ABSTRACT
Resistance in VanA-type vancomycin-resistant Enterococcus faecium (VREfm) is due to an inducible gene cassette encoding seven proteins (vanRSHAXYZ). This provides for an alternative peptidoglycan (PG) biosynthesis pathway whereby D-Ala–D-Ala is replaced by D-Ala–d-lactate (Lac), to which vancomycin cannot bind effectively. This study aimed to quantify cytoplasmic levels of normal and alternative pathway PG intermediates in VanA-type VREfm by liquid chromatography-tandem mass spectrometry before and after vancomycin exposure and to correlate these changes with changes in vanA operon mRNA levels measured by real-time quantitative PCR (RT-qPCR). Normal pathway intermediates predominated in the absence of vancomycin, with low levels of alternative pathway intermediates. Extended (18-h) vancomycin exposure resulted in a mixture of the terminal normal (UDP-N-acetylmuramic acid [NAM]–l-Ala–D-Glu–l-Lys–D-Ala–D-Ala [UDP-Penta]) and alternative (UDP-NAM–l-Ala–γ-D-Glu–l-Lys–D-Ala–D-Lac [UDP-Pentadepsi]) pathway intermediates (2:3 ratio). Time course analyses revealed normal pathway intermediates responding rapidly (peaking in 3 to 10 min) and alternative pathway intermediates responding more slowly (peaking in 15 to 45 min). RT-qPCR demonstrated that vanA operon mRNA transcript levels increased rapidly after exposure, reaching maximal levels in 15 min. To resolve the effect of increased van operon protein expression on PG metabolite levels, linezolid was used to block protein biosynthesis. Surprisingly, linezolid dramatically reduced PG intermediate levels when used alone. When used in combination with vancomycin, linezolid only modestly reduced alternative UDP-linked PG intermediate levels, indicating substantial alternative pathway presence before vancomycin exposure. Comparison of PG intermediate levels between VREfm, vancomycin-sensitive Enterococcus faecium, and methicillin-resistant Staphylococcus aureus after vancomycin exposure demonstrated substantial differences between S. aureus and E. faecium PG biosynthesis pathways.
IMPORTANCE VREfm is highly resistant to vancomycin due to the presence of a vancomycin resistance gene cassette. Exposure to vancomycin induces the expression of genes in this cassette, which encode enzymes that provide for an alternative PG biosynthesis pathway. In VanA-type resistance, these alternative pathway enzymes replace the D-Ala–D-Ala terminus of normal PG intermediates with D-Ala–D-Lac terminated intermediates, to which vancomycin cannot bind. While the general features of this resistance mechanism are well known, the details of the choreography between vancomycin exposure, vanA gene induction, and changes in the normal and alternative pathway intermediate levels have not been described previously. This study comprehensively explores how VREfm responds to vancomycin exposure at the mRNA and PG intermediate levels.
KEYWORDS: peptidoglycan, cell wall, Enterococcus faecium, VRE, vancomycin, antibiotic resistance, VanA, metabolomics, LC-MS/MS, mass spectrometry, metabolism
INTRODUCTION
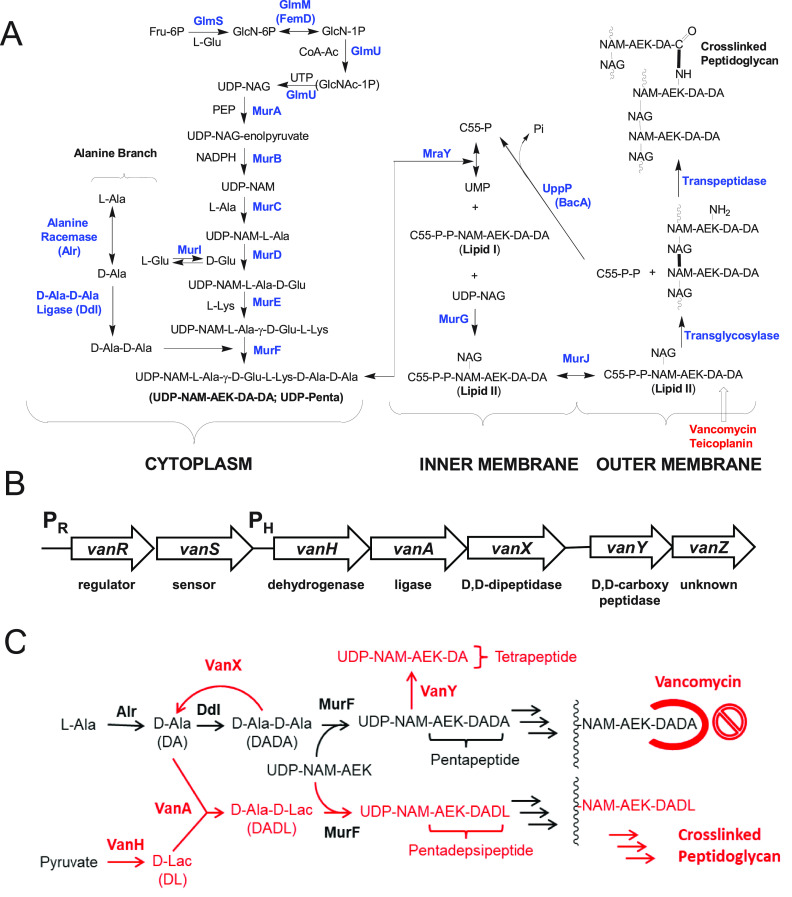
Vancomycin is an important agent for the treatment of Gram-positive bacterial infections that are resistant to other antibacterial agents, including vancomycin-sensitive enterococcus (VSE) and methicillin-resistant Staphylococcus aureus (MRSA) infections (1–3). Vancomycin resistance in enterococci is a serious public health issue, given the general resistance of these organisms to alternative agents (1, 4–8). Vancomycin acts by binding to D-Ala–D-Ala moieties in peptidoglycan (PG) precursors on the outer leaf of the cell membrane and blocking cell wall biosynthesis (CWB) (9–11) (Fig. 1). In the most clinically common VanA- and VanB-type vancomycin-resistant enterococcus (VRE) strains, D-Ala–D-Ala is replaced by D-Ala–d-lactate (Lac) (12–14). The gene cassette for vancomycin resistance encodes seven proteins (15) (Fig. 1B). VanH is a dehydrogenase that produces D-Lac from pyruvate, and VanA is a D-Ala–D-Lac ligase that links D-Lac to D-Ala (12, 16). VanX is a zinc dipeptidase that cleaves D-Ala–D-Ala (17, 18). VanY is a d,d-carboxypeptidase that cleaves the terminal D-Ala residue from UDP-N-acetylmuramic acid (NAM)–l-Ala–D-Glu–l-Lys–D-Ala–D-Ala (UDP-Penta) (13, 16) (Fig. 1C). VanR and VanS are regulatory proteins involved in vancomycin sensing and induction of vancomycin resistance (19). The function of the VanZ protein in VanA-type resistance to vancomycin is unknown, but it confers increased resistance to teicoplanin, a homolog of vancomycin (20, 21).
FIG 1.
(A) Generic PG biosynthesis process in Gram-positive bacteria. Note that in E. faecium a d-iAsp “bridging” residue is added to the ε-amino group of l-Lys (25), which is not included in this figure. (B) VanA-type resistance gene cluster (adapted from reference 15). (C) Alternative CWB pathway in VanA-type resistance. GlcN, glucosamine; GlcNAc, N-acetylglucosamine (NAG); GlcNAc-1P, N-acetylglucosamine-1-phosphate; PEP, phosphoenolpyruvate; AEK, L-Ala–γ-D-Glu–L-Lys.
An essential aspect of vancomycin resistance in enterococci is the shift from the vancomycin-naive state, with CWB based on normal D-Ala–D-Ala-based cell wall intermediates, to the vancomycin-exposed state, using alternative cell wall intermediates (15, 21) (Fig. 1C). This shift is poorly understood at the metabolite level, in part due to the lack of methods for the quantification of these PG intermediates. The cytoplasmic UDP-linked intermediates unique to vancomycin-resistant Enterococcus faecium (VREfm) resistance pathways (22) are UDP-NAM–l-Ala–γ-d-Glu–l-Lys–D-Ala–D-Lac (UDP-Pentadepsi), UDP-NAM–l-Ala–γ-d-Glu–l-Lys–D-Ala (UDP-Tetra), UDP-NAM–l-Ala–γ-d-Glu–l-Lys–(β-d-Asp)–D-Ala–D-Ala (UDP-Penta–d-isoAsp [UDP-Penta-D-iAsp]), UDP-NAM–l-Ala–γ-d-Glu–l-Lys–(β-d-Asp)–D-Ala–D-Lac (UDP-Pentadepsi–D-iAsp), and UDP-NAM–l-Ala–γ-d-Glu–l-Lys–(β-d-Asp)–D-Ala (UDP-Tetra-d-iAsp). This list contains several D-iAsp-containing PG intermediates. D-iAsp, subsequently α-amidated (23, 24), acts as a bridging residue for PG cross-linking reactions in E. faecium (25). It is preferentially added to the lipid I PG intermediate, with addition to UDP-Penta and UDP-Pentadepsi intermediates being observed in cells targeted with late-stage CWB inhibitors (22).
In recent studies, we have developed liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based quantification methods for cytoplasmic UDP-linked intermediates (26–28) and amine intermediates (l-Ala, D-Ala, and D-Ala–D-Ala) in S. aureus (27, 29, 30) and D-Ala–D-Lac in VRE (31). In this study, these methods are extended to the unique UDP-linked intermediates in VanA-type VREfm and are used to characterize how these metabolites respond to vancomycin exposure. To further support these observations, transcriptomic studies of VanA genes were also performed using real-time quantitative PCR (RT-qPCR).
RESULTS
LC-MS/MS method development.
In prior studies, the normal CWB pathway cytoplasmic intermediates in S. aureus were preparatively purified and LC-MS/MS methods for their quantification in bacterial extracts were developed (26). These normal CWB pathway intermediates are shared by vancomycin-sensitive Enterococcus faecium (VSEfm) and VREfm. VREfm also has several additional unique vancomycin resistance-associated intermediates (Fig. 1C) (22). Several of these intermediates (UDP-Pentadepsi, UDP-Tetra, and UDP-Pentadepsi-d-iAsp) were preparatively purified from vancomycin-treated VREfm cultures and were used to optimize their LC-MS/MS quantification (see Table S1 in the supplemental material) following previously described methods (26). UDP-Tetra-d-iAsp and UDP-Penta-d-iAsp lacked sufficient concentrations for purification, and their LC-MS/MS quantification parameters were adopted from those for UDP-Tetra and UDP-Penta, respectively. LC-MS/MS chromatograms are shown in Fig. S1. Serial dilutions of these intermediates in water and VREfm extract were linear over a detection range of 0.2 to 2,000 pmol with no apparent matrix effects, as observed in prior studies for the UDP-linked intermediates from S. aureus (26) (data not shown).
Comparison of sample preparation by centrifugation versus filtration for metabolite analysis.
Two different protocols for VREfm cytoplasmic intermediate isolation and extraction, i.e., filtration and centrifugation, were compared (27). Centrifugation showed slightly improved metabolite recovery (∼10%) (data not shown) and therefore was used for the remainder of this study.
Survey of vancomycin effects on CWB intermediates in E. faecium.
Data from VSEfm with or without vancomycin and VREfm with or without vancomycin are presented in Table 1. Values from MRSA with or without vancomycin are also included for comparison (28). A number of detailed comparisons can be made based on these data, which are presented in the supplemental material. Overall, VSEfm behaves very differently than MRSA in its response to vancomycin. UDP-linked intermediate levels increased remarkably in MRSA treated with vancomycin for 1.5 h, whereas VSEfm treated with vancomycin for 1.5 h showed only a modest overall increase in UDP-linked metabolite levels, indicating fundamental differences between E. faecium and MRSA PG biosynthesis pathways. A major difference between VSEfm treated with vancomycin for 1.5 h and VREfm treated with vancomycin for 1.5 h was that VREfm treated with vancomycin for 1.5 h showed a much lower level of UDP-Penta accumulation. In contrast, the total UDP-linked intermediate pool level (UDP-Sum) values were nearly the same for VSEfm treated with vancomycin for 1.5 h and VREfm treated with vancomycin for 1.5 h. The UDP-Sum value for VREfm treated with vancomycin for 18 h was similar to that for VREfm without vancomycin, indicating a complete return to normal levels of these key intermediates after prolonged vancomycin exposure.
TABLE 1.
Effect of vancomycin exposure on cytoplasmic PG intermediates in VSEfm and VREfm and comparison with MRSA (28)
| Concn (time zero concn) forb: |
|||||||
|---|---|---|---|---|---|---|---|
| VSEfm |
VREfm |
MRSA |
|||||
| Intermediatea | −Vm | +Vm (1.5 h) | −Vm | +Vm (1.5 h) | +Vm (18 h) | −Vm | +Vm (1.5 h) |
| UDP-linked intermediates (μM) | |||||||
| UDP-NAG | 940 (20) | 290 (20) | 590 (40) | 510 (30) | 470 (30) | 740 (60) | 77 (10) |
| UDP-NAM | 1,130 (60) | 740 (50) | 480 (20) | 740 (20) | 320 (30) | 1,250 (60) | 5,200 (900) |
| UDP-Mono | 113 (2) | 7.2 (0.5) | 80 (10) | 63 (4) | 35 (1) | 81 (7) | 2,900 (500) |
| UDP-Di | 55 (6) | 4.3 (1.5) | 95 (8) | 24 (2) | 47.8 (0.4) | 32 (6) | 110 (50) |
| UDP-Tri | 30 (5) | 97 (7) | 48 (7) | 43 (1) | 26 (4) | 10 (2) | 35 (8) |
| UDP-Penta | 590 (20) | 2,280 (120) | 356 (8) | 900 (40) | 190 (90) | 260 (30) | 62,000 (2,000) |
| UDP-Pentadepsi | ND | ND | 13.4 (0.4) | 730 (10) | 290 (50) | ND | ND |
| UDP-Tetra | ND | ND | 15.6 (0.4) | 774 (6) | 44 (5) | ND | ND |
| UDP-Penta-d-iAsp | ND | ND | ND | ND | 1.3 (0.2) | ND | ND |
| UDP-Pentadepsi-d-iAsp | ND | ND | 13.2 (0.9) | 46 (2) | 27 (2) | ND | ND |
| UDP-Tetra-d-iAsp | ND | ND | ND | ND | 2.1 (0.1) | ND | ND |
| UDP-Sum | 2,860 (60) | 3,500 (200) | 1,470 (130) | 3,820 (30) | 1,460 (60) | 2,380 (90) | 71,000 (1,900) |
| Amino acid and d-Ala–d-Ala intermediates (mM) | |||||||
| l-Ala | 52 (8) | 17.4 (0.6) | 29 (2) | 20.1 (0.8) | 15.2 (0.9) | 67 (7) | 64 (7) |
| d-Ala | 26 (3) | 5.2 (0.2) | 22 (2) | 12 (3) | 11 (1) | 48 (8) | 46 (16) |
| d-Ala–d-Ala | 2.8 (0.4) | 0.3 (0.1) | 0.71 (0.05) | 0.4 (0.1) | 0.12 (0.07) | 0.9 (0.3) | 17 (2) |
| d-Ala–d-Lac | ND | ND | 0.11 (0.01) | 2.8 (0.1) | 2.1 (0.3) | ND | ND |
| l-Glu | 126 (9) | 24.2 (0.6) | 81 (3) | 95 (2) | 57 (9) | 250 (30) | 210 (30) |
| d-Glu | 10.7 (0.2) | 4.7 (0.7) | 17.6 (0.3) | 51 (9) | 49 (2) | 200 (40) | 230 (50) |
| l-Lys | 11.1 (1.2) | 4.1 (0.4) | 11.8 (0.5) | 21 (1) | 12.6 (0.6) | 64 (5) | 79 (3) |
| l-Asp | 38 (4) | 9.3 (0.2) | 18.3 (0.4) | 21 (2) | 22 (3) | 96 (11) | 15 (8) |
| d-Asp | 22 (3) | 5 (1) | 14 (1) | 25.1 (0.5) | 15.7 (0.2) | 63 (9) | 15 (4) |
UDP-Mono, UDP-NAM–l-Ala; UDP-Di, UDP-NAM–l-Ala–d-Glu.
Values are mean ± SE (n = 3), Vm, vancomycin. UDP-NAG-enolpyruvate levels were not detectable (<0.5 μM). ND, not detectable.
Time dependence of vancomycin effects on metabolite pools in VREfm.
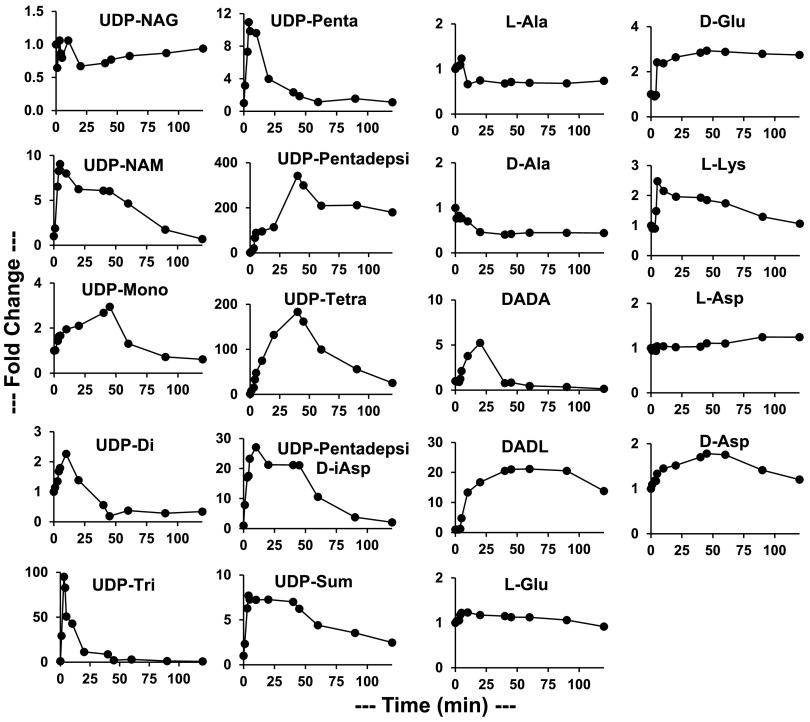
A time course experiment was performed to determine the rates at which PG intermediate levels changed in VREfm upon vancomycin exposure (Fig. 2) and in VSEfm (Fig. S2). These data (Fig. 2) revealed that most normal pathway VREfm PG metabolite levels changed rapidly in response to vancomycin exposure. UDP-NAM, UDP-NAM–l-Ala–D-Glu–l-Lys (UDP-Tri), and UDP-Penta peaked at 10 to 100 times their time zero levels in 3 to 10 min and then decreased back toward their time zero levels. UDP-Tri showed the largest fold increase, since its uninhibited level was low (37 μM) and the level increased nearly as high as the UDP-Penta level (1,080 μM for the peak UDP-Tri level versus 1,150 μM for the peak UDP-Penta level). D-Ala–D-Ala increased and spiked at a 5-fold increase at 20 min (3,800 μM), quite a bit lower (as a fold change) and slower than most of the normal UDP-linked intermediates; it then rapidly decreased to a low level. Key alternative UDP-linked PG pathway intermediates (UDP-Pentadepsi, UDP-Tetra, and D-Ala–D-Lac) peaked at 20 to 400 times their time zero levels at 30 to 45 min (peak levels of 4,450, 2,550, and 2,850 μM, respectively). UDP-Pentadepsi, the replacement for UDP-Penta in the alternative pathway, then plateaued at an ∼200-fold increase. D-Ala–D-Lac plateaued at a 15- to 20-fold increase, whereas UDP-Tetra continued to decline toward its time zero level. Interestingly, the D-Ala–D-Lac level appeared to show a distinct lag before beginning to increase at 5 min. UDP-Sum showed a sharp initial rise, peaking at 4 min, and then gradually decreased over the next 2 h. The rise of UDP-Sum was linear for 4 min after vancomycin exposure, with a slope of 2,800 μM/min. This represents a minimum value for the flux of metabolites through this pathway prior to vancomycin addition. Using the same approach, MRSA was found to have a minimum flux of 1,475 μM/min (28). The doubling times of MRSA and VREfm under logarithmic growth conditions are nearly the same (∼40 min), and the source of this difference in flux is presently unknown. UDP-Sum then plateaued until 40 min before gradually decreasing.
FIG 2.
Time course of VREfm PG intermediates in response to added vancomycin (16 μg/ml added at a culture OD600 of 0.5), plotted as fold changes versus the time zero values (shown in Table 1). UDP-Sum is the sum of all UDP-linked intermediates. DADA, D-Ala–D-Ala; DADL, D-Ala–D-Lac.
Time dependence of vanA gene induction.
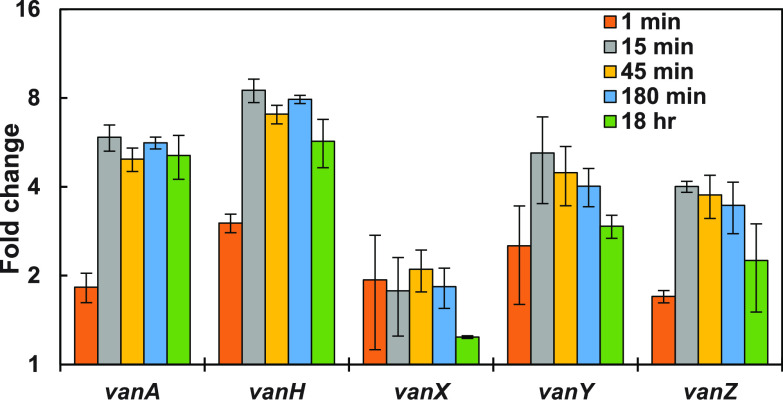
To assess the time dependence of the vanA gene induction, RT-qPCR was used to quantitatively amplify VanA-type mRNA transcripts for VanH, VanA, VanX, VanY, and VanZ proteins as a function of time after vancomycin exposure (Fig. 3). The response was rapid, as evident in the 1-min samples (collected immediately after vancomycin addition), which showed definite increases over control samples (collected immediately prior to vancomycin addition). mRNA levels peaked within 15 min after vancomycin exposure, and then decreased over time. These observations are consistent with prior studies (32–34).
FIG 3.
Time course of the effect of vancomycin (16 μg/ml) on RT-qPCR-determined mRNA levels in VREfm. Data are presented as mean ± SE (n = 3).
Vancomycin concentration dependence of VREfm metabolites and mRNA response.
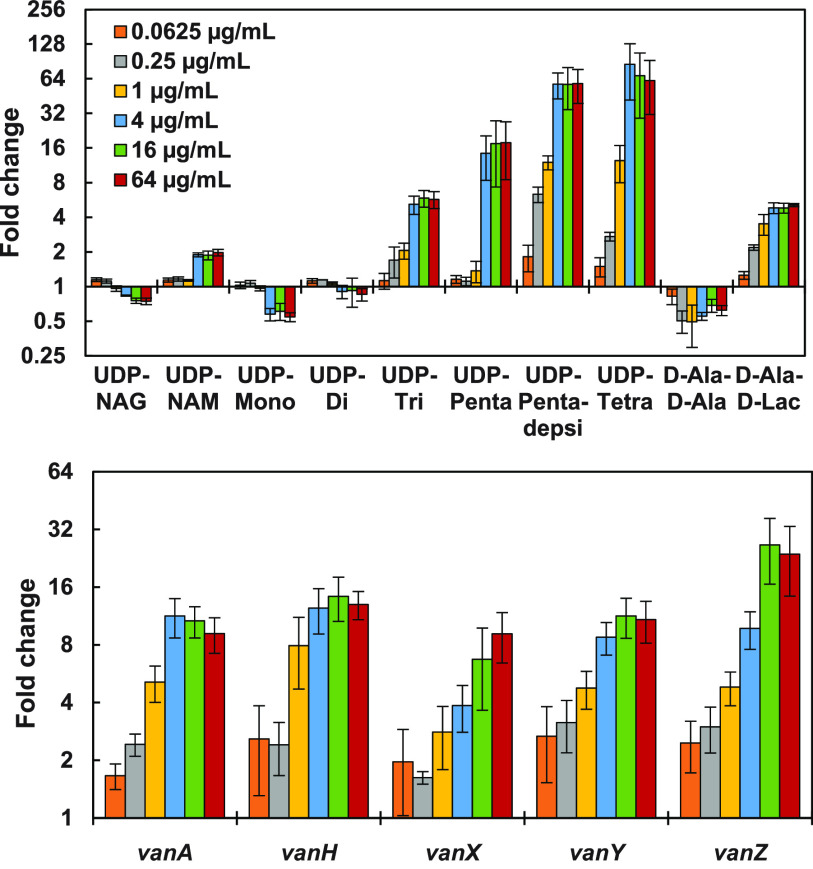
The effects of various concentrations of vancomycin on PG metabolite and mRNA levels after a 15-min exposure were also assessed (Fig. 4). The midpoints for the effects of vancomycin on the key PG metabolites (UDP-Penta, UDP-Pentadepsi, and D-Ala–D-Ala) and mRNA levels were 1 to 4 μg/ml vancomycin; they were possibly higher (4 to 16 μg/ml) for VanX, VanY, and VanZ protein mRNAs. The effects of increasing vancomycin concentrations were generally monotonic for most of these intermediates except for D-Ala–D-Ala, which initially decreased at low vancomycin concentrations and then approached normal levels at higher vancomycin concentrations. Late-stage intermediates (UDP-Tri, UDP-Penta, UDP-Tetra, and UDP-Pentadepsi) and D-Ala–D-Lac all increased substantially in response to increasing vancomycin concentrations.
FIG 4.
(Top) Fold changes (relative to no-vancomycin control) in key VREfm PG intermediate levels after 15-min exposure to different vancomycin concentrations (n = 4), shown with a semilogarithmic y axis. (Bottom) Corresponding fold changes in mRNA levels (n = 4).
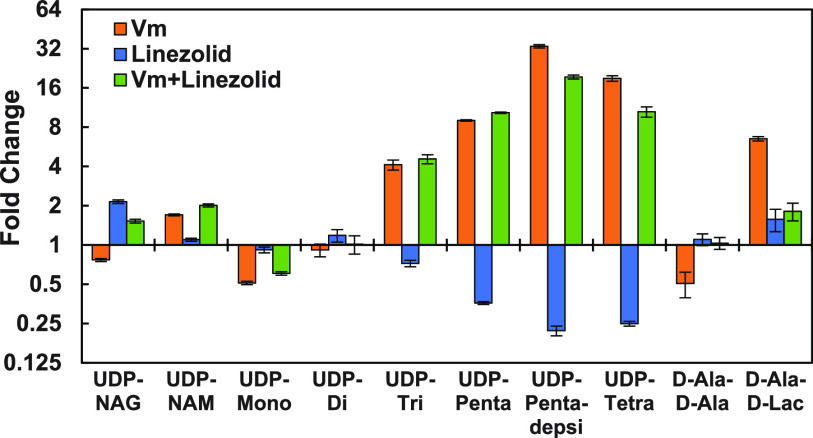
Effects of protein biosynthesis inhibition (by linezolid) on metabolite levels after vancomycin exposure.
To disentangle the effect of gene induction on metabolite levels in VREfm after vancomycin exposure, linezolid, a protein biosynthesis inhibitor effective against VREfm, was employed (Fig. 5). Linezolid alone increased UDP-N-acetylglucosamine (UDP-NAG) levels and decreased levels of later-stage intermediates (UDP-Penta, UDP-Tetra, and UDP-Pentadepsi).
FIG 5.
Fold changes (relative to no-vancomycin control) in key VREfm PG intermediate levels after 15-min exposure to vancomycin (Vm) and/or linezolid (n = 3), shown with a semilogarithmic y axis.
DISCUSSION
Bacterial PG biosynthesis is the target of many important antibacterial agents (35, 36). VREfm is particularly problematic, given its resistance to vancomycin and most other commonly used antibacterial agents (8, 37, 38). Vancomycin resistance in VRE is due to the presence of gene cassettes that encode alternative PG biosynthetic pathways (15). The most common of these clinically is VanA-type resistance, in which D-Ala–D-Ala is replaced by D-Ala–D-Lac (Fig. 1). The goals of this study were to investigate how cytoplasmic PG intermediate levels in VanA-type VREfm respond to vancomycin exposure and to compare this response to those of MRSA and VSEfm. The first step in this study was to extend LC-MS/MS assays for the normal PG pathway intermediates developed previously (27–31) to the unique intermediates responsible for vancomycin resistance in VREfm (Fig. 1). Purification of several of these intermediates from VREfm and ion-pairing (IP) LC-MS/MS method development were straightforward, based on the prior studies (see Fig. S1 and Table S1 in the supplemental material). Since vancomycin resistance involves induction of the alternative PG biosynthesis pathway, RT-qPCR was also used to provide complimentary data on the levels of VanA-type mRNA transcripts.
PG intermediate levels were first determined in both VSEfm and VREfm before and after vancomycin exposure (Table 1). Previously reported values for MRSA are included for comparison (28). Detailed comparisons within these data are provided in the supplemental material. Several significant features are apparent. VSEfm shows much less vancomycin exposure-associated UDP-linked intermediate accumulation (UDP-Sum) than does MRSA, demonstrating a substantial difference in this pathway between VSEfm and MRSA. In VSEfm, the substantial reduction in early intermediates indicates that reduced entry into the pathway is partially or wholly responsible for this effect. In VREfm, both normal and alternative pathway intermediates accumulate substantially after a 1.5-h vancomycin exposure (Table 1). After extended vancomycin exposure, levels of both normal and alternative pathway intermediates decrease considerably. Somewhat unexpectedly, UDP-Penta, the normal pathway terminal PG intermediate, is reduced to only about one-half the no-vancomycin level, equivalent to two-thirds the level of UDP-Pentadepsi, the analogous vancomycin resistance pathway intermediate. An early study of VanA-type resistance in vancomycin-resistant Enterococcus faecalis (VREfs) found a high UDP-Pentadepsi/UDP-Penta ratio of about 98:2 associated with a vancomycin MIC of 256 μg/ml (21), whereas a ratio of about 60:40 was observed in this study with a vancomycin MIC of 512 μg/ml. This observation indicates that a complete shift from D-Ala–D-Ala- to D-Ala–D-Lac-based intermediates is not required for high-level vancomycin resistance.
D-iAsp-containing intermediates were identified in E. faecium in prior studies (22, 25, 39). These intermediates (UDP-Penta-D-iAsp, UDP-Pentadepsi-D-iAsp, and UDP-Tetra-D-iAsp) were observed in VREfm (Table 1) but were undetectable in VSEfm. The step at which D-Asp is added to nascent PG has been unclear. Indications are that it is preferentially added to the lipid I PG intermediates, with addition to UDP-Penta and UDP-Pentadepsi intermediates being observed in cells targeted with late-stage CWB inhibitors (22). The results observed here are consistent with and support this interpretation.
Time course experiments for VREfm metabolite levels (Fig. 2) and mRNA levels (Fig. 3) were then performed. Similar data for VSEfm, overlaid with the VREfm data, are shown in Fig. S2 using a semi-square root x axis to expand and highlight early changes in metabolite levels. A key observation in VREfm is that normal pathway intermediates generally respond rapidly to vancomycin exposure, peaking in 3 to 5 min, whereas alternative pathway intermediates peak at around 30 to 45 min. PG intermediates in VSEfm initially respond similarly to those in VREfm but then begin to drop, relative to VREfm levels, after 3 to 5 min, with the exception of UDP-Penta in VSEfm, which increases more dramatically than in VREfm. This indicates that the PG pathway is more completely blocked in VSEfm than in VREfm, which is as expected. These observations also indicate that entry into the UDP-linked intermediate part of this pathway is substantially reduced in VSEfm, relative to VREfm, after vancomycin exposure. Similar time course experiments in MRSA show continuous increases in UDP-Penta and UDP-Sum accumulation for up to 120 min after vancomycin exposure (28), in stark contrast to VSEfm and VREfm, which show accumulation to peak levels between 5 and 20 min (Fig. 2; also see Fig. S2) and then decreasing levels of these intermediates. These observations indicate that entry into the UDP-linked intermediate part of this pathway is downregulated in E. faecium in response to vancomycin exposure, in contrast to MRSA, in which UDP-linked intermediates accumulate apparently unabated in response to vancomycin exposure.
A time course study of mRNA transcript levels for key vanA genes was also performed (Fig. 3). mRNA levels rose quickly, showing a significant jump even at 1 min and reaching a plateau within 15 min. This time frame precedes that over which alternative pathway intermediates (UDP-Pentadepsi, UDP-Tetra, and D-Ala–D-Lac) rise to maximum levels and then began to decrease (Fig. 2). It was also noted that there was a noticeable lag in D-Ala–D-Lac, UDP-Pentadepsi, and UDP-Tetra profiles (Fig. S2).
The effects of vancomycin concentration on metabolite and mRNA levels were also assessed after 15-min exposure (Fig. 4). A midpoint for the effect on metabolite levels was 0.25 to 1 μg/ml, whereas the midpoint for the effect on mRNA levels appeared slightly higher at 1 to 4 μg/ml. Similarly, the maximal effect on metabolite levels was at 4 μg/ml, whereas for mRNA it was at 16 μg/ml. UDP-Penta shows a sudden transition from no-vancomycin levels to plus-vancomycin levels at 4 μg/ml. Most other metabolites show a monotonic shift to the plus-vancomycin state.
To assess the role of gene induction in the observed metabolite pool changes, an experiment using linezolid to block new protein biosynthesis was performed (Fig. 5). Somewhat surprisingly, linezolid in the absence of vancomycin suppressed later UDP-linked intermediate (UDP-Penta, UDP-Pentadepsi, and UDP-Tetra) levels, increased the UDP-NAG level, and had no significant effects on D-Ala–D-Ala and UDP-Tri levels. This indicates that one of the enzymes in this pathway (MurA to MurE) may have a relatively short half-life, with activity loss contributing to the lower observed later metabolite levels and modest accumulation of the upstream UDP-NAG intermediate. Combination of linezolid with vancomycin resulted in a pattern of metabolite level changes nearly identical to that observed with vancomycin alone. This observation indicates that basal (uninduced) alternative pathway enzyme levels are sufficient for a substantial shift in UDP-linked intermediate levels toward the vancomycin-induced state even in the absence of new protein biosynthesis. It is also noteworthy that, in contrast to UDP-linked intermediate levels, new protein synthesis appears to be required for the vancomycin-induced shifts in D-Ala–D-Ala and D-Ala–D-Lac levels (Fig. 5).
A reasonable question is why did UDP-linked intermediate levels in the plus-vancomycin/plus-linezolid experiment not significantly decrease as in the plus-linezolid experiment? This observation can be explained as follows. In the plus-vancomycin experiment, levels of UDP-linked intermediates rise very quickly and remain substantially blocked at 15 min (Fig. 2). The action of linezolid on protein levels takes longer. In the plus-linezolid experiment, with continued but slowly diminishing flux through this pathway, decreasing key enzyme levels result in decreased UDP-linked metabolite levels. However, in the plus-vancomycin/plus-linezolid experiment, UDP-linked metabolite levels spike relatively quickly, followed by reduced flux through key enzyme depletion, such that high metabolite levels are established before enzyme depletion. These observations highlight the dynamic nature of this resistance mechanism.
MATERIALS AND METHODS
General.
VanA-type VREfm was a clinical isolate provided by Betty Herndon (University of Missouri-Kansas City, School of Medicine). VSE was obtained from the American Type Culture Collection (ATCC number BAA-2127). D-Ala, l-Ala, D-Ala–D-Ala, 13C-labeled D-Ala, UDP-glucuronic acid (UDP-GlcA), vancomycin, hemin, and triethylamine were from Millipore Sigma. N,N-Dimethylhexylamine (DMHA) was from Alfa Aesar. Other materials and reagents were generally as described previously (26, 27, 31). VRE growth medium, consisting of brain heart infusion (BHI) (37.5 g/liter), hemin (10 μg/ml), and NAD+ (10 μg/ml), was prepared following standard procedures. The forward and reverse primers for the vanH, vanA, vanX, vanY, and vanZ genes were from Integrated DNA Technologies (see Table S2 in the supplemental material). Diethyl pyrocarbonate (DEPC)-treated water was from Ambion, and the TRIzol Max bacterial RNA isolation kit and iTaq Universal SYBR green PCR kit were from Life Technologies and Bio-Rad, respectively. RT-qPCR was performed on a Bio-Rad CFX Connect real-time system. Bacterial cell densities were determined as the optical density at 600 nm (OD600) in a BioMate 3 spectrophotometer. Centrifugations were performed with a Sorvall RT 6000 centrifuge, a Beckman Coulter Avanti J-251 centrifuge, or an Eppendorf 5424 microcentrifuge. Marfey’s reagent (1-fluoro-2,4-dinitrophenyl-l-5-alanine amide) was from Novabiochem (a division of EMD Chemicals). LC-MS/MS analyses were performed with an AB Sciex 3200 Q-trap mass spectrometer coupled to a Shimadzu ultra-fast liquid chromatography (UFLC) system using electrospray ionization and run with Analyst v1.4.2 software (Sciex).
Purification of novel UDP-linked intermediates from VREfm for use as standards.
The alternative PG biosynthesis pathway in VanA-type VREfm uses several unique UDP-linked intermediates (Fig. 1). Several of these intermediates were purified from vancomycin (64 μg/ml)-treated VREfm cultures by preparative ion-pairing reverse-phase high-performance liquid chromatography (HPLC), as described previously for the isolation of the normal pathway UDP-linked intermediates from S. aureus (26). This provided purified samples of UDP-Pentadepsi, UDP-Pentadepsi-D-iAsp, and UDP-Tetra. These purified intermediates were used to optimize MS/MS parameters for their detection and quantification (see Table S1 and Fig. S1 in the supplemental material), and these detection parameters were combined with those for normal CWB pathway intermediates (26, 27) to provide a comprehensive method for both normal and alternative pathway UDP-linked intermediates in VREfm. Two unique VREfm UDP-linked intermediates (UDP-Penta-D-iAsp and UDP-Tetra-D-iAsp) were too low in abundance for purification, and for these intermediates the MS/MS detection parameters for UDP-Penta and UDP-Tetra, respectively, were used.
IP LC-MS/MS-based quantification of UDP-linked intermediates.
Analytical separations were performed on a Nucleodur 100-3 C18 column (125 by 2 mm; Macherey-Nagel) at 300 μl/min. Mobile phases were as follows: buffer A, 10 mM formic acid in water; buffer B, 10 mM formic acid in 70:30 acetonitrile-water; buffer C, 10 mM formic acid in acetonitrile; buffer D, 160 mM DMHA in water (adjusted to pH 3.0 with formic acid). The optimized elution gradient was 5% buffer D throughout (to provide 8 mM DMHA in the chromatography solvent stream) with 80% buffer A and 15% buffer B initially. After sample injection, a linear gradient to 75% buffer A and 20% buffer B over 10 min was used to elute analytes. MS/MS detection settings were as reported previously (26) for normal CWB pathway UDP-linked intermediates and as given in Table S1 for the VanA-type resistance-specific UDP-linked intermediates. Intracellular analyte concentrations were determined using purified UDP-linked intermediates as external standards, and intracellular concentrations were calculated using a culture to cell dry weight conversion factor of 533 mg of cell dry weight (liter of culture)−1 OD600−1 and an internal cell volume of 2 μl (mg of dry cells)−1, which were determined as described previously (27).
Amine and amino acid quantification.
Amino acids from bacterial extracts were diluted 5-fold with pure water, derivatized with Marfey’s reagent, and quantified by LC-MS/MS analysis in negative mode as described in detail elsewhere (27, 29–31).
Comparison of centrifugation versus filtration for metabolite extraction.
Bacterial cells for metabolomic studies can be collected by either filtration or centrifugation, with advantages and disadvantages for each. A comparison of these two approaches for VREfm was made to determine which strategy would be the best, as described previously for S. aureus (27). For filtration-based metabolite extraction, VREfm cultures were chilled on ice, and 10-ml samples were filtered through 47-mm-diameter 0.2-μm nylon membrane filters. Filters were quickly washed twice with 5 ml of ice-cold 0.9% saline and transferred to 15-ml centrifuge tubes containing 3 ml of ice-cold methanol-water-formic acid (66:33:1) with 100 μM 13C3-labeled D-Ala and 10 μM UDP-GlcA as internal standards. Samples were kept on ice for 5 min with regular vortex mixing and centrifuged at 3,000 × g at 4°C for 20 min, and the supernatants were collected. Filters and cell pellets were then extracted a second time with 2 ml of 67% methanol-water without any added internal standards and centrifuged at 3,000 × g at 4°C for 20 min, and the second supernatants were combined with the first. To the combined supernatants was added 2 ml of HPLC-grade water, which allowed the samples to be frozen prior to vacuum drying. The extracts were frozen at −80°C, dried in a vacuum centrifuge under strong vacuum (<50 μm Hg), and dissolved in 100 μl of 3.5% acetonitrile–24 mM DMHA–water. This level of DMHA in the samples facilitates UDP-linked intermediate binding to the C18 column for chromatographic analysis.
For centrifugation-based metabolite extraction, quadruplicate samples of 40 ml from each culture flask were transferred into chilled 50-ml centrifuge tubes on ice, and the cells were pelleted by centrifugation at 3,000 × g at 4°C for 20 min. Supernatants were discarded, and the cells were washed once by resuspension in 1 ml of ice-cold 0.9% saline and repelleting. Washed cells were resuspended in 1 ml of ice-cold 0.9% saline, and metabolites were extracted by adding 4 ml of the first extraction solvent (80% methanol with 100 μM 13C3-labeled D-Ala and 10 μM UDP-GlcA as internal standards). These samples were kept on ice for 5 to 15 min, the cell debris was pelleted at 3,000 × g at 4°C for 20 min, and the supernatants were collected into previously chilled 15-ml centrifuge tubes on ice. The pellets were then extracted a second time with 2 ml of the second extraction solvent (67% methanol-water without any internal standards added), using vortex mixing and pipetting to resuspend the samples. After 5 min on ice, the samples were centrifuged again at 3,000 × g at 4°C for 20 min, and the second sample extraction supernatants were removed and combined with the first sample extraction supernatants. This double extraction protocol maximizes both analyte and internal standard recovery and reduces variation. To the combined extracts, 2 ml of ice-cold pure water was added to allow freezing at −80°C, and the extracts were frozen in −80°C and then dried in a SpeedVac concentrator under strong vacuum (<50 μm Hg). The dried samples were then dissolved in 100 μl of 3.5% acetonitrile–24 mM DMHA and stored at −20°C prior to analysis. Centrifugation gave slightly higher levels of recovered metabolites than did filtration (∼10% overall) (data not shown). Since centrifugation was also more convenient, the remaining studies were performed using the centrifugation-based cell collection and extraction approach.
Standard growth and metabolite extraction procedures for VREfm vancomycin exposure experiments.
A 20-ml saturated overnight VREfm primary culture was grown in VRE medium and used to inoculate 1,300 ml of VRE medium in a baffled 2-liter flask (secondary culture) to an OD600 of 0.05. The cells were grown with good agitation at 35°C (doubling time of 45 min). When the secondary culture reached an OD600 of 0.5, 160-ml portions were transferred into separate 500-ml baffled flasks. Different concentrations of vancomycin were added to these flasks except for one (the no-vancomycin control), and the flasks were incubated with shaking for an appropriate time. The flasks were then rapidly chilled in an ice slush bath, and samples from individual flasks were collected in quadruplicate and stored on ice for up to 15 min prior to centrifugation and processing for metabolite extraction, as described above. Samples were analyzed for UDP-linked intermediates as described previously (28) with the expanded set of LC-MS/MS parameters for the alternative UDP-linked intermediates found in VREfm described above and for amine and amino acid intermediates using the Marfey’s derivatization-based approach with negative mode-based detection also as described previously (30, 31). This constituted a single experiment, which was replicated on separate days to provide three or four replications.
Experimental procedure for long-term vancomycin exposure.
A 20-ml saturated primary culture of VRE was grown in VRE medium with 16 μg/ml vancomycin. This primary culture was used to inoculate 100 ml of fresh VRE medium with 16 μg/ml vancomycin in a baffled flask (secondary culture). The culture was grown with good agitation at 35°C (doubling time of 45 min). When the secondary culture reached an OD600 of 0.5, triplicate samples of 15 ml were collected, and the cells were collected and extracted for LC-MS/MS analysis as described above.
Time dependence of cytoplasmic CWB intermediate levels in VREfm after vancomycin exposure.
This experiment was performed following the standard growth procedure described above. Cells were grown in the secondary culture (no vancomycin) to mid-log phase (OD600 of 0.5), and vancomycin (16 μg/ml) was added. At regular time points (0, 1, 2, 3, 4, 5, 10, 20, 40, 60, 90, 120, 180, and 240 min), triplicate samples of 15 ml were collected by centrifugation and prepared for analysis as described above.
Time dependence of VanA mRNA levels in VREfm after vancomycin exposure.
A 20-ml saturated overnight VREfm culture was grown in VRE medium and was used to inoculate 500 ml of VRE medium to an OD600 of 0.05. When the secondary culture reached an OD600 of 0.5, 16 μg/ml vancomycin was added to the flasks. Quadruplicate samples of 10 ml were collected at different time points after vancomycin addition (1, 15, 45, and 180 min and 18 h), chilled, and stored on ice prior to mRNA extraction. A sample was also collected immediately prior to vancomycin addition as the control (time zero) sample. mRNA was extracted from these samples using the TRIzol Max bacterial RNA isolation kit (Thermo Fisher Scientific) following the manufacturer’s protocol. The quality and concentration of mRNA were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific). RNA (100 ng) was then used as the template in RT-qPCR with primer pairs (Table S2) designed to amplify internal regions of VanH, VanA, VanX, VanY, and VanZ (40). RT-qPCR was performed with the comparative threshold cycle (CT) method (41). Measurements were performed on a Bio-Rad CFX Connect real-time system. RT-qPCR cycles were 10 s at 50°C, 5 min at 95°C, 30 s at 94°C, 30 s at 53.2°C, 30 s at 72°C (for 45 cycles), and 30 s at 25°C. The gene expression was internally normalized to the 16S rRNA gene. After initial results were obtained, mRNA levels were adjusted to give CT values of 18 to 29. CT values were used to calculate the fold change (FC) of van gene expression between control and vancomycin-treated cultures using the following formula:
| (1) |
where
| (2) |
The relative fold changes in vanA gene mRNA levels between control and vancomycin-exposed time course samples from three completely independent experiments were determined, and the independent experiment means and standard errors (SEs) are reported.
Vancomycin concentration dependence of VREfm mRNA levels.
Primary and secondary cultures were grown following the standard growth protocol. When the secondary culture reached an OD600 of 0.5, 50-ml aliquots were transferred to 250-ml baffled flasks and various concentrations of vancomycin (0.0625, 0.125, 1, 4, 16, and 64 μg/ml) were added. A no-vancomycin control flask was also included. The cultures were grown for 15 min, and quadruplicate samples of 10 ml were collected on ice. mRNA was isolated and quantified as described above. This experiment was repeated in pentaplicate.
Effect of linezolid on vancomycin-associated metabolite pool level changes.
Primary and secondary cultures were grown following the standard growth protocol. When the secondary culture reached an OD600 of 0.5, 50-ml portions were transferred into separate 250-ml baffled flasks. Different antibiotics (vancomycin at 64 μg/ml and linezolid at 8 μg/ml [4× MIC]) and their combination (vancomycin plus linezolid) were added to these flasks. A no-antibiotic flask was also included. The flasks were incubated at 35°C for 15 min with shaking, and 10-ml samples from individual flasks were collected in quadruplicate and processed for metabolite analysis as described above.
Effect of linezolid on vancomycin-associated mRNA level changes.
Primary and secondary cultures were grown following the standard growth protocol. When the secondary culture reached an OD600 of 0.5, 50-ml portions were transferred into separate 250-ml baffled flasks. Different antibiotics and their combination (vancomycin, linezolid, and vancomycin plus linezolid) were added to these flasks at 4× MIC. A no-antibiotic flask was also included. The flasks were incubated at 35°C for 15 min with shaking, and 10-ml samples from individual flasks were collected in quadruplicate and processed for metabolite analysis as described above.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R15-GM126502 (to W.G.G.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material is available online only.
Contributor Information
William G. Gutheil, Email: gutheilw@umkc.edu.
Michael J. Federle, University of Illinois at Chicago
REFERENCES
- 1.Wang JL, Hsueh PR. 2009. Therapeutic options for infections due to vancomycin-resistant enterococci. Expert Opin Pharmacother 10:785–796. 10.1517/14656560902811811. [DOI] [PubMed] [Google Scholar]
- 2.Koomanachai P, Crandon JL, Nicolau DP. 2009. Newer developments in the treatment of Gram-positive infections. Expert Opin Pharmacother 10:2829–2843. 10.1517/14656560903357491. [DOI] [PubMed] [Google Scholar]
- 3.Rivera AM, Boucher HW. 2011. Current concepts in antimicrobial therapy against select Gram-positive organisms: methicillin-resistant Staphylococcus aureus, penicillin-resistant pneumococci, and vancomycin-resistant enterococci. Mayo Clin Proc 86:1230–1243. 10.4065/mcp.2011.0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKenna M. 2013. Antibiotic resistance: the last resort. Nature 499:394–396. 10.1038/499394a. [DOI] [PubMed] [Google Scholar]
- 6.Nikolaidis I, Favini-Stabile S, Dessen A. 2014. Resistance to antibiotics targeted to the bacterial cell wall. Protein Sci 23:243–259. 10.1002/pro.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Tyne D, Gilmore MS. 2014. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol 68:337–356. 10.1146/annurev-micro-091213-113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller WR, Murray BE, Rice LB, Arias CA. 2020. Resistance in vancomycin-resistant enterococci. Infect Dis Clin North Am 34:751–771. 10.1016/j.idc.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nieto M, Perkins HR. 1971. Physicochemical properties of vancomycin and iodovancomycin and their complexes with diacetyl-l-lysyl-D-alanyl-D-alanine. Biochem J 123:773–787. 10.1042/bj1230773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barna JC, Williams DH. 1984. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu Rev Microbiol 38:339–357. 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- 11.Pootoolal J, Neu J, Wright GD. 2002. Glycopeptide antibiotic resistance. Annu Rev Pharmacol Toxicol 42:381–408. 10.1146/annurev.pharmtox.42.091601.142813. [DOI] [PubMed] [Google Scholar]
- 12.Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT. 1991. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30:10408–10415. 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- 13.Billot-Klein D, Gutmann L, Collatz E, van Heijenoort J. 1992. Analysis of peptidoglycan precursors in vancomycin-resistant enterococci. Antimicrob Agents Chemother 36:1487–1490. 10.1128/aac.36.7.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handwerger S, Pucci MJ, Volk KJ, Liu J, Lee MS. 1992. The cytoplasmic peptidoglycan precursor of vancomycin-resistant Enterococcus faecalis terminates in lactate. J Bacteriol 174:5982–5984. 10.1128/jb.174.18.5982-5984.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courvalin P. 2006. Vancomycin resistance in Gram-positive cocci. Clin Infect Dis 42(Suppl 1):S25–S34. 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 16.Arthur M, Molinas C, Courvalin P. 1992. Sequence of the vanY gene required for production of a vancomycin-inducible d,d-carboxypeptidase in Enterococcus faecium BM4147. Gene 120:111–114. 10.1016/0378-1119(92)90017-j. [DOI] [PubMed] [Google Scholar]
- 17.Wu Z, Wright GD, Walsh CT. 1995. Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 34:2455–2463. 10.1021/bi00008a008. [DOI] [PubMed] [Google Scholar]
- 18.Arthur M, Depardieu F, Cabanie L, Reynolds P, Courvalin P. 1998. Requirement of the VanY and VanX D,D-peptidases for glycopeptide resistance in enterococci. Mol Microbiol 30:819–830. 10.1046/j.1365-2958.1998.01114.x. [DOI] [PubMed] [Google Scholar]
- 19.Arthur M, Molinas C, Courvalin P. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol 174:2582–2591. 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arthur M, Depardieu F, Molinas C, Reynolds P, Courvalin P. 1995. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 154:87–92. 10.1016/0378-1119(94)00851-i. [DOI] [PubMed] [Google Scholar]
- 21.Arthur M, Depardieu F, Reynolds P, Courvalin P. 1996. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol 21:33–44. 10.1046/j.1365-2958.1996.00617.x. [DOI] [PubMed] [Google Scholar]
- 22.Billot-Klein D, Shlaes D, Bryant D, Bell D, Legrand R, Gutmann L, van Heijenoort J. 1997. Presence of UDP-N-acetylmuramyl-hexapeptides and -heptapeptides in enterococci and staphylococci after treatment with ramoplanin, tunicamycin, or vancomycin. J Bacteriol 179:4684–4688. 10.1128/jb.179.15.4684-4688.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staudenbauer W, Strominger JL. 1972. Activation of D-aspartic acid for incorporation into peptidoglycan. J Biol Chem 247:5095–5102. 10.1016/S0021-9258(19)44943-0. [DOI] [PubMed] [Google Scholar]
- 24.Mainardi JL, Legrand R, Arthur M, Schoot B, van Heijenoort J, Gutmann L. 2000. Novel mechanism of β-lactam resistance due to bypass of DD-transpeptidation in Enterococcus faecium. J Biol Chem 275:16490–16496. 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- 25.Ghuysen JM, Bricas E, Leyh-Bouille M, Lache M, Shockman GD. 1967. The peptide Nα-(l-alanyl-D-isoglutaminyl)-Nε-(D-isoasparaginyl)-l-lysyl-D-alanine and the disaccharide N-acetylglucosaminyl-β-1,4-N-acetylmuramic acid in cell wall peptidoglycan of Streptococcus faecalis strain ATCC 9790. Biochemistry 6:2607–2619. 10.1021/bi00860a044. [DOI] [PubMed] [Google Scholar]
- 26.Vemula H, Bobba S, Putty S, Barbara JE, Gutheil WG. 2014. Ion-pairing liquid chromatography-tandem mass spectrometry-based quantification of uridine diphosphate-linked intermediates in the Staphylococcus aureus cell wall biosynthesis pathway. Anal Biochem 465:12–19. 10.1016/j.ab.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Vemula H, Ayon NJ, Gutheil WG. 2016. Cytoplasmic peptidoglycan intermediate levels in Staphylococcus aureus. Biochimie 121:72–78. 10.1016/j.biochi.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 28.Vemula H, Ayon NJ, Burton A, Gutheil WG. 2017. Antibiotic effects on methicillin-resistant Staphylococcus aureus cytoplasmic peptidoglycan intermediate levels and evidence for potential metabolite level regulatory loops. Antimicrob Agents Chemother 61:e02253-16. 10.1128/AAC.02253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamindar D, Gutheil WG. 2010. A liquid chromatography-tandem mass spectrometry assay for Marfey's derivatives of l-Ala, D-Ala, and D-Ala–D-Ala: application to the in vivo confirmation of alanine racemase as the target of cycloserine in Escherichia coli. Anal Biochem 396:1–7. 10.1016/j.ab.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Ayon NJ, Sharma AD, Gutheil WG. 2019. LC-MS/MS-based separation and quantification of Marfey's reagent derivatized proteinogenic amino acid DL-stereoisomers. J Am Soc Mass Spectrom 30:448–458. 10.1007/s13361-018-2093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Putty S, Vemula H, Bobba S, Gutheil WG. 2013. A liquid chromatography-tandem mass spectrometry assay for D-Ala–D-Lac: a key intermediate for vancomycin resistance in vancomycin-resistant enterococci. Anal Biochem 442:166–171. 10.1016/j.ab.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 32.Wright GD, Molinas C, Arthur M, Courvalin P, Walsh CT. 1992. Characterization of VanY, a DD-carboxypeptidase from vancomycin-resistant Enterococcus faecium BM4147. Antimicrob Agents Chemother 36:1514–1518. 10.1128/aac.36.7.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panesso D, Abadia-Patino L, Vanegas N, Reynolds PE, Courvalin P, Arias CA. 2005. Transcriptional analysis of the vanC cluster from Enterococcus gallinarum strains with constitutive and inducible vancomycin resistance. Antimicrob Agents Chemother 49:1060–1066. 10.1128/AAC.49.3.1060-1066.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qureshi NK, Yin S, Boyle-Vavra S. 2014. The role of the staphylococcal VraTSR regulatory system on vancomycin resistance and vanA operon expression in vancomycin-resistant Staphylococcus aureus. PLoS One 9:e85873. 10.1371/journal.pone.0085873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bugg TD, Braddick D, Dowson CG, Roper DI. 2011. Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol 29:167–173. 10.1016/j.tibtech.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Silver LL. 2013. Viable screening targets related to the bacterial cell wall. Ann N Y Acad Sci 1277:29–53. 10.1111/nyas.12006. [DOI] [PubMed] [Google Scholar]
- 37.Barber KE, King ST, Stover KR, Pogue JM. 2015. Therapeutic options for vancomycin-resistant enterococcal bacteremia. Expert Rev Anti Infect Ther 13:363–377. 10.1586/14787210.2015.1001839. [DOI] [PubMed] [Google Scholar]
- 38.De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, Paterson DL, Walker MJ. 2020. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev 33:e00181-19. 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds PE, Snaith HA, Maguire AJ, Dutka-Malen S, Courvalin P. 1994. Analysis of peptidoglycan precursors in vancomycin-resistant Enterococcus gallinarum BM4174. Biochem J 301:5–8. 10.1042/bj3010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel R, Uhl JR, Kohner P, Hopkins MK, Cockerill FR, III.. 1997. Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J Clin Microbiol 35:703–707. 10.1128/JCM.35.3.703-707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong ML, Medrano JF. 2005. Real-time PCR for mRNA quantitation. Biotechniques 39:75–85. 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2, Fig. S1 and S2, and supplemental text. Download JB00230-21_Supp_1_seq5.pdf, PDF file, 0.4 MB (472.6KB, pdf)