Abstract
Transgenic neuromodulation tools have transformed the field of neuroscience over the past two decades by enabling targeted manipulation of neuronal populations and circuits with unprecedented specificity. Chemogenetic and optogenetic neuromodulation systems are among the most widely used and allow targeted control of neuronal activity through the administration of a selective compound or light, respectively. Innovative genetic targeting strategies are utilized to transduce specific cells to express transgenic receptors and opsins capable of manipulating neuronal activity. These allow mapping of neuroanatomical projection sites and link cellular manipulations with brain circuit functions and behavior. As these tools continue to expand knowledge of the nervous system in preclinical models, developing translational applications for human therapies is becoming increasingly possible. However, new strategies for implementing and monitoring transgenic tools are needed for safe and effective use in translational research and potential clinical applications. A major challenge for such applications is the need to track the location and function of chemogenetic receptors and opsins in vivo, and new developments in positron emission tomography (PET) imaging techniques offer promising solutions. The goal of this review is to summarize current research combining transgenic tools with PET for in vivo mapping and manipulation of brain circuits and to propose future directions for translational applications.
1. Introduction
The ability to manipulate neuronal activity with precise spatial and temporal control is essential for interrogating the neurobiological mechanisms underlying behavior and disease processes. Modern developments in transgenic neuromodulation technologies have transformed neuroscience research by enabling the manipulation of neuronal populations and circuits with incredible specificity in awake behaving subjects. Chemogenetic and optogenetic neuromodulation tools are among the most widely used in preclinical research for in vivo brain circuit manipulations. Innovative genetic targeting strategies (e.g., viral vector serotypes, cell-type specific promoter or enhancer sequences, and recombinase inducible transcription systems) can be used to transduce specific cell populations with transgenes encoding proteins whose expression can alter neuronal activity with administration of a selective compound or light. The ability to map highly specific neuroanatomical projections and manipulate neuronal activity in investigational settings has opened the door to explore novel clinical interventions (Assaf and Schiller, 2019; Curado et al., 2020; Iyer et al., 2016; Kätzel et al., 2014; Weir et al., 2017). However, safe and reliable strategies for delivering and monitoring transgenic tools are needed to enhance the translational potential of chemogenetic and optogenetic neuromodulation.
Chemogenetic neuromodulation is based on the premise that a neuronal system can be manipulated by introduction and expression of an exogenous transgenic receptor that responds selectively to an otherwise inert agonist. The ideal chemogenetic receptor construct would lack constitutive activity and would not be sensitive to endogenous neurotransmitters. Similarly, an ideal chemogenetic actuator ligand would be potent and selective for their chemogenetic effector and lack affinity for endogenous receptors to remain inert in the absence of chemogenetic receptors. Although some instances of constitutive activity have been observed and complete binding specificity has not yet been achieved (MacLaren et al., 2016; Saloman et al., 2016), several systems have demonstrated sufficient selectivity to allow effective chemogenetic applications (Armbruster et al., 2007; Magnus et al., 2019; Marchant et al., 2016; Vardy et al., 2015). These systems are well suited for sustained neuromodulation in freely moving subjects, with effects lasting hours and titratable depending on the chemogenetic compound dosage, route of administration, chemical half-life and duration of exposure (Grund et al., 2019; Magnus et al., 2019; Stachniak et al., 2014). Although these systems are ideal for use in untethered animal behavioral experiments, chemogenetic technologies lack the temporal control required to interrogate the neurobiological mechanisms of temporally precise brain functional behavioral couplings.
In contrast, optogenetic neuromodulation systems can be used to manipulate neuronal activity with exceptional temporal precision. These tools rely on the transduction and expression of light sensitive transmembrane opsins that open or close ion channels in response to light stimulation (Boyden et al., 2005; Zhang et al., 2006, 2007). When expressed in neurons, these transgenic opsins can promote or inhibit cell firing of action potentials, which can be scaled for spatiotemporally refined brain circuit manipulations. However, light delivery usually requires optical fiber implantation and tethering, which restricts the range of behavioral models that can be assessed during optogenetic manipulation of neuronal activity (Deisseroth, 2011; Tye and Deisseroth, 2012). In addition, overexpression of transgenic opsins can be damaging to cells and can affect endogenous receptors when light stimulation is administered at high intensities for long periods of time (Yizhar et al., 2011; Zimmermann et al., 2008). Moreover, the necessity of delivering light to target brain areas presents an obstacle for translational and clinical applications.
Despite their limitations, innovative applications of chemogenetic and optogenetic technologies have opened a new range of possibilities in preclinical research, and the potential for developing translational and clinical applications is becoming increasingly realizable. For example, chemogenetic and optogenetic tools have been proposed as potential treatments for sleep apnea (Curado et al., 2020), pain (Iyer et al., 2016; Weir et al., 2017), epilepsy (Kätzel et al., 2014), vision restoration (Gaub et al., 2015) and neurodegenerative disorders (Assaf and Schiller, 2019). However, new strategies for implementing and monitoring transgenic tools are needed for safe and effective use in humans. One major challenge is the fundamental need to track the location and function of chemogenetic receptors and opsins noninvasively and longitudinally in vivo, and we propose translational molecular imaging with positron emission tomography (PET) can be leveraged to safely monitor such transgenic constructs for this purpose. Recent studies demonstrate the use of PET for characterizing the functional effects of chemogenetic and optogenetic stimulation on brain activity, while development of selective PET-reporter molecules enable innocuous visualization of chemogenetic receptor location and occupancy (Bonaventura et al., 2019; Magnus et al., 2019; Nagai et al., 2020). In this review, we will summarize current research combining the use of transgenic tools with PET imaging for in vivo mapping and manipulation of brain circuits and outline future directions for translational applications.
2. Transgenic neuromodulation tools
2.1. Genetically engineered viral vector targeting
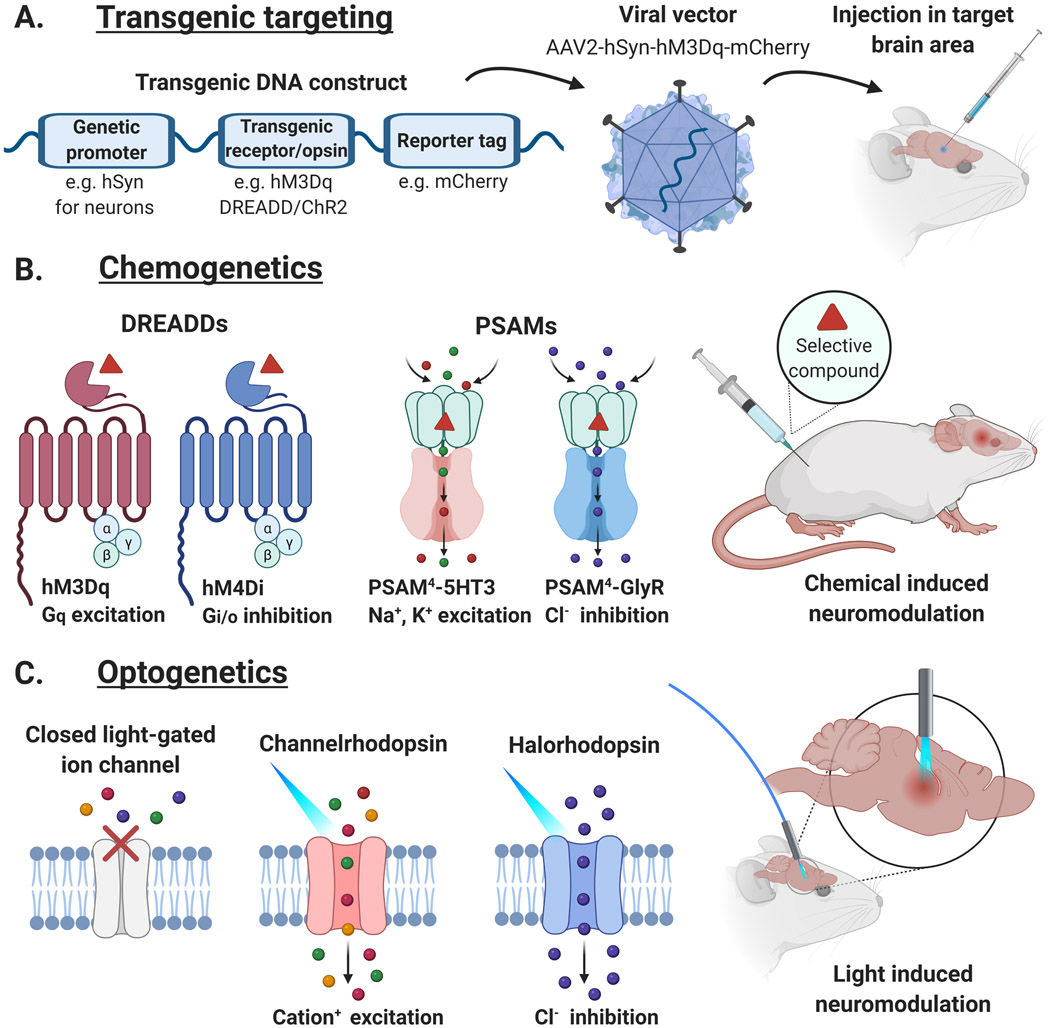
Recent advancements in genetic engineering have enabled the production of viral vectors carrying transgenes whose expression can be directed to specific neuronal populations and cell types, thereby enhancing the utility of transgenic neuromodulation tools (i.e., chemogenetics and optogenetics). Gene editing technologies allow precise engineering of DNA constructs that can be packaged into biological vectors (i.e., viral vectors with distinct serotypes and transduction properties) and delivered to target brain regions for transduction into cells (see Fig. 1A.). Most preclinical studies utilize recombinant adeno-associated viruses (AAVs) to package and deliver transgenes to cells because these vectors are relatively versatile and safe to use, although methods with lentiviral vectors have also been effectively implemented (Frecha et al., 2008; Taymans et al., 2007). Various targeting strategies can be used to direct transgene expression to specific cell populations based on anatomical projections, genetic identifiers, or inducible activation systems (Rothermel et al., 2013; Shevtosa et al., 2005; Tague et al., 2018). One commonly used approach is to employ distinct genetic promoter sequences as a practical and effective method for targeting specific cell types (Luo et al., 2008). Available promotor sequences include the human synapsin (hSyn) 1 promoter for pan-neuronal expression, CaMKII for targeting excitatory neurons, GAD67 and mDlx for GABAergic interneurons, and DRD1a for cells expressing dopamine D1 receptors (Dimidschstein et al., 2016; Durieux et al., 2011; Kügler et al., 2003; Watakabe et al., 2015).
Figure 1. Neuromodulation with chemogenetic and optogenetic tools.
A. Transgenic ssDNA combining a genetic promoter sequence (e.g., human synapsin 1 (hSyn) for neuronal expression) with transgenes for a neuromodulatory receptor/opsin (e.g., hM3Dq DREADD/Channelrhodopsin-2 (ChR2)) and reporter protein (e.g., mCherry fluorescent protein) is packaged into a viral vector (e.g., AAV serotype 2) and injected into a target brain area. B. Chemogenetic tools utilize genetically engineered receptors (e.g., DREADDs and PSAMs) that can alter brain activity following administration of a selective compound. C. Optogenetic tools rely on light-gated ion channels (e.g., channelrhodopsins and halorhodopsins) to modulate neuronal activity with light stimulation.
Additional strategies for targeted transgene expression include the use of inducible transcription systems and recombinase dependent systems (e.g., Cre-loxP, Dre-rox, Flp-recombinase). These systems can be employed through the development of transgenic animal lines such as mouse strains engineered to express a recombinase in specific cells. For example, in transgenic D1-Cre recombinase mice, an AAV containing DNA strands with two loxP sites flanking a gene of interest (e.g., chemogenetic or optogenetic transgenes) can be injected into a target brain region, such that expression of the transgene will only occur in cells that also express Cre recombinase under control of the dopamine D1 receptor promoter (Saunders et al., 2012). However, this approach is largely restricted to rodents because of breeding logistics necessary for developing transgenic animal lines.
Alternatively, inducible transcription systems can be applied through dual injection strategies such as the use of Cre recombinase and loxP containing vectors in multiple brain sites to target the projection areas of specific cell populations. For example, Oguchi et al., 2015 demonstrated a double injection technique with two different AAVs to express a transgene in the prefrontal network of macaques in a projection-specific manner. A local vector incorporated the Cre-On double floxed sequence while a retrograde vector containing Cre recombinase was injected at downstream axon terminals, therefore only cell bodies with projections to areas with the second vector expressed the given transgene. This latter intersectional approach is more widely applicable than the use of transgenic animal lines for translational purposes because it can be more readily implemented in non-transgenic animals and NHPs (O’Shea et al., 2018).
Nonetheless, conventional use of AAV vectors in the brain require intracranial injections, which are invasive and substantially restrict practical applications in humans. The development of AAVs capable of delivering genes through systemic or intravenous administration may provide a solution to this translational obstacle by circumventing the need for intracranial injections. Efforts are underway to develop new AAV capsid variants such as AAV-PHP capsids that can be used for noninvasive targeted gene delivery throughout the peripheral and central nervous system (Bedbrook et al., 2018; Challis et al., 2019; Chan et al., 2017). Altogether, genetic engineering technologies provide a wide array of targeted transduction approaches to express transgenic constructs (i.e., chemogenetic receptors and opsins) in specific cell populations and circuits, and advancements in this field aim to bolster the translational potential of neuromodulation with these tools.
2.2. Chemogenetic neuromodulation tools
The term chemogenetics encompasses the process of engineering macromolecules so they can interact with and respond to specific chemical stimuli. Chemogenetic neuromodulation techniques have mainly been used for interrogating the role of neuronal activity in preclinical research and are especially useful in animal behavioral models (Burnett and Krashes, 2016; Smith et al., 2016; Sternson and Roth, 2014). The most widely used chemogenetic tools rely on the transduction and expression of exogenous transgenic receptors (mostly G protein-coupled receptors (GPCRs) and ligand-gated ion channels (LGICs)) that can promote or inhibit neuronal activity upon administration of a highly potent and selective compound (Armbruster et al., 2007; Conklin et al., 2008; Wess et al., 2013). Although they lack the temporal control of other neuromodulation techniques such as optogenetics, these tools are well suited for sustained brain circuit modulations in freely behaving subjects (see Fig. 1B.).
One of the primary types of chemogenetic systems used in neuroscience research are designer receptors exclusively activated by designer drugs (DREADDs). This technology utilizes mutant versions of specific GPCRs that are activated by selective chemogenetic actuator ligands instead of endogenous ligands (Pei et al., 2008). Some DREADD systems utilize mutated human muscarinic acetylcholine receptors (Armbruster et al., 2007), while others adapt GPCRs such as opioid or adrenergic receptors (Conklin et al., 2008; Vardy et al., 2015). These tools can be used to excite or inhibit neuronal activity depending on the type of receptor-coupling used. For instance, activation of the hM3Dq receptor initiates Gq protein signaling which promotes intracellular calcium release and consequently increases neuronal activity. In contrast, the hM4Di receptor can decrease neuronal activity through the activation of inhibitory Gi/o proteins (Lee et al., 2014). DREADDs were first shown to be activated by clozapine-N-oxide (CNO) (Armbruster et al., 2007) and later in vivo by clozapine (an FDA approved drug) converted from the administered CNO (Gomez et al., 2017). Although multiple effective DREADD actuators exist, some have been shown to produce off-target effects at doses relevant for activating DREADDs (MacLaren et al., 2016), and therefore efforts to improve the potency and selectivity of DREADD ligands has been a continued focus in the field. A series of compounds for DREADDs was developed through testing the structure-activity relationships of CNO analogs such as C21 (Chen et al., 2015; Thompson et al., 2018), and various others have been identified as potent agonists with relatively favorable selectivities including deschloroclozapine (DCZ), fluorinated CNO analogs and the FDA approved drug olanzapine (Bonaventura et al., 2019; Nagai et al., 2020; Weston et al., 2019). Nonetheless, dose ranges need to be carefully considered to minimize off-target effects of such compounds, especially in NHPs (Upright and Baxter, 2020).
Aside from DREADDs, another form of chemogenetic system utilizes the ligand binding domain of the α7 nicotinic acetylcholine receptor (nAChR) coupled with the ion pore domains of other LGICs. These hybrid modular receptors are termed pharmacologically selective actuator modules (PSAMs), and they are activated through the binding of pharmacologically selective effector molecules (PSEMs) (Magnus et al., 2011). The latest generation of PSAMs (PSAM4) were recently developed through screening various mutations in the α7 nAChR ligand binding domain in search of enhanced channel interaction with the smoking cessation drug varenicline (Magnus et al., 2019). By combining these modified binding domains with various ion pore domains, chimeric LGICs were created to modulate neuronal activity in response to varenicline at concentrations much lower than the dosage used in clinical treatments targeting endogenous nicotinic receptors (Kaur et al., 2009; Rollema et al., 2010). The two most well characterized of these PSAMs, PSAM4-GlyR and PSAM4-5HT3, utilize the glycine receptor chloride channel (GlyR) and the serotonin cation channel (5-HT3R) and are intended to decrease or increase neuronal activity, respectively, although overall effects may vary if altering GABAergic inhibition (Magnus et al., 2019; Gantz et al., 2020). Like clozapine and olanzapine for DREADDs, the ability for selective activation of PSAMs using varenicline – an FDA approved drug – extends the translational potential of these chemogenetic tools for use in humans.
2.3. Optogenetic neuromodulation tools
In contrast to chemogenetic tools, optogenetic neuromodulation enables precise spatiotemporal manipulation of brain activity through the expression of transmembrane opsins with light sensitive ion channels that excite or inhibit neuronal cells in response to light (see Fig. 1C.) (Boyden et al., 2005; Zhang et al., 2006, 2007). To date, a variety of opsins with unique characteristics have been developed to permit excitation or inhibition of neuronal activity (e.g., channelrhodopsins and halorhodopsins). These opsins can be engineered to be permeable to different ions, distinct in their kinetics and sensitive to light at specific wavelengths (see reviews Duebel et al., 2015; Lin, 2011). Optogenetics can be used to interrogate precise neuronal and circuit functions with in vitro cells, in situ brain slices and in vivo experiments in anesthetized or awake behaving animals (Deisseroth, 2011; Mattis et al., 2012). Although optogenetic techniques allow excellent spatiotemporal control of neuronal manipulations, conventional light delivery usually necessitates hardware implantation and tethered animals. This restricts the range of applications that can be assessed during light-induced alterations in neuronal activity. In addition, the physical presence of implants or heat resulting from prolonged light stimulation can be damaging to cells (including those without opsins), and the need for targeted light delivery in brain tissue presents an obstacle for translational applications in larger subjects such as NHPs and in humans (Williams and Denison, 2013; Yizhar et al., 2011). Unsurprisingly, in vivo optogenetic technologies have predominately been developed in rodents thus far. Applications in NHPs have progressed more slowly due to a variety of practical obstacles, although a recently launched open resource platform should facilitate the ability of NHP researchers to compare and optimize methods (Bliss-Moreau et al., 2020; Tremblay et al., 2020). Despite the challenges, optogenetics has been effectively applied in numerous preclinical studies, and clinical use of opsins has already begun (Busskamp et al., 2012; RetroSense Clinical trial #NCT02556736). Continuing efforts are underway to improve translational potential by fine-tuning viral vector efficacy, enhancing the sensitivity of opsins at longer wavelengths, and developing wireless or skull permeant light sources (Büning and Srivastava, 2019; Chen et al., 2020; Wang et al., 2020).
2.4. Translational potential and obstacles for human applications
The rapid expansion and ongoing refinement of chemogenetic and optogenetic technologies presents an opportunity to develop transgenic neuromodulation applications for human therapies. Pathologies such as epilepsy, chronic pain, and Parkinson’s disease, which currently have poor therapeutic prognoses, are ideal candidates for such approaches (Alcacer et al., 2017; Iyer et al., 2016; Walker and Kullman, 2020). However, a major obstacle for practical use in translational models (i.e., NHPs) and in humans is the need to monitor the location and function of chemogenetic receptors and opsins in vivo noninvasively and longitudinally.
Conventional practices for confirming the expression of transgenic proteins in preclinical settings mainly rely on postmortem histology. When euthanasia is not desirable, such as in longitudinal experiments in NHPs, invasive in vivo techniques (e.g., calcium imaging, fiber photometry, implanted recording electrodes) can be used to indirectly assess transgene expression. However, aside from being intrusive, these indirect approaches are of low throughput and do not allow for assessing transgene expression directly, which is particularly important when studying novel transgenes or developing new vectors for transgene delivery. As such, the development of minimally invasive and longitudinal monitoring strategies is necessary to improve the translational potential and application of these tools in NHPs and humans. We suggest the versatile translational molecular imaging capabilities of PET may offer solutions to these challenges. Recent developments in the field demonstrate how PET imaging techniques can be leveraged to monitor chemogenetic receptor expression and function as well as functional effects of optogenetic stimulation for translational brain circuit mapping and manipulation. In the following sections, we describe various applications of PET imaging for in vivo mapping and manipulation of neuronal circuits with transgenic tools, highlighting recent advancements in the field that may facilitate chemogenetic and optogenetic applications in translational models and potentially human therapies.
3. PET applications for brain circuit mapping
3.1. Overview of PET imaging
PET is a translational molecular imaging modality with a diverse range of applications in neuroscience and medicine (Zimmer and Luxen, 2012). PET imaging relies on the injection of biologically relevant radiolabeled compounds (radiotracers) that produce gamma ray photons upon radioactive decay. When positrons emitted by the radiotracer collide with electrons in annihilation events, pairs of 511 keV photons are released at ~180° angles and detected by an array of photodetectors surrounding the subject, and these events can be computed to reconstruct three-dimensional volumetric and dynamic parametric images (Saha, 2005). The translational and repeatable nature of PET along with the extensive variety of compounds amenable for radiolabeling makes it a uniquely versatile tool for imaging neurobiological functions in vivo. However, radioactive decay of PET radiotracers necessitates their production be near the location of use, and therefore applications are restricted to select facilities. Radionuclides are primarily generated in particle accelerators called cyclotrons, and PET radiotracers typically incorporate [15O], [13N], [11C], or [18F], which are radioisotopes with half-lives of ~2-min, 10-min, 20-min, and 110-min, respectively. To date, over 200 PET radiotracers have been described for various types of brain imaging, with radiolabeled compounds available for examining brain activity patterns, neurotransmitter systems, neuroinflammation and other biomarkers used to characterize brain function and disease (Zimmer and Luxen, 2012), (see Table 1). The extensive utility of PET brain imaging applications is made possible by the existence of radiotracers with diverse chemical structures and biological relevance. The most optimal radiotracers have good blood-brain barrier penetration and exhibit high affinity and selectivity for their neurobiological targets. In addition, there are a variety of experimental designs and analytic approaches to interpret PET data in biologically meaningful ways, such as radioligand displacement (which can be used as a measure of receptor occupancy or neurotransmitter release) and a variety of kinetic modeling techniques for quantitative and parametric assessments. Overall, the adaptability of PET applications offers a unique and powerful approach for translational molecular imaging in the brain.
Table 1: PET applications for molecular brain imaging and examples of various PET radiotracers.
Brain activity: [15O] water for cerebral blood flow (Zhang et al., 2014) and [18F] fluorodeoxyglucose (FDG) for neurometabolic glucose use (Villien et al., 2014). Neurotransmitter receptors: Dopamine D2/D3 antagonists [11C]raclopride (Volkow et al., 1994) and [18F]fallypride (Slifstein et al., 2010). Serotonin 5- HT2A agonist [11C]Cimbi-36 (Ettrup et al., 2014). Neuroinflammation: Translocator protein (TSPO) tracer [18F]GE-180 (Vomacka et al., 2017). Neurodegeneration: Neurofibrillary tau protein tracer [18F]MK-6240 (Hostetler et al., 2016). Neuro-oncology: [18F]fluoroethyltyrosine (FET) and [18F]FDG for assessing brain tumors (Pauleit et al., 2009). Drug target engagement: Radiolabeled [11C]psilocin (Amatamey et al., 1998). Psilocybin displacement of [11C] Cimbi-36 at 5-HT2A receptors (Madsen et al., 2019). Tracking transgene expression: Enzyme-based HSV1-tk reporter [124I]2′-fluoro-2′-deoxy-1-β-D-arabinofuranosyl-5-iodouracil (FIAU) (Tjuvajev et al., 2002). Receptor based dopamine D2 reporter [18F]fluoroethylspiperone (FESP) (Liang et al., 2001).
| Applications | Description | Radiotracer examples |
|---|---|---|
| Brain activity | Measure cerebral blood flow or glucose metabolism as a proxy for brain activity | [15O] water (cerebral blood flow), [18F]FDG (glucose metabolism) |
| Neurotransmitter receptors and transporters | Evaluate neurotransmitter receptor availability and release using radioligands selective for endogenous receptors and transporters | Dopamine: [11C]raclopride and [18F]fallypride Serotonin (5-HT): [11C]Cimbi-36 |
| Neuroinflammation and Neurodegeneration | Assess neuroinflammation and neurodegeneration using selective radioligands to probe relevant biomarker proteins | Translocator protein (TSPO) tracers: [18F]GE-180 Neurofibrillary tau protein: [18F]MK-6240 |
| Neuro-oncology | Diagnose and monitor cancer in peripheral and central nervous system using relevant biomarkers | [18F] FET for tumors, [18F] FDG for abnormal glucose metabolism |
| Drug target engagement | Evaluate binding of radiolabeled drugs or blockade/displacement of receptor selective radioligands | Radiolabeled [11C]psilocin or displacement of [11C]Cimbi-36 at 5-HT2A receptors |
| Tracking transgene expression | Track transgene delivery and expression by co-packaging with PET-compatible reporter genes | Enzyme based: HSV1-tk reporter with [124I]FIAU. Receptor based: Dopamine D2 receptor reporter with [18F]FESP |
3.2. Measuring brain activity with FDG-PET
Two general approaches have been developed to utilize PET for measuring brain activity. These approaches take advantage of the oxidative energy demands required by ion pumps to restore and maintain ion gradients following neuronal activation. One of these methods relies on infusion of [15O]-labeled H2O to measure cerebral blood flow as a proxy for neuronal activity. The rapid half-life (~2-min) of the radiotracer enables multiple acquisition periods during a scan session, but it also limits scanning duration and restricts use to facilities with direct cyclotron access. Although useful in certain scenarios, this method has largely been replaced by functional magnetic resonance imaging (fMRI), which typically utilizes changes in blood oxygenation level dependent (BOLD) signal to characterize brain activity with better spatial and temporal resolution.
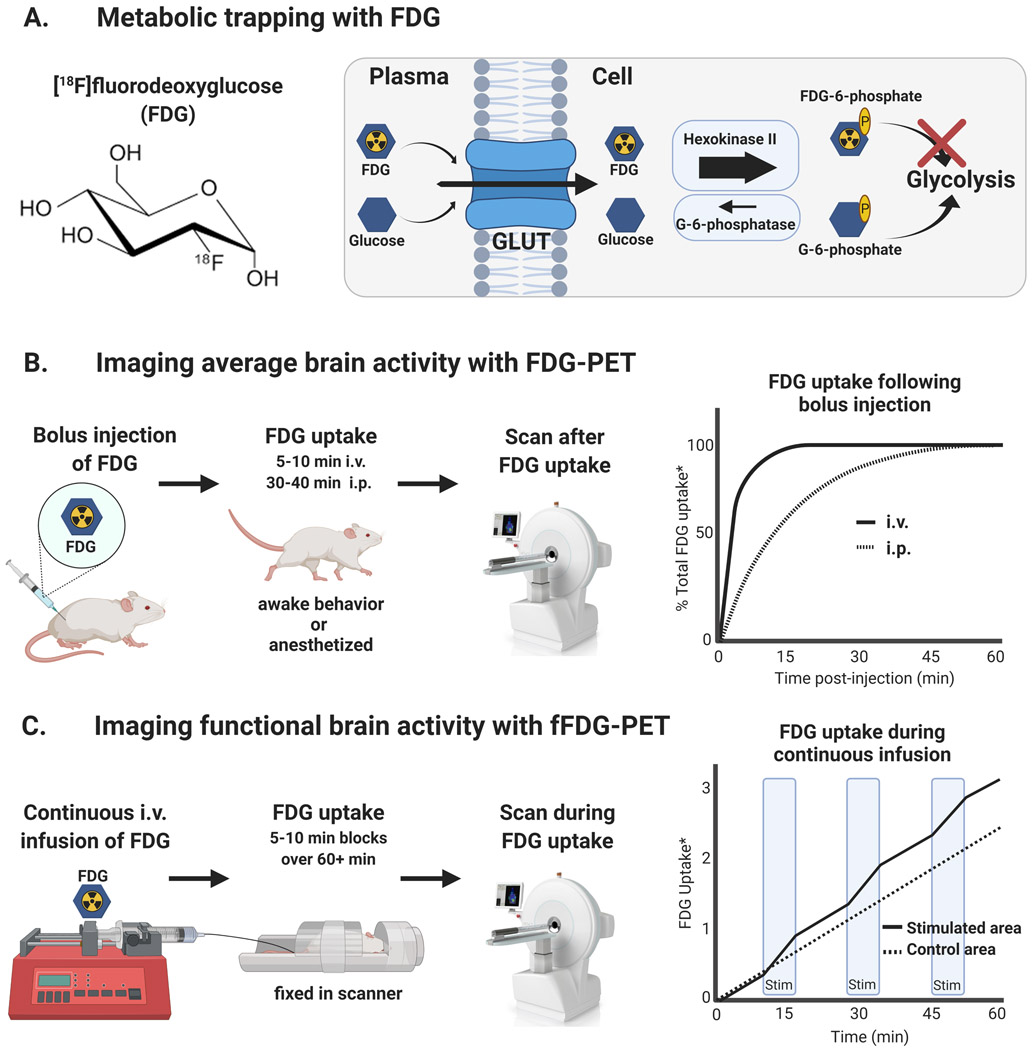
Aside from cerebral blood flow measures, brain activity can also be characterized using readout measures of glucose metabolism. The radiofluorinated glucose analog [18F]2-fluoro-2-deoxy-D-glucose (FDG) is among the most widely used radiotracers in clinical PET imaging with multiple indications (e.g., monitoring cancer and neuroinflammation) (Galldiks et al., 2019), and it can also be leveraged as a proxy measure of brain activity. Seminal autoradiography studies with the analogous [14C]-labeled and tritiated ([3H]) 2-deoxy-D-glucose (2-DG) (Alexander et al., 1981; Sokoloff et al., 1977) paved the way for the development of FDG-PET brain imaging techniques. These studies established a close link between glucose metabolism and brain activity, as ion pumps depend on oxidative metabolism of glucose to restore and maintain ion gradients following neuronal activation. Just as in the original 2-DG method, FDG is transported into active cells via glucose transporters (GLUT) and is then used as a readout measure of glucose metabolism (Reivich et al., 1979). After undergoing initial phosphorylation by hexokinase II, the converted FDG-6-phosphate cannot move on through glycolysis and is temporarily trapped inside cells (see Fig. 2A.). Importantly, this process of metabolic trapping allows brain activity to be imaged after it occurs as a cumulative average, which enables measuring whole-brain activity in awake and freely moving subjects. The FDG-6-phopshate undergoes radioactive decay and is eventually metabolized, allowing subjects to be imaged in a repeated and longitudinal manner. Standard uptake values (SUVs) normalizing for injected dose and body weight are often used to quantify FDG uptake, especially when serum concentration data are unavailable (Huang, 2000). In addition, PET based kinetic modeling of FDG uptake (e.g., arterial two-compartment or reference tissue modeling) enables quantitative characterizations of glucose transport and metabolism (Bentourkia and Zaidi, 2007).
Figure 2. FDG-PET for measuring brain activity.
A. Radiofluorinated glucose analog [18F]fluorodeoxyglucose (FDG) is taken up by cells through glucose transporters (GLUT) and trapped in active brain regions. B. Bolus injection of FDG can be used to image average brain activity after it occurs in anesthetized or awake and behaving animals (uptake period 5–10 min intravenous or 30–40 min intraperitoneal) (Aarons et al., 2012). C. Continuous infusion of FDG can be used to measure brain activity over longer durations with improved temporal resolution in functional FDG-PET (fFDG-PET) (Villien et al., 2014). *Model example data of FDG uptake for illustrative purposes.
The rate of FDG uptake into the brain is dependent on the route of radiotracer administration (i.e., intraperitoneal- i.p. or intravenous- i.v., bolus or continuous; see Fig. 2B-C.). Bolus injection of FDG can provide a cumulative snapshot of brain activity corresponding to the rate of local radiotracer uptake, and it is suited for assessing brain activity during freely moving behavior because brain activity can be imaged after it occurs. Intraperitoneal administration will result in a longer uptake period (30-40 min) than an i.v. injection (5-10 min) (Aarons et al., 2012). Thus, the route of administration can be adapted to obtain the temporal resolution necessary to examine the effect of interest. Although bolus techniques have the advantage of being able to image brain activity after it has occurred, they can only provide a measure of the average brain activity level that occurred during the 5-40 min uptake period. Alternatively, a continuous infusion of FDG can be administered throughout the scanning period to enable dynamic scans which are reconstructed as a series of snapshots to improve temporal resolution (e. g., 5-min time bins vs. 20-min average) and to extend uptake periods (40+ min). This method of continuous infusion has been referred to as functional FDG-PET (fFDG-PET) and can be used to image changes in brain activity throughout the scanning session (Villien et al., 2014), (see model data in Fig. 2C.). However, even with continuous fFDG-PET the temporal resolution (minutes) is limited in comparison to other whole-brain imaging methods (seconds/milliseconds) (e.g., fMRI, MEG, EEG), but new and continuing developments in scanner technology have enabled simultaneous use of PET with other imaging modalities such as fMRI to improve the interpretability of brain activity measurements (Musafargani et al., 2018; Sander et al., 2020).
3.3. Metabolic circuit mapping using FDG-PET with transgenic tools
FDG-PET can be applied as an effectively noninvasive approach for assessing the in vivo performance of chemogenetic and optogenetic constructs and their functional effects on brain activity patterns. The FDG radiotracer can be injected following the administration of a chemogenetic ligand or during light pulses in anesthetized or awake behaving animals to generate whole-brain metabolic maps of cell-specific functional circuits. One such strategy has been termed DREADD-assisted metabolic mapping (DREAMM) and can be used to derive functional brain maps induced by chemogenetic manipulation of distinct cell populations and circuits (Anderson et al., 2013; Klawonn et al., 2018; Mazzone et al., 2018; Michaelides et al., 2013; Michaelides and Hurd, 2015; Urban et al., 2015). For example, DREAMM has been applied to evaluate whole-brain effects of hM4Di activation in prodynorphin-expressing neurons in the rat periamygdaloid cortex and prodynorphin- and proenkephalin-expressing medium spiny neurons in the nucleus accumbens shell (Anderson et al., 2013; Michaelides et al., 2013). In one of these studies (Michaelides et al., 2013), it was shown that distinct brain networks were activated during conditions of awake vs. anesthetized chemogenetic activation. During anesthetized FDG uptake, chemogenetic activation led to changes in FDG accumulation in brain regions that were more proximally related to the anatomical circuits being manipulated, whereas during awake FDG uptake, chemogenetic activation led to changes in FDG accumulation in brain regions that extended beyond the anatomical circuit manipulated and were recruited as a function of the chemogenetic induced behavioral effects. These observations emphasize both the utility of this functional imaging approach as well as the importance of considering such factors in experimental design and interpretation when using FDG-PET (Cho et al., 2020; Michaelides et al., 2013).
FDG-PET has also been used to assess the in vivo efficacy of newly developed DREADD ligands. For example, FDG-PET was performed following i.p. administration of the fluorinated clozapine analog JHU37160 (J60) in transgenic D1-Cre mice expressing DREADDs under the control of the dopamine D1 receptor promoter. These scans showed distinct brain activation profiles of hM4Di and hM3Dq in DREADD- expressing mice compared to wildtype mice at doses of 0.1 mg/kg (Bonaventura et al., 2019). In another recent study, systemic injection of the highly potent DREADD actuator DCZ in NHPs with unilateral expression of hM3Dq in the amygdala produced dose-dependent increases of FDG uptake in this brain region with effects observed at doses as low as 1 μg/kg (Nagai et al., 2020). Importantly, this effect was not observed in the contralateral amygdala or in animals lacking hM3Dq, demonstrating the selectivity of DCZ by inducing activation only in regions expressing DREADDs.
Aside from imaging brain responses to chemogenetic neuromodulation, FDG-PET can also be combined with optogenetic neuromodulation to interrogate functional activation patterns among specific brain regions. This has been demonstrated in characterizing responses to optogenetic stimulation of neurons in the nucleus accumbens (NAc) of awake rats (Thanos et al., 2013). Increases in FDG uptake indicated enhanced activity in the stimulated region as well as the ipsilateral striatum, somatosensory cortex, periaqueductal gray (PAG), and multiple contralateral sites which were consistent with areas showing increased c-Fos expression. Decreased uptake in the retrosplenial cortex, cingulate gyrus and secondary motor cortex was also observed. In another study, FDG-PET was used to evaluate neurofunctional changes before and after optogenetic stimulation of the dorsal PAG (dPAG), a region implicated in the generation of panic disorders (He et al., 2019). Optogenetic stimulation of excitatory neurons in the dPAG of rats resulted in post-stimulation increases of FDG uptake in the dPAG and downstream areas such as the cingulate cortex, cerebellar lobule, and the septohypothalamic nucleus. Decreases in FDG uptake were also observed in the basal ganglia, frontal cortex, primary somatosensory and motor cortex post-stimulation (He et al., 2019). Overall, these studies demonstrate the utility of FDG-PET for validating the function of transgenic neuromodulation tools and for whole-brain metabolic mapping of cell-type specific functional networks.
3.4. Mapping DREADD expression with PET-reporters
Transgenic tools cannot be employed confidently without verifying their anatomical location and function. In most preclinical studies using chemogenetics or optogenetics, the location of transgene expression is verified through postmortem histology techniques and usually necessitates the inclusion of a gene for a reporter protein into the AAV-transgene construct (e.g., epitope tags or fluorescent proteins) which is transduced in addition to the transgenic receptor or opsin. Although usually not problematic, reporter proteins have the potential to compromise the function of the neuromodulatory protein and the viability of the cells that express it, underscoring the importance of functional validation and longitudinal monitoring (Galvan et al., 2019). In addition to genetic reporters for postmortem histology, in vivo functional measurements using technologies such as photometric biosensors, fiber photometry and electrophysiological recordings can be used for indirect monitoring of chemogenetic receptors and neuromodulatory opsins. However, these techniques are cumbersome and invasive, and they would not be widely adaptable for clinical applications. Additionally, they rely on expected functional effects and do not directly measure the presence of chemogenetic receptors or transgenic opsins.
A promising alternative strategy has emerged for direct and effectively noninvasive visualization of chemogenetic receptors. Since chemogenetic tools are based on receptor-binding ligands, using radiolabeled PET ligands that are selective and of high affinity for chemogenetic receptors can be suitable for in vivo mapping of chemogenetic receptor expression and the brain circuit(s) they occupy. One of the first demonstrations of this used [11C] radiolabeled clozapine-N-oxide (CNO) to localize DREADDs in mice expressing hM4Di (Ji et al., 2016), but low brain penetrance and affinity of CNO limits the sensitivity of this approach (Gomez et al., 2017; Raper et al., 2017). DREADDs have also been successfully visualized using [11C]clozapine and [11C]DCZ in rodents and NHPs (Gomez et al., 2017; Nagai et al., 2016; Nagai et al., 2020). While effective, these radiotracers have a relatively short half-life as they possess the 11C radionuclide (~20-min), which makes their application impractical at institutions without direct cyclotron access for radionuclide production. Although most clinical PET centers have cyclotron access, DREADD radioligands with longer half-lives could enhance accessibility for preclinical research labs and improve the ease of use in clinical settings.
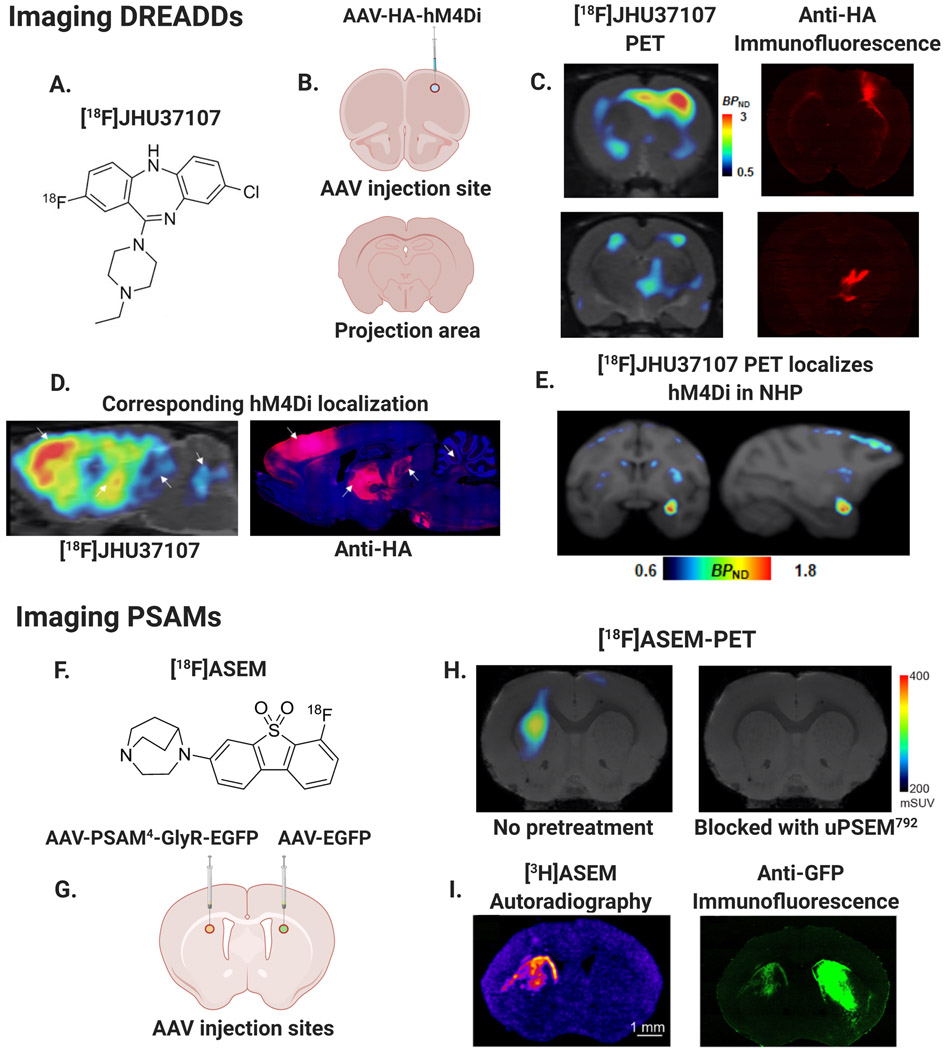
Therefore, to expand access to this approach, there is a push to develop selective fluorinated DREADD ligands that can be radiolabeled with 18F for a longer half-life (~110-min). For example, fluorinated compounds based on potent clozapine analogs (Chen et al., 2015) have recently been developed (Bonaventura et al., 2019; Hu et al., 2020), and of these compounds, the first high affinity and selective 18F labeled DREADD ligand – [18F]JHU37107 – was shown to successfully localize hM3Dq and hM4Di in rodents and NHPs (see Fig. 3A-E.). The use of [18F] JHU37107 to detect DREADD expression was tested in rats with unilateral expression of hM3Dq or hM4Di in the right motor cortex. DREADD specific binding of [18F]JHU37107 was observed both at the local AAV injection site and known anatomical projection sites including the contralateral motor cortex, striatum and motor thalamus (Fig. 3B-D.). This pattern was consistent with results from postmortem immunohistochemistry in brain slices (Fig. 3C-D.). In addition, a rhesus macaque expressing hM4Di in the right amygdala was imaged with [18F] JHU37107, and specific labelling of hM4Di receptors was observed unilaterally in this region (Fig. 3E.). This body of work demonstrated the first ever detection of DREADDs using an 18F-labled PET ligand in rodents and NHPs.
Fig. 3. PET-reporters for in vivo mapping of chemogenetic receptor expression.
A–E. Mapping DREADDs in rats and NHPs (modified from Bonaventura et al., 2019): A.) Chemical structure of DREADD radioligand [18F]-JHU37107. B.) AAV-HA-hM4Di injection site in rat right motor cortex (M1) and non-injected projection area. C.) Left: [18F]-JHU37107 PET reveals hM4Di expression at M1 injection site and putative projection sites, Right: Histological confirmation of hM4Di expression with hemagglutinin (HA) antibody immunofluorescence. D.) Anatomical localization of hM4Di from [18F]-JHU37107 PET in rats coincides with histological expression patterns of the HA-tagged DREADDs, arrows show corresponding anatomical sites of right M1 and motor cortical projection areas. E.) Localization of hM4Di in rhesus nonhuman primate (NHP) right amygdala with [18F]-JHU37107. F–I. Mapping PSAMs in mice (modified from Magnus et al., 2019): F.) Chemical structure of FDA approved radiotracer [18F]-ASEM capable of imaging PSAMs. G.) Schematic of AAV injection sites in mouse left dorsal striatum (AAV-PSAM4-GlyR-EGFP) and right dorsal striatum (AAV-EGFP control). H.) Left: [18F]-ASEM-PET showing left striatal PSAM4-GlyR expression, Right: [18F]-ASEM blocked following uPSEM792 pretreatment (1 mg/kg). I.) Left: Autoradiography of [3H]-ASEM binding in left dorsal striatum, Right: Immunofluorescence with anti-green fluorescent protein (anti- GFP) antibody showing PSAM4-GlyR-EGFP expression in left striatum and EGFP control in right striatum. Lack of PET and autoradiography binding in right striatum demonstrates specificity of ASEM for PSAM4-GlyR.
Aside from mapping the anatomical location of DREADD receptors, PET studies with radiotracers like [11C]clozapine, [18F]JHU37107 and [11C]DCZ enable validation of the in vivo target engagement of selective ligands at DREADD receptors through competitive binding experiments. For instance, dose-dependent reductions in [11C]clozapine occupancy of DREADDs were observed following pretreatment with various doses of CNO in NHPs expressing hM4Di in the rostromedial caudate (Nagai et al., 2016). This type of approach can facilitate the development of novel chemogenetic ligands by providing a metric for comparing the in vivo receptor occupancies of different actuator compounds. For example, [11C]clozapine-PET was used to quantify DREADD occupancies of fluorinated clozapine analogs JHU37152 (J52) and JHU37160 (J60) in comparison with the DREADD agonist C21 in rodents and NHPs (Bonaventura et al., 2019). Treatment with J52 and J60 occupied DREADDs (decreased [11C]clozapine binding) at doses 10-100 fold lower than those required by C21 (i.e., 0.1 mg/kg vs. 1-10 mg/kg), which was consistent with the superior potencies of J52 and J60 for producing DREADD-specific behavioral effects. The potent receptor occupancy of J60 was also confirmed by blocking the binding of [18F] JHU37107 with a 0.1 mg/kg dose of J60 in transgenic DREADD- expressing mice (Bonaventura et al., 2019; Hu et al., 2020). In another recent study, a highly potent clozapine analog − DCZ − was shown to exhibit greater selectivity for DREADDs (lower affinity for endogenous receptors) compared to clozapine in vitro, and was subsequently radiolabeled with [11C] to systematically compare dose-dependent hM4Di receptor occupancies of CNO, C21 and DCZ using [11C]DCZ-PET in NHPs (Nagai et al., 2020). The dose of DCZ required to reach 50% DREADD occupancy in vivo was ~20-fold lower than CNO and ~60-fold lower than C21. This was consistent with DCZ’s superior in vitro affinity and in vivo agonistic potency (~100-fold) compared to CNO and C21 and demonstrates that DCZ retains the high affinity and potency of clozapine while improving selectivity for DREADDs. Overall, these studies illustrate the utility of PET for monitoring DREADD expression at local and anatomical projection sites as well as comparing DREADD occupancy by different chemogenetic actuators.
3.5. Mapping PSAM4 expression
Pharmacologically selective actuator modules, or PSAMs, are another form of chemogenetic receptors with promising translational potential. As described in Section 2.2, they were developed by combining mutated versions of the α7 nAChR ligand binding domain with different ion pore domains. Like DREADDs, clinical use of PSAMs will require a way to track the expression of the transgenic receptors in vivo innocuously and longitudinally. An FDA approved PET ligand has recently been developed for visualizing α7 nAChRs in humans, namely the radiolabeled α7 nAChR antagonist [18F]ASEM (Horti et al., 2014). Just like [18F]JHU37107 and [11C]DCZ for DREADDs, [18F]ASEM can be used to localize the expression of PSAMs in rodents (Fig. 3F-I.) and potentially in NHPs and humans. For example, Magnus et al. (2019) demonstrated the use of [18F]-ASEM PET to visualize PSAM4-GlyR expression unilaterally in the mouse striatum. Expression areas identified by [18F]-ASEM PET in mice were consistent with expression areas of PSAM4-GlyR-IRES-EGFP in left dorsal striatum confirmed through [3H] ASEM autoradiography and ex vivo fluorescence microscopy in striatal brain slices (Fig. 3H-I.). Competitive binding PET experiments with [18F]ASEM were also used to characterize brain penetrance and PSAM4 receptor occupancy of newly developed ultrapotent PSEMs (uPSEMs) in vivo. [18F]ASEM binding was blocked by pretreatment with some of these novel uPSEMs including uPSEM792 and uPSEM817 at doses ranging from 0.3-1 mg/kg (Fig. 3H.). In contrast to [18F]JHU37107 and [11C] DCZ for DREADDs, the fact that [18F]ASEM has already been FDA approved for use as a PET radiotracer in humans makes it readily suitable for translational and clinical applications.
3.6. Other PET-reporters for mapping transgene expression
As described above, the use of selective PET-reporter compounds to image chemogenetic receptors in vivo is an effective technique for visualizing DREADD and PSAM expression and characterizing neuroanatomical projections in rodents and NHPs. However, not all transgenic tools have an available binding domain accessible to PET radiotracers. Therefore, there is a need to adapt other types of PET-reporter systems to monitor transgenic neuromodulatory proteins such as opsins.
A variety of PET-compatible transgene reporter systems have been developed over the past two decades with motivations for potential gene therapy applications. These systems typically utilize a reporter gene to express an enzyme, transporter protein or receptor that can selectively entrap or bind a specific PET radiotracer (Yaghoubi et al., 2012). The reporter genes are intended to be co-packaged with therapeutic transgenes and therefore provide a proxy confirmation of therapeutic transgene expression. One of the earliest enzyme-based transgene reporter systems was developed using herpes simplex virus thymidine kinase (HSV1-tk) in combination with radiolabeled substrates such as [124I]2′- fluoro-2′-deoxy-1-β-D-arabinofuranosyl-5-iodouracil (FIAU), which accumulates in cells expressing the transgene and can then be visualized with PET (Blasberg and Tjuvajev, 1999; Tjuvajev et al., 2002). The norepinephrine transporter has also been utilized as a reporter gene in combination with [131I]metaiodobenzylguanidine (Anton et al., 2004), and one of the first receptor-based transgene reporter systems was developed using a mutated dopamine D2 receptor in combination with [18F]-fluoroethylsisperone (FESP) (Liang et al., 2001). More recently, a PET reporter system using the PKM2 gene with the radiotracer [18F] DASA-23 was demonstrated in mice transfected using AAV9 (Haywood et al., 2019). Additionally, E. coli dihydrofolate reductase (ecDHFR) was utilized by Shimojo et al. (2020) for mapping transgenic protein expression in rodents and marmosets with radiolabeled analogs of the ecDHFR antagonist trimethoprim (i.e., [18F]FE-TMP). Importantly, they showed the ecDHFR-TMP system can be used in combination with transgenic neuromodulation tools such as DREADDs. Approaches like these can provide a proxy confirmation of the co-packaged transgene expression and can be adapted for identifying cells expressing a wide variety of transgenic constructs and neuromodulatory proteins.
Optogenetic technologies could greatly benefit from PET-compatible transgene reporters, because unlike chemogenetic receptors (which inherently possess a selective binding site), there is currently no available method to directly monitor opsin expression in a noninvasive and longitudinal manner. This limitation hinders the translational applications of optogenetics in NHPs and potential human applications because expression of opsins cannot be confirmed or localized in vivo, and therefore cannot be confidently used to manipulate discrete neuronal populations and projection sites without invasive or ex vivo procedures. Consequently, this presents an obstacle for clinical applications of optogenetics where medical decisions and patient outcomes would undoubtedly benefit from innocuous longitudinal tracking of opsin expression in targeted areas.
4. Discussion and Conclusions
4.1. Improving the translational capacity of transgenic tools
Modern advancements in transgenic neuromodulation technologies have revolutionized neuroscience research and clinical ambitions for gene therapies. However, to improve the translational capacity of these tools (i.e., chemogenetics and optogenetics), noninvasive methods for in vivo longitudinal monitoring are needed. The use of PET-compatible reporters is a promising strategy adaptable for this purpose, as selective radiotracers have been demonstrated to effectively localize chemogenetic receptors and other transgenic proteins in preclinical studies. Universal PET-reporter systems are necessary to expand translational applications of optogenetics and other transgenic technologies where exogenous proteins are introduced (e.g., CAR-T cells). Although PET imaging places some radioactive burden on subjects (albeit clinically safe), ongoing technological advancements in PET instrumentation are expected to increase the spatial resolution and sensitivity of this technique and therefore reduce subject radiation exposure by permitting lower doses of radiotracer (Hutton et al., 2018). Furthermore, the combination of PET with other modalities like MRI will enhance their utility for interrogating structural and functional neurobiology (Musafargani et al., 2018; Sander et al., 2020).
In addition to PET monitoring methods, the ability to safely deliver AAVs with high efficacy and precision is necessary for translational applications, and this will require innovations in genetic targeting technologies. Even with noninvasive methods for tracking transgenic tools in vivo, the challenge of transgene delivery remains, which in current practice requires intracranial injection of viral vectors into target brain regions. To overcome this challenge, efforts are underway to develop vectors that can be delivered systemically through intravenous injection and will pass through the blood-brain barrier to express in target regions of the brain. AAV capsid variants such as AAV-PHPs have been demonstrated to facilitate noninvasive, efficient, and trackable targeted gene delivery in rodent models (Challis et al., 2019; Seo et al., 2020). However, the efficacy of this technology has been shown to be strain-dependent in rodents and largely ineffective in NHPs, underscoring the need for further development in this area (Huang et al., 2019; Matsuzaki et al., 2018).
Another hurdle specific to practical applications of optogenetics is the challenge of delivering light to target areas. The development of new opsins with heightened sensitivity, especially at longer wavelengths (e. g., ChRmine), may provide some improvement because these wavelengths can penetrate deeper into tissue from an exogenous source (Chen et al., 2020; Duebel et al., 2015). Additionally, technologies are being developed to enable wireless and skull permeant light delivery into target brain areas (Kim et al., 2021; Wu et al., 2019). Although still an invasive technology, these advances may broaden the potential for translational optogenetic applications (Wang et al., 2020).
4.2. Conclusions
In this review, we summarized research demonstrating the utility of combining PET imaging with transgenic tools for in vivo mapping and manipulation of neuronal populations and circuits. We highlighted recent advances that improve translational applications of chemogenetics and optogenetics, and we propose PET as an ideal imaging modality for innocuous longitudinal tracking of transgenic constructs (i.e., chemogenetic receptors and neuromodulatory opsins). We also identified key domains where requisite innovations are needed to further develop these technologies in ways that enable safe and effective use for translational research applications and potentially human therapies (e.g., epilepsy, pain, vision restoration, Parkinson’s disease).
Highlights.
Transgenic neuromodulation tools enable targeted control of neuronal populations
Chemogenetics and optogenetics have translational utility and therapeutic potential
Translational and human applications require noninvasive longitudinal monitoring
PET enables in vivo imaging of chemogenetic receptor location and function
PET-reporter systems should be adapted to visualize expression of transgenic opsins
Acknowledgments
This work was funded in part by the National Institute on Drug Abuse Intramural Research Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: None
Figures 1-3 were created using BioRender.com.
Figure 3 was modified with permissions from Bonaventura et al. (2019) and Magnus et al. (2019).
References
- Aarons AR, Talan A, Schiffer WK (2012) Experimental Protocols for Behavioral Imaging: Seeing Animal Models of Drug Abuse in a New Light. In: Carter C, Dalley J (eds) Brain Imaging in Behavioral Neuroscience. Current Topics in Behavioral Neurosciences, vol 11. Springer, Berlin, Heidelberg, 10.1007/7854_2012_206 [DOI] [PubMed] [Google Scholar]
- Alcacer C, Andreoli L, Sebastianutto I, et al. (2017). Chemogenetic stimulation of striatal projection neurons modulates responses to Parkinson’s disease therapy. J Clin Invest. 127(2):720–734. 10.1172/JCI90132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander GM, Schwartzman RJ, Bell RD, 1981. Quantitative measurement of local cerebral metabolic rate for glucose utilizing tritiated 2-deoxyglucose. Brain Res. 223 (1), 59–67. 10.1016/0006-8993(81)90806-4. [DOI] [PubMed] [Google Scholar]
- Amatamey S, Vollenweider FX, Patt J, et al. (1998). 11C-Radiolabeling of hallucinogenic psilocin, a potential radioligand for studying the role of serotonin receptors in psychotic symptom formation. J. Labelled Cpd. Radiopharm, 41(7), 585–594. [DOI] [Google Scholar]
- Anderson SAR, Michaelides M, Zarnegar P, et al. (2013). Impaired periamygdaloid-cortex prodynorphin is characteristic of opiate addiction and depression. J Clin Invest. 123(12):5334–5341. 10.1172/JCI70395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton M, Wagner B, Haubner R, et al. (2004). Use of the norepinephrine transporter as a reporter gene for non-invasive imaging of genetically modified cells. J Genet Med. 6(1): 119–126. 10.1002/jgm.472 [DOI] [PubMed] [Google Scholar]
- Armbruster BN, Li X, Pausch MH, et al. (2007). Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. PNAS 104(12): 5163–5168 10.1073/pnas.0700293104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf F and Schiller Y (2019), A chemogenetic approach for treating experimental Parkinson's disease. Mov Disord., 34: 469–479. 10.1002/mds.27554 [DOI] [PubMed] [Google Scholar]
- Bedbrook CN, Deverman BE & Gradinaru V (2018) Viral strategies for targeting the central and peripheral nervous systems. Annu. Rev. Neurosci 41, 323–348. 10.1146/annurev-neuro-080317-062048 [DOI] [PubMed] [Google Scholar]
- Bentourkia M and Zaidi H (2007). Tracer kinetic modeling in PET. PET Clinics 2(2): 267–277. 10.1016/j.cpet.2007.08.003 [DOI] [PubMed] [Google Scholar]
- BioRender.com. Figures 1, 2 and 3 were created with BioRender.com
- Blasberg RG and Tjuvajev JG (1999). Herpes simplex virus thymidine kinase as a marker/reporter gene for PET imaging of gene therapy. Q J Nucl Med. 43(2):163–9. PMID: 10429512. https://pubmed.ncbi.nlm.nih.gov/10429512/ [PubMed] [Google Scholar]
- Bliss-Moreau E, Costa VD, Baxter MG (2020). A pragmatic reevaluation of the efficacy of nonhuman primate optogenetics. bioRxiv 2020.12.10.420331. 10.1101/2020.12.10.420331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventura J, Eldridge MAG, Hu F, et al. (2019). High-potency ligands for DREADD imaging and activation in rodents and monkeys. Nature Communications 10: 4627. 10.1038/s41467-019-12236-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, et al. (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neuroscience 8(9): 1263–1268. 10.1038/nn1525 [DOI] [PubMed] [Google Scholar]
- Büning H and Srivastava A (2019). Capsid modifications for targeting and improving the efficacy of AAV vectors. Molecular Therapy: Methods & Clinical Development, 12, 248–265. 10.1016/j.omtm.2019.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett CJ and Krashes MJ (2016). Resolving Behavioral Output via Chemogenetic Designer Receptors Exclusively Activated by Designer Drugs. Journal of Neuroscience, 36 (36) 9268–9282; 10.1523/JNEUROSCI.1333-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busskamp V, Picaud S, Sahel J et al. (2012). Optogenetic therapy for retinitis pigmentosa. Gene Ther, 19, 169–175. 10.1038/gt.2011.155 [DOI] [PubMed] [Google Scholar]
- Challis RC, Ravindra Kumar S, Chan KY et al. (2019). Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat Protoc 14, 379–414 10.1038/s41596-018-0097-3 [DOI] [PubMed] [Google Scholar]
- Chan KY, Jang MJ, Yoo BB, et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci 20, 1172–1179 (2017). 10.1038/nn.4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Gore F, Nguyen QA. et al. (2020). Deep brain optogenetics without intracranial surgery. Nat Biotechnol. 10.1038/s41587-020-0679-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Choo H, Huang X, et al. (2015). The first structure-activity relationship studies for designer receptors exclusively activated by designer drugs. ACS Chem Neurosci 6, 476–484 10.1021/cn500325v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Ryu S, Lee S et al. (2020).Optimizing clozapine for chemogenetic neuromodulation of somatosensory cortex. Sci Rep 10, 6001. 10.1038/s41598-020-62923-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin BR, Hsiao EC, Claeysen S, et al. (2008). Engineering GPCR signaling pathways with RASSLs. Nat Methods. 5(8):673–678. 10.1038/nmeth.1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curado TF, Pho H, Freire C, et al. (2020). DREADD approach to treatment of sleep disordered breathing. Am J Respir Crit Care Med (online ahead of print) 10.1164/rccm.202002-0321OC [DOI] [Google Scholar]
- Deisseroth K (2011). Optogenetics. Nat Methods 8(1): 26–29. 10.1038/nmeth.f.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimidschstein J, Chen Q, Tremblay R et al. (2016). A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat Neurosci 19, 1743–1749. 10.1038/nn.4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duebel J, Marazova K, & Sahel JA (2015). Optogenetics. Current opinion in ophthalmology, 26(3), 226–232. 10.1097/ICU.0000000000000140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux PF, Schiffmann SN, and de Kerchove d'Exaerde A (2011). Targeting neuronal populations of the striatum. Front. Neuroanatom 5(40) 10.3389/fnana.2011.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettrup A, da Cunha-Bang S, McMahon B, et al. (2014). Serotonin 2A receptor agonist binding in the human brain with [11C]Cimbi-36. Journal of cerebral blood flow and metabolism. J Cereb Blood Flow Metab, 34(7), 1188–1196. 10.1038/jcbfm.2014.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frecha C, Szecsi J, Cosset F, Verhoeyen E (2008). Strategies for targeting lentiviral vectors. Current Gene Therapy 8(6): 449–460. 10.2174/156652308786848003 [DOI] [PubMed] [Google Scholar]
- Galldiks N, Lohmann P, Albert NL, et al. (2019). Current status of PET imaging in neurooncology. Neuro-Oncology Advances, Volume 1, Issue 1, vdz010, 10.1093/noajnl/vdz010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan A, Raper J, Hu X, et al. (2019). Ultrastructural localization of DREADDs in monkeys. European Journal of Neuroscience 50(5): 2801–2813. 10.1111/ejn.14429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz SC, Oritz MM, Belilos AJ, Moussawi K (2020). Inhibitory ultrapotent chemogenetics activate dopamine D1 receptor-expressing medium spiny neurons. bioRxiv 2020.07.01.181925; 10.1101/2020.07.01.181925 [DOI] [Google Scholar]
- Gaub BM, Berry MH, Holt AE, et al. (2015). Optogenetic vision restoration using rhodopsin for enhanced sensitivity. Molecular Therapy 23(10): 1562–1571 10.1038/mt.2015.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez JL, Bonaventura J, Lesniak W, et al. (2017). Chemogenetics revealed: DREADD occupancy and activation via converted clozapine. Science 357(6350), 503–507 10.1126/science.aan2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund T, Tang Y, Benusiglio D, et al. (2019). Chemogenetic activation of oxytocin neurons: temporal dynamics, hormonal release, and behavioral consequences. Psychoneuroendocrinology 106: 77–84. 10.1016/j.psyneuen.2019.03.019 [DOI] [PubMed] [Google Scholar]
- Haywood T, Beinat C, Gowrishankar G, et al. (2019). Positron emission tomography reporter gene strategy for use in the central nervous system. PNAS 116(23) 11402–11407 10.1073/pnas.1901645116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Jin C, Ma M et al. (2019). PET imaging on neurofunctional changes after optogenetic stimulation in a rat model of panic disorder. Front. Med. 13, 602–609. 10.1007/s11684-019-0704-x [DOI] [PubMed] [Google Scholar]
- Horti AG, Gao Y, Kuwabara H, et al. (2014). 18F-ASEM, a radiolabeled antagonist for imaging the α7-nicotinic acetylcholine receptor with PET. Journal of nuclear medicine : official publication, Society of Nuclear Medicine, 55(4), 672–677. 10.2967/jnumed.113.132068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetler ED, Walji AM, Zeng Z, et al. (2016). Preclinical characterization of 18F-MK-6240, a promising pet tracer for in vivo quantification of human neurofibrillary tangles. Journal of Nuclear Medicine, 57 (10) 1599–1606; 10.2967/jnumed.115.171678 [DOI] [PubMed] [Google Scholar]
- Hu F, Morris PJ, Bonaventura J, et al. (2020). 18F-labeled radiotracers for in vivo imaging of DREADD with positron emission tomography, European Journal of Medicinal Chemistry 113047, ISSN 0223–5234, 10.1016/j.ejmech.2020.113047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Chan KY, Tobey IG, et al. (2019). Delivering genes across the blood-brain barrier: LY6A, a novel cellular receptor for AAV-PHP.B capsids. PLoS One, 14(11): e0225206. 10.1371/journal.pone.0225206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S (2000). Anatomy of SUV. Nuclear Medicine and Biology 27(7): 643–646. 10.1016/S0969-8051(00)00155-4 [DOI] [PubMed] [Google Scholar]
- Hutton BF, Erlandsson K and Thielemans K (2018). Advances in clinical molecular imaging instrumentation. Clin Transl Imaging 6, 31–45. 10.1007/s40336-018-0264-0 [DOI] [Google Scholar]
- Iyer SM, Vesuna S, Ramakrishnan C, et al. (2016). Optogenetic and chemogenetic strategies for sustained inhibition of pain. Scientific Reports 6:30570 10.1038/srep30570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji B, Kaneok H, Minamimoto T, et al. (2016) Multimodal imaging for DREADD-expressing neurons in living brain and their application to implantation of iPSC-derived neural progenitors. J Neurosci 7, 13605 10.1523/JNEUROSCI.1279-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur K, Kaushal S, Copra SC (2009). Varenicline for smoking cessation: a review of the literature. Current Therapeutic Research 70(1): 35–54. 10.1016/j.curtheres.2009.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kätzel D, Nicholson E, Schorge S, et al. (2014). Chemical-genetic attenuation of focal neocortical seizures. Nat. Commun 5, 3847. 10.1038/ncomms4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Ku MJ, Qazi R et al. (2021). Soft subdermal implant capable of wireless battery charging and programmable controls for applications in optogenetics. Nat Commun 12, 535. 10.1038/s41467-020-20803-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klawonn AM, Fritz M, Nilsson A, et al. (2018). Motivational valence is determined by striatal melanocortin 4 receptors. J Clin Invest. 128(7): 3160–3170. 10.1172/JCI97854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kügler S, Kilic E & Bähr M (2003). Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther 10, 337–347. 10.1038/sj.gt.3301905 [DOI] [PubMed] [Google Scholar]
- Lee H, Giguere PM, & Roth BL (2014). DREADDs: novel tools for drug discovery and development. Drug Discovery Today 19(4): 469–473. 10.1016/j.drudis.2013.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Satyamurthy N, Barrio JR, et al. (2001). Noninvasive, quantitative imaging in living animals of a mutant dopamine D2 receptor reporter gene in which ligand binding is uncoupled from signal transduction. Gene Ther 8, 1490–1498. 10.1038/sj.gt.3301542 [DOI] [PubMed] [Google Scholar]
- Lin JY (2011). A user's guide to channelrhodopsin variants: features, limitations and future developments. Experimental physiology, 96(1), 19–25. 10.1113/expphysiol.2009.051961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K (2008). Genetic dissection of neural circuits. Neuron, 57(5): 634–660. 10.1016/j.neuron.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLaren DA, Browne RW, Shaw JK, Krishnan Radhakrishnan S, Khare P, Espana RA, & Clark SD (2016). Clozapine N-oxide administration produces behavioral effects in Long–Evans rats: Implications for designing DREADD experiments. Eneuro, 3(5). 10.1523/ENEURO.0219-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen MK, Fisher PM, Burmester D et al. (2019). Psychedelic effects of psilocybin correlate with serotonin 2A receptor occupancy and plasma psilocin levels. Neuropsychopharmacol. 44, 1328–1334. 10.1038/s41386-019-0324-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus CJ, Lee PH, Atasoy D (2011). Chemical and genetic engineering of selective ion channel interactions. Science Vol. 333, Issue 6047, pp. 1292–1296 10.1126/science.1206606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus CJ, Lee PH, Bonaventura J, et al. (2019). Ultrapotent chemogenetics for research and potential clinical applications. Science 364 (6436): eaav5282. doi: 10.1126/science/aav5282 https://science.sciencemag.org/content/364/6436/eaav5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant N, Whitaker L, Bossert J et al. (2016). Behavioral and Physiological Effects of a Novel Kappa-Opioid Receptor-Based DREADD in Rats. Neuropsychopharmacol 41, 402–409. 10.1038/npp.2015.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Konno A, Mochizuki R, et al. (2018). Intravenous administration of the adeno-associated virus-PHP.B capsid fails to upregulate transduction efficiency in the marmoset brain. Neurosci Lett. 2018; 665: 182–188. 10.1016/j.neulet.2017.11.049 [DOI] [PubMed] [Google Scholar]
- Mattis J, Tye KM, Ferenczi EA, et al. , 2012. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat. Methods 9 (2), 159–172. 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone CM, Pati D, Michaelides M, et al. (2018). Acute engagement of Gq-mediated signaling in the bed nucleus of the stria terminalis induces anxiety-like behavior. Molecular psychiatry, 23(1), 143–153. 10.1038/mp.2016.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelides M, Anderson A, Ananth M et al. (2013). Whole-brain circuit dissection in free-moving animals reveals cell-specific mesocorticolimbic networks. J Clin Invest. 123(12):5342–50 10.1172/JCI72117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelides and Hurd. (2015). DREAMM: A biobehavioral imaging methodology for dynamic in vivo whole-brain mapping of cell type-specific functional networks. Neuropsychopharmacology 40: 239–240. 10.1038/npp.2014.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musafargani S, Ghosh KK, Mishra S et al. PET/MRI: a frontier in era of complementary hybrid imaging. European J Hybrid Imaging 2,(12). 10.1186/s41824-018-0030-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y, Kikuchi E, Lerchner W et al. (2016). PET imaging-guided chemogenetic silencing reveals a critical role of primate rostromedial caudate in reward evaluation. Nat Commun 7, 13605. 10.1038/ncomms13605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y, Miyakawa N, Takuwa H et al. (2020). Deschloroclozapine, a potent and selective chemogenetic actuator enables rapid neuronal and behavioral modulations in mice and monkeys. Nat Neurosci 23, 1157–1167. 10.1038/s41593-020-0661-3 [DOI] [PubMed] [Google Scholar]
- Oguchi M, Okajima M, Tanaka S, et al. (2015). Double virus vector infection to the prefrontal network of the macaque brain. PLOS ONE 10(7): e0132825. 10.1371/journal.pone.0132825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea DJ, Kalanithi P, Ferenczi EA, et al. (2018). Development of an optogenetic toolkit for neural circuit dissection in squirrel monkeys. Scientific Reports 8:6775 10.1038/s41598-018-24362-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauleit D, Stoffels G, Bachofner A, et al. (2009). Comparison of 18F-FET and 18F-FDG PET in brain tumors. Nuclear Medicine Biology, 36(7), 779–787. 10.1016/j.nucmedbio.2009.05.005 [DOI] [PubMed] [Google Scholar]
- Pei Y, Rogan SC, Yan F, Roth BL (2008). Engineered GPCRs as tools to modulate signal transduction. Physiology 23: 313–321. 10.1152/physiol.00025.2008 [DOI] [PubMed] [Google Scholar]
- Raper J, Morrison RD, Daniels JS, et al. (2017). Metabolism and distribution of clozapine-N-oxide: implications for nonhuman primate chemogenetics. ACS Chem. Neurosci. 8(7): 1570–1576. 10.1021/acschemneuro.7b00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reivich M, Kuhl D, Wolf A, Greenberg J, Phelps M, Ido T, et al. (1979). The [18F]fluorodeoxyglucose method for the measurement of local cerebral glucose utilization in man. Circ. Res. 44, 127–137. 10.1161/01.RES.44.1.127 [DOI] [PubMed] [Google Scholar]
- RetroSense Therapeutics Phase I/II Clinical Trial for RST-001; trial #NCT02556736. https://www.clinicaltrials.gov/ct2/show/NCT02556736
- Rollema H, Shrikhande A, Ward KM, et al. (2010). Pre-clinical properties of the alpha4beta2 nicotinic acetylcholine receptor partial agonists varenicline, cytisine and dianicline translate to clinical efficacy for nicotine dependence. Br J Pharmacol. 160(2): 334–345. 10.1111/j.1476-5381.2010.00682.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothermel M, Brunert D, Zabawa C, et al. (2013). Transgene expression in target-defined neuron populations mediated by retrograde infection with adeno-associated viral vectors. J of Neuroscience, 33(38): 15195–15206. 10.1523/JNEUROSCI.1618-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha GB (2005). Basics of PET Imaging: Physics, Chemistry and Regulations. Springer Science and Business Media, 10.1007/b138655 [DOI] [Google Scholar]
- Saloman JL, Scheff NN., Snyder LM., et al. (2016). Gi-DREADD Expression in Peripheral Nerves Produces Ligand-Dependent Analgesia, as well as Ligand-Independent Functional Changes in Sensory Neurons. Journal of Neuroscience, 36(42): 10769–10781. 10.1523/JNEUROSCI.3480-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander CY, Hansen HD, Wey H (2020). Advances in simultaneous PET/MR for imaging neuroreceptor function. Journal of Cerebral Blood Flow & Metabolism 40(6): 1148–1166 10.1177/0271678X20910038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Johnson CA, Sabatini BL, 2012. Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. Frontiers in Neurla Circuits 6 (47), 1–10. 10.3389/fncir.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JW, Ingham ES, Mahakian L et al. (2020). Positron emission tomography imaging of novel AAV capsids maps rapid brain accumulation. Nat Commun 11,(2102) 10.1038/s41467-020-15818-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtosa Z, Malik JMI, Michel U, et al. (2005). Promoters and serotypes: targeting of adeno-associated virus vectors for gene transfer in the rat central nervous system in vitro and in vivo. Experimental Physiology, 90: 53–59. 10.1113/expphysiol.2004.028159 [DOI] [PubMed] [Google Scholar]
- Shimojo M, Ono M, Takuwa H, et al. (2020). Genetically targeted reporter imaging of deep neuronal network in the mammalian brain. BioRxiv: 04.08.032870; 10.1101/2020.04.08.032870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifstein M, Kegeles LS, Xu X, et al. (2010). Striatal and extrastriatal dopamine release measured with PET and [18F] fallypride. Synapse, 64: 350–362. 10.1002/syn.20734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KS, Bucci DJ, Luikart BW, & Mahler SV (2016). DREADDS: Use and application in behavioral neuroscience. Behavioral Neuroscience, 130(2), 137–155. 10.1037/bne0000135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloff L, Reivich M, Kennedy C, Rosiers MHD, Patlak CS, Pettigrew KD, Sakurada O and Shinohara M (1977). The [14C]deoxyglucose method for the measurement of local cerebral glucose utilization: theory, procedure, and normal values in the conscious and anesthetized albino rat1. Journal of Neurochemistry, 28: 897–916. 10.1111/j.1471-4159.1977.tb10649.x [DOI] [PubMed] [Google Scholar]
- Stachniak TJ, Ghosh A, Sternson SM. (2014). Chemogenetic synaptic silencing of neural circuits localizes a hypothalamus-midbrain pathway for feeding behavior. Neuron 82: 797–808 10.1016/j.neuron.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternson SM, Roth BL. (2014). Chemogenetic tools to interrogate brain functions. Annu. Rev. Neurosci. 37:387–407 10.1146/annurev-neuro-071013-014048 [DOI] [PubMed] [Google Scholar]
- Taymans J, Vandenberghe LH, Van Den Haute C, et al. (2007). Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Human Gene Therapy 18(3): 195–206. 10.1089/hum.2006.178 [DOI] [PubMed] [Google Scholar]
- Tague EP, Dotson HL, Tunney SN, et al. (2018). Chemogenetic control of gene expression and cell signaling with antiviral drugs. Nat Methods 15, 519–522. 10.1038/s41592-018-0042-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos PK, Robison L, Nestler EJ, Kim R, Michaelides M, Lobo MK, & Volkow ND (2013). Mapping brain metabolic connectivity in awake rats with μPET and optogenetic stimulation. The Journal of neuroscience : the official journal of the Society for Neuroscience, 33(15), 6343–6349. 10.1523/JNEUROSCI.4997-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KJ, Khajehali E, Bradley SJ, et al. (2018). DREADD Agonist 21 is an effective agonist for muscarinic-based DREADDs in vitro and in vivo. ACS Pharmacol Transl Sci, 1: 61–72 10.1021/acsptsci.8b00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjuvajev JG, Doubrovin M, Akhurst T, et al. (2002). Comparison of radiolabeled nucleoside probes (FIAU, FHBG, and FHPG) for PET imaging of HSV1-tk gene expression. Journal of Nuclear Medicine, 43(8), 1072–1083. https://jnm.snmjournals.Org/content/43/8/1072.full [PubMed] [Google Scholar]
- Tremblay S, Acker L, Afraz A, et al. (2020). An open resource for non-human primate optogenetics. Neuron 108: 1075–1090. 10.1016/j.neuron.2020.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye KM, Deisseroth K (2012). Optogenetic investigation of neural circuits underlying brain disease in animal models. Nature Reviews Neuroscience 13: 251–266. 10.1038/nrn3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upright NA, Baxter MG Effect of chemogenetic actuator drugs on prefrontal cortex-dependent working memory in nonhuman primates. Neuropsychopharmacol. 45, 1793–1798 (2020). 10.1038/s41386-020-0660-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban DJ, Zhu H, Marcinkiewcz CA, et al. (2015). Elucidation of the behavioral program and neuronal network encoded by dorsal raphe serotonergic neurons. Neuropsychopharmacology 41(5): 1404–15. 10.1038/npp.2015.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy E, Robinson JE, Li C et al. (2015). A new DREADD facilitates the multiplexed chemogenetic interrogation of behavior. Neuron 86, 936–946 10.1016/j.neuron.2015.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villien M, Wey H, Mandeville JB, et al. (2014). Dynamic functional imaging of brain glucose utilization using FDG-PET. Neuroimage 100: 192–199 10.1016/j.neuroimage.2014.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang G-J, Fowler JS, et al. (1994), Imaging endogenous dopamine competition with [11C]raclopride in the human brain. Synapse, 16: 255–262. 10.1002/syn.890160402 [DOI] [PubMed] [Google Scholar]
- Vomacka L, Albert NL, Lindner S et al. (2017). TSPO imaging using the novel PET ligand [18F]GE-180: quantification approaches in patients with multiple sclerosis. EJNMMI Res 7, 89. 10.1186/s13550-017-0340-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MC, Kullman DM (2020). Optogenetic and chemogenetic therapies for epilepsy. Neuropsychopharmacology, 168:107751 10.1016/j.neuropharm.2019.107751 [DOI] [PubMed] [Google Scholar]
- Wang Y, Xie K, Yue H, et al. (2020). Flexible and fully implantable upconversion device for wireless optogenetic stimulation of the spinal cord in behaving animals. Nanoscale, 12, 2406–2414. 10.1039/C9NR07583F [DOI] [PubMed] [Google Scholar]
- Watakabe A, Ohtsuka M, Kinoshita M, et al. (2015). Comparative analyses of adeno-associated viral vector serotypes 1, 2, 5, 8 and 9 in marmoset, mouse and macaque cerebral cortex. Neuroscience Research, 93:144–157. 10.1016/j.neures.2014.09.002 [DOI] [PubMed] [Google Scholar]
- Weir G, et al. (2017). Using an engineered glutamate-gated chloride channel to silence sensory neurons and treat neuropathic pain at the source. Brain 140, 2570–2585. 10.1093/brain/awx201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wess J, Nakajima K, Jain S (2013). Novel designer receptors to probe GPCR signaling and physiology. Trends Pharamacol Sci. 34(7): 385–392 10.1016/j.tips.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston M, Kaserer T, Wu A, et al. (2019). Olanzapine: a potent agonist at the hM4D(Gi) DREADD amenable to clinical translation of chemogenetics. Sci. Adv 5(4), eaaw1567 . 10.1126/sciadv.aaw1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JC, Denison T (2013). From optogenetic technologies to neuromodulation therapies. Science Translational Medicine 5(177): 177ps6. 10.1126/scitranslmed.3003100 [DOI] [PubMed] [Google Scholar]
- Wu X, Zhu X, Chong P, et al. (2019). Sono-optogenetics facilitated by a circulation-delivered rechargeable light source for minimally invasive optogenetics. Proceedings of the National Academy of Sciences, 116 (52): 26332–26342. 10.1073/pnas.1914387116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaghoubi SS, Campbell DO, Radu CG, et al. (2012). Positron emission tomography reporter genes and reporter probes: gene and cell therapy applications. Theranostics 2(4): 374–391. 10.7150/thno.3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, et al. (2011). Optogenetics in neural systems. Neuron 71(1): 9–34. 10.1016/j.neuron.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang L, Boyden S, Deisseroth K (2006). Channelrhodopsin-2 and optical control of excitable cells. Nature Methods 3: 785–792. 10.1038/nmeth936 [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang L, Brauner M, et al. (2007). Multimodal fast optical interrogation of neural circuitry. Nature 446: 633–639. 10.1038/nature05744 [DOI] [PubMed] [Google Scholar]
- Zhang K, Herzog H, Mauler J, et al. (2014). Comparison of cerebral blood flow acquired by simultaneous [15O]water positron emission tomography and arterial spin labeling magnetic resonance imaging. Journal of Cerebral Blood Flow & Metabolism, 34, 1373–1380. 10.1038/jcbfm.2014.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer L, Luxen A (2012). PET radiotracers for molecular imaging in the brain: Past, present and future. Neuroimage 61: 363–370 10.1016/j.neuroimage.2011.12.037 [DOI] [PubMed] [Google Scholar]
- Zimmermann D, Zhou A, Kiesel M, et al. , 2008. Effects on capacitance by overexpression of membrane proteins. Biochem. Biophys. Res. Commun 369 (4), 1022–1026. 10.1016/j.bbrc.2008.02.153. [DOI] [PubMed] [Google Scholar]