Abstract
The Drosophila Pnut protein belongs to the family of septins, which are conserved GTPases participating in cytokinesis and many more other fundamental cellular processes. Because of their filamentous appearance, membrane association, and functions, septins are considered as the fourth component of the cytoskeleton, along with actin, microtubules, and intermediate filaments. However, septins are much less studied than the other cytoskeleton elements. We had previously demonstrated that the deletion of the pnut gene leads to mitotic abnormalities in somatic cells. The goal of this work was to study the role of the pnut in Drosophila spermatogenesis. We designed a construct for pnut RNA interference allowing pnut expression to be suppressed ectopically. We analyzed the effect of pnut RNA interference on Drosophila spermatogenesis. Germline cells at the earliest stages of spermatogenesis were the most sensitive to Pnut depletion: the suppression of the pnut expression at these stages leads to male sterility as a result of immotile sperm. The testes of these sterile males did not show any significant meiotic defects; the axonemes and mitochondria were normal. We also analyzed the effect of mutations in the Pnut’s conservative domains on Drosophila spermatogenesis. Mutations in the GTPase domain resulted in cyst elongation defects. Deletions of the C-terminal domain led to abnormal testis morphology. Both the GTPase domain and C-terminal domain mutant males were sterile and produced immotile sperm. To summarize, we showed that Pnut participates in spermiogenesis, that is, the late stages of spermatogenesis, when major morphological changes in spermatocytes occur.
Keywords: Drosophila, RNA interference, spermatogenesis, septin, peanut, Pnut
INTRODUCTION
Septins are group of conserved GTP-binding proteins that can be found in all eukaryotes, except the higher plants (Cao et al., 2007). The biological function of septins is connected with their ability to form heterooligomeric complexes which in turn form long polymers (filaments) (Sirajuddin et al., 2007; McMurray et al., 2011). Septins participate in cytokinesis, establishment and maintenance of cell polarity, the functioning of the microtubule and actin cytoskeleton, cell movement, vesicular transport, exocytosis, neurogenesis, and other processes (Saarikangas and Barral, 2011; Mostowy and Cossart, 2012). Septins share a conserved GTPase domain that, according to the latest studies, plays an important role in the dynamics of septin structures within the cell (Sirajuddin et al., 2009; Zent and Wittinghofer, 2014; Akhmetova et al., 2015). Many septins possess a C-terminal coiled-coil domain which is responsible for protein-protein interactions. In recent years, septins have come to be regarded as a new cytoskeleton component due to their filament organization and the functions they perform (Mostowy and Cossart, 2012).
It is known that Drosophila has five representatives of septin protein family: Pnut, Sep1, Sep2, Sep4, and Sep5. Earlier it was shown that septin, coded by the peanut (pnut) gene, participates in cytokinesis of somatic cells (Neufeld and Rubin, 1994). We showed that mutations in this gene, in addition to the defects of cytokinesis, also lead to abnormal chromosomal segregation, which causes ploidy abnormalities in somatic cells; however, proliferation of the germline cells in mutants remains unaffected (Akhmetova and Fedorova, 2011). In this work the role of a pnut gene product and its functional domains on the Drosophila spermatogenesis was studied.
EXPERIMENTAL
Hikone AW Drosophila melanogaster strain from the sector of the Сell cycle genetics of the Institute of Cytology and Genetics of Russian Academy of Sciences was used as a wild type. Fly strain w;T4+/CyO;bam-Gal4/VP16 was kindly provided by P.P. Laktionov from the Institute of molecular and cellular biology of the Siberian Division of the Russian Academy of Sciences. The remaining strains were obtained from the Bloomington Drosophila Stock Center, United States:
No. 5687: pnutXP/T(2;3)SM6a-TM6B,Tb1—null-allele of a peanut gene;
No. 4414: y1w*;P{Act5C-GAL4}25FO1/CyO,y+—ubiquitous Actin5C-GAL4 driver;
No. 5138: y1w*;P{w[+mC] = tubP-GAL4}LL7/TM3,Sb1Ser1—ubiquitous tubulin-GAL4 driver;
No. 4937: w1118;P{GAL4: VP16-nos.UTR}CG6325MVD1—nanos-GAL4 driver, specific for the germline cells;
No. 13134: w1118;P{GT1}chifBG02820a/CyO—chif-GAL4 driver.
Drosophila strains expressing the copy of a wild-type pnut gene and also the mutant gene forms (Table 1) were kindly provided by Dr. Chesnokov (Birmingham, United States) and described in the article of Akhmetova and colleagues (Akhmetova et al., 2015). All constructs contain 1.7 kb 5′ UTR of pnut gene (suggested promoter) and FLAG epitope in the 5′-end. The Pnut expression was verified for all transgenic strains using the western-blot analysis with anti-Pnut and anti-FLAG-epitope antibodies.
Table 1.
Transgenic fly strains used in this study
| Fly strain | Characteristic |
|---|---|
| FLAG-pmutWT | A full-size Pnut copy (1–539 amino acids) |
| FLAG-pmut(1–427) | Truncated Pnut copy (1–427 amino acids) |
| FLAG-pmut(1–460) | Truncated Pnut copy (1–460 amino acids) |
| FLAG-pmut(G4) | Pnut with mutation in the G4 motif of the GTPase domain |
| FLAG-pmut(G1, G3, G4) | Pnut with mutation in the G1, G3 and G4 motifs of the GTPase domain |
Obtaining a plasmid and transgenic flies to conduct pnut-gene RNA-interference was described earlier (Akhmetova et al., 2015).
Immunohistochemical staining and electron microscopic analysis were conducted as described earlier (Pertceva et al., 2010). Rabbit anti-Pnut (1 : 1000) (Huijbregts et al., 2009) primary antibodies; and secondary antibodies, goat anti-rabbit Alexa 488 fluor (Molecular Probes®, United States, 1 : 1000) were used. Microscopic analysis was conducted at the Center of Collective Use of Microscopic Analysis of Biological Objects at the Institute of Cytology and Genetics of the Siberian Division, Russian Academy of Sciences, using the Axioscope 2 plus (Zeiss) microscope. Ultrastructural analysis was conducted using the TEM Libra120 (Zeiss) microscope.
RESULTS
Analysis of Spermatogenesis Abnormalities Due to the Ectopic Suppression of the Pnut Gene Expression
The homozygotes carrying the null-allele of the pnut gene died in the late third instar larval stage. To study the functions of this gene in spermatogenesis, earlier we created the pUASP-pnut_RNAi plasmid to conduct RNA-interference of the pnut gene, and obtained transgenic Drosophila strain carrying this plasmid (Akhmetova et al., 2015). To estimate the pnut gene expression in wild type and while suppressing the expression of this gene using the RNA-interference plasmid created by us, the western-blot method with anti-Pnut, as well as anti-β-tubulin control antibodies, was applied. In Fig. 1 it is shown that using the ubiquitous driver tubulin-Gal4, the RNA-interference of a pnut gene led to a decrease in the amount of the gene product in testes; however, the amount of the control protein (β-tubulin) remained unchanged. Thus, the plasmid construct we created is functional.
Fig. 1.
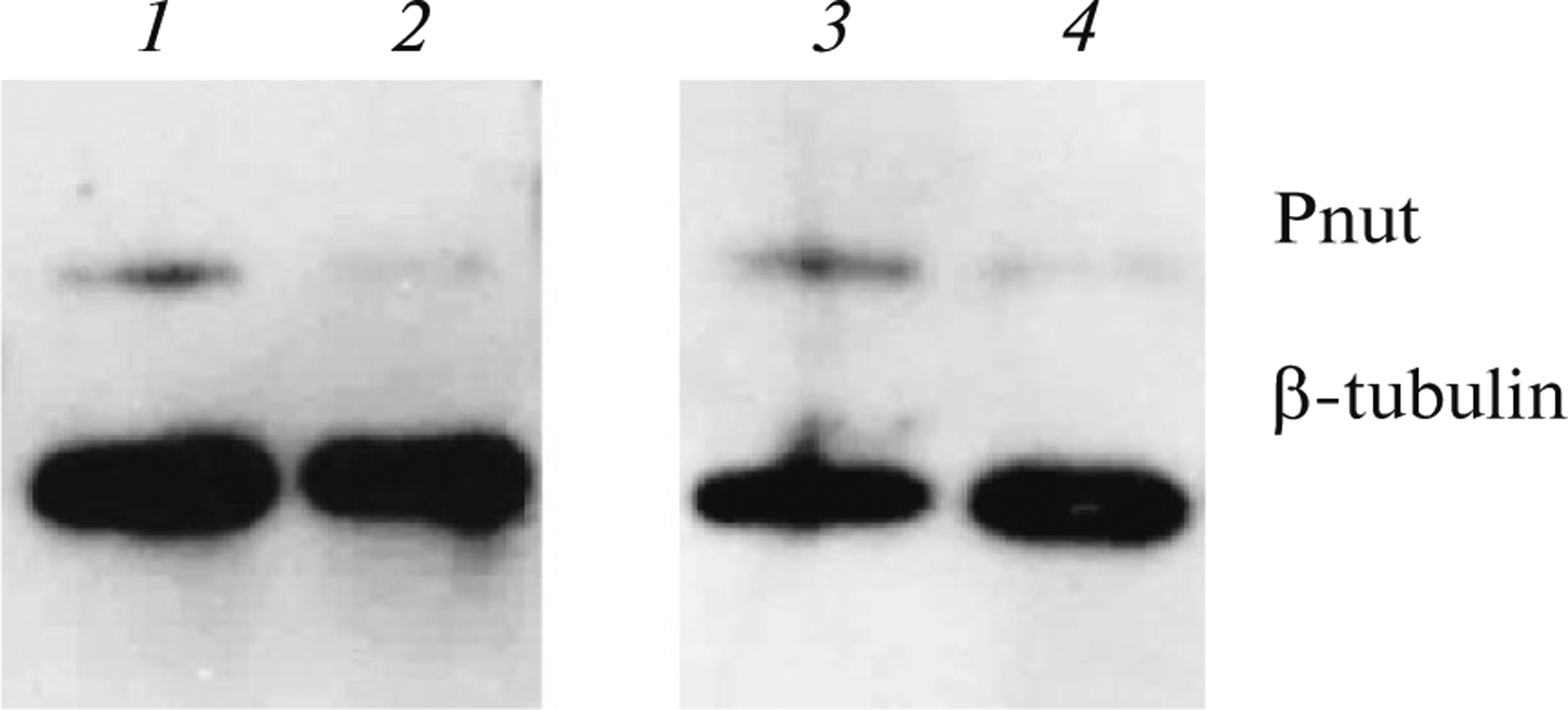
Analysis of the pnut gene expression level during RNA-interference. The results of western blot analysis of the testis extract are shown. Pnut (Huijbregts et al., 2009) antibodies and also β-tubulin control antibodies (E7, DSHB) were used. 1—control: tubulin-GAl4/Sb; 2—pUASP-W-pnut_RNAi/tubulin-GAl4; 3—control: pUASP-W-pnut_RNAi/Sb; 4—pUASP-W-pnut_RNAi/tubulin-GAl4.
The cytological and morphological analysis of Drosophila spermatogenesis in wild type and under the suppression of pnut gene expression using the Actin5C-GAL4 and tubulin-GAL4 ubiquitous drivers, as well as the nanos-GAL4 and bam-GAL4 drivers specific for germline cells, was conducted. When Actin5С-GAL4, tubulin-GAL4, and nanos-GAL4 drivers, expressed at all stages of the spermatogenesis, including the earlier ones, were used, an identical effect was observed: the sperm was immotile in approximately 60% of the seminal vesicles (Table 2). The suppression of pnut gene expression at the stage of premeiotic spermatocytes, when the main RNA accumulation for spermiogenesis takes place (bam-GAL4, chif-GAL4 drivers), did not lead to defects in the sperm motility. Thus the germ line cells at the earliest stages of spermatogenesis are the most sensitive to pnut gene RNA-interference; however, the effect of decreased expression manifests at the later stages of spermatogenesis, resulting in immotile sperm. When conducting the RNA-interference of the pnut gene, the cytological anomalies of spermatogenesis occurred with a low frequency and could not be the reason for the sperm’s sterility. To identify the reason of the immotile sperm due to the ectopic suppression of the pnut gene expression, electron-microscopy analysis of the testes from sterile males was conducted. Axoneme, mitochondria, and the basal body are important for sperm motility. We showed that the axoneme and mitochondria structure in mutants did not differ from control. However, the basal body structure of mutants and wild-type flies was different. The basal body is a modified centriole localized at the base of the axoneme and is needed for its assembly. During spermatid elongation, a collar-like dense electron structure called the centriolar adjunct is formed around the basal body. It resembles the pericentriolar material of centrosomes; however, the function of this structure remains unknown. In Fig. 2 it is seen that the centriolar adjunct in mutants has more blades (it resembles a firework), and its restructuring is delayed (in the control it gradually acquires a round shape and vacuolizes). At the spermiogenesis stages, when the nucleus has an elongated shape, in the pnut gene mutants, the centriolar adjunct continues to be elongated (Fig. 2b).
Table 2.
Sperm motility analysis at 24°С
| Genotype | Analyzed males | Sperm | ||
|---|---|---|---|---|
| Motile | Immotile | Partly motile + notes | ||
| nanos-GAL4/pnut-RNAi | 46 | 17 | 5 | 24, one seminal vesicle contains motile sperm and the other one contains immotile sperm |
| bam-GAL4/pnut-RNAi | 13 | 10 | 1 | 2, one seminal vesicle contains motile sperm and the other one contains immotile sperm |
| chif-GAL4/pnut-RNAi | 10 | 8 | 1 | 1, one seminal vesicle contains motile sperm and the other one contains immotile sperm |
| Actin5C-GAL4/pnut-RNAi | 30 | 12 | – | 8, one seminal vesicle contains motile sperm and the other one contains immotile sperm 10, partly motile sperm |
| Tubulin-GAL4/pnut-RNAi | 22 | 8 | 2 | 12, one seminal vesicle contains motile sperm and the other one contains immotile sperm |
| pnutXP; FLAG-pnut(1–427) | 24 | 4 | 7 | 13, impossible to estimate due to the absence of the contact of the testis with the seminal vesicle |
| pnutXP; FLAG-pnut(G1, G3, G4) | 9 | 2 | 7 | 0 |
Fig. 2.
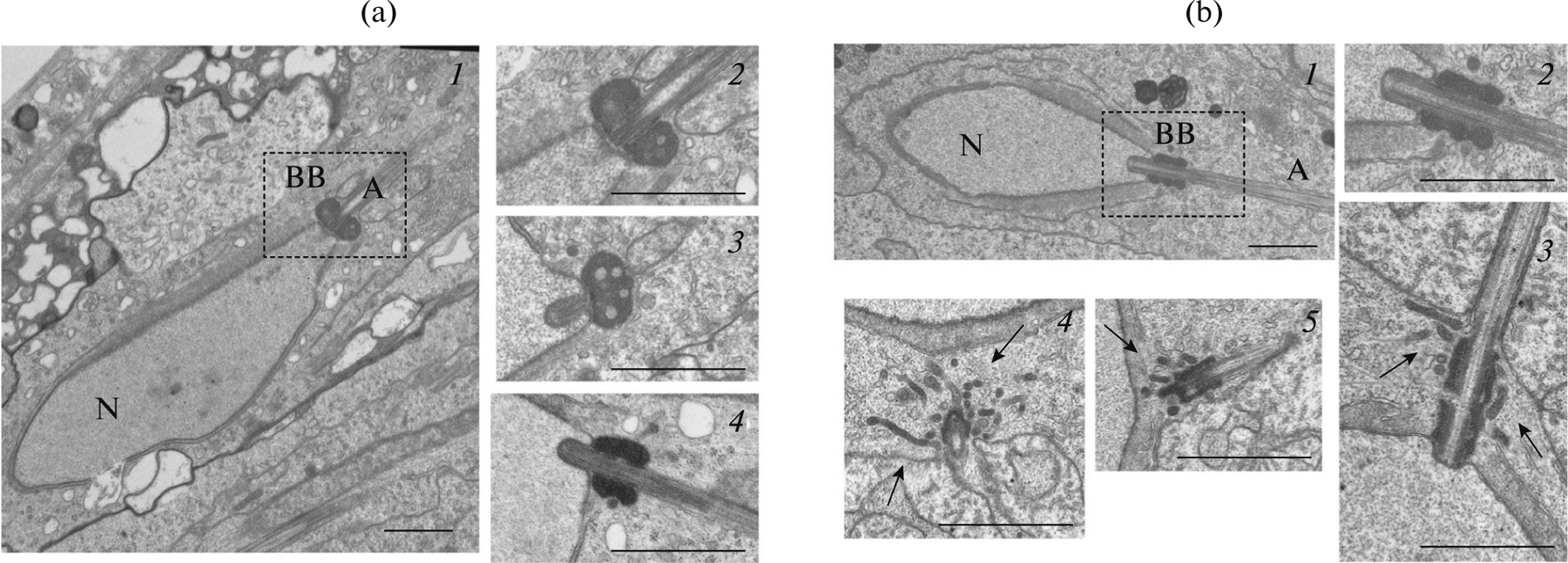
Electron microscopic (EM) analysis of the basal body of spermatids. (a) EM images of the cross section of wild-type spermatids: N is the nucleus of a spermatid; BB is the basal body; A—axoneme. 1, 2—longitudinal sections of the nucleus, slightly tangential to the axoneme (2—magnification of the dotted part); 3—cross section strongly tangential to the axoneme; 4—longitudinal section to the axoneme; the spermatid stage is earlier than shown in 1–3. Centriolar adjunct surrounding the basal body is round-shaped; inside the vacuoles are seen. The later the stage of spermatid development the more round the shape of the centriolar adjunct. (b) EM images of nanos-GAL4/pnut-RNAi spermatids cross sections: 1–3—longitudinal sections to the axoneme (2—magnification of 1); 4—cross section transverse to the axoneme; 5—cross section tangential to the axoneme. The centriolar adjunct is strongly extended, of an incorrect shape, surrounded by a large number of incorporations (arrows). Scale: 1 μm.
The role of Pnut Functional Domains
RNA-interference suppresses gene expression but does not allow us to obtain data about the functional domains of a product of the given gene. To identify the contribution of different Pnut domains, the mutations in conserved motifs and regions of the protein were used (Fig. 3). In this work, the single mutation in the G4 conserved motif of the Pnut GTPase domain—Pnut(G4), and also the triple mutant—Pnut(G1, G3, G4) were studied, with the conserved amino acids having been substituted by alanines. Moreover, the truncated forms of the Pnut protein, Pnut(1–427), and Pnut(1–460) with a deletions of the C-terminus were analyzed.
Fig. 3.
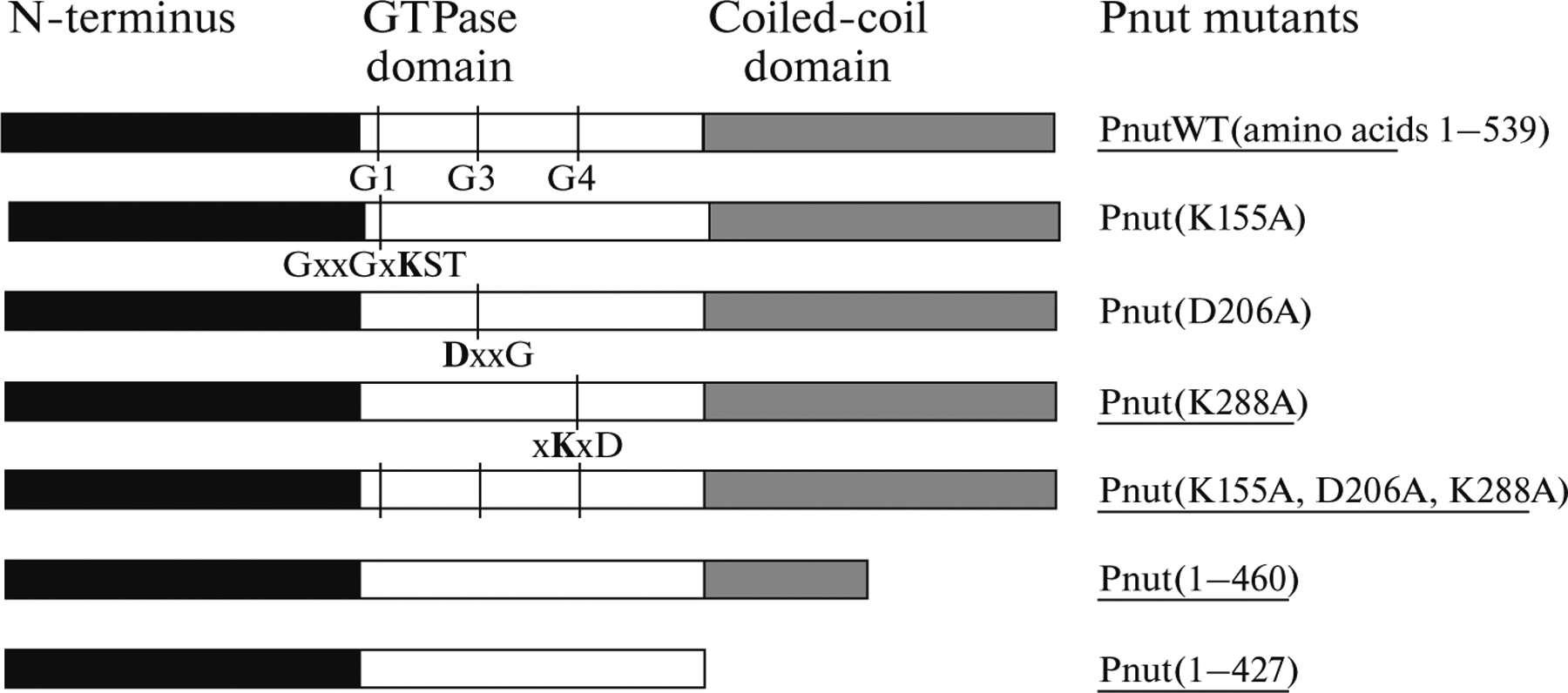
Domain structure of Pnut protein. On the left, the schematic view of a domain structure of Pnut is given; on the right, mutations in the conserved domains are shown. The conserved amino acids in the G1, G3, and G4 motifs of the GTPase domain (shown in bold letters) were substituted by alanines. The mutants used in the study are underlined.
It was shown earlier that the expression of the FLAG-pnutWT transgene, carrying a copy of a wild-type pnut gene, restores the viability of a pnutXP null-allele, which is lethal in the third larva instar. Introduction of the FLAG-pnut(1–427) or FLAG-pnut(1–460) transgenes, containing the deletion of the C-terminal part of the protein, as well as the GTPase deficient FLAG-pnut(G4) and FLAG-pnut(G1, G3, G4) transgenes did not rescue pnutXP deletion to the imago stage (Akhmetova et al., 2015). The described rescue experiments were conducted after the first balancing of the transgenic insertions just obtained, and no rescue with any of the mutant Pnut forms were detected. The transgenic pnutXP/Cy;FLAG-pnut strains were obtained. After several generations, as a result of the stabilization of the genetic background, an insignificant number of flies entering the late-pupa or even imago stage started to appear in the obtained stocks. This allowed us to study the effect of Pnut mutations on the process of Drosophila spermatogenesis.
The localization of the Pnut protein in the mutant testes using the Pnut antibodies was studied. In the wild type flies and pnutXP null-allele flies rescued with the wild-type FLAG-pnutWT transgene, Pnut is localized cortically, creating a dotted pattern (Fig. 4a). Both truncated forms of Pnut with the deletion of a C-terminal domain had a tendency to form aggregates that did not have characteristic Pnut localization pattern as demonstrated for the FLAG-pnut(1–427) (Fig. 4b). Mutations in the GTPase domain of Pnut disrupted the correct localization of the protein, membrane-specific dot-like staining was absent and the diffuse staining of the cytoplasm in the cells of the testes was observed (Fig. 4c, and also Akhmetova et al., 2015). Thus, it can be concluded that the C-terminal, as well as the GTPase domain, are important for the correct localization of protein in the testes of Drosophila in vivo.
Fig. 4.
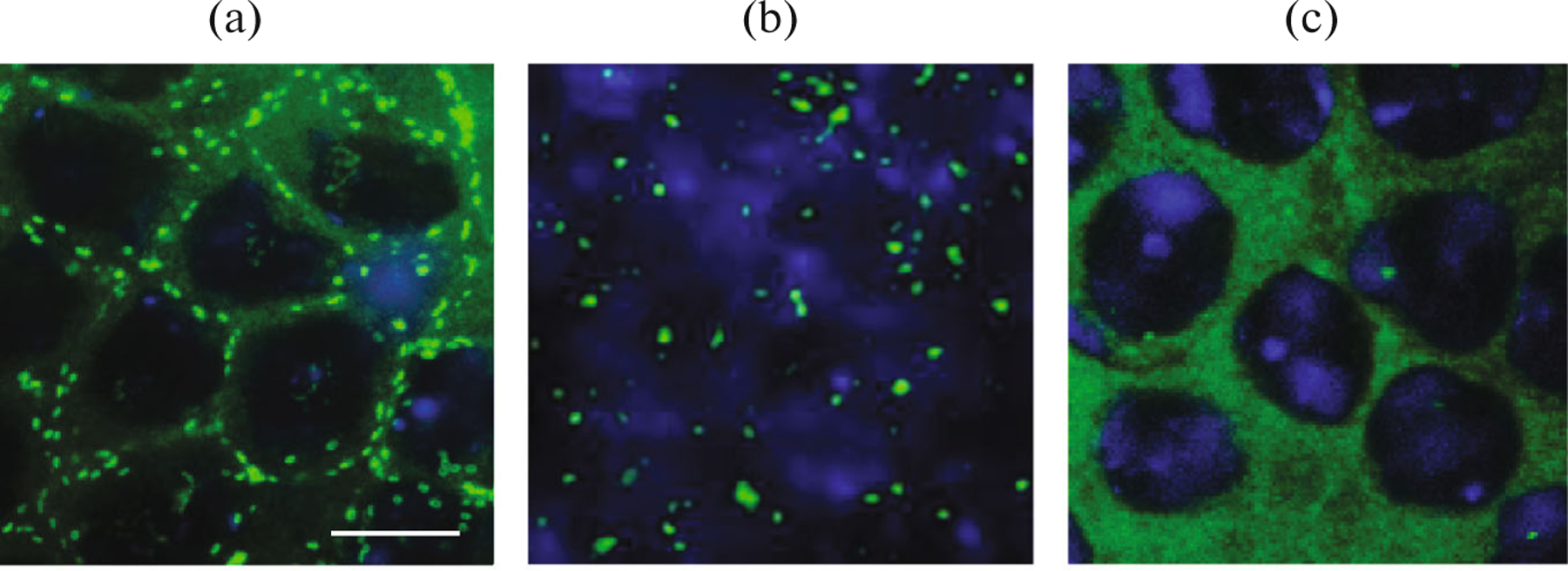
Mutations in the functional Pnut domains disrupt the correct protein localization in vivo: (a) dotted cortical distribution of a protein in the wild-type testis, stained with anti-Pnut antibodies; (b) disruption of a Pnut localization in mutants with deletion of a C-terminal domain (pnutXP; FLAG-pnut(1–427)); (c) diffuse Pnut localization in the testis of pnutXP; FLAG-pnut(G4) mutants defective in the GTP-binding domain. Green—Pnut staining, blue—DNA (DAPI staining). Scale: 10 μm.
The genetic tests for sterility showed that pnutXP males, expressing transgenes with FLAG-pnut(1–427) C-terminal deletion and also the FLAG-pnut(G1, G3, G4) GTPase mutant had an analogous immotile sperm phenotype as shown for the pnut RNA-interference (Table 2). Disruption of cytokinesis in meiosis was observed at a low frequency in both strains, however, these defects were not critical and could not lead to sterility in any of these cases.
In the spermatogenesis of deletion mutants pnutXP;FLAG-pnut(1–427) and FLAG-pnut(1–460) not only was the sperm immotile but also the shape of the testes was affected, which in 70–80% of the cases were underdeveloped, looked like the larval testes, and did not contact with the seminal vesicles (Fig. 5). Nevertheless, all processes specific for spermatogenesis could be observed in these round-shaped and mushroom-shaped testes: the germline cells were dividing and entering meiosis, followed by the start of spermiogenesis with the formation of the sperm that appeared normal. The motility of these sperm could not be estimated because the sperm from the seminal vesicle is used for motility analysis; however, in the case of mutants, it remained empty due to the absence of contact with the remaining part of the testis. In the spermatogenesis of these mutants, defects of cytokinesis also can be found, although they are not critical for fertility.
Fig. 5.
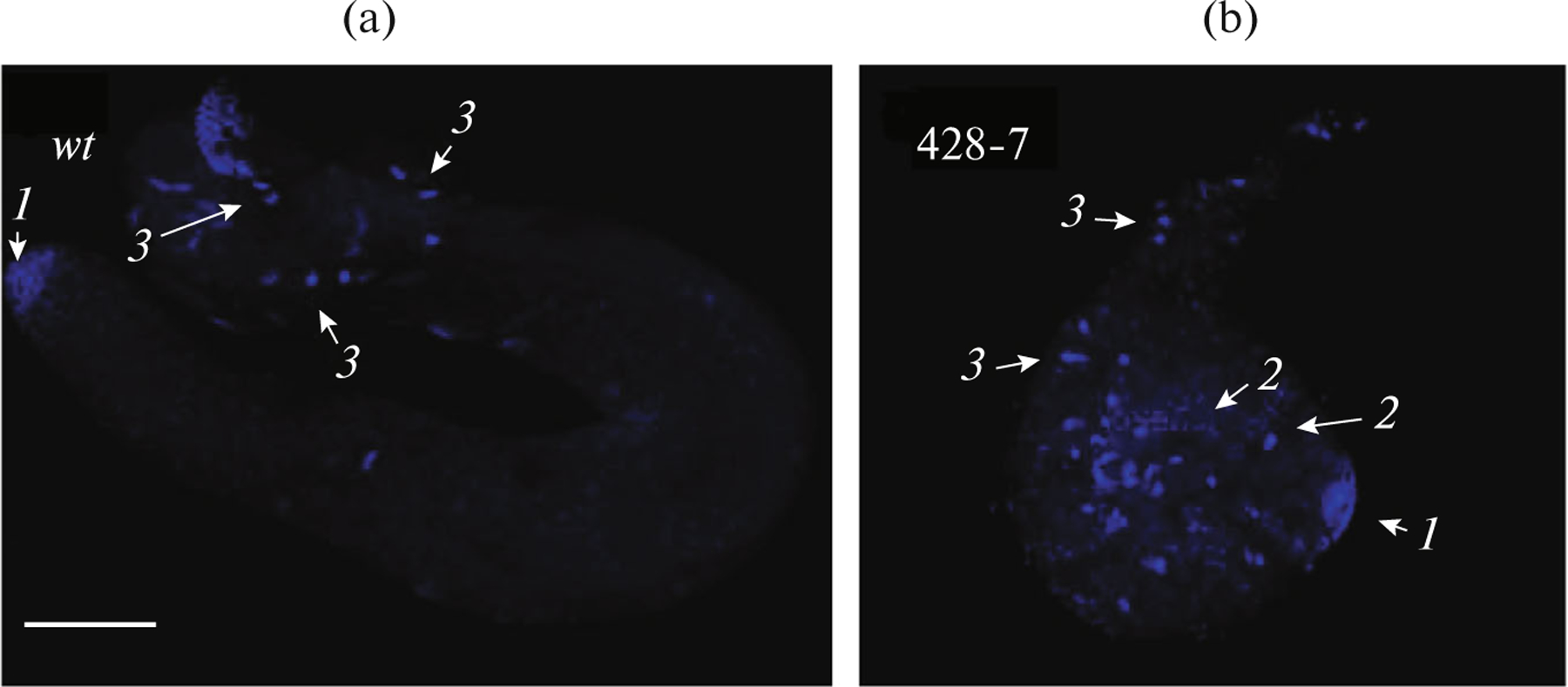
Defects of spermatogenesis in mutants with Pnut C-terminal domain deletion: (a) wild-type adult male testis; (b) round-shaped testis of mutant adult (pnutXP; FLAG-pnut(1–427) that is not in contact with the seminal vesicles. Marked with arrows: the mitotic divisions of the germline cells (arrow 1), meiosis (arrow 2), and spermiogenesis resulting in the formation of sperm (arrow 3). DNA was stained with DAPI. Scale: 100 μm.
Defects of cytokinesis were also observed in the spermatogenesis of the triple GTPase mutant pnutXP;FLAGpnut(G1, G3, G4); however, they were not critical. The most significant abnormalities were at the postmeiotic stages during the elongation of the spermatids. Very often mutants had disrupted elongation and/or polarization of the cysts that led to the formation of cysts with randomly scattered nuclei.
DISCUSSION
Conducting the RNA-interference of a pnut gene using the nanos-GAL4 driver specific to the germline cells, we found a high percentage of sterile males. The cytological analysis of spermatogenesis showed that about 60% of the sperm were immotile. While using other drivers, it was shown that germline cells at the early stages of spermatogenesis are sensitive to RNA-interference of the pnut gene and the first mitotic divisions are probably involved. However, the effect of decreased pnut gene expression was manifested later in the spermiogenesis and resulted in sperm immobility.
Similar studies were conducted on mammals. It was shown that in mammals proteins SEPT4 and SEPT7 (homologue of Pnut) are required for the formation of a specific ring-like membrane structure, the annulus, which divides the sperm tail into different compartments. In the SEPT4 knockout mice, the abnormal formation and absence of the annulus and disruption of an ultrastructural organization of the mitochondria leading to the immobility of the sperm and sterility were observed (Kissel et al., 2005; Lhuillier et al., 2009). However, in Drosophila sperm, an annulus-like structure was not found. Moreover, we found that when suppressing pnut gene expression in Drosophila, the ultrastructure of mitochondria and axoneme (the basic components, responsible for the sperm motility) remained intact. Besides the mitochondria and axoneme, the basal body (modified centriole) also provides sperm motility. After meiosis every haploid spermatid contains one basal body with a short axoneme. As it moves to the nucleus, it is associated with the microtubules (MT). After association with the nuclear membrane, the basal body serves as an MT-organizing center, taking part in assembling the axoneme of the sperm’s tail and also the perinuclear MT. The correct docking and maintenance of the association of a basal body with the nuclear membrane are critical for the formation of motile sperm in Drosophila. In the differentiation process of spermatid, when the basal body has already reached the nucleus, for some time it becomes surrounded by a dense electron structure, namely, the centriolar adjunct, which resembles the pericentriolar material of centrosomes. The centriolar adjunct is a very dynamic structure which contracts during nucleus elongation and takes the shape of a round collar, and then disappears. We found that the centriolar adjunct in the male testes with the RNA-interference of the pnut gene has morphological defects; moreover, its restructuring is delayed until the later stages of spermatogenesis. How the defects in this structure are connected with sperm immotility has not yet been elucidated. Probably the defects we observed in the centriolar adjunct actually reflect the anomalies of the basal body which directly ensures the motility of the sperm.
Mutants with the Pnut C-terminal deletion have an interesting phenotype. The testes of such males are underdeveloped and have a round- or mushroom-shaped form, similar to the testes of larva and, moreover, they do not contact with the seminal vesicles. In the literature, there is considerable information concerning the interaction of somatic niche cells with the germline cells, and the distribution of the morphogenes in the genital discs was studied in detail; however, there is no information about how these constitutive parts of a testis dock in the later embryogenesis or at the early larva stage. Probably, cell contacts and signal molecules are required for this. We assume that the cadherins can take part in the given processes. The Pnut C-terminal domain contains the coiled-coil involved in the interaction with other proteins, which is why abnormal Pnut localization can lead to the incorrect localization of the partner proteins of the given septin. As the Pnut is localized cortically in the cells of a testis, different components of cell contacts and also receptors can serve as partner proteins.
The mutants deficient in the Pnut GTPase domain have disrupted cyst polarization. Initially, a cyst consisting of 64 round haploid spermatids is not polarized. At the early elongation stages, the calcium channels are grouped next to the actin-rich areas in a cortical membrane at the distal (growing) end of the elongating spermatids; i.e., the polarization of cyst is taking place. We assume that Pnut is involved in the polarization of the cyst in the spermatogenesis due to the phospholipid level changes. Microscopy studies in real time of spermatids cultivated in vitro showed that for the correct and timely polarization the normal level of phosphatidylinositol 4,5-biphosphate (PIP2) on the plasma membrane is required (Fabian et al., 2010). A decrease in the membrane-associated PIP2 level due to the expression of the SigD protein, which is PIP2 phosphatase, leads to significant defects in the polarization of the cyst. The interaction with membrane phospholipids was shown for the majority of septins (Casamayor and Snyder, 2003). It was also shown that the SEPT2-SEPT6-SEPT7 human septin complex can modify the morphology of giant liposomes containing phosphoinositides (Tanaka-Takiguchi et al., 2009). The proposed hypothesis can be supported by the fact that the Pnut GTPase mutants are lacking in specific dotted structures in the cortical area. Probably, such mutant protein forms cannot interact with the membrane and as consequence cannot participate in the modification of the phospholipid level.
Thus, we showed that Pnut is involved in spermiogenesis—the final stages of spermatogenesis when the morphology of the spermatocytes is changed. Sterility caused by sperm immotility is a characteristic of Pnut mutants. Based on electron microscopy, we assume that the functioning of the basal body of mutants is disrupted. Other abnormalities of spermatogenesis in conservative Pnut domains, detected in the mutants, indicate that this protein has many functions in the spermatogenesis process; in particular, it is involved in cell signaling and also in cyst polarization.
ACKNOWLEDGMENTS
The work was supported by OPTEC grant no. 41/2014/51- Nvs “The role of septin Pnut and its functional domains in Drosophila spermatogenesis”, the Russian Fund for Basic Research, grant no. 14-04-31289, and also the federal budget, state project no. 0324-2015-0003.
Footnotes
CONFLICT OF INTEREST
The authors state that they do not have a conflict of interest.
REFERENCES
- Akhmetova KA and Fedorova SA, The peanut gene mutations effects on somatic and germ line cell division in Drosophila melanogaster, Russ. J. Genet.: Appl. Res, 2012, vol. 2, no. 3, pp. 229–234. [Google Scholar]
- Akhmetova K, Balasov M, Huijbregts RPH, and Chesnokov I, Functional insight into the role of Orc6 in septin complex filament formation in Drosophila, Mol. Biol. Cell, 2015, vol. 26, no. 1, pp. 15–28. doi 10.1091/mbc.E14-02-0734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmetova KA, Dorogova NV, Chesnokov IN, and Fedorova SA, Analysis of peanut gene RNAi in drosophila oogenesis, Russ. J. Genet, 2015, vol. 51, no. 9, pp. 847–854. [PMC free article] [PubMed] [Google Scholar]
- Cao L, Ding X, Yu W, Yang X, Shen S, and Yu L, Phylogenetic and evolutionary analysis of the septin protein family in metazoan, FEBS Lett., 2007, vol. 581, pp. 5526–5532. doi 10.1016/j.febslet.2007.10.032 [DOI] [PubMed] [Google Scholar]
- Casamayor A and Snyder M, Molecular dissection of a yeast septin: Distinct domains are required for septin interaction, localization, and function, Mol. Cell. Biol, 2003, vol. 23, no. 8, pp. 2762–2777. doi 10.1128/MCB.23.8.2762-2777.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian L, Wei HC, Rollins J, Noguchi T, Blanken-ship JT, Bellamkonda K, Polevoy G, Gervais L, Guichet A, Fuller MT, and Brill JA, Phosphatidylinositol 4,5-bisphosphate directs spermatid cell polarity and exocyst localization in Drosophila, Mol. Biol. Cell, 2010, vol. 21, no. 9, pp. 1546–1555. doi 10.1091/mbc.E09-07-0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbregts RP, Svitin A, Stinnett MW, Renfrow MB, and Chesnokov I, Drosophila Orc6 facilitates GTPase activity and filament formation of the septin complex, Mol. Biol. Cell, 2009, vol. 20, pp. 270–281. doi 10.1091/mbc.E08-07-0754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissel H, Georgescu MM, Larisch S, Manova K, Hunnicutt GR, and Steller H, The Sept4 septin locus is required for sperm terminal differentiation in mice, Dev. Cell, 2005, vol. 8, pp. 353–364. doi 10.1016/j.devcel.2005.01.021 [DOI] [PubMed] [Google Scholar]
- Lhuillier P, Rode B, Escalier D, Lorès P, Dirami T, Bienvenu T, Gacon G, Dulioust E, and Touré A, Absence of annulus in human asthenozoospermia: Case report, Hum. Reprod, 2009, vol. 24, no. 6, pp. 1296–1303. doi 10.1093/humrep/dep020 [DOI] [PubMed] [Google Scholar]
- McMurray MA, Bertin A, Garcia G, Lam L, Nogales E, and Thorner J, Septin filament formation is essential in budding yeast, Dev. Cell, 2011, vol. 20, no. 4, pp. 540–549. doi 10.1016/j.devcel.2011.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S and Cossart P, Septins: The fourth component of the cytoskeleton, Nat. Rev. Mol. Cell Biol, 2012, vol. 13, no. 3, pp. 183–194. doi doi 10.1038/nrm3284 [DOI] [PubMed] [Google Scholar]
- Neufeld TP and Rubin GM, The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins, Cell, 1994, vol. 77, pp. 371–379. doi 10.1016/0092-8674(94)90152-X [DOI] [PubMed] [Google Scholar]
- Pertceva JA, Dorogova NV, Bolobolova EU, Nerusheva OO, Fedorova SA, and Omelyanchuk LV, The role of Drosophila hyperplastic discs gene in spermatogenesis, Cell Biol. Int, 2010, vol. 10, pp. 991–996. doi 10.1042/CBI20100105 [DOI] [PubMed] [Google Scholar]
- Saarikangas J and Barral Y, The emerging functions of septins in metazoans, EMBO Rep, 2011, vol. 12, no. 11, pp. 1118–1126. doi 10.1038/embor.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Farkasovsky M, Hauer F, Kühlmann D, Macara IG, Weyand M, Stark H, and Wittinghofer A, Structural insight into filament formation by mammalian septins, Nature, 2007, vol. 449, pp. 311–315. doi 10.1038/nature06052 [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Farkasovsky M, Zent E, and Wittinghofer A, GTP-induced conformational changes in septins and implications for function, Proc. Natl. Acad. Sci. U.S.A, 2009, vol. 106, no. 39, pp. 16592–16597. doi 10.1073/pnas.0902858106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Takiguchi Y, Kinoshita M, and Takiguchi K, Septin-mediated uniform bracing of phospholipid membranes, Curr. Biol, 2009, vol. 19, no. 2, pp. 140–145. doi 10.1016/j.cub.2008.12.030 [DOI] [PubMed] [Google Scholar]
- Zent E and Wittinghofer A, Human septin isoforms and the GDP-GTP cycle, Biol. Chem, 2014, vol. 395, pp. 169–180. doi 10.1515/hsz-2013-0268 [DOI] [PubMed] [Google Scholar]


