Abstract
Extra-chromosomal genetic elements are important drivers of evolutionary transformations and ecological adaptations in prokaryotes with their evolutionary success often depending on their ‘utility’ to the host. Examples are plasmids encoding antibiotic resistance genes, which are known to proliferate in the presence of antibiotics. Plasmids carrying an essential host function are recognized as permanent residents in their host. Essential plasmids have been reported in several taxa where they often encode essential metabolic functions; nonetheless, their evolution remains poorly understood. Here we show that essential genes are rarely encoded on plasmids; evolving essential plasmids in Escherichia coli we further find that acquisition of an essential chromosomal gene by a plasmid can lead to plasmid extinction. A comparative genomics analysis of Escherichia isolates reveals few plasmid-encoded essential genes, yet these are often integrated into plasmid-related functions; an example is the GroEL/GroES chaperonin. Experimental evolution of a chaperonin-encoding plasmid shows that the acquisition of an essential gene reduces plasmid fitness regardless of the stability of plasmid inheritance. Our results suggest that essential plasmid emergence leads to a dose effect caused by gene redundancy. The detrimental effect of essential gene acquisition on plasmid inheritance constitutes a barrier for plasmid-mediated lateral gene transfer and supplies a mechanistic understanding for the rarity of essential genes in extra-chromosomal genetic elements.
Author summary
Mobile genetic elements have been extensively studied due to their role as agents of genetic innovation and rapid adaptation in prokaryotes. Specifically, prokaryotic plasmids have been the focus of investigation in the context of bacterial survival under growth limiting conditions with the prime example of resistance to antibiotics and heavy metals. In contrast, plasmids that encode for functions that are essential to their host viability are rarely described. We investigate the evolution of plasmids that encode for genes previously identified as essential for bacterial life. Our analysis of Escherichia isolates reveals only few plasmid-encoded essential genes, which likely function in the plasmid rather than the host life cycle. Following the evolution of plasmids encoding an essential gene in Escherichia coli in real time, we further find that the acquisition of a chromosomal essential gene may lead to plasmid loss. Our study supplies data and a mechanistic understanding on the rarity of essential genes in mobile genetic elements. We conclude that prokaryotic plasmids are rarely essential for their bacterial host.
Introduction
Plasmids are autonomously replicating genetic elements that are prominent in prokaryotes and have been studied extensively due to their contribution to lateral gene transfer. Plasmid invasion is often accompanied by the acquisition of novel traits that enable bacteria to survive under specific conditions or colonize specific ecological niches. Plasmids carrying beneficial genes, e.g., those that supply resistance against antibiotics (e.g., [1,2]) or heavy metals (e.g., [3,4]) enable their host to survive under transient selective conditions. A strong selection for the plasmid-encoded trait over many generations was shown to lead to co-adaptation of the plasmid and the host, and the evolution of stable plasmid inheritance [5–9]. Nonetheless, environmental conditions are rarely constant; a decrease in the strength of selection for plasmid-encoded beneficial genes–e.g., due to fluctuating abundance of growth limiting factors–may lead to plasmid loss and extinction. In contrast, plasmids that supply the host with essential functions, i.e., whose benefit to the host is less dependent on temporary environmental conditions, may persist over longer time scales and become an integral component of the lineage genome in the form of chromids [10]. The level of gene essentiality is defined as the extent to which a gene is required for the reproduction of an organism [11]. Borrowing that definition, plasmids that encode for essential genes may be thus considered as vital for the host proliferation. Examples include plasmids encoding for genes along the biosynthesis pathways of essential amino acids (e.g. [12,13]) and components of the ribosome machinery [14]. Nevertheless, it has been previously suggested that plasmids encoding essential genes should be rare in nature as their adaptation to the host is accompanied by reduction of host fitness due to plasmid loss [15]. Thus, the evolution of plasmids encoding essential genes may depend on the evolution of stable plasmid-host interactions.
In the absence of selection for plasmid-encoded genes, plasmid persistence largely depends on plasmid stability that comprises plasmid replication and segregation. Indeed, plasmids that have a negligible (i.e., neutral) effect on host fitness may evolve stable inheritance and consequently gain long-term persistence in the population–also in the absence of positive selection [16]. Stably inherited plasmids may serve as precursors for the evolution of larger plasmids carrying beneficial or essential genes. Notably, the essential plasmids reported in the literature so far are characterized by a stable vertical inheritance. The evolution of such essential plasmids may thus follow two alternative routes [17]: essentiality first, where the plasmid is initially essential to the host; a plasmid carrying an essential function may persist in the population over long timescales and eventually evolve a stable plasmid reproduction within the host. In the second route, termed stability first, a plasmid that initially evolved a stable reproduction cycle may subsequently evolve into an essential plasmid following an essential gene gain. Notwithstanding, as essential genes are typically encoded in the chromosomes [18], the presence of an essential gene on a plasmid may be comparable to the effect of gene duplication. In addition, the expression level of plasmid-encoded genes may be amplified following an increase in plasmid copy number (e.g., [19]). Therefore, the evolution of essential genes on plasmids is likely under selection pressure that is typical to gene duplication, such as dose effect (reviewed in [20]). Evidently, most genes (89 ± 8%) in prokaryotic genomes are found in a single copy [21].
Here we combine computational genomic analysis and experimental work to investigate the evolution of essential plasmids in the genus Escherichia. First, we investigate the frequency of shared genes between plasmids and chromosomes in Escherichia as well as the distribution of essential genes in Escherichia plasmids. Furthermore, we compare both the essentiality and stability first evolutionary scenarios using an evolution experiment in E. coli where we follow the evolution of stable and unstable plasmids encoding the chaperonin that is indispensable in E. coli.
Results
Plasmid gene content is rarely shared with the host chromosome
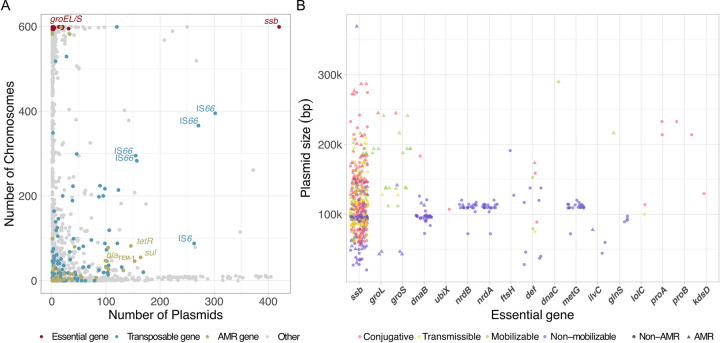
To study the distribution of chromosomal homologs to plasmid genes we examined isolates of Escherichia that are available as complete genome sequences at NCBI (S1 Table). Of the total 599 isolates, 416 isolates harbor between 1 and 10 plasmids. All 88,493 protein-coding genes encoded on plasmids were clustered into 4,780 proteins families by sequence similarity. Notably, we did not detect any universal plasmid gene–the largest plasmid protein family includes homologs from 41% (425/1035) plasmids. Additionally, we identified chromosomal homologs to the plasmid protein families using a sequence similarity search. This revealed that 2,682 (56%) of the plasmid protein families had no chromosomal homolog, while the remaining 2,098 (44%) families had a chromosomal homolog in at least one E. coli isolate. Previously it has been suggested that gene duplication via gene transfer may be disadvantageous in prokaryotes due to dose effect (e.g., [22,23]). Indeed, the distribution of protein family members on chromosomes and plasmids shows that the level of gene sharing between both replicon types is rather low (Fig 1A). A total of 383 (8%) protein families have a chromosomal homolog in a single isolate, and 544 (11%) of the plasmid protein families have a chromosomal homolog in 2–320 isolates (S1 Fig). Thus, in total 927 (19%) of the plasmid protein families have a chromosomal homolog in the same isolate (i.e., the plasmid host), of those, 133 (3%) families correspond to transposable elements (Fig 1A). The most abundant protein family among the transposable elements is that of IS66 that is found in varying copy numbers on 395 examined chromosomes and 302 plasmids.
Fig 1. The distribution of plasmid-encoded essential genes on plasmids and chromosomes.
A, The distribution of Escherichia protein families on plasmids in chromosomes (in grey). Essential genes (red) are found in most of the chromosomes and rarely on plasmids, with ssb as an exception. Transposable elements (blue) are frequent on both chromosomes and plasmids. IS66 is the most widely spread gene in Escherichia strains. IS66 was split into several protein families in our analysis, which are depicted by multiple data points. Antibiotic resistance (AMR) genes (yellow) are presented for comparison. The AMR genes can be roughly divided into two groups: the first group aligns along the y-axis hence it is more frequently found on chromosomes; those chromosome-encoded AMR genes are typically related to persistence and resilience functions. The second group aligns along the x-axis hence it is more frequently found on plasmids; those plasmid-encoded genes are typically related to antibiotics resistance functions (Wein et al. 2020). B, Distribution of plasmid size, mobility group and AMR for plasmids encoding an essential gene. Most of the ssb-coding plasmids are conjugative (red) while the majority of groEL/S coding plasmids are mobilizable (green). The remaining essential genes are encoded on plasmids that are often non-mobilizable (and non-AMR) (purple).
Essential genes are rarely encoded on plasmids
To study the evolution of essential genes encoded on plasmids, we examined the distribution of genes previously recognized as essential in E. coli on chromosomes and plasmids, where the extent of gene essentiality may depend on the environmental and nutritional conditions. The set of essential genes we tested here comprises 504 protein-coding genes (S2 Table); of those, 394 protein-coding genes were identified as essential in E. coli K-12 under standard laboratory conditions via gene knockout [24] or transposon mutagenesis [25]. Additional 110 protein-coding genes were identified as essential in a collection of 18 E. coli isolates by CRISPR interference of gene expression ([18]; only genes identified as essential in at least one out of the three tested growth media were included; S2 Table). Searching for plasmid-encoded homologs to the essential genes using sequence similarity revealed that only 17 (3%) of the essential genes had a homolog on plasmids (Fig 1A and Table 1). The 464 Escherichia plasmids encoding essential genes are typically of medium plasmid size (median: 106Kb), 32% of them are non-mobile and only 4% carry in addition an antibiotic resistance gene (Fig 1B). Importantly, all isolates where we identified a plasmid homolog of an essential gene also harbored the chromosomal copy of that gene (except one isolate carrying only metG on the plasmid).
Table 1. Data and phylogenetic analysis of essential genes encoded on plasmids.
Essential list indicates the source of essential genes: TKP is TraDIS-Keio-PEC, TP is TraDIS-PEC, T is TraDIS (Goodall et al. 2018); LB, M9 and GMM means this gene is essential in each of the three medias (Rousset et al. 2020). Relaxed selection shows evidence for relaxed selection on plasmid homologs’ branch was found using HyPhy-RELAX (Murrell et al. 2012). Tree topology shows the conclusions from the phylogenetic reconstruction of essential genes on plasmids and chromosomes: split between plasmids and chromosomes describe phylogenies that had a deep split between plasmid and chromosomal homologs (i.e., they are diverged); LGT events from chromosome to plasmid are labeled by transfer to plasmid; LGT events from plasmid to chromosome are labeled by transfer to chromosome; mixed is used to label phylogenies that contain deep divergence and LGT events. For gene trees with evidence for gene transfer (groEL, groES), we further tested the support in the plasmid/chromosome split while excluding the putatively transferred gene. In all tested cases the result showed that the constrained tree could not be rejected in a topology test, thus validating the gene transfer inference. All of the transfer event from plasmid to chromosome are better described as translocation of the plasmid homolog to chromosome.
| Gene | Product | Essential list | No. Plasmids | No. Isolates | Relaxed selection | Tree topology |
|---|---|---|---|---|---|---|
| ssb | single-stranded DNA-binding protein | TKP | 420 | 320 | n.a. | Mixed |
| groL | chaperonin GroEL | TP | 11 | 11 | On plasmid branch | One translocation to Chromosome |
| groS | co-chaperone GroES | TKP | 13 | 13 | No evidence | One translocation to Chromosome |
| dnaB | replicative DNA helicase | TKP | 31 | 31 | No evidence | Two translocations to Chromosome |
| ubiX | aromatic acid decarboxylase | T | 1 | 1 | n.a. | Split between plasmids and chromosomes |
| nrdB | ribonucleotide-diphosphate reductase subunit beta | TKP | 20 | 20 | No evidence | Split between plasmids and chromosomes |
| nrdA | ribonucleoside-diphosphate reductase subunit alpha | TKP | 19 | 19 | On plasmid branch | Split between plasmids and chromosomes |
| ftsH | ATP-dependent metalloprotease FtsH | TKP | 2 | 2 | n.a. | Three transfers to plasmid |
| def | peptide deformylase | TKP | 13 | 13 | No evidence | Split between plasmids and chromosomes |
| dnaC | DNA replication protein DnaC | TKP | 1 | 1 | n.a. | Single transfer to plasmid |
| metG | methionine—tRNA ligase | TKP | 19 | 19 | No evidence | Mixed |
| ilvC | ketol-acid reductoisomerase, NAD(P)-binding | M9 | 3 | 3 | No evidence | Split between plasmids and chromosomes |
| glnS | glutamine—tRNA ligase | TKP | 5 | 5 | n.a. | Five transfers to plasmid |
| lolC | lipoprotein-releasing ABC transporter permease subunit | TKP | 2 | 2 | n.a. | Split between plasmids and chromosomes |
| proB | gamma-glutamate kinase | M9 | 2 | 2 | n.a. | Two transfers to plasmid |
| proA | gamma-glutamylphosphate reductase | M9 | 2 | 2 | n.a. | Two transfers to plasmid |
| kdsD | D-arabinose 5-phosphate isomerase | LB, GMM | 1 | 1 | n.a. | Split between plasmids and chromosomes |
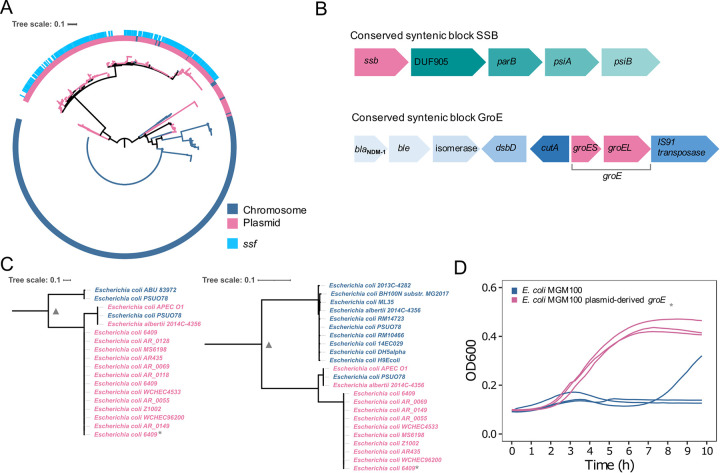
The most frequent essential gene on plasmids is a single-strand DNA binding protein (ssb), which is nearly ubiquitous in Escherichia plasmids (Table 1 and Fig 2A). Ssb functions in DNA replication and recombination and is important for the establishment of conjugative plasmids in the recipient host cell [26]. The chromosomal ssb gene function can be complemented by a plasmid borne ssb (termed ssf [27]) in E. coli when present on a high copy-number plasmid (Porter and Black 1991). To investigate whether the high abundance of ssb on plasmids is the result of frequent lateral transfer, we examined the genomic neighborhood of ssb on plasmids. This revealed that the ssb genomic neighborhood is conserved in 64% (270/420) of the plasmids, comprising five genes that include a parB gene encoding type II partitioning system and two genes annotated as inhibitors of SOS response to conjugation (psiA and psiB; Fig 2B and S3 Table). Indeed, many of the plasmids encoding ssb are conjugative (43%; Fig 1B). A phylogenetic reconstruction of the Escherichia Ssb evolutionary history reveals a deep divergence between the chromosomal and plasmid homologs (Fig 2A). Additionally, the phylogenetic tree suggests several events of lateral gene transfer (LGT) of ssb from plasmids to chromosomes as indicated by eight chromosomal ssb homologs that branch with plasmid homologs (Fig 2A). To further validate the LGT events, we compared the reconstructed tree likelihood to an alternative constrained topology having a split between chromosomal and plasmid ssb homologs. Since the alternative ssb topology was rejected (P = 0.0074, using AU test), we conclude that, indeed, the ssb evolution included multiple LGT events. According to the inferred ssb phylogeny, a recent translocation of the plasmid-encoded ssb to the chromosome occurred in the seven strains, all of them retained the original chromosomal copy and lacked a plasmid-encoded ssb; these seven isolates thus have two ssb copies encoded in their chromosome. These ssb LGT events correspond to the translocation of an essential plasmid gene to the chromosome.
Fig 2. Phylogeny of plasmid-encoded essential genes in Escherichia.
A, Phylogeny of the single-stranded DNA-binding protein Ssb. Ssb homologs encoded on plasmid are shown in pink and encoded on chromosome are shown in blue. The plasmid ssb gene termed ssf is most abundant across the plasmid Ssb homologs. B, The conserved neighborhood (conserved syntenic block, CSB) of ssb and the chaperone groE encoded on plasmids (S3 and S4 Tables). C, Phylogeny of the chaperonin GroES (left) and GroEL (right). The triangle symbol marks the branch split that was constrained in the test for an alternative tree topology. D, Growth measurements of E. coli MGM100 and MGM100 carrying the plasmid-derived GroE (marked by *). The exceptional growth behavior observed in one E. coli MGM100 replicate should be considered an outlier since we found no evidence for other explanations to that result.
Further phylogenetic reconstruction of the remaining plasmid homologs to essential genes revealed evidence for a recent transfer from the chromosome to the plasmid in six of the protein families including ftsH, dnaC, metG, glnS, proA and proB (Table 1 and S2–S7 Figs). The evolution of those gene families is evidence for rare duplication of essential genes onto a resident plasmid. In contrast, the phylogenetic trees of nine essential genes including groEL, groES, ubiX, nrdB, nrdA, def, ilvC, lolC and kdsD revealed a clear split between the chromosomal and plasmid homologs (Table 1 and Figs 2C and S8–S14). In two of these gene families—groEL and groES—we observed gene translocation from plasmid to chromosome (Table 1 and Fig 2C). The divergence between the plasmid and the chromosomal homologs in these gene families suggests that the plasmid homologs have undergone a sub- or neo-functionalization (i.e., similar to the plasmid Ssb).
Previous studies suggested that laterally transferred genes are often non-functionalized due to codon-usage incompatibility or the absence of regulatory elements [28]. To investigate whether plasmid-encoded homologs of essential genes are pseudogenes, we inferred the strength of selection pressure on the plasmid branch relative to chromosome branch in all relevant protein families. Whereas our results revealed the presence of relaxed selection on branches leading to plasmid homologs in groEL and nrdA (Table 1 and S15 and S16 Figs), we could not detect evidence for non-functionalization of those genes (e.g., frameshift or truncation). Our results thus suggest that the observed relaxation of selection is likely associated with sub- or neo-functionalization of the essential genes encoded on plasmids.
The plasmid-encoded chaperonin complements the chromosomal function
To further examine the evolution of plasmids encoding essential genes, we studied the implications of essential gene gain on a plasmid by focusing on the chaperonin genes groEL and groES. The chaperonin is universally encoded in eubacterial chromosomes with only rare exceptions (e.g., Mollicutes [29]) and plasmids encoding the chaperonin genes have been only reported in Rhizobium [30]. The groES and groEL genes are typically encoded within the groE operon. The GroE chaperonin plays a major role in the bacterial protein-folding pathway and is constitutively expressed in E. coli and essential for growth under all conditions [31]. An examination of the groE genomic neighborhood in Escherichia plasmids revealed a conserved neighborhood that includes, in addition to groEL and groES a blaNDM-1, ble (bleomycin binding protein Ble-MBL), phosphoribosylanthranilate isomerase, twin-arginine translocation (TAT) pathway signal sequence domain protein and cutA (divalent-cation tolerance protein) (Fig 2B and S4 Table). Indeed, a similar integron comprising groE has been previously reported in Escherichia [32,33]. Our results thus indicate that groE is likely not a pseudogene. To further test the function of the plasmid-derived groE, we cloned the gene sequence into a small model plasmid. The plasmid was then introduced into E. coli MGM100 (Fig 2D), which encodes the groE-operon under an inducible PBAD promoter, hence it is only viable in medium supplemented with arabinose (i.e., PBAD induction) or when it is complemented with groE [34]. In competition with E. coli MG1655, E. coli MGM100 has no measurable fitness disadvantage (S18 Fig). To test whether the plasmid-derived groE can complement the silenced groE in E. coli MGM100, we quantified the growth of this strain with and without supplemented arabinose. In addition, we recorded the overnight plasmid loss, as a plasmid that is essential to its host cannot be lost. Our results show that the plasmid can indeed complement the chromosomal GroE function, hence we conclude that the plasmid-encoded groE functions as a chaperonin.
Essential gene acquisition destabilizes plasmid inheritance
Our results so far show that essential genes are rarely found on plasmids with the plasmid-encoded GroE in Escherichia as a rare exception. Such plasmids that encode an essential gene may evolve via two possible routes–in the stability first a preliminary stably inherited plasmid gains an essential gene, while in the essentiality first a plasmid that encodes an essential gene gains by that a stable inheritance. To study the evolutionary consequences of the acquisition of an essential chromosomal gene by plasmids, we conducted an evolution experiment with E. coli K12 MG1655 carrying small plasmids encoding a chromosomal groE copy. To investigate the implications of plasmid stability versus essentiality in the evolution of essential plasmids, we compared the evolutionary dynamics of stable and unstable plasmids in two genetic host backgrounds.
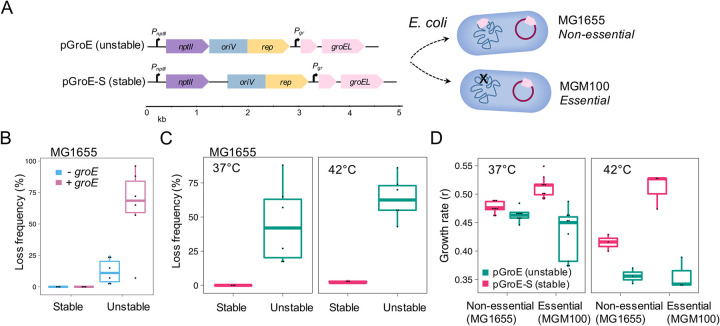
Our unstable model plasmid pCON originated from the pBBR1 backbone that is widely spread and associated with antibiotic resistance [16,35]. The plasmid pCON encodes nptII that is constitutively expressed and confers resistance to kanamycin; it has a negligible effect on the host fitness and is characterized by an unstable inheritance in the population [16]. Previously, we showed that pCON instability is caused by transcription-replication conflicts of the resistance gene transcription and plasmid replication. A stable inheritance of pCON is provided when transcription is silenced or when both machineries are physically separated, i.e., by a plasmid size increase [16]. Our stable model plasmid pCON-S is a derivative of pCON that is characterized by a stable inheritance due to a short DNA insertion between the origin of replication and the nptII gene; similarly to pCON, it has a negligible effect on the host fitness [36]. We equipped the unstable (pCON) and stable (pCON-S) plasmids with the chromosomal groE (including its native promoter) resulting in pGroE and pGroE-S (Fig 3A). The plasmids pGroE and pGroE-S were introduced into two host strains: E. coli MG1655 (wt) and E. coli MGM100 (Fig 3A). Thus, groE-encoding plasmids are essential in E. coli MGM100 while they are non-essential in E. coli MG1655.
Fig 3. Characterization of plasmids encoding an essential gene.
A, Genomic map of unstable pGroE and stable pGroE-S plasmids that were introduced into E. coli MG1655 (plasmid is non-essential) and E. coli MGM100 (plasmid is essential) hosts. B, Plasmid loss frequency of plasmids lacking (-) groE or encoding (+) groE in their genome in the host E. coli MG1655 (H0: L+>L-, P = 0.0247 using Wilcoxon test n = 6) C, Plasmid loss frequency of pGroE-S (stable) and pGroE (unstable) after incubation at 37°C or at 42°C (H0: L42>L37, P = 0.0091 using Wilcoxon test, n = 6). D, Growth rates of host strains E. coli MG1655 and E. coli MGM100 carrying pGroE (unstable) or pGroE-S (stable) after growth at 37°C and 42°C (H0: Gr42>Gr37, P = 0.051 using Wilcoxon test, n = 9).
To characterize the effect of GroE on plasmid stability, we quantified the loss frequency of pGroE (unstable) and pGroE-S (stable). Our results show that the pGroE loss frequency was significantly higher in comparison to the loss frequency of the plasmid pCON lacking GroE in E. coli MG1655 (Fig 3B, P = 0.0247 using Wilcoxon test, n = 6). Similar to pCON-S, the plasmid pGroE-S was stably maintained in E. coli MG1655 in an overnight incubation (Fig 3B). Both plasmids were maintained in the host strain E. coli MGM100, where the plasmid is essential for host survival. When the medium was supplemented with arabinose–rendering the plasmid dispensable–we observed a plasmid loss frequency of 21±3% (SE, n = 6) for the unstable pGroE and no loss of the stable pGroE-S (n = 6; S19 Fig). These results are in line with the loss frequency of pGroE in E. coli MG1655, in which the plasmid is dispensable. Our results show that the introduction of a redundant groE copy on an unstable plasmid further destabilizes the plasmid maintenance and may lead to an increase in plasmid loss.
In addition to variation in the extent of plasmid-encoded gene essentiality, the observed plasmid loss may be related to the chaperonin function. The chaperonin GroE is part of the heat shock regulon in E. coli hence its expression level is upregulated during growth at high temperature (42°C) [37,38]. The expression level of plasmid-encoded genes depends not only on their transcriptional regulation but also on the plasmid copy number (PCN). Variation in PCN is thus comparable to gene amplification where fluctuations in PCN lead to variation in the expression level of plasmid-encoded genes, which may enable the host to rapidly adapt to changing environmental conditions [19,39]. Thus, under heat stress bacteria that carry plasmid-encoded groE are expected to have an advantage over strains with chromosomally encoded groE due to a higher GroE abundance in the cell. To evaluate whether heat stress had an effect on plasmid persistence, we measured the plasmid loss frequency after overnight incubation at 42°C. Our results show no measurable plasmid loss for both plasmids when the plasmid is essential (i.e., in E. coli MGM100). When the plasmid is not essential (i.e., in E. coli MG1655), the pGroE-S remained stable, while the pGroE loss frequency was slightly increased in comparison to pGroE loss at 37°C (Fig 3C). This observation is in line with reduced growth rate of MG1655 at 42°C in comparison to 37°C (Fig 3C, P = 0.0091 using Wilcoxon test). Thus, the redundant plasmid-encoded GroE does not confer an advantage in E. coli MG1655 at 42°C. Nonetheless, a comparison of the growth rate (r) between E. coli MGM100 pGroE-S and MG1655 pGroE-S at 42°C revealed a growth advantage of E. coli MGM100 pGroE-S over MG1655 pGroE-S (Fig 3D, P = 0.051 using Wilcoxon test) and over the plasmid-free wildtype (rwt = 0.42±0.01, SE, n = 9). The growth advantage observed for expression E. coli MGM100 pGroE-S likely stems from differences in groE expression dynamics when it is expressed solely from the plasmid locus. Additionally, the growth rate of both strains carrying the unstable pGroE was decreased (Fig 3D, P = 0.0007 using Wilcoxon test). The reduced growth rate of E. coli MGM100 carrying the unstable pGroE is well explained by the occurrence of cell death following plasmid loss (S20 Fig).
We conclude that the presence of a redundant essential gene on an unstable plasmid may destabilize plasmid maintenance in the host regardless of growth conditions. The maintenance of stable plasmids can remain, however, unchanged in the presence of an essential gene. Furthermore, variation of the plasmid-encoded gene expression level due to fluctuations in the plasmid copy number may confer an advantage to the host under specific environmental conditions.
Essential plasmids persist over time regardless of their stability
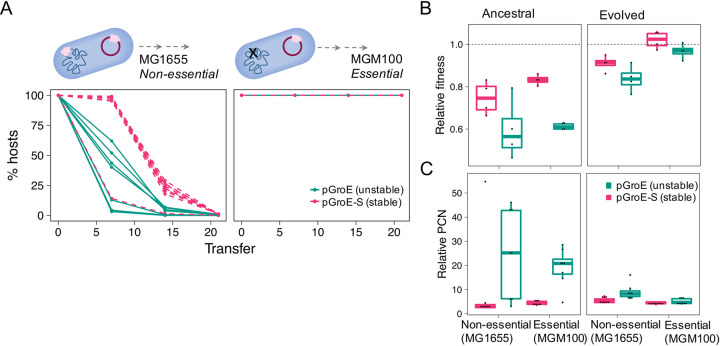
Constant positive selection for a beneficial gene is expected to result in gene transfer from the plasmid to the chromosome and consequently plasmid loss [40,41]. To test the effect of plasmid essentiality on plasmid evolution, we conducted an evolution experiment comparing both unstable (pGroE) and stable (pGroE-S) plasmids in host backgrounds where the plasmids are essential or non-essential. The experiment was performed with nine replicates of all plasmid and host combinations for approximately 320 generations in a serial batch transfer approach.
The results of the evolution experiment reveal that when the plasmids were essential for host survival, they were maintained over time regardless of their stability (Fig 4A). Sequencing a sample of the evolved populations, we did not detect evidence for groE transfer to the chromosome or mutations in the inducible PBAD-groE promoter (S5 Table). When the plasmid was non-essential to the host (i.e., in E. coli MG1655), the highly unstable pGroE was lost rapidly while the stable pGroE-S decreased in frequency after several transfers (Fig 4A). Thus, non-essential plasmids were lost regardless of their initial stability in the host (i.e., stable inheritance). To test whether plasmid loss in our experiment led to plasmid extinction, we exposed the evolved populations to antibiotics (kanamycin 25 μg/ml; S21 Fig), which was then followed by serial transfer under non-selective conditions (i.e., without antibiotics). This experiment showed that none of the plasmids went extinct over time. In addition, we observed a similar plasmid loss pattern, where the unstable plasmid was gradually lost from the population and the stable plasmid was maintained at a higher frequency (S21 Fig). Our results thus suggest that the decreased frequency of plasmid carrying hosts is not due to plasmid instability, rather is a result of host fitness differences between plasmid-carrying and plasmid-free cells.
Fig 4. Evolution of plasmids encoding an essential gene.
A, Evolution experiment of pGroE (unstable) and pGroE-S (stable) in the host strains E. coli MG1655 (non-essential, left) and MGM100 (essential, right). B, Relative fitness of plasmid-carrying ancestral and evolved populations. Pairwise competition experiments between the plasmid-carrying strains and tagged wild-type (Tmr). A significant negative fitness effect could be observed for all ancestral populations (w: 0.69, P = 1.526x10-5 using Wilcoxon test, n = 4 per population) while the fitness of evolved populations increased compared to the ancestral state (w: 0.93, P = 9.05x10-7 using Wilcoxon test, n = 4 per population). C, Plasmid copy number (PCN) of plasmid-carrying ancestral and evolved populations. The median pGroE PCN is significantly different between the ancestral and evolved populations (PMG1655 = 0.03, PMGM100 = 0.0016, using Wilcoxon test, n = 9 per population), with the PCN being lower in the evolved populations in both hosts.
To evaluate the relative fitness of the plasmid-carrying populations, we conducted competition experiments of the ancestral and evolved populations against the plasmid-free wildtype strain. Indeed, we observed a strong negative fitness effect of all groE-plasmids on host fitness (Fig 4B; w: 0.697, P = 1.52x10-5 using Wilcoxon test, n = 4 per population). Nonetheless, the negative impact on fitness decreased over time and the evolved populations revealed a higher relative fitness than their ancestors (Fig 4B, w: 0.93, P = 9.05x10-7 using Wilcoxon test, n = 4 per population). A reduction in the plasmid effect on the host fitness might have been associated with a decrease in PCN over time. Indeed, this is the case for the unstable plasmids in both host strains where the average PCN decreased about 10-fold (Fig 4C). Previously, we showed that a reduction of PCN for pCON plasmids is often a result of reduction in the plasmid tendency to form multimers (Wein et al., 2019; 2020). Moreover, the variance of pGroE PCN (in MG1655) among replicate populations decreased during the experiment (ancestral CVpGroE: 0.73, evolved CVpGroE: 0.35), and so did the loss frequency (S22 Fig), a result that is akin to the loss of plasmid multimers during the evolution experiment (as in [16,36]). Since we did not detect any high frequency chromosomal variants in the evolved populations (S5 Table; as previously observed for antibiotic resistance plasmids [42]), we conclude that the decrease in PCN is likely due to an efficient resolution of plasmid multimers in the evolved population.
Our results show that plasmid essentiality due to complementation of the chaperonin function leads to plasmid persistence that is accompanied by an increase in overall fitness of the evolved populations. Nonetheless, carrying a redundant chromosomal gene proved here to be disadvantageous for plasmid persistence regardless of plasmid stability. Consequently, we conclude that plasmids encoding an essential gene may have a higher chance to persist in the population over longer time scale if a chromosomal gene copy is either lacking or functionally divergent.
Discussion
The acquisition of an additional copy of single-copy chromosomal genes has been shown to be often deleterious, hence sensitivity to dose effect has been suggested as a barrier to gene acquisition by lateral gene transfer [23]. Newly acquired genes are often initially disconnected from the organisms’ metabolic network and their integration may take a long time [43]. Thus, additional copies of core essential genes can disrupt the cell physiology through protein dosage alterations that change metabolic fluxes (e.g., [44]). Regulatory networks in prokaryotes have evolved to respond to feedback loops that include monitoring of transcription level and substrate concentrations. Hence, the acquisition of a redundant gene copy may be deleterious due to indirect effect on the organism metabolism and cellular processes [22,45]. Notwithstanding, plasmid-mediated dose effect is not always deleterious to the host and may depend on the protein sequence characteristics and interactions with other proteins in the cell [46]. Our results for the chaperonin-encoding plasmid reveal that functional redundancy of an essential gene following plasmid acquisition can be highly disadvantageous for host fitness. In our experiment, the GroE abundance is associated with the PCN, hence the reduction in fitness effect of the plasmid in the evolved populations is likely explained by the decrease in PCN that is coupled with a decrease in GroE dose within the cell. An elevated expression level of the chaperonin has been shown to be detrimental for E. coli fitness under optimal growth conditions [47,48]. The fitness cost of GroE overexpression has been suggested to stem from the increased energetic investment in protein folding due to the high chaperonin availability [49]. The plasmid-mediated increase in GroE abundance may furthermore hamper the chaperonin assembly in the cell. Plasmids that gain a redundant essential gene are thus at risk of rapid loss and eventually extinction.
Based on our results, we suggest that the evolution of plasmids encoding essential genes is conditioned by either non-functionalization (or loss) of the chromosomal copy or its functional divergence, e.g., via neo-functionalization or sub-functionalization due to modification of the transcriptional regime. This constraint provides an explanation for the rarity of essential genes in Escherichia plasmids. Notably, two of the plasmid-encoded essential genes we report here–ssb and groE–seem to have undergone a sub-functionalization as part of the plasmid life cycle such that their function is no longer essential for the host. In other words, those essential genes were acquired by the plasmid and repurposed into essential plasmid genes that are required for persistence in the host. The Escherichia plasmids encoding ssb or groE are thus carrying homologs to essential genes but should not be considered essential plasmids. Hence, we predict that essential genes are rarely found on plasmids in other prokaryotic taxa and natural environments. The detrimental effect of essential gene acquisition on plasmid stability constitutes a barrier for plasmid-mediated lateral transfer of essential genes. Nonetheless, we cannot exclude that plasmids encoding an essential gene may become advantageous under specific environmental conditions. Our results show that the translocation of groE to a plasmid increased host fitness during growth at high temperature where groE expression is naturally upregulated [37,38]. Indeed, groE may be found on plasmids in nature as in the facultative plant symbiont Sinorhizobium meliloti, in which two out of five functional groE copies are encoded on the plasmid pSymA and one copy is encoded on the plasmid pSymB [50]. The chromosomal groE1 copy was shown to encode the major housekeeping chaperonin function, yet it could be functionally complemented by the pSymA-encoded groE2 [51]. The reason why multiple groE copies are retained in S. meliloti remains unknown. It is tenable to speculate that the facultative symbiotic lifestyle of S. meliloti entailed a sub-functionalization of the groE copies. An example for the translocation of an essential gene from the chromosome to a plasmid was reported in bacterial symbionts of lice that inhabit human, chimpanzee and gorilla (Riesia sp.). In these organisms, the B5 vitamin biosynthesis pathway is encoded on a small plasmid that is vital for the bacteria-louse symbiosis. In contrast, symbionts of lice that inhabit old world monkeys (Puchtella sp.) are lacking the plasmid and the B5 vitamin pathway is chromosomally encoded [52]. The phylogenetic relations of the two symbionts indicate that the chromosomal B5 copy is ancient and the gene translocation to the plasmid occurred during the lice adaptation to inhabit new-world monkeys [52]. We hypothesize that the fixation of the gene translocation on the plasmid may be related to the symbiotic Riesia lifestyle; a retention of the plasmid encoded gene copy may be advantageous under conditions that require a flexible gene expression level, which can be regulated rapidly by transient changes in plasmid copy number [39].
Considering the two possible routes for the evolution of essential genes on plasmids–our study supplies evidence that the essentiality first route is the more likely scenario. To better understand the difference between the two routes we shift our perspective to consider the fitness of the plasmid. Plasmid loss in our experimental system stems from two main reasons: plasmid instability of the unstable pGroE or fitness disadvantage of the stable pGroE-S-hosts; indeed, incomplete segregation and negative fitness effects are known causes of plasmid loss (e.g., [53,54]). The comparison between the loss dynamics of the unstable and stable plasmids (Fig 4A) indicates that plasmid loss due to intracellular processes–namely plasmid instability–may be more rapid in comparison to plasmid loss due to processes at the population level–where plasmid-hosts are outcompeted by non-hosts. Our results indicate that the acquisition of an essential chromosomal gene by a plasmid leads to a significant reduction in plasmid fitness, even when the plasmid was highly stable prior to the essential gene acquisition (as in the stability first route). Considering the constraint on the presence of chromosomal essential genes on plasmids, most plasmids are unlikely to be essential for their host.
Methods
Computational genomic analysis
All 599 completely sequenced genomes of Escherichia genus strains were downloaded from NCBI (version 2018) and analyzed as described in [36]. Briefly, plasmid-encoded protein coding genes were clustered into homologous protein families. Sequence similarity of plasmid-encoded protein sequences was assessed from the results of reciprocal best BLAST hits (rBBHs) applying a threshold of E-value ≤ 1×10−5 (using BLAST version 2.6.0+ [55]). Pairs of rBBHs were further compared by global alignment with needle (version 6.6.0.0; EMBOSS package [56]. Sequence pairs with ≥50% identical amino acids were clustered into protein families using the Markov clustering algorithm (MCL) (version 12–135) with the default parameters [57]. To identify chromosome-encoded homologs of plasmid genes, the sequences of all plasmid protein families were blasted against all the chromosome proteins using BLAST with a threshold of E-value ≤ 1×10−10. The resulting chromosomal hits were further compared by global alignment using needle; chromosome-encoded protein sequences were considered homologs using a threshold of ≥40% identical amino acids. AMR protein families were identified using Resistance Gene Identifier (version 5.1.0), with CARD database (version 3.0.7) [58]. Plasmid mobility was inferred depending on the presence of the Mob relaxase and T4SS encoding genes in the plasmid genomes (as listed in [59]). Plasmids were classified as mobilizable if they encode a Mob protein and as conjugative if they have at least 15 tra genes. The dataset of essential genes in E. coli includes 394 protein-coding genes included in the essential genome of E. coli K-12 [25]. These genes were identified manually in the genome of E. coli K-12 substr. MG1655 according to their gene symbol. This dataset was supplemented with additional 110 protein-coding genes identified as essential in a large-scale study that included multiple E. coli isolates and three growth media [18], conditions that the gene was essential in all 18 tested isolates in at least one of the tested media (LB, GMM, M9). These genes were identified manually in the genome of E. coli str. K-12 substr. MG1655 according to the gene symbol and locus tag. The final set included 504 protein-coding essential genes (listed in S2 Table). The protein sequences of chromosome-encoded essential genes and their plasmid homologs were aligned using MAFFT (version v7.123b) [60]. Maximum likelihood trees were reconstructed using IQ-Tree (version 1.5.5) [61] with LG model. The 17 alignments and trees are available as supplementary data. Trees were rooted using the midpoint criterion. The topology of trees comprising a deep plasmid-chromosome split and putative LGT events was further tested with IQ-TREE as following: a constrained tree topology assuming vertical evolution only (i.e., assuming a deep chromosome-plasmid split) was reconstructed; The likelihood of the resulting constrained topology was compared to the unconstrained topology (i.e., with the LGT event) using the approximately unbiased (AU) topology test. A rejection of the constrained topology thus supports the LGT inference. Codon-aware alignments were produced using PAL2NAL program [62]. For trees with a split formed by homologs on different replicons, selective strength on different branches were calculated using HyPhy-RELAX (version 2.5.15) [63]. The resulting trees were visualized using iTOL v5 (https://itol.embl.de/) [64]. Conserved syntenic blocks (i.e., gene order) were identified using CSBFinder-S [65] allowing ≥ 3 insertions.
Bacterial strains, plasmids and culture conditions
The strain Escherichia coli K-12 MG1655 was used as the model organism in all experiments (DSM No. 18039, German Collection of Microorganisms and Cell Cultures, DSMZ). In addition, we used the E. coli strain MGM100 that encodes the groE operon under an inducible PBAD promoter [34]. For the purpose of competition experiments, E. coli MG1655 equipped with a chromosomal mini-Tn7 insertion (attTn7:miniTn7(dhfrII) [16]) conferring resistance to trimethoprim (Tmr) was used. The strain E. coli DH5α was used during plasmid construction. All E. coli strains were routinely grown at 37°C in Luria Bertani (LB- Lennox) medium at 250 rpm shaking. 1% Arabinose was absent from the growth media, unless when stated otherwise. For molecular cloning and documentation purpose, the plasmid carrying strains were grown in LB supplemented with kanamycin (25 μg per ml).
The plasmids in this study are derived from the plasmids pCON and pCON-S that were constructed previously [16,36]. Their backbone is comprised of a pBBR1 origin (rep and oriV [35]) and they carry the antibiotic resistance gene nptII encoding for a neomycin phosphotransferase that confers resistance to kanamycin (including the natural Tn5 promoter [66]). The plasmids pGroE and pGroE-S were built by PCR amplification of pCON and pCON-S (for primers see S6 Table). The chromosomal operon groE, including its native promoter, was amplified from the genome of E. coli MG1655 by PCR using the primers groE_for/rev (S6 Table). Subsequently, the plasmids were assembled using the NEBuilder® protocol (New England Biolabs). The plasmid-derived GroE (WP_004201172.1, WP_004201176.1) was custom synthesized by Genscript and comprised the sequence of the strain E. coli 6409 (NZ_CP020056.1; Fig 2C). The sequence was introduced into pCON-S by PCR amplification (for primers see S6 Table) and NEBuilder® asssembly. All plasmids were extracted using the GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific) and DNA concentrations were measured using a NanoDrop (Thermo Fisher Scientific).
Fitness experiments
The relative fitness (w) [67] of plasmid-carrying versus ancestral plasmid-free strain (wt) was estimated by pairwise competition experiments, with four replicates per plasmid type. All competition experiments were initiated with a 1:1 mixture of 1:100 diluted plasmid-carrying and ancestral strain (E. coli MG1655 Tmr [16]) from overnight cultures, in a total volume of 1 ml of non-selective LB medium. The relative fitness of plasmid host strains was calculated by evaluating cell counts at time points 0 h and 24 h. The strains were distinguished through plating on non-selective (LB) and selective media (LB supplemented with trimethoprim 150 μg per ml and LB supplemented with kanamycin 25 μg per ml). The chromosomal integration (Tmr) as well as the plasmid pCON have no measurable impact on the fitness of E. coli MG1655 (previously shown in [16]).
Evolution experiment
Evolution experiments were conducted with plasmid-carrying strains under non-selective conditions. On the onset of the experiment, the host strains were plated on selective media to ensure plasmid carriage (kanamycin, 25 μg per ml). The experiments were founded by nine replicate colonies of each plasmid type which were inoculated in LB medium at 37°C constant shaking. After overnight growth all cultures were diluted 1:100,000 and transferred into 96-deep-well plates in a total volume of 1 ml. The diluted cultures were incubated at 37°C with constant shaking. The populations were transferred every 24 h by diluting the cultures 1:100,000 and the serial transfer was repeated over a total of 23 transfers for pGroE/pGroE-S populations. The number of generations was routinely measured by evaluating the cell number through plating directly after the dilution and before the next subsequent transfer. We observed a total number of ~320 generations for the pGroE/pGroE-S populations. During the evolution experiment, the frequency of plasmids in the population was estimated from the proportion of hosts, which was determined by replica plating (Lederberg and Lederberg 1952) every 7 transfers. Briefly, this was performed by first serially diluting the grown cultures followed by plating of ~500 cells on non-selective LB media. The number of plated cells was increased with decreasing plasmid frequency up to ~1000 cells. The plated populations were incubated for overnight growth. Colonies were counted and replicated using velvet cloth on selective media (LB supplemented with kanamycin 25 μg per ml). Colonies growing on the selective media were counted as plasmid hosts (for a detailed protocol see [68]).
To test for plasmid extinction and plasmid stability evolution, the evolved populations were transferred into a 96-deep-well plate containing selective media (LB supplemented with kanamycin, 25 μg per ml) and incubated for overnight growth (12 h). Thereafter, the cultures were transferred into non-selective conditions for a follow-up evolution experiment. Plasmid host frequency was monitored via replica plating.
Plasmid loss frequency assays
The plasmid loss frequency was estimated from the frequency of plasmid free cells occurring during overnight growth in non-selective media. To determine the loss frequency, cultures were inoculated from single colonies grown on selective media to ensure plasmid carriage. After 12 h growth in 37°C (approximately 8.5 generations), the cultures were serially diluted and plated on non-selective LB media. After overnight incubation the plates were replicated. Following overnight growth, colonies grow on replicated plates were counted as plasmid carrying. The loss frequency was calculated from plasmid-free cells (not resistant) and the total number of colonies tested.
Plasmid copy number determination
Plasmid copy number (PCN) was determined using quantitative real time PCR (qPCR) (as described in [16]). Bacterial cells were lysed by 10 min incubation at 98°C directly followed by 10 min at -20°C. The qPCR was conducted with primers targeting the chromosome and the plasmid DNA. The chromosomal primers were complementary to the idnT gene of E. coli (q_idnT_F/R) and the plasmid primers targeted the nptII gene (q_nptII_F/R). The qPCR reactions were conducted in volume of 10 μl containing 1x iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories), 100 nM of each primer (final concentration) and 1 μl sample. All qPCR reactions including positive and non-template controls were performed in technical replicates on a Real-Time PCR Detection System (Bio-Rad) using: 95°C for 3 min, and 40 cycles of 10 s at 95°C and 1 min at 59°C cycling conditions for all reactions. The ratio between the number of plasmid amplicons and chromosome amplicons is defined as the plasmid copy number (comparative CT (ΔΔCT) method) and was calculated while considering the amplification efficiencies of both primer pairs.
Population growth dynamics
The growth dynamics of E. coli populations carrying different plasmids were measured by determining the optical density at 600 nm (OD600) using a photo-spectrometer (Thermo Fisher Scientific) during growth at 37°C or 42°C for 24 h. Thereafter, the R package growthcurver [69], was used to fit a logistic growth model to the growth dynamics and estimate the growth parameters including the growth rate (r).
Bacterial viability assay
Bacterial viability was evaluated by staining E. coli cells with Propidium iodide (PI, Sigma). PI enters only compromised bacterial membranes and is therefore an indicator of membrane integrity (i.e., does not enter alive cells) and only stains dead cells. E. coli populations were inoculated in liquid media and the cultures were sampled after two hours (log cells) and ten hours (stationary cells) of growth (3 or 4 replicates per strain). The cells were washed in PBS (20 μl) and incubated with PI at a final concentration of 30 μM for 10 min in the dark at room temperature. Thereafter, 10 μl of the mixture were transferred to an agar-coated slide and the cells were visualized with an epifluorescence microscope (Zeiss Axio Imager 2, Plan-Apochromat 63×/1.40 Oil DIC M27 objective). For counting purposes at least 10 images were taken per sample at random locations. The cell number of stained versus non-stained cells were evaluated using the software ImageJ.
Sequencing analysis
Population sequencing of the ancestral and evolved populations (one or two per treatment) was used to detect genetic variants occurring either on the plasmid or the host chromosomes. Prior to the sequencing, cultures were grown in antibiotics to ensure plasmid presence (see S20 Fig). Total DNA was isolated from 1 ml culture using the Wizard Genomic DNA Purification Kit (Promega). Concentration and quality of the extracted plasmid and genomic DNA was assessed using the NanoDrop (Thermo Fisher Scientific) and Qubit (Invitrogen by Life Technologies). The sample libraries for Illumina sequencing were prepared using the Nextera Flex library kit (Illumina, Inc) and sequencing was performed with paired-end reads on the MiSeq platform (Illumina, Inc).
Sequencing reads were trimmed to remove Illumina specific adaptors and low quality bases using the program Trimmomatic v.0.35 [70] (parameters: ILLUMINACLIP:NexteraPE-PE.fa:2:30:10 CROP:250 HEADCROP:5 LEADING:20 TRAILING:20 SLIDINGWINDOW:4:20 MINLEN:36). As the reference we joined the E. coli MG1655 (GenBank acc. no. NC_000913.3) genome with the plasmid sequences (created in SnapGene software v.2.4 (GLS Biotech)). The sequencing reads were mapped to the reference genomes using BWA-MEM v.0.7.5a-r405 [71]. Mapping statistics were retrieved using BAMStats v.1.25 (https://sourceforge.net/projects/bamstats/files/). Subsequent indexing and local realignment of sequencing reads were performed using PICARD tools, SAMtools v.0.1.19 [72] and GATK v.3.6 [73] retaining only paired mapped reads with a minimum mapping quality of 20. SNPs were called using LoFreq v.2.1.2 [74].
Statistical analysis
All statistical tests and data analysis were performed in R version 3.5.1.
Supporting information
An inlay plot (in red dashed line) shows an enlargement of the top left corner. The distribution shows that 81% plasmid protein families have no chromosomal homolog in the same isolate. The remaining 19% plasmid protein families have a chromosomal homolog in at least one isolate.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes. MetG in Escherichia coli isolate 2014C-3307 is found only on a plasmid (i.e., it has no chromosomal homolog).
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Branches are colored according to the selection intensity parameter k, ranging between brown for k<1 (relaxed selection) and green for k>1 (intensified selection). Note that branch length information is excluded in this figure. Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Branches are colored according to the selection intensity parameter k, ranging between brown for k<1 (relaxed selection) and green for k>1 (intensified selection). Note that branch length information is excluded in this figure. Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
(TIFF)
(TIFF)
(TIFF)
Plasmid carrying populations were exposed to a transfer in antibiotics (kanamycin 25 μg/ml).
(TIFF)
The plasmid loss was measured after overnight incubation (n = 6).
(TIFF)
A total of 13 contigs were excluded from the analysis either because they better matched phage genomes, or were suspected as mis-assembly artifacts (i.e., suspected as chromosome sequences rather than plasmid sequences. Accession number are listed in the table).
(XLSX)
(XLSX)
The conserved genomic neighborhood comprises five genes including: ssb encoding single-strand DNA binding protein, parB encoding for a type II partitioning system as well as psiA and psiB that are known as inhibitors of the SOS response.
(XLSX)
The conserved genomic neighborhood comprises 7–8 genes that include the chaperonin complex (groEL and groES), blaNDM-1, ble (bleomycin binding protein Ble-MBL), a gene encoding a phosphoribosylanthranilate isomerase, a gene encoding twin-arginine translocation (TAT) pathway signal sequence domain protein and cutA (divalent-cation tolerance protein). In six out of ten instances the conserved neighborhood includes a gene encoding for a IS91 family transposase.
(XLSX)
Sequenced samples include ancestral populations as well as evolved strains carrying the plasmid pGroE or pGroE-S.
(TIFF)
(TIFF)
(GZ)
Acknowledgments
We thank Erik Brinks for his assistance with the genome sequencing. We thank Ana Garoña and Nils Hülter for critical comments on the manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files. All sequencing files are available from the SRA database (accession number PRJNA714291).
Funding Statement
TD received funding from Deutsche Forschungsgemeinschaft Grant No. DA1202/2-1 and GRK2501 (https://www.dfg.de/). Yiqing Wang was funded by the China Scholarship Council (CSC), Beijing, China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Heuer H, Schmitt H, Smalla K. Antibiotic resistance gene spread due to manure application on agricultural fields. Current Opinion in Microbiology. 2011;14: 236–243. doi: 10.1016/j.mib.2011.04.009 [DOI] [PubMed] [Google Scholar]
- 2.Blau K, Bettermann A, Jechalke S, Fornefeld E, Vanrobaeys Y, Stalder T, et al. The transferable resistome of produce. mBio. 2018;9: 464–15. doi: 10.1128/mBio.01300-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio. 2014;5: e1002158–10. doi: 10.1128/mBio.01918-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dziewit L, Pyzik A, Szuplewska M, Matlakowska R, Mielnicki S, Wibberg D, et al. Diversity and role of plasmids in adaptation of bacteria inhabiting the Lubin copper mine in Poland, an environment rich in heavy metals. Frontiers in Microbiology. 2015;6: 1–12. doi: 10.3389/fmicb.2015.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouma JE, Lenski RE. Evolution of a bacteria/plasmid association. Nature. 1988;335: 351–352. doi: 10.1038/335351a0 [DOI] [PubMed] [Google Scholar]
- 6.Heuer H, Fox RE, Top EM. Frequent conjugative transfer accelerates adaptation of a broad-host-range plasmid to an unfavorable Pseudomonas putida host. FEMS Microbiology Ecology. 2007;59: 738–748. doi: 10.1111/j.1574-6941.2006.00223.x [DOI] [PubMed] [Google Scholar]
- 7.Sota M, Yano H, Hughes JM, Daughdrill GW, Abdo Z, Forney LJ, et al. Shifts in the host range of a promiscuous plasmid through parallel evolution of its replication initiation protein. The ISME Journal. 2010;4: 1568–1580. doi: 10.1038/ismej.2010.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loftie-Eaton W, Bashford K, Quinn H, Dong K, Millstein J, Hunter S, et al. Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nature Ecology & Evolution. 2017;1: 1354–1363. doi: 10.1038/s41559-017-0243-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stalder T, Rogers LM, Renfrow C, Yano H, Smith Z, Top EM. Emerging patterns of plasmid-host coevolution that stabilize antibiotic resistance. Scientific Reports. 2017;7: 4853. doi: 10.1038/s41598-017-04662-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison PW, Lower RPJ, Kim NKD, Young JPW. Introducing the bacterial ‘chromid’: not a chromosome, not a plasmid. Trends in Microbiology. 2010;18: 141–148. doi: 10.1016/j.tim.2009.12.010 [DOI] [PubMed] [Google Scholar]
- 11.Rancati G, Moffat J, Typas A, Pavelka N. Emerging and evolving concepts in gene essentiality. Nature Reviews Genetics. 2018;19: 34–49. doi: 10.1038/nrg.2017.74 [DOI] [PubMed] [Google Scholar]
- 12.Gil R, Sabater-Muñoz B, Perez-Brocal V, Silva FJ, Latorre A. Plasmids in the aphid endosymbiont Buchnera aphidicola with the smallest genomes. A puzzling evolutionary story. Gene. 2006;370: 17–25. doi: 10.1016/j.gene.2005.10.043 [DOI] [PubMed] [Google Scholar]
- 13.diCenzo G, Milunovic B, Cheng J, Finan TM. The tRNAarg gene and engA are essential genes on the 1.7-Mb pSymB megaplasmid of Sinorhizobium meliloti and were translocated together from the chromosome in an ancestral strain. Journal of Bacteriology. 2012;195: 202–212. doi: 10.1128/JB.01758-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anda M, Ohtsubo Y, Okubo T, Sugawara M, Nagata Y, Tsuda M, et al. Bacterial clade with the ribosomal RNA operon on a small plasmid rather than the chromosome. Proceedings of the National Academy of Sciences. 2015;112: 14343–14347. doi: 10.1073/pnas.1514326112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tazzyman SJ, Bonhoeffer S. Why There Are No Essential Genes on Plasmids. Molecular Biology and Evolution. 2014; msu293–10. doi: 10.1093/molbev/msu293 [DOI] [PubMed] [Google Scholar]
- 16.Wein T, Hülter NF, Mizrahi I, Dagan T. Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nature Communications. 2019;10: 2595–13. doi: 10.1038/s41467-019-10600-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hülter N, Ilhan J, Wein T, Kadibalban AS, Hammerschmidt K, Dagan T. An evolutionary perspective on plasmid lifestyle modes. Current Opinion in Microbiology. 2017;38: 74–80. doi: 10.1016/j.mib.2017.05.001 [DOI] [PubMed] [Google Scholar]
- 18.Rousset F, Cabezas-Caballero J, Piastra-Facon F, Fernández-Rodríguez J, Clermont O, Denamur E, et al. The impact of genetic diversity on gene essentiality within the Escherichia coli species. Nature Microbiology. 2021;6: 301–312. doi: 10.1038/s41564-020-00839-y [DOI] [PubMed] [Google Scholar]
- 19.Nicoloff H, Hjort K, Levin BR, Andersson DI. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nature Microbiology. 2019;4: 504–514. doi: 10.1038/s41564-018-0342-0 [DOI] [PubMed] [Google Scholar]
- 20.Andersson DI, Jerlström-Hultqvist J, Näsvall J. Evolution of New Functions De Novo and from Preexisting Genes. Cold Spring Harbor Perspectives in Biology. 2015;7: a017996–19. doi: 10.1101/cshperspect.a017996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bratlie MS, Johansen J, Sherman BT, Huang DW, Lempicki RA, Drabløs F. Gene duplications in prokaryotes can be associated with environmental adaptation. BMC Genomics. 2010;11: 588. doi: 10.1186/1471-2164-11-588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: The complexity hypothesis. Proceedings of the National Academy of Sciences. 1999;96: 3801–3806. doi: 10.1073/pnas.96.7.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318: 1449–1452. doi: 10.1126/science.1147112 [DOI] [PubMed] [Google Scholar]
- 24.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular Systems Biology. 2006;2: 2460–11. doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodall ECA, Robinson A, Johnston IG, Jabbari S, Turner KA, Cunningham AF, et al. The essential genome of Escherichia coli K-12. mBio. 2018;9: 385–18. doi: 10.1128/mBio.02096-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcillán-Barcia MP, Alvarado A, de la Cruz F. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiology Reviews. 2011;35: 936–956. doi: 10.1111/j.1574-6976.2011.00291.x [DOI] [PubMed] [Google Scholar]
- 27.Chase JW, Merrill BM, Williams KR. F sex factor encodes a single-stranded DNA binding protein (SSB) with extensive sequence homology to Escherichia coli SSB. Proceedings of the National Academy of Sciences. 1983;80: 5480–5484. doi: 10.1073/pnas.80.18.5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao W, Golding GB. The fate of laterally transferred genes: Life in the fast lane to adaptation or death. Genome Research. 2006;16: 636–643. doi: 10.1101/gr.4746406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark GW, Tillier ERM. Loss and gain of GroEL in the Mollicutes. Biochemistry and Cell Biology. 2010;88: 185–194. doi: 10.1139/o09-157 [DOI] [PubMed] [Google Scholar]
- 30.Wallington EJ, Lund PA. Rhizobium leguminosarum contains multiple chaperonin (cpn60) genes. Microbiology. 1994;140: 113–122. doi: 10.1099/13500872-140-1-113 [DOI] [PubMed] [Google Scholar]
- 31.Fayet O, Ziegelhoffer T, Georgopoulos C. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol. 1989;171: 1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekizuka T, Matsui M, Yamane K, Takeuchi F, Ohnishi M, Hishinuma A, et al. Complete Sequencing of the blaNDM-1-Positive IncA/C Plasmid from Escherichia coli ST38 Isolate Suggests a Possible Origin from Plant Pathogens. PLoS ONE. 2011;6: e25334–7. doi: 10.1371/journal.pone.0025334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Feng Y, Zhou W, McNally A, Zong Z. Cryptic transmission of ST405 Escherichia coli carrying bla NDM-4 in hospital. Sci Rep. 2018;8: 390. doi: 10.1038/s41598-017-18910-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLennan N, Masters M. GroE is vital for cell-wall synthesis. Nature. 1998;392: 139–139. doi: 10.1038/32317 [DOI] [PubMed] [Google Scholar]
- 35.Antoine R, Locht C. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from Gram-positive organisms. Molecular Microbiology. 1992;6: 1785–1799. doi: 10.1111/j.1365-2958.1992.tb01351.x [DOI] [PubMed] [Google Scholar]
- 36.Wein T, Wang Y, Hülter NF, Hammerschmidt K, Dagan T. Antibiotics interfere with the evolution of plasmid stability. Current biology: CB. 2020;30: 3841–3847.e4. doi: 10.1016/j.cub.2020.07.019 [DOI] [PubMed] [Google Scholar]
- 37.Richmond C, Glasner JD, Mau R, Jin H, Blattner FR. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Research. 1999;27: 3821–3835. doi: 10.1093/nar/27.19.3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunasekera TS, Csonka LN, Paliy O. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. Journal of Bacteriology. 2008;190: 3712–3720. doi: 10.1128/JB.01990-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodríguez-Beltrán J, Hernandez-Beltran JCR, DelaFuente J, Escudero JA, Fuentes-Hernandez A, MacLean RC, et al. Multicopy plasmids allow bacteria to escape from fitness trade-offs during evolutionary innovation. Nature Ecology & Evolution. 2018; 1–11. doi: 10.1038/s41559-018-0529-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergstrom CT, Lipsitch M, Levin BR. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics. 2000;155: 1505–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison E, Brockhurst MA. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends in Microbiology. 2012;20: 262–267. doi: 10.1016/j.tim.2012.04.003 [DOI] [PubMed] [Google Scholar]
- 42.Wheatley R, Diaz Caballero J, Kapel N, de Winter FHR, Jangir P, Quinn A, et al. Rapid evolution and host immunity drive the rise and fall of carbapenem resistance during an acute Pseudomonas aeruginosa infection. Nature Communications. 2021;12: 2460. doi: 10.1038/s41467-021-22814-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lercher MJ, Pal C. Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Molecular Biology and Evolution. 2008;25: 559–567. doi: 10.1093/molbev/msm283 [DOI] [PubMed] [Google Scholar]
- 44.Baltrus DA. Exploring the costs of horizontal gene transfer. Trends in Ecology & Evolution. 2013;28: 489–495. doi: 10.1016/j.tree.2013.04.002 [DOI] [PubMed] [Google Scholar]
- 45.Cohen O, Gophna U, Pupko T. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Molecular Biology and Evolution. 2011;28: 1481–1489. doi: 10.1093/molbev/msq333 [DOI] [PubMed] [Google Scholar]
- 46.Acar Kirit H, Lagator M, Bollback JP. Experimental determination of evolutionary barriers to horizontal gene transfer. BMC Microbiology. 2020;20: 326–13. doi: 10.1186/s12866-020-01983-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fares MA, Ruiz-González MX, Moya A, Elena SF, Barrio E. GroEL buffers against deleterious mutations. Nature. 2002;417: 398–398. doi: 10.1038/417398a [DOI] [PubMed] [Google Scholar]
- 48.Sabater-Muñoz B, Prats-Escriche M, Montagud-Martínez R, López-Cerdán A, Toft C, Aguilar-Rodríguez J, et al. Fitness trade-offs determine the role of the molecular chaperonin GroEL in buffering mutations. Molecular Biology and Evolution. 2015;32: 2681–2693. doi: 10.1093/molbev/msv144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogumil D, Dagan T. Cumulative Impact of Chaperone-Mediated Folding on Genome Evolution. Biochemistry. 2012;51: 9941–9953. doi: 10.1021/bi3013643 [DOI] [PubMed] [Google Scholar]
- 50.Galibert F, Finan TM, Long SR, Pühler A, Abola P, Ampe F, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293: 668–672. doi: 10.1126/science.1060966 [DOI] [PubMed] [Google Scholar]
- 51.Bittner AN, Foltz A, Oke V. Only One of Five groEL Genes Is Required for Viability and Successful Symbiosis in Sinorhizobium meliloti. Journal of Bacteriology. 2007;189: 1884–1889. doi: 10.1128/JB.01542-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boyd BM, Allen JM, Nguyen N-P, Vachaspati P, Quicksall ZS, Warnow T, et al. Primates, lice and bacteria: speciation and genome evolution in the symbionts of hominid lice. Molecular Biology and Evolution. 2017;34: 1743–1757. doi: 10.1093/molbev/msx117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart FM, Levin BR. The population biology of bacterial plasmids: a priori conditions for the existence of conjugationally transmitted factors. Genetics. 1977;87: 209–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lau BTC, Malkus P, Paulsson J. New quantitative methods for measuring plasmid loss rates reveal unexpected stability. Plasmid. 2013;70: 353–361. doi: 10.1016/j.plasmid.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10: 421–9. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics. 2000;16: 276–277. doi: 10.1016/s0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- 57.Enright AJ, Dongen SV, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Research. 2002;30: 1575–1584. doi: 10.1093/nar/30.7.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Research. 2019;10: 226–9. doi: 10.1093/nar/gkz935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480: 241–244. doi: 10.1038/nature10571 [DOI] [PubMed] [Google Scholar]
- 60.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution. 2013;30: 772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Molecular Biology and Evolution. 2014;32: 268–274. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Research. 2006;34: W609–W612. doi: 10.1093/nar/gkl315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. Detecting Individual Sites Subject to Episodic Diversifying Selection. PLoS Genetics. 2012;8: e1002764–10. doi: 10.1371/journal.pgen.1002764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Research. 2019;47: W256–W259. doi: 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Svetlitsky D, Dagan T, Ziv-Ukelson M. Discovery of multi-operon colinear syntenic blocks in microbial genomes. Bioinformatics. 2020;36: i21–i29. doi: 10.1093/bioinformatics/btaa503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beck E, Ludwig G, Auerswald EA, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19: 327–336. doi: 10.1016/0378-1119(82)90023-3 [DOI] [PubMed] [Google Scholar]
- 67.Lenski RE. Quantifying Fitness and Gene Stability in Microorganisms. Assessing Ecological Risks of Biotechnology. Elsevier; 1991. pp. 173–192. doi: 10.1016/b978-0-409-90199-3.50015-2 [DOI] [PubMed] [Google Scholar]
- 68.Wein T, Stücker FT, Hülter NF, Dagan T. Quantification of plasmid-mediated antibiotic resistance in an experimental evolution approach. Journal of Visualized Experiments. 2019; e60749. doi: 10.3791/60749 [DOI] [PubMed] [Google Scholar]
- 69.Sprouffske K, Wagner A. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics. 2016;17: 172. doi: 10.1186/s12859-016-1016-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30: 2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25: 1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25: 2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20: 1297–1303. doi: 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilm A, Aw PPK, Bertrand D, Yeo GHT, Ong SH, Wong CH, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Research. 2012;40: 11189–11201. doi: 10.1093/nar/gks918 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An inlay plot (in red dashed line) shows an enlargement of the top left corner. The distribution shows that 81% plasmid protein families have no chromosomal homolog in the same isolate. The remaining 19% plasmid protein families have a chromosomal homolog in at least one isolate.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes. MetG in Escherichia coli isolate 2014C-3307 is found only on a plasmid (i.e., it has no chromosomal homolog).
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Branches are colored according to the selection intensity parameter k, ranging between brown for k<1 (relaxed selection) and green for k>1 (intensified selection). Note that branch length information is excluded in this figure. Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Branches are colored according to the selection intensity parameter k, ranging between brown for k<1 (relaxed selection) and green for k>1 (intensified selection). Note that branch length information is excluded in this figure. Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
Isolate names are colored according to gene location with red for plasmids and blue for chromosomes.
(TIF)
(TIFF)
(TIFF)
(TIFF)
Plasmid carrying populations were exposed to a transfer in antibiotics (kanamycin 25 μg/ml).
(TIFF)
The plasmid loss was measured after overnight incubation (n = 6).
(TIFF)
A total of 13 contigs were excluded from the analysis either because they better matched phage genomes, or were suspected as mis-assembly artifacts (i.e., suspected as chromosome sequences rather than plasmid sequences. Accession number are listed in the table).
(XLSX)
(XLSX)
The conserved genomic neighborhood comprises five genes including: ssb encoding single-strand DNA binding protein, parB encoding for a type II partitioning system as well as psiA and psiB that are known as inhibitors of the SOS response.
(XLSX)
The conserved genomic neighborhood comprises 7–8 genes that include the chaperonin complex (groEL and groES), blaNDM-1, ble (bleomycin binding protein Ble-MBL), a gene encoding a phosphoribosylanthranilate isomerase, a gene encoding twin-arginine translocation (TAT) pathway signal sequence domain protein and cutA (divalent-cation tolerance protein). In six out of ten instances the conserved neighborhood includes a gene encoding for a IS91 family transposase.
(XLSX)
Sequenced samples include ancestral populations as well as evolved strains carrying the plasmid pGroE or pGroE-S.
(TIFF)
(TIFF)
(GZ)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files. All sequencing files are available from the SRA database (accession number PRJNA714291).