Abstract
The spike protein of coronavirus is key target for drug development and other pharmacological interventions. In current study, we performed an integrative approach to predict antigenic sites in SARS-CoV-2 spike receptor binding domain and found nine potential antigenic sites. The predicted antigenic sites were then assessed for possible molecular similarity with other known antigens in different organisms. Out of nine sites, seven sites showed molecular similarity with 54 antigenic determinants found in twelve pathogenic bacterial species (Mycobacterium tuberculosis, Mycobacterium leprae, Bacillus anthracis, Borrelia burgdorferi, Clostridium perfringens, Clostridium tetani, Helicobacter Pylori, Listeria monocytogenes, Staphylococcus aureus, Streptococcus pyogenes, Vibrio cholera and Yersinia pestis), two malarial parasites (Plasmodium falciparum and Plasmodium knowlesi) and influenza virus A. Most of the bacterial antigens that displayed molecular similarity with antigenic sites in SARS-CoV-2 RBD (receptor binding domain) were toxins and virulent factors. Antigens from Mycobacterium that showed similarity were mainly involved in modulating host cell immune response and ensuring persistence and survival of pathogen in host cells. Presence of a large number of antigenic determinants, similar to those in highly pathogenic microorganisms, not merely accounts for complex etiology of the disease but also provides an explanation for observed pathophysiological complications, such as deregulated immune response, unleashed or dysregulated cytokine secretion (cytokine storm), multiple organ failure etc., that are more evident in aged and immune-compromised patients. Over-representation of antigenic determinants from Plasmodium and Mycobacterium in all antigenic sites suggests that anti-malarial and anti-TB drugs can prove to be clinical beneficial for COVID-19 treatment. Besides this, anti-leprosy, anti-lyme, anti-plague, anti-anthrax drugs/vaccine etc. are also expected to be beneficial in COVID-19 treatment. Moreover, individuals previously immunized/vaccinated or had previous history of malaria, tuberculosis or other disease caused by fifteen microorganisms are expected to display a considerable degree of resistance against SARS-CoV-2 infection. Out of the seven antigenic sites predicted in SARS-CoV-2, a part of two antigenic sites were also predicted as potent T-cell epitopes (KVGGNYNYL444-452 and SVLYNSASF366-374) against MHC class I and three (KRISNCVADYSVLYN356-370, DLCFTNVYADSFVI389-402, and YRVVVLSFELLHA508-520) against MHC class II. All epitopes possessed significantly lower predicted IC50 value which is a prerequisite for a preferred vaccine candidate for COVID-19.
Keywords: SARS-CoV-2, Mycobacterium, Yersinia, Listeria, Plasmodium, Bacillus, Borrelia, Vaccine, Antigenic variations, Antigenic drift, Antigenic shift
1. Introduction
The recent outbreak of novel coronavirus SARS-CoV-2 (earlier known as 2019-nCoV) in Wuhan city of China has led to serious health crisis as well as impacted socio-economic development globally. Alike previous SARS-CoV-1, the SARS-CoV-2 is also an enveloped “positive” single-stranded RNA virus. SARS-CoV-2’s genome encodes for 12 proteins that include non-structural proteins and four structural proteins such as S (spike), E (envelop), M (membrane) and N (nucleocapsid) proteins (Kuiken et al., 2003, Marra et al., 2003, Peiris et al., 2003, Rota et al., 2003). The ability of coronaviruses to enter into host cells and infect them is due to their ability to establish strong attachment with host cell receptor proteins such as integrins and others via their RGD motif present in their receptor binding domain of spike glycoproteins (Dakal, 2020, Li et al., 2005). The RGD motifs are well known in the field of cell biology, cell therapy and tissue engineering because of their remarkable cell-adhesive property (Bellis, 2011). Extracellular matrix proteins, such as fibronectin and laminin, possess RGD motif and are frequently coated onto the surface of biomaterials (petri-dishes, T-flasks) for facilitating human cells adhesion onto the surface of the biomaterials (Bellis, 2011). Bivalent ions such as Ca+2 & Mg+2 play indispensable role in promoting such cell adhesion and EDTA are used as a chelating agent for disaggregating Ca+2 mediated cell bondage. The RGD-integrin mediated virus-host attachment has been found to be dependent upon the calcium and other divalent ions’ concentration (Dakal, 2020).
Initially the spike proteins bind to cellular receptor angiotensin-converting enzyme 2 (ACE2). Following the cleavage of spike protein and with the help of proteases produce 2 domains of spike protein, S1 known as the receptor binding domain, which recognizes and bind to the RBD and S2 which fuse with the membrane following the entry of the virus into the host cell (Hoffmann et al., 2020). After that the viral RNA get exposed in the cytoplasm of the host cell. Virus infection in humans results in two major types of immune response. The first is an innate immune response, which is first line of defense and involves synthesis of proteins called interferons (key regulators of viral replication) and stimulation of a number of immune cells (Theofilopoulos et al., 2005, Wei et al., 2014). The viral infection is detected by innate immune system by pattern recognition receptors (PRRs) that recognize pathogen‐associated molecular patterns (PAMPs). The PRRs mainly include toll‐like receptor (TLR), RIG‐I‐like receptor (RLR), NOD‐like receptor (NLR), C‐type lectin‐like receptors (CLR), and free‐molecule receptors in the cytoplasm, such as cGAS, IFI16, STING, DAI, and so on (Thompson et al., 2011, Schock et al., 2017, Bottermann and James, 2018). After recognizing pathogenic features in the invading molecule, virus is delivered to macrophage and dendritic cells (DCs) that play crucial roles for viral destruction and immune response induction in mucosal-associated lymphoid tissues (MALT) (Spiegel et al., 2006, Spiegel and Weber, 2006, Frieman and Baric, 2008). Besides this, the host response is also regulated by a highly controlled network of cytokines (TNF-a, IL-6, IL-8, IP-10, MCP-1), chemokines (CXCL-1, CXCL-2, CCL-3 and CCL-5) and complement proteins (Frieman and Baric, 2008, Chen et al., 2010, van den Brand et al., 2014). Studies have also shown that activation of macrophages and DCs by SARS-CoV lead to excessive pro-inflammatory cytokine responses (Tseng et al., 2005, Mehta et al., 2020). On activation, an unleashed production of inflammatory cytokines and chemokines can be seen in the tissues and serum of the COVID-19 patients, a pathophysiological condition formally known as cytokine storm (Mehta et al., 2020, Huang et al., 2020). Also, the levels of IFN-γ, IL-1β, IL-2, IL-6, IL-7, IL-8, IL-10, IL-12, MCP-1, MIP1A, IP-10 and TNF-α are generally increased in the early infection; however, subsequently get lowered in the recovery stage (Huang et al., 2020). Upregulation of the inflammatory cytokines and chemokines like IL-1β, IFN-γ, IP-10, and MCP-1 may lead to activated T-helper-1 (Th1) cells response (Huang et al., 2020). However, it was also observed that SARS-CoV-2 patients secreted excessive IL-4 and IL-10 that may suppress inflammation via T-helper-2 (Th2) which makes the SARS-CoV-2 different from other previous coronavirus infections (Zhou and Zhao, 2020). The induction of interferons (IFN) along with anti-viral actions of macrophages and DCs at the sites of infection effectively hinders viral tropism in lung tissues/cells and eventually dampens virus’s efficiency to replicate and finally led to their elimination (Theofilopoulos et al., 2005, Garcia-Sastre and Biron, 2006, Seth et al., 2006, Versteeg et al., 2007).
In cases when the innate response is not enough to prevent viral infection, adaptive immunity comes into action, especially during the later stages of viral infection in which infection has already proceeded beyond the first few rounds of viral replication. As the part of adaptive immune response, antigen presenting cells such as B-cells, macrophages and dendritic cells (DCs) recognize and present viral antigens and trigger activation and proliferation of T-helper (Th) cells. The T-helper cells are required for the generation of the humoral response (the synthesis of virus-specific antibodies by B lymphocytes) and the cell-mediated response (recognition and targeted killing of virus infected antigen-displaying altered self-cells by cytotoxic T-cells). The respiratory dendritic cells (rDCs) that resides in lung epithelium acquire the invading virus or its antigens from infected lung cells and become activated (Peebles and Graham, 2011, Tognarelli et al., 2019). The rDCs process viral antigen and subsequently migrate to the draining lymph nodes (DLN), where the presentation of the processed antigen to naïve T cells takes place in the form of MHC/peptide complex (Braciale et al., 2012, Guilliams et al., 2013, Neyt and Lambrecht, 2013). Once the T cell receptor (TCR) get engaged with MHC/peptide complex and additional co-stimulatory signals, T cells get activated which in turn result into their rapid proliferation, differentiation and recruitment to the site of virus infection (Larsson et al., 2000, Norbury et al., 2002, Belz et al., 2004). At the site of virus infection or tropism, effector T cells produce antiviral pro-inflammatory cytokines and chemokines, most notably, IFN-γ, TNF-α, IL-2, CXCL-9, CXCL-10, and CXCL-11 as well as some cytotoxic molecules such as perforin and granzyme B (Wherry and Ahmed, 2004). Under the influence of IL-2, IL-15 and other cytokines and chemokines, natural killer (NK) cells also develop and target virus-infected cells using antibody dependent cell-mediated cytotoxicity (ADCC) (Cooper et al., 2009). These effector molecules (cytokines, chemokines and cytotoxic molecules) in a multi-faceted way inhibit viral replication and enhance antigen presentation leading to recruitment of additional immune cells (such as NK-cells) of innate and adaptive system at the site of viral infection for destroying infected epithelial cells (virus-infected altered self-cells) and eliminating virus (Cerwenka et al., 1999, Cerwenka et al., 1999, Roman et al., 2002, Swain et al., 2002, Saha et al., 2010).
The spike protein is the key target for drug and vaccine development for preventing SARS-CoV-2 infection and combating COVID-19 (Du et al., 2009). In particular, the receptor binding domain (RBD), which stretches from residue 330 to 583 within the spike protein, is the most important structural module (Dakal, 2020, Wong et al., 2004). Several researchers demonstrated that spike protein of SARS-CoV-1 plays a key role in eliciting potent T-cell responses and binds with neutralizing-antibodies (Prabakaran et al., 2006, Janice Oh et al., 2012). In particular, the S1 glycoprotein of SARS-CoV-1spike protein has been found be an important immunodominant epitope which induces a number of neutralizing antibodies (Tian et al., 2020).
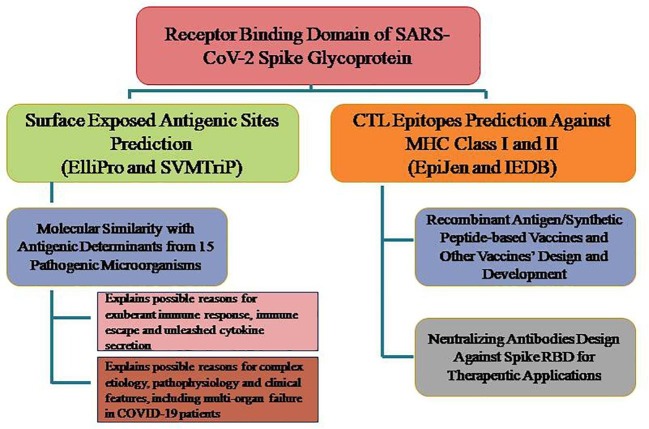
Of all structural proteins in SARS-CoV-2, spike proteins are the first to interact with the host cells, and therefore, the initial host cell immune response in COVID-19 patients is expected to be against the exposed antigenic epitopes in the spike proteins. This suggested that there is a strong need for exploration and prediction of antigenic sites and potent cytotoxic T-cell epitopes in spike RBD for rapid developments of novel vaccine candidates and neutralizing antibodies against SARS-CoV-2 infection. In current work, we have employed an integrated approach that comprises identification of potential antigenic sites and antigenic determinants in SARS-CoV-2’s RBD based on its primary sequence and 3D structure. These antigenic sites/determinants can trigger B-cells, T-cells and other immune cells mediated immune response and can explain the complex etiology, pathophysiology and other clinical features of COVID-19 patients. MHC molecules, also called as human leukocyte-associated (HLA) antigens, are cell surface glycoproteins that bind peptide fragments of proteins that either have been synthesized within the cell (class I MHC molecules) or that have been ingested by the cell and proteolytically processed (class II MHC molecules). Potent CTL epitopes against MHC class I/II have also been predicted. These CTL epitopes (peptides) can be used for development of DNA and recombinant vaccines as well as neutralizing antibodies against SARS-CoV-2. We believe that, it is most probable that SARS-CoV-2′s spike RBD-based vaccines will bear fruit in the near future, as they are expected to induce neutralizing antibodies to prevent viral entry into host cells and elicit long-term immune response against COVID-19.
2. Materials and methods
2.1. Retrieval of the SARS-CoV-2 proteome sequence data, sequence alignment and structural modeling
The complete proteome sequence of SARS-CoV-2 (YP_009724389, Wuhan, 12-2019) containing more than 7000 amino acids was downloaded from NCBI virus database (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/). The protein sequence of SARS-CoV-1 and SARS-CoV-2 coronaviruses were subjected to pair-wise sequence alignment using Clustal Omega using default setting (https://www.ebi.ac.uk/Tools/msa/clustalo/). The structural modeling was done using Chimera ver. 1.10.
2.2. Consensus antigenicity prediction
The consensus antigenicity of SARS-CoV-2 spike RBD was ascertained using AntigenPro (http://scratch.proteomics.ics.uci.edu/) (Magnan et al., 2010). The antigenic propensity was predicted using Antigenic Peptide tool of Universidad Complutense Madrid (imed.med.ucm.es/Tools/antigenic.pl).
2.3. Sequence- and Structure-based epitope prediction
We used the three-dimensional structure model of spike receptor binding domain (PB ID: 6LZG) (Wang et al., 2020) for the prediction of epitopes using ElliPro online server (http://tools.iedb.org/ellipro/) (Ponomarenko et al., 2008). We used an additional tool, namely SVMTriP, for prediction of linear epitopes in spike RBD of SARS-CoV-2 using its primary sequence (http://sysbio.unl.edu/SVMTriP/prediction.php). The tool employs support vector machine (SVM) in which the tripeptide similarity score and the propensity scores are combined to yield improved predictions (Yao et al., 2012).
2.4. Molecular similarity match
The predicted antigenic epitope sites were matched for molecular similarity with antigens present in other organisms using AntigenDB (https://webs.iiitd.edu.in/raghava/antigendb/epiquery.html) (Ansari et al., 2010). The antigenic sites were used as input in tripeptide format and output was obtained in the form of comprehensive information about experimentally verified epitopes from a wide range of known antigens.
2.5. Vaccine epitope prediction
The predicted antigenic sites in SARS-CoV-2 RBD were assessed for their ability to be processed and presented to cytotoxic T-cells by MHC class IB proteins. For this EpiJen (http://www.ddg-pharmfac.net/epijen/EpiJen/EpiJen.htm) was used for determining potential T-cell epitopes in SARS-CoV-2 spike RBD domain (Doytchinova et al., 2006). The prediction is based on known antigens that have been shown to be presented by MHC class I proteins in various experimental studies and are capable of inducing potent CTL immune responses. In addition, we also performed an analysis for prediction of peptides against the MHC class II alleles using NetMHCpan available in IEDB Analysis Resource of National Institute of Allergy and Infectious Diseases (NIH), USA (http://tools.iedb.org/mhci/) (Andreatta et al., 2015). Alleles of this HLA class are normally express on professional antigen-presenting cells (APCs) such as B lymphocytes, dendritic cells, mononuclear phagocytes, endothelial cells and thymic epithelial cells that are crucial in exacerbating immune responses in multi-faceted manner.
3. Results
3.1. SARS-CoV-2 spike RBD domain is highly antigenic
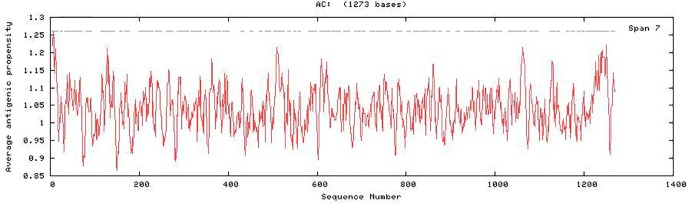
We performed pairwise sequence alignment of spike protein from SARS-CoV-1 and SARS-CoV-2 and found that the similarity between SARS-CoV-1 and SARS-CoV-2 spike receptor binding domain is approximately 70–80% (Dakal, 2020). The SARS-CoV-2 spike RBD was found to be highly antigenic with a predicted probability of antigenicity equal to 0.85, which is even more than the predicted probability of antigenicity of the complete SARS-CoV-2 spike protein (0.72). The average antigenic propensity of the SARS-CoV-2 is 1.0416 (Fig. 1 ). This showed that SARS-CoV-2 spike RBD possess highly antigenic sites that may activate immune cells and trigger immune response in multiple ways.
Fig. 1.
Antigenic plot for SARS-CoV-2 spike protein showing antigenic propensity for different antigenic sites predicted by Antigenic Peptide tool of Universidad Complutense Madrid (imed.med.ucm.es/Tools/antigenic.pl).
3.2. SARS-CoV-2 spike RBD domain possess nine potential antigenic sites
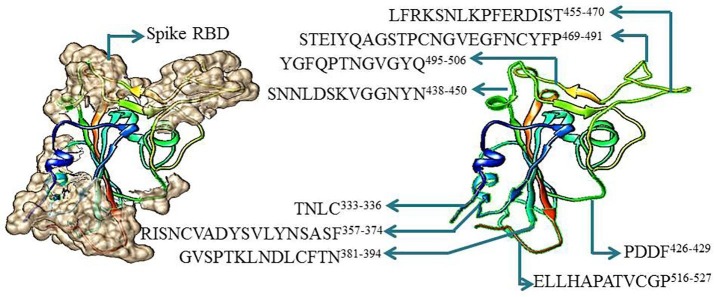
We subjected the 3D structure (PDB file) of SARS-CoV-2 spike RBD to epitope prediction using ElliPro and found that the RBD domain contains nine potential antigenic sites (Table 1 ). The length of these antigenic sites stretched from four amino acids to twenty three amino acids. The predicted antigenic sites were TNLC333-336, RISNCVADYSVLYNSASF357-374, GVSPTKLNDLCFTN381-394, PDDF426-429, SNNLDSKVGGNYN438-450, LFRKSNLKPFERDIST455-470, STEIYQAGSTPCNGVEGFNCYFP469-491, YGFQPTNGVGYQ495-506 and ELLHAPATVCGP516-527 (Table 1, Fig. 2 ). The antigenic sites prediction using sequence-based method (SVMTriP) also resulted in almost similar predictions with prediction of an additional antigenic site LFRKSNLKPFERDIST455-470 (Table 2 ). We performed structural analysis of spike RBD domain using Chimera and found that all the predicted antigenic sites are surface exposed loops of the spike RBD validating the ElliPro prediction (Fig. 2). The positions of predicted antigenic sites using Ellipro and SVMTrip in SARS-CoV-2 spike RBD domain have been also been represented in its sequence aligned with the spike proteins sequence from SARS-CoV-1 (Fig. 3 ).
Table 1.
Structure-based antigenic sites prediction in SARS-CoV-2 spike RBD using ElliPro.
| S. No. | Start | End | Peptide | Number of residues | Score |
|---|---|---|---|---|---|
| 1 | 469 | 491 | STEIYQAGSTPCNGVEGFNCYFP | 23 | 0.821 |
| 2 | 516 | 527 | ELLHAPATVCGP | 12 | 0.783 |
| 3 | 333 | 336 | TNLC | 4 | 0.773 |
| 4 | 438 | 450 | SNNLDSKVGGNYN | 13 | 0.689 |
| 5 | 495 | 506 | YGFQPTNGVGYQ | 12 | 0.674 |
| 6 | 381 | 394 | GVSPTKLNDLCFTN | 14 | 0.652 |
| 7 | 357 | 374 | RISNCVADYSVLYNSASF | 18 | 0.57 |
| 8 | 426 | 429 | PDDF | 4 | 0.505 |
| 9 | 455 | 470 | LFRKSNLKPFERDIST | 16 | 0.50 |
Fig. 2.
Structure model of SARS-CoV-2 receptor binding domain showing potential antigenic sites as predicted by ElliPro. All predicted sites are marked with their sequence and position.
Table 2.
Sequence-based antigenic sites prediction in SARS-CoV-2 spike RBD using SVMTriP. The potential sites with score >0.5 have been marked with asterisk * sign.
| S.No. | Site | Sequence | Score (0.5) |
|---|---|---|---|
| 1 | 571–590 | DTTDAVRDPQTLEILDITPC | 0.466 |
| 2 | 595–614 | VSVITPGTNTSNQVAVLYQD | 0.457 |
| 3 | 386–403 | KLNDLCFTNVYADSFVIR | 0.781* |
| 4 | 357–374 | RISNCVADYSVLYNSASF | 0.711* |
| 5 | 459–476 | SNLKPFERDISTEIYQAG | 0.581* |
| 6 | 357–372 | RISNCVADYSVLYNSA | 0.69* |
| 7 | 455–470 | LFRKSNLKPFERDIST | 0.605* |
| 8 | 388–403 | NDLCFTNVYADSFVIR | 0.544* |
| 9 | 356–369 | KRISNCVADYSVLY | 0.622* |
| 10 | 398–411 | DSFVIRGDEVRQIA | 0.496 |
| 11 | 457–470 | RKSNLKPFERDIST | 0.426 |
| 12 | 357–368 | RISNCVADYSVL | 0.727* |
| 13 | 356–365 | KRISNCVADY | 0.496 |
Fig. 3.
Pairwise sequence alignment of SARS-CoV-1 and SARS-CoV-2 showing the position of antigenic sites as predicted using sequence-based (SVMTriP) and structure-based (ElliPro) antigenic sites prediction tools. The site TNLC333-336 and PDDF426-429 sites are similar in both SARS-CoV-1 and SARS-CoV-2 while other six antigenic sites have sequence divergence. The antigenic site RISNCVADYSVLYNSASF357-374 of SARS-CoV-2 has been found to be quite unique as it is not present in SARS-CoV-1 as can be seen in alignment.
3.3. Predicted antigenic sites show molecular similarity with antigens from twelve pathogenic bacterial species, two Plasmodium parasites and influenza virus A
The predicted antigenic sites were then submitted to AntigenDB database for assessing their molecular similarity with other known antigens or antigenic determinants in different microorganisms. While using the whole antigenic site, we could not predict any similar antigenic determinant and as such the antigenic sites predicted in SARS-CoV-2 spike RBD appeared to be unique. However, when the same antigenic sequence was subjected for similarity search in small tripeptide fragments (with three amino acid residues at a time), for example the RISNCVADYSVLYNSASF357 antigenic sequence was used as RIS, ISN, SNC, NCV, CVA and so on, we could successfully predict antigenic sites’ similarity with other known antigens in different organisms.
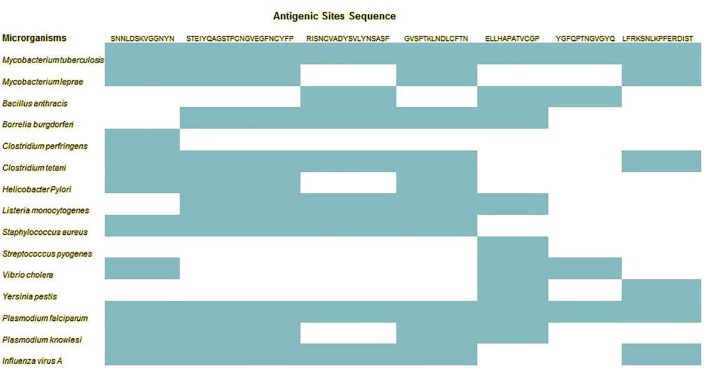
We subjected all nine antigenic sites’ sequence for similarity search in the form of tripeptide fragment and listed all the similar known antigens (Table 3 ). TNLC333-336 and PDDF426-429 showed no similarity with any known antigens from other organisms. Rest all other seven predicted antigenic sites showed molecular similarity with known antigens/antigenic determinants. To our surprise, the predicted antigenic sites showed similarity with antigenic determinants from twelve pathogenic bacterial species (Mycobacterium tuberculosis, Mycobacterium leprae, Bacillus anthracis, Borrelia burgdorferi, Clostridium perfringens, Clostridium tetani, Helicobacter Pylori, Listeria monocytogenes, Staphylococcus aureus, Streptococcus pyogenes, Vibrio cholera and Yersinia pestis), two malaria parasites (Plasmodium falciparum and Plasmodium knowlesi) and Influenza virus A. The antigenic determinants from both gram+ and gram− bacteria were predicted in spike RBD antigenic sites. All these microorganisms are well known for their pathogenesis and are associated with life threatening diseases in humans and in some animals as well. In total, out of nine sites, seven sites showed molecular similarity with 54 antigens from twelve pathogenic bacterial species, two malarial parasites and influenza virus A. The predicted molecular similarity between antigenic sites in SARS-CoV-2 and antigenic determinants from pathogenic organisms has three major implications: 1) the human immune system will recognize and activate the immune cells and trigger response in response to SARS-CoV-2 infection in the similar manner as the antigenic determinants could arouse after infection by these fifteen pathogenic organisms, 2) the pathophysiological outcomes and clinical features (symptoms) observed in COVID-19 patients are expected to be similar to the symptoms in any patient infected by any of these fifteen microorganisms (Huang et al., 2020, Qiu et al., 2020, Wang et al., 2020), and 3) the SARS-CoV-2 infection (due to unique antigenic features) is expected to have complex etiology and multiple pathophysiological features (symptoms), possibly multiple organ failure also.
Table 3.
List of antigenic proteins from different pathogenic microorganisms and virus showing sequence similarity with predicted antigenic sites’ sequence in SARS-CoV-2 spike RBD.
| S. No. | UniProt ID | Protein Name | Protein Description | Organisms |
|---|---|---|---|---|
| 1 | O33084 | ESXB_MYCLE | ESAT-6-like protein EsxB | Mycobacterium leprae (strain TN) |
| 2 | P01556 | CHTB_VIBCH | Cholera enterotoxin subunit B | Vibrio cholerae serotype O1 (strain ATCC 39315/El Tor Inaba N16961) |
| 3 | P01558 | ELTB_CLOPF | Heat-labile enterotoxin B chain | Clostridium perfringens |
| 4 | P02893 | CSP_PLAFA | Circumsporozoite protein | Plasmodium falciparum |
| 5 | P02894 | CSP_PLAKH | Circumsporozoite protein | Plasmodium knowlesi (strain H) |
| 6 | P03438 | HEMA_I000X | Hemagglutinin | Influenza A virus (strain A/X-31 H3N2) |
| 7 | P03449 | HEMA_I71A1 | Hemagglutinin | Influenza A virus (strain A/Memphis/1/1971 H3N2) |
| 8 | P03452 | HEMA_I34A1 | Hemagglutinin | Influenza A virus (strain A/Puerto Rico/8/1934 H1N1) |
| 9 | P03453 | HEMA_177AB | Hemagglutinin | Influenza A virus (strain A/USSR/90/1977 H1N1) |
| 10 | P03459 | HEMA_I34A0 | Hemagglutinin | Influenza A virus (strain A/Fowl plague virus/Rostock/8/1934 H7N1) |
| 11 | P03466 | NCAP_I34A1 | Nucleoprotein | Influenza A virus (strain A/Puerto Rico/8/1934 H1N1) |
| 12 | P04923 | CRA_PLAFA | Circumsporozoite protein-related antigen | Plasmodium falciparum |
| 13 | P04926 | EXP1_PLAFA | Malaria protein EXP-1 | Plasmodium falciparum |
| 14 | P04934 | MSP1_PLAFC | Merozoite surface protein 1 | Plasmodium falciparum (isolate Camp/Malaysia) |
| 15 | P04958 | TETX_CLOTE | Tetanus toxin | Clostridium tetani (strain Massachusetts/E88) |
| 16 | P08089 | M6B_STRPY | M protein, serotype 6 | Streptococcus pyogenes |
| 17 | P09621 | CH10_MYCTU | 10 kDa chaperonin | Mycobacterium tuberculosis (strain ATCC 25618/H37Rv) |
| 18 | P09621 | CH10_MYCTO | 10 kDa chaperonin | Mycobacterium tuberculosis (strain CDC 1551/Oshkosh) |
| 19 | P0A0L2 | ETXA_STAAU | Enterotoxin type A | Staphylococcus aureus |
| 20 | P0A4V2 | A85A_MYCTU | Diacylglycerolacyltransferase | Mycobacterium tuberculosis (strain ATCC 25618/H37Rv) |
| 21 | P0A4V2 | A85A_MYCTO | Diacylglycerolacyltransferase | Mycobacterium tuberculosis (strain CDC 1551/Oshkosh) |
| 22 | P0A550 | DPO1_MYCTU | DNA polymerase I | Mycobacterium tuberculosis (strain ATCC 25618/H37Rv) |
| 23 | P0A550 | DPO1_MYCTO | DNA polymerase I | Mycobacterium tuberculosis (strain CDC 1551/Oshkosh) |
| 24 | P0A564 | ESXA_MYCTU | 6 kDa early secretory antigenic target | Mycobacterium tuberculosis (strain ATCC 25,618 / H37Rv) |
| 25 | P0A564 | ESXA_MYCTO | 6 kDa early secretory antigenic tar | Mycobacterium tuberculosis (strain CDC 1551/Oshkosh) |
| 26 | P0A5J0 | LPQH_MYCTU | Lipoprotein LpqH | Mycobacterium tuberculosis (strain ATCC 25618/H37Rv) |
| 27 | P0A5J0 | LPQH_MYCTO | Lipoprotein LpqH | Mycobacterium tuberculosis (strain CDC 1551/Oshkosh) |
| 28 | P0A5Q4 | MP64_MYCTU | Immunogenic protein MPT64 | Mycobacterium tuberculosis (strain ATCC 25618/H37Rv) |
| 29 | P0A5Q4 | MP64_MYCTO | Immunogenic protein MPT64 | Mycobacterium tuberculosis (strain CDC 1551/Oshkosh) |
| 30 | P11089 | FLA1_BORBU | Flagellar filament 41 kDa core protein | Borrelia burgdorferi (strain ATCC 35210/B31/CIP 102532/DSM 4680) |
| 31 | P13423 | PAG_BACAN | Protective antigen | Bacillus anthracis |
| 32 | P13830 | RESA_PLAFF | Ring-infected erythrocyte surface antigen | Plasmodium falciparum (isolate FC27/Papua New Guinea) |
| 33 | P14013 | OSPA_BORBU | Outer surface protein A | Borrelia burgdorferi (strain ATCC 35210/B31/CIP 102532/DSM 4680) |
| 34 | P14013 | OSPA_BORBN | Outer surface protein A | Borrelia burgdorferi (strain N40) |
| 35 | P14013 | OSPA_BORBZ | Outer surface protein A | Borrelia burgdorferi (strain ZS7) |
| 36 | P14916 | URE23_HELPY | Urease subunit alpha | Helicobacter pylori (strain ATCC 700392/26695) (Campylobacter pylori) |
| 37 | P15712 | PSTS1_MYCTU | Phosphate-binding protein PstS 1 | Mycobacterium tuberculosis (strain ATCC 25618/H37Rv) |
| 38 | P15712 | PSTS1_MYCTO | Phosphate-binding protein PstS 1 | Mycobacterium tuberculosis (strain CDC 1551/Oshkosh) |
| 39 | P15917 | LEF_BACAN | Lethal factor | Bacillus anthracis |
| 40 | P16893 | TRAP_PLAFA | Thrombospondin-related anonymous protein | Plasmodium falciparum |
| 41 | P19214 | EBA1_PLAFC | Erythrocyte-binding antigen 175 | Plasmodium falciparum (isolate Camp/Malaysia) |
| 42 | P21171 | P60_LISMO | Probable endopeptidase p60 | Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679/EGD-e) |
| 43 | P23024 | TCPA_VIBCL | Toxin coregulated pilin | Vibrio cholera |
| 44 | P24301 | CH10_MYCLE | 10 kDa chaperonin | Mycobacterium leprae (strain TN) |
| 45 | P25893 | |||
| 46 | P26948 | CAF1_YERPE | F1 capsule antigen | Yersinia pestis |
| 47 | P31951 | A85B_MYCLE | Diacylglycerolacyltransferase | Mycobacterium leprae (strain TN) |
| 48 | P31952 | A85B_MYCTA | Diacylglycerolacyltransferase | Mycobacterium tuberculosis (strain ATCC 25177/H37Ra) |
| 49 | P31952 | A85B_MYCTU | Diacylglycerolacyltransferase | Mycobacterium tuberculosis (strain ATCC 25618/H37Rv) |
| 50 | P31952 | A85B_MYCTO | Diacylglycerolacyltransferase | Mycobacterium tuberculosis (strain CDC 1551/Oshkosh) |
| 51 | Q07337 | OSPC_BORBU | Outer surface protein C | Borrelia burgdorferi (strain ATCC 35210/B31/CIP 102532/DSM 4680) |
| 52 | Q25893 | Q25893_PLAFA | Liver stage antigen-1 | Plasmodium falciparum |
| 53 | Q9U0P0 | Q9U0P0_PLAFA | Liver stage antigen-3 | Plasmodium falciparum |
| 54 | Q60153 | TCPA_VIBCL | Toxin coregulatedpilin | Vibrio cholerae serotype O1 (strain ATCC 39315/El Tor Inaba N16961) |
3.4. All antigenic sequences in SARS-CoV-2 RBD had similarity with Mycobacterium tuberculosis and Plasmodium falciparum
All seven predicted antigenic sites in SARS-CoV-2 RBD had at least molecular similarity with antigenic determinants from Mycobacterium tuberculosis and Plasmodium falciparum (Fig. 4 ) suggesting that COVID-19 patients should have at least symptoms of these two diseases and the same can be useful for identification of SARS-CoV-2 infection. Antigenic sites in SARS-CoV-2 showed similarity with a number of proteins, enzymes, toxins and virulent factors with diverse functional role in pathogens. The role of proteins, enzymes, toxins and virulence factors has been presented here. Diacylglycerolacyltransferase fbpB from M. Tuberculosis have high affinity for extracellular matrix protein, fibronectin, which facilitates strong binding of M. tuberculosis to macrophages. The 10-kDa chaperonin (groS) of M. tuberculosis as found in M. Leprae is implicated for its virulence (Roberts et al., 2003). An in vitro study using recombinant M. tuberculosis showed that 10-kDa co-chaperonin (cpn10) led to bone weakness and fragility (Corrado et al., 2013). The 6 kDa early secretory antigenic target of M. tuberculosis, also known as esxA, is a secreted protein of M. tuberculosis which act as strong T-cell antigen and play key role in virus escape from host immune response. The lipoprotein Psts1 of M. tuberculosis codes for phosphate-binding protein that plays important role in phosphate uptake by M. tuberculosis and implicated in virulence (Peirs et al., 2005). Another lipoprotein Lpqh of M. tuberculosis induces T cell-mediated immunity and also acts as a TLR2 agonist as it downregulates antigen presentation to T-cells (Noss et al., 2001). The MPT64 (mpt64) of M. tuberculosis is involved in persistence and survival of pathogen in host cells and tissues (Kruh-Garcia et al., 2014). Sequence of DNA polymerase 1 of M. tuberculosis was also predicted similar to antigenic sites in SARS-CoV-2 RBD. In nutshell, the antigenic sites in SARS-CoV-2were similar to those found in pathogenic proteins in Mycobacterium having role in modulating the immune response for escaping immune recognition and ensuring persistence in host body, possibly in latent stage.
Fig. 4.
Similarity of antigenic sites in SARS-CoV-2 spike RBD with antigens predicted in different microorganisms, including bacteria, parasites and virus.
The spike RBD was also found to possess antigenic determinants as found in proteins from malarial parasites such as Plasmodium falciparum and Plasmodium knowlesi. The related proteins from malarial parasites were circumsporozoite protein (CSP), circumsporozoite protein-related antigen, malaria protein EXP-1, thrombospondin-related anonymous protein (TRAP), liver stage antigen-1 (LSA-1), liver stage antigen-3 (LSA-3), merozoite surface protein 1 (MSP1), erythrocyte-binding antigen 175 (EBA 175), and ring-infected erythrocyte surface antigen (RESA).All these proteins were found to be present in the sera of patients exposed natural to sporozoite or were immunized with it (Doolan et al., 2008). Naturally exposed individuals usually have blood stage antigens such as MSP1 (also MSP2, MSP4, MSP5, and MSP7), EXP1, LSA-3, EBA 175; while, CSP and TRAP were found to be present in sporozoite immunized individuals (Doolan et al., 2008). In addition, CSP and MSP1 have been found in both groups. Circumsporozoite proteins (CSPs) are important malarial sporozoite protein having role in liver cells invasion in humans (Doolan et al., 2008). The RESA is released by Plasmodium falciparum inside the RBCs on entry. The RESA migrates to the host cell membrane, where it binds to spectrin; however, the mechanism and type of binding and its pathological consequences are largely unknown yet (Pei et al., 2007).
3.5. Influenza virus A sequence having role in virus assembly were predicted in SARS-CoV-2 RBD
Antigenic determinants of two proteins from influenza virus A were predicted similar to SARS-CoV-2 RBD antigenic sites. The proteins are hemagglutinin (HA) (one of the three transmembrane protein of virus) and nucleoprotein (NP) (encapsulated in viral nucleocapsids). While HA is involved in viral assembly at host cell membrane, the viral nucleoproteins helps in incorporation of viral genetic material into newly budded virions (Zhang et al., 2000, Leser and Lamb, 2005). The nucleoproteins of influenza virus have RNA binding and protein interaction sites that may be important for their host cell functions such as stability and nuclear export of mRNA (Krug, 1993, Qian et al., 1994, Qiu and Krug, 1994) and activation of IRF3, STAT1 and NF-κB dependent pathways (Chien et al., 2004, Krug et al., 2003, Min et al., 2007).
3.6. Predicted similar antigenic sequences from bacteria were mainly toxins, virulent factors or other proteins that help invasion and survival of pathogens in host cells and tissues
Two antigenic determinants predicted from Bacillus anthracis were of protective antigen (pagA) and lethal factor. These proteins are two of the three proteins (PA, EF, and LF) that form anthrax toxin (Friebe et al., 2016). Five antigenic determinants predicted from spirochete Borrelia burgdorferi were of flagellar filament core protein (Fla1) and outer surface proteins A (OspA) and C (OspC). Fla1 protein is an immunodominant41kDaantigen present in the sera of Lyme-disease patients that allows bacteria to efficiently bore into the host cells for colonization and survival using outer surface proteins (Steere et al., 2004, Neelakanta et al., 2007). The antigenic sequences predicted from Clostridium perfringens and Clostridium tetani were also of the toxin proteins such as heat-labile enterotoxin B chain and tetanus toxin. Similarly, some other predicted antigenic determinants were also of well known toxins and virulent factor such as enterotoxin A (from Staphylococcus aureus), M protein (from Streptococcus pyogenes), cholera enterotoxin subunit B and toxin coregulated pilin (from Vibrio cholerae). The30kDa urease subunit alpha (ureA) from Helicobacter pylori acts as virulence factor and cause infection in stomach via host-pathogen interaction (Schoep et al., 2010) and mediate immune response in humans (Schoep et al., 2010). The endopeptidase p60 (Lm-p60) of Listeria monocytogenes whose sequence was also predicted in similar search is a highly conserved carbohydrate binding module which can be engineered for binding to peptidoglycans with high affinity (Yu et al., 2016). Mycobacterium leprae has two proteins (ESAT-6 like protein esxb and 10 kDa chaperonin groS) that showed molecular similarity with predicted antigens in SARS-CoV-2 RBD. These play role in bacterial virulence and act as chaperone to prevent membrane lysis in M. Leprae. F1 capsule of Yersinia pestis helps bacteria avoid up taken by macrophages (Levy et al., 2018).
3.7. Five peptide sequences predicted as novel vaccine candidates for COVID-19 treatment
The spike RBD of SARS-CoV-2 was subjected for T-cell epitope prediction using EpiJen which predicts peptides molecules that can be recognized by human T-cells after presentation by MHC class I proteins. In total fourteen peptide sequences were recognized as possible epitopes for T-cells (predicted against MHC class I) and only eight peptide sequences were predicted to be overlapping with the antigenic sites predicted (Table 4 ). However, the predicted IC50 value for six peptides was very high and thus these peptides failed to qualify the criteria of becoming a potent vaccine candidate. Finally, two peptides (KVGGNYNYL 444-452 andSVLYNSASF366-374) with low predicted IC50 value of 29.38 and 31.26 nM, respectively were considered suitable for designing DNA vaccine and recombinant vaccine and other vaccine type against COVID-19. Besides this, the receptor binding domain of the spike protein in SARS-CoV-2 has a stretch of sequence (TEIYQAGSTPCNGVEGF470-486) showing mismatch with SARS-CoV-1 and the same stretch has been found to be one of the antigenic sites and harboring peptide sequence for a potent vaccine candidate identified in the current study (Table 4). We have done an additional analysis in which prediction was done against the HLA class II alleles using NetMHCpan available in IEDB Analysis Resource of National Institute of Allergy and Infectious Diseases (NIH), USA (http://tools.iedb.org/mhci/). Alleles of this HLA class are normally found on professional antigen-presenting cells (APCs) such as B lymphocytes, dendritic cells, mononuclear phagocytes, endothelial cells and thymic epithelial cells. These cells are important in exacerbating immune responses in different ways. Herein, we predicted three potential peptides in SARS-CoV-2 RBD against MHC class II alleles (Table 5 ). These peptides are KRISNCVADYSVLYN356-370(IC50 = 2.6 nM), DLCFTNVYADSFVI389-402 (IC50 = 3.8 nM), and YRVVVLSFELLHA508-520 (IC50 = 3.8 nM).
Table 4.
Prediction of vaccine peptides in antigenic sites of SARS-CoV-2 RBD that can be presented to cytotoxic T-cells by MHC class I protein. The vaccine peptides are shown as bold and underlined in the antigenic sites predicted in SARS-CoV-2. The IC50 value has been marked with “asterisk” mark for the most potent vaccine candidates.
| S. No. | T-cell Epitopes | Antigenic Sites in SARS-CoV-2 | IC50 Value (nM) |
|---|---|---|---|
| 1 | CPFGEVFNA336-344 | TNLC333-336 | 331.13 |
| 2 | NCVADYSVL360-368 | RISNCVADYSVLYNSASF357-374 | 409.26 |
| 3 | CVADYSVLY361-369 | RISNCVADYSVLYNSASF357-374 | 579.43 |
| 4 | SVLYNSASF366-374 | RISNCVADYSVLYNSASF357-374 | 031.26* |
| 5 | NVYADSFVI394-402 | No overlap | 347.54 |
| 6 | KVGGNYNYL444-453 | SNNLDSKVGGNYN438-450 | 029.38* |
| 7 | GVEGFNCYF482-490 | STEIYQAGSTPCNGVEGFNCYFP469-491 | 294.44 |
| 8 | RVVVLSFEL509-517 | No overlap | 007.23 |
| 9 | VVVLSFELL510-518 | ELLHAPATVCGP516-527 | 010.64 |
| 10 | VVLSFELLH511-519 | ELLHAPATVCGP516-527 | 510.50 |
| 11 | LVKNKCVNF533-541 | No overlap | 079.25 |
| 12 | CVNFNFNGL538-546 | No overlap | 081.28 |
| 13 | LTESNKKFL533-541 | No overlap | 255.86 |
| 14 | ESNKKFLPF538-446 | No overlap | 115.35 |
Table 5.
Prediction of vaccine peptides in antigenic sites of SARS-CoV-2 RBD that can be presented to cytotoxic T-cells by MHC class II protein.
| S. No. | Start | End | Length | Core_peptide | Peptide | IC50 |
|---|---|---|---|---|---|---|
| 1 | 389 | 402 | 14 | FTNVYADSF | DLCFTNVYADSFVI | 2583.64 |
| 2 | 508 | 520 | 13 | VLSFELLHA | YRVVVLSFELLHA | 3827.15 |
| 3 | 356 | 370 | 15 | CVADYSVLY | KRISNCVADYSVLYN | 3851.20 |
4. Discussion
In brief, in current work, we have made an attempt to ascertain molecular similarity of antigenic sites predicted in SARS-CoV-2 spike protein with other proteins/antigens in other organisms. Molecular similarity can be defined as the theoretical explanation for sequence similarities (mainly in antigens) between two (or more) organisms. The concept of molecular or antigen similarity has been well presented and entrusted in literature in context to autoimmune disorders (Fujinami et al., 2006, Cusick et al., 2012, Pontes-de-Carvalho et al., 2013). In context to auto-immune pathogenesis, molecular similarity has been implicated between pathogenic organisms and humans self-antigens which results in generation of cross-reactive T-cells that targets human’s self-cells (Chodisetti et al., 2012). In context to current manuscript, the molecular similarity has been observed between antigenic sites in SARS-CoV-2 spike protein and similar antigenic determinants in fifteen microorganisms, including bacteria, parasites and viruses. We believe that the presence of tripeptide sequence(s) (displaying molecular similarity with highly potent antigenic determinants as present in fifteen pathogenic microorganisms) in the antigenic sites of SARS-CoV-2 spike proteins are sufficient to activate host immune cells (T or B cells or other immune cells) in similar as the fifteen microorganisms can do. Some researchers showed that even a tripeptide motif (predicted as antigenic determinants in current study) can activate CD4+ T-cells and can induce immune response (Hemmer et al., 2000). Additionally, other researchers demonstrated how an antigenic tripeptide motif in the amino acid sequence of a protein in pathogens can also differentially activate one of the two immune cells, either B-cell or T-cell (Yao et al., 2012). The molecular similarity of spike RBD with antigenic determinants as found in different pathogenic microorganisms is expected to induce exuberant innate and adaptive immune responses leading to excessive secretion of cytokines that is expected to adversely affect several vital organs leading to multi-organ failure in COVID-19 patients (Fig. 5 ). Such an exacerbated innate and adaptive immune response is attributed to hyper-activation of B-cells, T-cells (both CTL and Th-cells), DCs, NK-cells and macrophage/monocyte lineage cells (Fig. 5). Since, T-cell play critical role in controlling immune response and one can expect deregulated immune response and dysregulated cytokine secretion in aged and immune-compromised patients who have less number of T-cells in their body (Kim et al., 2007, Palm and Medzhitov, 2007). There is also a correlation between the secretion of some cytokines and chemokines, such as IL-6, IL-8, and MCP-1 and IP-10, with higher mortality and severity of disease (Reghunathan et al., 2005).
Fig. 5.
The study explains the complex etiology and pathophysiology of COVID-19 and sheds light on implications and future prospects.
We predicted nine potential surface exposed antigenic sites in SARS-CoV-2 spike RBD. We also predicted CTL epitopes that can be presented with MHC class I and class II and found five peptides in spike RBD of SARS-CoV-2 (KVGGNYNYL 444-452 andSVLYNSASF366-374 against MHC class I; KRISNCVADYSVLYN356-370, DLCFTNVYADSFVI389-402, and YRVVVLSFELLHA508-520 against MHC class II) with low predicted IC50 values, which confirms their suitability for vaccine design against COVID-19. Some researchers used RVDFCGKGY peptide (CTL epitope) to design a vaccine against SARS-CoV-1 and experimental trials showed the vaccine to be effective as well on different animal models (Choy et al., 2004). Finally, the results obtained in current study are also in congruence with previous experimental studies in which SARS-CoV-specific CD4+ and CD8+ T cell epitopes were found in C57BL/6 and BALB/C mice (Zhi et al., 2005, Huang et al., 2007, Zhao et al., 2010).
Presence of antigenic sites in SARS-CoV-2 with similarity with antigenic determinants found in pathogenic bacterial, malarial and viral species is seriously alarming as their presence makes the SARS-CoV-2 more pathogenic than any other previously known coronavirus. Especially, antigenic determinants those are unique to SARS-CoV-2. The antigenic patches from pathogenic microorganisms in SARS-CoV-2 could be traced only as small tripeptide motifs and this makes their origin uncertain. However, most of the antigenic determinants found in pathogenic microorganisms were related to antigenic proteins having established function as toxin and proven role in virulence and pathogenicity. Besides this, other antigenic determinants found similar to Mycobacterium antigens having role in persistence and survival of pathogen in host cells. Some antigenic determinants from HA and NP of influenza virus A were also in antigenic sites of SARS-CoV-2 and these antigens from influenza virus have role in assembly of newly budded virions in case of influenza virus A. Due to the presence of antigenic sites with molecular similarity with antigens from different pathogenic organisms, parasites and virus, it can be ruled out that COVID-19 patients may possibly suffer multi-organ failure as these pathogenic antigens are well known to cause serious damages to different vital organs of human body such as liver, kidney, blood, stomach, heart and bones. This is also clearly evident from complex etiology and pathophysiological outcomes (clinical features or symptoms) as observed in COVID-19 patients. Common in all seven antigenic sites predicted in SARS-CoV-2 was the presence of antigenic determinants from Mycobacterium and Plasmodium antigens suggesting that antimalarial and anti-TB drugs and vaccines could be a good treatment options for COVID-19. Besides this, anti-leprosy, anti-lyme, anti-plague, anti-anthrax drugs/vaccine etc are also expected to be beneficial in COVID-19 treatment. Moreover, individuals previously immunized/vaccinated or had previous history of malaria, tuberculosis or other disease caused by fifteen microorganisms are expected to display a considerable degree of resistance against SARS-CoV-2 infection. The possible explanation is that the memory B or T cells previously generated by the microorganisms (in context here) would get activated again upon SARS-CoV-2 infection (long-lived immunity to reinfection) because of the similar antigenic specificity and due to presence of common antigenic determinants in both. For some of the pathogenic antigens, whose antigenic determinants were predicted similar to SARS-CoV-2′s antigenic sites, very limited information was available in literature regarding their function.
As such, nothing can be explicitly stated regarding the presence of antigenic determinants as found in this current study. We speculate that viruses may be doing this as a genetic trick for tackling genetic constraints imposed by host cells to ensure their persistence and survival. There are two explanations for high divergence between spike RBD in SARS-CoV-1 and SARS-CoV-2. The SARS-CoV-2 might have changed or switched the molecular composition of its spike protein using a process called as antigenic variation (Smith, 2004, Chibo and Birch, 2006, de Jong et al., 2007). Antigenic variations are genetic tricks using which some viruses, such as influenza virus and coronavirus etc., evade protective immune system by altering their immunodominant epitopes that otherwise could be recognized by the host adaptive immune system leading to their elimination (Bidokhti et al., 2013, Lewis et al., 2014). Antigenic variations are brought up by periodic and random genetic mutations, generally in viral surface antigens, so as to temporarily camouflage host immune cells and to prevent clearance (Smith, 2004, de Jong et al., 2007). Antigenic variation that occurs in the HA and NA antigens of influenza virus A are known. The antigenic alteration can occur in two different ways: 1) genetic drift that causes subtle changes in amino acids, and 2) antigenic shift that causes major alterations in the antigenic properties of the protein (Smith, 2004, de Jong et al., 2007, Ren et al., 2015). In latter case, the acquisition of new antigenic determinants in virus would render virus no longer identifiable (similarly as “eclipsed antigens” do) by host immune cells and neutralizing antibodies (Smith, 2004, de Jong et al., 2007). Such antigenic-shifted strains of virus generate periodically when genes encoding structural proteins are acquired from viruses that infect animal hosts. These antigenic-shifted strains are known to cause global pandemic and recurring acute infections (Smith, 2004, de Jong et al., 2007).
Currently, computational biology approaches for epitope prediction, identification and analysis are well-developed and have been proved highly successful to predict & identify both weak and strong antibody epitopes, some of them are often experimentally ignored (Jespersen et al., 2017). Antigenic properties of spike glycoprotein of SARS-CoV-2, especially of the receptor binding domain (RBD), were well appreciated by many experimental researchers including myself in current study (Baruah and Bose, 2020, Lucchese, 2020). Most of the vaccines such as virus vector and protein subunit vaccine developed or under clinical trails have been developed using full length spike protein or RBD of spike protein of SARS-CoV-2. The identified sequence in current study are part of all vaccine under clinical or pre-clinical development. (Baruah and Bose, 2020). The strategy to find antigenic sites in spike protein of SARS-CoV-2 is also known (Ren et al., 2003). Some new antibody epitopes have also been found in SARS-CoV-2 spike protein that dominate the antigenicity of spike protein in SARS-CoV-2 as compared to other coronaviruses (Zheng and Song, 2020). Potential antigenic cross-reactivity of SARS-CoV-2 with dengue virus has also been observed (Lustig et al., 2020).
This study provides the first evidence in favor of antigenic variations in spike RBD of SARS-CoV-2 that succour virus to undergo adaptive evolution in order to infect humans for their survival and persistence. These antigenic variations not only explain the complex immunological aspects, etiology and pathophysiology of the disease but also suggest different therapeutics (anti-malarial, anti-TB, anti-leprosy, anti-plague, anti-lyme, anti-anthrax, anti-cholera etc.), including drugs, medicines, antibodies and vaccines, for their promising role in inactivating SARS-CoV-2, which is thought to mutate quickly. Besides this, our body also requires B-cells, CD8+ and CD4+ T-helper cells and other immune cells based innate and adaptive immunity and antibodies for specifically targeting infectious cells (Janice Oh et al., 2012, Choy et al., 2004). We envisage that several lines of research endeavors are still required towards understanding the multifaceted mechanisms of immunomodulation, host-virus interaction and SARS-CoV-2 infection.
Author contributions
TCD conceived the idea and designed the experimental strategy and designed methodology, performed the analyses, wrote the manuscript, revised the manuscript.
Funding
The work was conducted in absence of any funding source.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imbio.2021.152091.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. (2011)3(6):920–940. [DOI] [PMC free article] [PubMed]
- Andreatta M., Karosiene E., Rasmussen M., Stryhn A., Buus S., Nielsen M. Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics. 2015;67(11-12):641–650. doi: 10.1007/s00251-015-0873-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari HR, Flower DR, Raghava GP. AntigenDB: an immunoinformatics database of pathogen antigens. Nucleic Acids Res. (2010) 38:D847-853. [DOI] [PMC free article] [PubMed]
- Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J. Med. Virol. 2020;92(5):495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellis S.L. Advantages of RGD peptides for directing cell association with biomaterials. Biomaterials. 2011;32(18):4205–4210. doi: 10.1016/j.biomaterials.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belz G.T., Smith C.M., Kleinert L., Reading P., Brooks A., Shortman K., et al. Distinct migrating and nonmigrating dendritic cell populations are involved in MHC class I-restricted antigen presentation after lung infection with virus. Proc. Natl. Acad. Sci. USA. 2004;101:8670–8675. doi: 10.1073/pnas.0402644101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidokhti MR, Tråvén M, Krishna NK, Munir M, Belák S, Alenius S, et al. Evolutionary dynamics of bovine coronaviruses: natural selection pattern of the spike gene implies adaptive evolution of the strains. J. Gen.Virol. (2013) 94:2036–2349. [DOI] [PubMed]
- Bottermann M., James L.C. Intracellular antiviral immunity. Adv. Virus Res. 2018;100:309–354. doi: 10.1016/bs.aivir.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braciale T.J., Sun J., Kim T.S. Regulating the adaptive immune response to respiratory virus infection. Nat. Rev. Immunol. 2012;12(4):295–305. doi: 10.1038/nri3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerwenka A., Morgan T.M., Harmsen A.G., Dutton R.W. Migration kinetics and final destination of type 1 and type 2 CD8 effector cells predict protection against pulmonary virus infection. J. Exp. Med. 1999;189:423–434. doi: 10.1084/jem.189.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerwenka A., Morgan T.M., Dutton R.W. Naive, effector, and memory CD8 T cells in protection against pulmonary influenza virus infection: homing properties rather than initial frequencies are crucial. J. Immunol. 1999;163:5535–5543. doi: 10.4049/jimmunol.163.10.5535. [DOI] [PubMed] [Google Scholar]
- Chen J., Lau Y.F., Lamirande E.W., Paddock C.D., Bartlett J.H., Zaki S.R., Subbarao K. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J. Virol. 2010;84:1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibo, D., Birch, C. (2006) Analysis of human coronavirus 229E spike and nucleoprotein genes demonstrates genetic drift between chronologically distinct strains. J. Gen. Virol. 87:1203–1208. [DOI] [PubMed]
- Chien C.Y., Xu Y., Xiao R., Aramini J.M., Sahasrabudhe P.V., Krug R.M., et al. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry. 2004;43(7):1950–1962. doi: 10.1021/bi030176o. [DOI] [PubMed] [Google Scholar]
- Chodisetti S.B., Rai P.K., Gowthaman U., Pahari S., Agrewala J.N. Potential T cell epitopes of Mycobacterium tuberculosis that can instigate molecular mimicry against host: implications in autoimmune pathogenesis. BMC Immunol. 2012;13:13. doi: 10.1186/1471-2172-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy W.Y., Lin S.G., Chan P.K., Tam J.S., Lo Y.M., Chu I.M., et al. Synthetic peptide studies on the severe acute respiratory syndrome (SARS) coronavirus spike glycoprotein: perspective for SARS vaccine development. Clin. Chem. (2004) 50(6):1036–1042. [DOI] [PMC free article] [PubMed]
- Cooper M.A., Colonna M., Yokoyama W.M. Hidden talents of natural killers: NK cells in innate and adaptive immunity. EMBO Rep. 2009;10:1103–1110. doi: 10.1038/embor.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrado A., Neve A., Macchiarola A., Gaudio A., Marucci A., Cantatore F.P. RANKL/OPG ratio and DKK-1 expression in primary osteoblastic cultures from osteoarthritic and osteoporotic subjects. J. Rheumatol. 2013;40:684–694. doi: 10.3899/jrheum.120845. [DOI] [PubMed] [Google Scholar]
- Cusick M.F., Libbey J.E., Fujinam R.S. Molecular mimicry as a mechanism of autoimmune disease. Clin. Rev. Allerg. Immu. 2012;42:102–111. doi: 10.1007/s12016-011-8294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakal T.C. (2020) SARS - CoV - 2 attachment to host cells is possibly mediated via RGD-integrin interaction in a calcium-dependent manner. OSF Preprints. doi:10.31219/osf.io/chvsa. [DOI] [PMC free article] [PubMed]
- de Jong J.C., Smith D.J., Lapedes A.S., Donatelli I., Campitelli L., Barigazzi G., et al. Antigenic and Genetic Evolution of Swine Influenza A (H3N2) Viruses in Europe. J. Virol. 2007;81(8):4315–4322. doi: 10.1128/JVI.02458-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolan D.L., Mu Y., Unal B., Sundaresh S., Hirst S., Valdez C., et al. (2008). Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 8:4680–4694. 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed]
- Doytchinova I.A., Guan P., Flower D.R. EpiJen: a server for multistep T cell epitope prediction. BMC Bioinf. 2006;7:131. doi: 10.1186/1471-2105-7-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe S., van der Goot F.G., Bürgi J. (2016). The ins and outs of anthrax toxin. Toxins (Basel). 8(3):pii E69. 10.3390/toxins8030069. [DOI] [PMC free article] [PubMed]
- Frieman M., Baric R. Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation. Microbiol. Mol. Biol. R. 2008;72(4):672–685. doi: 10.1128/MMBR.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinami R.S., von Herrath M.G., Christen U., Whitton J.L. Molecular mimicry, bystander activation, or viral persistence: infections and autoimmune disease. Clin. Microbiol. Rev. 2006;19(1):80–94. doi: 10.1128/CMR.19.1.80-94.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A., Biron C.A. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Guilliams M., Lambrecht B.N., Hammad H. Division of labor between lung dendritic cells and macrophages in the defense against pulmonary infections. Mucosal Immunol. 2013;6(3):464–473. doi: 10.1038/mi.2013.14. [DOI] [PubMed] [Google Scholar]
- Hemmer B., Kondo T., Gran B., Pinilla C., Cortese I., Pascal J., et al. (2020) Minimal peptide length requirements for CD4+ T cell clones—implications for molecular mimicry and T cell survival. Int. Immunol. 12(3):375–383. [DOI] [PubMed]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Cao Y., Du J., Bu X., Ma R., Wu C. Priming with SARS CoV S DNA and boosting with SARS CoV S epitopes specific for CD4+ and CD8+ T cells promote cellular immune responses. Vaccine. 2007;25(39–40):6981–6991. doi: 10.1016/j.vaccine.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.i., Zhang L.i., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L.i., Xie J., Wang G., Jiang R., Gao Z., Jin Q.i., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janice Oh H.-L., Ken-En Gan S., Bertoletti A., Tan Y.-J. Understanding the T cell immune response in SARS coronavirus infection. Emerg. Microbes Infect. 2012;1:1–6. doi: 10.1038/emi.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen M.C., Peters B., Nielsen M., Marcatili P. (2017). BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 45:W24–W29. [DOI] [PMC free article] [PubMed]
- Kim K.D., Zhao J., Auh S., Yang X., Du P., Tang H., Fu Y.X. Adaptive immune cells temper initial innate responses. Nat. Med. 2007;13:1248–1252. doi: 10.1038/nm1207-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug R.M. The regulation of export of mRNA from nucleus to cytoplasm. Curr. Opin. Cell Biol. 1993;5(6):944–949. doi: 10.1016/0955-0674(93)90074-z. [DOI] [PubMed] [Google Scholar]
- Krug R.M., Yuan W., Noah D.L., Latham A.G. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology. 2003;309(2):181–189. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Kruh-Garcia N.A., Lisa M., Wolfe L.M., Chaisson L.H., Worodria W.O., Nahid P., et al. (2014). Detection of Mycobacterium tuberculosis peptides in the exosomes of patients with active and latent M. tuberculosis infection using MRM-MS. PLoS One. 9(7):e103811. 10.1371/journal.pone.0103811. [DOI] [PMC free article] [PubMed]
- Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson M., Messmer D., Somersan S., Fonteneau J.F., Donahoe S.M., Lee M., et al. Requirement of mature dendritic cells for efficient activation of influenza A-specific memory CD8+ T cells. J. Immunol. 2000;165:1182–1190. doi: 10.4049/jimmunol.165.3.1182. [DOI] [PubMed] [Google Scholar]
- Leser G.P., Lamb R.A. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342(2):215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Levy Y., Vagima Y., Tidhar A., Aftalion M., Gur D., Nili U., Chitlaru T., Zauberman A., Mamroud E. Targeting of the Yersinia pestis F1 capsular antigen by innate-like B1b cells mediates a rapid protective response against bubonic plague. NPJ Vacc. 2018;3(1) doi: 10.1038/s41541-018-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis N.S., Anderson T.K., Kitikoon P., Skepner E., Burke D.F., Vincent A.L. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. J. Virol. 2014;88(9):4752–4763. doi: 10.1128/JVI.03805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Lucchese G. Epitopes for a 2019-nCoV vaccine. Cell Mol. Immunol. 2020;17(5):539–540. doi: 10.1038/s41423-020-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig Y., Keler S., Kolodny R., Ben-Tal N., Atias-Varon D., Shlush E., et al. Potential antigenic cross-reactivity between SARS-CoV-2 and Dengue viruses. Clin. Infect. Dis. 2020:ciaa1207. doi: 10.1093/cid/ciaa1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnan C.N., Zeller M., Kayala M.A., Vigil A., Randall A., Felgner P.L., et al. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics. 2010;26:2936–2943. doi: 10.1093/bioinformatics/btq551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J.-Y., Li S., Sen G.C., Krug R.M. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology. 2007;363(1):236–243. doi: 10.1016/j.virol.2007.01.038. [DOI] [PubMed] [Google Scholar]
- Neelakanta G., Li X., Pal U., Liu X., Beck D.S., DePonte K., Fish D., Kantor F.S., Fikrig E., Schneewind O. Outer surface protein B is critical for Borrelia burgdorferi adherence and survival within Ixodes ticks. PLoS Pathog. 2007;3(3):1–11. doi: 10.1371/journal.ppat.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyt K., Lambrecht B.N. The role of lung dendritic cell subsets in immunity to respiratory viruses. Immunol. Rev. 2013;255(1):57–67. doi: 10.1111/imr.12100. [DOI] [PubMed] [Google Scholar]
- Norbury C.C., Malide D., Gibbs J.S., Bennink J.R., Yewdell J.W. Visualizing priming of virus-specific CD8+ T cells by infected dendritic cells in vivo. Nat. Immunol. 2002;3:265–271. doi: 10.1038/ni762. [DOI] [PubMed] [Google Scholar]
- Noss E.H., Pai R.K., Sellati T.J., Radolf J.D., Belisle J., Golenbock D.T., Boom W.H., Harding C.V. Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19-kDa lipoprotein of Mycobacterium tuberculosis. J. Immunol. 2001;167(2):910–918. doi: 10.4049/jimmunol.167.2.910. [DOI] [PubMed] [Google Scholar]
- Palm N.W., Medzhitov R. Not so fast: adaptive suppression of innate immunity. Nat. Med. 2007;13(10):1142–1144. doi: 10.1038/nm1007-1142b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles R.S., Graham B.S. Viruses, dendritic cells and the lung. Respir. Res. 2011;2:245–249. doi: 10.1186/rr63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei X., Guo X., Coppel R., Bhattacharjee S., Haldar K., Gratzer W., et al. The ring-infected erythrocyte surface antigen (RESA) of Plasmodium falciparum stabilizes spectrin tetramers and suppresses further invasion. Blood. 2007;110:1036–1042. doi: 10.1182/blood-2007-02-076919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirs P., Lefèvre P., Boarbi S., Wang X.-M., Denis O., Braibant M., et al. Mycobacterium tuberculosis with disruption in genes encoding the phosphate binding proteins PstS1 and PstS2 is deficient in phosphate uptake and demonstrates reduced in vivo virulence. Infect. Immun. 2005;73:1898–1902. doi: 10.1128/IAI.73.3.1898-1902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko J., Bui H.-H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9(1) doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes-de-Carvalho L., Mengel J., Figueiredo C.A., Social Changes, Asthma and Allergy in Latin America (SCAALA)’s Study Group, Alcântara-Neves N.M. (2013). Antigen mimicry between infectious agents and self or environmental antigens may lead to long-term regulation of inflammation. Front. Immunol. 4:314.10.3389/fimmu.2013.00314. [DOI] [PMC free article] [PubMed]
- Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X., Ji X., Dimitrov D.S. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281(23):15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X.Y., Alonso-Caplen F., Krug R.M. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 1994;68(4):2433–2441. doi: 10.1128/jvi.68.4.2433-2441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Krug R.M. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J. Virol. 1994;68(4):2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Wu J., Hong L., Luo Y., Song Q., Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet. 2020;20(6):689–696. doi: 10.1016/S1473-3099(20)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reghunathan R., Jayapal M., Hsu L.-Y., Chng H.-H., Tai D., Leung B.P., et al. (2005). Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 6:2. [DOI] [PMC free article] [PubMed]
- Ren L., Zhang Y., Li J., Xiao Y., Zhang J., Wang Y., Chen L., Paranhos-Baccalà G., Wang J. Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Sci. Rep. 2015;5(1) doi: 10.1038/srep11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Zhou Z., Liu J., Lin L., Li S., Wang H., Xia J.i., Zhao Z., Wen J., Zhou C., Wang J., Yin J., Xu N., Liu S. A strategy for searching antigenic regions in the SARS-CoV spike protein. Genomics Proteomics Bioinformatics. 2003;1(3):207–215. doi: 10.1016/S1672-0229(03)01026-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M.M., Coker A.R., Fossati G., Mascagni P., Coates A.R.M., Wood S.P. Mycobacterium tuberculosischaperonin 10 heptamers self-associate through their biologically active loops. J. Bacteriol. 2003;185:4172–4185. doi: 10.1128/JB.185.14.4172-4185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman E., Miller E., Harmsen A., Wiley J., Von Andrian U.H., Huston G., Swain S.L. CD4 effector T cell subsets in the response to influenza: heterogeneity, migration, and function. J. Exp. Med. 2002;196:957–968. doi: 10.1084/jem.20021052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogel J.P., et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Saha B., JyothiPrasanna S., Chandrasekar B., Nandi D. Gene modulation and immunoregulatory roles of interferon gamma. Cytokine. 2010;50:1–14. doi: 10.1016/j.cyto.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Schock S.N., Chandra N.V., Sun Y., Irie T., Kitagawa Y., Gotoh B., et al. Induction of necroptotic cell death by viral activation of the RIG-I or STING pathway. Cell Death Differ. 2017;24:615–625. doi: 10.1038/cdd.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoep T.D., Fulurija A., Good F., Lu W., Himbeck R.P., Schwan C., Choi S.S., Berg D.E., Mittl P.R.E., Benghezal M., Marshall B.J., Ahmed N. Surface properties of Helicobacter pylori urease complex are essential for persistence. PLoS One. 2010;5(11):e15042. doi: 10.1371/journal.pone.0015042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Chen Z.J. Antiviral innate immunity pathways. Cell Res. 2006;16(2):141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- Smith D.J. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305(5682):371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- Spiegel M., Schneider K., Weber F., Weidmann M., Hufert F.T. Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J. Gen. Virol. 2006;87:1953–1960. doi: 10.1099/vir.0.81624-0. [DOI] [PubMed] [Google Scholar]
- Spiegel M., Weber F. Inhibition of cytokine gene expression and induction of chemokine genes in non-lymphatic cells infected with SARS coronavirus. Virol. J. 2006;3:17. doi: 10.1186/1743-422X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere A.C., Coburn J., Glickstein L. The emergence of Lyme disease. J. Clin. Invest. 2004;113(8):1093–1101. doi: 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain S.L., Agrewala J.N., Brown D.M., Roman E. Regulation of memory CD4 T cells: generation, localization and persistence. Adv. Exp. Med. Biol. 2002;512:113–120. doi: 10.1007/978-1-4615-0757-4_15. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos A.N., Baccala R., Beutler B., Kono D.H. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognarelli E.I., Bueno S.M., González P.A. Immune-modulation by the human respiratory syncytial virus: focus on dendritic cells. Front. Immunol. 2019;10:810. doi: 10.3389/fimmu.2019.00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.-T., Perrone L.A., Zhu H., Makino S., Peters C.J. Severe acute respiratory syndrome and the innate immune responses: modulation of effector cell function without productive infection. J. Immunol. 2005;174(12):7977–7985. doi: 10.4049/jimmunol.174.12.7977. [DOI] [PubMed] [Google Scholar]
- van den Brand J.M., Haagmans B.L., van Riel D., Osterhaus A.D., Kuiken T. The pathology and pathogenesis of experimental severe acute respiratory syndrome and influenza in animal models. J. Comp. Pathol. 2014;151(1):83–112. doi: 10.1016/j.jcpa.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A., Bredenbeek P.J., van den Worm S.H.E., Spaan W.J.M. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology. 2007;361(1):18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B.o., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Wang S., Chen Q., Chen Y., Chi X., Zhang L., Huang S., Gao G.F., Chen J.-L., Gack M.U. Suppression of interferon lambda signaling by SOCS-1 results in their excessive production during influenza virus infection. PloS Pathog. 2014;10(1):e1003845. doi: 10.1371/journal.ppat.1003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., Ahmed R. Memory CD8 T-cell differentiation during viral infection. J. Virol. 2004;78:5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B., Zhang L., Liang S., Zhang C., Porollo A. SVMTriP: A method to predict antigenic epitopes using support vector machine to integrate tri-peptide similarity and propensity. PLoS One. 2012;7(9):e45152. doi: 10.1371/journal.pone.0045152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Yang J., Guo M. Is the LysM domain of L. monocytogenes p60 protein suitable for engineering a protein with high peptidoglycan binding affinity? Bioengineered. 2016;7:406–410. doi: 10.1080/21655979.2016.1200772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Pekosz A., Lamb R.A. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 2000;74(10):4634–4644. doi: 10.1128/JVI.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Perlman S. (2010) T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice ? J. Virol. 84:9318–9325. 10.1128/JVI.01049-1010. [DOI] [PMC free article] [PubMed]
- Zheng M., Song L. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell Mol. Immunol. 2020;17(5):536–538. doi: 10.1038/s41423-020-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi Y., Kobinger G.P., Jordan H., Suchma K., Weiss S.R., Shen H., Schumer G., Gao G., Boyer J.L., Crystal R.G., Wilson J.M. Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein. Virology. 2005;335(1):34–45. doi: 10.1016/j.virol.2005.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Zhao Q.i. Perspectives on therapeutic neutralizing antibodies against the Novel Coronavirus SARS-CoV-2. Int. J. Biol. Sci. 2020;16(10):1718–1723. doi: 10.7150/ijbs.45123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.