This work dissects the role of different KIF1A tail domains in motor activity and cargo binding. Based on the results, the authors propose a mechanism for the regulation of KIF1A cargo transport in hippocampal neurons.
Abstract
Intracellular transport in neurons is driven by molecular motors that carry many different cargos along cytoskeletal tracks in axons and dendrites. Identifying how motors interact with specific types of transport vesicles has been challenging. Here, we use engineered motors and cargo adaptors to systematically investigate the selectivity and regulation of kinesin-3 family member KIF1A–driven transport of dense core vesicles (DCVs), lysosomes, and synaptic vesicles (SVs). We dissect the role of KIF1A domains in motor activity and show that CC1 regulates autoinhibition, CC2 regulates motor dimerization, and CC3 and PH mediate cargo binding. Furthermore, we identify that phosphorylation of KIF1A is critical for binding to vesicles. Cargo specificity is achieved by specific KIF1A adaptors; MADD/Rab3GEP links KIF1A to SVs, and Arf-like GTPase Arl8A mediates interactions with DCVs and lysosomes. We propose a model where motor dimerization, posttranslational modifications, and specific adaptors regulate selective KIF1A cargo trafficking.
Introduction
Molecular motors use the cellular cytoskeleton to transport cargos throughout the cell. Transport of cargos and other building blocks is crucial for many cellular functions and is especially important in highly complex cells such as neurons. In neurons, cargos (such as protein complexes, membranous vesicles, organelles, and mRNA) are often generated in the cell body and transported over long distances into the correct compartments (Bentley and Banker, 2016). Kinesin, dynein, and myosin are molecular motors that transport these cargos into the axon and dendrites. Tight regulation of the transport machinery is critically important to ensure that cargo is picked up from and delivered to the right place at the right time (Hirokawa et al., 2010; van den Berg and Hoogenraad, 2012). Disruption of the transport machinery leads to neuronal disfunction, and genetic mutations in motor proteins have been linked to various neurological diseases in humans (Millecamps and Julien, 2013; Franker and Hoogenraad, 2013). For example, mutations in the molecular motor KIF1A, a kinesin-3 family member, have been reported to be involved in hereditary spastic paraplegias (Gabrych et al., 2019).
KIF1A is a dimeric motor that drives transport to both axons and dendrites. In neurons, KIF1A exists in an inactive, autoinhibited state, in which the C-terminal tail domain folds back on the N-terminal motor domain (Hammond et al., 2009). Autoinhibition is a common regulation mechanism for kinesin motor proteins (Verhey and Hammond, 2009). Several mechanisms have been found to release autoinhibition, among which are binding of cargo vesicles (Coy et al., 1999; Soppina et al., 2014) or binding to adaptor proteins (Siddiqui et al., 2019; Fu and Holzbaur, 2013). KIF1A and UNC104, its orthologue in Caenorhabditis elegans and Drosophila, were originally identified as the primary motors for axonal transport of synaptic vesicles (SVs; Hall and Hedgecock, 1991; Okada et al., 1995). This was supported by the observation that overactivation of KIF1A/UNC104 leads to an increase in the number of SVs at the synapse, whereas depletion of KIF1A/UNC104 leads to a decrease in SVs and defects in synapse maturation (Chiba et al., 2019; Zhang et al., 2016). Later, KIF1A was also found to be the primary anterograde motor for transport of dense core vesicles (DCVs; Zahn et al., 2004; Lo et al., 2011). Unlike SVs, DCVs are transported from the Golgi apparatus to the plasma membrane in both axons and dendrites (de Wit et al., 2006). Furthermore, in nonneuronal cells, KIF1A has been implicated in transport of organelles, such as lysosomes (Guardia et al., 2016). While KIF1A drives trafficking of several selective vesicle types, it remains unclear how KIF1A specificity is achieved for each cargo.
The interaction between motor protein and cargo is often mediated by adaptor proteins (Hirokawa et al., 2010). Different adaptor proteins have been proposed to mediate the KIF1A–cargo interaction. For example, liprin-α has long been suggested as an adaptor protein for KIF1A-mediated SV transport (Shin et al., 2003; Miller et al., 2005). In addition, tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 (TANC2) was found to be a KIF1A interacting partner. However, it was recently shown that liprin-α and TANC2 might not function as classic adaptors, but rather as signposts recruiting KIF1A-driven cargo to dendritic synapses (Stucchi et al., 2018). The Arf-like small G protein, Arl8, has also been implicated in transport of SVs, and its loss of function decreases axonal trafficking of SVs and accumulates the cargo in the proximal axon (Klassen et al., 2010). GTP-Arl8 bound to SVs releases UNC-104 autoinhibition, thereby promoting cargo transport (Niwa et al., 2016). Mammalian Arl8 has two isoforms, Arl8A and Arl8B, which in nonneuronal cells localize on lysosomes (Hofmann and Munro, 2006). Furthermore, Arl8B was found to mediate lysosomal transport by kinesin-1 family motors in both nonneuronal and neuronal cells (Rosa-Ferreira and Munro, 2011; Farías et al., 2017).
Differentially expressed in normal and neoplastic cells/MAP kinase activating death domain (DENN/MADD or Rab3-GEP, further referenced as MADD) is a protein that has multiple roles, for example in neurotransmission (Miyoshi and Takai, 2004) and SV trafficking (Tanaka et al., 2001). It was also found to be an essential factor in transport of Rab3-containing SVs by KIF1Bβ and KIF1A (Niwa et al., 2008). Thus, several KIF1A adaptor proteins are candidates for selective cargo binding. However, a systematic and in-depth analysis of how KIF1A–adaptor interactions control selective cargo recognition is currently lacking.
To better understand how KIF1A interacts with different types of cargos, we used engineered motors to identify the mechanism of the KIF1A interaction with DCVs, lysosomes, and SVs. Using an inducible assay, we mapped the region in the KIF1A tail domain that is involved in the association with different cargos and found that the KIF1A PH and CC3 domain are required for interaction with all three vesicle types. These findings were further validated using KIF1A depletion and rescue experiments. We also show that KIF1A motor activity is regulated by autoinhibition via CC1 as well as motor dimerization via CC2. In addition, we find that KIF1A-cargo binding depends on phosphorylation events by calmodulin-dependent protein kinase II (CaMKII). Finally, we show that Arl8A acts as an adaptor for KIF1A mediating the interaction with DCVs and lysosomes, whereas MADD is an adaptor for SVs, suggesting a model in which KIF1A-cargo selectivity is achieved by specific adaptors.
Results
Engineering motors to identify cargo for KIF1A in neurons
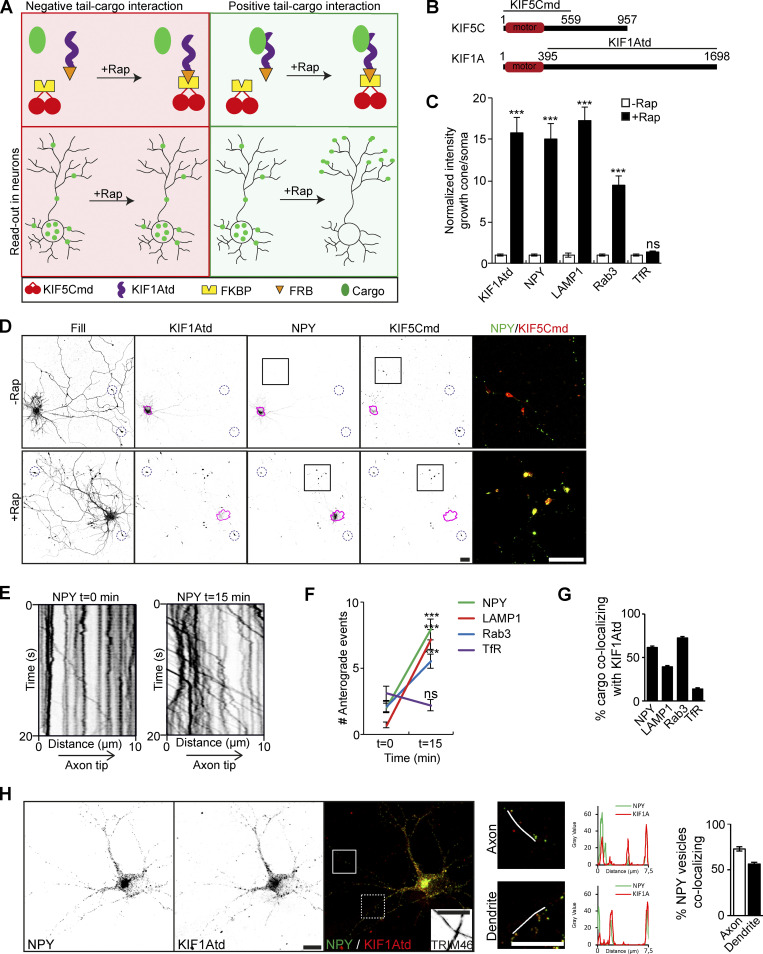
Engineered motors can be used to identify kinesin–cargo interactions. Here, we used the motor domain of the kinesin-1 family member KIF5C (KIF5Cmd) fused to a FKBP domain and the tail domain of KIF1A (KIF1Atd) to a FKBP12–rapamycin-binding (FRB) domain (Fig. 1, A and B). Addition of the chemical dimerizer AP21968, a rapamycin analogue (rapalog), leads to linkage of the FKBP and FRB domain (Hoogenraad et al., 2003; Kapitein et al., 2010a), thereby generating a full kinesin protein with motor and cargo binding functionalities. The constitutively active KIF5Cmd traffics selectively into the axon (Jacobson et al., 2006), which provides a clear readout of cargo relocalization into axonal tips after addition of rapalog in case of an interaction between KIF1Atd and cargo (Fig. 1 A). Validation of the assay was done by coexpressing FKBP-mRFP-KIF5Cmd and FRB-3myc-KIF1Atd constructs in hippocampal neurons. KIF5Cmd was visualized by mRFP immunofluorescence and KIF1Atd by immunostaining with myc antibodies. Addition of rapalog leads to a clear relocalization of KIF1Atd into axonal tips, where it colocalizes with KIF5Cmd (Fig. S1 A). This was quantified by the ratio of KIF1Atd intensity in a distal tip over KIF1Atd intensity in the soma (Fig. 1 C).
Figure 1.
Identification of KIF1A cargo in neurons. Identification of KIF1A cargos using engineered motors and evaluation of KIF1A–cargo interaction in neuronal compartments. (A) Schematic depiction of the engineered motor assay in case of a negative (left) or positive (right) tail–cargo interaction. Upper panels show components of the assay, and lower panels show the readout in a neuron. (B) Schematic depiction of the structure of KIF5C and KIF1A motors. (C) Quantification of the normalized ratio of cargo intensity in axonal tips to that of the soma of hippocampal neurons coexpressing FRB-3myc-KIF1Atd, cargo-GFP, and FKBP-mRFP-KIF5Cmd without rapalog or with addition of 1 µM rapalog (N = 1, n = 15). (D) Representative images of hippocampal neurons coexpressing FRB-3myc-KIF1Atd, NPY-GFP, and FKBP-mRFP-KIF5Cmd without (upper panels) or with (lower panels) addition of 1 µM rapalog. Pink lines mark the cell soma. Blue dotted circles indicate examples of axonal tips. Zooms of the boxed regions are shown as a merge on the right. (E) Representative kymographs showing movement of NPY vesicles in the AIS before (left) and 15 min after (right) addition of 1 µM rapalog. (F) Quantification of the axonal entries of cargo before and 15 min after addition of 1 µM rapalog in neurons expressing FRB-3myc-KIF1Atd, cargo-GFP, and FKBP-mRFP-KIF5Cmd (N = 3, n = 17–27). (G) Quantification of the percentage of cargo vesicles colocalizing with KIF1Atd puncta (N = 3, n = 81–108). (H) Representative image of a neuron coexpressing KIF1Atd and GFP-NPY. White boxes on the merged image mark the axon (continuous line) and a dendrite (dotted line). TRIM46 staining in the axon is shown in the merged image. Merged zooms of the boxes are depicted on the right. Graphs show the NPY (green) and KIF1A (red) intensity along the line marked in the zooms. Quantification of the percentage of NPY colocalizing with KIF1A in the axon and dendrites is shown on the right (N = 3, n = 36 axons and 72 dendrites). Data are displayed as mean ± SEM. Mann–Whitney U test; ***, P < 0.001. Scale bars, 20 µm (D) and 10 µm (H).
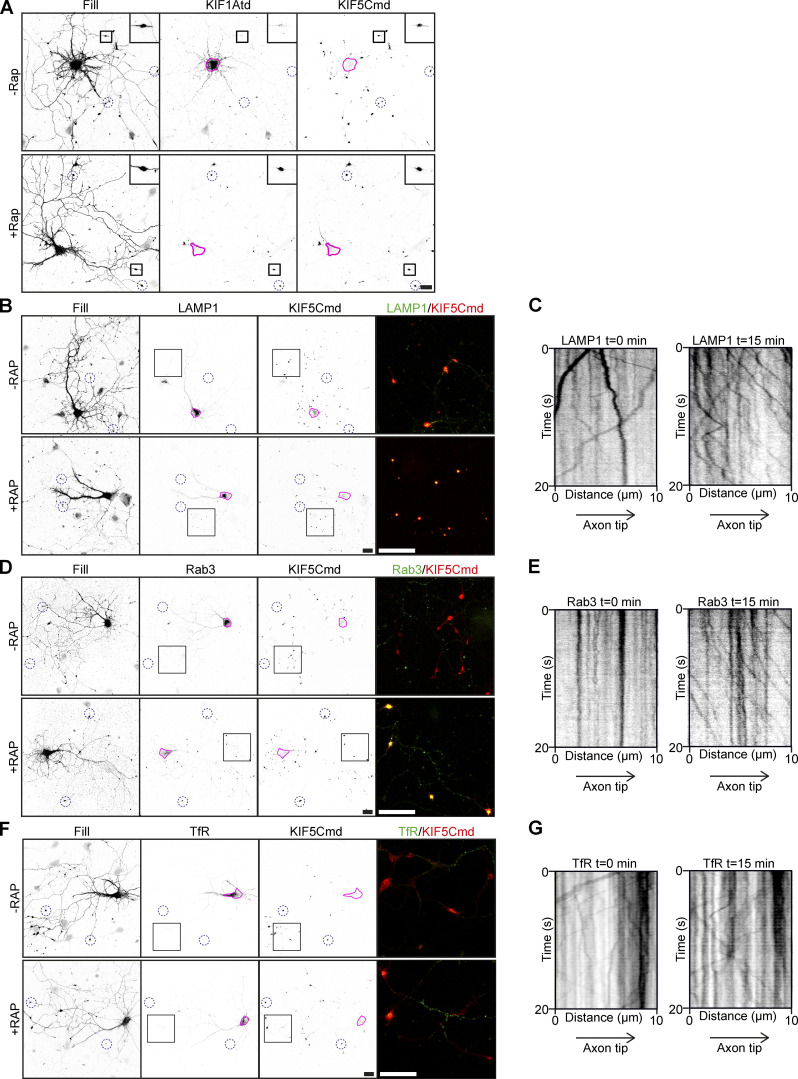
Figure S1.
KIF1A–cargo interactions in the engineered motor assay. Related to Fig. 1. Images validating the engineered motor assay and showing lysosome, SV, and endosomal vesicle localization and motility in the assay. (A) Representative images of hippocampal neurons coexpressing FRB-3myc-KIF1Atd and FKBP-mRFP-KIF5Cmd without (upper panels) or with (lower panels) addition of 1 µM rapalog. Pink lines mark the cell soma. Blue dotted circles indicate examples of axonal tips. Zooms of the boxed regions are shown in the top right of each image. (B, D, and F) Representative images of hippocampal neurons coexpressing FRB-3myc-KIF1Atd, FKBP-mRFP-KIF5Cmd, and LAMP1-GFP (B), Rab3-GFP (D), or TfR-GFP (F) without (upper panels) or with (lower panels) addition of 1 µM rapalog. Pink lines mark the cell soma. Blue dotted circles indicate examples of axonal tips. Zooms of the boxed regions are shown as a merge on the right. (C, E, and G) Representative kymographs showing movement of LAMP1 (C), Rab3 (E), or TfR (G) vesicles in the AIS before (left) and 15 min after (right) addition of 1 µM rapalog. Scale bars, 20 µm.
Next, we used these engineered motors to screen for an interaction of different vesicle populations with KIF1A. Previously, DCVs, lysosomes and SVs have been described as cargo for KIF1A (Lo et al., 2011; Guardia et al., 2016; Hall and Hedgecock, 1991). For each vesicle type a GFP-tagged marker protein was used to label the vesicle population: neuropeptide Y (NPY) for DCVs (Kwinter et al., 2009), lysosomal-associated membrane protein 1 (LAMP1) for lysosomes (Chen et al., 1985), and Rab3 for SVs (Fischer von Mollard et al., 1990). To identify an association between these vesicles and KIF1A, hippocampal neurons were cotransfected with FKBP-mRFP-KIF5Cmd, FRB-3myc-KIF1Atd, and GFP-tagged marker proteins. Addition of rapalog led to relocalization of all three cargos into axonal tips (Fig. 1 D; and Fig. S1, B and D). Relocalization was not observed for endosomal vesicles present in the somatodendritic region, which were labeled using GFP-tagged transferrin receptor (TfR; Fig. S1 F; Burack et al., 2000), indicating that these vesicles do not interact with KIF1A. Relocalization of cargos was clear from quantifications, which show that the ratio of marker intensity in the axonal tip to that of the soma increases ∼15-fold for DCVs and lysosomes and ∼10-fold for SVs (Fig. 1 C). We further verified these interactions by live-cell imaging. For this, we imaged the axon initial segment (AIS), where in case of an interaction between kinesin and cargo, an increase in vesicle entry into the axon will be observed upon addition of rapalog. Kymographs show that already 15 min after rapalog addition, axonal vesicle entry increased for DCVs (Fig. 1 E), lysosomes (Fig. S1 C), and SVs (Fig. S1 E), but not dendritic vesicles (Fig. S1 G), which was confirmed by quantifications (Fig. 1 F). Together, these data show that in hippocampal neurons, KIF1A interacts with DCVs, lysosomes, and SVs, but not with TfR-positive endosomal vesicles.
Association of KIF1A with different cargo is not restricted to neuronal compartments
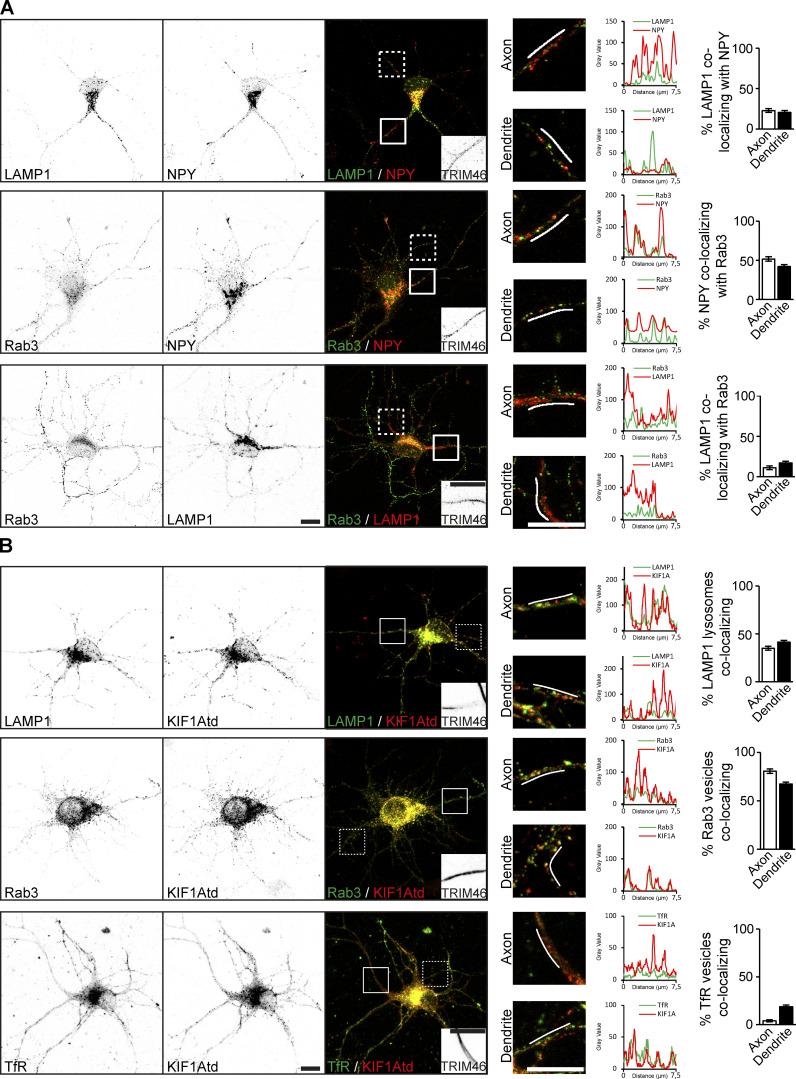
KIF1A interacts with three distinct vesicle populations in hippocampal neurons, raising the question of how specificity for a cargo is achieved. One possibility is that KIF1A associates with different vesicle populations in different neuronal compartments. First, we investigated the colocalization of our marker proteins on vesicles. We found that there was almost no colocalization between LAMP1-containing lysosomes and either NPY or Rab3 vesicles. However, we did observe that ∼50% of the Rab3 vesicles also contained NPY in both axonal and dendritic compartments, suggesting that there is some overlap between these vesicle types (Fig. S2 A). Next, we looked at colocalization of KIF1Atd with different vesicle types in specific neuronal compartments. We found that KIF1Atd colocalizes with DCVs, lysosomes and SVs in both the axon and dendrites (Fig. 1, G and H; and Fig. S2 B). Further analysis revealed that DCVs and SVs show a slightly higher colocalization in the axon, whereas lysosomes colocalize slightly more with KIF1Atd in dendrites (Fig. 1 H; and Fig. S2 B). However, there is no indication of any preference for KIF1A colocalization with a specific cargo in a specific compartment. Low colocalization was observed for KIF1Atd with TfR-containing endosomal vesicles in dendrites, but not in axons (Fig. S2 B). These results confirm the specificity of KIF1A for DCVs, lysosomes, and SVs and demonstrate that KIF1A specificity for a cargo is not restricted to axons or dendrites.
Figure S2.
Colocalization of cargo vesicles and KIF1A. Related to Fig. 1. Representative images and quantifications showing colocalization of vesicle markers and KIF1A colocalization with vesicles. (A) Representative images of hippocampal neurons coexpressing different combinations of vesicle markers. White boxes on the merged image mark the axon (continuous line) and a dendrite (dotted line). TRIM46 staining in the axon is shown in the merged image. Merged zooms of the boxes are depicted on the right. Graphs show the intensity of cargo along the line marked in the zooms. Quantifications of the percentage of vesicle markers colocalizing with each other in the axon and dendrites are shown on the right (N = 3, n = 29–33 axons and 87–99 dendrites). (B) Representative images of hippocampal neurons coexpressing KIF1Atd and GFP-tagged LAMP1, Rab3, or TfR. White boxes on the merged image mark the axon (continuous line) and a dendrite (dotted line). TRIM46 staining in the axon is shown in the merged image. Merged zooms of the boxes are depicted on the right. Graphs show the cargo (green) and KIF1A (red) intensity along the line marked in the zooms. Quantifications of the percentage of vesicle markers colocalizing with KIF1A in the axon and dendrites are shown on the right (N = 3, n = 27–36 axons and 54–72 dendrites). Scale bars, 10 µm.
KIF1A interacts with vesicles via its PH and CC3 domains
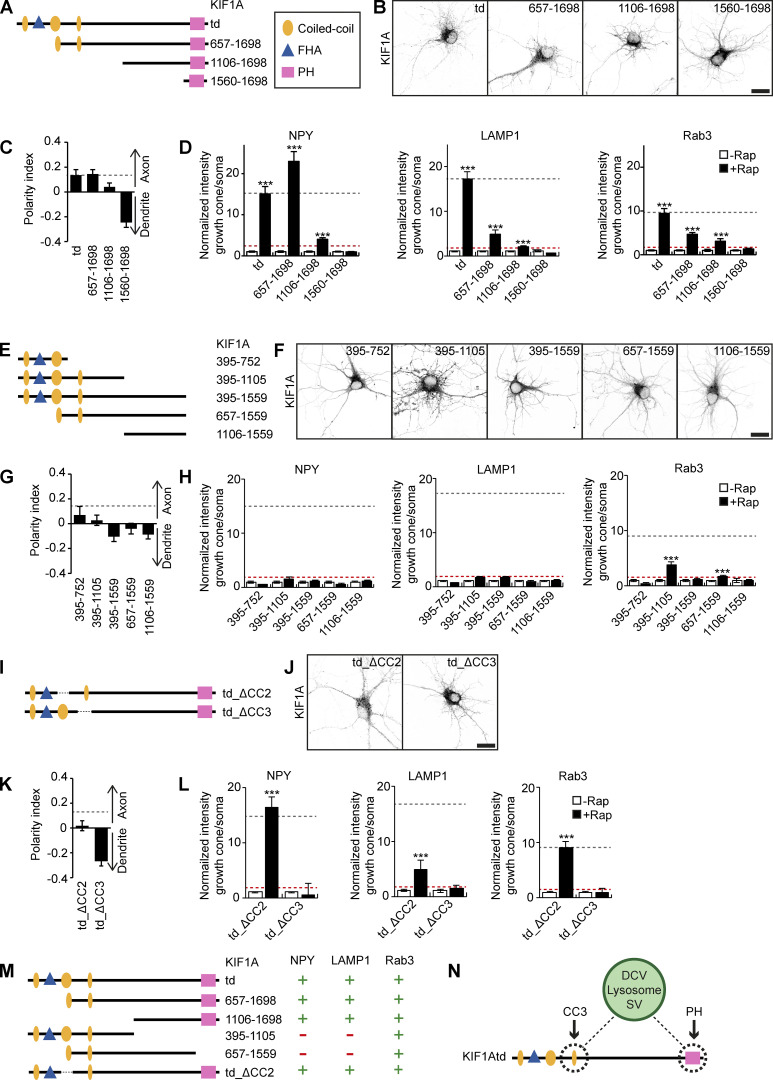
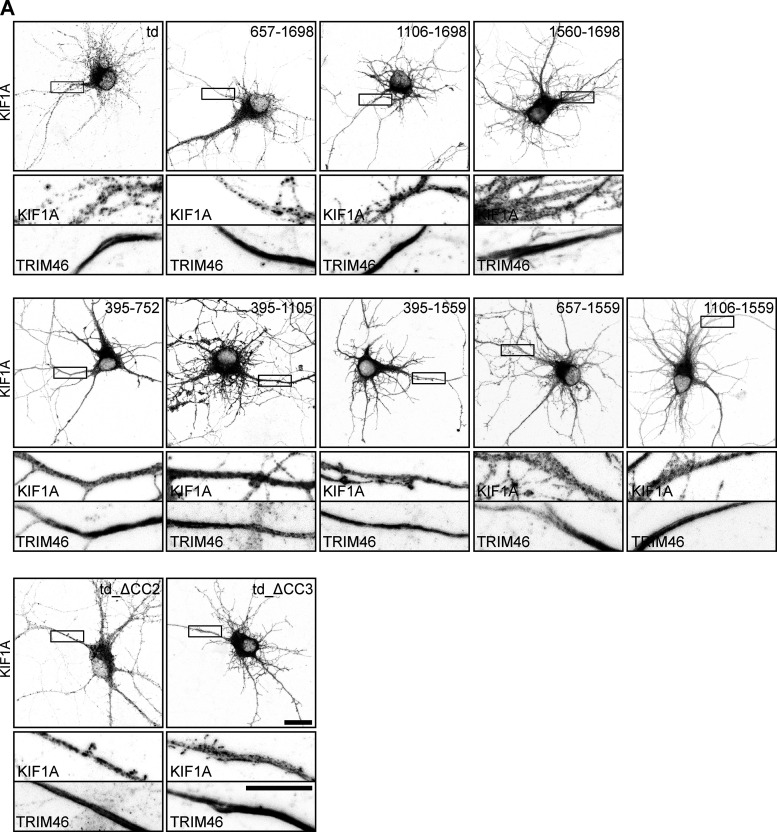
As specificity of KIF1A for a cargo is not achieved by localization in different neuronal compartments, we wondered whether different vesicles interact with different domains of the KIF1A motor as a mechanism to achieve selectivity. The KIF1A tail domain consists of three coiled-coil (CC) domains with a forkhead-associated (FHA) domain between CC1 and CC2 at the N-terminal side and a PH domain at the C-terminal side (Fig. 2 A). We generated several fragments that lack the N-terminus of KIF1Atd (Fig. 2 A). These were expressed in hippocampal neurons to visualize their localization pattern and subjected to our engineered motor assay to identify cargo interactions. Expression of KIF1A_657–1698 and KIF1A_1106–1698 in hippocampal neurons showed a punctate more axonal localization comparable to KIF1Atd, whereas KIF1A_1560–1698 showed a diffuse localization that was more dendritic (Fig. 2, B and C; and Fig. S3 A). This suggests that KIF1A_657–1698 and KIF1A_1106–1698 associate with vesicles, but the PH domain alone is not sufficient for cargo interaction. Each fragment was then screened in our motor assay for an interaction with DCVs, lysosomes, and SVs. We used the following baseline criterion to identify positive cargo interactions: the ratio of cargo in distal tips over the soma is ≥5% of the KIF1Atd signal. This showed that KIF1A_657–1698, lacking only the CC1 and FHA domain, was able to bind all three cargos. The ratio of NPY intensity in the axonal tips to that of the soma increased for KIF1A_657–1698 compared with KIF1Atd, whereas for LAMP1 and Rab3, the ratio decreased for KIF1A_657–1698 with respect to KIF1Atd (Fig. 2 D). KIF1A_1106–1698, having a punctate localization pattern, was found to interact with DCVs, lysosomes, and SVs, although the association with each cargo was decreased strongly when compared with KIF1Atd (Fig. 2 D). The shortest fragment, KIF1A_1560–1698, containing only the PH domain, was unable to bind any of the cargo (Fig. 2 D). This was expected from its diffuse distribution and confirms that the PH domain alone is not sufficient for cargo interaction.
Figure 2.
Identification of the minimum KIF1A domain required for cargo interaction. Overview of different KIF1Atd fragments, their localization, and interactions with cargo. (A, E, and I) Schematic depiction of the structure of truncated KIF1Atd constructs. (B, F, and J) Representative images of hippocampal neurons expressing KIF1Atd truncations (depicted in A, E, and I, respectively). (C, G, and K) Quantification of the polarity index of KIF1Atd truncations (depicted in A, E, and I, respectively). Dotted gray line marks the polarity index of KIF1Atd (N = 3, n = 32–40). (D, H, and L) Quantification of the normalized ratio of cargo intensity in axonal tips to that of the soma of hippocampal neurons coexpressing FRB-3myc–fused truncated KIF1Atd constructs (depicted in A, E, and I, respectively), marker-GFP, and FKBP-mRFP-KIF5Cmd, without or with addition of 1 µM rapalog. Dotted gray line marks the normalized intensity for KIF1Atd with rapalog treatment. Dotted red line marks the baseline of 5% of the KIF1Atd signal (N = 1, n = 7–15). (M) Overview of the identified interactions between truncated KIF1Atd and cargo. (N) Schematic depiction showing the interaction between KIF1A CC3 and PH domain with DCVs, lysosomes, and SVs. Data are displayed as mean ± SEM. Mann–Whitney U test; ***, P < 0.001. Scale bars, 20 µm.
Figure S3.
Expression of KIF1Atd truncation constructs in hippocampal neurons. Related to Fig. 2. (A) Representative images of hippocampal neurons expressing KIF1A deletion constructs (shown in Fig. 2, B, F, and J) including zooms of the axon, marked by the boxes. TRIM46 staining and the KIF1A channel in the axon are shown below. Scale bars, 20 µm (upper images) and 10 µm (lower zooms).
To further identify the minimum binding domain of the KIF1A tail that interacts with cargo, we then examined truncations of KIF1Atd lacking the PH domain. KIF1Atd was truncated from the C-terminus or from both the N- and C-termini to generate five KIF1A fragments: KIF1A_395–752, KIF1A_395–1105, KIF1A-395–1559, KIF1A_657–1559, and KIF1A_1106–1559 (Fig. 2 E). The distribution pattern of these constructs was diffuse, with the latter three constructs having a more dendritic localization compared with KIF1Atd (Fig. 2, F and G; and Fig. S3 A). The screen from our motor assay shows that none of these constructs can associate with DCVs or lysosomes. For SVs, positive interactions were detected with KIF1A_395–1105 and KIF1A_657–1559 (Fig. 2 H), suggesting that two sites might be involved in the interaction, one site in KIF1A_395–1105 and one site in KIF1A_1106–1698. Lastly, two constructs lacking either CC2 or CC3 (td_ΔCC2 and td_ΔCC3; Fig. 2 I) were generated. Whereas KIF1Atd_ΔCC2 showed a similar distribution pattern as KIF1Atd, KIF1Atd_ΔCC3 exhibited a diffuse somatodendritic localization (Fig. 2, J and K; and Fig. S3 A). The engineered motor assay shows that KIF1Atd_ΔCC2 associates with all three vesicle populations, where an interaction comparable to KIF1Atd was observed for DCVs and SVs and a slightly weaker binding for lysosomes. In contrast, KIF1Atd_ΔCC3 could not interact with any of the cargos (Fig. 2 L). In summary, these results suggest that both the PH and CC3 domains of KIF1Atd are necessary for interaction with DCVs, lysosomes, and SVs (Fig. 2, M and N).
The KIF1A PH and CC3 domains are required for cargo trafficking
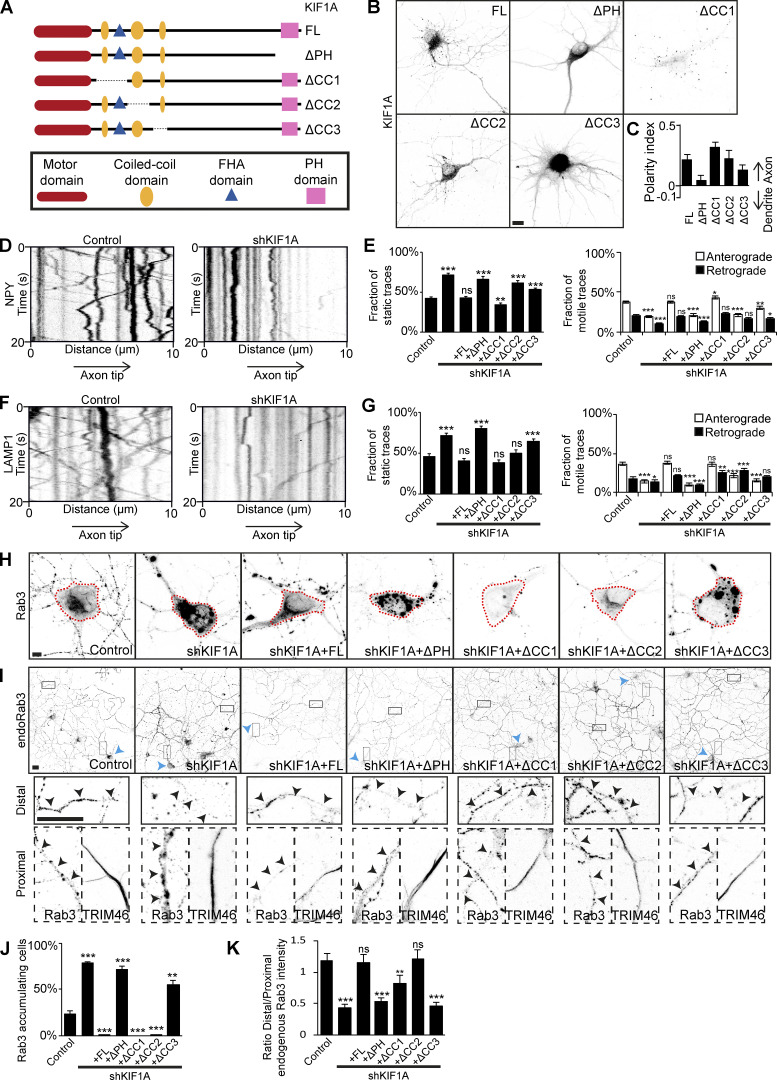
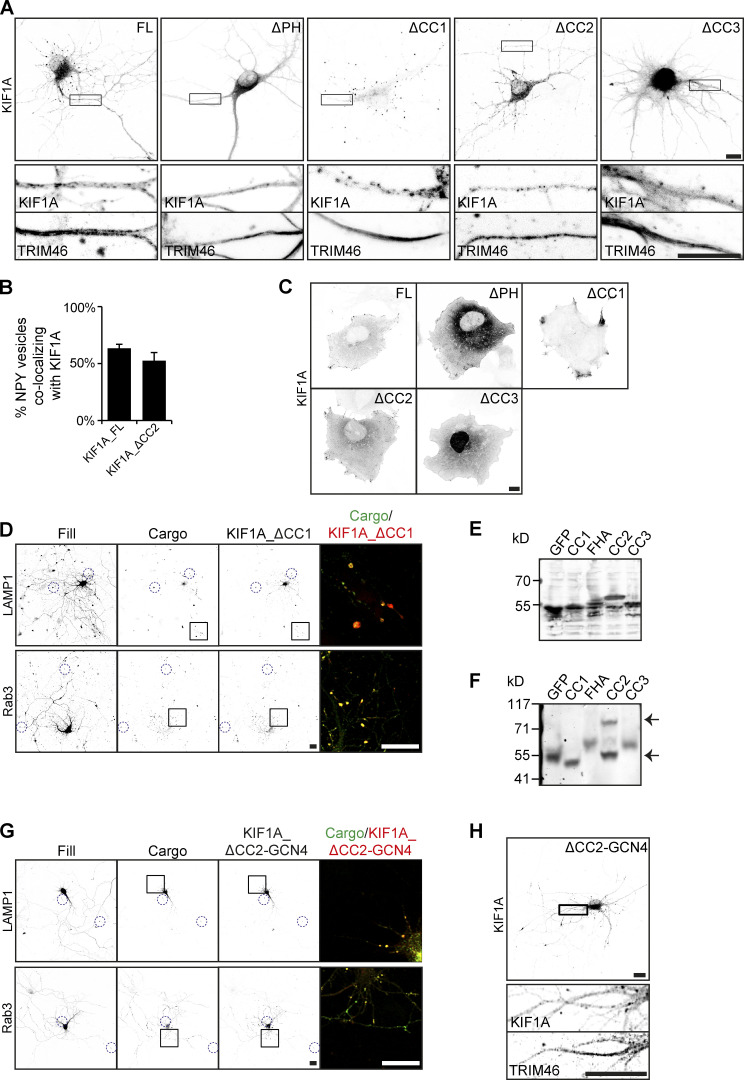
To verify the importance of the PH and CC3 domains of KIF1A for cargo interaction, we performed KIF1A knockdown (KD) and rescue experiments. First, we generated KIF1A deletion constructs lacking the PH, CC1, CC2, or CC3 domain (KIF1A_ΔPH, KIF1A_ΔCC1, KIF1A_ΔCC2, and KIF1A_ΔCC3; Fig. 3 A). Comparison of the localization and polarity indices of these deletion mutants to full-length KIF1A (KIF1A_FL), showed that KIF1A_ΔCC1 and KIF1A_ΔCC2 have a similar vesicular-like pattern, whereas KIF1A_ΔPH, and KIF1A_ΔCC3 have a diffuse unpolarized pattern (Fig. 3, B and C; and Fig. S4 A), suggesting that the latter two are not associated with vesicles. KD of KIF1A in hippocampal neurons gave different phenotypes for each vesicle population. Depletion of KIF1A resulted in decreased axonal motility of DCVs (Fig. 3 D). Quantifications show that there is an increase in static traces and a decrease in both anterograde and retrograde traces (Fig. 3 E). The decreased vesicle motility phenotype was rescued by KIF1A_FL and KIF1A_ΔCC1, but not by KIF1A_ΔPH, KIF1A_ΔCC2, or KIF1A_ΔCC3 (Fig. 3 E). Lysosomes showed a similar decreased axonal motility phenotype upon KIF1A depletion (Fig. 3, F and G). This phenotype was rescued by KIF1A_FL, KIF1A_ΔCC1, and KIF1A_ΔCC2, but not by KIF1A_ΔPH or KIF1A_ΔCC3 (Fig. 3 G). For SVs, we found two phenotypes upon depletion of KIF1A. SVs, visualized by overexpression of marker Rab3, accumulate in the cell soma in KIF1A KD (Fig. 3, H and J). This was rescued by KIF1A_FL, KIF1A_ΔCC1, and KIF1A_ΔCC2, but not by KIF1A_ΔPH or KIF1A_ΔCC3 (Fig. 3, H and J). Accordingly, endogenous Rab3 vesicles were less transported along the axon upon KIF1A depletion (Fig. 3 I). The ratio of Rab3 intensity in the distal over the proximal axon indicates a decrease in SVs distributed in the distal axon in KIF1A KD compared with control (Fig. 3 K). The axonal transport of endogenous Rab3 vesicles was rescued by KIF1A_FL, KIF1A_ΔCC1, and KIF1A_ΔCC2, but not by KIF1A_ΔPH or KIF1A_ΔCC3 (Fig. 3, I and K). Thus, the PH and CC3 domains are required for KIF1A-mediated trafficking of DCVs, lysosomes, and SVs.
Figure 3.
Validation of the KIF1A CC3 and PH domains as minimal domains for cargo interaction. KIF1A KD and rescue experiments to verify the requirement of KIF1A CC3 and PH domains in cargo association. (A) Schematic depiction of the structure of KIF1A deletion constructs. (B) Representative images of hippocampal neurons expressing KIF1A deletion constructs. (C) Quantification of the polarity index of KIF1A deletion constructs (N = 3, n = 39–45). (D and F) Representative kymographs showing movement of NPY (D) or LAMP1 (F) vesicles in the AIS in control or when transfected with KIF1A shRNA. (E and G) Quantification of the fraction of static, anterograde, and retrograde NPY (E) or LAMP1 (G) traces in control, KIF1A KD, and rescue with KIF1A deletion constructs (N = 3, n = 40–45; except NPY in KD + ΔPH, N = 2, n = 25). (H) Representative images of the soma of hippocampal neurons expressing GFP-Rab3 in control, cotransfected with KIF1A shRNA, or with KIF1A shRNA and KIF1A deletion constructs. Dotted red line marks the cell soma. (I) Representative images of hippocampal neurons immunostained for Rab3 in control situation, when transfected with KIF1A shRNA, or with KIF1A shRNA and KIF1A deletion constructs. Blue arrowheads indicate the soma of a transfected cell, closed boxes mark part of the distal axon, and dotted boxes mark the proximal axon. Zooms of the boxes are shown underneath, including TRIM46 staining in the proximal axon. Black arrowheads indicate the axon of the transfected neuron. (J) Quantification of the number of cells with Rab3 accumulation in the cell soma in control, KIF1A KD, and rescue with KIF1A deletion constructs (N = 4, n = 370–400). (K) Quantification of the ratio of endogenous Rab3 intensity in the distal axon to that of the proximal axon in control, KIF1A KD, and rescue with KIF1A deletion constructs (N = 3, n = 38–40). Data are displayed as mean ± SEM. Unpaired t test (E) and Mann–Whitney U test (G, J, and K); *, P < 0.05; **, P < 0.01; ***, P < 0.001. Scale bars, 10 µm (B), 5 µm (H), and 20 µm (I).
Figure S4.
Expression of KIF1A constructs and colocalization with cargo. Related to Figs. 3 and 4. Example images of cells expressing KIF1A constructs in neurons and in COS7 cells or together with cargo in neurons and dimerization analysis on Western blot. (A) Representative images of hippocampal neurons expressing KIF1A deletion constructs (shown in Fig. 3 B) including zooms of the axon, marked by the boxes. TRIM46 staining and the KIF1A channel in the axon are shown below. (B) Quantification of the percentage of NPY vesicles colocalizing with KIF1A during live-imaging experiments (N = 7, n = 15 for KIF1A_FL; N = 4, n = 15 for KIF1A_ΔCC2). (C) Representative images of COS7 cells expressing different KIF1A constructs. (D) Representative images of hippocampal neurons coexpressing KIF1A_ΔCC1 and LAMP1 or Rab3. Blue dotted circles indicate examples of distal tips. Merged image of the boxed region is shown on the right. (E and F) Normal (E) and native (F) SDS-PAGE analysis of GFP-fused KIF1A domains expressed in HEK293T cell lysates. Arrows (F) mark the bands of monomeric and dimeric CC2 domain. (G) Representative images of hippocampal neurons coexpressing KIF1A_ΔCC2-GCN4 and LAMP1 or Rab3. Blue dotted circles indicate examples of distal tips. Merged image of the boxed region is shown on the right. (H) Representative image of a hippocampal neuron expressing KIF1A_ ΔCC2-GCN4 (shown in Fig. 4 H) including zoom of the axon, marked by the box. TRIM46 staining and the KIF1A channel in the axon are shown below. Scale bars, 10 µm (A, C, D, and G) and 20 µm (H).
Interaction of KIF1A with cargo via PH and CC3 is needed for motor activity
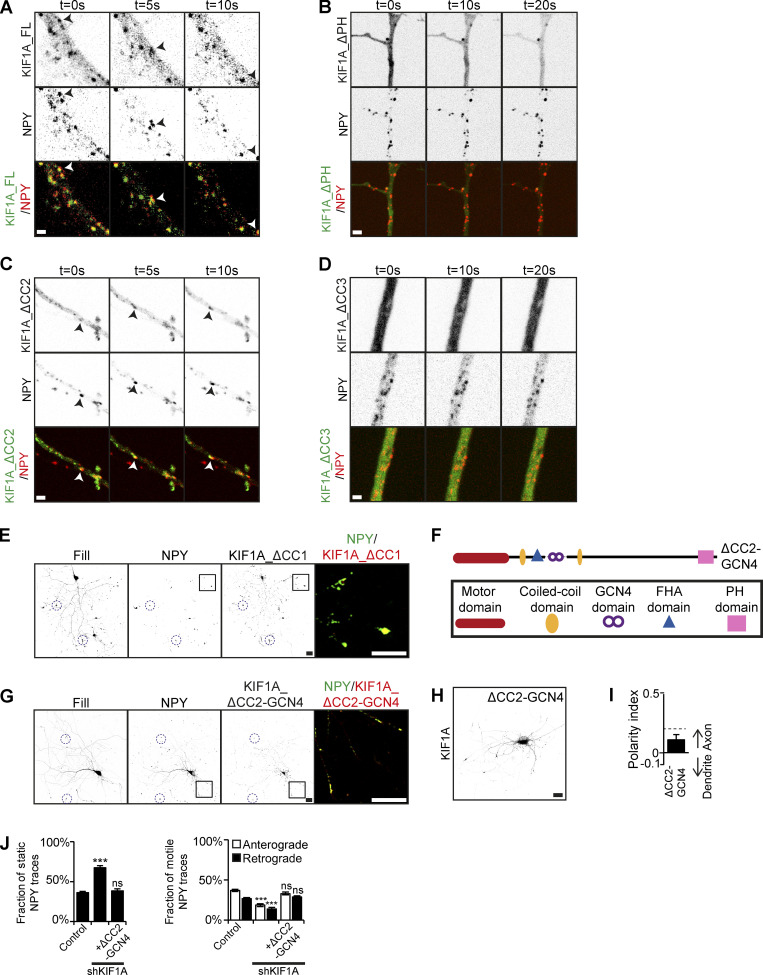
To further show the role of the PH and CC3 domain of KIF1A for vesicle interaction, we investigated live movement of KIF1A deletion constructs together with DCVs. KIF1A_FL colocalizes and moves together with NPY-containing DCVs (Fig. 4 A), and similar comovement was observed for KIF1A_ΔCC2 (Fig. 4 C). During live imaging, ∼50–60% of NPY vesicles colocalized with KIF1A_FL and KIF1A_ΔCC2 motors (Fig. S4 B). In contrast, KIF1A_ΔPH, and KIF1A_ΔCC3 showed diffuse localization and did not colocalize with DCVs. Interestingly, we observed that upon their expression, DCV movements were abolished, indicating that they might act as dominant negatives (Fig. 4, B and D). As the CC1 is involved in regulation of motor activity, removal of CC1 results in a constitutively active kinesin motor, which accumulates in distal tips, making it challenging to live image its movement (Huo et al., 2012). We did observe that NPY vesicles are still motile when coexpressed with KIF1A_ ΔCC1. Furthermore, DCVs, lysosomes, and SVs accumulate and colocalize with KIF1A_ ΔCC1 in distal tips, indicating an interaction between the cargos and KIF1A_ ΔCC1 (Figs. 4 E and S4 D). When we expressed KIF1A deletion constructs in COS7 cells, we observed that KIF1A_FL, KIF1A_ΔCC1, and KIF1A_ΔCC2 translocate into the cell periphery, indicating that they are active motors, which is consistent with our live-imaging experiments in neurons. For KIF1A_ΔCC1, we observed a large accumulation of the motor in the cell periphery, confirming its high motor activity. In contrast, KIF1A_ΔPH and KIF1A_ΔCC3 showed a diffuse localization pattern (Fig. S4 C). Together, these results suggest that interaction of the KIF1A PH and CC3 domains with cargo is crucial for KIF1A motor activity.
Figure 4.
KIF1A vesicle association and motor dimerization are required for vesicle transport. Comovement of KIF1A with cargo and investigation of the involvement of KIF1A dimerization in cargo transport. (A–D) Stills showing movement of KIF1A_FL (A), KIF1A_ΔPH (B), KIF1A_ΔCC2 (C), or KIF1A_ΔCC3 (D) and NPY vesicles in the axon. Arrowheads mark a moving NPY vesicle colocalizing with KIF1A_FL (A) or KIF1A_ΔCC2 (C). (E) Representative image of a hippocampal neuron coexpressing KIF1A_ΔCC1 and NPY. Blue dotted circles indicate examples of distal tips. Merged image of the boxed region is shown on the right. (F) Schematic depiction of the structure of the KIF1A_ΔCC2-GCN4 motor. (G) Representative image of a hippocampal neuron coexpressing KIF1A_ΔCC2-GCN4 and NPY. Blue dotted circles indicate examples of distal tips. Merged image of the boxed region is shown on the right. (H) Representative image of a hippocampal neuron expressing KIF1A_ΔCC2-GCN4. (I) Quantification of the polarity index of KIF1A_ΔCC2-GCN4 motor (N = 3, n = 38). (J) Quantification of the fraction of static, anterograde, and retrograde NPY traces in control, KIF1A KD, and rescue with KIF1A_ΔCC2-GCN4 (N = 3, n = 35). Data are displayed as mean ± SEM. Unpaired t test, ***, P < 0.001. Scale bars, 1 µm (A–D) and 20 µm (E, G, and H).
Dimerization of KIF1A via its CC2 domain is important for cargo transport
Previously, the CC2 domain of KIF1A has been implicated in motor dimerization (Hammond et al., 2009). In our KD and rescue experiments, we observed that KIF1A_ ΔCC2 was not able to rescue the reduced NPY motility in KIF1A KD, and we speculated that this might be due to a reduced ability of monomer KIF1A to interact with vesicles. To investigate the involvement of different KIF1A domains in dimerization, we expressed GFP-fused domain constructs in HEK293T cells and analyzed cell lysates by normal and native SDS-PAGE. Results show an additional band for the CC2 domain, which is twice its normal size on native gel, suggesting that the domain is dimerized (Fig. S4, E and F). This was not observed for any of the other domains, suggesting that the CC2 domain of KIF1A is responsible for motor dimerization. Next, we generated a construct in which we replaced the CC2 domain with a GCN4 leucine zipper domain (O’Shea et al., 1991), thereby artificially dimerizing the motor (Fig. 4 F). Expression of the artificially dimerized KIF1A motor in neurons showed that it accumulates in distal tips, while maintaining a slightly axonal polarity comparable to KIF1A_FL (Fig. 4, H and I; and Fig. S4 H). When we expressed KIF1A_ΔCC2-GCN4 together with GFP-labeled cargo, we observed that the motor colocalized with vesicles in distal tips, suggesting that the motor interacts with DCVs, lysosomes, and SVs (Figs. 4 G and S4 G). Next, we performed KIF1A KD experiments and found that dimerized KIF1A could rescue the NPY motility phenotype (Fig. 4 J). These data show that KIF1A dimerization by CC2 is important for the regulation of cargo transport.
KIF1A-cargo binding is regulated by phosphorylation
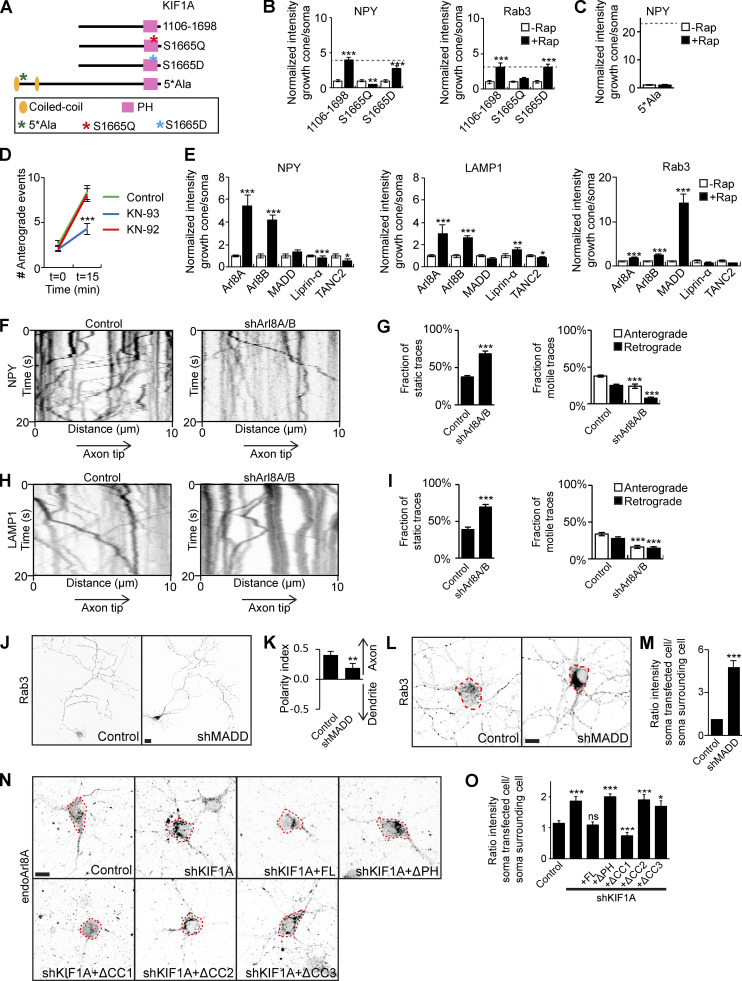
KIF1A-associated neurological disorder is a neurodegenerative disorder characterized by symptoms including spastic paraplegia, ataxia, and intellectual disability. Several mutations in the KIF1A gene were found to underlie the neurodegenerative symptoms, most of which are found in the KIF1A motor domain. Five mutations in the human KIF1Atd have been described, one of them being the mutation of serine 1758 to glutamine, which localizes in the PH domain (Gabrych et al., 2019; Rivière et al., 2011). We reasoned that this mutation might disrupt the KIF1A–cargo interaction. Human S1758 corresponds to S1665 in our mouse cDNA, and we therefore mutated S1665 to Q1665 in KIF1A_1106–1698, the minimal KIF1A fragment that associates with cargo (Fig. 5 A). The interaction of KIF1A_S1665Q with DCVs and SVs was then assessed in our motor assay, and we found that it was unable to transport DCV or SVs, indicating that this mutation indeed interferes with KIF1A-cargo binding (Fig. 5 B). As the mutation involved the change from a serine, a potential phosphorylation site, to a glutamine, a phosphodeficient site, we wondered whether a phosphorylation mechanism might play a role in the KIF1A–cargo interaction. Therefore, we generated KIF1A_S1665D, containing a phosphomimetic aspartic acid residue. Using this construct, we observed that transport of DCVs and SVs was restored to levels comparable to KIF1A_1106–1698, suggesting that phosphorylation of serine 1665 is required for an interaction between KIF1A and cargo.
Figure 5.
Identification of KIF1A adaptors for specific cargo transport. Identification of a phosphorylation mechanism in KIF1A-cargo binding and Arl8A and MADD as adaptors regulating KIF1A specificity for DCVs and lysosomes or SVs, respectively. (A) Schematic depiction of the structure of KIF1A_1106–1698 mutated constructs and KIF1A_5*Ala. (B and C) Quantification of the normalized ratio of cargo intensity in axonal tips to that of the soma of hippocampal neurons coexpressing FRB-3myc–fused mutated KIF1A_1106–1698 (B) or KIF1A_5*Ala (C), marker-GFP, and FKBP-mRFP-KIF5Cmd, without or with addition of 1 µM rapalog. Dotted gray line marks the normalized intensity for KIF1A_1106–1698 (B) or KIF1A_657–1698 (C) with rapalog treatment (N= 1, n = 15). (D) Quantification of the axonal entries of NPY vesicles before and 15 min after addition of 1 µM rapalog in neurons expressing FRB-3myc-KIF1Atd, NPY-GFP, and FKBP-mRFP-KIF5Cmd in control or after treatment with 1 µM KN-93 or 1 µM KN-92 for 20 h (N = 3, n = 30). (E) Quantification of the normalized ratio of cargo intensity in axonal tips to that of the soma of hippocampal neurons coexpressing FRB-3myc–fused adaptors, cargo-GFP, and FKBP-mRFP-KIF5Cmd, without or with addition of 1 µM rapalog (N = 1, n = 10–15). (F and H) Representative kymographs showing movement of NPY (F) or LAMP1 (H) vesicles in the AIS in control or when transfected with Arl8A/B shRNA. (G and I) Quantification of the fraction of static, anterograde, and retrograde NPY (G) or LAMP1 (I) traces in control or when transfected with Arl8A/B shRNA (N = 3, n = 29–30). (J) Representative images of hippocampal neurons expressing GFP-Rab3 in control or when cotransfected with MADD shRNA. (K) Quantification of the polarity index of GFP-Rab3 in control or MADD KD (N = 3, n = 29–30). (L) Representative images of the soma of hippocampal neurons immunostained for Rab3 in control or when cotransfected with MADD shRNA. Dotted red lines mark the cell soma. (M) Quantification of the ratio of endogenous Rab3 intensity in a transfected cell to that of a surrounding cell in control or MADD KD (N = 3, n = 29–30). (N) Representative images of the soma of hippocampal neurons immunostained for Arl8A in control, when cotransfected with KIF1A shRNA, or with KIF1A shRNA and KIF1A deletion constructs. Dotted red lines mark the cell soma. (O) Quantification of the ratio of endogenous Arl8A intensity in a transfected cell to that of a surrounding cell in control, KIF1A KD, and rescue with KIF1A deletion constructs (N = 3, n = 26–30). Data are displayed as mean ± SEM. Unpaired t test (G, I, and K) and Mann–Whitney U test (B–E, M, and O), *, P < 0.05; **, P < 0.01; ***, P < 0.001. Scale bars, 10 µm (L and N) and 20 µm (J).
Previously, it was shown that KIF1A has a calmodulin (CaM) binding site and that activation of the motor is calcium dependent (Stucchi et al., 2018). Therefore, we hypothesized that binding of CaM activates Ca2+/CaMKII, which then phosphorylates KIF1A. To test this, we generated KIF1A_5*Ala (Fig. 5 A), which contains five alanine substitutions in the CaM-binding domain of KIF1A_657–1698, thereby preventing the interaction of KIF1A with CaM (Stucchi et al., 2018). In our motor assay, KIF1A_5*Ala was unable to interact with DCVs (Fig. 5 C), suggesting that the interaction of KIF1A with CaM is needed for vesicle association. We then investigated NPY axonal vesicle entry in our motor assay in the presence of KN-93, a specific CaMKII inhibitor. When cells were treated with KN-93, there was only a small increase in axonal NPY entry after addition of rapalog. In contrast, the negative control, treated with KN-92, behaved like the untreated control condition (Fig. 5 D). Altogether, these results suggest that phosphorylation of KIF1A by CaMKII, potentially in the PH domain, is an important step in vesicle association.
Identification of adaptors for specific cargo types
Next, we turned our focus to different KIF1A adaptors. The main adaptors described for KIF1A are liprin-α, TANC2, Arl8A/B, and MADD, and they have been implicated in transport of DCVs, lysosomes, and SVs. Using our engineered motor platform, we generated FRB-3myc-adaptor constructs and expressed these in neurons together with FKBP-mRFP-KIF5Cmd and GFP-tagged markers. This allowed assessment of the interaction of each adaptor with DCVs, lysosomes, and SVs. Results show that Arl8A and Arl8B both interact with DCVs and lysosomes, whereas MADD clearly interacts only with SVs (Fig. 5 E). Liprin-α and TANC2 did not interact with any of the vesicle types in our motor assay (Fig. 5 E).
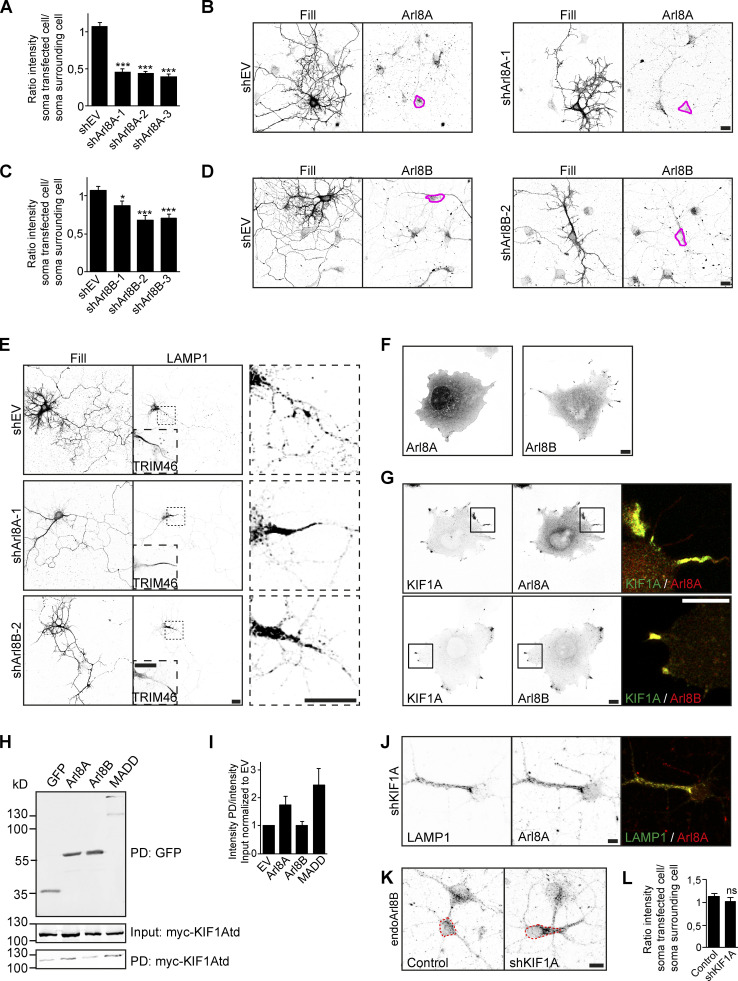
We further focused our attention on Arl8A/B and MADD and depleted these adaptors in hippocampal neurons. Arl8A and Arl8B shRNA constructs were validated using immunofluorescence (Fig. S5, A–D), and we used shArl8A-1 and shArl8B-2 for further experiments. When coexpressing these shRNA constructs together with LAMP1, we observed that LAMP1 is prevented from entry into the axon and accumulates in the proximal axon (Fig. S5 E). To circumvent potential redundancy between the two Arl8 proteins, we then performed an Arl8A/B double KD and found a decrease in both NPY and LAMP1 motility (Fig. 5 F–I), supporting a role for Arl8 in transport of these vesicles. When we knocked down MADD using a previously validated shRNA (Niwa et al., 2008), we observed changes in the localization pattern of overexpressed Rab3-GFP. In control situations, it has a punctate pattern, but it becomes diffuse upon MADD depletion, suggesting loss of its vesicle association (Fig. 5 J). In addition, polarity index quantifications show that Rab3-GFP localizes more somatodendritically when MADD is depleted (Fig. 5 K). We also looked at endogenous Rab3 and found that it accumulates in the cell soma upon MADD KD (Fig. 5, L and M). These findings show that Arl8A/B are adaptors for DCVs and lysosomes, and MADD is an adaptor for SVs.
Figure S5.
Identification of Arl8A and MADD as specific KIF1A adaptors. Related to Fig. 5. Validation of Arl8 shRNAs and investigation of Arl8A and MADD as specific KIF1A adaptors. (A and C) Quantification of the ratio of endogenous Arl8A (A) or Arl8B (C) intensity in a transfected cell to that of a surrounding cell in control and with different shRNAs against Arl8A (A) or Arl8B (C; N = 4, n = 40–35). (B and D) Representative images of hippocampal neurons expressing shEV, shArl8A-1 (B), or shArl8B-2 (D), immunostained for Arl8A (B) or Arl8B (D). Pink lines mark the cell soma of the transfected cell. (E) Representative images of hippocampal neurons coexpressing LAMP1 together with shEV, shArl8A-1, or shArl8B-2. TRIM46 staining in the dotted box is shown in the bottom left to mark the axon. Zooms of the dotted boxes are shown on the right. (F) Representative images of COS7 cells expressing Arl8A or Arl8B. (G) Representative images of COS7 cells coexpressing KIF1A together with Arl8A or Arl8B. Merged image of the boxed region is shown on the right. (H) Pull-down experiment showing the interaction between KIF1A_395–1105 and bioGFP-adaptors in HEK293T cell lysates. (I) Quantification of the ratio of KIF1A_395–1105 intensity in the pull-down fraction to that of the input fraction normalized to bioGFP-EV (N = 3). (J) Representative image of a hippocampal neuron coexpressing shKIF1A and LAMP1-GFP and immunostained for Arl8A. (K) Representative images of the soma of hippocampal neurons immunostained for Arl8B in control or when transfected with KIF1A shRNA. Dotted red lines mark the cell soma. (L) Quantification of the ratio of endogenous Arl8B intensity in a transfected cell to that of a surrounding cell in control or KIF1A KD (N = 3, n = 29–30). Data are displayed as means ± SEM. Unpaired t test (A) and Mann–Whitney U test (C and L), *, P < 0.05; ***, P < 0.001. Scale bars, 20 µm (B, D, and E) and 10 µm (F, G, J, and K).
Arl8A and MADD mediate the interaction between KIF1A CC3 and specific cargo
We further explored the interaction between the adaptors and KIF1A. First, we expressed Arl8A and Arl8B in COS7 cells and found that Arl8A is diffuse over the cell, whereas Arl8B is mainly diffuse but also has some accumulation in the cell periphery (Fig. S5 F). When we coexpressed these adaptors together with KIF1A_FL, a clear relocalization of Arl8A into the cell periphery was observed, where it colocalized with KIF1A. As Arl8B already localized in the cell periphery, we could not confirm that the observed colocalization with KIF1A was due to an interaction with the motor (Fig. S5 G). To further investigate the interaction between KIF1A and adaptors, we performed pull-down experiments where we used the adaptors to pull down KIF1A_395–1105. Western blot analysis confirmed an interaction of KIF1A_395–1105 with Arl8A and MADD, whereas no interaction was seen with Arl8B (Fig. S5, H and I). These data point to a role for the CC3 domain in the interaction with adaptors. To confirm the observation that Arl8A, but not Arl8B, is an adaptor for KIF1A-mediated DCV and lysosome transport, the localization of Arl8A was analyzed in KIF1A-depleted neurons, and we observed that endogenous Arl8A accumulates in the cell body in KIF1A KD. Rescue experiments using different KIF1A deletion constructs (Fig. 3 A) show that this phenotype was rescued by KIF1A_FL and KIF1A_ΔCC1, but not by KIF1A_ΔPH, KIF1A_ΔCC2, or KIF1A_ΔCC3 (Fig. 5, N and O). This effect resembles the pattern observed in rescue experiments of the NPY motility phenotype (Fig. 3 E), suggesting that Arl8A is an adaptor for KIF1A-mediated DCV transport. In addition, when LAMP1 was coexpressed in KIF1A-depleted neurons, we observed that the Arl8A accumulation colocalizes with LAMP1-containing lysosomes, supporting a role for Arl8A in lysosomal transport by KIF1A (Fig. S5 J). In contrast, no changes in endogenous Arl8B localization were observed upon KIF1A depletion (Fig. S5, K and L). Altogether, these data suggest that Arl8A and MADD interact with KIF1A via its CC3 domain and that Arl8A specifically is involved in the association between KIF1A and DCVs and lysosomes, whereas MADD specifically mediates the interaction between KIF1A and SVs.
Discussion
The three KIF1A CC domains play distinct roles in regulating motor activity
Kinesin motors contain different structural domains that contribute to their regulation and function. The stalk domain of KIF1A contains three CC domains and an FHA domain between CC1 and CC2. Previous studies have described different roles for these domains in motor regulation. For instance, CC1 was found to regulate motor activity by inhibition of the motor domain (Ren et al., 2018; Huo et al., 2012). In addition, both CC1 and CC2 have been implicated in motor dimerization (Huo et al., 2012; Hammond et al., 2009). Also, the FHA domain was found to be involved in the regulation of motor activity, suggesting that the central CC1-FHA-CC2 region might function as a negative regulatory center for KIF1A motor activity (Lee et al., 2004; Huo et al., 2012). Here, we found that removal of CC1 results in a very processive motor, which in neurons accumulates in distal tips. This localization is comparable to the localization of only the KIF1A motor domain (Huang and Banker, 2012), thereby supporting a role for CC1 in motor inhibition. Furthermore, we demonstrate that CC2 is involved in motor dimerization. Although removal of CC2 does not completely abolish cargo trafficking, we find that artificially dimerized KIF1A accumulates rapidly in distal tips, suggesting that dimerization of KIF1A is important for its activity. Lastly, our results show that the KIF1A CC3 is required for vesicle interaction. This is in line with previous work, which has suggested that the stalk region of KIF1A is critically important for cargo interaction (Klopfenstein and Vale, 2004; Xue et al., 2010; Guardia et al., 2016). Interestingly, removal of the CC3, thereby abolishing cargo interaction, resulted in a nonprocessive motor, suggesting that kinesin–cargo interactions could be required for motor activity. Overall, our data point to a role for all three CC domains in motor activity, however via various mechanisms.
KIF1A PH domain binding to vesicular cargo is regulated by phosphorylation
The C-terminal PH domain is a unique feature of KIF1A and KIF1Bβ motors and has been implicated in linking these motors to cargo. The PH domain of KIF1A has been shown to bind to PIP2 on vesicle membranes and mediates efficient vesicle transport in different cell types (Klopfenstein et al., 2002; Klopfenstein and Vale, 2004; Xue et al., 2010). Here, we confirm that the PH domain of KIF1A associates with DCVs, lysosomes, and SVs in living neurons, indicating that cargo binding to the PH domain is a conserved mechanism across cell types. We identified S1665 in the PH domain as a crucial residue for cargo interaction, and by using phosphodeficient S1665Q and phosphomimetic S1665D mutants, we showed that phosphorylation of this residue is essential. One could speculate that phosphorylation of a serine residue enhances binding with phosphoinositides on the cargo membrane. We also found that cargo interaction is dependent on CaMKII activity. Interestingly, Ca2+/CaM-regulated kinesin motors have been engineered, and they revealed that Ca2+ levels regulate kinesin motor activity (Shishido and Maruta, 2012). In addition, phosphorylation-dependent cargo loading and trafficking has been described for other kinesin motors, such as KIF3A (Ichinose et al., 2015), and a Ca2+ regulation mechanism was found for mitochondrial trafficking by kinesin-1 (Wang and Schwarz, 2009). Furthermore, it has been shown that KIF1A activity is regulated by CaM binding, as Ca2+/CaM-dependent modulation of KIF1A enables transport of DCVs (Stucchi et al., 2018). It is tempting to speculate on a phosphorylation-dependent mechanism for regulating KIF1A-cargo transport, where binding of CaM to KIF1A activates CaMKII to phosphorylate the PH domain and allow cargo interaction. This model fits well with our findings that phosphorylation events are critical steps in cargo binding and in regulating KIF1A cargo trafficking.
Cargo binding is stabilized by interaction with specific adaptors
Cargo-adaptor proteins are known to regulate kinesin–cargo interactions (Hirokawa et al., 2010). Several adaptors have been described for KIF1A, including liprin-α, TANC2, Arl8A/B, and MADD. Recently, our laboratory described that liprin-α and TANC2 do not act as classic motor-cargo adaptors but rather as signposts to recruit KIF1A-cargo complexes at synaptic sites (Stucchi et al., 2018). Using engineered motors, we show that liprin-α and TANC2 do not interact with vesicles, thereby supporting their role as signposts rather than classic cargo adaptors. Arl8A and Arl8B have been implicated in transport of lysosomes and SVs across different cell types (Niwa et al., 2016; Farías et al., 2017; Rosa-Ferreira and Munro, 2011). Furthermore, a direct interaction between the KIF1A CC3 domain and Arl8A was established by affinity chromatography and yeast two-hybrid assays (Wu et al., 2013). MADD has been described as an adaptor for KIF1A- and KIF1Bβ-mediated transport of SVs, and it was shown to interact with the KIF1Bβ stalk domain (Niwa et al., 2008). Our findings that KIF1A_395–1105 binds to Arl8A and MADD support a role of the stalk domain in the interaction between KIF1A and its adaptors. Interestingly, KIF1A_657–1105 was also identified as the binding region for the nonclassic adaptors liprin-α and TANC2 (Stucchi et al., 2018), suggesting that adaptors with different functions associate with the same region in KIF1A. Based on our data and previous literature, we propose a general mechanism for KIF1A-mediated cargo trafficking. In its autoinhibited state, inactive KIF1A exists as a dimer (Hammond et al., 2009). This dimeric conformation is established by the CC2 domain. An increase in the local Ca2+ level activates CaM and induces its binding to KIF1A, thereby diminishing KIF1A autoinhibition by CC1 and opening the motor domain. In parallel, CaM activates CaMKII, which phosphorylates the PH domain of active KIF1A and enables cargo binding. The KIF1A-cargo association is stabilized by specific adaptors, including Arl8A and MADD, which interact with both the cargo and the KIF1A CC3 domain. KIF1A transports the cargo until it reaches an adaptor signpost, such as liprin-α and TANC2, which outcompetes the classic adaptor for binding with the KIF1A stalk domain. This decreases the strength of the KIF1A–cargo interaction and results in release of the vesicle in the designated area.
In summary, we have dissected the role of different CC domains in KIF1A motor activity and show that CC1 regulates autoinhibition, CC2 regulates motor dimerization, and CC3 is involved in cargo interaction. Furthermore, we propose a two-step model for KIF1A-cargo binding and transport in neuronal cells. First, phosphorylation of S1665 in the KIF1A PH domain may enable binding to any cargo vesicle, whether it is DCVs, lysosomes, or SVs. Specificity for a cargo is achieved in a second step in which a specific adaptor stabilizes the interaction of KIF1A with a certain cargo. The adaptor interacts with both the cargo and the CC3 domain of KIF1A. We identified Arl8A as specific adaptor for DCVs and lysosomes and MADD as a specific adaptor for SVs. This mechanism fits well in the model for specific KIF1A-mediated cargo transport and delivery. As many similarities have been found in KIF1A vesicle transport in different cell types and between KIF1A and KIF1Bβ, the described mechanism may be a generic model for KIF1-mediated cargo trafficking.
Materials and methods
Animals
All animal experiments were approved by the Dutch Animal Experiments Committee and were performed in line with guidelines of Utrecht University, Dutch law (Wet op de Dierproeven, 1996), and European regulations (Guideline 86/609/EEC). In this study, neurons obtained from embryonic day 18 stage embryos of both genders from female pregnant Wistar rats (Janvier) were used. Pregnant rats were at least 10 wk old and not involved in previous experiments. Animals were housed in transparent Plexiglas cages with wood-chip bedding and paper tissue and provided with unrestricted access to food and water. Adult rats were kept with a companion and kept in a 12-h light–dark cycle with a temperature of 22 ± 1°C.
Primary hippocampal neuron cultures and transfections
Primary hippocampal neuron cultures were prepared from embryonic day 18 rat brains according to protocols described previously (Kapitein et al., 2010c). Cells were plated at a density of 100K neurons per well in a 12-well plate on coverslips coated with poly-L-lysine (37.5 µg/ml; Sigma-Aldrich) and laminin (1.25 µg/ml; Roche) and grown in Neurobasal medium (NB; Gibco) supplemented with 2% B27 (Gibco), 0.5 mM L-glutamine (Gibco), 15.6 µM glutamate (Sigma-Aldrich), and 1% penicillin/streptomycin (Gibco) at 37°C and 5% CO2.
Hippocampal neurons were transfected at the indicated time points using Lipofectamine 2000 (Invitrogen). In short, DNA (1.8 µg/well) was mixed with Lipofectamine 2000 (3.3 µl/well) in 200 µl NB, incubated for 30 min, and added to neurons in transfection medium (NB supplemented with 0.5 mM glutamine) for 45 min at 37°C and 5% CO2. Neurons were then washed with NB and transferred to their original medium at 37°C and 5% CO2 until fixation at indicated time points. For engineered motor experiments, neurons were transfected at 7 days in vitro (DIV7). When used for analysis in fixed cells, rapalog (final concentration of 1 µM) was added directly after transfection, and neurons were fixed on DIV8. When used for live-imaging experiments, neurons were imaged on DIV8. For KIF1A-cargo or cargo-cargo colocalization experiments, neurons were transfected on DIV3 and fixed or imaged on DIV4. To calculate the polarity indices, neurons were transfected on DIV7 and fixed on DIV8. For KD experiments, neurons were transfected on DIV4 and fixed or imaged on DIV8.
Cultured cells and transfections
HEK293T and COS7 cells were cultured in 50/50 DMEM/Ham’s F10 (Lonza) medium supplemented with 10% FCS (Sigma-Aldrich) and 1% penicillin/streptomycin (Sigma-Aldrich). HEK293T cells were transfected using MaxPEI (Polysciences) in a ratio of 3:1 PEI:DNA, according to the manufacturer’s protocol. COS7 cells were plated on 18-mm glass coverslips 1 d before transfection with DNA constructs using FuGENE6 (Roche) and following the manufacturer’s protocol.
DNA and shRNA constructs
The following DNA constructs used in this study were described before: pβ-actin-HA-β-galactosidase (Hoogenraad et al., 2005), pGW2-NPY-GFP and pGW2-NPY-RFP (Schlager et al., 2010), GFP-LAMP1 and LAMP1-RFP (Farías et al., 2017), GFP-Rab3C (van Vlijmen et al., 2008), TfR-GFP (Burack et al., 2000), and BirA coding vector and pebioGFP (van der Vaart et al., 2013). GW1-KIF5C_1–559-mRFP-(FKBP)2 was cloned by PCR-based strategy from pBa-Kif5C_1–559-GFP (plasmid 45059; Addgene) using GW1-PEX-mRFP-(FKBP)2 (Kapitein et al., 2010a) as backbone. GW1-FRB-3myc-KIF5A_375-1032 was cloned using a PCR-based Gibson Assembly strategy with the GW1-HA expression vector as backbone and GW1-GFP-FRB (Kapitein et al., 2010b) and GW1-GFP-KIF5A to generate the GW1-FRB-3myc backbone. GW1-FRB-3myc-KIF1A_395-1698 (KIF1Atd) was cloned using GW1-HA-KIF1A (Kevenaar et al., 2016) as PCR template into the GW1-FRB-3myc backbone. The following KIF1Atd fragments were generated by PCR using GW1-FRB-3myc-KIF1A_395-1698 as template and were cloned into the GW1-FRB-3myc backbone: KIF1A_657–1698, KIF1A_1106–1698, KIF1A_1560–1698, KIF1A_395–752, KIF1A_395–1106, KIF1A_395–1559, KIF1A_657–1559, KIF1A_1106–1559, KIF1A_395–600_701–1698 (KIF1A_ΔCC2), and KIF1A_395–780_850–1698 (KIF1A_ΔCC3). Full-length KIF1A (KIF1A_FL) and deletion constructs (KIF1A_ΔPH, KIF1A_ΔCC1, KIF1A_ΔCC2, KIF1A_ΔCC3, and KIF1A_ ΔCC2-GCN4) were cloned with PCR-based strategies using GW1-HA-KIF1A as template and ligated into GW1-HA and pGW1-GFP (Jaworski et al., 2009) expression vectors. GFP-KIF1A domain constructs (CC1, 410–480; FHA, 500–580; CC2, 600–700; and CC3, 781–850) were generated using PCR-based strategies with GW1-HA-KIF1A as template and pGW1-GFP as backbone. Adaptors were cloned into the GW1-FRB-3myc, bioGFP, and pGW1-GFP backbone using PCR-based strategies with Arl8A (plasmid 67403; Addgene), Arl8B (plasmid 67404; Addgene), and MADD (IMAGE clone 5271798) as templates.
For KD experiments, shRNAs were inserted into the pSuper vector (Brummelkamp et al., 2002), which contains a scrambled sequence. The shRNA for KIF1A has been described before (Kevenaar et al., 2016). For Arl8A and Arl8B shRNAs, the following targeting sequences were used: shArl8A-1, 5′-GCGTCAGGACAGTTCAATG-3′; shArl8A-2, 5′-TCCACAACCTGCTAGACAA-3′; sh-Arl8A-3, 5′-ACCGAGAGATCTGCTGCTA-3′; shArl8B-1, 5′-ACCGAGAGATCTGCTGCTA-3′; shArl8B-2, 5′-AGAGGCGTCAATGCAATTG-3′; and shArl8B-3, 5-GAACCTGTCTGCTATTCAA-3′. The sequence used for shMADD (5′-ATCCTGCCAGAACCGGAAT-3′) has been described before (Niwa et al., 2008).
Antibodies and reagents
The following antibodies and dilutions were used in this study for immunofluorescence experiments: mouse anti-myc (1:200, #SC-40; Bio Connect), chicken anti–β-galactosidase (1:2,500, BGL-1040; Aveslab), mouse anti-Rab3 (1:200, 610379; BD Biosciences), rabbit anti-Arl8A (1:200, 17060-1-AP; Proteintech), rabbit anti-Arl8B (1:200, 13049-1-AP; Proteintech), rabbit anti-TRIM46 serum (1:500, described before [van Beuningen et al., 2015]), goat anti-chicken Alexa Fluor 405 (1:400, ab175675; Abcam), goat anti-rabbit Alexa Fluor 405 (1:400, A31556; Thermo Fisher Scientific), goat anti-mouse Alexa Fluor 488 (1:400, A11029; Thermo Fisher Scientific), goat anti-rabbit Alexa Fluor 488 (1:400, A11034; Thermo Fisher Scientific), goat anti-mouse Alexa Fluor 568 (1:400, A11031; Thermo Fisher Scientific), goat anti-chicken Alexa Fluor 568 (1:400, A11041, Thermo Fisher Scientific), and goat anti-mouse Alexa Fluor 647 (1:400, A21236; Thermo Fisher Scientific). For live-imaging analysis, NF-CF555 (Farías et al., 2016) was used. For Western blot, the following antibodies were used: mouse anti-myc (1:200, SC-40; Bio Connect), rabbit anti-GFP (1:10,000, ab290; Abcam), goat anti-mouse IRDye800CW (1:15,000, 926-32210; LI-COR), and goat-anti-rabbit IRDye680LT (1:20,000, 926-68021; LI-COR). Reagents used in this study are rapalog (AP21967, 635056; TaKaRa), KN-93, and KN-92.
Immunofluorescence staining and imaging
After transfection, neurons or COS7 cells were fixed for 10 min with 4% formaldehyde/4% sucrose in PBS at RT. Cells were washed three times in PBS-CM (PBS, 1 mM MgCl2, and 0.1 mM CaCl2), permeabilized in 0.2% Triton X-100 for 15 min, washed once with PBS-CM, and incubated with 0.2% gelatin for 30 min at 37°C. Primary antibodies were diluted in 0.2% gelatin and incubated for 30 min at 37°C. Next, coverslips were washed three times in PBS-CM, incubated with secondary antibody diluted in 0.2% gelatin for 30 min at 37°C, washed three times in PBS-CM, and mounted in Fluoromount (Invitrogen).
Fixed cells were imaged on (1) a Carl Zeiss LSM 700 confocal laser scanning microscope running ZEN2011 software, using a Plan-Apochromat 40×/1.30 oil differential interference contrast objective or a Plan-Apochromat 63×/1.40 oil differential interference contrast objective; or (2) a Nikon Eclipse 80i upright widefield fluorescence microscope, equipped with a Photometrics CoolSNAP HQ2 charge-coupled device camera and Nikon NIS Br software, using a Plan Fluor 40× NA/1.30 oil objective. For quantitative experiments, image settings were kept identical for all images within one experiment.
Pull-down experiments and Western blotting
For pull-down experiments, HEK293T cells were diluted and plated into six-well plates 1 d before transfection with with pCl-Neo-BirA, GW1-FRB-3myc-KIF1A_395–1105, and bioGFP-tagged constructs. 24 h after transfection, cells were washed with ice-cold PBS and lysed in lysis buffer (100 mM Tris HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, and protease inhibitors [Roche]) on ice for 30 min. Lysates were cleared by centrifugation for 30 min at 13.2 krpm at 4°C. For pull-down experiments, supernatants were incubated for 1.5 h at 4°C with blocked (incubation for 30 min at RT in 50 mM Tris HCl, pH 7.5, 150 mM KCl, and 0.2 µg/µl chicken egg albumin) Streptavidin Dynabeads M-280 (Invitrogen), after which beads were washed five times with washing buffer (100 mM Tris HCl, pH 7.5, 250 mM NaCl, and 0.5% Triton X-100). Proteins were eluted from beads by boiling for 10 min at 95°C in 2× DTT + sample buffer (20% glycerol, 4% SDS, 200 mM DTT, 100 mM Tris HCl, pH 6.8, and bromophenol blue), and protein samples were run on 10% SDS-PAGE gels. For SDS-PAGE experiments, lysates from HEK293T cells expressing GFP-KIF1A domain constructs were made as described above. Lysates were either boiled at 95°C in 1× DTT + sample buffer and run on 12% SDS-PAGE gels or run on 12% mini-protean TGX precast protein gels (Bio-Rad) without boiling in 1× DTT − sample buffer. Proteins were then transferred from the gel to nitrocellulose membranes (Bio-Rad) by semidry blotting at 16 V for 1 h, and membranes were blocked by incubation for 1 h at RT in 3% BSA in PBST (PBS supplemented with 0.02% Tween 20). Next, membranes were incubated overnight at 4°C with primary antibodies in 3% BSA-PBST and washed three times with PBST, followed by incubation with secondary antibody in 3% BSA-PBST for 1 h at RT, and washed three times with PBST. Membranes were scanned using an Odyssey Infrared Imaging system (LI-COR Biosciences), and blots were acquired at 680 and 800 nm.
Image analysis and quantification
Engineered motor assay
For quantification of transport in the engineered motor assay, confocal images of neurons transfected with HA-β-galactosidase, FKBP-mRFP-KIF5Cmd, FRB-3myc-constructs, and cargo were acquired. For each cell, fluorescence intensity of the cargo was measured in five growth cones and the cell soma (regions of interest [ROIs] determined from β-galactosidase staining). The ratio of fluorescence intensity in a growth cone to that of the cell soma was calculated and normalized to the ratio of the condition without rapalog.
Vesicle colocalization
To investigate cargo colocalization or KIF1Atd-cargo colocalization, confocal images of neurons expressing different cargo combinations or KIF1Atd and GFP-labeled cargo were acquired. For each cell, a line of 20–30-µm length was drawn in the axon (identified from TRIM46 staining) and two dendrites. For cargo colocalization, first the total amount of puncta of one cargo along a line was counted and then the amount of colocalizing puncta of the other cargo was counted, which allowed calculation of the percentage of colocalization. For KIF1Atd-cargo colocalization, the total amount of KIF1Atd puncta along a line was counted. Next, the amount of colocalizing cargo particles was counted, and the percentage of colocalization was calculated.
Polarity indices
For calculation of the polarity index, neurons expressing HA-β-galactosidase and different KIF1A constructs were imaged on the upright fluorescent microscope. A line of 5-µm width and 40–90-µm length was drawn in the axon (identified from TRIM46 staining) and in three dendrites (ROIs determined from β-galactosidase staining). The same ROIs were positioned in a near area where no neurons were present, and this background intensity was subtracted from raw measurements. The intensity in the axon, Ia, and the average intensity in three dendrites, Id, were then used to calculate the polarity index with the formula PI = (Ia − Id)/(Ia + Id).
Rab3 accumulation
For calculation of Rab3-accumulating cells, neurons expressing Rab3-GFP, shKIF1A, and indicated rescue constructs were visualized on the upright fluorescent microscope. For each condition, ≤100 neurons were counted and classified as Rab3 accumulating or not Rab3 accumulating, which enabled quantification of the percentage of Rab3-accumulating cells.
Quantification of fluorescence intensity ratio in distal to proximal axons
To quantify axonal fluorescent Rab3 intensity, confocal images of neurons expressing indicated shRNAs and rescue constructs, stained for Rab3 and TRIM46, were acquired. A line of 5-µm width and 15–40-µm length was drawn in the proximal axon (identified from TRIM46 staining), and the same ROIs were positioned in a near area where no neurons were present. The same was done in a distal part of the proximal axon. Background intensities were subtracted from raw measurements, and the ratio of fluorescence intensity in distal to proximal axons was calculated.
Quantification of fluorescence intensity in cell soma
To quantify the fluorescence intensity in the cell soma, confocal images of neurons expressing HA–β-galactosidase and the indicated shRNAs and stained for the protein of interest were acquired. An ROI was drawn around the soma of a transfected cell (determined from β-galactosidase staining) and then positioned in a near area where no neurons were present. The same was done for two surrounding untransfected cells. Background intensities were subtracted from raw measurements, and the ratio of fluorescence intensity in the transfected cell to the average intensity in the surrounding cells was calculated.
Live-cell imaging and analysis
Live-cell imaging experiments were performed using an inverted Nikon Eclipse Ti-E confocal microscope equipped with a perfect focus system (Nikon), a CSU-X1-A1 Spinning Disc unit (Yokogawa) and a Photometrics Evolve 512 electron-multiplying charge-coupled device camera (Roper Scientific) with a Plan Apo VC 100× NA 1.40 oil objective. For imaging, coverslips were mounted in a Ludin chamber (Life Imaging Services) and maintained in culture medium at 37°C and 5% CO2 in a stage incubator (Tokai Hit) during image acquisition.
Imaging of axonal vesicle entry in the engineered motor assay
Neurons expressing FKBP-mRFP-KIF5Cmd, FRB-3myc-KIF1Atd, and GFP-tagged markers were incubated for 30 min with NF-CF555 before imaging. Neurons were then visualized in the axon (identified by NF-CF555 staining), and videos in the GFP channel were acquired as stream acquisitions at 10 frames/s for 20 s. Videos were taken before addition of rapalog and 15 min after addition of rapalog (1 µM final concentration). For analysis, a line along the AIS was drawn and straightened, and kymographs were generated using Kymoreslicewide plugin for Fiji (https://github.com/ekatrukha/KymoResliceWide). An area of 10 µM on the kymograph was selected, and all anterograde events were traced and counted for quantification of axonal vesicle entry.
Imaging of cargo motility
Neurons expressing indicated shRNAs, rescue constructs, and GFP-tagged markers were incubated for 30 min with NF-CF555 before imaging. Neurons were then visualized in the axon (identified by NF-CF555), and videos were acquired in the GFP channel as stream acquisitions at 10 frames/s for 20 s. For analysis, a line along the AIS was drawn, and kymographs were generated. An area of 10 µM on the kymograph was selected, and all static, anterograde, and retrograde events were traced and counted for quantification.
Imaging of comovement of motor and cargo
Neurons expressing the indicated GFP-tagged KIF1A constructs and NPY-RFP were visualized, and axons were determined based on morphology. Videos in the distal axon were acquired in the GFP and RFP channels as stream acquisitions at 10 frames/s for 20 s. Videos were processed using Fiji. To quantify colocalization of KIF1A and NPY during live imaging, stills of the videos at 1 and 5 s were made. In these stills, the total amount of KIF1A puncta and colocalizing NPY particles were counted in the imaged axon, and the percentage of colocalization was calculated.
Statistical analysis
Statistical analyses were performed using GraphPad 5. Data distribution was tested for normality using a D’Agostino and Pearson omnibus normality test. Normally distributed datasets were compared using unpaired t tests. Datasets that did not have a normal distribution were compared using Mann-Whitney U tests. Tests were two-tailed. Statistical tests used for each experiment are detailed in the figure legends. All error bars are displayed as mean ± SEM. The following P value conventions are used throughout the paper: ns, P > 0.05; *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Online supplemental material
Fig. S1 shows images that validate the engineered motor assay and lysosome, SV, and endosomal vesicle localization and motility in the assay. Fig. S2 contains representative images and quantifications of the colocalization of different cargos with KIF1Atd. Fig. S3 compares the expression patterns of different KIF1Atd constructs in hippocampal neurons. Fig. S4 shows example images of neurons and COS7 cells expressing KIF1A constructs and dimerization analysis on Western blot. Fig. S5 shows the validation of Arl8 shRNAs and investigates Arl8A and MADD as specific KIF1A adaptors.
Supplementary Material
Acknowledgments
We thank Ginny Farias, Robin Buijs, and Robbelien Kooistra for discussion, feedback, and advice on experimental design.
This work was supported by the Netherlands Organisation for Scientific Research (NWO-ALW-VICI to C.C. Hoogenraad), the Foundation for Fundamental Research on Matter (to C.C. Hoogenraad), which is part of the Netherlands Organisation for Scientific Research, the Netherlands Organization for Health Research and Development (ZonMW-TOP to C.C. Hoogenraad), and the European Research Council (ERC-consolidator: 617050, C.C. Hoogenraad).
C.C. Hoogenraad is an employee of Genentech, Inc., a member of the Roche group. The authors declare no additional competing financial interests.
Author contributions: J.J.A. Hummel designed, performed, and analyzed the experiments, processed data for representation in the figures, and wrote the manuscript. C.C. Hoogenraad supervised the research, coordinated the study, and edited the manuscript.
References
- Bentley, M., and Banker G.. 2016. The cellular mechanisms that maintain neuronal polarity. Nat. Rev. Neurosci. 17:611–622. 10.1038/nrn.2016.100 [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards R., and Agami R.. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2:243–247. 10.1016/S1535-6108(02)00122-8 [DOI] [PubMed] [Google Scholar]
- Burack, M.A., Silverman M.A., and Banker G.. 2000. The role of selective transport in neuronal protein sorting. Neuron. 26:465–472. 10.1016/S0896-6273(00)81178-2 [DOI] [PubMed] [Google Scholar]
- Chen, J.W., Murphy T.L., Willingham M.C., Pastan I., and August J.T.. 1985. Identification of two lysosomal membrane glycoproteins. J. Cell Biol. 101:85–95. 10.1083/jcb.101.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, K., Takahashi H., Chen M., Obinata H., Arai S., Hashimoto K., Oda T., McKenney R.J., and Niwa S.. 2019. Disease-associated mutations hyperactivate KIF1A motility and anterograde axonal transport of synaptic vesicle precursors. Proc. Natl. Acad. Sci. USA. 116:18429–18434. 10.1073/pnas.1905690116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy, D.L., Hancock W.O., Wagenbach M., and Howard J.. 1999. Kinesin’s tail domain is an inhibitory regulator of the motor domain. Nat. Cell Biol. 1:288–292. 10.1038/13001 [DOI] [PubMed] [Google Scholar]
- de Wit, J., Toonen R.F., Verhaagen J., and Verhage M.. 2006. Vesicular trafficking of semaphorin 3A is activity-dependent and differs between axons and dendrites. Traffic. 7:1060–1077. 10.1111/j.1600-0854.2006.00442.x [DOI] [PubMed] [Google Scholar]
- Farías, G.G., Britt D.J., and Bonifacino J.S.. 2016. Imaging the polarized sorting of proteins from the Golgi complex in live neurons. Methods Mol. Biol. 1496:13–30. 10.1007/978-1-4939-6463-5_2 [DOI] [PubMed] [Google Scholar]
- Farías, G.G., Guardia C.M., De Pace R., Britt D.J., and Bonifacino J.S.. 2017. BORC/kinesin-1 ensemble drives polarized transport of lysosomes into the axon. Proc. Natl. Acad. Sci. USA. 114:E2955–E2964. 10.1073/pnas.1616363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard, G., Mignery G.A., Baumert M., Perin M.S., Hanson T.J., Burger P.M., Jahn R., and Südhof T.C.. 1990. rab3 is a small GTP-binding protein exclusively localized to synaptic vesicles. Proc. Natl. Acad. Sci. USA. 87:1988–1992. 10.1073/pnas.87.5.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franker, M.A.M., and Hoogenraad C.C.. 2013. Microtubule-based transport - basic mechanisms, traffic rules and role in neurological pathogenesis. J. Cell Sci. 126:2319–2329. 10.1242/jcs.115030 [DOI] [PubMed] [Google Scholar]
- Fu, M.M., and Holzbaur E.L.F.. 2013. JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J. Cell Biol. 202:495–508. 10.1083/jcb.201302078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrych, D.R., Lau V.Z., Niwa S., and Silverman M.A.. 2019. Going too far is the same as falling short: Kinesin-3 family members in hereditary spastic paraplegia. Front. Cell. Neurosci. 13:419. 10.3389/fncel.2019.00419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardia, C.M., Farías G.G., Jia R., Pu J., and Bonifacino J.S.. 2016. BORC functions upstream of kinesins 1 and 3 to coordinate regional movement of lysosomes along different microtubule tracks. Cell Rep. 17:1950–1961. 10.1016/j.celrep.2016.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, D.H., and Hedgecock E.M.. 1991. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 65:837–847. 10.1016/0092-8674(91)90391-B [DOI] [PubMed] [Google Scholar]
- Hammond, J.W., Cai D., Blasius T.L., Li Z., Jiang Y., Jih G.T., Meyhofer E., and Verhey K.J.. 2009. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 7:0650–0663. 10.1371/journal.pbio.1000072 [DOI] [PMC free article] [PubMed]
- Hirokawa, N., Niwa S., and Tanaka Y.. 2010. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 68:610–638. 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- Hofmann, I., and Munro S.. 2006. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 119:1494–1503. 10.1242/jcs.02958 [DOI] [PubMed] [Google Scholar]
- Hoogenraad, C.C., Milstein A.D., Ethell I.M., Henkemeyer M., and Sheng M.. 2005. GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat. Neurosci. 8:906–915. 10.1038/nn1487 [DOI] [PubMed] [Google Scholar]
- Hoogenraad, C.C., Wulf P., Schiefermeier N., Stepanova T., Galjart N., Small J.V., Grosveld F., de Zeeuw C.I., and Akhmanova A.. 2003. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J. 22:6004–6015. 10.1093/emboj/cdg592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.F., and Banker G.. 2012. The translocation selectivity of the kinesins that mediate neuronal organelle transport. Traffic. 13:549–564. 10.1111/j.1600-0854.2011.01325.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo, L., Yue Y., Ren J., Yu J., Liu J., Yu Y., Ye F., Xu T., Zhang M., and Feng W.. 2012. The CC1-FHA tandem as a central hub for controlling the dimerization and activation of kinesin-3 KIF1A. Structure. 20:1550–1561. 10.1016/j.str.2012.07.002 [DOI] [PubMed] [Google Scholar]
- Ichinose, S., Ogawa T., and Hirokawa N.. 2015. Mechanism of Activity-Dependent Cargo Loading via the Phosphorylation of KIF3A by PKA and CaMKIIa. Neuron. 87:1022–1035. 10.1016/j.neuron.2015.08.008 [DOI] [PubMed] [Google Scholar]
- Jacobson, C., Schnapp B., and Banker G.A.. 2006. A change in the selective translocation of the Kinesin-1 motor domain marks the initial specification of the axon. Neuron. 49:797–804. 10.1016/j.neuron.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Jaworski, J., Kapitein L.C., Gouveia S.M., Dortland B.R., Wulf P.S., Grigoriev I., Camera P., Spangler S.A., Di Stefano P., Demmers J., et al. 2009. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 61:85–100. 10.1016/j.neuron.2008.11.013 [DOI] [PubMed] [Google Scholar]
- Kapitein, L.C., Schlager M.A., Kuijpers M., Wulf P.S., van Spronsen M., MacKintosh F.C., and Hoogenraad C.C.. 2010a. Mixed microtubules steer dynein-driven cargo transport into dendrites. Curr. Biol. 20:290–299. 10.1016/j.cub.2009.12.052 [DOI] [PubMed] [Google Scholar]
- Kapitein, L.C., Schlager M.A., van der Zwan W.A., Wulf P.S., Keijzer N., and Hoogenraad C.C.. 2010b. Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys. J. 99:2143–2152. 10.1016/j.bpj.2010.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein, L.C., Yau K.W., and Hoogenraad C.C.. 2010c. Microtubule dynamics in dendritic spines. Methods Cell Biol. 97:111–132. 10.1016/S0091-679X(10)97007-6 [DOI] [PubMed] [Google Scholar]
- Kevenaar, J.T., Bianchi S., van Spronsen M., Olieric N., Lipka J., Frias C.P., Mikhaylova M., Harterink M., Keijzer N., Wulf P.S., et al. 2016. Kinesin-binding protein controls microtubule dynamics and cargo trafficking by regulating kinesin motor activity. Curr. Biol. 26:849–861. 10.1016/j.cub.2016.01.048 [DOI] [PubMed] [Google Scholar]
- Klassen, M.P., Wu Y.E., Maeder C.I., Nakae I., Cueva J.G., Lehrman E.K., Tada M., Gengyo-Ando K., Wang G.J., Goodman M., et al. 2010. An Arf-like small G protein, ARL-8, promotes the axonal transport of presynaptic cargoes by suppressing vesicle aggregation. Neuron. 66:710–723. 10.1016/j.neuron.2010.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein, D.R., and Vale R.D.. 2004. The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans. Mol. Biol. Cell. 15:3729–3739. 10.1091/mbc.e04-04-0326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein, D.R., Tomishige M., Stuurman N., and Vale R.D.. 2002. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 109:347–358. 10.1016/S0092-8674(02)00708-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwinter, D.M., Lo K., Mafi P., and Silverman M.A.. 2009. Dynactin regulates bidirectional transport of dense-core vesicles in the axon and dendrites of cultured hippocampal neurons. Neuroscience. 162:1001–1010. 10.1016/j.neuroscience.2009.05.038 [DOI] [PubMed] [Google Scholar]
- Lee, J.R., Shin H., Choi J., Ko J., Kim S., Lee H.W., Kim K., Rho S.H., Lee J.H., Song H.E., et al. 2004. An intramolecular interaction between the FHA domain and a coiled coil negatively regulates the kinesin motor KIF1A. EMBO J. 23:1506–1515. 10.1038/sj.emboj.7600164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, K.Y., Kuzmin A., Unger S.M., Petersen J.D., and Silverman M.A.. 2011. KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci. Lett. 491:168–173. 10.1016/j.neulet.2011.01.018 [DOI] [PubMed] [Google Scholar]
- Millecamps, S., and Julien J.P.. 2013. Axonal transport deficits and neurodegenerative diseases. Nat. Rev. Neurosci. 14:161–176. 10.1038/nrn3380 [DOI] [PubMed] [Google Scholar]
- Miller, K.E., DeProto J., Kaufmann N., Patel B.N., Duckworth A., and Van Vactor D.. 2005. Direct observation demonstrates that Liprin-α is required for trafficking of synaptic vesicles. Curr. Biol. 15:684–689. 10.1016/j.cub.2005.02.061 [DOI] [PubMed] [Google Scholar]
- Miyoshi, J., and Takai Y.. 2004. Dual role of DENN/MADD (Rab3GEP) in neurotransmission and neuroprotection. Trends Mol. Med. 10:476–480. 10.1016/j.molmed.2004.08.002 [DOI] [PubMed] [Google Scholar]
- Niwa, S., Lipton D.M., Morikawa M., Zhao C., Hirokawa N., Lu H., and Shen K.. 2016. Autoinhibition of a neuronal kinesin UNC-104/KIF1A regulates the size and density of synapses. Cell Rep. 16:2129–2141. 10.1016/j.celrep.2016.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, S., Tanaka Y., and Hirokawa N.. 2008. KIF1Bbeta- and KIF1A-mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD. Nat. Cell Biol. 10:1269–1279. 10.1038/ncb1785 [DOI] [PubMed] [Google Scholar]
- O’Shea, E.K., Klemm J.D., Kim P.S., and Alber T.. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 254:539–544. 10.1126/science.1948029 [DOI] [PubMed] [Google Scholar]
- Okada, Y., Yamazaki H., Sekine-Aizawa Y., and Hirokawa N.. 1995. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 81:769–780. 10.1016/0092-8674(95)90538-3 [DOI] [PubMed] [Google Scholar]
- Ren, J., Wang S., Chen H., Wang W., Huo L., and Feng W.. 2018. Coiled-coil 1-mediated fastening of the neck and motor domains for kinesin-3 autoinhibition. Proc. Natl. Acad. Sci. USA. 115:E11933–E11942. 10.1073/pnas.1811209115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière, J.B., Ramalingam S., Lavastre V., Shekarabi M., Holbert S., Lafontaine J., Srour M., Merner N., Rochefort D., Hince P., et al. 2011. KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am. J. Hum. Genet. 89:219–230. 10.1016/j.ajhg.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Ferreira, C., and Munro S.. 2011. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell. 21:1171–1178. 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager, M.A., Kapitein L.C., Grigoriev I., Burzynski G.M., Wulf P.S., Keijzer N., de Graaff E., Fukuda M., Shepherd I.T., Akhmanova A., and Hoogenraad C.C.. 2010. Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J. 29:1637–1651. 10.1038/emboj.2010.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, H., Wyszynski M., Huh K.H., Valtschanoff J.G., Lee J.R., Ko J., Streuli M., Weinberg R.J., Sheng M., and Kim E.. 2003. Association of the kinesin motor KIF1A with the multimodular protein liprin-α. J. Biol. Chem. 278:11393–11401. 10.1074/jbc.M211874200 [DOI] [PubMed] [Google Scholar]
- Shishido, H., and Maruta S.. 2012. Engineering of a novel Ca2+-regulated kinesin molecular motor using a calmodulin dimer linker. Biochem. Biophys. Res. Commun. 423:386–391. 10.1016/j.bbrc.2012.05.135 [DOI] [PubMed] [Google Scholar]
- Siddiqui, N., Zwetsloot A.J., Bachmann A., Roth D., Hussain H., Brandt J., Kaverina I., and Straube A.. 2019. PTPN21 and Hook3 relieve KIF1C autoinhibition and activate intracellular transport. Nat. Commun. 10:2693. 10.1038/s41467-019-10644-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina, V., Norris S.R., Dizaji A.S., Kortus M., Veatch S., Peckham M., and Verhey K.J.. 2014. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc. Natl. Acad. Sci. USA. 111:5562–5567. 10.1073/pnas.1400759111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucchi, R., Plucińska G., Hummel J.J.A., Zahavi E.E., Guerra San Juan I., Klykov O., Scheltema R.A., Altelaar A.F.M., and Hoogenraad C.C.. 2018. Regulation of KIF1A-driven dense core vesicle transport: Ca2+/CaM controls DCV binding and Liprin-α/TANC2 recruits DCVs to postsynaptic sites. Cell Rep. 24:685–700. 10.1016/j.celrep.2018.06.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M., Miyoshi J., Ishizaki H., Togawa A., Ohnishi K., Endo K., Matsubara K., Mizoguchi A., Nagano T., Sato M., et al. 2001. Role of Rab3 GDP/GTP exchange protein in synaptic vesicle trafficking at the mouse neuromuscular junction. Mol. Biol. Cell. 12:1421–1430. 10.1091/mbc.12.5.1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beuningen, S.F.B., Will L., Harterink M., Chazeau A., van Battum E.Y., Frias C.P., Franker M.A.M., Katrukha E.A., Stucchi R., Vocking K., et al. 2015. TRIM46 controls neuronal polarity and axon specification by driving the formation of parallel microtubule arrays. Neuron. 88:1208–1226. 10.1016/j.neuron.2015.11.012 [DOI] [PubMed] [Google Scholar]
- van den Berg, R., and Hoogenraad C.C.. 2012. Molecular motors in cargo trafficking and synapse assembly. In Synaptic Plasticity: Dynamics, Development and Disease. Kreutz M.R., and Sala C., editors. Springer Vienna, Vienna, Switzerland. 173–196. 10.1007/978-3-7091-0932-8_8 [DOI] [PubMed] [Google Scholar]
- van der Vaart, B., van Riel W.E., Doodhi H., Kevenaar J.T., Katrukha E.A., Gumy L., Bouchet B.P., Grigoriev I., Spangler S.A., Yu K.L., et al. 2013. CFEOM1-associated kinesin KIF21A is a cortical microtubule growth inhibitor. Dev. Cell. 27:145–160. 10.1016/j.devcel.2013.09.010 [DOI] [PubMed] [Google Scholar]
- van Vlijmen, T., Vleugel M., Evers M., Mohammed S., Wulf P.S., Heck A.J.R., Hoogenraad C.C., and van der Sluijs P.. 2008. A unique residue in rab3c determines the interaction with novel binding protein Zwint-1. FEBS Lett. 582:2838–2842. 10.1016/j.febslet.2008.07.012 [DOI] [PubMed] [Google Scholar]
- Verhey, K.J., and Hammond J.W.. 2009. Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777. 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Wang, X., and Schwarz T.L.. 2009. The mechanism of Ca2+ -dependent regulation of kinesin-mediated mitochondrial motility. Cell. 136:163–174. 10.1016/j.cell.2008.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y.E., Huo L., Maeder C.I., Feng W., and Shen K.. 2013. The balance between capture and dissociation of presynaptic proteins controls the spatial distribution of synapses. Neuron. 78:994–1011. 10.1016/j.neuron.2013.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, X., Jaulin F., Espenel C., and Kreitzer G.. 2010. PH-domain-dependent selective transport of p75 by kinesin-3 family motors in non-polarized MDCK cells. J. Cell Sci. 123:1732–1741. 10.1242/jcs.056366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn, T.R., Angleson J.K., MacMorris M.A., Domke E., Hutton J.F., Schwartz C., and Hutton J.C.. 2004. Dense core vesicle dynamics in Caenorhabditis elegans neurons and the role of kinesin UNC-104. Traffic. 5:544–559. 10.1111/j.1600-0854.2004.00195.x [DOI] [PubMed] [Google Scholar]
- Zhang, Y.V., Hannan S.B., Stapper Z.A., Kern J.V., Jahn T.R., and Rasse T.M.. 2016. The Drosophila KIF1A homolog unc-104 is important for site-specific synapse maturation. Front. Cell. Neurosci. 10:207. 10.3389/fncel.2016.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.