Trillet et al. report that the glycoprotein GP130 orchestrates the availability of the G protein–coupled receptor for apelin APLNR at the plasma membrane of glioblastoma stem-like cells, thereby modulating APLN signaling toward self-renewal. By modulating ELMOD1 expression, GP130 ultimately tunes ARF-mediated endovesicular internalization and recycling.
Abstract
Glioblastoma is one of the most lethal forms of adult cancer, with a median survival of ∼15 mo. Targeting glioblastoma stem-like cells (GSCs) at the origin of tumor formation and relapse may prove beneficial. In situ, GSCs are nested within the vascular bed in tight interaction with brain endothelial cells, which positively control their expansion. Because GSCs are notably addicted to apelin (APLN), sourced from the surrounding endothelial stroma, the APLN/APLNR nexus has emerged as a druggable network. However, how this signaling axis operates in gliomagenesis remains underestimated. Here, we find that the glycoprotein GP130 interacts with APLNR at the plasma membrane of GSCs and arbitrates its availability at the surface via ELMOD1, which may further impact on ARF-mediated endovesicular trafficking. From a functional standpoint, interfering with GP130 thwarts APLNR-mediated self-renewal of GSCs ex vivo. Thus, GP130 emerges as an unexpected cicerone to the G protein–coupled APLN receptor, opening new therapeutic perspectives toward the targeting of cancer stem cells.
Introduction
Glioblastoma multiforme (GBM), an aggressive adult primary brain tumor, remains one of the most pernicious malignancies. In defiance of intrusive treatments, including neurosurgery and combined radiation and chemotherapy with temozolomide, median survival has leveled off at ∼15 mo since the introduction of the so-called Stupp protocol more than a decade ago (Stupp et al., 2005). However, most patients inexorably relapse and shortly succumb to their disease. The intratumor and intertumor heterogeneity of GBM surfaced upon systematic profiling of patient samples (Verhaak et al., 2010; Lathia et al., 2015; Jin et al., 2017; Gimple et al., 2019). Within these multifarious masses exists a population of rare cells with stem-like properties termed glioblastoma stem-like cells (GSCs; Lathia et al., 2015; Gimple et al., 2019). Rather than possessing a molecular identity based on gene signature (i.e., expression and mutations), GSCs are phenotypically defined by their ability to self-renew and expand in vitro as nonadherent spheroid culture in mitogen-defined, serum-free medium. GSCs can also differentiate, escape from chemotherapy- and radiation-provoked damages, and initiate and propagate tumors upon serial implantations in immunocompromised mice (Singh et al., 2004; Lathia et al., 2015; Gimple et al., 2019). The expression of classical stem markers, such as NESTIN, sex-determining Y-box2, octamer binding transcription factor 4, and nanog homeobox, is nonetheless frequently associated with the GSC population. Because GSCs are believed to be at the origin of gliomagenesis, tumor expansion, and treatment failure, they represent an interesting opportunity for targeted therapies.
Comparable to neural stem cells, GSCs reside, at least in part, in a confined and protective microenvironment, which provides essential cues to maintain their self-renewal and dictates their fate (Shen et al., 2004; Calabrese et al., 2007; Gilbertson and Rich, 2007; Prager et al., 2019; Day et al., 2019). The neural stem cell niche notably integrates the blood vasculature, as neo-neurons arise along vessels in adulthood, and employs intermingled vacular endothelial growth factor (VEGF) signaling (Bao et al., 2006; Oka et al., 2007; Batchelor et al., 2007; Treps et al., 2016). Likewise, GSCs interact with endothelial cells and pericytes from the tumor vasculature (Calabrese et al., 2007; Cheng et al., 2013; Harford-Wright et al., 2017). Seminal work highlighted that NESTIN-positive brain tumor cells physically interface with endothelial cells (Calabrese et al., 2007). In the same vein, the endothelial secretome recapitulated in vitro the supportive action of endothelial cells on GSC subsistence (Galan-Moya et al., 2011; Harford-Wright et al., 2017; Jacobs et al., 2017). In this scenario, the localization of GSCs, in close proximity to endothelial cells, facilitates communication between these cells, delivering a paracrine supply of angiocrine factors essential to nurture GSCs. Few factors, such as basic fibroblast growth factor, have been definitively implicated in the endothelial-ignited communication toward GSCs (Takahashi et al., 1991; Batchelor et al., 2007; Loilome et al., 2009). Our recent mass spectrometry analysis of brain endothelial cell conditioned media identified the vasoactive peptide apelin (APLN) as critical for GSC maintenance and growth (Harford-Wright et al., 2017). Indeed, APLN could maintain GSC identity in the absence of all other mitogens, while the pharmacological blockade of the apelin receptor (APLNR; also known as APJ) drastically reduced tumor growth in experimental animal models, and sensitized GSCs to temozolomide treatment in vitro (Harford-Wright et al., 2017). This endogenous peptide, which may emanate from endothelial cells throughout diverse organs and tissues, engages the G protein–coupled receptor APLNR, and conducts a large repertoire of physiological, protective and regenerative actions, as exemplified in heart and skeletal muscles (Yang et al., 2015). In the context of GBM, APLN functions may deviate in order to guard the pool of GSCs. The potent role of APLN in tumorigenesis has been further elucidated in diverse experimental tumor models (Uribesalgo et al., 2019). Moreover, endothelial cell–produced APLN contributes to hematopoietic stem cell repopulation, which in turn supports vessel integrity, further emphasizing the intertwined connection between stem cells and vessels (Chen et al., 2019). A pillar of this reciprocal communication may thus be APLN.
Based on the essential role we recently ascribed to endothelial APLN in glioma progression via its cognate receptor, APLNR, we explored in depth the molecular mechanisms involved in APLN-based signaling in GBM. A recent study demonstrated that APLNR is linked to the Janus kinase (JAK) pathway in cancer cells and therefore suggests that APLN might prime atypical signaling, beyond G-protein coupling (Patel et al., 2017). Here, we report that the glycoprotein GP130, the transducing receptor for JAK-associated cytokine receptors, interacts with APLNR and orchestrates its availability at the plasma membrane for further ligand-dependent activation in GSCs. Our results help to better understand the molecular basis for tumor perversion of the physiological APLN/APLNR signaling axis in the course of glioblastoma progression.
Results
APLNR interacts with GP130 at membrane microdomains at the surface of GSCs
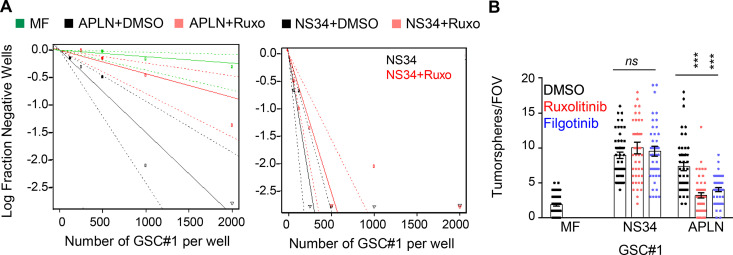
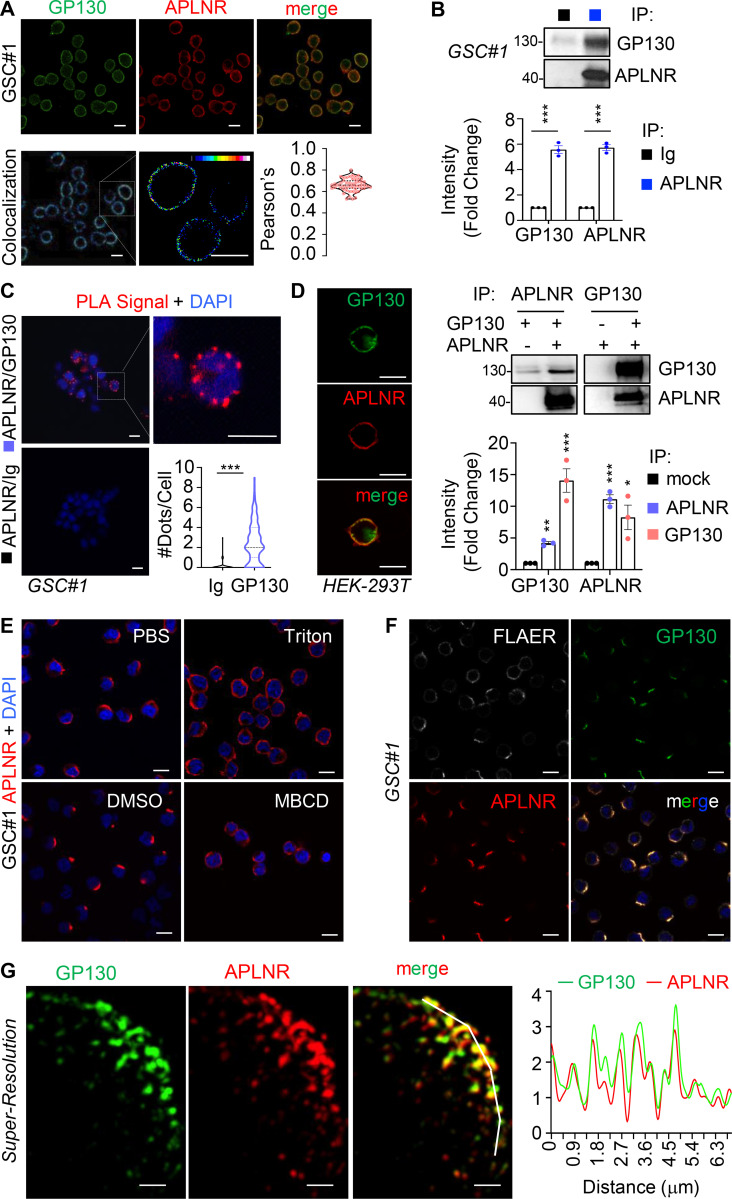
We and others recently showed that GSCs are addicted to APLN secreted by surrounding endothelial cells, thereby placing the APLN/APLNR nexus as a druggable network in glioblastoma (Harford-Wright et al., 2017; Uribesalgo et al., 2019; Mastrella et al., 2019). Of note, the nonreceptor tyrosine kinase JAK1 that bridges the transducer cytokine receptor GP130 to the activation of the STAT (signal transducer and activator of transcription) transcription factors was found bound to a mutated version of APLNR in tumor cells (Patel et al., 2017). Although none of these recently identified APLNR mutations were found in the genomic DNA of three different glioblastoma patient–derived cells (see Materials and methods), the implication of GP130 in maintaining GSCs (Jacobs et al., 2017; Shi et al., 2017) prompted us to examine the connection between APLNR and GP130. First, we found that blocking JAK activity with ruxolitinib largely impaired self-renewal and sphere formation of GSCs in APLN-only mitogen-free (MF) medium (Fig. S1, A and B). This was however not the case in mitogen-supplemented medium (NS34; Fig. S1, A and B), suggesting a specific involvement of JAK in APLN-ignited signaling rather than a selective yet toxic effect of this drug. Similar results were obtained with filgotinib, an additional JAK inhibitor (Fig. S1 B). In line with this functional connection between the JAK pathway and the APLN cascade, confocal analysis revealed that APLNR and GP130 coalesce at the plasma membrane of GSCs (Fig. 1 A). Moreover, coimmunoprecipitation experiments demonstrated that GP130 binds to APLNR (Fig. 1 B). This was further confirmed with a proximity ligation assay, which highlighted that the interaction between GP130 and APLNR occurs within a maximum 30–40 nm distance (Fig. 1 C). Of note, this GP130/APLNR association was recapitulated in ectopic expression system, using transfected HEK293T cells (Fig. 1 D).
Figure S1.
JAK inhibitors impaired APLN-based GSC maintenance. (A) Patient-derived GSCs (mesenchymal GSC#1) were cultured in MF conditions (green) or MF containing APLN (1 µM) plus either vehicle (APLN+DMSO, black) or ruxolitinib (APLN+Ruxo, 1 µM, pink). Linear regression plot of in vitro LDA is shown. Alternatively, LDA assays were done in mitogen-containing medium plus vehicle (NS34+DMSO, black) and NS34 plus ruxolitinib (NS34+Ruxo, pink). Data are representative of two independent experiments. (B) Tumorspheres per FOV were manually, single-blindly counted in similarly treated cells cultured in MF medium, mitogen-containing medium (NS34), or MF containing APLN (1 µM, APLN). DMSO (black) and the JAK inhibitors ruxolitinib (1 µM, red) and filgotinib (1 µM, blue) were also added as indicated. Data are presented as the mean ± SEM from three independent experiments. Each dot (n > 25) represents one sample count. All data are representative of at least three independent experiments. ***, P < 0.001 using ANOVA tests.
Figure 1.
APLNR interacts with GP130 at membrane microdomains at the surface of GSCs. (A) Patient-derived GSCs (mesenchymal GSC#1) were fixed, permeabilized and analyzed by confocal microscopy for GP130 (green) and APLNR (red). Merge images are shown, colocalization were visualized using Fiji software. Scale bars, 10 µm. Pearson’s correlation factor was measured using the Fiji Coloc2 analysis tool in >50 cells from at least three independent experiments. Data are presented as violin plot, and dashed line delineates the mean. (B) Protein lysates from patient-derived GSC#1 were processed from immunoprecipitation using control isotype Ig (black) or anti-APLNR antibody (blue). Samples were resolved on Tris-glycine SDS gels for Western blot using the indicated antibodies. Densitometry analysis was performed in three independent experiments and expressed as the mean fold change ± SEM. (C) Proximity ligation assay (PLA) was performed in GSC#1 using either APLNR alone (Ig/APLNR) or APLNR/GP130 antibodies. PLA signal is indicated in red. Nuclei are shown in blue (DAPI). Scale bars, 10 µm. The number of dots per cell was counted in single-blinded images from three independent experiments (n = 63 in Ig, n = 145 in GP130). Dashed lines delineate the mean. (D) HEK293T cells were transfected with GP130 and/or APLNR and then fixed, permeabilized, and analyzed by confocal microscopy for GP130 (green) and APLNR (red). Merge images are shown. Images are representative of three independent experiments. Scale bars, 10 µm. Similarly, protein lysates from transfected cells were processed for immunoprecipitation with APLNR (blue) or GP130 (pink) antibodies. IP fractions were analyzed by Western blot, as indicated. Densitometry analysis was performed in three independent experiments and expressed as the mean fold change ± SEM. (E) GSC#1 were fixed (PBS) and further permeabilized (Triton) before analysis by confocal microscopy for APLNR (red) and nuclei (DAPI, blue). Alternatively, cells were pretreated with vehicle (DMSO 1%, 30 min) and MBCD (10 mM, 30 min). Scale bars, 10 µm. (F) GSC#1 cells were fixed and analyzed by confocal microscopy for GPI-enriched domains (FLAER, gray), GP130 (green), and APLNR (red). Merge images are shown. Scale bars, 10 µm. (G) Structured illumination microscopy was deployed to image nanoclusters of APLNR and GP130 in superresolution. Scale bars, 1 µm. Graph shows the quantification of staining intensity alongside the white broken line in the merge panel. All data are representative of at least three independent experiments. ***, P < 0.001; **, P < 0.01; *, P < 0.05 using ANOVA tests. IP, immunoprecipitation.
To gain insights into the distribution of the two receptors at the plasma membrane, confocal analysis was next performed upon manipulation of the plasma membrane topology. Strikingly, we observed that APLNR and GP130 staining were restricted to a crescent-shape portion of the plasma membrane in the absence of nonionic detergents (i.e., triton, saponin, and digitonin; Fig. 1, E and F; and Fig. S2). Likewise, cholesterol depletion using methyl-β-cyclodextrin (MBCD) resulted in a more homogeneous distribution of APLNR around the plasma membrane (Fig. 1 E). These membrane microdomains segregated in zones also enriched with glycophosphatidylinositol (GPI) anchors, as detected with fluorescein-labeled proaerolysin (FLAER), further reinforcing the idea of membrane clustering (Fig. 1 F). Finally, structured-illuminated microscopy detailed the intrabiomembrane organization of APLNR/GP130 nanoclusters (Fig. 1 G). The superresolution images showed that APLNR and GP130 indeed coalesce at a confined region at the surface of GSCs.
Figure S2.
Effects of nonionic detergents on APLNR localization. GSC#1 was fixed (PBS) and further permeabilized (Triton, saponin, and digitonin) before analysis by confocal microscopy for APLNR (red) and nuclei (DAPI, blue). Scale bars, 10 µm. All data are representative of at least three independent experiments.
GP130 is required for APLN-mediated GSC expansion
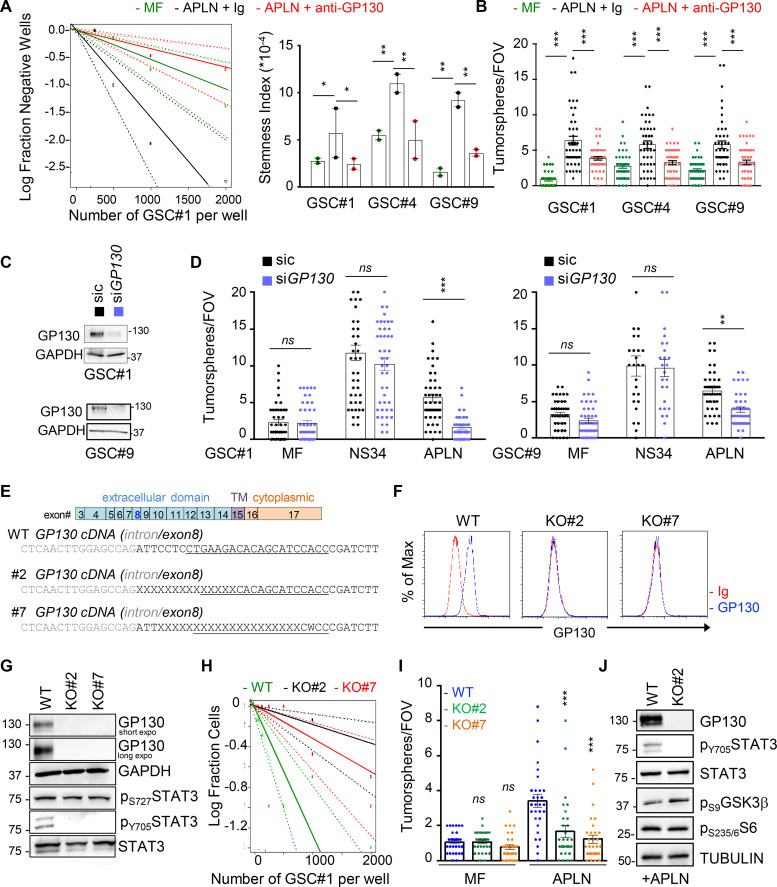
We next assessed the functional impact of the GP130/APLNR association on the stem properties of GSCs. As expected (Harford-Wright et al., 2017), APLN alone promotes self-renewal of patient-derived GSCs, as monitored via limiting dilution assays (LDAs) and secondary spheroid assays (tumorspheres; Fig. 2, A and B). However, this was reduced upon neutralization of GP130 signaling and expression, with blocking antibodies and RNA interference, respectively (Fig. 2, B–D). To delve into the role of GP130 in APLN supporting action on patient-derived GSCs, GP130 was knocked out via CRISPR/Cas9-based gene editing (Fig. 2 E). Single clones were selected by fluorescence-activated cell sorting and genomic DNA was sequenced to identify bi-allelic mutants in the GP130 gene (Fig. 2 E). GP130 knockout (KO) was confirmed at the protein level on two independent clones (Fig. 2, F and G). In agreement with the obliteration of GP130-mediated signaling, the two KO clones exhibited a sharp reduction in STAT3 Y705 phosphorylation (Fig. 2 G; Guanizo et al., 2018), which reflects canonical JAK-mediated activation of this transcription factor downstream of GP130, in growing GSCs. It is noteworthy that STAT3 S727 phosphorylation was not affected, as this posttranslational modification might be caused by an alternative signaling cascade, such as mTOR (mechanistic target of rapamycin) and MAPK (Guanizo et al., 2018; Fig. 2 G). To next determine the importance of GP130 expression in APLN signaling, self-renewal capabilities were estimated with LDAs and spheroid assays (Fig. 2, H and I). We found that depletion of GP130 expression strongly impaired APLN-dependent tumorsphere formation. Previous findings had highlighted the contribution of the glycogen synthase kinase 3 β (GSK3β) activity in gliomagenesis and more precisely that GSK3β inactivation by S9 phosphorylation was associated with APLN inhibition (Zhou et al., 2016; Harford-Wright et al., 2017). In line with this, the level of inactive S9 phosphorylation was increased in GP130 KO cells, as compared with parental cells grown in APLN-only MF medium (Fig. 2 J). S6 phosphorylation engaged upon mTOR activation was, however, intact (Figs. 2 J and S3 A). Thus, GP130 expression licenses APLN to operate in GSCs.
Figure 2.
GP130 is required for APLN-mediated GSC expansion. (A) Patient-derived GSCs (mesenchymal GSC#1, mesenchymal GSC#4, and classical GSC#9) were cultured in MF conditions (green), MF medium containing APLN (1 µM) plus control isotype Ig (2 µg/µl, black), and MF containing APLN (1 µM) plus anti-GP130 antibody (2 µg/µl, red). Linear regression plot of in vitro LDA is shown for GSC#1. Data are representative of two independent experiments. Stemness frequency was calculated from LDA in GSC#1, GSC#4, and GSC#9. Data are presented as the mean ± SEM on two independent experiments. (B) Tumorspheres per FOV were manually, single-blindly counted in GSC#1, GSC#4, and GSC#9 treated as in A. Data are presented as the mean ± SEM of three independent experiments. Each dot (n > 25) represents one sample count. (C) Silencing efficiency is shown by Western blot for GSC#1 and GSC#4 transfected with nonsilencing (sic, black) or GP130 targeting siRNA duplexes (siGP130, blue). (D) Tumorspheres per FOV were manually, single-blindly counted in GSC#1 and GSC#9, as treated in C, in MF medium, in mitogen-containing medium (NS34) and MF containing APLN (1 µM, APLN). Data are presented as the mean ± SEM of three independent experiments. Each dot (n > 25) represents one sample count. (E) Schematic representation of the GP130 (IL6ST) gene, with positioning of the CRISPR sequence guide at the junction between intro and exon8 (underlined sequence), that encodes for the N-terminal part of the extracellular domain. Genomic sequences in WT and two bi-allelic clones (KO#2 and #7) are shown. TM, transmembrane domain. (F) Flow cytometry analysis in WT and GP130 KO (#2 and #7) GSC#1. Histogram plots are represented with isotype Ig (red) and GP130 staining (blue). (G) Western blot analysis of total protein lysates from WT and GP130 KO (#2 and #7) GSC#1 using the indicated antibodies. Both GAPDH and STAT3 serve as loading controls. (H) LDA were performed in WT (green) and GP130 KO (#2, black, and #7, red) GSC#1 in APLN-only containing medium. Plot is representative of two independent experiments. (I) Tumorspheres per FOV were manually, single-blindly counted in WT and GP130 KO (#2 and #7) GSC#1, cultured in MF and APLN-only medium. Data are presented as the mean ± SEM of three independent experiments. Each dot (n > 25) represents one sample count. (J) Western blot analysis of total protein lysates from WT and GP130 KO#2 cells in APLN-only containing medium (2 d) using the indicated antibodies. Both TUBULIN and STAT3 serve as loading controls. All data are representative of at least three independent experiments, unless otherwise stated. ***, P < 0.001; **, P < 0.01; *, P < 0.05 using ANOVA tests.
Figure S3.
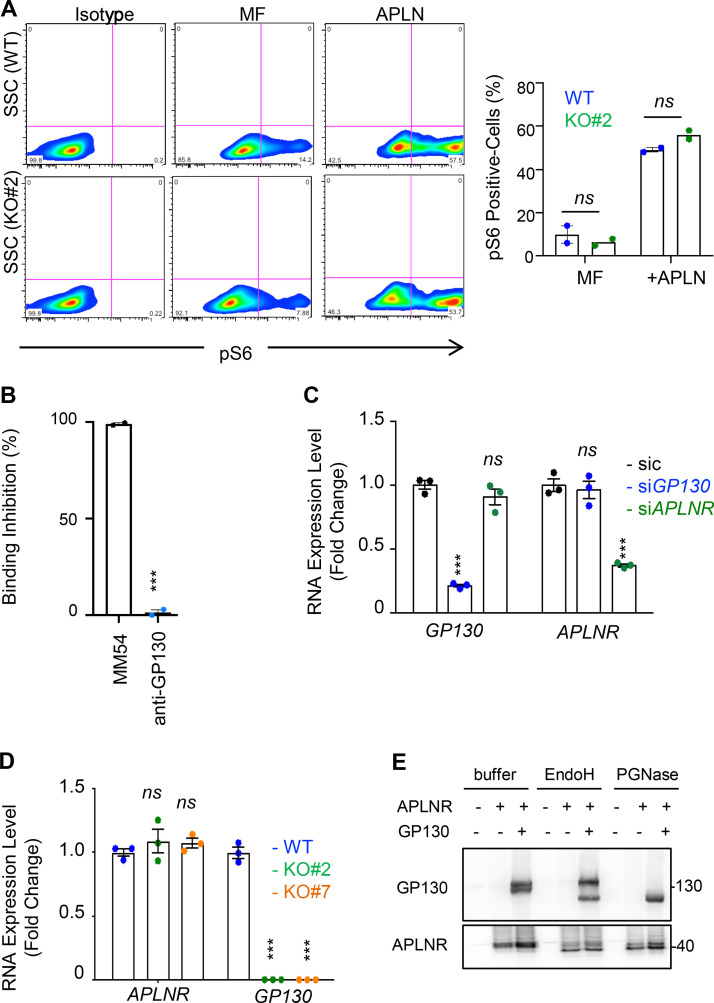
GP130 does not affect all APLN/APLNR signaling activities. (A) S6 phosphorylation was monitored by flow cytometry in WT (blue) and GP130 KO#2 (green) patient-derived GSCs (mesenchymal GSC#1) cultured in MF conditions and MF containing APLN only (1 µM). Isotype staining is also shown as a control for gating. Histogram represents the mean ± SEM percentage of phospho S6 ribosomal protein (pS6)–positive cells in two independent experiments. SSC, side scatter. (B) A radioligand binding assay of APLN to APLNR was measured in the presence of the either competitive APLNR antagonist (MM54, black) or anti-GP130 antibodies (blue) in APLNR–stably expressing cells. Data are expressed as the mean ± SEM percentage of binding inhibition on technical duplicates. (C) Patient-derived GSCs (mesenchymal GSC#1) transfected with nonsilencing (sic, black) and either GP130 or APLNR targeting siRNA duplexes (siGP130, blue and siAPLNR, green) were analyzed by qPCR for APLNR and GP130. Data are presented as the mean ± SEM fold change of three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. (D) qPCR was performed in WT (blue) and GP130 KO (#2, green; and #7, orange) GSC#1. Data are presented as the mean ± SEM fold change of three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. (E) HEK293T cells were transfected with GP130 and/or APLNR encoding plasmids. Protein lysates were incubated with buffer, EndoH or PGNase, and further analyzed by Western blot, as indicated. All data are representative of at least three independent experiments. ***, P < 0.001 using ANOVA tests.
GP130 contributes to APLNR availability at the plasma membrane
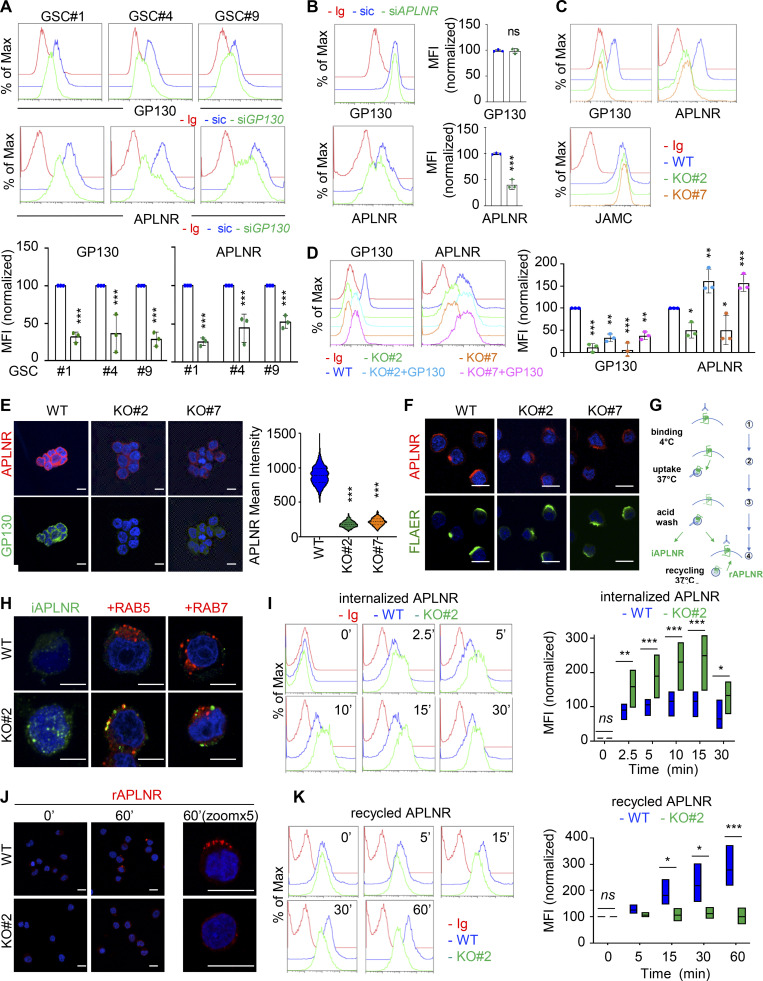
Because GP130 interacts with APLNR and is coopted in APLN signaling, we wondered whether GP130 could directly modulate APLN binding to its receptor in GSCs. However, while the competitive antagonist of APLNR, namely MM54, precluded APLN binding, anti-GP130 antibodies failed to do so (Fig. S3 B; Harford-Wright et al., 2017). We therefore inferred that presentation of APLNR at the cell surface could be the mechanisms by which its downstream signaling is modulated. Supporting this hypothesis, silencing of GP130 expression decreased the membrane expression of APLNR, as monitored by flow cytometry, in three different patient-derived GSCs (Fig. 3 A). Conversely, interfering with APLNR expression did not mitigate the level of GP130 detected at the plasma membrane, therefore suggesting an unidirectional relationship within the APLNR–GP130 complex (Fig. 3 B). Furthermore, GSC#1, with CRISPR/Cas9-mediated KO of GP130 gene expression, phenocopied transient knockdown cells in terms of diminished APLNR membrane expression (Fig. 3 C). Of note, GP130 deletion did not impinge on the expression of JAMC (junctional adhesion molecule C), an alternate membrane receptor expressed in GSCs, ruling out a broad action of GP130 expression on protein presentation at the plasma membrane (Fig. 3 C). This effect was most likely posttranslational, as neither APLNR silencing nor GP130 knockdown and KO altered the global level of expression of the partner receptor (Fig. S3, C and D). One nonexclusive alternative mechanism might be that GP130 cotraffics with APLNR from the ER along the secretory pathway. To challenge this hypothesis, APLNR was ectopically expressed in HEK293T together or not with GP130 and further treated with Endoglycosidase H (endo H) and peptide/glycosidase (PGNase), which removes asparagine-linked high mannose glycans from immature and mature glycoproteins, respectively. The proportion of EndoH-sensitive and EndoH-resistant APLNR was not overly modified with GP130 expression (Fig. S3 E). Importantly, the reintroduction of GP130, with GP130 episomal expression in KO cells, was sufficient to restore APLNR membrane availability (Fig. 3 D). Echoing flow cytometry analysis, the measurement of mean intensity of APLNR-conjugated fluorescent staining by high-content microscopy revealed a drastic reduction in APLNR signal when GP130 expression was abolished (Fig. 3 E). This most likely results from the dispersion and/or processing of the APLNR immunosignal into intracellular compartments. Of note, FLAER staining of GPI-enriched membrane microdomains was not modified, and remaining surface-exposed APLNR localized within this zone (Fig. 3 F). We next challenged APLNR trafficking from and to the plasma membrane (i.e., internalization and recycling, respectively) in WT and KO cells (Fig. 3 G). Tracking the internalized fraction of APLNR demonstrated an enlarged proportion of APLNR-positive intracellular puncta within RAB5-stained (i.e., early endosome), RAB7-negative (i.e., late endosome) vesicles in GP130 KO GSCs (Fig. 3 H). Flow measurements identified an increase in anti-APLNR antibody uptake in GP130 KO clones, as compared with WT (Fig. 3 I). Conversely, APLNR recycled back at the plasma membrane solely in WT cells 1 h following antibody uptake (Fig. 3 J). The receptor failed to do so in GP130 KO cells (Fig. 3 J). This was further quantified with flow cytometry analysis over time (Fig. 3 K). Altogether, our data support the notion that GP130 expression dictates the level, but not the clustering, of APLNR surface presentation.
Figure 3.
GP130 contributes to APLNR availability at the plasma membrane. (A) Flow cytometry analysis of APLNR and GP130 in patient-derived GSCs (mesenchymal GSC#1, mesenchymal GSC#4, and classical GSC#9) transfected with nonsilencing (sic, blue) and GP130 targeting siRNA duplexes (siGP130, green). Ig control staining plots are shown (red). Histograms present the mean fluorescence intensity (MFI) normalized to respective sic conditions for GP130 and APLNR staining as indicated. Data are presented as the mean ± SEM of three independent experiments. (B) Flow cytometry analysis of APLNR and GP130 in GSC#1 transfected with nonsilencing (sic, blue) and APLNR targeting siRNA duplexes (siAPLNR, green). Ig control staining plots are shown (Ig, red). Data are representative of three independent experiments. (C) Flow cytometry analysis of GP130, APLNR and JAMC in WT (blue) and GP130 KO (#2, green, and #7, orange) GSC#1. Ig control staining plots are shown (Ig, red). Data are representative of three independent experiments. (D) Flow cytometry analysis of GP130 and APLNR in GSC#1 WT (parental, blue), GP130 KO (#2, green, and #7, orange), and GP130 KO reconstituted with GP130 cDNA (#2+GP130, light blue, and #7+GP#130, pink). Ig control staining plots are shown (Ig, red). Data are representative of three independent experiments. Histograms present the MFI normalized to respective sic conditions for GP130 and APLNR staining as indicated. Data are presented as the mean ± SEM on three independent experiments. (E) Confocal analysis of GP130 (green), APLNR (red) and nuclei (DAPI, blue) in WT and GP130 KO (#2 and #7) permeabilized GSC#1. Scale bars, 10 µm. Fluorescence mean intensity for APLNR signal (arbitrary unit) was quantified by high-content microscopy in WT and GP130 KO (#2 and #7) GSC#1 and represented as violin diagram. Lines delineate the mean. Data are representative of three independent experiments, with n > 950 cells. (F) WT and GP130 KO (#2 and #7) GSC#1 were fixed and analyzed by confocal microscopy for GPI-enriched domains (FLAER, green) and APLNR (red). Nuclei are shown in blue (DAPI). Scale bars, 10 µm. Data are representative of at least three independent experiments. (G) Schematic diagram for the anti-APLNR uptake to analyze internalization (iAPLNR) and further recycling (rAPLNR). (H) Anti-APLNR antibody uptake (iAPLNR, green) was assessed in WT and GP130 KO#2 cells after 15 min at 37°C. Following acid wash and fixation, cells were stained for RAB5 (red) or RAB7 (red) and analyzed by confocal microscopy. Nuclei are shown in blue (DAPI). Merge images are shown. Scale bars, 5 µm. Data are representative of three independent experiments. (I) Similarly, anti-APLNR antibody uptake was assessed by flow cytometry after incubation at 37°C at the indicated times (0, 2.5, 5, 10, 15, and 30 min) in WT (blue) and GP130 KO#2 (green) GSC#1. Ig control staining plots are shown (red). Data are representative of three independent experiments. Boxplots depict the APLNR uptake in WT (blue) and GP130 KO#2 (green) GSC#1, calculated from normalized MFI at the indicated time points (minutes) following 37°C incubation. Line delineates the mean, and boxes show upper and lower quartiles. (J) Anti-APLNR antibody recycling (rAPLNR, red) was evaluated in WT and GP130 KO#2 cells after a 60-min chase at 37°C (following a 15-min pulse at 37°C and acid washes, as depicted in G). Cells were fixed and not permeabilized to analyze recycling APLNR (red) by confocal microscopy. Nuclei are shown in blue (DAPI). Merge images are shown. Scale bars, 10 µm. (K) Similarly, anti-APLNR antibody recycling was assessed by flow cytometry after incubation at 37°C at the indicated times (0, 5, 15, 30, and 60 min) in WT (blue) and GP130 KO#2 (green) GSC#1. Ig control staining plots are shown (red). Data are representative of three independent experiments. Boxplots depict the APLNR recycling in WT (blue) and GP130 KO#2 (green) GSC#1, calculated from normalized MFI at the indicated time points (minutes) following 37°C chasing incubation. Line delineates the mean, and boxes show upper and lower quartiles. All data are representative of at least three independent experiments. ***, P < 0.001; **, P < 0.01; *, P < 0.05 using ANOVA tests.
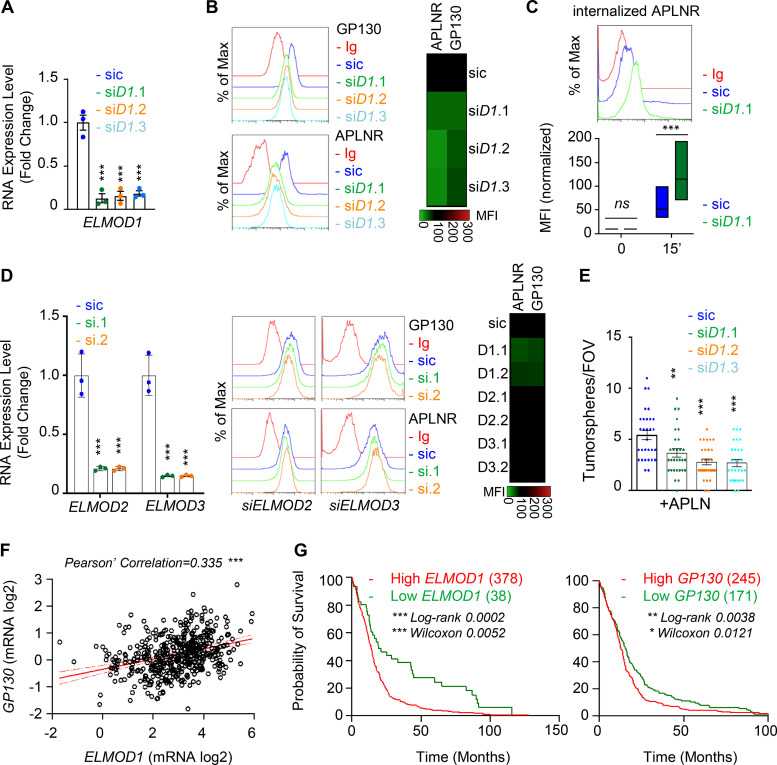
The expression of the GTPase activating protein ELMOD1 is modulated by GP130
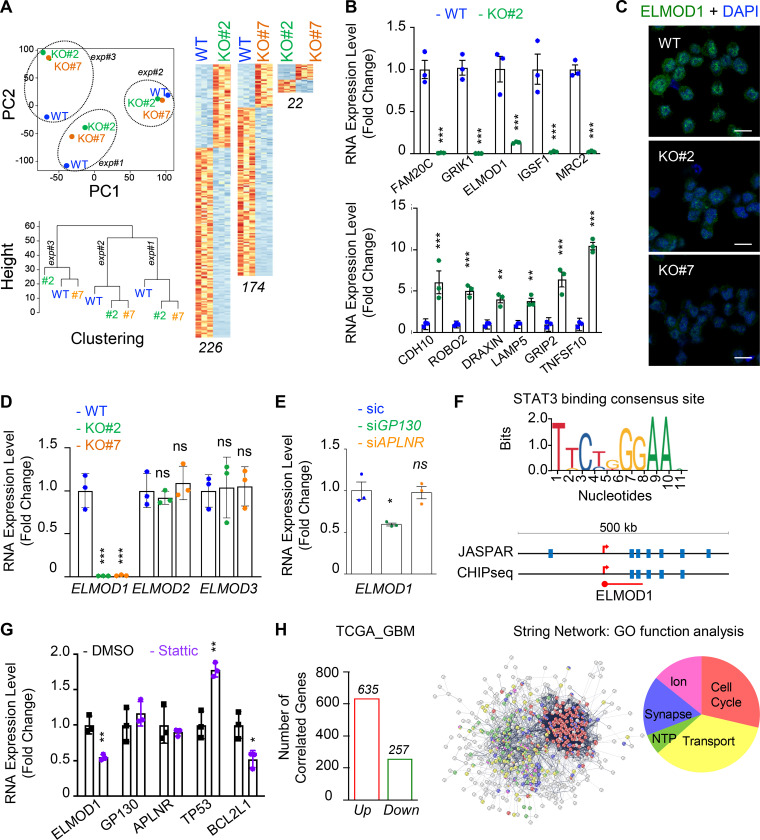
To gain further mechanistic insights into GP130-orchestrated APLN-based signaling, whole-transcriptome shotgun sequencing was performed in WT and GP130 KO GSC#1 (Fig. 4 A). Principal-component analysis and unsupervised clustering indicated a minimal culture drift between independent samples (Fig. 4 A), with a sparse set of genes whose expression was modified upon GP130 deletion, rather than massive differences between WT and GP130 KO clones. RNA sequencing (RNA-seq) data were next confirmed by qPCR on a panel of 11 differentially expressed genes (5 down and 6 up) shared between the two KO clones (Fig. 4 B). Among those was ELMOD1 (engulfment and cell motility proteins [ELMO] domain containing 1), a protein with GTPase-activating protein (GAP) for the ARF (ADP-ribosylation factor) family of small G proteins (Zhang et al., 1998; Donaldson and Jackson, 2011; Ivanova et al., 2014; Sztul et al., 2019). Consistent with the observed decrease in mRNA level, ELMOD1 protein abundance was also reduced when analyzed by immunostaining in GP130 KO cells (Fig. 4 C). Of note, this was not the case for ELMOD2- and ELMOD3-related members (Fig. 4 D). Likewise, transient silencing of GP130, but not APLNR, reiterated the reduction in ELMOD1 mRNA (Fig. 4 E). GP130 signaling culminates in STAT3 Y705 phosphorylation, a signaling event, which is abolished in GP130 KO clones (Fig. 2 G). Supporting the idea that ELMOD1 expression is controlled by a GP130/STAT3 axis, our analysis of publicly available datasets unveiled putative STAT3 binding sites around ELMOD1 gene, similar to the ones identified in the ELMOD2 and ELMOD3 regions (Figs. 4 F and S4). In keeping with this idea, inhibiting STAT3 in GSC#1 led to a reduction in ELMOD1 transcription (Fig. 4 G), along with the expected increase and decrease in the level of TP53 and BCL2L1, respectively, used as internal controls (Louault et al., 2019). Conversely, neither GP130 nor APLNR mRNA levels were modified upon STAT3 pharmacological blockade (Fig. 4 G). The Cancer Genome Atlas (TCGA) was then interrogated for the correlated genes associated with ELMOD1 expression in primary GBM patients. This reveals 635 up-regulated and 257 down-regulated genes, positively and negatively correlated with ELMOD1 expression, respectively (Fig. 4 H). Interestingly, gene ontology function analysis partitions the ELMOD1-associated signature to “transport,” “cell cycle,” “synapse,” “ion homeostasis,” and “NTP,” which agrees with reported functions of ELMODs (Ivanova et al., 2014; Sztul et al., 2019). Collectively, these results suggest that GP130 expression governs the expression of ELMOD1 in GSCs.
Figure 4.
ELMOD1 is modulated by GP130 expression. (A) RNA was extracted from three independent batches of WT (blue) and GP130 KO (#2, green; and #7, orange) patient-derived GSCs (mesenchymal GSC#1) and analyzed by RNA-seq. Representation of principal-component (PC) analysis and nonsupervised clustering show are shown. “Up” (orange) and “down” (blue) differentially expressed genes are also represented by pairs. n (226, 174, and 22) indicates the number of differentially expressed genes for each comparison. (B) qPCR analysis of up- and down-regulated hits in WT (blue) and GP130 KO#2 (green) GSC#1. Data are presented as the mean ± SEM fold change of three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. (C) Confocal analysis of ELMOD1 (green) and nuclei (DAPI, blue) in WT and GP130 KO (#2 and #7). Data are representative of three independent experiments. Scale bars, 10 µm. (D) qPCR analysis of ELMOD1, ELMOD2, and ELMOD3 in WT (blue) and GP130 KO (#2, green; and #7, orange). Data are presented as the mean ± SEM fold change on three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. (E) qPCR analysis of ELMOD1 in GSC#1 transfected with nonsilencing (sic, green) and either GP130 (green) or APLNR (orange) targeting siRNA duplexes. Data are presented as the mean ± SEM fold change of three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. (F) STAT3 consensus binding sequence was obtained from JASPAR, while putative sites were predicted around ELMOD1 gene using JASPAR and ENCODE (transcription factor ChIP-seq database). (G) qPCR analysis of the ELMOD1, APLNR, and GP130 and known STAT3 target genes (TP53 and BCL2L1) in GSC#1 treated with DMSO (1%, overnight, black) and STAT3 inhibitor (Stattic, 8 μM, overnight, purple). Data are presented as the mean ± SEM fold change of three independent experiments, using ACTB and HPRT1 as housekeeping genes for normalization. (H) Number of coregulated genes (635 “Up,” red, and 257 “Down,” green) with ELMOD1 expression in 489 patients from the cancer genome atlas for glioblastoma (TCGA_GBM) database. Search tool for the retrieval of interacting genes/proteins (STRING) analysis of the coregulated genes is shown, together with the main gene ontology (GO) functions. All data are representative of at least three independent experiments. ***, P < 0.001; **, P < 0.01; *, P < 0.05 using ANOVA tests.
Figure S4.
Prediction of STAT3 consensus binding sites on the three loci surrounding ELMOD1, ELMOD2, and ELMOD3 genes. Putative STAT3 consensus binding sites were predicted using JASPAR and ENCODE (transcription factor ChIP-seq database), in a 500-kb region on chromosome (chr) 11, 4, and 2, surrounding ELMOD1, ELMOD2, and ELMOD3 genes, respectively.
ELMOD1 is involved in the GP130-dependent trafficking of APLNR
We then wondered whether ELMOD1 drives the phenotype of GP130 KO cells. First, ELMOD1 silencing in parental GSC#1 attenuated APLNR level at the cell surface (Fig. 5, A and B). Likewise, a portion of GP130 was withdrawn from the plasma membrane in ELMOD1-knockdown GSCs, while APLNR internalization was enhanced (Fig. 5, B and C). Favoring a specific involvement of GP130 in APLNR trafficking via ELMOD1, ELMOD2 and ELMOD3 silencing did not affect APLNR surface presentation (Fig. 5 D). At the functional level, APLN-mediated secondary sphere formation was reduced when ELMOD1 mRNA was silenced (Fig. 5 E). Finally, this correlation between ELMOD1 and GP130 was discernible at the level of their expression in clinical GBM samples (Fig. 5 F). Data mining of the TCGA further unveiled that the high levels of ELMOD1 and, to a lesser extent, GP130 mRNA were associated with a worsen prognosis in primary GBM (Fig. 5 G), supporting the notion that impairing plasma membrane signaling could be of therapeutic interest.
Figure 5.
ELMOD1 is involved in the GP130-dependent trafficking of APLNR. (A) qPCR analysis of ELMOD1 in GSC#1 transfected with nonsilencing (sic, dark blue) and three independent ELMOD1 targeting siRNA duplexes (siD1.1, siD1.2, and siD1.3, in green, orange, and light blue, respectively). Data are presented as the mean ± SEM fold change of three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. (B) Flow cytometry analysis of APLNR and GP130 in sic (dark blue) and siELMOD1 (seq.1, 0.2, and 0.3, in green, orange, and light blue, respectively). Ig control staining plots are shown (red). Data are representative of at least three independent experiments. Heatmap recapitulates the mean MFI for GP130 and APLNR staining as indicated, normalized to sic, in three independent experiments. (C) Flow cytometry analysis of anti-APLNR uptake in sic (blue) and siELMOD1 (green) GSC#1 upon 15-min incubation at 37°C. Ig control staining plots are shown (red). Boxplots depict the uptake of the anti-APLNR uptake at the indicated time points (15 min) following 37°C incubation. Line delineates the mean, and boxes show upper and lower quartiles. Data are representative of four independent experiments. (D) qPCR analysis of ELMOD2 and ELMOD3 in GSC#1 transfected with nonsilencing (sic, dark blue) and ELMOD2 or ELMOD3 targeting siRNA duplexes (seq.1, and 0.2, in green and orange, respectively). Data are presented as the mean ± SEM fold change of three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. Flow cytometry analysis of APLNR and GP130 in sic (blue) and siELMOD2 or D3 targeting siRNA duplexes (seq.1 and 0.2, in green and orange, respectively). Ig control staining plots are shown (red). Heatmap recapitulates the MFI for GP130 and APLNR staining as indicated, normalized to sic, in three independent experiments. (E) Tumorspheres per FOV were manually, single-blindly counted in similarly transfected cells with sic (dark blue), ELMOD1 targeting duplexes, and siELMOD1 (seq.1, 0.2, and 0.3, in green, orange, and light blue, respectively) cultured in MF APLN-only medium. Data are presented as the mean ± SEM of three independent experiments. Each dot (n > 25) represents one sample count. (F) Scatterplot shows the linear correlation between GP130 and ELMOD1 RNA level in 489 GBM patients from TCGA_GBM database. (G) Kaplan–Meier survival curves validate the prognostic value of GP130 and ELMOD1 genes when overexpressed in 416 GBM patients. All data are representative of at least three independent experiments. ***, P < 0.001; **, P < 0.01; *, P < 0.05 using ANOVA tests, unless specified.
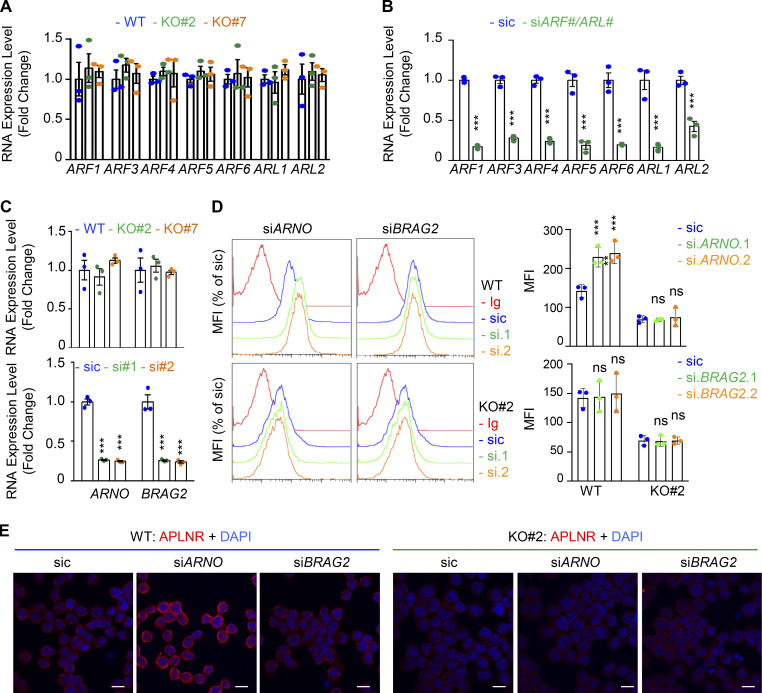
ARF GTPases contribute to APLNR internalization
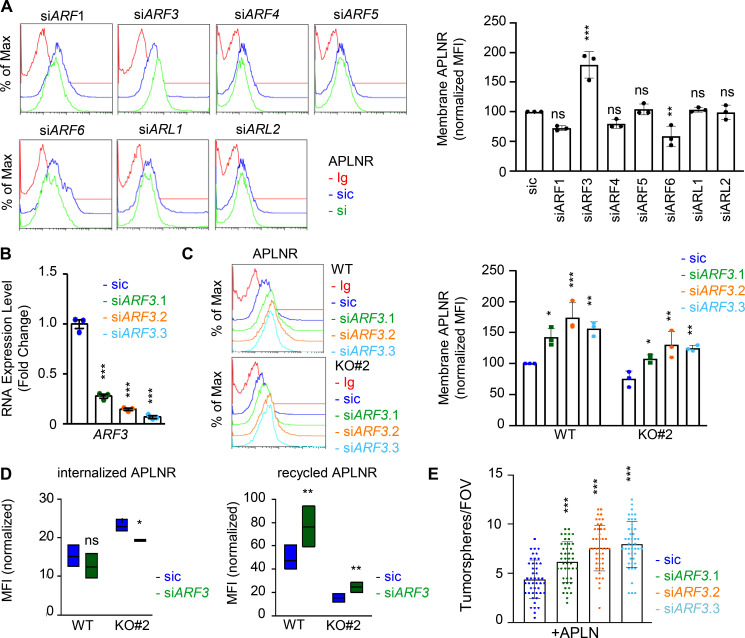
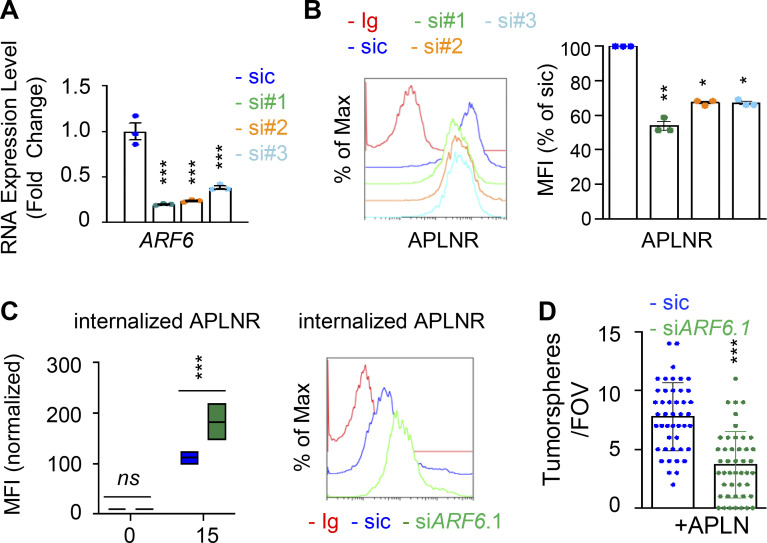
Because of the prominent role of ARFs in receptor trafficking and intracellular dynamic processes (Donaldson and Jackson, 2011; Sztul et al., 2019), we next examined whether GP130 extinction modifies the expression of ARF family members. However, the RNA expression for the seven members tested here, namely ARF1, ARF3, ARF4, ARF5, ARF6, ARL1, and ARL2, remained even in both WT and GP130 KO cells (Fig. S5, A and B). Because ARF enzyme activity could be controlled by the GAP activity of ELMOD1 (Schweitzer et al., 2011; Ivanova et al., 2014), we then challenged whether interfering with the expression of two ARF G-protein exchange factors (GEFs), namely BRAG2 (IQSEC1) and ARNO (CYTH2; Donaldson and Jackson, 2011; Sztul et al., 2019; Peurois et al., 2017), modulates APLNR membrane expression (Fig. S5, C and D). We observed that ARNO, but not BRAG2, silencing indeed increased APLNR plasma membrane staining in GSC#1, suggesting an ARF-dependent control of APLNR surface expression (Fig. S5, C and D). This augmentation was not, however, detected in GP130 KO cells (Fig. S5 D). Although we cannot exclude complex, parallel mechanisms, it is tempting to speculate that ARFs cannot be further inactivated in the GP130 KO background as a consequence of the loss of ELMOD1 expression. It is therefore conceivable that the functions of ARFs in vesicular trafficking are altered in GP130 KO cells. To test this hypothesis, seven ARF members were individually knocked down in GSC#1 (Fig. 6 A; Sztul et al., 2019). Interestingly, ARF3 silencing boosted APLNR expression at the plasma membrane and was the only tested ARF, whose silencing inversely mirrored the phenotype observed in GP130- and ELMOD1-silenced cells (Fig. 6 A). Indeed, ARF3 silencing increases APLNR membrane presentation in GSC#1 (Fig. 6, B and C), which might be explained rather via an increased recycling activity than APLNR endocytosis in WT cells (Fig. 6 D). Importantly, ARF3 knockdown was sufficient to moderately rescue the GP130 KO phenotype on APLNR membrane trafficking (Fig. 6, B and C). In these cells, where APLNR was less directed toward the plasma membrane (Fig. 3), ARF3 silencing partially opposed to the effects induced by GP130 deletion (Fig. 6 D). Internalization was slightly but significantly reduced, while recycling was increased, suggesting that ARF3 silencing may in part normalize APLNR flux in GP130 KO cells. However, this partial effect suggested the coexistence of combined and paralleled mechanisms. For instance, it is noteworthy that ARF6 silencing negatively affects the level of membrane-exposed APLNR (Figs. 6 A and S6), suggesting counteracting action between ARF3 and ARF6 in regulating APLNR membrane. From a functional standpoint, ARF3 knockdown enhanced APLN-mediated secondary sphere formation, again inversely mirroring ELMOD1 and GP130 silencing, as well as ARF6 silencing phenotype (Figs. 6 E and S6). This complex phenotype might result from a combined, stepwise involvement of multiple ARFs cooperating in parallel and in a timely manner (Figs. 6 A and S6).
Figure S5.
Modulation of APLNR membrane expression by ARF-related GEFs. (A) qPCR analysis of ARFs1-6 and ARLs1-2 in WT (blue) and GP130 KO (#2, green; and #7, orange) GSC#1. Data are presented as the mean ± SEM fold change of three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. (B) qPCR analysis of the indicated targets in GSC#1 transfected with nonsilencing (sic) and indicated siRNA duplexes. Data are presented as the mean ± SEM fold change of three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. (C) qPCR analysis of ARNO (CYTH2) and BRAG2 (IQSEC1) GSC#1 transfected with nonsilencing (sic) and respective siRNA duplexes. Data are presented as the mean ± SEM fold change of three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. (D) Flow cytometry analysis of APLNR in WT (blue) and GP130 KO#2 cells transfected with sic (blue) and ARNO and BRAG2 siRNA duplexes (green and orange, respectively). Ig control staining plots are shown (red). Histograms present the MFI normalized to respective sic conditions for APLNR staining as indicated. Data are presented as the mean ± SEM of three independent experiments. (E) Similarly treated cells were fixed (PBS) and further permeabilized (Triton) before analysis by confocal microscopy for APLNR (red) and nuclei (DAPI, blue). Scale bars, 10 µm. All data are representative of at least three independent experiments. ns, P > 0.05; ***, P < 0.001 using ANOVA tests.
Figure 6.
ARF GTPases controls APLNR availability at the plasma membrane. (A) Flow cytometry analysis of APLNR in sic (blue) and siRNA duplexes (green) targeting the indicated ARFs. Ig control staining plots are shown (red). Histograms present the MFI normalized to respective sic conditions for APLNR staining in the indicated ARF/ARL-silenced cells. Data are presented as the mean ± SEM of three independent experiments. (B) qPCR analysis of ARF3 in GSC#1 transfected with nonsilencing (sic, dark blue) and ARF3 targeting siRNA duplexes (seq.1, 0.2, and 0.3, in green, orange, and light blue, respectively). Data are presented as the mean ± SEM fold change of three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. (C) Flow cytometry analysis of APLNR in sic (blue) and siARF3 (seq.1, 0.2, and 0.3, in green, orange, and blue, respectively) in WT and GP130 KO#2. Graph recapitulates the normalized MFI for APLNR staining as indicated in three independent experiments. (D) Boxplots depict the internalization and recycling of the anti-APLNR in sic (blue) and siARF3 (green) WT and GP130 KO#2 cells at 15-min pulse time (iAPLNR), followed by a 60-min chasing at 37°C incubation (rAPLNR). Line delineates the mean, and boxes show upper and lower quartiles. Data are representative of three independent experiments. (E) Tumorspheres per FOV were manually, single-blindly counted in similarly transfected cells cultured in MF APLN-only medium. Data are presented as the mean ± SEM of three independent experiments. Each dot (n > 25) represents one sample count. All data are representative of at least three independent experiments. ***, P < 0.001; **, P < 0.01; *, P < 0.05 using ANOVA tests.
Figure S6.
ARF GTPases contribute to APLNR availability at the plasma membrane. (A) qPCR analysis of ARF6 in GSC#1 transfected with nonsilencing (sic, dark blue) and three independent ARF6 targeting siRNA duplexes (si#1, si#2, and si#3, in green, orange, and light blue, respectively). Data are presented as the mean ± SEM fold change of three independent experiments using ACTB and HPRT1 as housekeeping genes for normalization. (B) Flow cytometry analysis of APLNR in sic (blue) and siARF6 (seq#1, #2, and #3, in green, orange, and blue, respectively). Ig control staining plots are shown (red). Histograms present the MFI normalized to respective sic conditions for APLNR staining as indicated. Data are presented as the mean ± SEM of three independent experiments. (C) Boxplots depict the uptake of the anti-APLNR uptake, in sic (blue) and siARF6 (green) GSC#1, at the indicated time points (15 min) following 37°C incubation. Line delineates the mean, and boxes show upper and lower quartiles. Data are representative of four independent experiments. Flow cytometry analysis of anti-APLNR uptake in sic (blue) and siARF6 (green) GSC#1 upon 15-min incubation at 37°C. Ig control staining plots are shown (red). Data are representative of four independent experiments. (D) Tumorspheres per FOV were manually, single-blindly counted in similarly transfected cells, cultured in APLN-only medium. Data are presented as the mean ± SEM of three independent experiments. Each dot (n > 25) represents one sample count. All data are representative of at least three independent experiments. ***, P < 0.001; **, P < 0.01; *, P < 0.05 using ANOVA tests.
Thus, GP130 not only interacts with APLNR but also contributes to their copresentation at the plasma membrane by transcriptionally enabling ELMOD1 expression, which in turn involves a fine-tuned balance between endocytosis/recycling ARF-based activities (Donaldson and Jackson, 2011; Sztul et al., 2019; Schweitzer et al., 2011). This could be attributed to the modulation of receptor membrane trafficking at the plasma membrane and/or coreceptor clustering, as previously reported for VEGFR2 and EGFR (Zhu et al., 2017; Kulasekaran et al., 2021). Our data therefore suggest that this endocytic–recycling pathway may constitute an interesting process to target in cancer cells dependent on APLN supply.
Discussion
Here, we found that the glycoprotein GP130 commissions APLN-mediated action on GSC self-renewal. From a molecular standpoint, interfering with GP130 lowers APLNR availability at the plasma membrane, thereby dampening APLN signaling. In fact, GP130 dictates APLNR surface presentation by enabling the expression of ELMOD1. This might ultimately detune ARFs, which are instrumental molecular switches of vesicular trafficking (Donaldson and Jackson, 2011; Sztul et al., 2019). In line with previous reports pointing to the role of endovesicular trafficking in GBM cell vitality (Shingu et al., 2017; Jacobs et al., 2020; Kulasekaran et al., 2021), our study reports on how modulating receptor dynamics might limit nononcogene signaling involved in tumor progression.
The role of the APLN peptide has initially been investigated in the cardiovascular system, highlighting its protective actions against ischemia–reperfusion injury, elevated blood pressure, and aging (Yang et al., 2015). Further involvement of the apelinergic system in metabolic disorders, such as diabetes and obesity, has been unmasked (Chaves-Almagro et al., 2015). This hormonal peptide operates through the G protein–coupled receptor APLNR, which is widely distributed throughout the body, with a preponderant expression in smooth muscle cells and endothelial cells. The canonical Gi/G0-coupling toward cAMP inhibition conducts the main functions of APLN (Chaves-Almagro et al., 2015). Another layer of complexity is added by the fact that APLNR coalesces with other G protein–coupled receptors, including angiotensin, bradykinin, opioid, and neurotensin receptors (Chaves-Almagro et al., 2015). Recent findings had further highlighted the role of the vasoactive peptide APLN in the paracrine communication between endothelial cells and their neighboring cells in the tumor ecosystem (Harford-Wright et al., 2017; Uribesalgo et al., 2019; Chen et al., 2019; Mastrella et al., 2019). Indeed, genetic and pharmacological targeting of APLNR abrogates APLN- and endothelial-mediated expansion of tumor cells in vitro and suppresses tumor growth in vivo (Harford-Wright et al., 2017; Uribesalgo et al., 2019; Mastrella et al., 2019). Functionally, selective competitive antagonists of APLNR are safe and effective in reducing tumor growth and lengthening the survival of intracranially xenografted mice (Harford-Wright et al., 2017). Therefore, the APLN–APLNR signaling nexus may operate as a paracrine signal that sustains tumor cell expansion and progression. Our study shows that GP130 controls the magnitude of APLNR expression at the cell surface of GSCs. It does so by interfering with ligand-independent APLNR endocytosis, although alternate mechanisms such as cotrafficking from the ER secretory pathway might also contribute. Future work will delineate the contribution of GP130 in the other facets of APLNR functions besides GSCs.
APLN appears to modulate proliferation, survival and migration in varied cell types (Chaves-Almagro et al., 2015). In line with the discovery of these multiple cellular actions sparked by APLN, the downstream mechanisms have been extended to the activation of intracellular kinase pathways, such as MAPK and phosphoinositide 3-kinase/AKT (Chaves-Almagro et al., 2015). Our data suggest that APLN increases the phosphorylation of the ribosomal protein pS6, a typical target of mTOR (mechanistic target of rapamycin), constitutively active in most glioma and GSCs (Galan-Moya et al., 2011). However, GP130 deletion did not impair either the constitutive activation of the mTOR pathway or in response to APLN, as pS6 phosphorylation was left intact in both conditions. Recent works also demonstrated the importance of the GSK3β activation in GSCs, notably in the response to APLN (Zhou et al., 2016; Harford-Wright et al., 2017). Likewise, the inhibitory phosphorylation of GSK3β was elevated in GP130 KO cells upon APLN exposure. Thus, in the absence of GP130, APLN stimulation can no longer maintain GSK3β activity and its effect on GSC self-renewal. Moreover, GP130 KO interfered with the constitutive STAT3 activation in proliferative GSCs. Pharmacological blockade of JAK–STAT signaling mimics the impact of GP130 deletion on APLN-driven self-renewal of GSCs (Harford-Wright et al., 2017; Jacobs et al., 2017). However, our ongoing observations suggest that the level of STAT3 phosphorylation is not altered in APLN-stimulated GP130 KO cells, indicating that STAT3 activation is not a direct consequence of APLN stimulation. Rather, it is more likely due to an autocrine stimulation of the GP130 receptor via IL-6 (Wang et al., 2009; Shi et al., 2017). Thus, GP130-dependent activation of JAK/STAT in GSCs might impact the capacity of APLNR to conduct downstream molecular signaling via a bystander effect. Hence, GP130 deletion selectively opposes APLN downstream signaling (i.e., GSK3β inhibition), but not mTOR activation. Although the decisional mechanism involved is still unclear, it might be linked to the observed defined localization of APLNR at the cell membrane, enriched in FLAER-positive zones. Because the GPI-enriched nanodomains integrate signaling platforms and sort signaling hubs (Helms and Zurzolo, 2004), these detergent-resistant membrane domains most likely contribute to compartmentalize APLNR signaling and trafficking. The restricted, polarized localization of APLNR occurs in a ligand-independent manner, at steady-state in GSCs. However, whether this localization guides APLN signaling is still unclear. Interestingly, our data showed that APLNR was addressed, at a slow pace, to such lipid domains of the plasma membrane, even in the absence of GP130 expression. Therefore, GP130 might modulate APLNR surface presentation rather than its guided delivery toward these domains. How GP130 further controls APLNR membrane availability might be a multi-modal mechanism, such as internalization and/or recycling, cotrafficking through the ER secretory pathway, and, membrane stability. While we cannot entirely rule out any of these latter dynamic processes, our data point toward the regulation of APLNR ligand-independent internalization via the modulation of the endocytosis/recycling machinery, as illustrated by anti-APLNR antibody uptake experiments.
In keeping with this idea of GP130 modulating APLNR membrane presentation, our RNA-seq analysis in GP130 KO GSCs unveils changes in ELMOD1 expression, a putative negative regulator of ARF GTPases (Donaldson and Jackson, 2011; Ivanova et al., 2014; Sztul et al., 2019). The ELMO family consists of six paralogs in mammals, with ELMOD1 exhibiting in vitro GAP activity toward ARFs (Donaldson and Jackson, 2011; Ivanova et al., 2014; Sztul et al., 2019). Our experimental results corroborate the analysis of TCGA clinical data showing a positive correlation between GP130 and ELMOD1 gene expression. In addition, the ELMOD1 loci harbor putative STAT3-binding sites, which may in turn account for the decreased level of ELMOD1 mRNA in GP130 KO cells. Likewise, pharmacological blockade of STAT3 reduces ELMOD1 expression. ELMOD1 is highly expressed within the brain, marking stereotyped neuronal structures within the hippocampus (Johnson et al., 2012). ELMOD1 has been reported to contribute to the stabilization of apical structures in vestibular cells during development, while mutations in the human gene have been linked with mental retardation and autism disorder syndromes (Johnson et al., 2012; Krey et al., 2018; Miryounesi et al., 2019). Although ELMOD1 mutations have not been documented in cancer patients, TCGA analysis predicts that a low level of its transcripts suggests a better prognosis. It is therefore tempting to speculate here that ELMOD1 conveys permissive nononcogene signaling, at least via its action on APLNR in GSCs. Whether ELMOD1 acts on the magnitude of other membrane receptors will require further investigation. Additionally, it will be important to decipher whether ELMOD1 operates directly on ARFs in GSCs, alone or in conjunction with the GEF ARNO (CYTH2) but possibly other GEFs besides BRAG2 (IQSec1; Peurois et al., 2017; Sztul et al., 2019). In this context, our siRNA screening of seven ARF members unveils that at least two of them, namely ARF3 and ARF6, exert opposing effects on APLNR trafficking from and to the plasma membrane. Alternatively, such ARFs might operate in sequence. Yet, how exactly ELMOD1 controls APLNR surface expression requires further investigation, as ELMOD1 might target ARF GTPases either by its GAP activity and/or binding, reminiscent of SAT (suppressors of Arf ts) identified in yeast (Zhang et al., 1998). Overall, this suggests a complex modulation in endovesicular dynamics, which recently emerged as a potent checkpoint for tumor cell expansion in GBM cells (Shingu et al., 2017; Jacobs et al., 2020; Kulasekaran et al., 2021).
In conclusion, we report that GP130 interacts with APLNR and guides its surface presentation and availability for ligand binding. This supports the notion that impairing membrane signaling might be achieved in multiple ways, including blocking ligand binding and camouflaging the receptor from agonists, and therefore offers new opportunities for anticancer therapeutic options.
Materials and methods
Ethics statement
Informed consent was obtained from all patients before sample collection for diagnostic purposes. This study was reviewed and approved by the institutional review boards of Sainte Anne Hospital (Paris, France) and Laennec Hospital (Nantes, France) and performed in accordance with Helsinki protocol.
Cell culture, siRNA, and DNA transfection
HEK293T cells (ATCC) were cultured as per the ATCC’s recommendation. Three different patient-derived GSCs, namely GSC#1 (mesenchymal, 68-yr-old male), GSC#4 (mesenchymal, 76-yr-old female), and GSC#9 (classical, 68-yr-old female) were isolated and grown as spheres in NS34 medium (DMEM-F12 with N2, G5, and B27 supplements, plus GlutaMAX and antibiotics; Life Technologies), as described earlier (Harford-Wright et al., 2017). Genomic DNA from three different patient-derived GSCs was sequenced for APLNR. Sequences used for PCR amplification and sequencing of human APLNR gene were as follows: forward1, 5′-GGCAGAATCAGGGGACAGT-3′; reverse1, 5′-GTGGCCACGGCAGTTCTT-3′; forward2, 5′-CACGGTGGTGGACGAGAC-3′; reverse2, 5′-TTGCAGAGTGGGTGACAGAG-3′. None presented any mutations as reported previously (Patel et al., 2017), and transcripts were identical to the reference one (GenBank accession no. NM_005161.4): 5′-CTAGTCAACCACAAGGGTCTCCTGGCTGTAGGGGATGGATTTCTCGTGCATCTGTTCTCCACCCTTGCCCATGTTGGGGCCGGGCCCCTGGCTGTGCCCCGAAGAGTAGCTGGCTGACTTCTCCCCACTGCTGCTGTGGGAGGTGCCTGCGCACCTGCTCTGGCCACAGCAGAGCATGGAGGTGCAGGCCTGGCGGAAGCGGGGGTCGAAAAAGGCATAGAGGAAGGGGTTGAGGCAGCTGTTGACGTAGCTGATGCAGGTGCAGTAGGGGAAGATGTTCATGAGGAAGAGGTCAAAGTCACAGGGCCAGTGCAGCAGGCTGCCCAGCATGTACAGCGTCTTCACCAGGTGGTAGGGCATCCAGCACAGGGCAAAGGTCACCACCAGCACCACGATGATGCTGAGCAGCCGGCGCCGCTTCCGCAGGCCCTCGATGCGTTCCTTGCGGAAGTGGCCAGCGATGGTTTGGGCGATGAAGAAGTAACAGGTCAGCATGATGGTGAAGGGCACCACAAAGCCCACGGTGGTGGACGAGACCCCAAGGCCCACCTCCCAGGCCCACTCTGAGCTCACAGTGGCCACCATGGAGTAGTCCATGTAGCACTGCACCTTAGTGGTGTTCTCCAAGTCCCCGGTGGTGCGTAACACCATGACAGGCATGGCCAGGAGGGCGGCCAGCACCCAAAGAACTGCCGTGGCCACGGCCCCGCTGACCCGCAGCCTCAGCCGAGCATTGGCCACTGGCCTCACGATGGCCAGGTAGCGGTCGAAGCTGAGGCCGGTGAGGCAGAAGACGCTGGCGTACATGTTGACGAAGATGAGGTAGCTGCTGAGCTTGCAGAAGAAGGTCCCAAAGGGCCAGTCATAGTCCCGGTACGTGTAGGTAGCCCACAGGGGCAGCGTCACCACGAAGGTCAGGTCAGCCACCGCCAGGCTAGCAATGAAGATATCAGCTGAGCGCCTCTTCTCCCGGCTGCTCCGAAACACGGTCCAGAGCACCAGACCGTTKCCCGTGGTGCCCAGGAGGAAGACCAACATGTAGATGGCAGGGATGAGGGCCCCCGAGGATTTCCAGTCTGTGTACTCACACTCAGACTGGTTGTCTGCCCCATAGTAGTTGTCAAAATCACCACCTTCCTCCA-3′.
Stealth nonsilencing control duplexes (low-GC 12935111; Life Technologies), and siRNA duplexes (Stealth RNAi; Life Technologies) were transfected using RNAiMAX Lipofectamine (Life Technologies). The following duplexes targeting the respective human genes were used: GP130, 5′-CUCACUUGCAACAUUCUUA-3′; APLNR, 5′-CCAUCAUGCUGACCUGUUACUUCUU-3′; ELMOD1 HSS124561 (si.1), HSS124562 (si.2), and HSS124563 (si.3), ELMOD2 5′-AUAUAAAUUCUGUUGAAUG-3′ (si.1) and 5′-UCAAUUUAAUACAAAUACC-3′ (si.2), ELMOD3 HSS130688 (si.1) and HSS130687 (si.2), ARF1 HSS141267, ARF3 5′-CCAUUGGGUUCAAUGUGGAGACAGU-3′ (si.1), 5′-UGUGGAGACAGUGGAGUAU-3′ (si.2) and 5′-ACAGGAUCUGCCUAAUGCU-3′ (si.3), ARF4 5′-GAUAGCAACGAUCGUGAA-3′, ARF5 5′-UGAGCGAGCUGACUGACAA-3′, ARF6 HSS100646 (si.1), HSS180192 (si.2) and HSS180193 (si.3), ARL1 5′-AAGAAGAGCUGAGAAAAGCCA-3′, ARL2 5′-CAACCAUCCUGAAGAAGUU-3′, ARNO/CYTH2 HSS145078 (si.1) and HSS145079 (si.2), BRAG2/IQSEC1 HSS115015 (si.1) and HSS115016 (si.2).
pORF-GP130 (OriGene) and HA-tagged APLNR (Chun et al., 2008) plasmid DNA were introduced in GSCs and HEK-293T using the Neon electroporation system and Lipofectamine, respectively, according to the manufacturer’s instructions (Life Technologies).
CRISPR
Single gRNAs (sgRNAs) targeting GP130 were chosen in the sgRNA library and cloned into a lentiviral lentiCRISPRv2 (GeCKO; ZhangLab) backbone (Sanjana et al., 2014; Shalem et al., 2014). The sgRNA used was 5′-GTGGATGCTGTGTCTTCAGG-3′. For infections, lentiviral particles were produced in HEK293T cells by cotransfection of the various constructs together with pVSV-G and psPAX2 plasmids. Supernatants were collected after 2 d and applied on GSC#1 during a 1,000 g centrifugation for 90 min in the presence of 8 µg/ml polybrene (Santa Cruz Biotechnology). Cells were cultured with 1 µg/ml puromycin to select infected cells and sorted by fluorescence-activated cell sorting (anti-GP130; Abcam; and ab34324, BD FACS Aria III; BD Biosciences). Viable single clones were tested by flow cytometry to control for GP130-negative staining, and GP130 KO was confirmed by genomic DNA sequencing (forward, 5′-CAAGGCTGTGCCTCAAAAA-3′; reverse, 5′-CCCACTTGCTTCTTCACTCC-3′). Two bi-allelic KO clones were further selected.
Antibodies and reagents
The following primary antibodies were used (dilution 1:1,000 for Western blot, 1:200 for immunofluorescence and flow cytometry): GAPDH (mouse SC-25778; Santa Cruz Biotechnology), GP130 (rabbit SC-656 for Western blot, mouse SC-376280 for immunofluorescence; Santa Cruz Biotechnology; and mouse ab34324 for flow cytometry; Abcam), APLNR (mouse FAB8561R and MAB856; R&D Systems), JAMC (mouse FAB11891A; R&D Systems), STAT3 (rabbit 9132; Cell Signaling), pS727-STAT3 (rabbit 9134; Cell Signaling), pY705-STAT3 (rabbit 9145; Cell Signaling), pS235/S236-S6 (rabbit 2211 for Western blot and rabbit 5316 for flow cytometry; Cell Signaling), pS9-GSK3β (rabbit 9336; Cell Signaling), ELMOD1 (rabbit ab127541; Abcam), RAB5 (rabbit ab218624; Abcam) and RAB7 (rabbit 9367; Cell Signaling). HRP-conjugated secondary antibodies (anti-rabbit, mouse Ig, mouse IgG1, mouse IgG2a, and mouse IgG2b) were purchased from Southern Biotech. Alexa-conjugated secondary antibodies were from Life Technologies. FLAER (Alexa Fluor 488–conjugated proaerolysin) was purchased from Cedarlane, MBCD was from Sigma-Aldrich, and Stattic, ruxolitinib, and filgotinib were all from Selleckchem. [Pyr1]-Apelin-13 was resuspended and aliquoted according to the supplier’s recommendation (Phoenix).
Exploration of public datasets
The STAT3 consensus binding sequence was obtained from the JASPAR database, together with the list of matching sites across the genome. A closer look was then applied to the 500-kb region surrounding the ELMOD1, ELMOD2, and ELMOD3 genes. They were further compared with STAT3 transcription factor chromatin immunoprecipitation sequencing (ChIP-seq) database from ENCODE. TCGA was explored in Agilent databases via the Gliovis platform (http://gliovis.bioinfo.cnio.es/; Bowman et al., 2017). Optimal cutoffs were set. All subtypes were included, and histology was the only selective criteria.
Tumorsphere formation
To analyze tumorsphere formation, GSCs (100 cells/µl) were seeded in triplicate in NS34, MF media and MF media supplemented with Pyr-APLN (1 µM; Phoenix) as previously described (Harford-Wright et al., 2017). Cells were dissociated manually each day to reduce aggregation influence and maintained at 37°C 5% CO2 until day 5 (day 4 for siRNA). Tumorsphere formation was assessed in response to anti-GP130 (2 µg/ml, rabbit ab234105; Abcam) or control antibodies (swap70, 2 µg/ml, rabbit ab228846; Abcam). Tumorspheres per field of view (FOV) were calculated by a manual, single-blinded count of the total number of tumorspheres in five random FOVs for each well.
LDAs
To evaluate the self-renewal of GSCs, LDAs were performed as previously described (Tropepe et al., 1999). GSCs were plated in a 96-well plate using serial dilution ranging from 2,000 to 1 cell per well with eight replicates per dilution and treated as indicated. After 14 d, each well was binary evaluated for tumorsphere formation. Stemness frequency was then calculated using ELDA software (Hu and Smyth, 2009). The mean stemness frequency for each treatment was calculated by averaging across two independent experiments.
Flow cytometry
Flow cytometry analyses were performed on FACSCalibur (BD Biosciences and Cytocell) and processed using FlowJo software. Staining was performed on living cells (incubation, 1 h at 4°C). Intracellular flow cytometry experiments were performed using BD Cytofix/Cytoperm Kit (BD Biosciences) following the manufacturer’s protocol.
Immunostaining
Cells were fixed in PBS-paraformaldehyde 4% and processed for immunostaining with or without permeabilization with Triton X-100 (0.05%, 5 min). Immunofluorescence was performed using Alexa Fluor 488–, Alexa Fluor 568–, and Alexa Fluor 640–conjugated secondary antibodies (dilution 1:500; Life Technologies). Samples were mounted in DAPI-containing mounting medium (Prolong gold anti-fade reagent; Life Technologies). APLNR–GP130 interaction was visualized through the Duolink in situ kit following the manufacturer’s instructions (Sigma-Aldrich). All images were acquired on confocal Nikon A1 RSi using a 60× oil-immersion lens with NIS-Element Software at the Nikon Excellence Center (Micropicell, SFR François Bonamy, Nantes, France). Signal intensity quantification was performed on Imaris software (Oxford Instruments). Structured illumination microscopy images were acquired with a Nikon N-SIM microscope. Z-stacks of 0.12 µm were performed using a 100× oil-immersion lens with a 1.49 aperture and reconstructed in 3D using the NIS-Element Software. Reconstructed images validated with the SIMcheck plugin 1.2. All images were analyzed using the Image J software.
Antibody uptake and recycling experiment
To investigate endocytosis, cells were first placed at 4°C for 30 min to stop endocytosis processes and stained with anti-APLNR AF647-coupled antibody (mouse IgG1, 1 h, 4°C, FAB8561R, R&D Systems). After three ice-cold PBS washes, cells were transferred into prewarmed NS34 at 37°C and harvested into ice-cold PBS at different time points (0, 2.5, 5, 10, 15, and 30 min). An acid wash (50 mM glycine and 100 mM NaCl, pH 3, for 10 min at 4°C) stripped membrane-bound antibody and left intact intracellular staining to be processed for flow cytometry or confocal microscopy. To track recycling processes, a similar procedure was deployed using uncoupled anti-APLNR antibody (mouse IgG1, 1 h, 4°C, MAB8561; R&D Systems). After 15 min of endocytosis in prewarmed NS34 at 37°C (pulse), membrane-bound antibody was stripped using the acid wash solution described above. Then, cells were placed back at 37°C and harvested into ice-cold PBS at different time points (0, 5, 10, 15, 30, and 60 min, chase). APLNR membrane staining was revealed with Alexa-conjugated secondary anti-mouse IgG1 antibodies (1 h, 4°C; Life Technologies) followed by three ice-cold PBS washes. Samples were analyzed by flow cytometry and confocal microscopy.
Glycosylation status
Total protein lysates from GP130- and/or APLNR-transfected HEK293T cells were denaturated (55°C, 10 min) and treated overnight with EndoH and for 1 h with PGNase (New England Biolabs) according to the manufacturer’s instructions.
Immunoblotting and immunoprecipitation
Briefly, cells were lysed at 4°C in TNT lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% Igepal, and 2 mM EDTA, supplemented with protease inhibitor) for 30 min and cleared by centrifugation at 8,000 g. For immunoprecipitation, samples were precleared by a 30-min incubation with Protein G agarose, and then incubated for 2 h at 4°C with Protein G agarose and 5 µg of indicated antibodies. Protein concentrations were determined by bicinchoninic acid. Equal amount of 5–10 µg proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were revealed using a chemiluminescent HRP substrate and visualized using the Fusion imaging system (Vilber-Lourmat).
RNA-seq analysis
Parental and GP130 KO GSC#1 were harvested in three independent experiments and snap-frozen on dry ice. RNA extraction (all RNA integrity number >9.0), library preparation, RNA-seq and bioinformatics analysis was performed at Active Motif (Carlsbad, CA). Briefly, 2 µg of total RNA were isolated using the Qiagen RNeasy Mini Kit and further processed in Illumina’s TruSeq Stranded mRNA Library kit. Libraries are sequenced on Illumina NextSeq 500 as paired-end 42-nt reads. Sequence reads are analyzed with the STAR alignment – DESeq2 software pipeline described in the Data Explanation document. The list of differentially expressed genes from DESeq2 output was selected based on 10% adjusted P value level and false discovery rate of 0.1. Gene ontology and KEGG pathway enrichment analysis was done using DAVID bioinformatics resources portal.
qPCR
RNA extraction was done using Qiagen RNeasy kit on three biological replicates (3.106 GSC#1). Equal amounts of RNA were reverse transcribed using the Maxima Reverse transcription kit, and 30 ng of the resulting cDNA was amplified by qPCR using PerfeCTa SYBR Green SuperMix Low ROX. Data were analyzed using the 2-ΔΔCt method and normalized by the housekeeping genes ACTB and HPRT1. The following primers were used: ACTB forward, 5′-GGACTTCGAGCAAGAGATGG-3′; ACTB reverse, 5′-AGCACTGTGTTGGCGTACAG-3′; HPRT1 forward, 5′-TGACACTGGCAAAACAATGCA-3′; HPRT1 reverse, 5′-GGTCCTTTTCACCAGCAAGCT-3′; GP130 forward, 5′-TGTAGATGGCGGTGATGGTA-3′; GP130 reverse, 5′-CCCTCAGTACCTGGACCAAA-3′; APLNR forward, 5′-GAGTGCTGGGAAGGACTCTG-3′; APLNR reverse, 5′-ACTGGTTGTCTGCCCCATAG-3′; FAM20C forward, 5′-CCTCAGCTCCCAGAGGATCA-3′; FAM20C reverse, 5′-TCCCTCGTTTGTTTCATGGGT-3′; GRIK1 forward, 5′-CTCCCTGCAGTGCCATAGAC-3′; GRIK1 reverse 5′-TCCCAATCTTTTCAGTTCCTTCC-3′; ELMOD1 forward, 5′-GGAAGGCGGAGATGAAGACC-3′; ELMOD1 reverse, 5′-GGATCAACATTCTCAGGAAGTGC-3′; ELMOD2 forward, 5′-GGAAGGCGGAGATGAAGACC-3′; ELMOD2 reverse, 5′-TGTCGCCTTCTGTAAAACCTTA-3′; ELMOD3 forward, 5′-GCTCAGTGTGGCCTGGATAG-3′; ELMOD3 reverse, 5′-TCGGCAAGGTCTTTGAGTCC-3′; IGSF1 forward, 5′-GCCAGCAGGCTATCTCACC-3′; IGSF1 reverse, 5′-GCAGAGAAGGGCCGTGACTA-3′; MRC2 forward, 5′-CGGACACCAAACTCCGGTAT-3′; MRC2 reverse, 5′-CGGGTTCTGATTCACCGAAGA-3′; CDH10 forward, 5′-ATGGCAAGGGACCCAGATTC-3′; CDH10 reverse, 5′-GGGATTGTTGATTTCAGCAGCA-3′; ROBO2 forward, 5′-TGCAGACTTGCCAAGAGGAA-3′; ROBO2 reverse, 5′-AAACTGTGGGGGAGCAACAG-3′; DRAXIN forward, 5′-CTGCCACAGAAGAGTCCCTG-3′; DRAXIN reverse, 5′-ACCGCGGTGTTTCTCTTTCT-3′; LAMP5 forward, 5′-TCTTTGTAAAGGAAAGCCACAACAT-3′; LAMP5 reverse, 5′-CAGAGAGGTGGTGCGAGTTG-3′; GRIP2 forward, 5′-TAGCGAACATGCTGTGTGGG-3′; GRIP2 reverse, 5′-CCCTCGGAACTCCTCTGGAA-3′; TNFSF10 forward, 5′-GCAGCTCACATAACTGGGACC-3′; TNFSF10 reverse, 5′-TTGCTCAGGAATGAATGCCC-3′; BCL2L1 forward, 5′-TTCAGTGACCTGACATCCCA-3′; BCL2L1 reverse, 5′-TCCACAAAAGTATCCCAGCC-3′; TP53 forward, 5′-GGAGCACTAAGCGAGCACTG-3′; TP53 reverse, 5′-TCTCGGAACATCTCGAAGCG-3′; ARF1 forward, 5′-GGGCCGCAGAGACGTT-3′; ARF1 reverse, 5′-TCCACGTTGAAGCCTATGG-3′; ARF3 forward, AGGAAAACCGTCCGAGGAG-3′; ARF3 reverse, 5′-TGGTCCCTGGTATGGAAGACT-3′; ARF4 forward, 5′-AGCTGCAGAAAATGCTTCTGGT-3′; ARF4 reverse, 5′-TGACAGCCAGTCAAGTCCTTC-3′; ARF5 forward, 5′-CGTGTCCGCGCTCTTTTC-3′; ARF5 reverse, 5′-CGCATCCAAGCCAACCATGA-3′; ARF6 forward, 5′-GTGACCACCATTCCCACTGT-3′; ARF6 reverse, 5′-GCCCACATCCCATACGTTGA-3′; ARL1 forward, 5′-CAGTCTGTTTGGAACTCGGGA-3′; ARL1 reverse, 5′-ACATCTCCAGTATGGCCTGA-3′; ARL2 forward, 5′-CTGGAGCACCGAGGATTCAAG-3′; ARL2 reverse, 5′-AAAGATGAGGAGGGTTGCTCC-3′; ARNO/CYTH2 forward, 5′-AGGACGGCGTCTATGAACC-3′; ARNO/CYTH2 reverse, 5′-CGATCTCCTCGGGTGTGTTC-3′; BRAG2/IQSEC1 forward, 5′-CATAGAGGCGTTCAGCCAGC-3′; BRAG2/IQSEC1 reverse, 5′-CTCACGGGGAATGTCCTCAC-3′.
Statistics
Data are representative of at least three independent experiments, unless otherwise stated. Statistical analysis was performed with GraphPad Prism8 using one-way and two-way ANOVA. For each statistical test, a P value of <0.05 was considered significant. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows that JAK inhibitors impair APLN-based GSC maintenance. Fig. S2 reports the effects of nonionic detergents on APLNR membrane localization. Fig. S3 shows that GP130 does not affect all APLN/APLNR signaling activities in terms of downstream S6 phosphorylation, APLN binding to APLNR, and APLNR expression and glycosylation. Fig. S4 documents STAT3 predictive consensus binding sites on the three loci surrounding ELMOD1, ELMOD2, and ELMOD3 genes. Fig. S5 shows the modulation of APLNR membrane expression by the ARF-related GEFs ARNO and BRAG2. Fig. S6 highlights that the ARF GTPase ARF6 contributes to modulate APLNR availability at the plasma membrane.
Supplementary Material
Acknowledgments
We thank Signaling in Oncogenesis, Angiogenesis, and Permeability team members, especially Tiphaine Douanne, Yanis Mace, Thomas Maurice, Margot Machu, and Margo Fernandez (Université de Nantes, Institut National de la Santé et de la Recherche Médicale [INSERM], Centre National de la Recherche Scientifique, Nantes, France). We are grateful to Thomas Bonneaud (Université de Nantes, INSERM, Centre National de la Recherche Scientifique, Nantes, France) for providing the STAT3 inhibitor and technical advice, as well as Eric Chevet (INSERM, Rennes, France) for helpful discussion. We acknowledge Micropicell and Cytocell core facilities (Structure Fédérative de Recherche Santé François Bonamy, Nantes, France).
This research was funded by Fondation pour la Recherche Médicale (Equipe Labellisée DEQ20180339184); Fondation ARC contre le Cancer (J. Gavard; PJA20171206146 and PJA20191209477); Ligue Nationale contre le Cancer Comités de Loire-Atlantique, Maine et Loire, Vendée, Ille-et-Vilaine (J. Gavard and N. Bidère); and Région Pays de la Loire et Nantes Métropole under Connect Talent Grant (J. Gavard). K.A. Jacobs received Nantes Métropole and Fondation ARC contre le Cancer PhD fellowships. The team is part of the SIRIC ILIAD (INCA-DGOS-INSERM_12558).
The authors declare no competing financial interests.
Author contributions: K. Trillet and K.A. Jacobs: conceptualization, formal analysis, investigation, methodology, visualization, and writing (review and editing). G. André-Grégoire, A. Thys, C. Maghe, J. Cruard, and S. Gonzalez Diest: formal analysis, investigation, and visualization. J. Cruard and S. Minvielle: resources, visualization. G. Montagnac: formal analysis, resources, and methodology. N. Bidère: conceptualization, formal analysis, funding acquisition, and writing (review and editing). J. Gavard: conceptualization, formal analysis, resources, funding acquisition, and writing (original draft). All authors approved the manuscript.
References
- Bao, S., Wu Q., Sathornsumetee S., Hao Y., Li Z., Hjelmeland A.B., Shi Q., McLendon R.E., Bigner D.D., and Rich J.N.. 2006. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 66:7843–7848. 10.1158/0008-5472.CAN-06-1010 [DOI] [PubMed] [Google Scholar]
- Batchelor, T.T., Sorensen A.G., di Tomaso E., Zhang W.-T., Duda D.G., Cohen K.S., Kozak K.R., Cahill D.P., Chen P.-J., Zhu M., et al. 2007. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 11:83–95. 10.1016/j.ccr.2006.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, R.L., Wang Q., Carro A., Verhaak R.G.W., and Squatrito M.. 2017. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro-oncol. 19:139–141. 10.1093/neuonc/now247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese, C., Poppleton H., Kocak M., Hogg T.L., Fuller C., Hamner B., Oh E.Y., Gaber M.W., Finklestein D., Allen M., et al. 2007. A perivascular niche for brain tumor stem cells. Cancer Cell. 11:69–82. 10.1016/j.ccr.2006.11.020 [DOI] [PubMed] [Google Scholar]
- Chaves-Almagro, C., Castan-Laurell I., Dray C., Knauf C., Valet P., and Masri B.. 2015. Apelin receptors: From signaling to antidiabetic strategy. Eur. J. Pharmacol. 763(Pt B):149–159. 10.1016/j.ejphar.2015.05.017 [DOI] [PubMed] [Google Scholar]
- Chen, Q., Liu Y., Jeong H.-W., Stehling M., Dinh V.V., Zhou B., and Adams R.H.. 2019. Apelin+ Endothelial Niche Cells Control Hematopoiesis and Mediate Vascular Regeneration after Myeloablative Injury. Cell Stem Cell. 25:768–783.e6. 10.1016/j.stem.2019.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L., Huang Z., Zhou W., Wu Q., Donnola S., Liu J.K., Fang X., Sloan A.E., Mao Y., Lathia J.D., et al. 2013. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 153:139–152. 10.1016/j.cell.2013.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun, H.J., Ali Z.A., Kojima Y., Kundu R.K., Sheikh A.Y., Agrawal R., Zheng L., Leeper N.J., Pearl N.E., Patterson A.J., et al. 2008. Apelin signaling antagonizes Ang II effects in mouse models of atherosclerosis. J. Clin. Invest. 118:3343–3354. 10.1172/JCI34871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, B.W., Lathia J.D., Bruce Z.C., D’Souza R.C.J., Baumgartner U., Ensbey K.S., Lim Y.C., Stringer B.W., Akgül S., Offenhäuser C., et al. 2019. The dystroglycan receptor maintains glioma stem cells in the vascular niche. Acta Neuropathol. 138:1033–1052. 10.1007/s00401-019-02069-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, J.G., and Jackson C.L.. 2011. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12:362–375. 10.1038/nrm3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan-Moya, E.M., Le Guelte A., Lima Fernandes E., Thirant C., Dwyer J., Bidere N., Couraud P.-O., Scott M.G.H., Junier M.-P., Chneiweiss H., and Gavard J.. 2011. Secreted factors from brain endothelial cells maintain glioblastoma stem-like cell expansion through the mTOR pathway. EMBO Rep. 12:470–476. 10.1038/embor.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson, R.J., and Rich J.N.. 2007. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat. Rev. Cancer. 7:733–736. 10.1038/nrc2246 [DOI] [PubMed] [Google Scholar]
- Gimple, R.C., Bhargava S., Dixit D., and Rich J.N.. 2019. Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 33:591–609. 10.1101/gad.324301.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guanizo, A.C., Fernando C.D., Garama D.J., and Gough D.J.. 2018. STAT3: a multifaceted oncoprotein. Growth Factors. 36:1–14. 10.1080/08977194.2018.1473393 [DOI] [PubMed] [Google Scholar]
- Harford-Wright, E., Andre-Gregoire G., Jacobs K.A., Treps L., Le Gonidec S., Leclair H.M., Gonzalez-Diest S., Roux Q., Guillonneau F., Loussouarn D., et al. 2017. Pharmacological targeting of apelin impairs glioblastoma growth. Brain. 140:2939–2954. 10.1093/brain/awx253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms, J.B., and Zurzolo C.. 2004. Lipids as targeting signals: lipid rafts and intracellular trafficking. Traffic. 5:247–254. 10.1111/j.1600-0854.2004.0181.x [DOI] [PubMed] [Google Scholar]
- Hu, Y., and Smyth G.K.. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347:70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Ivanova, A.A., East M.P., Yi S.L., and Kahn R.A.. 2014. Characterization of recombinant ELMOD (cell engulfment and motility domain) proteins as GTPase-activating proteins (GAPs) for ARF family GTPases. J. Biol. Chem. 289:11111–11121. 10.1074/jbc.M114.548529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, K.A., Harford-Wright E., and Gavard J.. 2017. Neutralizing gp130 interferes with endothelial-mediated effects on glioblastoma stem-like cells. Cell Death Differ. 24:384. 10.1038/cdd.2016.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, K.A., André-Grégoire G., Maghe C., Thys A., Li Y., Harford-Wright E., Trillet K., Douanne T., Alves Nicolau C., Frénel J.-S., et al. 2020. Paracaspase MALT1 regulates glioma cell survival by controlling endo-lysosome homeostasis. EMBO J. 39:e102030. 10.15252/embj.2019102030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, X., Kim L.J.Y., Wu Q., Wallace L.C., Prager B.C., Sanvoranart T., Gimple R.C., Wang X., Mack S.C., Miller T.E., et al. 2017. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat. Med. 23:1352–1361. 10.1038/nm.4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, K.R., Longo-Guess C.M., and Gagnon L.H.. 2012. Mutations of the mouse ELMO domain containing 1 gene (Elmod1) link small GTPase signaling to actin cytoskeleton dynamics in hair cell stereocilia. PLoS One. 7:e36074. 10.1371/journal.pone.0036074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey, J.F., Dumont R.A., Wilmarth P.A., David L.L., Johnson K.R., and Barr-Gillespie P.G.. 2018. ELMOD1 Stimulates ARF6-GTP Hydrolysis to Stabilize Apical Structures in Developing Vestibular Hair Cells. J. Neurosci. 38:843–857. 10.1523/JNEUROSCI.2658-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekaran, G., Chaineau M., Piscopo V.E.C., Verginelli F., Fotouhi M., Girard M., Tang Y., Dali R., Lo R., Stifani S., and McPherson P.S.. 2021. An Arf/Rab cascade controls the growth and invasiveness of glioblastoma. J. Cell Biol. 220:e202004229. 10.1083/jcb.202004229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia, J.D., Mack S.C., Mulkearns-Hubert E.E., Valentim C.L.L., and Rich J.N.. 2015. Cancer stem cells in glioblastoma. Genes Dev. 29:1203–1217. 10.1101/gad.261982.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loilome, W., Joshi A.D., ap Rhys C.M.J., Piccirillo S., Vescovi A.L., Gallia G.L., and Riggins G.J.. 2009. Glioblastoma cell growth is suppressed by disruption of Fibroblast Growth Factor pathway signaling. J. Neurooncol. 94:359–366. 10.1007/s11060-009-9885-5 [DOI] [PubMed] [Google Scholar]
- Louault, K., Bonneaud T.L., Séveno C., Gomez-Bougie P., Nguyen F., Gautier F., Bourgeois N., Loussouarn D., Kerdraon O., Barillé-Nion S., et al. 2019. Interactions between cancer-associated fibroblasts and tumor cells promote MCL-1 dependency in estrogen receptor-positive breast cancers. Oncogene. 38:3261–3273. 10.1038/s41388-018-0635-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrella, G., Hou M., Li M., Stoecklein V.M., Zdouc N., Volmar M.N.M., Miletic H., Reinhard S., Herold-Mende C.C., Kleber S., et al. 2019. Targeting APLN/APLNR Improves Antiangiogenic Efficiency and Blunts Proinvasive Side Effects of VEGFA/VEGFR2 Blockade in Glioblastoma. Cancer Res. 79:2298–2313. 10.1158/0008-5472.CAN-18-0881 [DOI] [PubMed] [Google Scholar]
- Miryounesi, M., Bahari S., Salehpour S., Alipour N., and Ghafouri-Fard S.. 2019. ELMO Domain Containing 1 (ELMOD1) Gene Mutation Is Associated with Mental Retardation and Autism Spectrum Disorder. J. Mol. Neurosci. 69:312–315. 10.1007/s12031-019-01359-z [DOI] [PubMed] [Google Scholar]
- Oka, N., Soeda A., Inagaki A., Onodera M., Maruyama H., Hara A., Kunisada T., Mori H., and Iwama T.. 2007. VEGF promotes tumorigenesis and angiogenesis of human glioblastoma stem cells. Biochem. Biophys. Res. Commun. 360:553–559. 10.1016/j.bbrc.2007.06.094 [DOI] [PubMed] [Google Scholar]
- Patel, S.J., Sanjana N.E., Kishton R.J., Eidizadeh A., Vodnala S.K., Cam M., Gartner J.J., Jia L., Steinberg S.M., Yamamoto T.N., et al. 2017. Identification of essential genes for cancer immunotherapy. Nature. 548:537–542. 10.1038/nature23477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peurois, F., Veyron S., Ferrandez Y., Ladid I., Benabdi S., Zeghouf M., Peyroche G., and Cherfils J.. 2017. Characterization of the activation of small GTPases by their GEFs on membranes using artificial membrane tethering. Biochem. J. 474:1259–1272. 10.1042/BCJ20170015 [DOI] [PubMed] [Google Scholar]
- Prager, B.C., Xie Q., Bao S., and Rich J.N.. 2019. Cancer Stem Cells: The Architects of the Tumor Ecosystem. Cell Stem Cell. 24:41–53. 10.1016/j.stem.2018.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana, N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer, J.K., Sedgwick A.E., and D’Souza-Schorey C.. 2011. ARF6-mediated endocytic recycling impacts cell movement, cell division and lipid homeostasis. Semin. Cell Dev. Biol. 22:39–47. 10.1016/j.semcdb.2010.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem, O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., and Zhang F.. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q., Goderie S.K., Jin L., Karanth N., Sun Y., Abramova N., Vincent P., Pumiglia K., and Temple S.. 2004. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 304:1338–1340. 10.1126/science.1095505 [DOI] [PubMed] [Google Scholar]
- Shi, Y., Zhou W., Cheng L., Chen C., Huang Z., Fang X., Wu Q., He Z., Xu S., Lathia J.D., et al. 2017. Tetraspanin CD9 stabilizes gp130 by preventing its ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells. Cell Death Differ. 24:167–180. 10.1038/cdd.2016.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingu, T., Ho A.L., Yuan L., Zhou X., Dai C., Zheng S., Wang Q., Zhong Y., Chang Q., Horner J.W., et al. 2017. Qki deficiency maintains stemness of glioma stem cells in suboptimal environment by downregulating endolysosomal degradation. Nat. Genet. 49:75–86. 10.1038/ng.3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., and Dirks P.B.. 2004. Identification of human brain tumour initiating cells. Nature. 432:396–401. 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- Stupp, R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J.B., Belanger K., Brandes A.A., Marosi C., Bogdahn U., et al. National Cancer Institute of Canada Clinical Trials Group . 2005. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352:987–996. 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- Sztul, E., Chen P.-W., Casanova J.E., Cherfils J., Dacks J.B., Lambright D.G., Lee F.S., Randazzo P.A., Santy L.C., Schürmann A., et al. 2019. ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol. Biol. Cell. 30:1249–1271. 10.1091/mbc.E18-12-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, J.A., Fukumoto M., Kozai Y., Ito N., Oda Y., Kikuchi H., and Hatanaka M.. 1991. Inhibition of cell growth and tumorigenesis of human glioblastoma cells by a neutralizing antibody against human basic fibroblast growth factor. FEBS Lett. 288:65–71. 10.1016/0014-5793(91)81004-R [DOI] [PubMed] [Google Scholar]
- Treps, L., Edmond S., Harford-Wright E., Galan-Moya E.M., Schmitt A., Azzi S., Citerne A., Bidère N., Ricard D., and Gavard J.. 2016. Extracellular vesicle-transported Semaphorin3A promotes vascular permeability in glioblastoma. Oncogene. 35:2615–2623. 10.1038/onc.2015.317 [DOI] [PubMed] [Google Scholar]
- Tropepe, V., Sibilia M., Ciruna B.G., Rossant J., Wagner E.F., and van der Kooy D.. 1999. Distinct neural stem cells proliferate in response to EGF and FGF in the developing mouse telencephalon. Dev. Biol. 208:166–188. 10.1006/dbio.1998.9192 [DOI] [PubMed] [Google Scholar]
- Uribesalgo, I., Hoffmann D., Zhang Y., Kavirayani A., Lazovic J., Berta J., Novatchkova M., Pai T.-P., Wimmer R.A., László V., et al. 2019. Apelin inhibition prevents resistance and metastasis associated with anti-angiogenic therapy. EMBO Mol. Med. 11:e9266. 10.15252/emmm.201809266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak, R.G.W., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Cancer Genome Atlas Research Network . 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 17:98–110. 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Lathia J.D., Wu Q., Wang J., Li Z., Heddleston J.M., Eyler C.E., Elderbroom J., Gallagher J., Schuschu J., et al. 2009. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells. 27:2393–2404. 10.1002/stem.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., Maguire J.J., and Davenport A.P.. 2015. Apelin, Elabela/Toddler, and biased agonists as novel therapeutic agents in the cardiovascular system. Trends Pharmacol. Sci. 36:560–567. 10.1016/j.tips.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C.J., Cavenagh M.M., and Kahn R.A.. 1998. A family of Arf effectors defined as suppressors of the loss of Arf function in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273:19792–19796. 10.1074/jbc.273.31.19792 [DOI] [PubMed] [Google Scholar]
- Zhou, A., Lin K., Zhang S., Chen Y., Zhang N., Xue J., Wang Z., Aldape K.D., Xie K., Woodgett J.R., and Huang S.. 2016. Nuclear GSK3β promotes tumorigenesis by phosphorylating KDM1A and inducing its deubiquitylation by USP22. Nat. Cell Biol. 18:954–966. 10.1038/ncb3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W., Shi D.S., Winter J.M., Rich B.E., Tong Z., Sorensen L.K., Zhao H., Huang Y., Tai Z., Mleynek T.M., et al. 2017. Small GTPase ARF6 controls VEGFR2 trafficking and signaling in diabetic retinopathy. J. Clin. Invest. 127:4569–4582. 10.1172/JCI91770 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.