We tested the role of endothelial dysfunction in the development of COPD/emphysema. We found that restoration of lung endothelial cells and blockade of endothelial-derived leucine-rich α-2-glycoprotein perturbs the pathogenesis of COPD/emphysema.
Abstract
Chronic obstructive pulmonary disease (COPD) is marked by airway inflammation and airspace enlargement (emphysema) leading to airflow obstruction and eventual respiratory failure. Microvasculature dysfunction is associated with COPD/emphysema. However, it is not known if abnormal endothelium drives COPD/emphysema pathology and/or if correcting endothelial dysfunction has therapeutic potential. Here, we show the centrality of endothelial cells to the pathogenesis of COPD/emphysema in human tissue and using an elastase-induced murine model of emphysema. Airspace disease showed significant endothelial cell loss, and transcriptional profiling suggested an apoptotic, angiogenic, and inflammatory state. This alveolar destruction was rescued by intravenous delivery of healthy lung endothelial cells. Leucine-rich α-2-glycoprotein-1 (LRG1) was a driver of emphysema, and deletion of Lrg1 from endothelial cells rescued vascular rarefaction and alveolar regression. Hence, targeting endothelial cell biology through regenerative methods and/or inhibition of the LRG1 pathway may represent strategies of immense potential for the treatment of COPD/emphysema.
Introduction
Chronic obstructive pulmonary disease (COPD) is a heterogeneous lung disease that is associated with long-term cigarette smoking and represents the third-leading cause of death worldwide (Quaderi and Hurst, 2018). The cellular and molecular determinants underpinning the pathogenesis of COPD remain to be fully understood, but the current thought is that long-term inflammation serves as a driver of remodeling and parenchymal destruction in the proximal airways and distal lung tissue leading to the disease states of chronic bronchitis and emphysema (Hogg et al., 2004). Current disease-modifying therapies are limited in their ability to halt the progression of COPD and in relieving symptoms of dyspnea and airflow obstruction.
Recent investigations have uncovered the functional role for vascular endothelial cells (ECs) in organ regeneration and repair in multiple model systems (Rafii et al., 2016; Augustin and Koh, 2017). ECs maintain highly adaptable cellular functions that promote the development of organ-specific vascular niches that are critical to the maintenance of tissue homeostasis. Through the release of growth factors, known as angiocrine factors, vascular ECs coordinate propagation, patterning, and behavior of adjacent parenchymal and mesenchymal cells within a given tissue type (Rafii et al., 2016; Cao et al., 2014; Ding et al., 2011; Cao et al., 2017; Ding et al., 2014). By contrast, maladaptive ECs that emerge from environments of chronic cellular stresses and injury drive the development of fibrosis or tumorigenesis (Cao et al., 2014). Lung-specific endothelium has been shown to encourage alveologenesis following injury through the release of known angiocrine factors, such as metalloprotease-14 (MMP-14) and bone morphogenetic protein 4 (BMP4; Ding et al., 2011; Lee et al., 2014). As such, harnessing the regenerative potential of the endothelium through delivery of healthy ECs has therapeutic potential in diseased lungs.
The notion that pulmonary vascular health is linked to the parenchymal destruction and functional loss of alveolar surface area observed in COPD/emphysema was first described by Liebow (1959). Further morphometric analyses demonstrated a reduction in capillary length and density in COPD patients relative to healthy human subjects (Wiebe and Laursen, 1998). Endothelial dysfunction, assessed by monitoring nitric oxide–mediated relaxation in pulmonary artery rings, was identified in COPD samples at both early and late disease states and correlated with airflow obstruction (Peinado et al., 1998; Dinh-Xuan et al., 1991). Several studies have used ultrasound-based flow-mediated vasodilation to correlate endothelial dysfunction with a reduction in 6-min walk and forced expiratory volume in 1 s (FEV1), and increased emphysema (Minet et al., 2012; Vukic Dugac et al., 2015; Moro et al., 2008; Eickhoff et al., 2008; Barr et al., 2007). Additionally, pruning of the peripheral vasculature and a reduction in lung perfusion in COPD lungs has been directly shown by computed tomography, wherein abundant microvasculature loss, decreased capillary length, and reduced capillary density are observed in areas with reduced alveolar surface area (Estépar et al., 2013; Iyer et al., 2016). The paucity of endothelium is attributed to EC death, which is associated with vascular endothelial growth factor receptor-2 (VEGFR2; Kdr, Flk1) inhibition and the development of emphysema in murine models (Kasahara et al., 2000). Despite these tantalizing associations, it is not clear that endothelial dysfunction drives COPD pathophysiology or is simply the consequence of damaged alveolar surface area.
We queried the role of EC biology in the development of COPD/emphysema through human tissue analysis and the use of purified ECs from the elastase-induced murine model of emphysema. In human COPD samples, substantial loss of the EC signature reflected more advanced clinical disease. EC loss and dysfunction were hallmarks of emphysematous lungs harvested from elastase-treated mice. These phenotypes were reversed by intravenous delivery of healthy lung ECs. Additionally, elevation of leucine-rich α-2-glycoprotein-1 (LRG1) was found in human COPD tissue and in the purified endothelium of elastase-treated mice. We identified LRG1 as a driver of emphysema when deletion of endothelial Lrg1 rescued the phenotype.
Results
Loss of endothelial markers in human COPD tissue
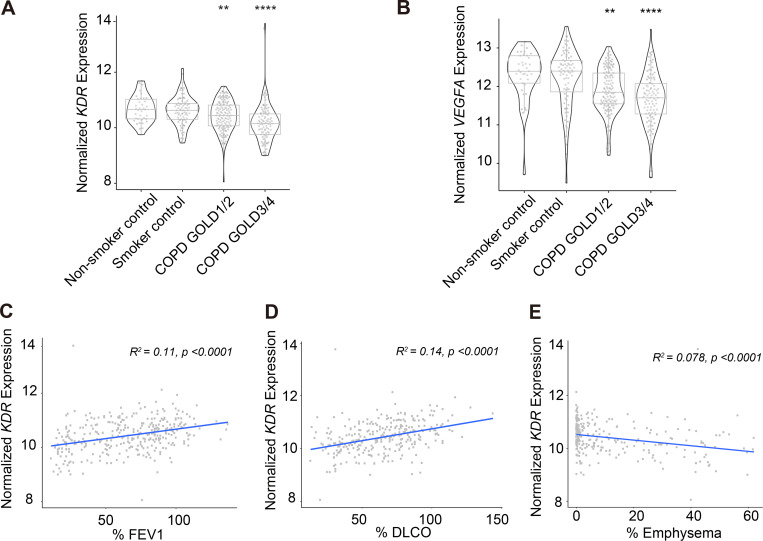
In this study, in silico analyses of microarray and RNA sequencing (RNA-seq) datasets from the Lung Genomics Research Consortium (LGRC) identified transcriptional changes in common EC markers within whole lung homogenates obtained from patients with severe COPD/emphysema (GOLD 3/4), mild COPD (GOLD 1/2), no COPD (smoker control), and no smoke exposure (nonsmoker control; Fig. 1 and Table 1). We used generalized linear models to assess changes in gene expression as they relate to lung function and disease severity. Significant loss of expression of KDR and VEGF-A was observed in lungs isolated from patients with both mild and severe COPD defined by GOLD stage criteria (Fig. 1, A and B). We found a direct relationship between the loss of expression of several endothelial markers and a reduction in lung function, defined as an increase in airway obstruction (decrease in FEV1) and decreased alveolar surface area (diffusing capacity of the lungs for carbon monoxide [DLCO]; Fig. 1, C and D; and Table 1). Loss of the lung EC signature also reflected increased severity of radiographical evidence of advanced emphysematous changes, suggesting that endothelium dysfunction underscores the loss of competent lung parenchyma observed in cases of severe emphysema (Fig. 1 E and Table 1). We questioned if the loss of EC marks reflected a global phenomenon of lung-specific cellular loss through an examination of the epithelial transcriptional signature. Expression of key epithelial markers, namely CDH1, SFTPD, and AQP5, was much less impacted by the presence of either mild (GOLD 1/2) or severe (GOLD 3/4) disease states (Fig. S1). Additionally, alterations of epithelial gene expression patterns did not appear to correlate with changes in FEV1 or radiographical evidence of emphysema (Table 1). The observed changes in the EC signature in these human COPD lung samples extend previous work and emphasize the association between lung EC dysfunction and the pathogenesis of this disease (Kasahara et al., 2001). We hypothesized that dysfunctional ECs could comprise maladaptive lung vascular ECs that help drive the development of COPD/emphysema. A testable corollary to this hypothesis is that reestablishing a healthy vasculature—by either intravenous delivery of normal lung ECs or reversal of aberrant EC signaling—will encourage repair and regeneration of damaged lung tissue.
Figure 1.
Loss of expression of key endothelial cell marks is a hallmark of COPD in human lung tissue and correlates with disease severity.(A and B) In silico analyses of microarray and RNA-seq datasets from the LGRC of VEGFR2/KDR (A) and VEGFA expression (B) in nonsmoker control, smoker control, COPD GOLD1/2 (mild), and COPD GOLD3/4 (severe) lungs. (C and D) Correlation between the expression of VEGFR2/KDR and physiological parameters % FEV1 (C) and % DLCO (D). (E) Correlation between VEGFR2/KDR expression and severity of emphysema determined by chest computed tomography (% emphysema). **, P < 0.01; ****, P < 0.0001.
Table 1. Summary of the correlation of expression of several endothelial- and epithelial-specific factors with physiological markers of COPD (% FEV1, % DLCO, and % emphysema).
| % FEV1 adjusted for age, sex, library | % DLCO adjusted for age, sex, library | % Emphysema adjusted for age, sex, library | ||||
|---|---|---|---|---|---|---|
| R 2 | P valuea | R 2 | P valuea | R 2 | P valuea | |
| Endothelial markers | ||||||
| KDR | 0.11 | 5.5 × 10−9 | 0.136 | 1.01 × 10−10 | 0.078 | 4.74 × 10−5 |
| VEGFA | 0.09 | 4.7 × 10−9 | 0.152 | 3.9 × 10−12 | 0.014 | 0.035 |
| ICAM2 | 0.022 | 0.0031 | 0.06 | 6.17 × 10−6 | 0.009 | 0.08 |
| TEK | 0.019 | 0.013 | 0.06 | 2.46 × 10−5 | 0.01 | 0.023 |
| TIE1 | 0.047 | 5.7 × 10−6 | 0.095 | 1.7 × 10−8 | 0.051 | 5.8 × 10−5 |
| Epithelial cell markers | ||||||
| CDH1 | 0.007 | 0.1 | 0.018 | 0.0167 | 0.0016 | 0.24 |
| SFTPD | 2.12 × 10−6 | 0.8 | 0.023 | 0.0049 | 0.0018 | 0.346 |
| AQP5 | 0.011 | 0.02 | 1.6 × 10−5 | 0.97 | 0.0027 | 0.153 |
| SFTPC | 0.0078 | 0.149 | 0.03 | 0.003 | 0.0045 | 0.3 |
Generalized linear model Wald statistics were used to assess the significance of coefficients in relevant models.
Figure S1.
The expression of key epithelial marks is maintained in human COPD lung tissue. In silico analysis from LGRC of expression of CDH1, AQP5, SFTPD, and SFTPC mRNA expression in nonsmoker control, smoker control, COPD GOLD1/2, and COPD GOLD3/4 lungs. Data are presented as median (line) ± SD (box). Generalized linear model Wald statistics were used to assess significance of coefficients in relevant models.
Reduced and disrupted EC population following elastase treatment
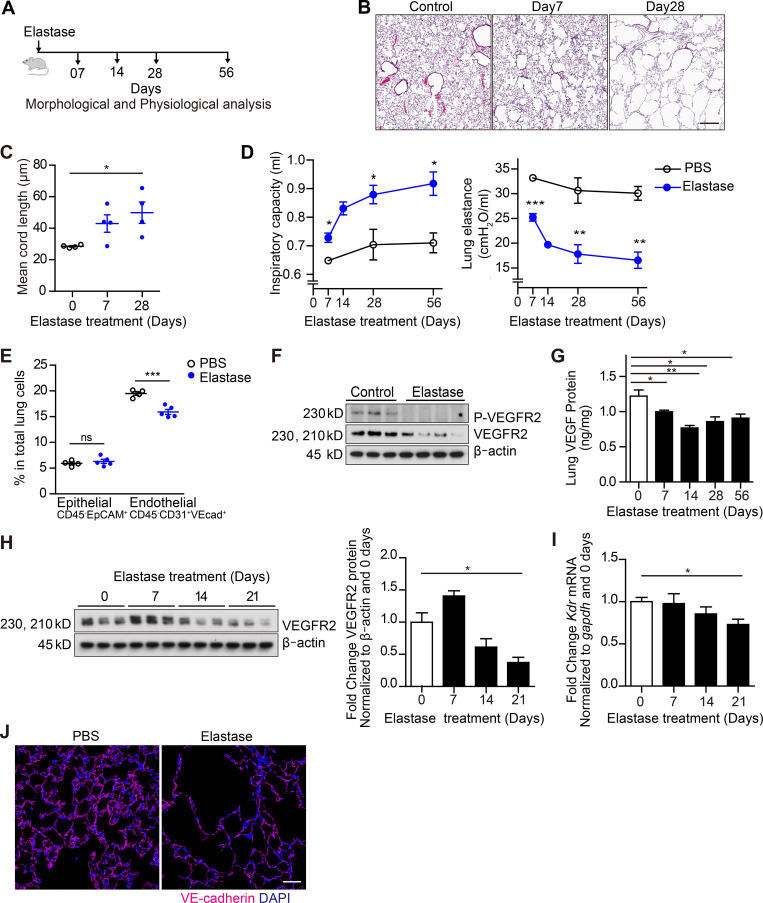
To test this hypothesis, we used an elastase-induced murine model of COPD (Suki et al., 2017; Fig. 2 A). Histological analyses of lung tissue following intratracheal instillation of elastase showed alveolar enlargement evident by day 7, progressing to diffuse parenchymal/alveolar destruction by day 28 (Fig. 2 B) that continued through day 56 (data not shown). Lung injury was characterized by destruction of alveolar walls, loss of alveolar surface area, and increased alveolar air space enlargement quantified by comparing mean cord length—a measure of mean free distance between air spaces (Mitzner, 2008)—in elastase- and vehicle-treated samples (Fig. 2 C). These morphological emphysematous changes were associated with increased inspiratory capacity and reduced pulmonary elastance consistent with a functional COPD/emphysema phenotype (Fig. 2 D).
Figure 2.
Dysfunctional lung vasculature in the murine elastase-induced emphysema model. (A) Schematic of the experimental design for results in B–D and G. (B) Representative hematoxylin and eosin staining of lung sections 7 and 28 d after elastase instillation and 28 d after PBS instillation (control). Scale bar, 300 µm. (C) Mean cord length quantification 7 and 28 d after elastase instillation and 28 d after PBS instillation (control; n = 4 per group). (D) Inspiratory capacity (left) and lung elastase (right) quantification 7, 14, 28, and 56 d after PBS or elastase instillation. n = 3 (PBS), n = 4 (elastase) per time point. (E) Proportion of EpCAM+ epithelial cells (CD45−CD31−VEcadherin−EpCAM+) and ECs (CD45−CD31+VEcadherin+) in total lung 14 d after PBS or elastase instillation (n = 5 per group) measured by FACS. (F) Total levels of phospho (P)-VEGFR2 and VEGFR2 (immunoblot) in whole lung lysates 28 d after elastase instillation. (G) Total VEGF levels in whole lung measured by ELISA (n = 4). (H) Total VEGFR2 levels in purified lung ECs. Lung ECs (CD45−CD31+) were purified using magnetic sorting (n = 3; left). Quantification of VEGFR2/β-actin (right) in purified lung ECs. (I) Loss of Kdr expression in purified lung ECs (right) measured by RT-qPCR (n = 3). (J) Representative immunofluorescent images of frozen lung sections stained for VE-cadherin (intravital labeling, magenta) and DAPI (blue) 21 d after elastase instillation. Scale bar, 50 µm. Data in C–E and G–I are represented as mean ± SEM. P values were determined by an unpaired Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with PBS). In F and H, the experiments were independently repeated at least two times with similar results. VEcad+, VE-cadherin+.
We next examined the effects of intratracheal elastase on murine lung vascular ECs through the use of a previously established protocol of intravital labeling of ECs followed by FACS (Fig. 2 E and Fig. S2; Nolan et al., 2013). In elastase-treated tissue, the number of purified ECs (CD45− CD31+ VE-cadherin+) was reduced compared with control mice (Fig. 2 E and Fig. S2). In contrast, the total number of epithelial cells (CD45− EpCAM+) appeared to be unchanged in the elastase-damaged lungs (Fig. 2 E). Additionally, a reduction of phosphorylated-VEGFR2, total VEGFR2, and total VEGF-A levels in total lung homogenates was observed in elastase-treated mice (Fig. 2, F and G). Isolated ECs also exhibited less VEGFR2 levels at days 14 and 21 following elastase treatment (Fig. 2, H and I), showing that elastase appeared to disrupt this key endothelial signaling pathway. We also noted that by 14 d after elastase treatment, a drastic loss of intricate alveolar capillary networks within lung tissue was observed (Fig. 2 J). Therefore, a depleted—and potentially dysregulated—endothelial population exists in lung following elastase treatment. These observations manifest as severe air space enlargement and respiratory dysfunction akin to the clinical disease entity of emphysema.
Figure S2.
Example of flow gating strategy for ECs (CD31+VEcadherin+ cells) and epithelial cells (EpCAM+).(A) 14 d after elastase instillation, ECs were intravitally labeled with anti–VEcadherin–Alexa Fluor 647. Then, lungs were enzymatically digested and quantified with FACS. ECs are defined as CD45−CD31+VEcadherin+ cells, whereas epithelial cells are defined as CD45−CD31−VEcadherin−EpCAM+ cells. Transcriptome analysis of lung ECs following elastase instillation. (B) Heatmaps of top 10 up-regulated and down-regulated genes for elastase-treated lungs at days 7, 14, and 21. (C) GSEA using vascular and angiocrine gene set (upper panel), as well as blood vessel development and angiogenesis (lower panel). Enrichment plot for vascular and angiogenesis (upper) and blood vessel development (lower). DN, down; Neg., negative; SSC-A, side scatter A.
Loss of endothelial marks and increased angiogenesis following elastase
Microvascular dysfunction and vascular remodeling are known features of emphysema, and the loss of substantial capillary bed surface area is thought to be linked to dysregulated angiogenesis and endothelial apoptosis (Kropski et al., 2018; Kasahara et al., 2001). To uncover the temporal molecular mechanisms driving elastase-induced vascular EC injury and how they may recapitulate the pathogenesis of emphysema, in particular as it relates to endothelial alterations, we compared the transcriptome of purified lung vascular ECs isolated from elastase-treated and control mice at days 7, 14, and 21 (Fig. 3 A). Elastase treatment induced distinct transcriptional changes in lung endothelium at each time point. Comparisons between ECs exposed to elastase versus control at each time point identified 74, 101, and 45 differentially expressed genes (false discovery rate [FDR] < 5%) at days 7, 14, and 21 after elastase treatment, respectively (Fig. 3 B, Fig. S2 B, and Table S1). Supervised gene ontology analyses of the differentially expressed genes identified pathways of blood vessel development and angiogenesis, TGF-β signaling, and extracellular matrix signaling as markedly altered following exposure to elastase (Fig. 3 C). Gene set enrichment analyses (GSEAs) highlighted the robust activation of the blood vessel development and angiogenesis pathway in ECs isolated from elastase-treated lungs at day 7 (enrichment score: 1.33, P value: 0.007), day 14 (enrichment score: 1.381, P value: 0.002), and day 21 (enrichment score: 1.347, P value: 0.001; Fig. 3 D, Fig. S2 C, and Table S1). Additionally, in these same samples, loss of EC vascular marks and the angiocrine gene signature also correlated with elastase treatment, most notably at days 7 and 14 (Fig. S2 C and Table S1). We next queried the cellular damage and death mechanism induced in lung ECs through elastase treatment (Table S1). Elastase-treated lung endothelium exhibited a pro-apoptotic gene signature at day 7 and a pro-inflammatory state at days 14 and 21 (Table S1). Transcription of additional processes of autophagy, necroptosis, and senescence were not significantly affected by elastase treatment (Table S1).
Figure 3.
Transcriptome profiling of lung ECs identifies a state with loss of prototypical endothelial marks and enhanced features of angiogenesis. (A) Schematic of murine EC isolation with FACS-assisted sorting at 7, 14, and 21 d after elastase treatment. Anti-VEcadherin–Alexa Fluor 647 was retro-orbitally injected. 8 min after injection, mice were sacrificed, and the lungs were enzymatically digested. ECs were isolated as CD45− CD31+VEcadherin+ cells and sorted directly into TRIzol. Total RNA was isolated, and paired-end RNA-seq was performed. (B) Volcano plots illustrating differentially expressed genes (FDR < 0.05, fold change [FC] > 1.5) in ECs following elastase treatment at days 7, 14, and 21 compared with control (PBS). Red and blue genes represent significantly up-regulated and down-regulated genes, respectively. Black lines indicate threshold cutoffs for FDR and FC. (C) Gene ontology analysis of differentially expressed genes in ECs treated with elastase compared with control at different time points using biological process pathways (BP5). Top 15 pathways with FDR < 0.05 are illustrated. Bar graphs (in gray) represent number of genes occurring within the indicated biological pathway. Red line indicates −log10 (P value) of the hypergeometric distribution test used in our analyses. Pathways are listed in order of significance (top to bottom). (D) Heatmaps of top 20 variant genes within the biological pathway “blood vessel development and angiogenesis” identified from ECs following treatment with elastase at days 7, 14, and 21 compared with control. Red and blue genes represent significantly up-regulated and down-regulated genes, respectively. Reg, regulation; SSC-A, side scatter A; VEcad, VEcadherin.
Previous studies have highlighted the importance of the cross-talk between lung ECs and epithelial cells as a key feature of lung homeostasis and physiology (DeLisser et al., 2006). Likewise, dysregulated angiogenesis and inflammation are proposed to encourage the small airway epithelium remodeling observed in the COPD/emphysema pathogenesis (Carmeliet, 2005; Wang et al., 2017; Chung and Adcock, 2008). Here, we find that the endothelium isolated from elastase-treated lung samples manifests features of apoptosis, inflammation, and angiogenesis associated with loss of prototypical endothelial marks and angiocrine factor expression. We take these features to denote a potentially dysfunctional state that drives the development of emphysema.
Under normal physiological conditions, ECs exhibit highly adaptable cellular and functional plasticity to support organ homeostasis and regeneration, whereas maladaptive ECs are drivers of fibrosis and tumorigenesis. Recent studies show that intravenous delivery of healthy ECs serves as primers to instruct regeneration and repair (Kusumbe et al., 2016; Poulos et al., 2017). Therefore, we postulated that intravenous delivery of lung ECs following installation of elastase may thwart the elastase effect and facilitate lung repair and regeneration. To test this hypothesis, we used FACS to obtain purified lung vascular ECs as previously described and transduced them with adenoviral E4ORF1, a gene product that permits long-term survival, expansion, and sustained pro-regenerative functions of ECs (Seandel et al., 2008; Butler et al., 2010).
Intravenous delivery of lung ECs reverses emphysema
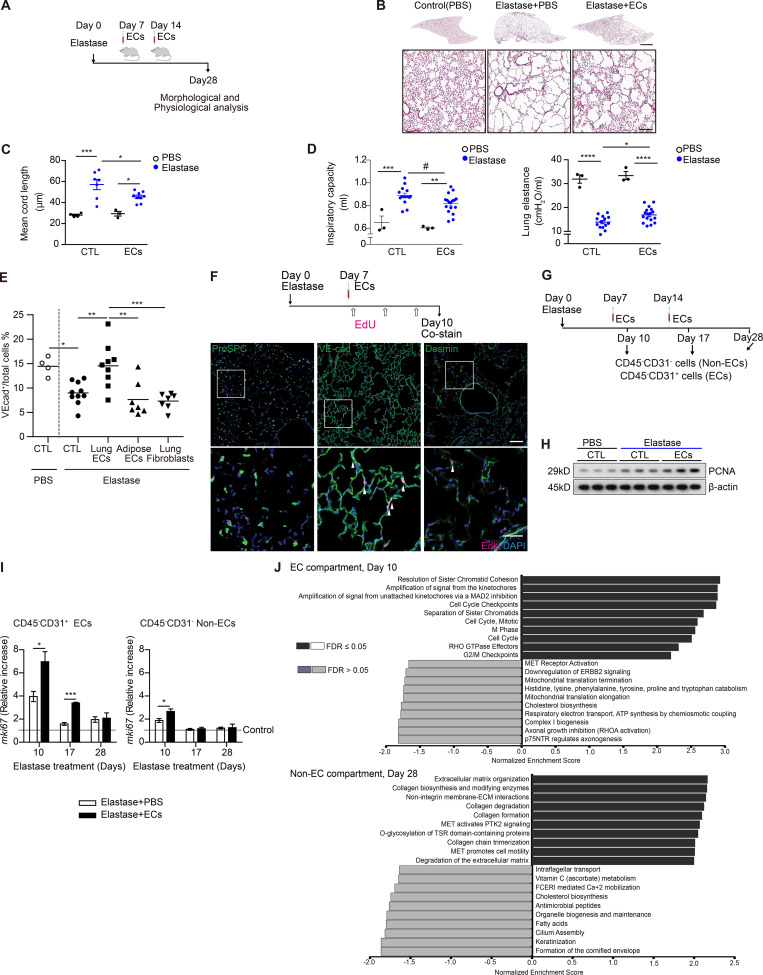
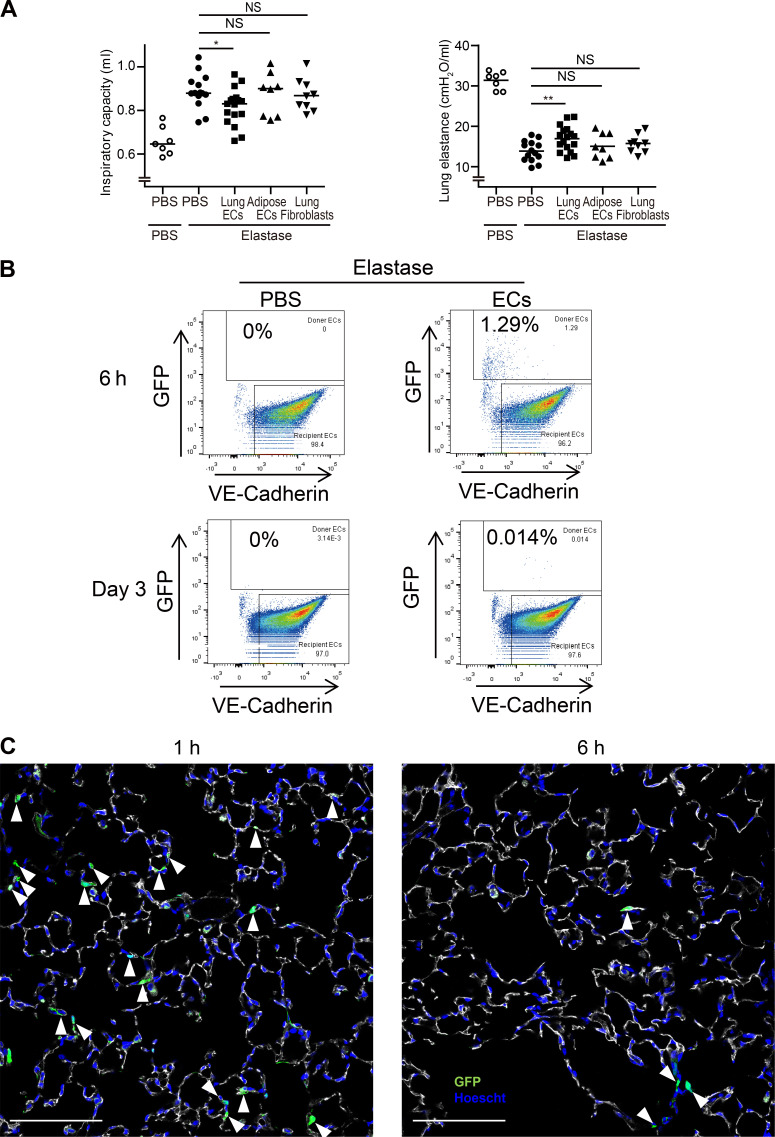
We delivered 5 × 105 GFP-labeled lung ECsE4ORF1 systemically through retro-orbital or jugular vein injections at days 7 and 14 after elastase instillation (Fig. 4 A). Notably, mice treated with intravenous injection of lung ECsE4ORF1 at these time points after elastase treatment (for 28 d) showed significantly less parenchymal destruction, observed histologically as a reduction in mean cord length, than mice treated with elastase only (Fig. 4, B and C). The lung ECE4ORF1–treated mice also had improved lung function with a reduced inspiratory capacity and an increase in lung elastance compared with mice that did not receive lung ECsE4ORF1 (Fig. 4 D). Additionally, the observed protective effect appeared to be dependent on the delivery of healthy lung ECs (Fig. 4 E and Fig. S3 A). Elastase-induced EC loss (VE-cadherin+ cells) was mitigated by injection of lung ECsE4ORF1, but not lung fibroblasts or, importantly, nonlung ECs, such as adipose-derived ECs (Fig. 4 E). Likewise, only delivery of lung ECsE4ORF1 perturbed the increase in inspiratory capacity (Fig. S3 A) and reduction in lung elastance (Fig. S3 A) observed in mice treated with elastase only. These findings support previous reports that underscored the importance of organ-specific ECs in tissue repair and function (Ding et al., 2014; Ding et al., 2011). Notably, despite the robust phenotype following lung ECsE4ORF1 injection, we found <2% of lung ECs to be GFP-positive (within 6 h) following EC injection (Fig. S3 B). Histologically, we were able to observe GFP-positive cells in lung parenchyma 1 h after EC treatment, but less so at the 6-h time point (Fig. S3 C). Hence, it appears that even transient healthy lung EC transplantation confers an acute and persistent impact on the surrounding parenchyma even after clearance of the injected cells.
Figure 4.
Intravenous delivery of lung ECsE4ORF1 ameliorates the development of emphysema following elastase treatment. (A) Schematic of experiments presented in B–E. Intravenous delivery of lung ECsE4ORF1 (ECs) occurred at days 7 and 14 after elastase treatment. (B) Representative images of lung sections at 28 d after elastase instillation with (Elastase + ECs) or without (Elastase + PBS) injections of lung ECsE4ORF1 at days 7 and 14. Scale bars, 2 mm (upper panel), 300 µm (lower panel). (C) Mean cord length quantification of lungs isolated from mice 28 d after elastase instillation with (ECs) or without (CTL) injections of lung ECsE4ORF1. PBS (n = 4 [CTL], n = 3 [ECs]) and elastase (n = 7 [CTL], n = 9 [ECs]). (D) Inspiratory capacity (left) and lung elastance (right) quantification 28 d after elastase instillation with (EC) or without (CTL) lung ECE4ORF1 delivery. PBS (n = 3 [CTL], n = 3 [ECs]) and elastase (n = 14 [CTL], n = 17 [ECs]). Data are from two independent experiments. (E) Proportion of ECs (VE-cadherin+) in total lung cells 28 d after elastase instillation, followed by treatment with PBS (CTL, n = 10), lung ECsE4ORF1 (n = 9), adipose ECsE4ORF1 (n = 7), and lung fibroblasts (n = 7) at day 7 and day 14. ECs (VE-cadherin+) were intravitally labeled with anti–VE-cadherin (VEcad)–Alexa Fluor 647, and the proportion of VE-cadherin–positive cells was quantified with FACS. Represented are biologically independent samples. (F) Schematic of EdU labeling experiments (upper panel). Representative immunofluorescent images of frozen lung sections stained for ProSPC, VE-cadherin, Desmin (green), EdU (magenta), and DAPI (blue). Scale bar, 100 µm (upper panel), 40 µm (lower panel). Arrowheads indicate EdU-positive cells. (G–I) Isolation of EC (CD45−CD31+) and Non-EC (CD45−CD31−) cellular fractions was performed using magnetic sorting. Schematic of the experiment (G). Representative immunoblots of PCNA levels in purified lung ECs 10, 17, and 28 d after elastase treatment with (ECs) or without (CTL) lung ECE4ORF1 injection (n = 3 per group; H). mRNA expression level of mKi67 in cellular fractions 10 d after elastase treatment with (ECs) or without (CTL) lung ECE4ORF1 injection (n = 3 per group). Relative increase compared with control (PBS, dotted line; I). (J) GSEA of bulk RNA-seq obtained from the lung endothelial and non–EC compartments of elastase mice with EC infusion compared with control samples at day 10 (EC compartment) and day 28 (non-EC compartment). Gene ontology and Reactome databases were used to perform unsupervised analyses using log fold change values. Data in C, D, and I are presented as mean ± SEM. P values were determined by one-way ANOVA in C–E or by unpaired Student’s t test in I (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; #, not significant in one-way ANOVA; P = 0.0222 in unpaired Student’s t test). In F and H, the experiments were independently repeated at least two times with similar results. GTPase, guanosine triphosphatase.
Figure S3.
Intravenous delivery of lung-specific ECs is required to recover physiological pulmonary function following elastase treatment.(A) Inspiratory capacity and lung elastance quantification at day 28 after elastase instillation. PBS or 0.5 × 106 of cells described (see below) was delivered at days 7 and 14 after elastase instillation. PBS (n = 14), Lung ECsE4ORF1 (n = 17), adipose ECsE4ORF1 (n = 8), and lung fibroblasts (n = 9) were delivered at stated time points. Data are from two independent experiments. Data are presented as mean. P values were determined by unpaired Student’s t test (*, P < 0.05; **, P < 0.01; NS, not significant compared with PBS). (B) 1 × 106 of GFP tagged ECs was injected through the retro-orbital cavity into elastase-treated mice. Following intravital labeling with anti–VEcadherin-Alexa647, total lungs were harvested and digested as previously described. FACS analysis was performed to identify the percentage of GFP-positive/VE-cadherin+ cells (Donor cells) in the total lung EC population. Percentages were quantified at 6 h and 3 d after injection. Recipient cells represent the endogenous lung ECs. (C) Representative immunofluorescent images of GFP (green) in frozen lung sections stained for VE-cadherin (white) and DAPI (blue). 1 × 106 of GFP-tagged lung ECsE4ORF1 was injected from the retro-orbital cavity into elastase-treated mice. 1 and 6 h after ECs implantation, mice were sacrificed after intravital labeling with anti–VE-cadherin. Arrowheads show GFP-positive cells, indicating that implanted GFP-tagged lung ECsE4ORF1 home to endothelium. Scale bars, 100 µm.
Given our observation that the murine elastase model appears to reproduce, at least in part, the vascular loss and dysregulation observed in emphysematous tissue, we aimed to further understand the pathways affected by EC infusion following elastase. With the marked histological and functional improvements observed following injection of lung ECsE4ORF1 in our murine elastase model, we looked for changes in cellular growth through the labeling of proliferating cells using 5-ethynyl-2′-deoxyuridine (EdU). Mice treated with elastase with or without intravenous injection of lung ECsE4ORF1 were administered EdU 0–2 d after systemic delivery of ECs (Fig. 4 F). We costained lung tissue with cell-specific antibodies to define endothelial, epithelial, and mesenchymal cell populations. VE-Cadherin+ cells stained most readily with EdU, suggesting a highly proliferative state in the EC population (Fig. 4 G). To a lesser extent, mesenchymal cells (Desmin+) were also found to costain with EdU, whereas epithelial cells (Spc) were found to be minimally stained (Fig. 4 F). Further evidence of EC proliferation was illustrated by a rise in PCNA and Ki67 in ECs harvested from animals treated with elastase followed by lung ECsE4ORF1 injection than in mice treated with elastase alone (Fig. 4, G–I). The non-EC fraction isolated from lung ECE4ORF1–treated mice did not appear to exhibit the same increase in Ki67 levels as the lung EC compartment (Fig. 4 I, left panel).
We pursued the molecular response to EC delivery further through bulk RNA-seq of both the endothelial and nonendothelial lung cellular fractions at 10 and 28 d after elastase treatment following EC delivery (Fig. 4 J). We did not observe significant differences between EC- and control (PBS infusion)–treated samples using multidimensional scaling. We proceeded to perform unsupervised GSEA using gene ontology and Reactome databases to identify enriched pathways within the various conditions (Fig. 4 J). A highly significant feature of both the EC and non-EC fractions at 10 d following elastase treatment was a marked increase in processes linked to cellular proliferation, suggesting that EC infusion may encourage this state in both compartments (Fig. 4 J). Notably, at 28 d following elastase treatment, markers of extracellular matrix were greatly enhanced in the non–EC fraction preparations but were not in the EC population (Fig. 4 J). We took these data to support the notion that delivery of healthy lung ECs appears to diminish emphysema and or promote repair and regeneration through possible encouragement of proliferation and tissue remodeling.
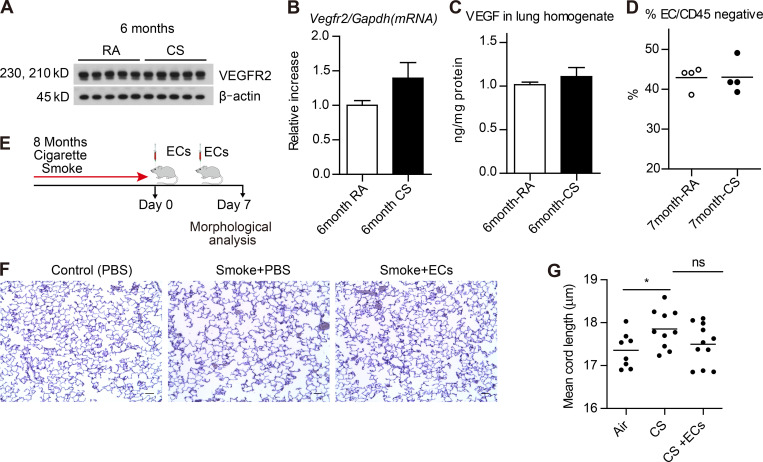
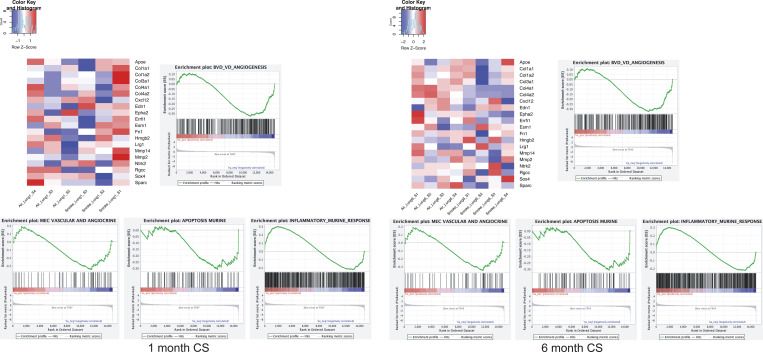
We extended our RNA-seq and EC delivery studies to the cigarette smoke (CS) murine model, where mice were exposed to either CS or room air (RA) for 1 mo or 6–8 mo. Using this model, we found that VEGFR2 and total VEGF protein levels were similar between lungs from mice exposed to CS or RA (Fig. 5, A–C). Likewise, the EC population appeared relatively stable between control mice and mice exposed to 7 mo of CS (Fig. 5 D). We then isolated lung ECs, as described in Fig. 3, from mice exposed to either 1 mo or 6 mo of CS and control mice at RA. Transcriptional profiles in lung ECs following either 1 mo or 6 mo of CS were less distinct than those observed in the elastase model (Fig. S4). There were a total of 17 and 12 differentially expressed genes identified in lung ECs isolated from 1 mo and 6 mo CS exposed mice, respectively (Table S2). We queried the impact of CS exposure on vascular, cell death, and inflammatory pathways that are known to be altered in COPD/emphysema (Kasahara et al., 2001; Kasahara et al., 2000). At both time points, angiogenic and apoptosis genes trended downward, while inflammatory signals were up-regulated (Fig. S4 and Table S2). The observed persistence of the EC population and lack of apoptosis in the murine smoke model is a notable distinction between the data obtained from the elastase mouse model (Fig. 2) and human COPD/emphysema lung tissue (Fig. 1; Kasahara et al., 2000; Kasahara et al., 2001).
Figure 5.
CS exposure does not alter murine lung ECE4ORF1 populations but EC transplant may reverse murine smoke-induced emphysema.(A–C) VEGFR2 protein by immunoblotting and ELISA (A and C) and mRNA (B) levels in whole lung tissue homogenates of mice exposed to RA or CS for 6 mo (n = 5 per group). (D) Proportion of ECs (CD45−CD31+VEcadherin+) in CD45− lung cells of mice exposed to RA or CS for 7 mo (n = 4 per group). (E) Schematic of the timeline of lung ECE4ORF1 (ECs) transplantation in mice exposed to CS. (F and G) Modified Gill’s immunohistochemical stain (F) and mean cord length of inflated whole lung sections from mice exposed to 8 mo of whole-body CS followed by EC transplant (500,000 ECs injected retro-orbitally) 0 and 3 d after smoking cessation (8 mo RA, n = 8; 8 mo smoke alone, n = 10; 8 mo smoke with EC transplant, n = 11). Scale bars, 100 µm. All data are mean ± SEM. *, P < 0.05 by unpaired t test.
Figure S4.
GSEA of RNA-seq profiles in purified lung ECs isolated from 1-mo– and 6-mo–CS-exposed mice. Analyses were performed using log fold change for gene sets of blood vessel development, EC and angiocine markers, apoptosis, and inflammation. Heatmaps for blood vessel development and angiogenesis gene set are shown. Enrichment score and P value are displayed.
Despite the lack of profound EC changes in the smoke-induced COPD model, we probed the impact of healthy lung EC delivery on damaged lungs obtained from animals exposed to CS compared with RA controls. We assessed the degree of emphysema in mice exposed to 8 mo of CS and found mild lung disease compared with RA control mice (P < 0.03; Fig. 5, E–G). Notably, CS-exposed mice who underwent EC infusion twice within 1 wk of smoking cessation appeared to have less emphysema, by mean cord length, compared with the CS group, but statistical significance was not achieved (Fig. 5 G; CS vs. CS + EC). We repeated these experiments at different time intervals, but the mild nature of the emphysema and the spontaneous recovery that occurs following cessation of CS limited our ability to assess the impact of endothelial infusion on damaged smoked lungs (Fig. 5; Canning and Wright, 2008).
We pursued the molecular response to EC delivery further through bulk RNA-seq of both the endothelial and nonendothelial lung cellular fractions at 10 and 28 d after elastase treatment following EC delivery (Fig. 4 J). We did not observe significant differences between EC- and control (PBS infusion)–treated samples using multidimensional scaling. We proceeded to perform unsupervised GSEA using gene ontology and Reactome databases to identify enriched pathways within the various conditions (Fig. 4 J). A highly significant feature of both the EC and non-EC fractions at 10 d following elastase treatment was a marked increase in processes linked to cellular proliferation, suggesting that EC infusion may encourage this state in both compartments (Fig. 4 J). Notably, at 28 d following elastase treatment, markers of extracellular matrix were greatly enhanced in the non–EC fraction preparations but were not in the EC population (Fig. 4 J). We took these data to support the notion that delivery of healthy lung ECs appears to diminish emphysema and or promote repair and regeneration through possible encouragement of proliferation and tissue remodeling.
LRG1 is up-regulated in emphysema lung-derived ECs and human COPD tissues
We next aimed to further understand the mechanism of endothelial dysfunction observed in the elastase murine model. RNA-seq (Fig. 3, B and D) revealed a subset of common, differentially expressed genes across all three time points (7, 14, and 21 d after elastase treatment). LRG1, a known glycoprotein that binds to the TGF-β accessory receptor (endoglin) and modulates the smad 1/5/8 signaling pathway promoting angiogenesis in ECs, was up-regulated in the EC population following elastase at all three time points (Fig. 3, B and D; and Fig. S3). LRG1 is known to mediate neovascularization through TGF-β1/Alk1 signaling in several disease processes, including ocular disease and diabetic nephropathy (Wang et al., 2013; Hong et al., 2019). LRG1 levels are up-regulated in the plasma of patients with ovarian and pancreatic cancer and in the urine of patients with bladder and non–small cell lung cancer (Andersen et al., 2010; Ding et al., 2011; Lindén et al., 2012). Recently, a link between LRG1 and the promotion of apoptosis and autophagy in ischemia and reperfusion injury was also made (Jin et al., 2019). In these cases, the potential for LRG1 levels to serve as a biomarker and its role in promoting pathogenesis through neo-angiogenesis have been proposed. Notably, global loss of LRG1 in a murine model was also shown to be protective against bleomycin-induced lung fibrosis, suggesting a role for this molecule in chronic lung disease (Honda et al., 2017). However, its role in the vascular-driven pathogenesis of emphysema has not been described.
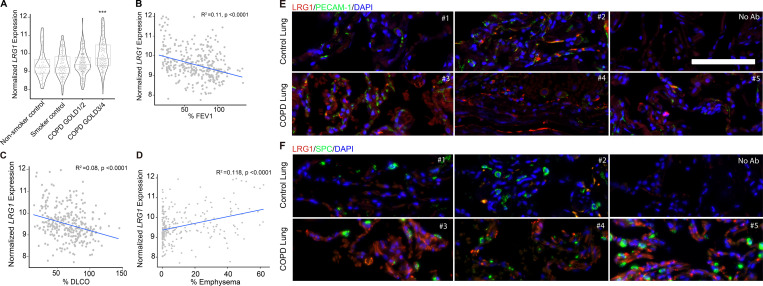
We examined the potential role of LRG1 in the development of COPD by following changes in expression in human lung from the LGRC datasets. The expression of LRG1 was up-regulated in human COPD samples, and these changes directly correlated with severity of the COPD phenotype (GOLD 3/4; Fig. 6 A). Additionally, marked airflow obstruction and decline in lung function (decrease in FEV1 and DLCO; Fig. 6, B and C), as well as severity of emphysematous lung parenchymal changes, also directly correlated with increased levels of LRGI (Fig. 6 D). We compared protein levels of LRG1 in human COPD lung tissue samples with control tissue and found substantially higher levels in COPD tissue. Costaining of lung tissue samples with PECAM-1 (CD31), a marker of ECs, and LRG1 revealed colocalization, suggesting that the increase in LRG1 observed in COPD tissue was particular to the EC population (Fig. 6, E and F; and Fig. S5 A). This colocalization was not observed in the type II epithelial population (labeled as SPC; Fig. 5 F and Fig. S5 A). Thus, these data point to a potential link between endothelial LRG1 accumulation and the development and progression of COPD.
Figure 6.
LRG1 levels are up-regulated in human COPD tissue and directly correlate with severity of disease and lung function decline. (A) In silico analysis from LGRC of expression of LRG1 mRNA in nonsmoker control, smoker control, COPD GOLD1/2, and COPD GOLD3/4 lungs. Data are presented as mean ± SD. Generalized linear model Wald statistics were used to assess significance of coefficients in relevant models (***, P < 0.001). (B–D) Correlation between the expression of LRG1 and DLCO, FEV1, and severity of emphysema observed on chest computed tomography. Generalized linear model Wald statistics were used to assess the significance of coefficients in relevant models. (E and F) Representative immunofluorescent staining images of formalin-fixed, paraffin-embedded human COPD lung tissue obtained from LTRC (as described in Materials and methods). Individual control and COPD patient samples were stained for total levels of LRG1 (red), PECAM-1 (top panel, green), DAPI (blue), and SPC (bottom panel, green). Scale bar, 100 µm.
Figure S5.
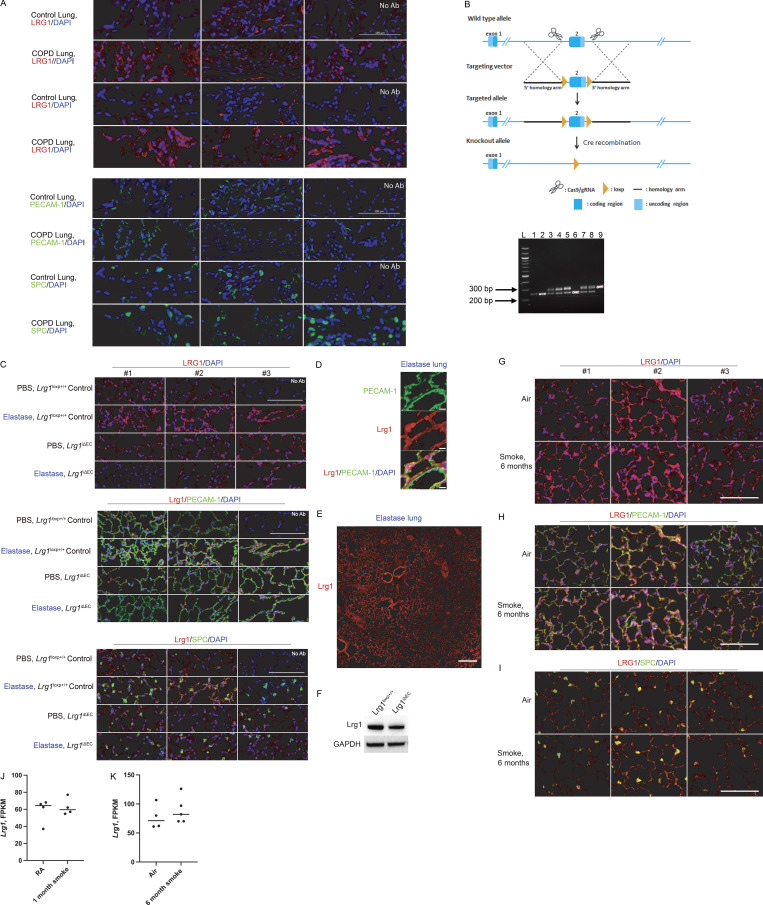
LRG1, PECAM-1, SPC, and DAPI staining in control and COPD human lungs. (A) Top: LRG1 and DAPI staining in control and COPD tissue used in bottom panel. Bottom: PECAM-1 or SPC, and DAPI in control and COPD tissue. No Ab panel represents tissue sample without primary antibody staining. Scale bar is 100 µm. (B) Generation of EC-specific Lrg1 knockout mouse line. Lrg1loxP/+ mice were obtained from Nanjing University. Top: Schematic of the strategy used to generate Lrg1loxP/+ mice using CRISPR Cas9 technology. Bottom: Genotyping of littermates generated from Lrg1loxP/+ × Lrg1loxP/+ breeding. Lanes 1, 2, and 6 represent wild-type mice with PCR product at 255 bp. Lanes 3–5, 7, and 8 represent heterozygous mice with product at 255 bp (wt) and product at 289 bp (following insertion of LoxP site). Lane 9 represents a homozygous condition, where LoxP sites were inserted at both alleles (single 289-bp PCR product). (C) Representative images of immunofluorescence staining with LRG-1 (red), PECAM-1 or SPC (green), and DAPI (blue) in control and elastase-treated lung obtained from control mice and Lrg1iΔEC. Scale bars, 100 µm. (D) Representative images of immunofluorescence staining highlighting colocalization of LRG1 and PECAM-1. Scale bars, 10 µm. (E) Representative images of immunofluorescence staining of Lrg1 in whole lung following elastase. Scale bar, 200 µm. (F) Lrg1 levels in MACS-isolated ECs obtained from control mice and Lrg1iΔEC. (G–I) Immunofluorescence staining with LRG-1 (red), PECAM-1 or SPC (green), and DAPI (blue) in mouse lung following 6 mo of CS or air exposure. Scale bars, 200 μm. (J and K) FPKM values of transcript level of Lrg-1 levels following 1 mo (J) and 6 mo (K) of smoke. gRNA, guide RNA; MACS, magnetic-activated cell sorting.
Endothelial LRG1 drives COPD/emphysema phenotype in elastase-treated mice
To explore the role of endothelial LRG1 in the development of emphysema in our elastase model, we used an EC-specific inducible gene deletion strategy. VE-cadherin-CreERT2 mice, in which a tamoxifen responsive–Cre is expressed under the control of the EC-specific VE-cadherin promoter, were crossed with mice with floxed Lrg1 that was generated using CRISPR Cas9 technology (Fig. 7 A and Fig. S5 B). Treatment of the resulting mice (VE-cadherin-CreERT2+/− Lrg1LoxP/LoxP; referred to as Lrg1iΔEC) with tamoxifen selectively ablated Lrg1 in ECs of adult mice (Fig. 7 A). VE-cadherin-CreERT2−/−Lrg1LoxP/LoxP (referred to as Lrg1LoxP/LoxP Control) was used as the control group.
Figure 7.
Loss of EC LRG1 blocks the development of emphysema in the murine elastase model. (A) Schematic of generation of mouse line with EC-specific loss of Lrg1 expression. Transgenic mice in which VE-cadherin promoter drives expression of tamoxifen-responsive CreERT2 (VE-Cad–CreERT2 mice) were crossed with Lrg1loxP/loxP mice and treated with tamoxifen to induce EC-specific deletion of LRG1. (B) Representative immunofluorescent images of lung sections stained for LRG1 (red), DAPI (blue), SPC (green, middle panel), and PECAM-1 (green, right panel). Scale bar, 100 µm. (C) Representative images of lung sections at 28 d after elastase instillation in conditions where EC expression of Lrg1 is either intact (Lrg1loxp+/+) or lost (Lrg1iΔEC). Scale bars, 4 mm (upper panel), 300 µm (lower panel). (D) Mean cord length quantification 28 d after elastase instillation in conditions where EC expression of Lrg1 is either intact (Lrg1loxp+/+) or lost (Lrg1iΔEC). PBS (n = 8 [Lrg1loxp+/+], n = 8 [Lrg1iΔEC]) and elastase (n = 25 [Lrg1loxp+/+], n = 25 [Lrg1iΔEC]). Combined data are from at least three independent experiments. **, P < 0.01; ****, P < 0.0001. VE-Cad, VE-cadherin.
LRG1 levels increased following elastase treatment in control mice (Fig. 7 B and Fig. S5, B–F). We localized the increase to the endothelium, as opposed to type II epithelial cells (PECAM-1 and SPC, respectively; Fig. 7 B and Fig. S5 C). Additionally, we noted heterogeneous staining of LRG1 across the entire lung following elastase treatment (Fig. S5 E). In contrast to both the human COPD and our elastase data, changes in LRG1 levels were less clear in our CS model. At the protein level, areas where LRG1 appeared increased in response to CS were identified, but there were also areas lacking this increase. Likewise, Lrg1 transcript levels trended upward in ECs following 6 mo of smoke (Fig. S5, G–K).
In Lrg1iΔEC mice, we observed a substantial reduction in lung LRG1 levels following elastase treatment compared with control tissues (Fig. 7 B and Fig. S5 C). This decrease is consistent with the down-regulation of LRG1 observed in the Lrg1iΔEC mice treated with tamoxifen (Fig. S5 F). Notably, Lrg1iΔEC mice treated with elastase were protected from the severe parenchymal destruction observed in control animals, as determined by histological assessment and morphometric analysis (Fig. 7, C and D). Hence, EC LRG1 is a critical factor involved in promoting the development of the maladaptive lung vasculature observed in our murine elastase model of COPD.
Discussion
To further decipher the role of EC biology in the pathogenesis of COPD, we queried expression and physiological data from COPD patients enrolled in the LGRC dataset and used the elastase-induced murine model of COPD/emphysema. Our data strongly suggest the critical role of EC function in mediating the pathogenesis of COPD/emphysema. Human COPD tissue reflects a state where markers of healthy endothelium are depleted (e.g., VEGFR2 and VEGFA) and markers of pathogenic angiogenesis (e.g., LRG1) are up-regulated. Loss of endothelial marks correlated with the progression of pulmonary function decline and increased emphysema. These data extend and support prior work linking endothelial apoptosis and dysfunction to severity of COPD (Kasahara et al., 2001; Liebow, 1959). Given the significant role of airway epithelial dysfunction and inflammation in the pathogenesis of COPD, we also investigated alterations in the epithelial cell compartment in human COPD tissues. Notably, expression of several epithelial marks did not appear to be altered in severe COPD tissues compared with control samples. The significance of the pulmonary ECs to the development and regeneration of the epithelial cell compartment has been highlighted in multiple studies (Ding et al., 2011; Lee et al., 2014). Loss of EC marks and angiogenic capabilities impaired proper alveolarization in multiple animal models (DeLisser et al., 2006; Jakkula et al., 2000). Additionally, endothelial-derived angiocrine factor signaling, through VEGF-A, FGF, MMP14, and BMP4 pathways, has been shown to support the differentiation, proliferation, and repair of alveolar epithelial progenitor cells (Ding et al., 2011; Lee et al., 2014). We propose that the unexpected stability of epithelial-specific marks in human COPD tissues suggests that the observed dysfunctional endothelial state may serve as a driver of tissue malfunctions found in the COPD disease state.
We used the elastase-induced murine model of COPD/emphysema to further decipher the role of EC biology in the development of functional and histological COPD/emphysema. We found substantial loss of lung ECs following treatment with elastase. Additionally, transcriptional profiling of highly purified elastase-treated lung ECs defined a pro-angiogenic state that appears to underscore the development of emphysema (Fig. 3 C, Fig. S2 C, and Fig. 6, E and F). The significance of EC function in the development of COPD was further stressed when intravenous delivery of healthy lung ECs to mice previously exposed to elastase rescued the lung phenotype. The regeneration and repair of tissue through the delivery of organ-specific ECs has been shown in previous work (Poulos et al., 2017; Ding et al., 2011). Our studies extend this work and highlight the regenerative potential of lung EC administration to patients with advanced COPD/emphysema.
ECs isolated from elastase-treated lungs were enriched for elevated levels of LRG1, a glycoprotein protein known to be involved in the progression of several disease states (Honda et al., 2017; Hong et al., 2019; Wang et al., 2013). Further analysis of the transcriptomic data from the human LGRC datasets revealed a significant increase in LRG1 levels in patients with severe COPD, reduced lung function, and significant emphysema. Therefore, EC-derived LRG1 contributes to the pathogenesis of COPD. To further clarify the role of EC LRG1 in the pathogenesis of COPD, we used Lrg1iΔEC mice in our elastase model. Lrg1iΔEC mice were protected from the development of COPD/emphysema following elastase, suggesting that a rise in EC LRG1 is key to the development of COPD/emphysema. Additionally, we identified LRG1 as a key driver of COPD/emphysema pathogenesis in both human COPD samples and our murine model.
Our findings give credence to the “vascular hypothesis” postulated by previous studies assessing EC function in the COPD/emphysema disease state (Voelkel, 2018). We propose that EC dysfunction supports a maladaptive pulmonary vasculature that perpetuates the development of COPD/emphysema through autocrine and paracrine effectors. This paradigm is supported by the observations that delivery of healthy lung ECs or endothelial-specific deletion of LRG1 thwarts the development of COPD/emphysema (Fig 4, B and C; Fig 7, C and D; and Fig. 6, E and F). Hence, uncovering both the perfusion-dependent and perfusion-independent reparative functions of the lung vasculature sets forth a new potential therapeutic target either through cell therapeutics or inhibition of factors to revert the pathogenic vascular environment into regenerative modalities.
Materials and methods
Mice
C57BL/6J mice were purchased from The Jackson Laboratory. Lrg1loxP/+ mice were obtained from Nanjing University. To generate mice with tamoxifen-inducible deletion of Lrg1 specifically in ECs, Lrg1loxP/loxP mice were crossed with VE-cadherin-CreERT2 mice. VE-cadherin-CreERT2 mice were provided by Ralf H. Adams (Max Planck Institute for Molecular Biomedicine, Munster, Germany). Lrg1loxP/loxP mice were crossed with VE-cadherin-CreERT2 to generate VE-cadherin-CreERT2 Lrg1loxP/loxP mice. To induce deletion of Lrg1 by CreERT2, intraperitoneal injections of tamoxifen (100 mg/kg/dose; Sigma-Aldrich; T5648) prepared using sunflower seed oil (Sigma-Aldrich; S5007) were administered on 3 d consecutively for a total of three times (total of nine injections) starting at 4 wk of age (Cao et al., 2017).
Source of human lung COPD and control tissues
The LGRC provides molecular and quantitative phenotype data for human subjects in the National Heart, Lung, and Blood Institute’s Lung Tissue Research Consortium (LTRC) biorepository (https://biolincc.nhlbi.nih.gov/studies/ltrc/). Human samples obtained from LTRC used in Fig. 5 were tissues obtained from open lung biopsies described previously (Cloonan and Choi, 2016). Lung tissue samples were collected as part of the routine care of patients and were submitted with a standardized series of questions. Deidentified formalin-fixed paraffin-embedded tissues from a subset of LTRC patient samples were used for immunofluorescence studies.
Elastase-induced murine emphysema model
8-wk-old C57BL/6J mice were treated with oropharyngeal instillation of either 0.25 IU of porcine pancreatic elastase (Sigma-Aldrich; E7885) or PBS in 50 µl using Microsprayer (Penn-Century), after anesthesia with intraperitoneal ketamine (100 mg/kg) and xylazine (10 mg/kg). Pulmonary function was measured, and mice were euthanized at different time points after elastase instillation for lung harvest as outlined in the figure legends (Suki et al., 2017).
CS exposure
Age- and sex-matched C57/BL6 mice were selected at random and exposed to total body CS or RA conditions for 6–8 mo. CS exposures (mainstream and sidestream smoke) were performed using the TE-10 inhalation exposure apparatus (Teague Enterprises) using 3R4F cigarettes (University of Kentucky Center for Tobacco Reference Products) and exposure to an average total particulate matter of 150 mg/m3 for 2 h per day, 5 d per week for 6–8 mo. For animals subjected to CS exposure, early death was used as an exclusion criterion. At the end of the exposure regimen, mice were euthanized by CO2 narcosis.
Pulmonary function test
Pulmonary function tests were performed as previously described (Siempos et al., 2018). In brief, mice were anesthetized with an intraperitoneal injection of pentobarbital (70 mg/kg). The trachea was exposed, and a 19-gauge cannula was inserted and stabilized in the trachea lumen. A rodent computer-controlled piston ventilator (Flexivent; SCIREQ) was connected to the trachea cannula. Lungs were recruited by inflation with up to 30 cm H2O pressure, and lung respiratory function parameters (including lung parenchymal tissue elastance and inspiratory capacity) were measured using the forced oscillation technique operated by FlexiWare 7 software.
Morphometric analysis of lung sections
Tissue preparation
Left lungs were inflated by gravity with 4% paraformaldehyde (PFA) and held at a pressure of 30 cm H2O for 15 min. Right lungs were immediately frozen for subsequent analysis. Left lungs were fixed with 4% PFA for 24 h at 4°C and then transferred to 70% ethanol at 4°C. Lungs were transferred to tissue cassettes (Tissue-Tek), and then lungs were embedded in paraffin. Lung sections were stained with modified Gill’s staining.
We quantified air space enlargement using the mean linear intercept (chord) length method (Chen et al., 2010; Smith and Mitzner, 1980). Randomized images were acquired as black-and-white TIFF files using a microscope, a 10× objective, and a camera and software that can acquire high-quality digital images. 5–10 images (10× magnification) were captured per mouse in a randomized manner, with the observer blinded to the experimental condition, avoiding underinflated areas of the lung (at 10× magnification). Lung sections were blinded by taping the slide identifiers before acquiring images and before data analysis. Random number generations using Excel, and a gridded coverslip were used to choose the sample image area for acquisition in all cases.
Mean chord length measurements
This protocol measures mean alveolar chord length and alveolar area on paraffin-embedded lung sections stained with Gill’s stain. We used a technique previously described (Cloonan and Choi, 2016). Morphometry software quantified the length of chords (vertical lines within a grid) within areas identified as airspace (Laucho-Contreras et al., 2015; Cloonan and Choi, 2016). Using this method, it is possible to measure the size of the alveoli in all parts of the lung in a standardized and relatively automated manner (Cloonan and Choi, 2016; Laucho-Contreras et al., 2015). Large airways, blood vessels, and other nonalveolar structures such as macrophages were manually removed from the images. Alveolar chord length for each image was calculated, and the average mean chord length ± SEM for each mouse was calculated (Cloonan and Choi, 2016).
Immunofluorescence
We prepared and stained formalin-fixed, paraffin-embedded, 10-μm-thick lung sections using standard procedures. Human COPD tissues obtained from LTRC were deidentified. Both human and murine paraffin-embedded lung tissues underwent deparaffinization (washes with graded alcohol and PBS) and rehydration processing as an initial step. Heat-activated antigen retrieval was performed using Sodium Citrate Buffer (10 mM Sodium Citrate and 0.05% Tween 20, pH 6.0) and 30-min heat using a steamer. Lung sections were incubated in primary antibodies overnight at 4°C: anti-LRG1 (Abcam; 178698, 1:100, human), anti–PECAM-1 (ARP; 24–5175-MSM9, 1:100, human), anti–PECAM-1 (Dianova; DIA-310, 1:100, mouse), anti-SPC (Life Biosciences; LS-B10952-200, 1:100, human and mouse), and anti-LRG1 (Abcam; 231188, mouse, 1:100). The negative control used substituted PBS for primary antibody. Following incubation with fluorophore-conjugated secondary antibodies (2.5 µg/ml; Jackson ImmunoResearch), a Zeiss Cell Observer SD confocal with a Yokagawa CSU-X1 spinning disk, Plan-Apochromat 63×/1.4, 40×/1.4, or 20×/0.8 numerical aperture objectives paired with a 1.2× adapter to a Photometrics Evolve 512 EMCCD camera was used for image acquisition with Zen 2 acquisition software.
EdU labeling
Lung cell proliferation was measured in vivo by EdU labeling. Briefly, 100 µg of EdU in 200 µl PBS was intraperitoneally injected once a day from day 7 to day 9. At day 10, lungs were fixed with 4% PFA and embedded in optimum cutting temperature formulations (Tissue-Tek). 14-µm-thick cryosections were incubated with PBS containing 5% normal donkey serum (Jackson ImmunoResearch) and 0.15% Triton X-100 (Sigma-Aldrich) for blocking and permeation. EdU was developed with Click-iT Plus EdU Imaging Kits (Molecular Probes; C10640), and sequentially, sections were incubated with diluted primary antibodies in a humidified chamber overnight at 4°C. Primary antibodies against Prosurfactant Protein C (Abcam; ab90716, 1:800), Desmin (Abcam; ab15200, 1:200), and VE-cadherin (Biolegend; BV13, 1:100) were used. 24 h later, the cryosections were incubated with secondary antibody (Thermo Fisher Scientific) for 1 h under ambient temperature. The slides were mounted using Prolong Gold antifade solution (Invitrogen). Images were captured on an AxioVert LSM710 confocal microscope (Zeiss).
Generation of lung ECsE4ORF1 from mouse lungs
25 μg of VE-Cadherin Alexa Fluor 647 (Biolegend; BV13) and 100 µg of Isolectin GSIB4 Alexa Flour 488 (Invitrogen/Molecular Probes) was injected retro-orbitally under anesthesia 8 min before sacrifice (Nolan et al., 2013). Lungs and subcutaneous fat were minced and incubated with Collagenase A (2.5 mg/ml; Sigma-Aldrich), Dispase II (1.0 mg/ml; Sigma-Aldrich), and DNase (80 U/ml; Sigma-Aldrich) at 37°C for 30 min. Cells were filtered through a 40-µm filter, and VE-Cadherin+IsolectinGSIB4+ cells were isolated as lung ECs. Primary ECs were sorted with a BD FACS aria II, at 25 p.s.i., using a 100-µm nozzle. The FACS setup and gating strategy for EC isolations are depicted in Fig. 3 A.
Primary ECs were cultured on the fibronectin-coated plates with EC-media (advanced DMEM/F12 media, 50 µg/ml EC supplement, 20% fetal bovine serum, 1× antibiotic-antimycotic solution, 10 mM Hepes, 5 µM SB431542 small molecule, 10 U/ml heparin, 1× Glutamax solution, 1× non-essential amino acid solution, 20 ng/ml FGF-2, and 10 ng/ml VEGF). Primary ECs were transduced with lentiviral vector expressing the E4ORF1 gene and maintained in the X-VIVO medium for 2 d to purify E4ORF1-transduced ECs (Seandel et al., 2008). Further, cells were stained with VE-Cadherin Alexa Fluor 647, CD31 PE/Cy7 (Biolegend; MEC13.3), and CD45-FITC (Biolegend; 30-F11), and ECs were purified as VE-Cadherin+CD31+CD45− cells. ECs were passaged in EC media, used for experiment between passages 10 and 20.
FACS analysis of EC compartment in mouse lung
14 d after elastase instillation, 25 µg of VE-Cadherin Alexa Fluor 647 (Biolegend; BV13) was injected retro-orbitally under anesthesia 8 min before sacrifice. Lungs were minced and incubated with Collagenase A (2.5 mg/ml; Sigma-Aldrich), Dispase II (1.0 mg/ml; Sigma-Aldrich), and DNase (80 U/ml; Sigma-Aldrich) at 37°C for 30 min. Single cells were stained with CD45-BV421 (Biolegend; 30-F11), CD31-PE/Cy7 (Biolegend; MEC13.3), EpCAM-FITC (Biolegend; G8.8), and DAPI and quantified with BD FACS aria II. ECs are defined as CD45−VEcadherin+CD31+cells, whereas EpCAM+ epithelial cells are defined as CD45−VEcadherin−CD31−EpCAM+ cells. Proportion is defined as a ratio of either endothelial or epithelial cell count to total cells counted in lung. For analysis of EC compartment after intravenous delivery of lung ECs, VEcadherin (intravital labeling) cells were quantified with FACS analysis 28 d after elastase instillation.
Intravenous delivery of healthy lung ECsE4ORF1
Mice were anesthetized with Isoflurane or an intraperitoneal injection of pentobarbital (70 mg/kg). 500,000 lung ECsE4ORF1 or other cells (lung fibroblasts, adipose ECsE4ORF1) in 50 µl PBS were injected via retro-orbital cavity or jugular vein at days 7 and 14 after elastase treatment. At 28 d after elastase instillation, either pulmonary functions were measured or mice were sacrificed for histological analysis.
Isolation of ECs from mouse lung with magnetic sorting
ECs were isolated from a single-cell suspension described above (collagenase and dispase) with magnetic sorting, according to the manufacturer’s instructions (Miltenyi Biotec). CD45−CD31+ cells were collected as ECs, and CD45−CD31− cells were collected as non-ECs.
Immunoblotting
Radio immunoprecipitation assay buffer with protease inhibitor cocktail was used to prepare the lysates. Equal amounts of protein were resolved by NuPAGE 4–12% Bis-Tris gels and transferred to polyvinylidene difluoride membranes. The following primary antibodies were used to detect VEGFR2 (Cell Signaling; #9698), Phospho-VEGFR2 (Cell Signaling; #2478), PCNA (Abcam; ab29), and β-actin (Sigma-Aldrich; A2228). The densitometry of the bands was measured using FIJI running ImageJ software (version 1.52b).
RT-quantitative PCR (qPCR)
cDNA was prepared by using the high-capacity cDNA RT kit (Applied Biosystems). Relative qPCR was performed on a 7500 Fast Real-Time PCR System using primers for Kdr (forward 5′-TTTGGCAAATACAACCCTTCAGA-3′, reverse 5′-GCAGAAGATACTGTCACCACC-3′); mki67 (forward 5′-CTGCCTGTTTGGAAGGAGTAT-3′, reverse 5′-TGCCTCTTGCTCTTTGACTT-3′); and GAPDH (forward 5′-TCAACAGCAACTCCCACTCTTCCA-3′, reverse 5′-ACCCTGTTGCTGTAGCCGTATTCA-3′), and SYBR Green Master Mix (Applied Biosystems).
Lrg1loxP/loxP genotyping
Genotyping of Lrg1loxP/+ × Lrg1loxP/+ breeding was performed to generate Lrg1loxP/loxP. Forward 5′-AGGGGCTGCTTTGTGAGAATGAA-3′ and reverse 5′-CTGGAGGCTGAGGTTAGTGAA-3′ were used to identify insertion of the LoxP site. Bands at 255 bp represent wild-type allele, whereas bands at 289 represent insertion of LoxP site. Both bands signify animals heterozygous for both the LoxP and wild-type allele.
ELISA
Concentrations of analytes in lung lysate were quantified with Duoset ELISAs (R & D) according to the manufacturer’s instructions.
Transcriptomic microarray expression analysis of human LGRC data
Normalized gene expression profiling of chronic lung disease for the LGRC was downloaded from the Gene Expression Omnibus, under accession no. GSE47460. After analyzing clinical metadata, generalized linear models were created for select gene expression and clinical variables, including GOLD status as ordered factors, FEV percentage, DLCO percentage, Emphysema percentage, patient age, sex, and the microarray platform using the function glm in the R stats package. Boxplots and scatterplots were made using ggplot2 R package. Generalized linear model Wald statistics were used to assess the significance of coefficients in relevant models. Genomic data, as well as associated clinical data, are available for download on the LGRC website (https://biolincc.nhlbi.nih.gov/studies/ltrc/).
Isolation of ECs for RNA-seq
7, 14, and 21 d after either elastase or PBS instillation, 25 µg of VE-Cadherin Alexa Fluor 647 (Biolegend; BV13) was injected retro-orbitally under anesthesia 8 min before sacrifice. Lungs were minced and incubated with Collagenase A (2.5 mg/ml; Sigma-Aldrich), Dispase II (1.0 mg/ml; Sigma-Aldrich), and DNase (80 U/ml; Sigma-Aldrich) at 37°C for 30 min. Cells were filtered through a 40-µm filter, and single cells were stained with CD45-FITC (Biolegend; 30-F11), CD31-PE/Cy7 (Biolegend; MEC13.3). CD45−VEcadherin+CD31+ cells were isolated as ECs. ECs were sorted with a BD FACS aria II, at 25 p.s.i., using a 100-µm nozzle. The FACS setup and gating strategy for EC isolations are depicted in Fig. 3 A.
Transcriptome data analysis
RNA library preparation and sequencing
At least 100 ng of total RNA from freshly harvested cells was isolated (phenol-chloroform separation of TRIzol LS) and purified using Qiagen’s RNeasy Mini Kit. RNA quality was verified using an Agilent Technologies 2100 Bioanalyzer before sequencing. RNA library preps were generated and multiplexed using Illumina’s TruSeq RNA Library Preparation Kit v2 (nonstranded and poly-A selection). 10 nM of cDNA was used as input for high-throughput sequencing via Illumina’s HiSeq 4000, producing 51 bp paired-end reads.
Next generation sequencing data processing and statistical analysis for bulk RNA-seq described in Fig. 3, Fig. S3, and Table S1
Sequencing reads were de-multiplexed (bcl2fastq v2.17), checked for quality (FastQC v0.11.5), and trimmed/filtered when appropriate (Trimmomatic v0.36). The resultant high-quality reads were mapped (TopHat2 v2.1.0; Bowtie2 v2.2.6) to the transcriptome sequence reference of the University of California Santa Cruz mm10 genome build. Gene counts were quantified using the Python package HTSeq (v0.11.1). Transcript abundance measures (fragments per kilobase of transcript per million mapped reads [FPKM] values) were quantified using Cufflinks (v2.2.1). Gene-level differential expression analysis was performed using Bioconductor R package DESeq2 (v1.22.2).
Transcriptomic data analyses are summarized in the forms of heatmaps, volcano plots, and gene set enrichment plots. Heatmaps were generated using the CRAN R package pheatmap (v1.0.12). GSEA plots were generated using the R scripts available from the Broad Institute (GSEA v1.0). Volcano plots were generated using the built-in R commands.
Next generation sequencing data processing and statistical analysis for bulk RNA-seq described in Fig. 4 and Fig. S4
Sequence files were checked for quality (FastQC v0.11.5) and processed using the Digital Expression Explorer 2 workflow (Ziemann et al., 2019). Adapter trimming was performed with Skewer (v0.2.2; Jiang et al., 2014). Further adapter quality control was done with Minion, part of the Kraken package (Davis et al., 2013). The resultant filtered reads were mapped to mouse reference genome GRCm38 using STAR aligner, and gene-wise expression counts were generated using the “-quantMode GeneCounts” parameter (Dobin et al., 2013). After further filtering and quality control, R package edgeR (Robinson et al., 2010) was used to calculate trimmed mean of M values library normalized FPKM and Log2 counts per million matrices as well as to perform differential expression analysis. Principal component analysis was performed using Log2 counts per million values. GSEA preranked pathway analysis was run with the University of California San Diego and Broad Institute java application (v4.1.0; Subramanian et al., 2005; Mootha et al., 2003). Gene ontology and Reactome databases were used to perform unsupervised GSEA by log fold change.
Statistics
All data are represented as mean ± SEM. Unpaired Student’s t test was used for the comparisons between two groups, and one-way ANOVA was used for multigroup comparisons. The detailed statistical analyses for RNA-seq data are described in the Materials and methods details of RNA-seq. A two-sided P value <0.05 was statistically significant. All analyses were performed using GraphPad Prism version 5.0 (GraphPad Software). For human studies, generalized linear model Wald statistics were used to assess significance of coefficients in relevant models.
Study approval
All animal experiments and procedures were approved by the Institutional Animal Care and Use Committee at Weill Cornell Medicine. Human tissue samples were obtained from the LTRC repository as described in the Materials and methods section. The LTRC project received approval from the Mayo Clinic institutional review board (IRB number 1640–04). Informed consent for this study was covered under the initial enrollment into the LTRC database.
Online supplemental material
Fig. S1 shows the expression of key epithelial marks is maintained in human COPD lung tissue. Fig. S2 provides an example of the flow gating strategy for ECs (CD31+VEcadherin+ cells) and epithelial cells (EpCAM+). Fig. S3 shows that intravenous delivery of lung-specific ECs is required to recover physiological pulmonary function following elastase treatment. Fig. S4 presents GSEA of RNA-seq profiles in purified lung ECs isolated from 1-mo– and 6-mo–CS-exposed mice. Fig. S5 shows LRG1, PECAM-1, SPC, and DAPI staining in control and COPD human lungs. Table S1 summarizes the transcriptomic analysis of elastase: differential expression analysis, gene ontology, and GSEA. Table S2 summarizes the transcriptomic analysis of CS: differential expression analysis and GSEA.
Supplementary Material
summarizes the transcriptomic analysis of elastase.
summarizes the transcriptomic analysis of CS.
Acknowledgments
The authors thank Nanjing University for the gift of the Lrg1loxP/+ mice and Ralf H. Adams for the VE-Cadherin-CreERT2. The authors thank Deebly Chavez and Freddy Reyes for animal husbandry and also thank the Genomic Resources Core Facility and the Translational Research Program at Pathology Laboratory Medicine for assistance and support in this study.
This work was supported by National Institutes of Health grants R01HL132198, P01Hl114501 (to A.M.K. Choi), R00 HL125899 (to S.M. Cloonan), 5T32HL134629 (to A.C. Racanelli), R35 HL150809, U01 AI 138329, RC2 DK11477 (to S. Rafii), CRIM (to M. Ginsberg and D.J. Nolan), T32 HD060600, and RFA#0906290930 (to B. Kunar), Leukemia and Lymphoma Society (to J.M. Scandura), the Cancer Research and Treatment Fund (to J.M. Scandura, P. Kermani), the Taub Foundation (to J.M. Scandura, P. Kermani), the National Heart, Lung, and Blood Institute (to J.M. Scandura, P.Kermani), the Daedalus Fund for Innovation from Weill Cornell Medicine, and the Tri-Institutional Stem Cell Initiatives (TRI-SCI 2019-029 to S. Rafii).
Author contributions: S. Hisata, A.C. Racanelli, S.M. Cloonan, A.M.K. Choi, and S. Rafii conceived and designed the study. S. Hisata, A.C. Racanelli, B. Palikuqi, B. Kunar, A. Zhou, P. Kermani, D.J. Nolan, M. Ginsberg, and B.-S. Ding performed experiments. K. McConn analyzed histology to obtain mean linear intercept (chord) length data. R. Schreiner provided technical support and expertise in image acquisition. B. Kunar performed analysis for mouse RNA-seq data. D. Redmond and S. Houghton performed analyses of microarray and RNA-seq datasets obtained from human LTRC and non-EC and EC fractions and smoke RNA-seq. R. Schreiner performed imaging of human and mouse lung tissues. A. Capili performed data analysis. S. Hisata, A.C. Racanelli, B. Palikuqi, B. Kunar, P. Kermani, D. Redmond, D.J. Nolan, M. Ginsberg, B.-S. Ding, F.J. Martinez, J.M. Scandura, S.M. Cloonan, S. Rafii, and A.M.K. Choi provided critical discussions for data interpretation. A.C. Racanelli, S. Hisata, S.M. Cloonan, A.M.K. Choi, and S. Rafii wrote the manuscript, and all coauthors reviewed and approved the final manuscript.
Data availability
Both raw and processed sequencing data generated in this study are available under Gene Expression Omnibus accession number GSE140413 as FASTQ and .TSV files. In addition, publicly available datasets that were reanalyzed for the purpose of this study are available under Gene Expression Omnibus accession number GSE47460.
References
- Andersen, J.D., Boylan K.L., Jemmerson R., Geller M.A., Misemer B., Harrington K.M., Weivoda S., Witthuhn B.A., Argenta P., Vogel R.I., and Skubitz A.P.. 2010. Leucine-rich alpha-2-glycoprotein-1 is upregulated in sera and tumors of ovarian cancer patients. J. Ovarian Res. 3:21. 10.1186/1757-2215-3-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin, H.G., and Koh G.Y.. 2017. Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science. 357:eaal2379. 10.1126/science.aal2379 [DOI] [PubMed] [Google Scholar]
- Barr, R.G., Mesia-Vela S., Austin J.H., Basner R.C., Keller B.M., Reeves A.P., Shimbo D., and Stevenson L.. 2007. Impaired flow-mediated dilation is associated with low pulmonary function and emphysema in ex-smokers: the Emphysema and Cancer Action Project (EMCAP) Study. Am. J. Respir. Crit. Care Med. 176:1200–1207. 10.1164/rccm.200707-980OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, J.M., Nolan D.J., Vertes E.L., Varnum-Finney B., Kobayashi H., Hooper A.T., Seandel M., Shido K., White I.A., Kobayashi M., et al. 2010. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 6:251–264. 10.1016/j.stem.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning, B.J., and Wright J.L.. 2008. Animal models of asthma and chronic obstructive pulmonary disease. Pulm. Pharmacol. Ther. 21:695. 10.1016/j.pupt.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Cao, Z., Ding B.S., Guo P., Lee S.B., Butler J.M., Casey S.C., Simons M., Tam W., Felsher D.W., Shido K., et al. 2014. Angiocrine factors deployed by tumor vascular niche induce B cell lymphoma invasiveness and chemoresistance. Cancer Cell. 25:350–365. 10.1016/j.ccr.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Z., Ye T., Sun Y., Ji G., Shido K., Chen Y., Luo L., Na F., Li X., Huang Z., et al. 2017. Targeting the vascular and perivascular niches as a regenerative therapy for lung and liver fibrosis. Sci. Transl. Med. 9:eaai8710. 10.1126/scitranslmed.aai8710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet, P.2005. Angiogenesis in life, disease and medicine. Nature. 438:932–936. 10.1038/nature04478 [DOI] [PubMed] [Google Scholar]
- Chen, Z.H., Lam H.C., Jin Y., Kim H.P., Cao J., Lee S.J., Ifedigbo E., Parameswaran H., Ryter S.W., and Choi A.M.. 2010. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc. Natl. Acad. Sci. USA. 107:18880–18885. 10.1073/pnas.1005574107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, K.F., and Adcock I.M.. 2008. Multifaceted mechanisms in COPD: inflammation, immunity, and tissue repair and destruction. Eur. Respir. J. 31:1334–1356. 10.1183/09031936.00018908 [DOI] [PubMed] [Google Scholar]
- Cloonan, S.M., and Choi A.M.. 2016. Mitochondria in lung disease. J. Clin. Invest. 126:809–820. 10.1172/JCI81113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, M.P., van Dongen S., Abreu-Goodger C., Bartonicek N., and Enright A.J.. 2013. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods. 63:41–49. 10.1016/j.ymeth.2013.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisser, H.M., Helmke B.P., Cao G., Egan P.M., Taichman D., Fehrenbach M., Zaman A., Cui Z., Mohan G.S., Baldwin H.S., et al. 2006. Loss of PECAM-1 function impairs alveolarization. J. Biol. Chem. 281:8724–8731. 10.1074/jbc.M511798200 [DOI] [PubMed] [Google Scholar]
- Ding, B.S., Nolan D.J., Guo P., Babazadeh A.O., Cao Z., Rosenwaks Z., Crystal R.G., Simons M., Sato T.N., Worgall S., et al. 2011. Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell. 147:539–553. 10.1016/j.cell.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B.S., Cao Z., Lis R., Nolan D.J., Guo P., Simons M., Penfold M.E., Shido K., Rabbany S.Y., and Rafii S.. 2014. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature. 505:97–102. 10.1038/nature12681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh-Xuan, A.T., Higenbottam T.W., Clelland C.A., Pepke-Zaba J., Cremona G., Butt A.Y., Large S.R., Wells F.C., and Wallwork J.. 1991. Impairment of endothelium-dependent pulmonary-artery relaxation in chronic obstructive lung disease. N. Engl. J. Med. 324:1539–1547. 10.1056/NEJM199105303242203 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff, P., Valipour A., Kiss D., Schreder M., Cekici L., Geyer K., Kohansal R., and Burghuber O.C.. 2008. Determinants of systemic vascular function in patients with stable chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 178:1211–1218. 10.1164/rccm.200709-1412OC [DOI] [PubMed] [Google Scholar]
- Estépar, R.S., Kinney G.L., Black-Shinn J.L., Bowler R.P., Kindlmann G.L., Ross J.C., Kikinis R., Han M.K., Come C.E., Diaz A.A., et al. COPDGene Study . 2013. Computed tomographic measures of pulmonary vascular morphology in smokers and their clinical implications. Am. J. Respir. Crit. Care Med. 188:231–239. 10.1164/rccm.201301-0162OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg, J.C., Chu F., Utokaparch S., Woods R., Elliott W.M., Buzatu L., Cherniack R.M., Rogers R.M., Sciurba F.C., Coxson H.O., and Paré P.D.. 2004. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N. Engl. J. Med. 350:2645–2653. 10.1056/NEJMoa032158 [DOI] [PubMed] [Google Scholar]
- Honda, H., Fujimoto M., Serada S., Urushima H., Mishima T., Lee H., Ohkawara T., Kohno N., Hattori N., Yokoyama A., and Naka T.. 2017. Leucine-rich α-2 glycoprotein promotes lung fibrosis by modulating TGF-β signaling in fibroblasts. Physiol. Rep. 5:e13556. 10.14814/phy2.13556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Q., Zhang L., Fu J., Verghese D.A., Chauhan K., Nadkarni G.N., Li Z., Ju W., Kretzler M., Cai G.Y., et al. 2019. LRG1 Promotes Diabetic Kidney Disease Progression by Enhancing TGF-β-Induced Angiogenesis. J. Am. Soc. Nephrol. 30:546–562. 10.1681/ASN.2018060599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, K.S., Newell J.D. Jr., Jin D., Fuld M.K., Saha P.K., Hansdottir S., and Hoffman E.A.. 2016. Quantitative Dual-Energy Computed Tomography Supports a Vascular Etiology of Smoking-induced Inflammatory Lung Disease. Am. J. Respir. Crit. Care Med. 193:652–661. 10.1164/rccm.201506-1196OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakkula, M., Le Cras T.D., Gebb S., Hirth K.P., Tuder R.M., Voelkel N.F., and Abman S.H.. 2000. Inhibition of angiogenesis decreases alveolarization in the developing rat lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 279:L600–L607. 10.1152/ajplung.2000.279.3.L600 [DOI] [PubMed] [Google Scholar]
- Jiang, H., Lei R., Ding S.W., and Zhu S.. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 15:182. 10.1186/1471-2105-15-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J., Sun H., Liu D., Wang H., Liu Q., Chen H., Zhong D., and Li G.. 2019. LRG1 Promotes Apoptosis and Autophagy through the TGFβ-smad1/5 Signaling Pathway to Exacerbate Ischemia/Reperfusion Injury. Neuroscience. 413:123–134. 10.1016/j.neuroscience.2019.06.008 [DOI] [PubMed] [Google Scholar]
- Kasahara, Y., Tuder R.M., Taraseviciene-Stewart L., Le Cras T.D., Abman S., Hirth P.K., Waltenberger J., and Voelkel N.F.. 2000. Inhibition of VEGF receptors causes lung cell apoptosis and emphysema. J. Clin. Invest. 106:1311–1319. 10.1172/JCI10259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara, Y., Tuder R.M., Cool C.D., Lynch D.A., Flores S.C., and Voelkel N.F.. 2001. Endothelial cell death and decreased expression of vascular endothelial growth factor and vascular endothelial growth factor receptor 2 in emphysema. Am. J. Respir. Crit. Care Med. 163:737–744. 10.1164/ajrccm.163.3.2002117 [DOI] [PubMed] [Google Scholar]
- Kropski, J.A., Richmond B.W., Gaskill C.F., Foronjy R.F., and Majka S.M.. 2018. Deregulated angiogenesis in chronic lung diseases: a possible role for lung mesenchymal progenitor cells (2017 Grover Conference Series). Pulm. Circ. 8:2045893217739807. 10.1177/2045893217739807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumbe, A.P., Ramasamy S.K., Itkin T., Mäe M.A., Langen U.H., Betsholtz C., Lapidot T., and Adams R.H.. 2016. Corrigendum: Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature. 539:314. 10.1038/nature19782 [DOI] [PubMed] [Google Scholar]
- Laucho-Contreras, M.E., Taylor K.L., Mahadeva R., Boukedes S.S., and Owen C.A.. 2015. Automated measurement of pulmonary emphysema and small airway remodeling in cigarette smoke-exposed mice. J. Vis. Exp. (95):52236. 10.3791/52236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H., Bhang D.H., Beede A., Huang T.L., Stripp B.R., Bloch K.D., Wagers A.J., Tseng Y.H., Ryeom S., and Kim C.F.. 2014. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell. 156:440–455. 10.1016/j.cell.2013.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebow, A.A.1959. Pulmonary emphysema with special reference to vascular changes. Am. Rev. Respir. Dis. 80:67–93. [DOI] [PubMed] [Google Scholar]
- Lindén, M., Lind S.B., Mayrhofer C., Segersten U., Wester K., Lyutvinskiy Y., Zubarev R., Malmström P.U., and Pettersson U.. 2012. Proteomic analysis of urinary biomarker candidates for nonmuscle invasive bladder cancer. Proteomics. 12:135–144. 10.1002/pmic.201000810 [DOI] [PubMed] [Google Scholar]
- Minet, C., Vivodtzev I., Tamisier R., Arbib F., Wuyam B., Timsit J.F., Monneret D., Borel J.C., Baguet J.P., Lévy P., and Pépin J.L.. 2012. Reduced six-minute walking distance, high fat-free-mass index and hypercapnia are associated with endothelial dysfunction in COPD. Respir. Physiol. Neurobiol. 183:128–134. 10.1016/j.resp.2012.06.017 [DOI] [PubMed] [Google Scholar]
- Mitzner, W.2008. Use of mean airspace chord length to assess emphysema. J Appl Physiol (1985). 105:1980–1981. 10.1152/japplphysiol.90968.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha, V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Moro, L., Pedone C., Scarlata S., Malafarina V., Fimognari F., and Antonelli-Incalzi R.. 2008. Endothelial dysfunction in chronic obstructive pulmonary disease. Angiology. 59:357–364. 10.1177/0003319707306141 [DOI] [PubMed] [Google Scholar]
- Nolan, D.J., Ginsberg M., Israely E., Palikuqi B., Poulos M.G., James D., Ding B.S., Schachterle W., Liu Y., Rosenwaks Z., et al. 2013. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell. 26:204–219. 10.1016/j.devcel.2013.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado, V.I., Barbera J.A., Ramirez J., Gomez F.P., Roca J., Jover L., Gimferrer J.M., and Rodriguez-Roisin R.. 1998. Endothelial dysfunction in pulmonary arteries of patients with mild COPD. Am. J. Physiol. 274:L908–L913. 10.1152/ajplung.1998.274.6.L908 [DOI] [PubMed] [Google Scholar]
- Poulos, M.G., Ramalingam P., Gutkin M.C., Llanos P., Gilleran K., Rabbany S.Y., and Butler J.M.. 2017. Endothelial transplantation rejuvenates aged hematopoietic stem cell function. J. Clin. Invest. 127:4163–4178. 10.1172/JCI93940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaderi, S.A., and Hurst J.R.. 2018. The unmet global burden of COPD. Glob. Health. Epidemiol. Genom. 3:e4. 10.1017/gheg.2018.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii, S., Butler J.M., and Ding B.S.. 2016. Angiocrine functions of organ-specific endothelial cells. Nature. 529:316–325. 10.1038/nature17040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seandel, M., Butler J.M., Kobayashi H., Hooper A.T., White I.A., Zhang F., Vertes E.L., Kobayashi M., Zhang Y., Shmelkov S.V., et al. 2008. Generation of a functional and durable vascular niche by the adenoviral E4ORF1 gene. Proc. Natl. Acad. Sci. USA. 105:19288–19293. 10.1073/pnas.0805980105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siempos, I.I., Ma K.C., Imamura M., Baron R.M., Fredenburgh L.E., Huh J.-W., Moon J.-S., Finkelsztein E.J., Jones D.S., Lizardi M.T., et al. 2018. RIPK3 mediates pathogenesis of experimental ventilator-induced lung injury. JCI Insight. 3:e97102. 10.1172/jci.insight.97102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.C., and Mitzner W.. 1980. Analysis of pulmonary vascular interdependence in excised dog lobes. J. Appl. Physiol. 48:450–467. 10.1152/jappl.1980.48.3.450 [DOI] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suki, B., Bartolák-Suki E., and Rocco P.R.M.. 2017. Elastase-Induced Lung Emphysema Models in Mice. Methods Mol. Biol. 1639:67–75. 10.1007/978-1-4939-7163-3_7 [DOI] [PubMed] [Google Scholar]
- Voelkel, N.F.2018. Cigarette Smoke Is an Endothelial Cell Toxin. Am. J. Respir. Crit. Care Med. 197:274. 10.1164/rccm.201706-1123LE [DOI] [PubMed] [Google Scholar]
- Vukic Dugac, A., Ruzic A., Samarzija M., Badovinac S., Kehler T., and Jakopovic M.. 2015. Persistent endothelial dysfunction turns the frequent exacerbator COPD from respiratory disorder into a progressive pulmonary and systemic vascular disease. Med. Hypotheses. 84:155–158. 10.1016/j.mehy.2014.11.017 [DOI] [PubMed] [Google Scholar]
- Wang, X., Abraham S., McKenzie J.A.G., Jeffs N., Swire M., Tripathi V.B., Luhmann U.F.O., Lange C.A.K., Zhai Z., Arthur H.M., et al. 2013. LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature. 499:306–311. 10.1038/nature12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Xu Z., Chen B., He W., Hu J., Zhang L., Liu X., and Chen F.. 2017. The Role of Vascular Endothelial Growth Factor in Small-airway Remodelling in a Rat Model of Chronic Obstructive Pulmonary Disease. Sci. Rep. 7:41202. 10.1038/srep41202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebe, B.M., and Laursen H.. 1998. Lung morphometry by unbiased methods in emphysema: bronchial and blood vessel volume, alveolar surface area and capillary length. APMIS. 106:651–656. 10.1111/j.1699-0463.1998.tb01395.x [DOI] [PubMed] [Google Scholar]
- Ziemann, M., Kaspi A., and El-Osta A.. 2019. Digital expression explorer 2: a repository of uniformly processed RNA sequencing data. Gigascience. 8:giz022. 10.1093/gigascience/giz022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
summarizes the transcriptomic analysis of elastase.
summarizes the transcriptomic analysis of CS.
Data Availability Statement
Both raw and processed sequencing data generated in this study are available under Gene Expression Omnibus accession number GSE140413 as FASTQ and .TSV files. In addition, publicly available datasets that were reanalyzed for the purpose of this study are available under Gene Expression Omnibus accession number GSE47460.