Abstract
Dermatofibrosarcoma protuberans (DFSP) is a spindle cell neoplasm of the skin and superficial soft tissue with a tendency for locally aggressive behavior; metastatic potential coincides with fibrosarcomatous transformation. The vast majority of DFSPs harbor the t(17;22) translocation resulting in a COL1A1-PDGFB fusion that drives autocrine growth stimulation via PDGFB overexpression. Here, we examined the utility of PDGFB RNA chromogenic in situ hybridization (CISH) for the diagnosis of DFSP. A total of 337 tumors represented in whole tissue sections and tissue microarrays, including 37 cases of DFSP and 300 histologically similar spindle cell tumors, were subjected to PDGFB RNA CISH using commercially available probes. PDGFB overexpression was observed by light microscopy in 24 of 26 conventional DFSPs (92%) and 11 of 11 fibrosarcomatous DFSPs (100%). One of two DFSPs negative for PDGFB by RNA CISH was found to harbor an uncommon alternative rearrangement involving PDGFD. All examined cases of histologic mimics were negative for PDGFB overexpression; limited PDGFB expression, not reaching an empirical threshold of greater than 5 puncta or one aggregate of chromogen in more than 25% of cells, was observed in 7 of 300 mimics (2.3%), including desmoplastic melanoma, malignant peripheral nerve sheath tumor, angiosarcoma, and pleomorphic dermal sarcoma. Vascular PDGFB expression was seen in several tumor types. We conclude that PDGFB RNA CISH, with careful interpretation and the use of appropriate thresholds, may serve as a surrogate marker of PDGFB rearrangement and a useful ancillary tool for the diagnosis of DFSP.
Keywords: PDGFB, dermatofibrosarcoma protuberans, in situ hybridization, fusion gene, sarcoma, soft tissue tumors
INTRODUCTION
Dermatofibrosarcoma protuberans (DFSP) is a superficial, fibroblastic spindle cell neoplasm of intermediate biologic potential manifesting with locally aggressive behavior. Clinically, DFSP presents as a slow-growing nodular cutaneous or soft tissue mass, most commonly affecting the trunk and proximal extremities, and tends to occur in young to middle-aged adults, with a slight male predominance [1]. Histologically, these neoplasms are characterized by a dermal-based proliferation of uniform spindle cells with infiltration into the subcutis, a storiform growth pattern, minimal cytologic atypia, and infrequent mitoses.
Less commonly, DFSP can have a prominent myxoid matrix and lose its storiform architecture, potentially simulating other myxoid mesenchymal neoplasms [2]. More rarely, tumors can entrap pigmented dendritic melanocytes (Bednar tumor) or exhibit nodules of myoid differentiation [3, 4]. DFSP can also transform into a sarcoma (fibrosarcomatous DFSP) with a capacity for more aggressive behavior, including metastatic spread [5, 6]. In the fibrosarcomatous variant of DFSP, tumor cells are arranged as highly cellular fascicles in a distinct herringbone pattern, and nuclear atypia and mitotic activity are more pronounced [5, 7].
Establishing the diagnosis of DFSP and excluding benign and malignant histologic mimics may require a combination of immunohistochemical and molecular studies, particularly in the setting of a limited biopsy sample or unusual morphology. By immunohistochemistry, most cases of conventional DFSP are diffusely positive for CD34 expression [8, 9]. However, CD34 has low specificity, and up to half of cases of fibrosarcomatous DFSP lose expression of CD34 [5, 7]. Other widely available immunohistochemical markers are consistently negative in these tumors.
Most cases of DFSP harbor the recurrent unbalanced chromosome translocation t(17;22)(q21;q13), commonly in the form of a supernumerary ring chromosome, resulting in a COL1A1-PDGFB chimeric gene [10]. Rare cryptic fusions and alternative rearrangements involving the related PDGFD gene have also been reported [11, 12]. In clinical practice, the molecular confirmation of DFSP often relies on DNA fluorescence in situ hybridization (FISH), reverse transcription-polymerase chain reaction (RT-PCR), or next generation sequencing (NGS). Although these methods have been shown to be sensitive and specific for the detection of COL1A1-PDGFB in DFSP [13–16], they do not allow for analysis with conventional light microscopy and each typically takes several days to perform in most clinical laboratories. Here, we examine the diagnostic utility of an RNA-based chromogenic in situ hybridization (CISH) approach targeting PDGFB to aid in the histologic identification of DSFP. Using formalin-fixed paraffin-embedded (FFPE) archival specimens and commercially available probes, we show that PDGFB RNA CISH represents a histologic tool to support the diagnosis of DFSP.
MATERIALS AND METHODS
Cases were retrieved from the surgical pathology archives of Stanford Hospital and Brigham and Women’s Hospital under an Institutional Review Board-approved protocol. Representative hematoxylin and eosin (H&E) stained slides were reviewed to confirm the diagnosis. A combination of whole tissue sections (WTS) and tissue microarrays (TMA) were used to evaluate 337 cases altogether: DFSP (8 WTS, 18 TMA); fibrosarcomatous DFSP (2 WTS, 9 TMA); dermatofibroma (1 WTS, 13 TMA); cellular dermatofibroma (1 WTS, 10 TMA); atypical fibrous histiocytoma (4 TMA);angiomatoid fibrous histiocytoma (2 TMA); superficial acral fibromyxoma (3 TMA); solitary fibrous tumor (2 WTS, 13 TMA); atypical fibroxanthoma (8 TMA); pleomorphic dermal sarcoma (8 TMA); Kaposi sarcoma (3 TMA); angiosarcoma (12 TMA); desmoid fibromatosis (28 TMA); nodular fasciitis (8 TMA); leiomyosarcoma (57 TMA); monophasic synovial sarcoma (13 TMA); desmoplastic melanoma (9 TMA); malignant peripheral nerve sheath tumor (65 TMA); and neurofibroma (41 TMA). The TMAs were constructed using a tissue arrayer (Beecher Instruments, Silver Spring, MD) as previously described [17]. Tissues were evaluated as at least 0.6-mm cores taken from representative areas of each FFPE block. Cores were not considered if targeted tissue was not included on the array, as assessed morphologically for each core.
PDFGB RNA CISH was performed using the RNAscope 2.5 high definition assay using probe sets targeting bases 1995–3047 (designated PDGFB-1) or 665–2037 (PDGFB-2) of the human PDGFB gene (catalog numbers 441578 and 406708, respectively; Advanced Cell Diagnostics, Hayward, CA). These bases are distal to the fusion breakpoint and are conserved in COL1A1-PDGFB rearrangements. Hybridization was completed manually on 4-μm histologic sections of FFPE tissues according to the manufacturer’s recommended protocol. For all examined cases, histologic sections were cut from the tissue block less than 4 weeks prior to hybridization with the exception of the PDGFD-rearranged DFSP. A case of DFSP with known COL1A1-PDGFB rearrangement served as an external positive control for PDGFB expression. The housekeeping gene peptidylprolyl isomerase B (PPIB) provided control for RNA integrity.
Conventional light microscopy was used to visualize mRNA expression manifesting as red (Fast Red) intracellular puncta or aggregates as previously described [18]. Tumors were considered positive for PDGFB overexpression if greater than 25% of neoplastic cells exhibited greater than 5 puncta or at least one aggregate. Any expression below this threshold, present in greater than 1% of neoplastic cells, was considered limited. In parallel, a semiquantitative scoring system was used to assess the degree of PDGFB expression according to the lowest objective magnification at which chromogen was visualized (2× or 4×: 3+, 10×: 2+, 20× or 40×: 1+), as in previous studies [19].
RESULTS
The 26 conventional DFSPs included in this study affected 15 men and 11 women with a median age of 39 years (range 3 to 73 y). The tumors arose in the trunk (n=14), extremities (n=8), head/neck (n=3), and breast (n=1). Characteristic morphologic features of DFSP, including storiform architecture and bland cytomorphology with limited mitotic activity, were present in 23 cases; two additional cases were characterized by abundant myxoid stroma and one by pigmentation (Bednar tumor). Immunohistochemistry for CD34 was performed in 22 cases, 20 of which exhibited strong, diffuse expression; CD34 expression was heterogeneous in the other two cases. The diagnosis of DFSP was established by morphology and immunohistochemistry in the majority of cases (n=20). In 6 cases, the diagnosis was informed by identification of a PDGFB translocation using conventional cytogenetics (n=2) or break-apart FISH (n=4).
PDGFB overexpression was observed by RNA CISH in 24 of 26 conventional DFSPs (92%) using the PDGFB-1 probe set (Figure 1, Table 1). Diffuse overexpression (more than 5 puncta or a single aggregate of chromogen in greater than 90% of neoplastic cells) was evident in 21 of 24 positive tumors (88%); at a minimum, expression was seen in 50% of cells in one case. While the majority of tumors exhibited diffuse intracellular chromogen aggregates (e.g., Figure 1A–B), PDGFB overexpression manifested as scattered discrete cytoplasmic puncta in a subset (e.g., Figure 1C–D). Using a semiquantitative scoring system based on the lowest magnification required for visual chromogen detection, 20 of 24 PDGFB-positive DFSPs (83%) received a score of 3+; all other positive cases received a score of 2+. Strong and diffuse PDGFB overexpression was identified in 4 cases with confirmation of PDGFB rearrangement by break-apart FISH and in 2 cases with t(17;22) by conventional cytogenetics. Overexpression was seen in the two DFSPs with prominent myxoid stroma and the pigmented DFSP.
Figure 1.
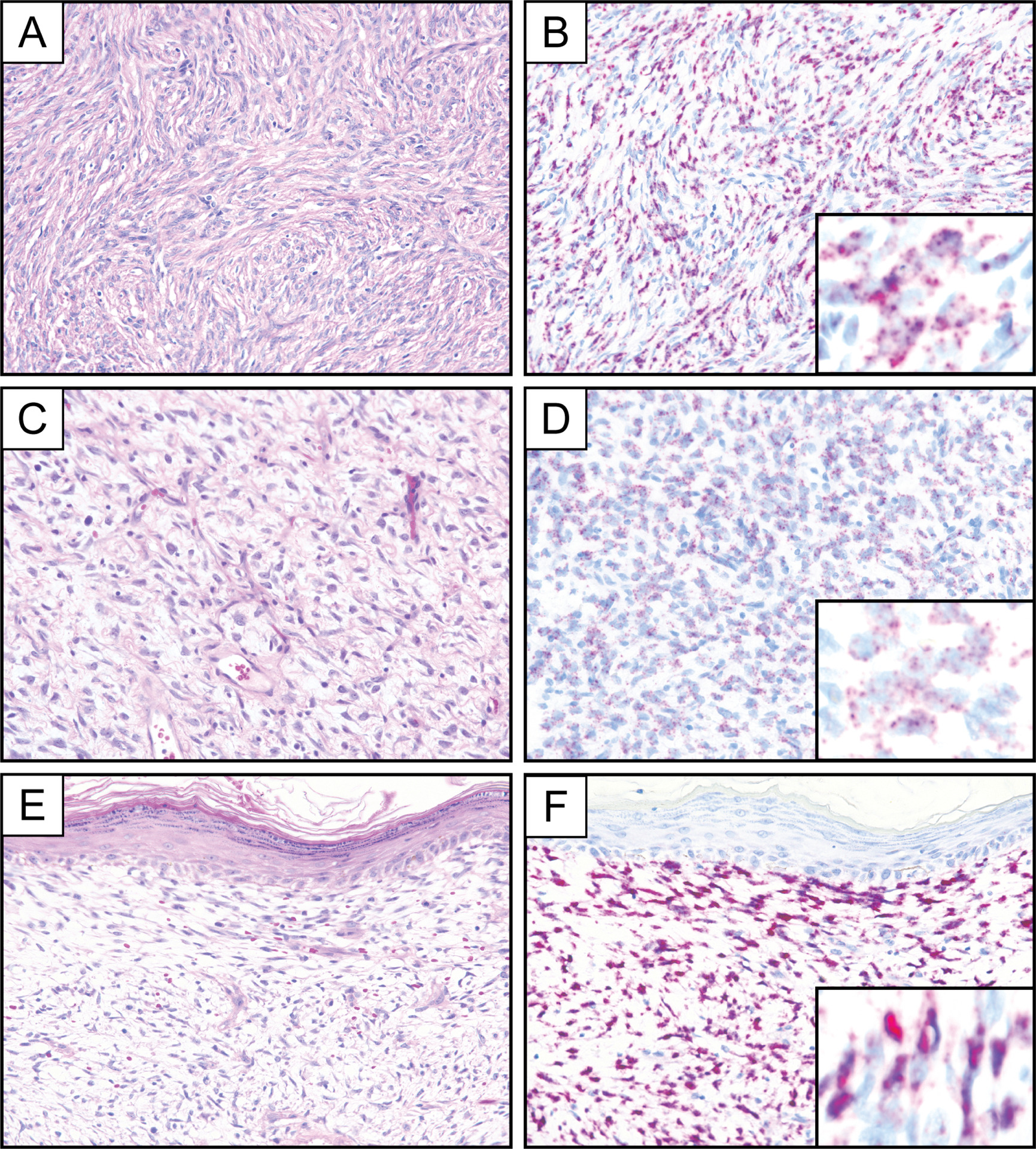
Representative photomicrographs of hematoxylin and eosin stain and PDGFB RNA chromogenic in situ hybridization (PDGFB-1 probe set) demonstrating diffuse and strong PDGFB expression in a conventional DFSP with characteristic storiform architecture (A, B). PDGFB expression in DFSP can manifest by RNA chromogenic in situ hybridization as numerous small puncta (C, D) or as larger subcellular aggregates (E, F). Insets represent PDGFB RNA chromogenic in situ hybridization at high magnification.
Table 1.
Summary of PDGFB expression by RNA chromogenic in situ hybridization
| Tumor type | Total cases | PDGFB overexpression (%) | PDGFB limited (%) | PDGFB negative (%) |
|---|---|---|---|---|
| Conventional DFSP | 26 | 24 (92) | 0 (0) | 2 (8) |
| PDGFD-rearranged DFSP | 1 | 0 | 0 (0) | 1 (100%) |
| Fibrosarcomatous DFSP | 11 | 11 (100) | 0 (0) | 0 (0) |
| Dermatofibroma | 14 | 0 (0) | 0 (0) | 14 (100) |
| Cellular dermatofibroma | 11 | 0 (0) | 0 (0) | 11 (100) |
| Atypical fibrous histiocytoma | 4 | 0 (0) | 0 (0) | 4 (100) |
| Angiomatoid fibrous histiocytoma | 2 | 0 (0) | 0 (0) | 2 (100) |
| Superficial acral fibromyxoma | 3 | 0 (0) | 0 (0) | 3 (100) |
| Atypical fibroxanthoma | 8 | 0 (0) | 0 (0) | 8 (100) |
| Pleomorphic dermal sarcoma | 8 | 0 (0) | 2 (25) | 6 (75) |
| Solitary fibrous tumor | 15 | 0 (0) | 0 (0) | 15 (100) |
| Synovial sarcoma (monophasic) | 13 | 0 (0) | 0 (0) | 13 (100) |
| Angiosarcoma | 12 | 0 (0) | 1 (8) | 11 (92) |
| Kaposi sarcoma | 3 | 0 (0) | 0 (0) | 3 (100) |
| Melanoma (desmoplastic) | 9 | 0 (0) | 2 (22) | 7 (78) |
| Nodular fasciitis | 8 | 0 (0) | 0 (0) | 8 (100) |
| Desmoid fibromatosis | 28 | 0 (0) | 0 (0) | 28 (100) |
| Leiomyosarcoma (extrauterine) | 57 | 0 (0) | 0 (0) | 57 (100) |
| Neurofibroma | 41 | 0 (0) | 0 (0) | 41 (100) |
| MPNST | 65 | 0 (0) | 2 (3) | 63 (97) |
DFSP, dermatofibrosarcoma protuberans; MPNST, malignant peripheral nerve sheath tumor
Eleven additional fibrosarcomatous DFSPs, each exhibiting characteristic fascicular architecture, were strongly positive for PDGFB overexpression using the PDGFB-1 probe set (11/11, 100%). This group of fibrosarcomatous DFSPs included 2 cases with COL1A1-PDGFB confirmed by targeted hybrid capture-based RNA-sequencing [20, 21]. All fibrosarcomatous DFSPs showed diffuse PDGFB overexpression with a score of 3+, which was qualitatively stronger on average than that seen in conventional DFSP (Figure 2). In comparison, there was at least partial loss of CD34 expression by immunohistochemistry in 7 of 11 fibrosarcomatous DFSPs.
Figure 2.
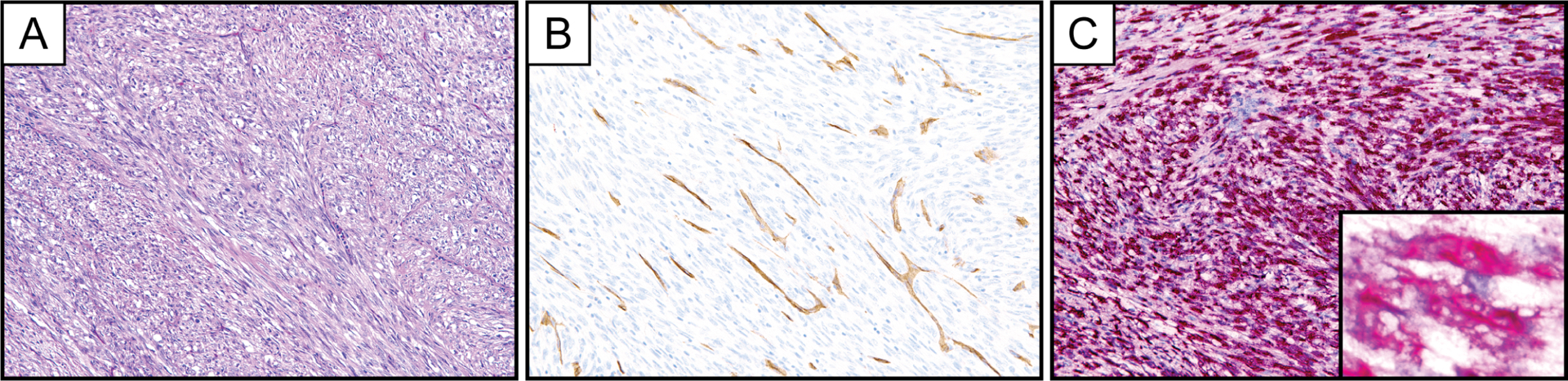
Representative photomicrographs of hematoxylin and eosin stain (A), CD34 immunohistochemistry (B), and PDGFB RNA chromogenic in situ hybridization (C, PDGFB-1 probe set) in a fibrosarcomatous DFSP with diffuse and strong PDGFB expression. The COL1A1-PDGFB translocation was identified in this case by targeted next generation sequencing. Inset represents PDGFB RNA chromogenic in situ hybridization at high magnification.
A second probe set (PDGFB-2) exhibited a staining pattern similar to that of PDGFB-1 in 12 conventional DFSPs and 3 fibrosarcomatous DFSPs (Figure 3). Although the staining patterns of these two probe sets did not differ in terms of either the percentage of tumor cells staining positive for PDGFB or the semiquantitative assessment of staining intensity, the PDGFB-1 probe set qualitatively exhibited somewhat more intense staining in both conventional and fibrosarcomatous DFSP.
Figure 3.
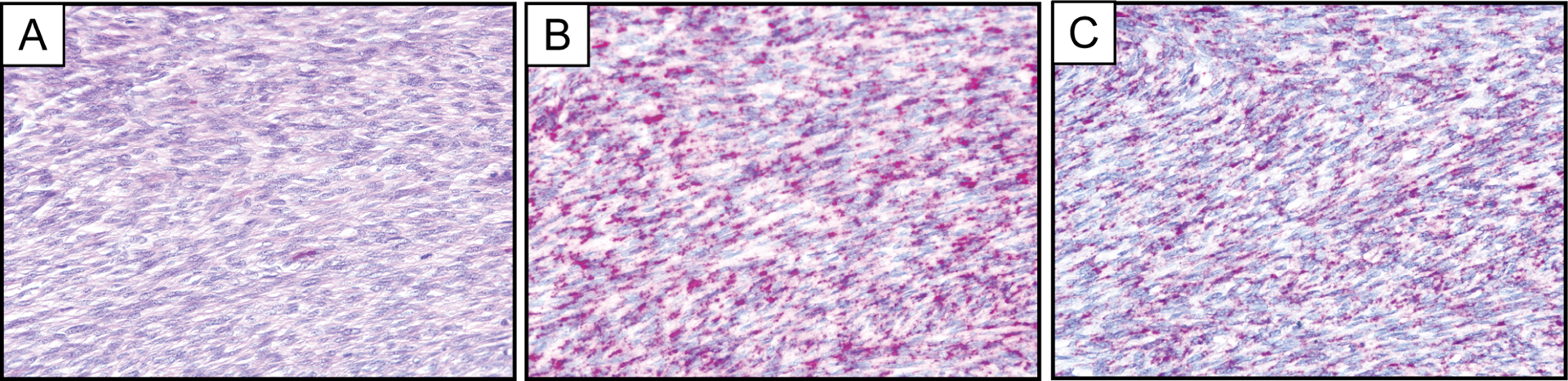
Representative photomicrographs of hematoxylin and eosin stain (A) and PDGFB RNA chromogenic in situ hybridization in a fibrosarcomatous DFSP comparing the PDGFB-1 (B) and PDGFB-2 (C) probe sets.
Both conventional DFSP cases lacking PDGFB expression by RNA CISH using the PDGFB-1 probe set manifested histologically as spindle cell neoplasms exhibiting characteristic storiform architecture and diffuse CD34 expression. One case, which involved the breast of a female patient, was found to harbor a PDGFD rearrangement (Figure 4). This observation suggests that the finding of PDGFB overexpression by RNA CISH using the PDGFB-1 probe set may distinguish DFSPs bearing PDGFB rearrangements from the much rarer tumors exhibiting PDGFD rearrangement. Genetic characterization of the second PDGFB RNA CISH-negative case was not performed. This case, which presented as a superficial mass on the trunk of a 70-year-old male patient, retained PPIB expression, indicating generally preserved RNA integrity. The case was evaluated in a single iteration of RNA CISH using the PDGFB-1 probe set with several other DFSPs in the same histologic section of the TMA serving as positive control.
Figure 4.
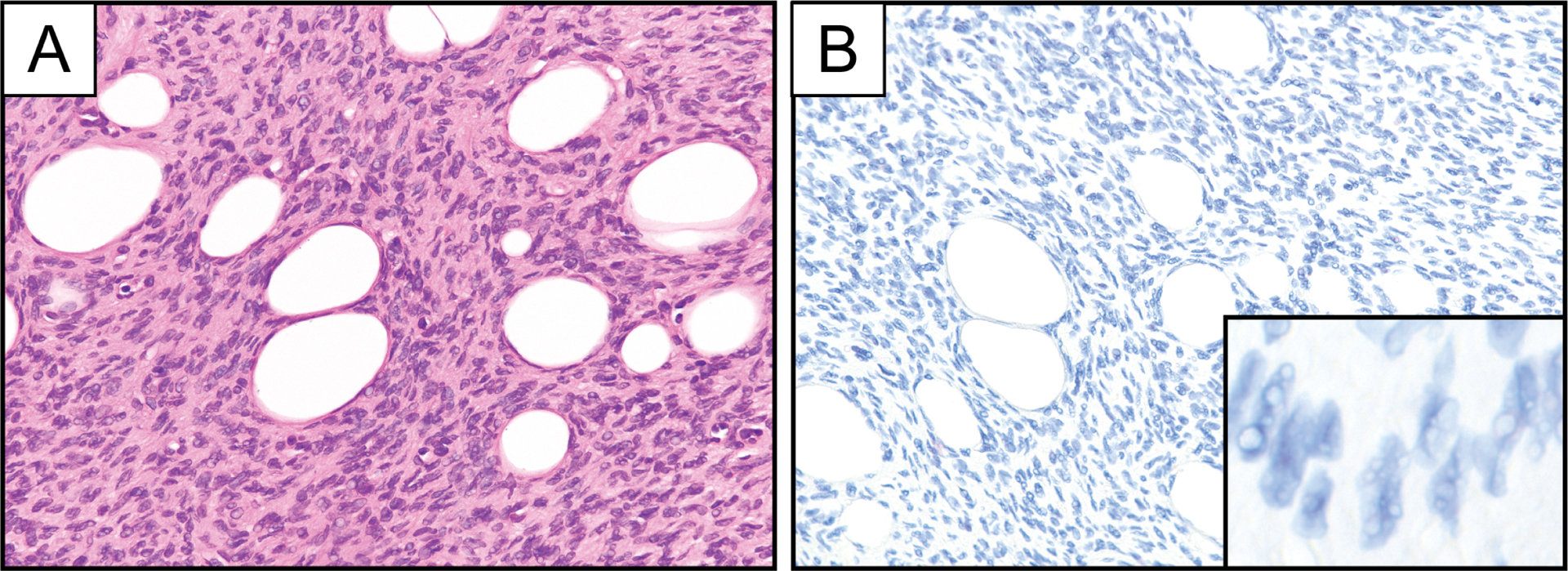
Representative photomicrographs of hematoxylin and eosin stain (A) and PDGFB RNA chromogenic in situ hybridization (B; PDGFB-1 probe set) in a case of PDGFD-rearranged DFSP. Inset represents PDGFB RNA chromogenic in situ hybridization at high magnification.
All tumors in a broad morphologic differential diagnosis of both conventional and fibrosarcomatous DFSP were negative for PDGFB overexpression when a threshold of more than 5 puncta or at least one chromogen aggregate in greater than 25% of neoplastic cells was used to determine positivity (Figure 5, Table 1). Minimal PDGFB expression (fewer than 5 puncta in 20–50% of cells) was observed in 2 cases each of desmoplastic melanoma, malignant peripheral nerve sheath tumor, and pleomorphic dermal sarcoma (Figure 6). In addition, PDGFB expression was observed within the vasculature of several tumors, including dermatofibroma, angiomatoid fibrous histiocytoma, desmoplastic melanoma, malignant peripheral nerve sheath tumor, leiomyosarcoma, desmoid fibromatosis, solitary fibrous tumor, and pleomorphic dermal sarcoma/atypical fibroxanthoma (Figure 5). Vascular PDGFB staining was often strong, frequently exceeding 5 puncta or one aggregate within an individual endothelial cell. Despite this level of expression in tumor-associated vessels, the angiosarcomas examined in this study lacked PDGFB overexpression, with one case of angiosarcoma showing limited expression manifesting as 1–2 puncta in approximately 15% of tumor cells.
Figure 5.
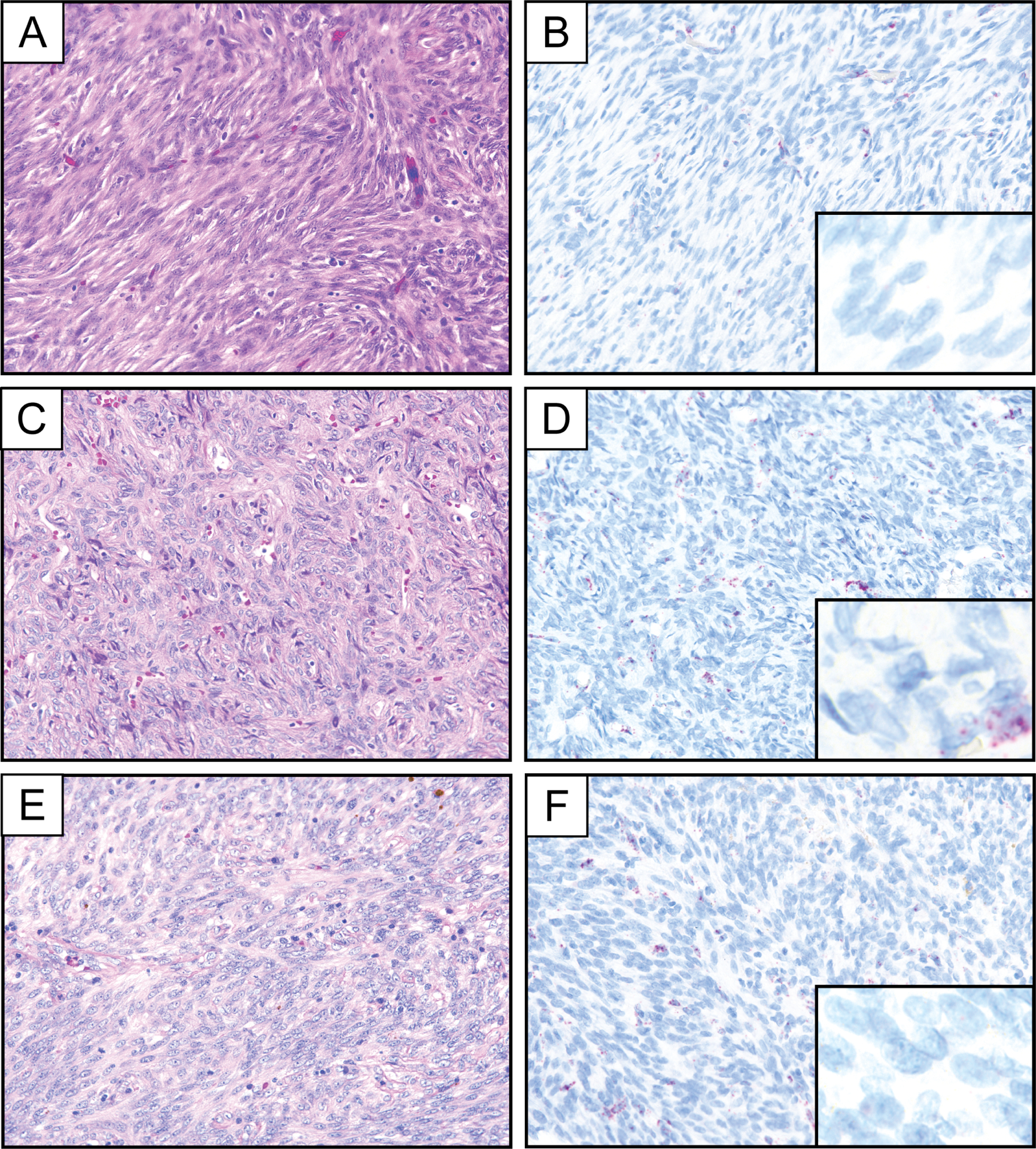
Representative photomicrographs of hematoxylin and eosin stain and PDGFB RNA chromogenic in situ hybridization demonstrating a lack of PDGFB expression in cellular dermatofibroma (A, B), solitary fibrous tumor (C, D), and angiomatoid fibrous histiocytoma (E, F). The pictured cases exhibit vascular PDGFB expression in the background. Insets represent PDGFB RNA chromogenic in situ hybridization at high magnification.
Figure 6.
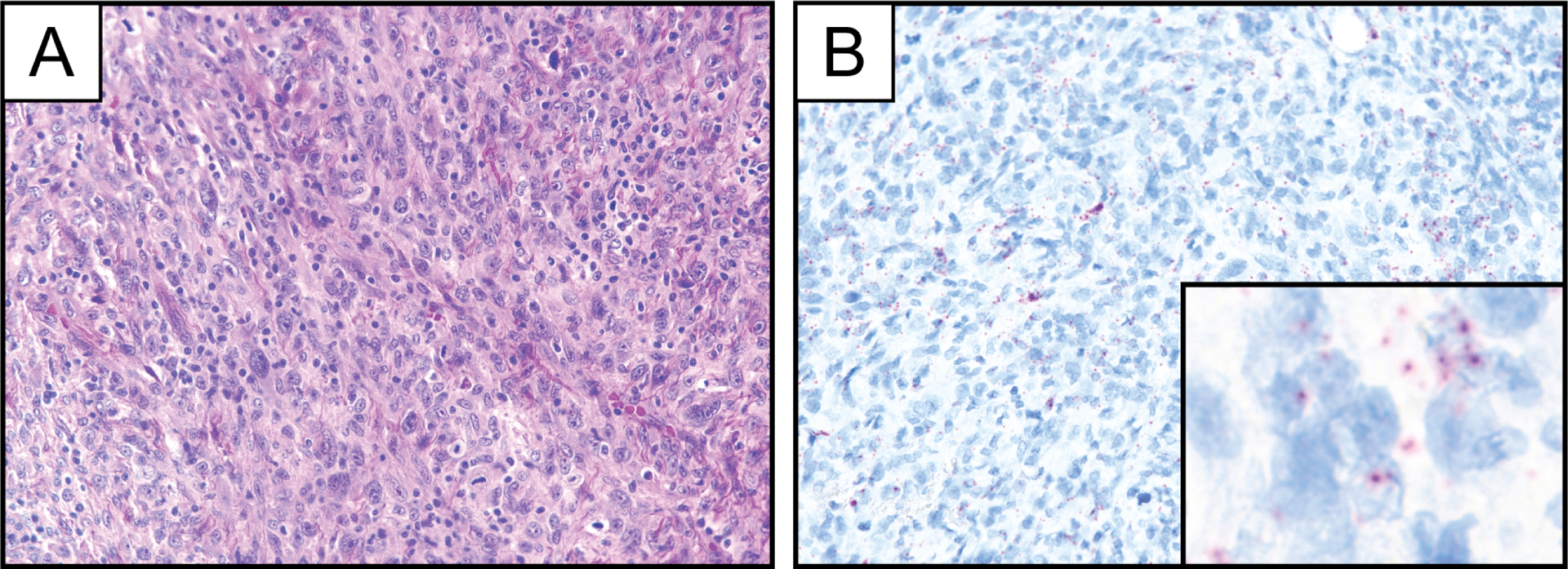
Representative photomicrographs of hematoxylin and eosin stain (A) and PDGFB RNA chromogenic in situ hybridization (B) demonstrating limited PDGFB expression in a case of pleomorphic dermal sarcoma. Inset represents PDGFB RNA chromogenic in situ hybridization at high magnification.
DISCUSSION
In this study, PDGFB RNA CISH was found to be a sensitive and specific tool for the diagnosis of DFSP when implemented with careful interpretation of staining patterns and appropriate thresholds for determining positivity. Although we expect that conventional morphological and immunohistochemical assessment will be sufficient for the diagnosis of DFSP in the majority of cases, an ancillary diagnostic tool such as PDGFB RNA CISH may be useful in the evaluation of scant biopsy specimens and tumors exhibiting variant histology, such as fibrosarcomatous transformation or extensive myxoid change. Indeed, for the majority of cases in our cohort of DFSPs, morphology and immunohistochemistry enabled accurate classification in agreement with PDGFB RNA CISH results, with molecular-cytogenetic findings informing the diagnosis in a minor subset. While PDGFB RNA CISH is strongly and diffusely positive in the fraction of fibrosarcomatous DFSPs with loss of CD34 expression by immunohistochemistry, a rare subpopulation of DFSP with PDGFD rearrangement, may be identified by CD34 immunohistochemistry, but not by PDGFB RNA CISH. Therefore, CD34 in particular remains an important histologic biomarker in the assessment of dermal and subcutaneous spindle cell neoplasms.
In addition to PDGFB RNA CISH, more direct approaches to detect an underlying PDGFB rearrangement, such as FISH and RT-PCR, also continue to be important adjunctive techniques for tumor classification, which have been shown to be highly sensitive and specific for the diagnosis of DFSP [13–16]. In comparison to the RNA CISH approach presented here, FISH and RT-PCR do not enable detection of tumor cells using conventional light microscopy and ordinarily have longer turnaround time. Thus, access to the resources required for implementation of these molecular-cytogenetic techniques, such as a fluorescence microscope and appropriate staff for FISH studies, will likely dictate the relative utility of PDGFB RNA CISH in pathology laboratories. In this regard, the utility of RNA CISH for PDGFB in DFSP is somewhat analogous to that of immunohistochemistry for the components of chimeric fusion oncoproteins, such as STAT6 in solitary fibrous tumor [22–24], CAMTA1 in epithelioid hemangioendothelioma [25, 26], or DDIT3 in myxoid liposarcoma [27]. While implementation of RNA CISH currently is limited relative to immunohistochemistry, the advent of RNA CISH assays for use with FFPE samples on automated immunohistochemistry platforms is expected to enable more widespread adoption of this technology.
We anticipate that PDGFB RNA CISH may have particular utility in the diagnosis of fibrosarcomatous DFSP, given its less distinctive morphological features and the possibility of CD34 loss of expression in a substantial proportion of cases. In our study, fibrosarcomatous DFSP unexpectedly showed qualitatively stronger PDGFB expression than conventional DFSP, a feature that may reflect genomic gains of COL1A1-PDGFB known to occur with fibrosarcomatous transformation [28]. Although our quantification of PDGFB staining based on visual metrics did not enable distinction of conventional and fibrosarcomatous DFSP, it remains possible that computational RNA CISH image analysis or alternative methods for mRNA quantification (RT-qPCR, RNA-seq, etc.) may offer new approaches to the diagnosis of fibrosarcomatous transformation based on the level of PDGFB expression.
A notable limitation of PDGFB-targeted RNA CISH for the diagnosis of DFSP is its inherent specificity for tumors driven by PDGFB rearrangement. Although this approach is useful for detecting the PDGFB overexpression seen in the vast majority (~95%) of conventional and fibrosarcomatous DFSPs, a rare subset of DFSPs known to harbor PDGFD rearrangements will go undetected, accounting for a low “false negative” rate. As PDGFD-rearranged DFSPs appear to exhibit a distinctive anatomic predilection for the female breast [12], particular attention should be paid to this limitation in the evaluation of breast lesions. Based on the proof-of-concept in our studies of PDGFB, it is feasible that one could employ an alternative PDGFD-specific probe set to diagnose PDGFD-rearranged DFSPs using RNA CISH.
The instability of mRNA warrants special consideration of sample quality when undertaking diagnostic RNA CISH assays. To limit errors of interpretation due to signal degradation, we generally performed studies on freshly cut (less than one month-old) tissue sections and routinely assessed expression of the housekeeping gene PPIB in conjunction with PDGFB. Anecdotally, in our studies of PDGFB on histologic sections stored at room temperature, we have observed limited signal loss at 1 year, appreciable signal loss at 2 years, and complete signal loss at 5 years. On the other hand, we have observed robust PDGFB staining in freshly cut sections from FFPE blocks stored at room temperature for up to 6 years. Strong chromogen was also observed in DFSP cases from fresh sections of FFPE blocks stored at 4° C for as much as 26 years. In practice, RNA degradation is expected to be less of a concern as most tissues will have been newly obtained.
There are additional barriers to the clinical implementation of RNA CISH that may limit the utility of this assay in practice, at least in the near-term. Currently, the “cost per slide” of the RNAscope assay, while favorable in comparison to a more labor-intensive assay such as FISH, substantially exceeds that of immunohistochemistry. Additionally, although RNA CISH can be performed on an automated clinical platform analogous to immunohistochemistry, many clinical laboratories may not currently have such a platform, and this capital investment must be considered in the overall cost and practicality of adopting this methodology for routine practice.
A separate challenge of the RNA CISH approach relates to the difficulty of interpreting equivocal staining patterns in a subset of cases, most notably those with minimal expression. Because the branched-chain RNA CISH assay employed by RNAscope is designed to be highly sensitive for the detection of RNA molecules, even low levels of expression can manifest some amount of staining. Thus, as in the case of the PDGFB RNA CISH approach described here, empirically defined thresholds of “positivity” must be employed to ensure appropriate sensitivity and specificity. Nonetheless, based on our experience with the cohort of DFSPs examined in this study, the robust PDGFB expression driven by the oncogenic translocation manifests with distinctive strong and diffuse RNA CISH staining in the vast majority of specimens, enabling unequivocal, fast interpretation. We also envision that in time RNA CISH will become more widespread in surgical pathology and that the interpretation of such thresholds will become more engrained in our practice.
A shortcoming of this study is the use of TMAs to interrogate PDGFB expression in the majority of cases, potentially limiting evaluation of tissue heterogeneity. In this study, the restricted tumor sampling inherent to the use of TMAs may lead to an underestimation of the RNA CISH assay’s sensitivity or an overestimation of its specificity. One might reasonably anticipate that heterogeneity is less of a concern for a translocation-driven tumor, such as DFSP, generally characterized by uniform histology. Indeed, we observed relatively homogeneous PDGFB RNA CISH staining in the subset of DFSPs studied using whole tissue sections and the CISH assay exhibited high sensitivity among conventional and fibrosarcomatous DFSPs represented on TMAs (26/27; 96%). Still, with regard to the issue of specificity in particular, caution is advised when implementing PDGFB RNA CISH as a diagnostic tool for use on whole tissue sections.
We found evidence of PDGFB expression by RNA CISH in the blood vessels of several tumors. This observation is in keeping with an extensive body of research that has defined a role for PDGFB signaling in developmental and tumor angiogenesis; in particular, PDGFB produced by endothelial cells appears to be required for the recruitment and proliferation of pericytes and vascular smooth muscle cells in newly formed blood vessels [29]. Overall, PDGFB expression by blood vessels did not pose a challenge in terms of diagnostic interpretation of RNA CISH, despite the fact that expression within individual endothelial cells was occasionally rather strong, as the expression could be localized to vessels morphologically. We saw substantial variability in vascular PDGFB expression even within a given tumor subtype, suggesting that angiogenic PDGFB signaling may be dynamic or even tumor specific. Given this variability of expression, we expect that tumor-associated vessels will be an inconsistent source of internal positive control for PDGFB RNA CISH.
We conclude that RNA CISH serves as a useful technique for the detection of PDGFB overexpression, seen in the vast majority of DFSPs as a consequence of a recurrent COL1A1-PDGFB rearrangement. Our study adds to the growing list of applications for RNA CISH in surgical pathology, including the detection of MDM2 in liposarcoma [30], MYB in adenoid cystic carcinoma [18], RANKL in chondroblastoma [19], and CSF1 in tenosynovial giant cell tumor [31], among others. The highly sensitive nature of RNA CISH for the analysis of gene expression demands careful interpretation with the use of empirically defined criteria or thresholds, as in the case of PDGFB. Further study and adoption of RNA CISH in surgical pathology will help to refine methods for quantification and rules for interpretation, which may be unique for each target gene. We would expect that variability inherent to a histologic technique such as RNA CISH will demand that individual laboratories evaluate staining criteria as a component of assay validation, similar to immunohistochemical approaches. Ultimately, the ability to evaluate histologically a continuum of gene expression changes holds the promise of moving beyond largely binary readouts to obtain enhanced diagnostic insight based on a more granular assessment of relative expression levels.
FUNDING
G.W.C. is supported in part by the Stanford University School of Medicine Clinical and Translational Science Award Program (National Center for Advancing Translational Sciences, KL2TR003143).
Footnotes
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was performed in accordance with research policies approved by the Institutional Review Boards of Stanford University and the Brigham and Women’s Hospital.
DATA AVAILABILITY
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
DECLARATION OF CONFLICTING INTERESTS
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- 1.WHO Classification of Tumours Editorial Board. Soft Tissue and Bone Tumours. (Lyon: International Agency for Research on Cancer, 2020). [Google Scholar]
- 2.Reimann JDR, Fletcher CDM. Myxoid dermatofibrosarcoma protuberans: a rare variant analyzed in a series of 23 cases. Am J Surg Pathol 31, 1371–1377 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Dupree WB, Langloss JM, Weiss SW. Pigmented dermatofibrosarcoma protuberans (Bednar tumor). A pathologic, ultrastructural, and immunohistochemical study. Am J Surg Pathol 9, 630–639 (1985). [DOI] [PubMed] [Google Scholar]
- 4.Calonje E, Fletcher CD. Myoid differentiation in dermatofibrosarcoma protuberans and its fibrosarcomatous variant: clinicopathologic analysis of 5 cases. J Cutan Pathol 23, 30–36 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Mentzel T, Beham A, Katenkamp D, Dei Tos AP, Fletcher CDM. Fibrosarcomatous (‘High-Grade’) Dermatofibrosarcoma Protuberans: Clinicopathologic and Immunohistochemical Study of a Series of 41 Cases With Emphasis on Prognostic Significance. Am J Surg Pathol 22, 576–587 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Bowne WB, Antonescu CR, Leung DH, Katz SC, Hawkins WG, Woodruff JM, et al. Dermatofibrosarcoma protuberans: A clinicopathologic analysis of patients treated and followed at a single institution. Cancer 88, 2711–2720 (2000). [PubMed] [Google Scholar]
- 7.Goldblum JR, Reith JD, Weiss SW. Sarcomas arising in dermatofibrosarcoma protuberans: a reappraisal of biologic behavior in eighteen cases treated by wide local excision with extended clinical follow up. Am J Surg Pathol 24, 1125–1130 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Goldblum JR, Tuthill RJ. CD34 and factor-XIIIa immunoreactivity in dermatofibrosarcoma protuberans and dermatofibroma. Am J Dermatopathol 19, 147–153 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Abenoza P, Lillemoe T. CD34 and factor XIIIa in the differential diagnosis of dermatofibroma and dermatofibrosarcoma protuberans. Am J Dermatopathol 15, 429–434 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Sirvent N, Maire G, Pedeutour F. Genetics of dermatofibrosarcoma protuberans family of tumors: from ring chromosomes to tyrosine kinase inhibitor treatment. Genes Chromosomes Cancer 37, 1–19 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Dadone-Montaudié B, Alberti L, Duc A, Delespaul L, Lesluyes T, Pérot G, et al. Alternative PDGFD rearrangements in dermatofibrosarcomas protuberans without PDGFB fusions. Mod Pathol 31, 1683–1693 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Dickson BC, Hornick JL, Fletcher CDM, Demicco EG, Howarth DJ, Swanson D, et al. Dermatofibrosarcoma protuberans with a novel COL6A3-PDGFD fusion gene and apparent predilection for breast. Genes Chromosomes Cancer 57, 437–445 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahira T, Oda Y, Tamiya S, Higaki K, Yamamoto H, Kobayashi C, et al. Detection of COL1A1-PDGFB fusion transcripts and PDGFB/PDGFRB mRNA expression in dermatofibrosarcoma protuberans. Mod Pathol 20, 668–675 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Patel KU, Szabo SS, Hernandez VS, Prieto VG, Abruzzo LV, Lazar AJ, et al. Dermatofibrosarcoma protuberans COL1A1-PDGFB fusion is identified in virtually all dermatofibrosarcoma protuberans cases when investigated by newly developed multiplex reverse transcription polymerase chain reaction and fluorescence in situ hybridization assays. Hum Pathol 39, 184–193 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Segura S, Salgado R, Toll A, Martín-Ezquerra G, Yébenes M, Sáez A, et al. Identification of t(17;22)(q22;q13) (COL1A1/PDGFB) in dermatofibrosarcoma protuberans by fluorescence in situ hybridization in paraffin-embedded tissue microarrays. Hum Pathol 42, 176–184 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Karanian M, Pérot G, Coindre JM, Chibon F, Pedeutour F, Neuville A. Fluorescence in situ hybridization analysis is a helpful test for the diagnosis of dermatofibrosarcoma protuberans. Mod Pathol 28, 230–237 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4, 844–847 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Rooper LM, Lombardo KA, Oliai BR, Ha PK, Bishop JA. MYB RNA In Situ Hybridization Facilitates Sensitive and Specific Diagnosis of Adenoid Cystic Carcinoma Regardless of Translocation Status. Am J Surg Pathol Published Ahead of Print, (2021). [DOI] [PubMed] [Google Scholar]
- 19.Suster DI, Kurzawa P, Neyaz A, Jarzembowski JA, Lozano-Calderon S, Raskin K, et al. Chondroblastoma Expresses RANKL by RNA In Situ Hybridization and May Respond to Denosumab Therapy. Am J Surg Pathol 44, 1581–1590 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Nohr E, Kunder CA, Jones C, Sutton S, Fung E, Zhu H, et al. Development and clinical validation of a targeted RNAseq panel (Fusion-STAMP) for diagnostic and predictive gene fusion detection in solid tumors. bioRxiv 870634 (2019). [Google Scholar]
- 21.He J, Abdel-Wahab O, Nahas MK, Wang K, Rampal RK, Intlekofer AM, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 127, 3004–3014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyle LA, Vivero M, Fletcher CD, Mertens F, Hornick JL. Nuclear expression of STAT6 distinguishes solitary fibrous tumor from histologic mimics. Mod Pathol 27, 390–395 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Yoshida A, Tsuta K, Ohno M, Yoshida M, Narita Y, Kawai A, et al. STAT6 immunohistochemistry is helpful in the diagnosis of solitary fibrous tumors. Am J Surg Pathol 38, 552–559 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Cheah AL, Billings SD, Goldblum JR, Carver P, Tanas MZ, Rubin BP. STAT6 rabbit monoclonal antibody is a robust diagnostic tool for the distinction of solitary fibrous tumour from its mimics. Pathology 46, 389–395 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Doyle LA, Fletcher CDM, Hornick JL. Nuclear Expression of CAMTA1 Distinguishes Epithelioid Hemangioendothelioma From Histologic Mimics. Am J Surg Pathol 40, 94–102 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Shibuya R, Matsuyama A, Shiba E, Harada H, Yabuki K, Hisaoka M. CAMTA1 is a useful immunohistochemical marker for diagnosing epithelioid haemangioendothelioma. Histopathology 67, 827–835 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Scapa JV, Cloutier JM, Raghavan SS, Peters-Schulze G, Varma S, Charville GW. DDIT3 Immunohistochemistry Is a Useful Tool for the Diagnosis of Myxoid Liposarcoma. Am J Surg Pathol 45, 230–239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbott JJ, Erickson-Johnson M, Wang X, Nascimento AG, Oliveira AM. Gains of COL1A1-PDGFB genomic copies occur in fibrosarcomatous transformation of dermatofibrosarcoma protuberans. Mod Pathol 19, 1512–1518 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev 22, 1276–1312 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkarni AS, Wojcik JB, Chougule A, Arora K, Chittampalli Y, Kurzawa P, et al. MDM2 RNA In Situ Hybridization for the Diagnosis of Atypical Lipomatous Tumor: A Study Evaluating DNA, RNA, and Protein Expression. Am J Surg Pathol 43, 446–454 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Mastboom MJL, Hoek DM, Bovée JVMG, van de Sande MAJ, Szuhai K. Does CSF1 overexpression or rearrangement influence biological behaviour in tenosynovial giant cell tumours of the knee? Histopathology 74, 332–340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


