Abstract
The lung is inhabited by a diverse microbiome that originates from the oropharynx by a mechanism of micro-aspiration. Its bacterial biomass is usually low; however, this condition shifts in lung cancer (LC), chronic obstructive pulmonary disease (COPD) and interstitial lung disease (ILD). These chronic lung disorders (CLD) may coexist in the same patient as comorbidities and share common risk factors, among which the microbiome is included. We characterized the microbiome of 106 bronchoalveolar lavages. Samples were initially subdivided into cancer and non-cancer and high-throughput sequenced for the 16S rRNA gene. Additionally, we used a cohort of 25 CLD patients where crossed comorbidities were excluded. Firmicutes, Proteobacteria and Bacteroidetes were the most prevalent phyla independently of the analyzed group. Streptococcus and Prevotella were associated with LC and Haemophilus was enhanced in COPD versus ILD. Although no significant discrepancies in microbial diversity were observed between cancer and non-cancer samples, statistical tests suggested a gradient across CLD where COPD and ILD displayed the highest and lowest alpha diversities, respectively. Moreover, COPD and ILD were separated in two clusters by the unweighted UniFrac distance (P value = 0.0068). Our results support the association of Streptoccocus and Prevotella with LC and of Haemophilus with COPD, and advocate for specific CLD signatures.
Subject terms: Respiratory tract diseases, Microbiome
Introduction
The human lung was originally thought to be a sterile organ; however, it is now accepted to harbor a complex community of microorganisms referred to as the lung microbiome1,2. Notably, the lung microbiome has been shown to be remarkably similar to the oropharyngeal microbiota due to the physiological mechanism of micro-aspiration, which facilitates bacterial dissemination from the upper airways into the lower respiratory tract2,3. In healthy lungs there is indeed a bidirectional flow of microbes with a steady equilibrium between immigration and elimination through mucociliary clearance2–5. Although bacterial biomass in the lung is maintained at a low concentration, it displays a remarkable microbiological diversity2,4. According to different studies, the lung microbiome is dominated by the phyla Firmicutes, Proteobacteria and Bacteroidetes, and the genera Prevotella, Veillonella and Streptococcus2,4. Bacterial burden, however, frequently fluctuates in chronic lung disorders (CLD), particularly during acute disease stages (exacerbations) and life-threatening complications (e.g., septicemia)4–6.
CLD encompass several airway pathologies, such as chronic obstructive pulmonary disease (COPD), interstitial lung disease (ILD) and lung cancer (LC). COPD is a common CLD and a leading cause of morbidity and mortality worldwide, and is associated mainly with cigarette smoking as well as several indoor and outdoor hazards7,8. Nowadays, COPD is characterized by persistent respiratory symptoms and airflow limitation that is not fully reversible as assessed by lung function tests (spirometry). Small airways obstruction (e.g., bronchitis and bronchiolitis) and loss of lung parenchyma (emphysema) are major underlying causes of COPD and usually coexisting at different scales7,8. The COPD microbiome shows high heterogeneity during stable phases and undergoes notable shifts toward Proteobacteria (mostly Moraxella and Haemophilus) during exacerbations and advanced disease stages9,10.
ILD comprises a wide group of disorders sharing common features of enhanced fibrosis. ILD affects primarily the lung interstitium and can be triggered by a plethora of environmental and/or immunological exposures2,11. Idiopathic pulmonary fibrosis (IPF) represents a paradigmatic example of ILD in which lung architecture is seriously compromised by the accumulation of extensive scar tissue of unknown etiology. Other less prevalent and scrutinized ILD include sarcoidosis, a systemic disorder characterized by idiopathic appearance of granuloma that affects predominantly the lung, and hypersensitivity pneumonitis (HP) a complex syndrome resulting from a negative reaction to antigen inhalation (e.g., non-tuberculosis mycobacteria).
Concerning the ILD microbiome, most of the available data is related to IPF. Those studies showed that in stable patients there is a two-fold increase in bacterial load coupled with diversity loss mostly due to an overgrowth of potentially pathogenic genera (Streptococcus, Neisseria and Haemophilus)2,12. In addition, during acute IPF exacerbations, microbial abundance was found to increase and, as in COPD, a boost in Proteobacteria prevalence was also observed13.
Finally, LC is the most commonly diagnosed and lethal of all cancers. Like COPD, LC is also directly correlated with the tobacco epidemic and several air pollutants (e.g., asbestos and biomass burning)14,15. LC is classified into different histological types being the most prevalent the non-small cell lung cancer (NSCLC), which can be further subdivided into distinct carcinomas where the most common are the adenocarcinoma (ADC) and the squamous cell carcinoma (SCC). Few microbiome studies have discriminated between LC subtypes, but overall, they seem to reveal a reduction in microbial diversity coupled with significant changes in some bacterial genera (e.g., Streptococcus and Veillonella enrichment) during LC. Remarkably, those alterations seem to be perceptible not only in tumor sites, but also in distant non-cancerous regions of the lung16–18. Moreover, according to a recent study, lung microbiota seems to differentially impact SCC patient survival, either because bacteria (Enterobacteriaceae) cause non-cancer complications of infectious nature, or because they enhance inflammatory pathways and carcinogenic events19.
In LC as well as in COPD and ILD, inflammatory processes are often upregulated20,21. It is thought that microbiome dysbiosis may play a role in the activation and perpetuation of inflammatory processes, which ultimately may impact biological networks and disease progression4,5,22. Furthermore, these CLD are proposed to be linked by common mechanisms of pathogenesis, where pulmonary emphysema and fibrosis have been recognized as critical lung injuries often preceding malignant transformation. Moreover, COPD and ILD may coexist with LC in the same individual as comorbidities, which leads to worse outcomes of the disease20,23,24.
Therefore, to understand the etiology of CLD, it is crucial to disentangle the contribution of the lung microbiome to each disorder, in particularly to LC, which is far less studied when compared to COPD and ILD. Furthermore, it is also fundamental to assess whether lung microbiotas are influenced by different risk factors, such as smoking history, patient age, gender or even disease type and their overlap. To this end, we have combined 16S rRNA amplicon next-generation sequencing with bioinformatics to first characterize the lung microbiome of 89 Portuguese individuals with and without LC, regardless of their histological type and COPD or ILD co-occurrence. Additionally, we have compared the lung microbiomes of 25 CLD patients diagnosed with LC, COPD or ILD and controlled for the absence of crossed comorbidities.
Material and methods
Ethics approval and consent to participate
Sample collection for research purposes was authorized by the ethical committees of participating institutions: Comissão de Ética para a Saúde (CES) Centro Hospitalar São João (#95_14); CES Centro Hospitalar Baixo Vouga (#054031), and the Ethics Committees from the Centro Hospitalar Lisboa Norte and the National Health Institute Dr. Ricardo Jorge (DIRCLN-8ABR2014-130). Informed consent was obtained from all participants and patient samples and data were treated anonymously. The study was conducted in accordance with ethical guidelines and regulations for Human research and the Helsinki Declaration.
Samples
Bronchoalveolar lavage fluid (BALF) samples were collected by pulmonologists from individuals subjected to a bronchoscopy for evaluation of lung disease at three hospitals in Portugal: Centro Hospitalar São João (CHSJ), Porto; Centro Hospitalar Baixo Vouga (CHBV), Aveiro; and Hospital Pulido Valente—Centro Hospitalar Universitário Lisboa Norte (CHULN), Lisbon. Sample collection targeted affected lung segments and was carried out as previously described19,25. Briefly, BALF samples had a minimum volume of 15 mL (0.9% saline solution) and were initially stored by pulmonologists at − 20 to 4 °C according to the facilities available at the participating hospitals. Samples were then transported on ice to research centers where they were stored at − 80 °C until needed. Overall, we collected 106 samples to address two main goals: (1) compare the lung microbiome of LC cases with other non-cancerous patients and (2) contrast the lung microbiome of LC patients with those of COPD and ILD patients. Towards the first goal, we sampled 49 patients with a positive cancer diagnosis (LC) (regardless of histological type and the presence of other known comorbidities such as COPD or ILD), and 40 patients with a negative cancer diagnosis (non-LC; Supplementary Table 1). Moreover, we did not include in the non-LC group any subject with a primary diagnosis of COPD or ILD. No healthy controls were collected due to bronchoscopy invasiveness and risk of complications.
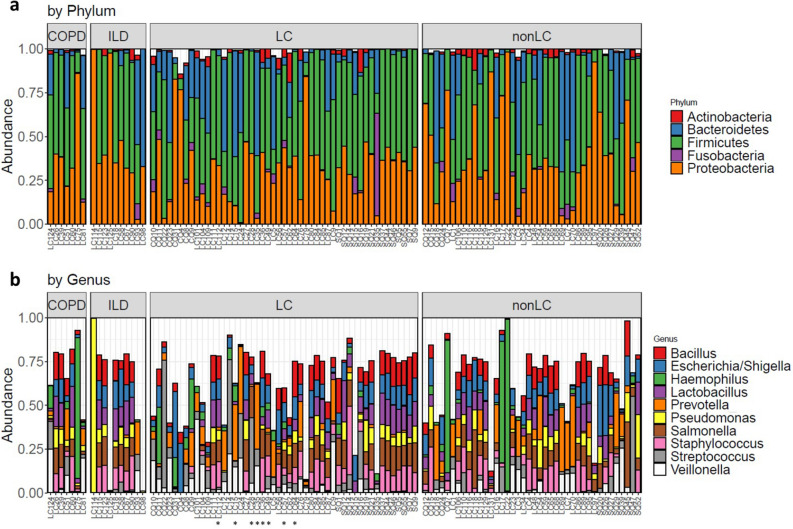
To address our second goal, we selected three homogenous patient groups with a single CLD diagnosis (controlled for other comorbidities): LC (N = 8), COPD (N = 7) and ILD (N = 10). LC patients were included in the comparison above (Supplementary Table 1; Table 2). For simplicity, this subset of LC samples will be designated from this point forward as LC*. This is also indicated in Fig. 1. To our knowledge, none of the patients included in this study had a record of acute exacerbations at the time of sampling.
Table 2.
Demographic and clinical data of the comorbidity-controlled dataset.
| Variables | LC* (N = 8) |
COPD (N = 7) |
ILD (N = 10) |
P value* |
|---|---|---|---|---|
| Age (yrs. ± SD) | 58.5 ± 14.1 | 56.7 ± 13.9 | 62.9 ± 12.3 | NS |
| Sex, men N (%) | 5 (62.5) | 6 (85.7) | 8 (80.0) | NS |
| Smoking status N (%) | ||||
| Smoker | 3 (37.5) | 2 (28.6) | 2 (20.0) | NS |
| Former smoker | 2 (25.0) | 3 (42.6) | 6 (60.0) | |
| Non-smoker | 3 (37.5) | 2 (28.6) | 2 (20.0) | |
| Pack Per Year | 37.6 ± 18.2 | 64.5 ± 37.4 | 35.7 ± 17.7 | NSa |
| LC Diagnosis N (%) | ||||
| NSCLC | 7 (87.5) | NA | ||
| ADC | 5 (62.5) | |||
| Unknown | 2 (25.0) | |||
| SCLC | 1 (12.5) | |||
| COPD Diagnosis N (%) | ||||
| GOLD 2 | 5 (71.4) | NA | ||
| Unknown | 2 (28.6) | |||
| ILD Diagnosis N (%) | ||||
| HP | 3 (30.0) | NA | ||
| Sarcoidosis | 2 (20.0) | |||
| IPF | 1 (10.0) | |||
| Other | 4 (40.0) | |||
*P values based on Kruskal–Wallis test or chi-square (normal distributed or categorical variables, respectively) for the comparison of the three groups, lung cancer (LC), chronic obstructive pulmonary disease (COPD) and interstitial lung disease (ILD). HP- Hypersensitivity pneumonitis; NS—Non-significant (p value > 0.05); NA—Not applicable. a—results based in pairwise Welch's t-tests.
Figure 1.
Microbial profiles of most abundant (> 1%) phyla (a) and genera (b) per individual BALF sample. Comorbidity controlled groups: COPD (N = 7) and ILD (N = 10), and LC (N = 49) and non-LC (N = 40) samples are indicated. Samples included in the LC* (N = 8) controlled group are marked with asterisks (*).
Lung microbiota 16S rRNA screening and analysis
DNA extraction from BALF (200–250 µL) was performed using DNA Mini kit (Qiagen) according to manufacturer’s instructions for capturing bacterial DNA in body fluids. We amplified and sequenced a fragment of ~ 250 bp of the 16S rRNA gene covering the V4 region using the dual-index sequencing strategy described in Kozich et al.26. Sequencing was performed using the next-generation sequencing Illumina MiSeq platform at the GWSPH Genomics Core Facility. We sequenced both negative controls and mock communities (reference samples with a known composition) to detect potential contaminating microbial DNA in reagents and measure sequencing error rate. No evidence of contamination was found and our sequencing error rate was low. Sequence data have been deposited in GenBank under BioProject PRJNA742244.
16S rRNA–V4 amplicon sequence variants (ASV) in each sample were inferred using dada2 version 1.1627. Exact sequence variants provide a more accurate and reproducible description of amplicon-sequenced communities than is possible with operational taxonomic units (OTUs) defined at a constant level (97% or other) of sequence similarity27. Reads were filtered using standard parameters, with no uncalled bases, maximum of 2 expected errors, and truncating reads at a quality score of 2 or less. Forward and reverse reads were truncated after 225 and 100 bases, respectively. The standard dada2 pipeline was then applied to perform ASV inference, merge paired reads and identify chimeras. Taxonomic assignment was performed against the Silva v132 database using the implementation of the RDP naive Bayesian classifier available in the dada2 R package28,29. ASV sequences were aligned using MAFFT30 and used to build a tree with FastTree31. The resulting ASV tables and phylogenetic tree were imported into phyloseq32 for further analysis.
We normalized our samples using the negative binomial distribution as recommended by McMurdie and Holmes33 and implemented in the Bioconductor package DESeq234. This approach simultaneously accounts for library size differences and biological variability and it has increased sensitivity in small and homogeneous datasets with less than 20 samples per group35. Microbial normalized counts generated this way are referred to as taxon abundances throughout the text. Taxonomic and phylogenetic alpha-diversity were estimated using Chao richness and Shannon, ACE, Simpson, Fisher and Phylogenetic (Faith’s) diversity indices. Beta-diversity was estimated using phylogenetic Unifrac (unweighted and weighted), Bray–Curtis and Jaccard distances. Dissimilarity between samples was explored using principal coordinates analysis (PCoA).
Significant associations between alpha-diversity indices and taxon abundances and lung disorders and covariables (clinical history, age and sex) were assessed using the Mann–Whitney-Wilcoxon Test. Beta-diversity indices were compared using permutational multivariate analysis of variance (adonis) as implemented in the vegan R package36. We applied the Benjamini–Hochberg method at alpha = 0.05 to correct for multiple hypotheses testing37,38. Effect sizes were calculated using Cohen’s ds estimator for unequal group sizes39. All the analyses above were performed in R40 and RStudio41.
Results
Subjects biodemographic and clinical characteristics
In our study LC patients averaged 65.6 years of age, 41 (83.7%) were men and 67.3% were reported as former or current smokers. NSCLC was the most prevalent cancer (51%) among these patients, with ADC and SCC subtypes representing 34.7% and 10.2% of cases, respectively. A small fraction of LC subjects was diagnosed with SCLC or with other rarer cancers types (14.3%) and for the remaining samples no cancer type classification was available (Table 1; Supplementary Table 1). Non-LC individuals were younger and averaged 59.5 years of age, 27 (54%) were men and 50% described as former or current smokers (Table 1; Supplementary Table 1). A heterogeneous array of respiratory conditions was reported for non-LC subjects, including many benign findings (22.5%) and several lung abnormalities such as hemoptysis and atelectasis (Table 1; Supplementary Table 1).
Table 1.
Demographic and clinical data of the extended BALF dataset.
| Variables | LC (N = 49) |
Non-LC (N = 40) |
P value* |
|---|---|---|---|
| Age (yrs. ± SD) | 65.6 ± 11.4 | 59.5 ± 12.7 | 0.01805 |
| Sex, men N (%) | 41 (83.7) | 27 (54.0) | NS |
| Smoking status N (%) | |||
| Smoker | 16 (32.7) | 10 (25.0) | NS |
| Former smoker | 17 (34.7) | 10 (25.0) | |
| Non-smoker | 9 (18.4) | 15 (37.5) | |
| Unknown | 7 (14.3) | 5 (12.5) | |
| LC Diagnosis N (%) | |||
| NSCLC | 25 (51.0) | NA | |
| ADC | 17 (34.7) | ||
| SCC | 5 (10.2) | ||
| LCC | 1 (2.0) | ||
| Unknown | 2 (4.1) | ||
| SCLC | 5 (10.2) | ||
| Others | 2 (4.1) | ||
| Unknown | 17 (34.7) | ||
| Non-LC Diagnosis N (%) | |||
| Hemoptysis | 5 (12.5) | NA | |
| Atelectasis | 3 (7.5) | ||
| Unaffected | 2 (5.0) | ||
| Benign findings | 9 (22.5) | ||
| Asthma | 1 (2.5) | ||
| Hamartoma | 1 (2.5) | ||
| Other | 7 (17.5) | ||
| Unknown | 14 (35.0) | ||
*P values based on Welch's t-test or chi-square (normal distributed or categorical variables, respectively) for the comparison lung cancer (LC) and non-LC groups. NS—non-significant (p value > 0.05). NA—not applicable.
In the CLD comorbidity-controlled groups, the LC subset (LC*) averaged 58.5 years of age and comprised 7 NSCLC (5 ADC) and 1 SCLC types; five were men, and five had a history of heavy smoking (20–63 packs per year; PPY). The COPD group (mean age 56.7 years) included only moderate disease cases (GOLD 2), a single woman and four heavy smokers (38–120 PPY). Finally, the ILD group (mean age 62.9 years) included 3 HP, 2 sarcoidosis and a single IPF case, 7 patients were men and 6 were former smokers (Supplementary Table 1; Table 2).
Taxonomic characterization
In general, the microbiome analysis of BALF samples consistently showed Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria as the prevalent phyla across the five groups (Table 3; Fig. 1A). Similarly, the results obtained at the genus level indicated that abundant bacteria such as Prevotella, Staphylococcus, Veillonella, Pseudomonas and Streptococcus were also shared by the different groups (Table 3; Fig. 1B). Some inter-individual variability in microbial composition could be detected as suggested by a few outlier samples dominated by single genera (Fig. 1B). Interestingly, those samples were all subjected to microbiological culture testing, one being classified as negative (LC114), two positive and concordant with 16S rRNA results (LC75 with Haemophilus and LC125 with Serratia) and another positive but discordant (LC98; Supp. Table 1).
Table 3.
Mean relative proportions of dominant phyla and genera (> 1%) identified in the different groups.
| Taxon | Extended dataset | Comorbidity controlled dataset | |||
|---|---|---|---|---|---|
| LC | Non-LC | LC* | COPD | ILD | |
| Phyla | |||||
| Firmicutes | 47.11 | 40.30 | 48.75 | 49.69 | 39.30 |
| Proteobacteria | 31.35 | 37.94 | 30.17 | 34.71 | 45.06 |
| Bacteroidetes | 15.52 | 17.61 | 17.34 | 13.42 | 12.82 |
| Actinobacteria | 2.80 | 1.95 | 2.40 | < 1.00 | 1.50 |
| Genera | |||||
| Prevotella | 6.09 | 8.32 | 9.47 | 4.63 | 2.14 |
| Escherichia/Shigella | 8.80 | 8.96 | 8.58 | 6.05 | 7.91 |
| Staphylococcus | 7.27 | 7.02 | 7.33 | 6.95 | 8.40 |
| Lactobacillus | 6.41 | 6.75 | 6.37 | 8.86 | 9.03 |
| Bacillus | 7.66 | 6.79 | 8.51 | 7.72 | 7.53 |
| Salmonella | 7.40 | 7.60 | 10.43 | 6.66 | 7.83 |
| Veillonella | 6.00 | 4.63 | 5.69 | 8.29 | 1.48 |
| Haemophilus | 3.21 | 7.06 | 2.90 | 14.28 | < 1.00 |
| Pseudomonas | 3.56 | 5.09 | 2.63 | 5.16 | 14.18 |
| Streptococcus | 7.45 | 3.94 | 8.74 | 4.29 | 1.85 |
Taxa proportions with significant differences are highlighted in bold (P < 0.05).
Escherichia/Shigella, Bacillus, Streptococcus and Salmonella displayed the largest mean abundances in LC cases (Table 3). However, only Streptococcus diverged between LC and non-LC groups (Wilcoxon rank sum test; p value = 0.03852; Cohen’s dS = 0.30). Streptococcus, Prevotella, Salmonella and Escherichia/Shigella were found as the most prevalent taxa in the comorbidity-controlled LC* group (Table 3), whereas Prevotella proportions separated LC* from ILD cases (Wilcoxon rank sum test;p value = 0.04405; Cohen’s dS = 0.65).
Conversely, in the ILD group the most common taxa according to their mean abundances were Pseudomonas, Lactobacillus, Staphylococcus, and Escherichia/Shigella, (Table 3). Besides Prevotella (ILD vs. LC*), Haemophilus also varied significantly in the ILD versus COPD comparison (Wilcoxon rank sum test; p value = 0.005107; Cohen’s dS = 0.74).
Finally, in the COPD controlled group Haemophilus, Lactobacillus, Veillonella and Bacillus comprised the most prevalent taxa (Table 3). No statistically significant differences were observed between COPD and LC* groups at the genus level.
Given that a strict definition of a common shared microbiome could not be applied to CLD comorbidity-controlled groups, we used instead a less constrained threshold, in which taxa were considered as common if present in at least 80% of the samples. With this approach Enterobacteriaceae (Escherichia/Shigella and Salmonella), Staphylococcus, Streptococcus, Lactobacillus, Listeria and Bacillus were recognized as members of a stable bacterial community shared across LC*, COPD and ILD.
Microbiota diversity
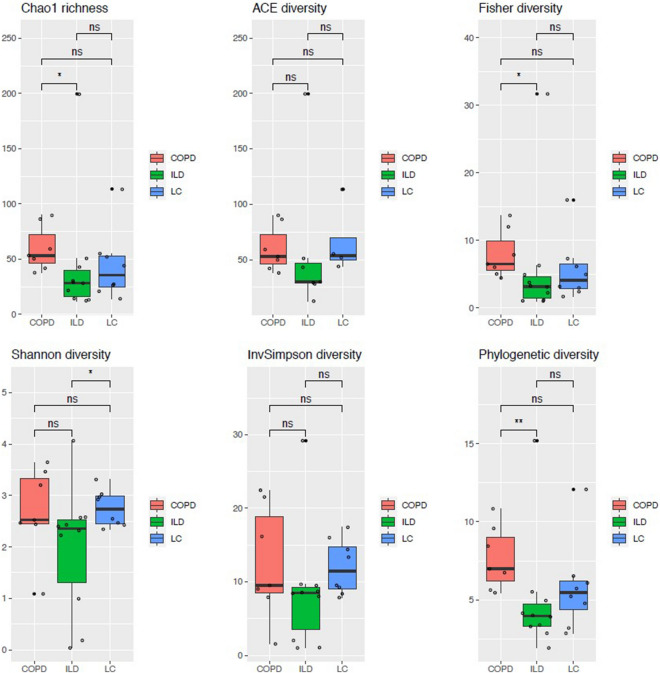
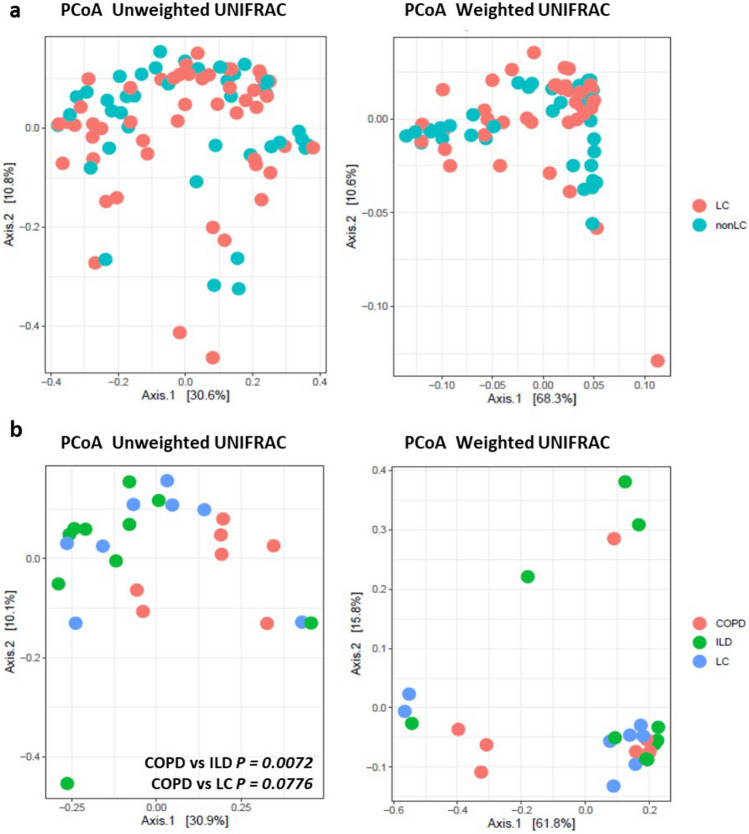
Alpha-diversity indices did not vary significantly between LC and non-LC groups (Supplementary Fig. 1; Supplementary Table 2). In contrast, CLD groups were found to differ, with COPD showing higher diversity than LC* and ILD. Statistically significant results were observed in Chao richness, Fisher and Phylogenetic diversity indices for COPD versus ILD (PChao = 0.0250; PFisher = 0.0185; and PPD = 0.0068) and in Shannon diversity index for LC* versus ILD (P value = 0.0476; Fig. 2; Supplementary Table 2). PCoA plots did not reveal microbial structure (beta-diversity) for LC and non-LC groups, as suggested by the overlap of samples and non-significance of the adonis tests (Fig. 3A; Supplementary Fig. 2). On the other hand, among CLD types, PCoA plots showed some dissimilarities (Fig. 3B, Supplementary Fig. 2), with COPD versus ILD yielding significant differences in the unweighted Unifrac distance (adonis test P = 0.0072) and with COPD versus LC* showing a borderline, yet non-significant p value for the same statistic (P = 0.0776; Fig. 3B). In general, alpha- and beta-diversity were not affected by analyzed co-variables (data not shown), except for the LC* versus ILD comparison, in which smoking history could be associated with statistically significant differences in Bray–Curtis and Jaccard indices (P = 0.027 and P = 0.025, respectively; Supplementary Fig. 3).
Figure 2.
Alpha diversity of the CLD comorbidity-controlled dataset COPD (N = 7), ILD (N = 10) and LC* (N = 8) groups. Displayed estimates: Chao richness and Shannon, ACE, Inversed Simpson, Fisher and Phylogenetic (Faith’s) diversity indices. Significant p-values for pairwise group comparisons are indicated as (*) for p < 0.05 and (**) for p < 0.01. ns: non-significant.
Figure 3.
Beta diversity as shown by principle coordinate analysis of unweighted UniFrac distances and weighted UniFrac distances. (a) Extended dataset comprising LC (N = 49) and non-LC groups (N = 40); (b) Comorbidity controlled dataset including COPD (N = 7), ILD (N = 10) and LC* (N = 8) groups.
Discussion
Currently, few studies have attempted to simultaneously analyze the microbiome of distinct CLD considering their frequent co-occurrence in a single individual. In this study, we perform a characterization of BALF samples first stratified into LC and non-LC cases and then into three CLD groups (LC*, COPD and ILD) controlled for the absence of crossed comorbidities. Although, we found lung microbiome to be relatively stable among the studied groups, significant differences in the proportions of certain taxa were detected, suggesting a possible role for bacteria in the onset, progression and eventual outcome of distinct CLD. A recent comparison of bacterial communities from LC cases with an assorted group of lung disorders and with healthy controls (BALF samples also), presented no distinct profiles in alpha-diversity18. After stratifying samples into cancer and non-cancer types, the same study detected significant differences in beta-diversity tests18. We, however, found no significant differences in alpha- or beta-diversities between LC and non-LC groups, which could be related to the heterogeneous nature of the 49 cancerous samples compared, which comprise only 34.7% of ADC and 10.5% of SCC subtypes. Contrarily, Tsay et al. (2018) studied mostly ADC and SCC, representing 56.4% and 25.6% of the 39 cases analyzed, respectively18. Nonetheless, we did observe significant differences between other CLD groups. When controlling for comorbidities and comparing strict CLD phenotypes, COPD versus ILD displayed a remarkable divergence across both alpha- and beta- diversity indices (but not against LC*), indicating some community structuring by disease. According to our results, COPD communities are generally the most diverse and composed by a larger number of low-abundance taxa as suggested by the Chao index results. On the other hand, ILD cases show the lowest bacterial richness and diminished phylogenetic diversity. Interestingly, LC* samples, which overlapped with both COPD and ILD groups in most alpha indices, exhibited a stronger phylogenetic relatedness with ILD cases, as uncovered by the unweighted Unifrac distances.
Concerning bacteria differential abundances, Streptococcus was identified as significantly increased in the LC group compared to non-LC, whereas in the comorbidity-controlled dataset Prevotella was identified to be augmented in LC* when contrasted with ILD. In addition, in the COPD group, Haemophilus proportions were found to be higher than in ILD. All these genera, typically associated with the oral microbiome, have already been reported as prevalent taxa in affected lungs of CLD patients12,16,18,42.
Notably, Streptococcus and Prevotella proportions, which discriminate our LC cases from other assorted pathologies (LC vs. non-LC and LC* vs ILD groups, respectively), replicated to some extent Tsay et al. (2018) findings, where the same taxa were identified as good predictors of LC18. Therefore, our results may also support the association between the high prevalence of these microbes and lung carcinogenesis. To be more accurate, Tsay et al. (2018) demonstrated by means of in vitro studies that Streptococcus and Prevotella are able to induce the up-regulation of PI3K (phosphoinositide 3-kinase) and ERK (extracellular signal–regulated kinase) signaling pathways, which are associated with cancer transformation18. Importantly, Streptococcus is also a well-known pneumonia agent (Streptococcus pneumoniae), particularly among LC subjects. Moreover, Streptococcus has been shown to raise cytokine levels and promote diverse inflammatory responses through the activation of Toll-like receptors and by the degradation of extracellular matrix elements16,43.
Conversely, a high content of Prevotella in the airways has been correlated with enhanced concentrations of interleukin 17 (IL17), among other cytokines, and T helper 17 cells (Th17), underlying a status of subclinical lung inflammation seen also among healthy individuals5,44,45. Furthermore, in a recent study using bleomycin-induced mouse models of lung fibrosis, it was shown that a dysbiotic microbiome enriched in Prevotella could activate multiple pro-inflammatory and pro-fibrotic genes. These, in turn, were found to promote both lung immune cell infiltration and massive extracellular matrix deposition, ultimately leading to animal death in an IPF-like phenotype46. Once again, IL17 and Th17 cells were pinpointed as key drivers of inflammatory networks induced by Prevotella in mice46. In our study, we observed a decreased prevalence of Prevotella in the ILD cohort, but a higher prevalence in LC*. This may indicate a potential interaction between Prevotella and Th17 cells, which were hitherto shown to promote lung tumorigenesis47.
The detection of a higher proportion of Haemophilus in our COPD cases, a taxon frequently associated with acute exacerbations (Haemophilus influenzae), supports previous evidence for an early dysbiosis caused by this genus that can be observed even in stable phases of the disease9,10. Interestingly, Haemophilus has been described to provoke a more aggressive inflammatory response than Prevotella, as depicted by a fold increase in IL10, IL12 and IL23 cytokines48,49. In addition, it was correlated with the activation in the airways of the nuclear factor kappa beta (NF-KB) pathway and other inflammatory markers, such as IL1B and IL6, myeloperoxidase, and CXC-chemokine ligand 8. Moreover, Haemophilus is also capable of triggering other host responses that might be correlated with COPD pathogenesis, including the production of reactive oxygen species (ROS) and the formation of extracellular protease networks traps by both neutrophil and macrophage cells50,51.
The hypothesis of the microbiome fulfilling a pivotal role in CLD seems quite plausible if considering the negative effects of the aforementioned bacteria in lung biology, as well as, the diversity differences observed between COPD and ILD. For example, the increased prevalence of Haemophilus compared to Prevotella and Streptococcus in COPD may contribute to a pro-inflammatory and protease enriched microenvironment that promotes the airflow obstruction by inflating and filing the bronchi with mucus (bronchitis) and/or by destroying extracellular matrix and pulmonary parenchyma (emphysema). Although, we could not establish a link between any taxa and a pro-fibrotic stimulus in ILD, its microbial structure was distinct from that of COPD. The genera Pseudomonas and Staphylococcus previously described as associated with a worse IPF prognosis42,52,53 tended to be higher in our ILD cases compared to COPD and LC*, but this was not significant.
Oddly, although COPD has been shown to increase the risk of LC development 2- to fourfold21, the LC* microbiome appears to be more closely related to ILD than COPD. This finding may then question whether the lung microbiome takes part in cancer transformation among COPD patients, particularly when our cases are essentially moderate ones (GOLD 2) and microbial diversity tends to decrease along with disease progression to advanced stages—very severe COPD (GOLD4), reducing the abundances of the potentially carcinogenic genera Prevotella and Veillonella9.
Although less frequently than in COPD, subjects with IPF (ILD) were also reported to be at risk of progressing to cancer54,55, suggesting the similarity of LC* and ILD microbiomes as a predisposing factor for cancer occurrence. However, this hypothesis appears to be contradicted by the low prevalence in the ILD group of the cancer associated taxa Prevotella, Streptococcus and Veillonella 18. On the other hand, the decay of microbial diversity registered from COPD to ILD may be correlated with the severity or life-expectancy of each CLD, in which pulmonary fibrosis tends to have the worse prognosis17. In support of this conjecture are former reports of reduced diversity levels in LC and severe COPD and the findings in IPF (ILD) of an association between bacterial burden and patient survival9,56–58. If proven true, microbiome studies might be clinically useful to identify patients at risk of cancer complications and to predict disease outcomes.
Even though our study supports previous microbial associations with CLD (e.g., Haemophilus and COPD) and provides some evidence for a disease differentiation based in microbiome diversity, it is worth noticing that our comorbidity-controlled groups have a small sample size. Moreover, there is also a large variability in microbiome composition across COPD and LC patients, where some sub-phenotypes (or endotypes) were already connected with specific microbial signatures9,19,59–61. Furthermore, in the absence of a healthy group as a control, we could not assess the extent to which the lung microbiome is altered by each CLD. To the best of our knowledge, our work represents a first attempt to consider crossed comorbidities as a factor to characterize the large microbiome heterogeneity in lung cancer cases.
Conclusions
No clear cut divergence was observed between LC and non-LC cases, aside from the previously recognized Streptococcus link to lung cancer. Nonetheless, we uncovered several differences across CLD microbiomes: COPD, ILD and LC* varied not only in microbial composition and evenness, but also in the proportions of Prevotella and Haemophilus. Altogether, our findings point out to the presence of distinct microbiome hallmarks specific to each CLD subtype that should be further explored in larger cohorts of COPD, LC and ILD cases.
Supplementary Information
Acknowledgements
We would like to thank all patients for donating their samples and for collaborating in this study. IPATIMUP integrates the i3S Research Unit, which is partially supported by the Portuguese Foundation for Science and Technology (FCT). This work was supported by Norte Portugal Regional Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (FEDER)—project NORTE-01-0145-FEDER-000029. This work was also financed by FEDER funds through COMPETE 2020 (Operacional Programme for Competitiveness and Internationalization— POCI) and by private funds through the Young Investigator Prize Francisco Augusto da Fonseca Dias and Maria José Melenas da Fonseca Dias to P.I.M. FCT supports P.I.M. through a post-doctoral fellowship (SFRH/BPD/120777/2016), financed by the Portuguese State Budget of the Ministry for Science, Technology and High Education and from the European Social Fund (Programa Operacional do Capital Humano—POCH). M.P-L was partially supported by FCT under the Programa Operacional Potencial Humano—Quadro de Referência Estratégico Nacional funds from the European Social Fund and Portuguese State Budget of the Ministry for Science, Technology and High Education (IF/00764/2013; RX: IF/00359/2015).
Author contributions
S.S. wrote the manuscript; S.S. and M.P-L conceived the study; S.G. and P.I.M. performed laboratorial work; A.R.K., and M.P.L. carried out bioinformatics and statistical analyses; M.S., C.S., L.V.R., G.T., P.P., T.T.A., C.B., J.S., L.M., A.S.C., R.M. provided samples and managed clinical data, P.I.M. and M.P.L. critically revised the manuscript. All authors approved the final version of the manuscript.
Data availability
The data used in this study is included in the manuscript and in supplementary material files. Sequence data used to generate microbiome analyses is deposited in GenBank under BioProject PRJNA742244.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-94468-y.
References
- 1.Dickson RP, Huffnagle GB. The lung microbiome: New principles for respiratory bacteriology in health and disease. PLoS Pathog. 2015;11:e1004923. doi: 10.1371/journal.ppat.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Lung Microbiome. (2019).
- 3.Dickson RP, Erb-Downward JR, Huffnagle GB. Towards an ecology of the lung: New conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir. Med. 2014;2:238–246. doi: 10.1016/s2213-2600(14)70028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ubags NDJ, Marsland BJ. Mechanistic insight into the function of the microbiome in lung diseases. Eur. Respir. J. 2017;50:1602467. doi: 10.1183/13993003.02467-2016. [DOI] [PubMed] [Google Scholar]
- 5.Huffnagle GB, Dickson RP, Lukacs NW. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal. Immunol. 2017;10:299–306. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laroumagne S, et al. Bronchial colonisation in patients with lung cancer: A prospective study. Eur. Respir. J. 2013;42:220–229. doi: 10.1183/09031936.00062212. [DOI] [PubMed] [Google Scholar]
- 7.Vogelmeier CF, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report: GOLD executive summary. Eur. Respir. J. 2017 doi: 10.1183/13993003.00214-2017. [DOI] [PubMed] [Google Scholar]
- 8.Celli BR, et al. Standards for the diagnosis and treatment of patients with COPD: A summary of the ATS/ERS position paper. Eur. Respir. J. 2004;23:932–946. doi: 10.1183/09031936.04.00014304. [DOI] [PubMed] [Google Scholar]
- 9.Mayhew D, et al. Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax. 2018 doi: 10.1136/thoraxjnl-2017-210408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, et al. Lung microbiome dynamics in COPD exacerbations. Eur. Respir. J. 2016;47:1082–1092. doi: 10.1183/13993003.01406-2015. [DOI] [PubMed] [Google Scholar]
- 11.Mikolasch TA, Garthwaite HS, Porter JC. Update in diagnosis and management of interstitial lung disease. Clin. Med. (Lond.) 2016;16:s71–s78. doi: 10.7861/clinmedicine.16-6-s71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molyneaux PL, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2014;190:906–913. doi: 10.1164/rccm.201403-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molyneaux PL, et al. Changes in the respiratory microbiome during acute exacerbations of idiopathic pulmonary fibrosis. Respir. Res. 2017;18:29. doi: 10.1186/s12931-017-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 15.Hardavella, G. & Sethi, T. In Lung Cancer (eds Dingemans, A. M. C., Reck, M. & Westeel, V.) 285 (2015).
- 16.Liu HX, et al. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int. J. Cancer. 2018;142:769–778. doi: 10.1002/ijc.31098. [DOI] [PubMed] [Google Scholar]
- 17.Yu G, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17:163. doi: 10.1186/s13059-016-1021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsay JJ, et al. Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am. J. Respir. Crit. Care Med. 2018;198:1188–1198. doi: 10.1164/rccm.201710-2118OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomes S, et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci. Rep. 2019;9:12838. doi: 10.1038/s41598-019-49195-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meiners S, Eickelberg O, Konigshoff M. Hallmarks of the ageing lung. Eur. Respir. J. 2015;45:807–827. doi: 10.1183/09031936.00186914. [DOI] [PubMed] [Google Scholar]
- 21.Vermaelen K, Brusselle G. Exposing a deadly alliance: Novel insights into the biological links between COPD and lung cancer. Pulm. Pharmacol. Ther. 2013;26:544–554. doi: 10.1016/j.pupt.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Dickson RP, et al. The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am. J. Respir. Crit. Care Med. 2018;198:497–508. doi: 10.1164/rccm.201711-2180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank AL, Kreuter M, Schwarzkopf L. Economic burden of incident interstitial lung disease (ILD) and the impact of comorbidity on costs of care. Respir. Med. 2019;152:25–31. doi: 10.1016/j.rmed.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Lung Cancer. (2015).
- 25.Carvalho AS, et al. Bronchoalveolar lavage proteomics in patients with suspected lung cancer. Sci. Rep. 2017;7:42190. doi: 10.1038/srep42190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMurdie PJ, Holmes S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss S, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 38.Cook RD. Detection of influential observation in linear regression. Technometrics. 1977;19:15–18. doi: 10.1080/00401706.1977.10489493. [DOI] [Google Scholar]
- 39.Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2. Hillsdale: Lawrence Earlbaum Associates; 1988. [Google Scholar]
- 40.Team, R. D. C. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2008).
- 41.RStudio, R. T. Integrated development for R. RStudio, IncBoston, MA (2015).
- 42.Han MK, et al. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: An analysis of the COMET study. Lancet Respir. Med. 2014;2:548–556. doi: 10.1016/s2213-2600(14)70069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim GL, Seon SH, Rhee DK. Pneumonia and Streptococcus pneumoniae vaccine. Arch. Pharm. Res. 2017;40:885–893. doi: 10.1007/s12272-017-0933-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segal LN, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segal LN, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 2016;1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, D. et al. Dysregulated lung commensal bacteria drive interleukin-17b production to promote pulmonary fibrosis through their outer membrane vesicles. Immunity50, 692–706 e697. 10.1016/j.immuni.2019.02.001 (2019). [DOI] [PubMed]
- 47.Marshall EA, et al. Emerging roles of T helper 17 and regulatory T cells in lung cancer progression and metastasis. Mol. Cancer. 2016;15:67. doi: 10.1186/s12943-016-0551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mika M, et al. Microbial and host immune factors as drivers of COPD. ERJ Open Res. 2018;4:00015–02018. doi: 10.1183/23120541.00015-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larsen JM, et al. Divergent pro-inflammatory profile of human dendritic cells in response to commensal and pathogenic bacteria associated with the airway microbiota. PLoS ONE. 2012;7:e31976. doi: 10.1371/journal.pone.0031976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King PT, Sharma R. The lung immune response to nontypeable haemophilus influenzae (lung immunity to NTHi) J. Immunol. Res. 2015;2015:706376. doi: 10.1155/2015/706376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.King PT, et al. Nontypeable Haemophilus influenzae induces sustained lung oxidative stress and protease expression. PLoS ONE. 2015;10:e0120371. doi: 10.1371/journal.pone.0120371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newton CA, Molyneaux PL, Oldham JM. Clinical genetics in interstitial lung disease. Front. Med. (Lausanne) 2018;5:116. doi: 10.3389/fmed.2018.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y, et al. Microbes are associated with host innate immune response in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2017;196:208–219. doi: 10.1164/rccm.201607-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Opron K, et al. Lung microbiota associations with clinical features of COPD in the SPIROMICS cohort. NPJ. Biofilms Microbiomes. 2021;7:14. doi: 10.1038/s41522-021-00185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Idiopathic Pulmonary Fibrosis. (2016).
- 56.Invernizzi R, et al. Bacterial burden in the lower airways predicts disease progression in idiopathic pulmonary fibrosis and is independent of radiological disease extent. Eur. Respir. J. 2020 doi: 10.1183/13993003.01519-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abe Y, et al. A severe pulmonary complication in a patient with COL4A1-related disorder: A case report. Eur. J. Med. Genet. 2016 doi: 10.1016/j.ejmg.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 58.Jin J, et al. Diminishing microbiome richness and distinction in the lower respiratory tract of lung cancer patients: A multiple comparative study design with independent validation. Lung Cancer. 2019;136:129–135. doi: 10.1016/j.lungcan.2019.08.022. [DOI] [PubMed] [Google Scholar]
- 59.Barnes PJ. Inflammatory endotypes in COPD. Allergy. 2019;74:1249–1256. doi: 10.1111/all.13760. [DOI] [PubMed] [Google Scholar]
- 60.Dima E, et al. The lung microbiome dynamics between stability and exacerbation in chronic obstructive pulmonary disease (COPD): Current perspectives. Respir. Med. 2019;157:1–6. doi: 10.1016/j.rmed.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 61.Ghebre, M. A. et al. Biological exacerbation clusters demonstrate asthma and chronic obstructive pulmonary disease overlap with distinct mediator and microbiome profiles. J. Allergy Clin. Immunol.141, 2027–2036 e2012. 10.1016/j.jaci.2018.04.013 (2018). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study is included in the manuscript and in supplementary material files. Sequence data used to generate microbiome analyses is deposited in GenBank under BioProject PRJNA742244.