Abstract
Thermophilic bacteria, especially from the genus Bacillus, constitute a huge potential source of novel enzymes that could be relevant for biotechnological applications. In this work, we described the cellulose and hemicellulose-related enzymatic activities of the hot spring Bacillus aerius CCMM B940 from the Moroccan Coordinated Collections of Microorganisms (CCMM), and revealed its potential for hemicellulosic biomass utilization. Indeed, B940 was able to degrade complex polysaccharides such as xylan and lichenan and exhibited activity towards carboxymethylcellulose. The strain was also able to grow on agriculture waste such as orange and apple peels as the sole carbon source. Whole-genome sequencing allowed the reclassification of CCMM B940 previously known as B. aerius into Bacillus paralicheniformis since the former species name has been rejected. The draft genome reported here is composed of 38 contigs resulting in a genome of 4,315,004 bp and an average G + C content of 45.87%, and is an important resource for illuminating the molecular mechanisms of carbohydrate metabolism. The annotated genomic sequences evidenced more than 52 genes encoding glycoside hydrolases and pectate lyases belonging to 27 different families of CAZymes that are involved in the degradation of plant cell wall carbohydrates. Genomic predictions in addition to in vitro experiments have revealed broad hydrolytic capabilities of the strain, thus reinforcing its relevance for biotechnology applications.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02919-0.
Keywords: Bacillus paralicheniformis, Whole genome, Hemicellulose, Glycoside hydrolase, Lichenan, Xylan
Genome research reports
Lignocellulosic biomass, mainly composed of cellulose, hemicelluloses and lignin, is the most abundant complex biopolymer in the nature. It is of major interest for biotechnology as this feedstock is used for the production of renewable biofuels and other high-value chemicals, through an enzymatic saccharification process (Zhang et al. 2018). However, depolymerization of lignocellulose is challenging and it requires high-performance enzymes that act in synergy (Houfani et al. 2020). Therefore, enzymes produced by microorganisms, such as bacteria, are used for the deconstruction of the complex polysaccharides of cellulose and hemicelluloses which are tightly bound together in plant cell walls (Zeng et al. 2017).
Xylan is the main component of hemicelluloses and consists of a β-1,4-d-xylan backbone with short side chains of O-acetyl, β-l-arabinofuranosyl, d-α-glucuronic acid and phenolic acid (Biely et al. 2016). Other polysaccharides such as xyloglucan, lichenan and mannans are part of the hemicellulolytic fraction of plant cell walls. Xylanases, xyloglucanases, lichenases, and endoglucanases are necessary to completely degrade these polysaccharides into simple sugars to allow glycolytic fermentation to produce energy. These enzymes belong to one of the glycoside hydrolase (GH) families, a subgroup of the carbohydrate-active enzymes (CAZymes) (Lombard et al. 2014) and very often associated with carbohydrate-binding modules (CBM) that facilitate adhesion of enzymes to cellulose or hemicelluloses.
Most microorganisms living in extreme environmental habitats such as hot springs have been considered attractive producers of lignocellulolytic enzymes for industrial bioconversion processes (Thapa et al. 2020). Morocco has more than 20 hot springs distributed in different regions (Cidu and Bahaj 2000; Bouchaou et al. 2017) which are suitable habitats for thermophilic microorganisms growing optimally between 55 and 80 °C. In addition, thermophilic bacteria are known for their ability to produce thermostable enzymes of biotechnological interest (Knapik et al. 2019). Previously, Aanniz et al. (2015) investigated the diversity of thermophilic bacteria in 4 different hot springs in Morocco, which enabled the isolation of 79 bacterial strains belonging to the genus Bacillus (www.ccmm.ma). Some of these strains were identified as Bacillus aerius. They grow optimally at 55 °C and exhibit amylolytic, proteolytic or cellulolytic activity (Aanniz et al. 2015). However, strains belonging to B. aerius species should be reclassified as this species name has been rejected since 2015 (Dunlap 2015). For this reason, we chose the CCMM B940 strain as the representative of the B. aerius group isolated by Aanniz et al. (2015) from a hot spring for biological characterization of its potential hemicellulolytic and cellulolytic activities and whole-genome sequencing analysis.
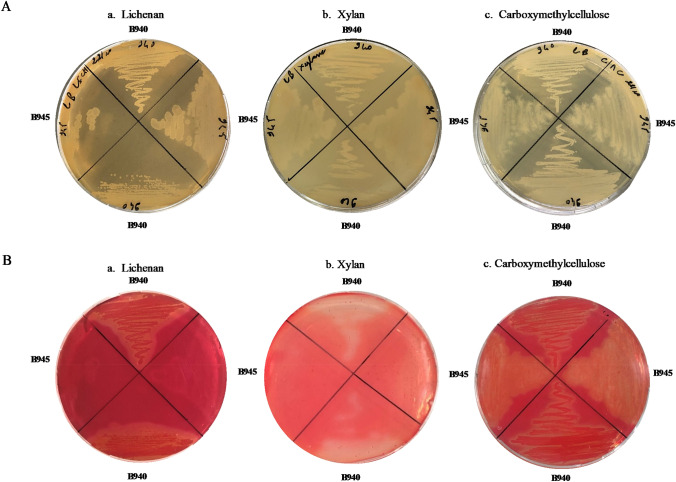
B940 strain was grown twice on Luria–Bertani (LB) agar medium (BD Difco, Sparks, Maryland, USA) for 18 h at 37 °C under aerobic conditions. Individual colonies were then picked from LB plates and cultivated at 37 °C for 18 h in LB agar medium supplemented with 0.5% Icelandic moss Lichenan (Megazyme, Wicklow, Irlande), beechwood Xylan (Megazyme, Wicklow, Irlande), or medium viscosity Carboxymethylcellulose (CMC, Sigma, MO, USA), respectively. A second bacterial strain (CCMM B945) from the same B. aerius group, known for inability to degrade lichenan and xylan (data not shown) was used as a negative control for degradation of hemicellulose and following the same experimental protocol as for CCMM B940. Both strains grew well on LB agar medium supplemented with purified fibres (Fig. 1A).
Fig. 1.
Growth A and detection of lichenase (a), xylanase (b) and carboxymethylcellulase (c) activities B of Bacillus CCMM B940 and CCMM B945 strains cultivated in LB agar streak plates supplemented with 0.5% (w/v) lichenan, xylan or carboxymethylcellulose (CMC), respectively. Plates were incubated 18 h at 37 °C. Congo red staining was performed as described in Patrascu et al. (2017). Clear halos around the streaks indicate the degradation of the corresponding polysaccharide.
To reveal whether the strains were able to degrade the complex polysaccharides added into the medium, the plates were stained with 0.1% Congo red as previously described (Patrascu et al. 2017). Clear halos around the streaks indicated that B940 strain can degrade all three polysaccharides, and thus capable of producing the appropriate enzymes. As expected, lichenases and xylanases were only produced by the CCMM B940 strain (Fig. 1B). Strain CCMM B945 exhibited significant activity against CMC, indicating production of endoglucanases, similar to the CCMM B940 strain.
The Congo red test demonstrated that live cells of the B940 strain exhibited lichenase and xylanase activities and also produced endoglucanases for glucan-polymer degradation. However, to evaluate biomass utilization potential of this strain for biotechnology applications, we searched for the presence of those enzymes in the rather easily accessible B940 strain supernatants. For this purpose, the strain was cultivated in the basal salt solution (BSS), a chemically defined medium modified from Parab et al. (2017), with a lower salt concentration (NaCl 6 g L−1). The medium was supplemented with fruits peels (orange or apple) as the sole source of carbon and then sterilized 15 min at 121 °C. Fruit peels are a rich source of cellulose, hemicellulose, lignin and pectin (Rivas et al. 2008; Joglekar et al. 2019). Orange peel is rich in structural polysaccharides, including cellulose (18–20%), hemicellulose (14–16%), pectin (20–22%) and lignin (5–7%) (de la Torre et al. 2019). Apple residues typically contain 7.2–43.6% cellulose, 4.3–24.4% hemicellulose, 15.2–23.5% lignin and 3.5–14.32% pectin (Dhillon et al. 2013). Both substrates were chosen because of their abundance as industrial waste around the world (de la Torre et al. 2019).
A single colony of CCMM B940 and CCMM B945 from LB agar medium was picked to inoculate 5 mL of LB Broth (Difco) and incubated overnight at 37 °C (with continuous stirring at 170 rpm), followed by second culture in 5 mL of BSS medium supplemented with 0.5% apple or orange peels and inoculated at 1%. Before adding them in the medium, apple and orange peels were dried for 24 h at 60 °C and crumbled finely using an electric grinder. After 48 h of incubation at 37 °C with stirring (170 rpm), both strains grew well in the BSS supplemented with fruit peels representing agro-industrial wastes as shown in Figure S1. Additionally, this test confirmed the ability of CCMM B940 and CCMM B945 strains to break down fibres from the peels and use the degradation products.
Protein samples from B940 and B945 strains cultivated for 48 h at 37 °C in 30 mL BSS medium supplemented with 0.5% orange and apple peels were prepared to detect and quantify hemicellulolytic and cellulolytic activities in the extracellular fluids. Briefly, cultures were centrifuged for 15 min at 5000×g and 4 °C, supernatants were filtered through a 0.22 μm Millipore filter, and then concentrated 25-fold in a Spin-X UF10K concentrator following the supplier’s protocol (Corning B.V., Amsterdam, The Netherlands).
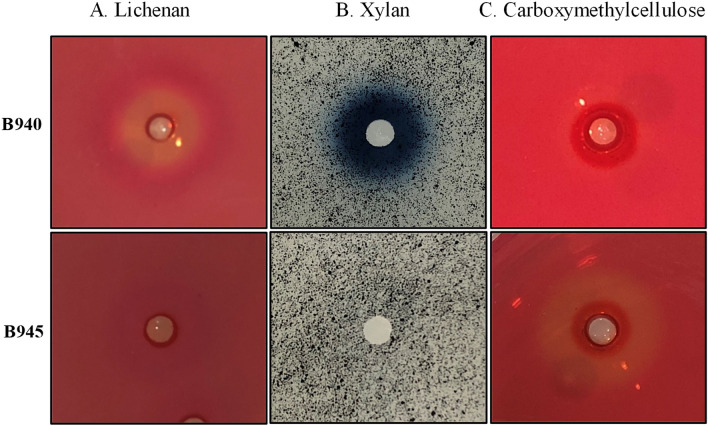
Agar-well plate enzymatic assays were performed to detect xylanase, lichenase and endoglucanase activities using concentrated extracellular proteins. Concentrated extracellular proteins from CCMM B940 and control strain CCMM B945 were prepared in the same way. The samples were loaded (60 μL/well) in Petri dishes containing BSS medium supplemented with either 0.1% chromogenic AZCL-Xylan (Megazyme, Wicklow, Ireland), 0.5% non-chromogenic CMC (Sigma-Aldrich, MO, USA) or lichenan (Megazyme) polysaccharides. The plates were incubated overnight at 37 °C and those with non-chromogenic polysaccharide were stained with Congo red as described above. Clear and blue halos around the wells indicated that the CCMM B940 cultivated with apple peels was able to produce lichenase (Fig. 2A), endoglucanase (Fig. 2C, although weakly in the tested conditions), and xylanase (Fig. 2B). The same results were obtained with orange peels (not shown).
Fig. 2.
Detection of lichenase A, xylanase B and carboxymethylcellulase C activities of Bacillus CCMM B940 and B945 strains, using agar-well plate assays with concentrated extracellular proteins of BSS cultures with 0.5% apple. Agar plate BSS medium was supplemented with 0.5% (w/v) lichenan, carboxymethylcellulose (CMC), or 0.1% AZCL beechwood xylans. Plates were incubated overnight at 37 °C. Clear and blue halos around the wells indicate a positive sample for the corresponding glycoside hydrolase activity
The CCMM B940 strain is able to grow at 55 °C in Tryptic Soy broth (Aanniz et al. 2015) and we also observed that the strain is capable of growing in BSS medium supplemented with lichenan and xylan at the same temperature (data not shown), thus reinforcing the interest of its characteristics for industrial applications requiring thermostable enzymes.
In contrast to CCMM B940 strain results, the control strain CCMM B945 was not able to produce xylanase or lichenase when cultivated in the presence of apple or orange peels (Fig. 2).
Moreover, extracellular hemicellulolytic activities of the strain CCMM B940 were quantified using the dinitrosalicylic acid (DNS) spectrophotometric method for reducing sugars as described by Miller (1959). Protein concentration was determined by the Bradford protocol (Bradford 1976) with bovine serum albumin (BSA) as the standard. The amount of protein samples and the incubation time were chosen to provide assay conditions in which the measured activity was proportional to these two parameters. The enzymatic assays were all carried out at 55 °C to quantify the activities under conditions close to those of industrial processes using thermostable enzymes. Reducing sugars that were released after enzyme–substrate incubations were quantified in triplicates using glucose or xylose as a standard. One international unit (IU) was defined as the amount of enzyme, which produced 1 µmol of reducing sugars in 1 min. The xylanase and lichenase activities of the extracellular proteins from BSS cultures with 0.5% apple and orange peels are listed in Table 1. Strain CCMM B945 was again used as a control. No reducing sugars was detectable for strain CCMM B945 proteins incubated with xylan or lichenan. The lichenase activity of CCMM B940 was approximately two times higher than the xylanase activity in the cultures with natural peels. Both activities were higher in apple peel cultures than in orange peels, suggesting that the produced enzymes were either more numerous or more active in metabolizing carbohydrates in apple peels. This could be due to the nature of fibres present in the peel (sugar composition and types of glycosidic bonds) and/or to differential induction of glycoside hydrolases encoding gene expression. Strain CCMM B940 exhibited xylanase activities approximately twofold and over tenfold higher in apple peel cultures than thermophilic strains B. licheniformis 2D55 (Kazeem et al. 2017) and Bacillus sp. 275 (Gong et al. 2017), respectively.
Table 1.
Lichenase and xylanase specific activities of extracellular proteins from Bacillus CCMM B940 and CCMM B945 cultivated in BSS medium supplemented with 0.5% apple and orange peels
| Strains | Apple | Orange | ||
|---|---|---|---|---|
| CCMM B940 | CCMM B945 | CCMM B940 | CCMM B945 | |
| Lichenase activity (IU/mg) | 4.06 ± 1.1 | nd | 1.6 ± 0.2 | nd |
| Xylanase activity (IU/mg) | 2.39 ± 0.29 | nd | 0.98 ± 0.2 | nd |
Specific activities are given as µmol of glucose (incubation with lichenan) or xylose (incubation with xylans) equivalent produced per min and per mg of protein. Each value is the mean of three different assays (± SD)
nd not detectable
To further investigate the putative functions of the CCMM B940 strain and to achieve its reclassification within the genus Bacillus, we sequenced and examined the whole genome of this strain. Thus, genomic DNA was extracted with Wizard® Genomic DNA Purification Kit (Promega, Charbonnières-les-Bains, France). The concentration and quality were determined using Nanodrop and Qubit spectrophotometers. Genomic DNA was sequenced at Eurofins Genomics (Cologne, Germany) using Illumina HiSeq technology. The library was prepared with 2 × 150-bp paired-end read length, including DNA fragmentation, adapter ligation, amplification, and size selection. All the steps were performed according to Eurofins Genomics protocols, producing 7,612,190 reads.
The raw reads were then trimmed using Trimmomatic v0.39 (Bolger et al. 2014) with the following parameters: SLIDINGWINDOW:4:20; ILLUMINACLIP: TruSeq3-PE.fa:2:30:20:2: LEADING:3, TRAILING:3 and MINLEN:60. We verified the presence/absence of phix sequences using bowtie2 (Langmead and Salzberg 2012) with the following parameters: –trim-to 80 –end-to-end –no-unal –no-sq –no-head –p 12. Processed reads were then assembled using the SPAdes de novo assembler v3.13.1 (Bankevich et al. 2012) testing assemblies with k-mer values: 21,33,55,77 using the parameter: –careful. Depending on the size of insert, scaffolds smaller than 350 bp were filtered out. Assembly quality was assessed using in-house script and QUAST v5.0.0 (Gurevich et al. 2013). We estimated genome completeness at 99.59% and checked that no contamination was detected using CheckM v1.1.3 (Parks et al. 2015).
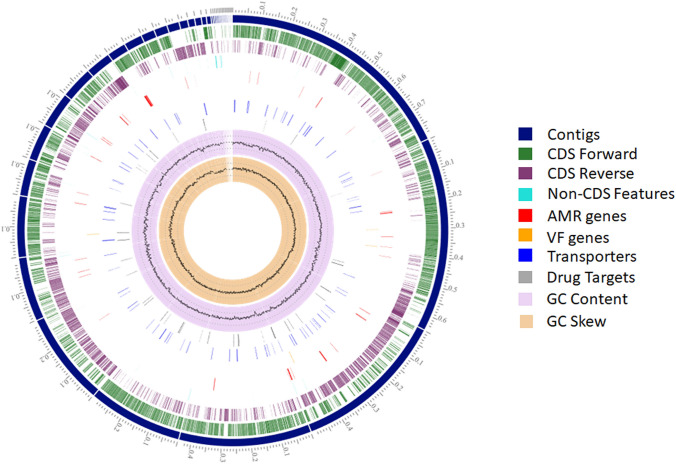
After eliminating low coverage and small contigs (< 350 bp), we obtained a total of 38 contigs and 34 scaffolds resulting in a genome of 4,315,004 bp (NCBI accession number JADRJL000000000) and an average G + C content of 45.87% which is close to other genomes from the Bacillus subtilis group (Rey et al. 2004; Du et al. 2019).The N50 of the draft genome is 440,121 bp, with an average of 126,912 bp and a notable four gap region (Fig. 3). Species identity was confirmed by comparing the genome reference sequences of B. licheniformis and Bacillus paralicheniformis available in the NCBI database (https://www.ncbi.nlm.nih.gov) using the BLAST service deployed on local servers with the genome obtained sequences and fastANI (Jain et al. 2018). The draft genome was most closely related to B. paralicheniformis Bac84 (accession number ASM299392v1) living in sea environment (Othoum et al. 2019) with an ANI value of 98.94%, and shared only 94.36% identity with B. licheniformis DSM 13T (NC_006270.3). B. aerius CCMM B940 was, therefore, reclassified as B. paralicheniformis CCMM B940, placing it in the B. subtilis group; a group known for its wide array of uses in biotechnology mainly through species like B. subtilis and B. licheniformis (Fan et al. 2017). Taxonomic identity of the CCMM B940 strain was also confirmed using the GTDB-Tk tool for genome classification (Chaumeil et al. 2020) with 98.95% identity to the B. paralicheniformis KJ-16 genome (accession number ASM104248v2). The genome was then annotated using the default parameters of Prokka 1.14 (Seemann 2014) showing 4259 protein-coding genes, 51 tRNAs and 4 complete rRNAs.
Fig. 3.
Circular representation of the B. paralicheniformis CCMM B940 chromosome using PATRIC based on 34 assembled scaffolds. From outer to inner ring—contigs (scale—x1Mbp), CDS on the forward strand, CDS on the reverse strand, RNA genes, homologous CDS to known antimicrobial resistance (AMR) genes, CDS with homology to known virulence factors (VF), GC content and GC skew
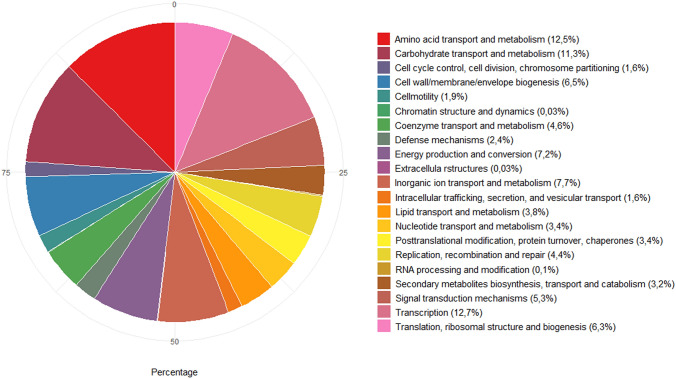
We further used eggNOG-mapper v2.0.1 (Huerta-Cepas et al. 2017) to assign proteins sequences into functional categories. More than 76% of the predicted proteins were attributed to functional subsystems. Proportions of genes with known functions (3047 CDS) were assigned to each category as presented in Fig. 4. Interestingly, a high proportion of CCMM B940 genes was assigned to the carbohydrate transport and metabolism (11.3%) as well as amino-acid transport and metabolism (12.5%) proteins. In a study conducted on Bacillus velenzis, Bacillus safensis and Bacillus altitudinis genomes, only 6.1% of the predicted proteins were assigned to carbohydrate transport and metabolism (Datta et al. 2020), while in Bacillus amyloliquefaciens and Bacillus siamensis, it represents a maximum of 8% in the pan and the core genomes (Chun et al. 2019). B. paralicheniformis has not been thoroughly studied for carbohydrate metabolism yet, but is phylogenetically very close to the B. licheniformis that has been widely used in the fermentation industry for production of enzymes, antibiotics and other chemicals (Madslien et al. 2012).
Fig. 4.
Distribution of biological functions of B. paralicheniformis CCMM B940 based on eggNOG analysis. Colors show different gene features categories and their proportions. Percentages of genes with known functions are given within parentheses
Hereafter, we focused on CCMM B940 glycoside hydrolases related to hemicellulose, cellulose and pectin degradation, comparing their protein sequences to the sequences of GH and pectate lyase (PL) of seven strains of B. paralicheniformis (A4-3, ATCC 9945a, BL-09, CBMAI 1303, FA6, MDJK30, ZAP17) available in the CAZY database (Lombard et al. 2014; database release 22 November 2020). Blast results was obtained using default parameters. Best hits with almost 90% amino-acid identity and 50% length alignment were selected and illustrated in Table 2 (the corresponding sequences are provided in Fig S2). Among 74 GH and PL sequences from CCMM B940 showed homology to other strains of B. paralicheniformis, 52 are dedicated to the degradation of carbohydrates in plant cell walls, representing 27 different CAZymes families according to the classification of B. Henrissat (Lombard et al. 2014). Thirty-two sequences of GH1, GH3, GH5, GH8, GH11, GH16, GH26, GH30, GH31, GH32, GH42, GH43, GH51, GH53, GH68, GH73 were linked to the degradation of hemicellulose, 4 sequences of GH5, GH9, GH12, GH48 for cellulose metabolism, 1 sequence (GH5) either for hemicellulose or cellulose degradation, and 11 sequences of GH (GH28, GH105) and PL (PL1, PL3, PL9, PL11, PL2) were attributed to pectin degradation (Table 2). Four sequences corresponded to enzymes (GH1, GH4) involved in glycolytic pathways. The remaining sequences were mainly related to the hydrolysis of starch and peptidoglycan (not shown).
Table 2.
Annotated genes encoding lignocellulose-degrading enzymes in B. paralicheniformis CCMM B940
| Enzymes | Accession number | Activity | CAZyme family | Reference genes | Identity (%) | Subject sequence length (bp) | Alignment length (bp) | |
|---|---|---|---|---|---|---|---|---|
| Accession number | Species | |||||||
| Hemicellulose related | EOOONOAK_01749 | Aryl-phospho-beta-d-glucosidase BglC | GH1 | ARA87766.1 | Bacillus paralicheniformis MDJK30 | 99.78 | 472 | 472 |
| EOOONOAK_03335 | Aryl-phospho-beta-d-glucosidase BglH | GH1 | AGN35231.1 | Bacillus paralicheniformis ATCC 9945a | 100 | 100 | 100 | |
| EOOONOAK_03002 | Aryl-phospho-beta-d-glucosidase BglC | GH1 | AJO16530.1 | Bacillus paralicheniformis BL-09 | 100 | 491 | 478 | |
| EOOONOAK_01773 | Aryl-phospho-beta-d-glucosidase BglH | GH1 | QEO05069.1 | Bacillus paralicheniformis A4-3 | 99.57 | 469 | 469 | |
| EOOONOAK_03715 | Endo-1,4-beta-xylanase A | GH11 | QEO05734.1 | Bacillus paralicheniformis A4-3 | 100 | 213 | 213 | |
| EOOONOAK_02553 | Beta-glucanase | GH16 | ARA86799.1 | Bacillus paralicheniformis MDJK30 | 99.59 | 243 | 243 | |
| EOOONOAK_03322 | Mannan endo-1,4-beta-mannosidase | GH26 | QFY39913.1 | Bacillus paralicheniformis FA6 | 98.89 | 360 | 360 | |
| EOOONOAK_03205 | Beta-hexosaminidase | GH3 | QEO05378.1 | Bacillus paralicheniformis A4-3 | 99.69 | 644 | 643 | |
| EOOONOAK_02186 | Glucuronoxylanase XynC | GH30 | AJO20016.1 | Bacillus paralicheniformis BL-09 | 100 | 420 | 420 | |
| EOOONOAK_01418 | Oligosaccharide 4-alpha-d-glucosyltransferase | GH31 | QFY38704.1 | Bacillus paralicheniformis FA6 | 99.75 | 802 | 802 | |
| EOOONOAK_02225 | Alpha-xylosidase | GH31 | AJO19971.1 | Bacillus paralicheniformis BL-09 | 99.61 | 769 | 769 | |
| EOOONOAK_02112 | Levanbiose-producing levanase | GH32 | ARA87316.1 | Bacillus paralicheniformis MDJK30 | 99.8 | 515 | 495 | |
| EOOONOAK_01584 | Sucrose-6-phosphate hydrolase | GH32 | AJO20434.1 | Bacillus paralicheniformis BL-09 | 99.37 | 478 | 478 | |
| EOOONOAK_01729 | Sucrose-6-phosphate hydrolase | GH32 | AJO20595.1 | Bacillus paralicheniformis BL-09 | 98.98 | 492 | 492 | |
| EOOONOAK_00085 | Levanase | GH32CBM66 | QFY38018.1 | Bacillus paralicheniformis FA6 | 100 | 677 | 677 | |
| EOOONOAK_02224 | Hypothetical protein | GH3CBM6 | AJO19972.1 | Bacillus paralicheniformis BL-09 | 100 | 980 | 980 | |
| EOOONOAK_03904 | Beta-galactosidase BglY | GH42 | ARA84401.1 | Bacillus paralicheniformis MDJK30 | 99.42 | 690 | 690 | |
| EOOONOAK_04173 | Beta-galactosidase YesZ | GH42 | QFY39293.1 | Bacillus paralicheniformis FA6 | 100 | 665 | 665 | |
| EOOONOAK_01837 | Beta-galactosidase GanA | GH42 | QFY40673.1 | Bacillus paralicheniformis FA6 | 99.56 | 684 | 684 | |
| EOOONOAK_03858 | Beta-xylosidase | GH43 | ARA84797.1 | Bacillus paralicheniformis MDJK30 | 99.62 | 532 | 532 | |
| EOOONOAK_03859 | Non-reducing end alpha-l-arabinofuranosidase | GH43 | ARA84798.1 | Bacillus paralicheniformis MDJK30 | 99.03 | 515 | 515 | |
| EOOONOAK_03489 | Extracellular endo-alpha-(1- > 5)-l-arabinanase 1 | GH43 | ARA85201.1 | Bacillus paralicheniformis MDJK30 | 100 | 316 | 316 | |
| EOOONOAK_00089 | Beta-xylosidase | GH43 | ARA86580.1 | Bacillus paralicheniformis MDJK30 | 100 | 533 | 533 | |
| EOOONOAK_01777 | Extracellular endo-alpha-(1- > 5)-l-arabinanase 2 | GH43 | ARA88110.1 | Bacillus paralicheniformis MDJK30 | 99.79 | 469 | 469 | |
| EOOONOAK_02586 | Extracellular endo-alpha-(1- > 5)-l-arabinanase 1 | GH43 | QFY37786.1 | Bacillus paralicheniformis FA6 | 99.69 | 320 | 320 | |
| EOOONOAK_02185 | Arabinoxylan arabinofuranohydrolase | GH43CBM6 | QFY37223.1 | Bacillus paralicheniformis FA6 | 100 | 515 | 515 | |
| EOOONOAK_01157 | hypothetical protein | GH5 | ARA85656.1 | Bacillus paralicheniformis MDJK30 | 99.82 | 560 | 560 | |
| EOOONOAK_02594 | Intracellular exo-alpha-(1- > 5)-l-arabinofuranosidase | GH51 | ARA86758.1 | Bacillus paralicheniformis MDJK30 | 99.80 | 502 | 502 | |
| EOOONOAK_01836 | Arabinogalactan endo-beta-1,4-galactanase | GH53 | AGN38579.1 | Bacillus paralicheniformis ATCC 9945a | 98.58 | 437 | 424 | |
| EOOONOAK_02113 | Levansucrase | GH68 | ARA87315.1 | Bacillus paralicheniformis MDJK30 | 100 | 481 | 481 | |
| EOOONOAK_04229 | Hypothetical protein | GH73 | QEO06161.1 | Bacillus paralicheniformis A4-3 | 98.95 | 570 | 570 | |
| EOOONOAK_03432 | Reducing-end xylose-releasing exo-oligoxylanase Rex8A xylanase probable | GH8 | QFY39429.1 | Bacillus paralicheniformis FA6 | 99.77 | 434 | 434 | |
| Cellulose-related | EOOONOAK_03622 | Endoglucanase S | GH12 | ARA86997.1 | Bacillus paralicheniformis MDJK30 | 100 | 261 | 261 |
| EOOONOAK_01156 | Exoglucanase-2 | GH48 | AJO18128.1 | Bacillus paralicheniformis BL-09 | 100 | 717 | 704 | |
| EOOONOAK_01385 | Endoglucanase | GH5, CBM3 | AJO18373.1 | Bacillus paralicheniformis BL-09 | 99.81 | 518 | 518 | |
| EOOONOAK_01155 | Endoglucanase A | GH9CBM3 | AJO18127.1 | Bacillus paralicheniformis BL-09 | 100 | 653 | 653 | |
| Glucose metabolism | EOOONOAK_00372 | 6-Phospho-beta-glucosidase GmuD | GH1 | AJO18923.1 | Bacillus paralicheniformis BL-09 | 100 | 471 | 471 |
| EOOONOAK_01723 | 6-Phospho-beta-glucosidase GmuD | GH1 | AJO20590.1 | Bacillus paralicheniformis BL-09 | 99.16 | 478 | 478 | |
| EOOONOAK_03054 | Putative 6-phospho-beta-glucosidase | GH4 | ARA84298.1 0 | Bacillus paralicheniformis MDJK30 | 99.55 | 444 | 444 | |
| Hemicellulose or cellulose related | EOOONOAK_01655 | Putative 6-phospho-beta-glucosidase | GH4 | ARA87677.1 | Bacillus paralicheniformis MDJK30 | 100 | 442 | 442 |
| EOOONOAK_01158 | Hypothetical protein | GH5 | QEO07050.1 | Bacillus paralicheniformis A4-3 | 100 | 395 | 395 | |
| Pectin degradation | EOOONOAK_04163 | Unsaturated rhamnogalacturonyl hydrolase YesR | GH105 | AJO17709.1 | Bacillus paralicheniformis BL-09 | 99.71 | 344 | 344 |
| EOOONOAK_02457 | Unsaturated rhamnogalacturonyl hydrolase YteR | GH105 | AJO19596.1 | Bacillus paralicheniformis BL-09 | 99.73 | 373 | 373 | |
| EOOONOAK_02256 | Exo-poly-alpha-d-galacturonosidase | GH28 | QFY37288.1 | Bacillus paralicheniformis FA6 | 100 | 436 | 434 | |
| EOOONOAK_02563 | Hypothetical protein | PL1 | QFY37765.1 | Bacillus paralicheniformis FA6 | 100 | 494 | 494 | |
| EOOONOAK_04252 | Pectate lyase | PL1 | AJO17744.1 | Bacillus paralicheniformis BL-09 | 99.53 | 428 | 428 | |
| EOOONOAK_01685 | Pectate trisaccharide-lyase | PL1 | QEO04986.1] | Bacillus paralicheniformis A4-3 | 99.71 | 341 | 341 | |
| EOOONOAK_04170 | Rhamnogalacturonan exolyase YesX | PL11 | QEO06607.1 | Bacillus paralicheniformis A4-3 | 100 | 628 | 628 | |
| EOOONOAK_04168 | Rhamnogalacturonan endolyase YesW | PL11 | QEO08401.1 | Bacillus paralicheniformis A4-3 | 100 | 622 | 622 | |
| EOOONOAK_04174 | Hypothetical protein | PL26 | QEO06610.1 | Bacillus paralicheniformis A4-3 | 99.07 | 886 | 863 | |
| EOOONOAK_02080 | Pectate lyase C | PL3 | ARA87346.1 | Bacillus paralicheniformis MDJK30 | 99.55 | 222 | 221 | |
| EOOONOAK_01392 | Hypothetical protein | PL9 | QEO07258.1 | Bacillus paralicheniformis A4-3 | 100 | 468 | 468 | |
The accession numbers correspond to the CDS sequences annotated with Prokka as indicated in the manuscript. The corresponding sequences are provided in Figure S2
These predictions first confirmed the experimental observations in this study for the degradation of xylan, lichenan and CMC as the CCMM B940 genome carries glycoside hydrolases from GH5, GH9, GH11 and GH16 families of CAZYmes known to target these substrates. They also provided additional information on the carbohydrate metabolism of CCMM B940 such as pectinase activity as well as the presence of carbohydrate-binding modules (CBM3, CBM6, CBM66) associated with GHs helping them to bind to cellulose or hemicellulose polysaccharides to improve their efficiency. A few other studies (Wang et al. 2017; Chen et al. 2018) examined the genomic sequences of B. paralicheniformis and predicted their CAZymes, but without the experimental validation. To our knowledge, this is the first report that combines in silico genomic analysis and in vitro experiments to study the carbohydrate metabolism of B. paralicheniformis. These combined analyses reinforce the practical and potential use of CCMM B940 strain for industrial purposes. Indeed, Bacillus species are known to produce 60% of commercially available enzymes, mostly being homologous proteins that are naturally secreted in the growth medium (Westers et al. 2004). The strain CCMM B940 exhibits cellulolytic and hemicellulolytic activities at high temperature and produces extracellular thermostable enzymes.
Based on whole-genome sequencing, we demonstrated that the B. aerius strain CCMM B940 could be reclassified as B. paralicheniformis. Genomic analysis confirmed the fibrolytic potential of the strain CCMM B940 observed in vitro using purified and natural substrates and provided information on its ability to degrade pectins. This statement reinforces the strain's interest especially in agro-waste bioconversion. To our knowledge, this is the first report of lichenase and xylanase activities by a thermophilic strain of B. paralicheniformis isolated from a hot spring. The CCMM B940 strain is part of the large Coordinated Moroccan Collections of Microorganisms (CCMM), which comprise more than 1800 isolates of bacteria, yeasts and fungi. Many of these microorganisms have also been isolated from extremophile environments (Berrada et al. 2012; Aanniz et al. 2015) and they may harbor beneficial attributes such as production of heat-resistance enzymes as already observed in B. amyloliquefaciens (Beladjal et al. 2018) and B. subtilis (den Besten et al. 2017) strains. For this purpose, complete genome sequencing not only enables accurate identification and reclassification of CCMM isolates, i.e. the thermophilic strains of the B. aerius group, but also opens the door to the field of biotechnological applications such as production of second- and third-generation biofuels from raw or pre-treated biomass, medical and nutraceutical fields and food processing (Benedetti et al. 2019), in the favor of the fibrolytic potential (CAZymes) of these strains. Enzymatic activities of these strains should be further studied to determine the most suitable markets in biotechnology (agro-waste bioconversion, depollution, health, pharmaceuticals, etc.…).
Accession numbers
The assembled genome and all relevant sequences were deposited in NCBI's GenBank on November 24, 2020 under the following accession numbers, BioProject PRJNA680612; BioSample SAMN16895636; Genome JADRJL000000000.
Supplementary Information
Below is the link to the electronic supplementary material.
Fig. S1 Cultures of the CCMM B940 and CCMM B945 Bacillus strains in BSS medium supplemented with 0.5% orange or apple peels for 48h at 37°C. a. negative control without bacteria; b. growth (turbidity) is observed for cultures on both substrates (DOCX 1356 kb)
Fig.S2 CDS annotation of the genome of B.paralicheniformis CCMM B940 for lignocellulose-degrading enzymes (TXT 4743 kb)
Acknowledgements
Funding for this research was provided by Belgian Cooperation FRAB/CCMM2, the National Center for Scientific and Technical Research (CNRST) and the National Research Institute for Agriculture, Food and Environment (INRAE). Maski Soufiane is a PhD student at Faculty of Science, Mohammed V University, Rabat, Morocco. Serigne Inssa Ngom is a recipient of a grant for doctoral research allocated by the French ministry of higher education and research (doctoral school 569, University of Paris-Saclay). The authors would like to thank Mrs Zehra Esra Ilhan for English improvement of the manuscript.
Author contributions
MA, EE, MB and CBM conceived the study. SM, SIN, MA, EE, CBM designed the experiments. SM, SIN did genomic and enzymatic analyses, respectively. SIN, BR, TC prepared the samples and extracted DNA. SM performed nucleic Blast, the assembly and the annotation of the genome, and performed the map of functional categories. SIN performed all the enzymatic assays. MA, EE, MB, BR, CBM analyzed the data. SM, SIN, CBM, MA wrote the manuscript with the help of all authors. All authors have read and approved the final manuscript.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Contributor Information
Soufiane Maski, Email: soufianemaski1@gmail.com.
Serigne Inssa Ngom, Email: serigneinssa.ngom@inrae.fr.
Bahia Rached, Email: rached@cnrst.ma.
Taha Chouati, Email: chouati@cnrst.ma.
Elmostafa El Fahime, Email: elfahime@cnrst.ma.
Mohamed Amar, Email: m.amar.cnrst@gmail.com.
Christel Béra-Maillet, Email: christel.maillet@inrae.fr.
References
- Aanniz T, Ouadghiri M, Melloul M, Swings J, Elfahime E, Ibijbijen J, Ismaili M, Amar M, et al. Thermophilic bacteria in Moroccan hot springs, salt marshes and desert soils. Braz J Microbiol. 2015;46(2):443–453. doi: 10.1590/S1517-838246220140219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beladjal L, et al. Life from the ashes: survival of dry bacterial spores after very high temperature exposure. Extremophiles. 2018;22(5):751–759. doi: 10.1007/s00792-018-1035-6. [DOI] [PubMed] [Google Scholar]
- Benedetti M, et al. Green production and biotechnological applications of cellwall lytic enzymes. Appl Sci. 2019;9(23):5012. doi: 10.3390/app9235012. [DOI] [Google Scholar]
- Berrada I, et al. Diversity of culturable moderately halophilic and halotolerant bacteria in a marsh and two salterns a protected ecosystem of lower Loukkos (Morocco) Afr J Microbiol Res. 2012;6(10):2419–2434. doi: 10.5897/AJMR11.1490. [DOI] [Google Scholar]
- Biely P, Singh S, Puchart V. Towards enzymatic breakdown of complex plant xylan structures: state of the art. Biotechnol Adv. 2016;34(7):1260–1274. doi: 10.1016/j.biotechadv.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchaou L, Warner NR, Tagma T, Hssaisoune M, Vengosh A. The origin of geothermal waters in morocco: multiple isotope tracers for delineating sources of water–rock interactions. Appl Geochem. 2017;84:244–253. doi: 10.1016/j.apgeochem.2017.07.004. [DOI] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1–2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics. 2020;36(6):1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, et al. Comparative genome analysis of Bacillus velezensis reveals a potential for degrading lignocellulosic biomass. 3 Biotech. 2018;8(5):253. doi: 10.1007/s13205-018-1270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun BH, Kim KH, Jeong SE, Jeon CO. Genomic and metabolic features of the bacillus amyloliquefaciens group—B. Amyloliquefaciens, B. Velezensis, and B. Siamensis—revealed by pan-genome analysis. Food Microbiol. 2019;77:146–157. doi: 10.1016/j.fm.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Cidu R, Bahaj S. Geochemistry of thermal waters from Morocco. Geothermics. 2000;29(3):407–430. doi: 10.1016/S0375-6505(00)00007-9. [DOI] [Google Scholar]
- Datta S, Saha D, Chattopadhyay L, Majumdar B. Genome comparison identifies different bacillus species in a bast fibre-retting bacterial consortium and provides insights into pectin degrading genes. Sci Rep. 2020;10(1):1–15. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre I, et al. Utilisation/upgrading of orange peel waste from a biological biorefinery perspective. Appl Microbiol Biotechnol. 2019;103(15):5975–5991. doi: 10.1007/s00253-019-09929-2. [DOI] [PubMed] [Google Scholar]
- den Besten HMW, Berendsen EM, Wells-Bennik MHJ, Straatsma H, Zwietering MH. Two complementary approaches to quantify variability in heat resistance of spores of Bacillus subtilis. Int J Food Microbiol. 2017;253:48–53. doi: 10.1016/j.ijfoodmicro.2017.04.014. [DOI] [PubMed] [Google Scholar]
- Dhillon GS, Kaur S, Brar SK. Perspective of apple processing 401 wastes as low-cost substrates for bioproduction of high value products: a review. Renewable 402 and sustainable energy reviews. Pergamon. 2013 doi: 10.1016/j.rser.2013.06.046. [DOI] [Google Scholar]
- Du Y, et al. Comparative genomic analysis of Bacillus paralicheniformis MDJK30 with its closely related species reveals an evolutionary relationship between B. paralicheniformis and B. licheniformis. BMC Genom. 2019;20(1):283. doi: 10.1186/s12864-019-5646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap CA. The status of the species Bacillus aerius. Request for an opinion. Int J Syst Evol Microbiol. 2015;65(7):2341. doi: 10.1099/ijs.0.000271. [DOI] [PubMed] [Google Scholar]
- Fan B, Blom J, Klenk H-P, Borriss R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an ‘Operational Group B. amyloliquefaciens’ within the B. subtilis species Complex. Front Microbiol. 2017;8:22. doi: 10.3389/fmicb.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, et al. Complete genome sequence of Bacillus sp. 275, producing extracellular cellulolytic, xylanolytic and ligninolytic enzymes. J Biotechnol. 2017;254:59–62. doi: 10.1016/j.jbiotec.2017.05.021. [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houfani AA, et al. Insights from enzymatic degradation of cellulose and hemicellulose to fermentable sugars: a review. Biomass Bioenergy. 2020;134:105481. doi: 10.1016/j.biombioe.2020.105481. [DOI] [Google Scholar]
- Huerta-Cepas J, et al. Fast genome-wide functional annotation through orthology assignment by EggNOG-mapper. Mol Biol Evol. 2017;34(8):2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C, et al. High-throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar SN, Pathak PD, Mandavgane SA, Kulkarni BD. Process of fruit peel waste biorefinery: a case study of citrus waste biorefinery, its environmental impacts and recommendations. Environ Sci Pollut Res. 2019;26(34):34713–34722. doi: 10.1007/s11356-019-04196-0. [DOI] [PubMed] [Google Scholar]
- Kazeem MO, Shah UKM, Baharuddin AS, AbdulRahman NA. Prospecting agro-waste cocktail: supplementation for cellulase production by a newly isolated thermophilic B. licheniformis 2D55. Appl Biochem Biotechnol. 2017;182(4):1318–1340. doi: 10.1007/s12010-017-2401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapik K, Becerra M, González-Siso MI. Microbial diversity analysis and screening for novel xylanase enzymes from the sediment of the lobios hot spring in Spain. Sci Rep. 2019;9(1):11195. doi: 10.1038/s41598-019-47637-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, et al. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madslien EH, Olsen JS, Granum PE, Blatny JM. Genotyping of B. licheniformis based on a novel multi-locus sequence typing (MLST) scheme. BMC Microbiol. 2012;12(1):230. doi: 10.1186/1471-2180-12-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GL. Determination of reducing sugar by DNS method. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Othoum G, et al. Comparative genomics study reveals Red Sea Bacillus with characteristics associated with potential microbial cell factories (MCFs) Sci Rep. 2019;9(1):23955–24900. doi: 10.1038/s41598-019-55726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parab P, Khandeparker R, Amberkar U, Khodse V. Enzymatic saccharification of seaweeds into fermentable sugars by xylanase from marine Bacillus sp. strain BT21. 3 Biotech. 2017;7(5):296. doi: 10.1007/s13205-017-0921-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, et al. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrascu O, et al. A fibrolytic potential in the human ileum mucosal microbiota revealed by functional metagenomic. Sci Rep. 2017;7:40248. doi: 10.1038/srep40248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey MW, et al. complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biol. 2004;5(10):R77. doi: 10.1186/gb-2004-5-10-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas B, et al. Submerged citric acid fermentation on orange peel autohydrolysate. J Agric Food Chem. 2008;56(7):2380–2387. doi: 10.1021/jf073388r. [DOI] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Thapa S, et al. Microbial cellulolytic enzymes: diversity and biotechnology with reference to lignocellulosic biomass degradation. Rev Environ Sci Biotechnol. 2020;19(3):621–648. doi: 10.1007/s11157-020-09536-y. [DOI] [Google Scholar]
- Wang Y, et al. Complete genome sequence of Bacillus paralicheniformis MDJK30, a plant growth-promoting rhizobacterium with antifungal activity. Genome Announc. 2017;5(25):577–594. doi: 10.1128/genomeA.00577-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westers L, Westers H, Quax WJ. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochimica Et Biophysica Acta (molecular Cell Research) 2004;1694(1–3 SPEC.ISS.):299–310. doi: 10.1016/j.bbamcr.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Himmel ME, Ding SY. Visualizing chemical functionality in plant cell walls mike himmel. Biotechnol Biofuels. 2017;10(1):263. doi: 10.1186/s13068-017-0953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-Y, et al. Complete genome sequence of Bacillus velezensis ZY-11 reveals the genetic basis for its hemicellulosic/cellulosic substrate-inducible xylanase and cellulase activities. 3 Biotech. 2018;8(11):465. doi: 10.1007/s13205-018-1490-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Cultures of the CCMM B940 and CCMM B945 Bacillus strains in BSS medium supplemented with 0.5% orange or apple peels for 48h at 37°C. a. negative control without bacteria; b. growth (turbidity) is observed for cultures on both substrates (DOCX 1356 kb)
Fig.S2 CDS annotation of the genome of B.paralicheniformis CCMM B940 for lignocellulose-degrading enzymes (TXT 4743 kb)