Abstract
Huntington disease (HD) is a devastating monogenic autosomal dominant disorder. HD is caused by a CAG expansion in exon 1 of the gene coding for huntingtin, placed in the short arm of chromosome 4. Despite its well-defined genetic origin, the molecular and cellular mechanisms underlying the disease are unclear and complex. Here, we review some of the currently known functions of the wild-type huntingtin protein and discuss the deleterious effects that arise from the expansion of the CAG repeats, which are translated into an abnormally long polyglutamine tract.
Also, we present a modern view on the molecular biology of HD as a representative of the group of polyglutamine diseases, with an emphasis on conformational changes of mutant huntingtin, disturbances in its cellular processing, and proteolytic stress in degenerating neurons. The main pathogenetic mechanisms of neurodegeneration in HD are discussed in detail, such as autophagy, impaired mitochondrial biogenesis, lysosomal dysfunction, organelle and protein transport, inflammation, oxidative stress, and transcription factor modulation. However, other unraveling mechanisms are still unknown. This practical and brief review summarizes some of the currently known functions of the wild-type huntingtin protein and the recent findings related to the mechanisms involved in HD pathogenesis.
Keywords: Huntington disease, Mechanisms, Pathophysiology
1. Introduction to Huntington's disease: genetics, pathology and clinical stages
HD is a devastating autosomal dominant disease, clinically characterized by motor, behavioral, cognitive and neuropsychiatric symptoms. The mean age at onset (AO) is 35 to 44 years [1,2].
HD is characterized by a general shrinkage of the brain and degeneration of the striatum (caudate nucleus and putamen), with specific loss of efferent medium spiny neurons (MSNs). Although the striatum appears to be the most affected region of the brain, a regionally-specific thinning of the cortical ribbon has been found in patients with HD. This regionally-selective cortical degeneration may explain the heterogeneity of clinical expression in HD.
In recent years, a large amount of evidence has supported a premotor onset many years before motor manifestations. Lately, HD has been mentioned as a neurodevelopmental disorder, with a wide range of AO, and a wide phenotypic variability. In this sense, a new definition of AO in HD is required [3,4].
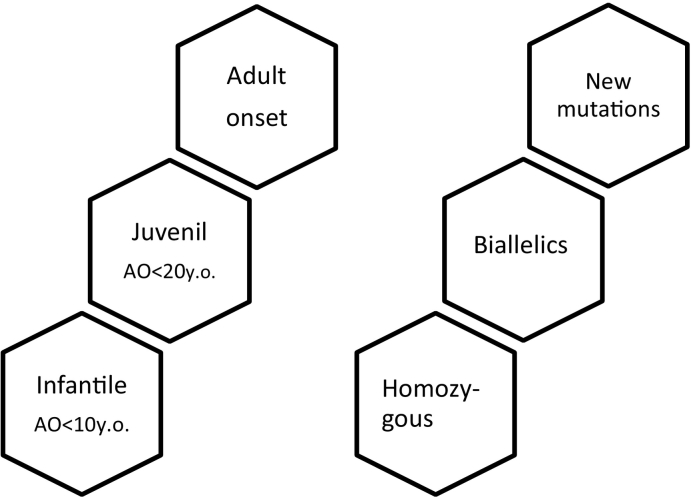
Juvenile HD is defined by an AO under 20 years; however, new evidence suggests two additional sub-categories: early infantile onset (AO < 10 years old) and juvenile onset (AO < 20 years old) [5].
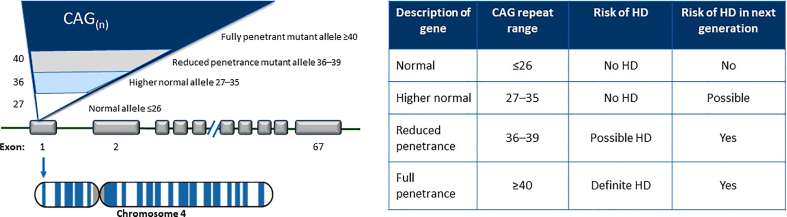
HD is a monogenic disease caused by a CAG expansion in the gene coding for htt, located at the short arm of chromosome 4. This expansion induces a polyglutamine stretch (polyQ) at the N-terminus.
A CAG repeat length ≥ 40 represents an expanded allele, inducing complete penetrance with full clinical expression. Those individuals with a range between 36 and 39 CAG repeats are associated with reduced penetrance, older age at onset, and a variable or absent clinical expression.
Intermediate alleles comprise those individuals with 27–35 CAG repeats; controversial data report a mild cognitive impairment and behavioral involvement [6,7]. The major concern in this population is the expansion during meiosis increasing the nCAG repeats in the offspring [8] (Table 1).
Table 1.
Classification of disease status in HD.
New pathogenic mutation and homozygous expansions are rare [9]. Homozygous is classically defined as individuals with >36 CAG repeats in both alleles. More recently, the term biallelic HD (B-HD) has been introduced to describe those individuals with 1 mutated allele (≥40 CAG repeats) and another non-identical CAG expanded allele (≥27).
The fact remains under discussion whether biallelic and homozygous HD patients constitute a more severe phenotype, since there may be a gain of function in both mutated alleles [1,2,7,10,11,12] (Fig. 1).
Fig. 1.
Special HD populations.
The age at onset (AO) of HD is inversely correlated to CAG repeat length; this correlation has been found to account for between 42% and 79% of the variation in AO. It is important to identify additional factors modifying AO of HD in order to understand the mechanisms of disease onset, probably revealing possible targets for treatment that could delay onset of symptoms.
New genetic technologies have identified a number of genes that modulate the expression of the HTT gene. These genes have been identified as HD genetic modifiers. Recently, at least three loci in chromosomes 8 and 15 have been involved in motor onset age [13,14].
Some possible modulator genes include nearby genes MTMR10, FAN1, and pseudogene HERC2P10 at the chr15 locus, and RRM2B and UBR5 genes at the chr8 locus. All these genes appear associated with DNA repair pathways, modulating the pathogenicity of the CAG repeats in HD [13,14].
Recently, the overexpression of the FAN1 gene (a coding gene for a DNA repair enzyme) has been associated with delayed AO and slower progression of HD. R. Goold et al. [15] described a protective effect of FAN1 mediated by restriction of CAG repeat expansion in HD medium spiny neurons (MSNs) at the dividing and post-mitotic stages, opening a new therapeutic pathway [15].
TRACK-HD and Predict-HD longitudinal studies have provided interesting clinical and biomarker information, which support HD as a neurodegenerative and neurodevelopmental disorder [16,17]. Biological and pathological changes take place a long time before symptom onset. In this sense, reliable biomarkers confer a unique window of opportunity to identify underlying biological processes before clinical manifestations.
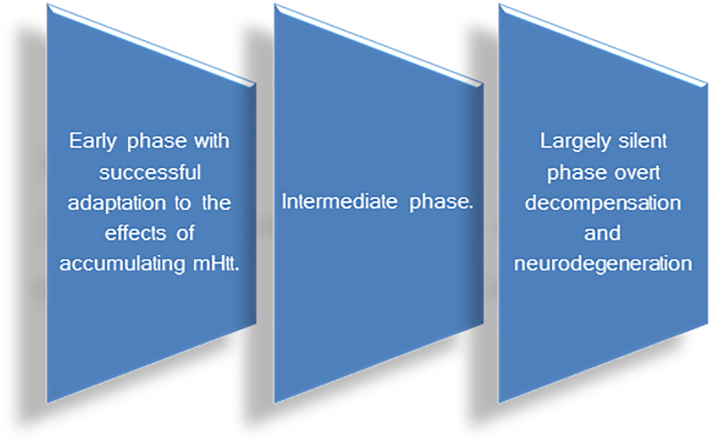
HD stages comprise (Fig. 2)
-
1.
Prenatal stage: defined by molecular findings.
-
2.
Presymptomatic stage: defined by a biological process including neurodevelopmental and neurodegenerative commitment.
-
3.
Prodromal stage: characterized by lack of insight, Stroop Word Reading, Symbol digit modality test (SDMT), verbal fluency, grasping, tapping and total motor score impairment.
-
4.
Early and moderate manifest stage: motor, cognitive, neurobehavioral and neuropsychiatric classical manifestations
Fig. 2.
HD stages.
The early stage (previously mentioned as prenatal and presymptomatic stages) is characterized by a successful adaptation to reduce the damage of accumulating mutant huntingtin (mhtt); but when this balance is broken, the next two stages, intermediate and last stage, occur. The last one is characterized by the end of the largely silent phase with the beginning of the overt decompensation and neurodegenerative stage. As in other neurodegenerative disorders, an extensive preclinical stage is present in HD [18].
In this sense, Stephan von Hörsten et al. have reported the presence of behavioral abnormalities before mhtt aggregation in a transgenic rat model [19]. Recently, the same authors have confirmed an htt developmental role supported by behavioral, cellular and molecular postnatal changes that could be reversed by a histone deacetylase inhibitor [18].
Other experimental studies that support the neurodevelopmental hypothesis:
-
1.
Wild htt expression in preimplantation stages of the embryo and htt is necessary for brain development; htt expression loss is lethal to the embryo. Cell adhesion mediated by htt may play a key role during embryonic neurogenesis.
-
2.
mhtt-induced changes affect striatal development, ergo, striatonigral and corticostriatal projections [20].
-
3.
4-week-old transgenic HD (tgHD) rats and R6/2 mice developed behavioral, cellular, and metabolic abnormalities, while 14-day-old Hdh-Q250 mice showed changes in myelination [21].
-
4.
Moreover, htt is required for ciliogenesis and neurogenesis in experimental models [22].
-
5.
mhtt-associated developmental impairments in neurogenesis contribute to regional cellular vulnerabilities to late-life stressors leading to dysfunction and culminating in cell death.
Several studies conducted in iPSC neuronal cells showed early transcriptional proteomics and transcriptional changes in affected cells in order with the developmental hypothesis [23].
In summary, developmental aberrations may play important roles in HD pathogenesis and subsequent progression.
Selective exposure to the pathogenic protein during development recapitulates characteristic features of HD. Early impairments could induce a neuronal vulnerability that could rend these neurons (particularly medium spiny neurons) susceptible to late-life stressors leading to dysfunction and apoptosis [24].
2. Wild-type huntingtin: structure and functions
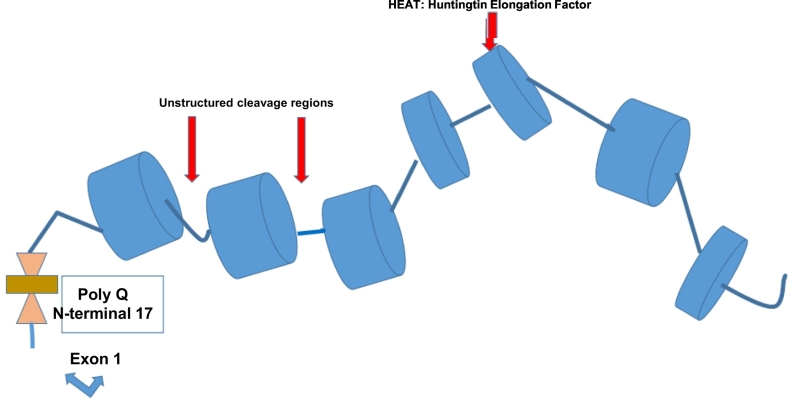
Huntingtin (htt) is an alpha-helix, 380 Kilo-Dalton protein made up by a sequence of several consensus areas called HEAT [Huntingtin, Elongation factor 3 (EF3), protein phosphatase 2A (PP2A), yeast kinase TOR1] repeats.
The HEAT sequences are resistant to the proteolytic cleavage providing a scaffold function to the protein. Exon 1 corresponds to 90 AA of HTT and comprises a N-terminal 17 amino-acid (N17) segment with α-helical structure, the PolyQ tract and a 51-residue proline-rich domain (PRD) [25].
Proteolytic and post-translational changes occur at non-HEAT consensus regions, leading to htt conformational and protein-protein modifications [26]. A number of caspases, calpains and endopeptidases contribute to the cleavage, providing a variety of N-terminal fragments, including a short sequence encoding exon 1 of the protein (Fig. 3).
Fig. 3.
Huntingtin protein schema: HEAT regions and cleavage regions, polyQ expansion at N-terminal -17.
Although the complete functions of htt are still under investigation, an extensive number of functions have been characterized.
HTT is involved in:
-
1.
brain development, with a crucial role in the formation of cortical and striatal excitatory synapses and signaling
-
2.
regulation of the transcriptional process, providing neurotrophic support and neurotrophin receptor balance
-
3.
balance of histone acetylation/deacetylation and glial activation
-
4.
mitochondrial function surveillance and biogenesis
-
5.
axonal transport of organelles by microtubules
-
6.
regulation of signaling pathways
-
7.
multimerization of mhtt
- 8.
3. Mechanisms of pathogenesis in HD
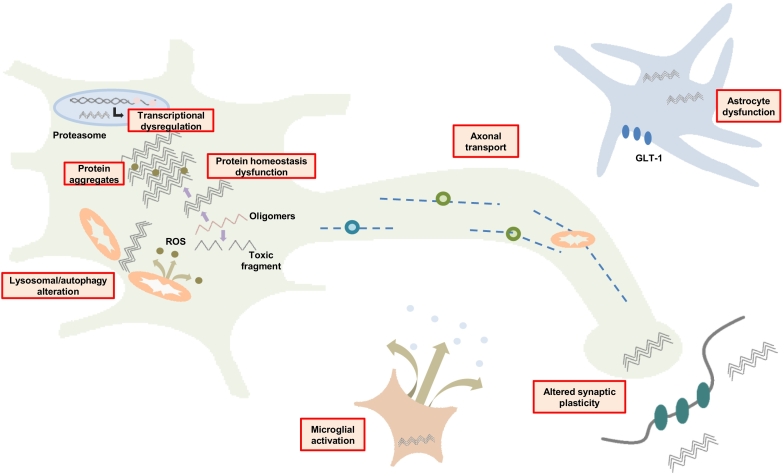
Several steps have been involved in the pathogenic mechanism of HD: mhtt affects the regulation of transcription factors, impairs mitochondrial energy pathways, alters protein homeostasis, increases the presence of aggregates that repress many factors, affects vesicular, organelle and neurotransmitter axonal trafficking, induces synaptic plasticity failure and glial activation) (Fig. 4).
Fig. 4.
Scheme of target areas affected by mHTT.
3.1. Alterations in gene expression beyond transcription: epigenetics and non-coding RNAs
We previously mentioned Mendelian genetic modifiers of HD. Epigenetic factors designate a series of inherited and non-inherited conditions capable of modifying the clinical expression of HD. They include DNA nucleotide modifications, histone protein involvement and other regulator factors including non-coding RNAs [30].
-
1.
Chromatin is a complex of DNA and protein macromolecules. Its basic structural unit is the nucleosome, composed by a DNA segment and histone protein. During cell division, it turns into a chromosome. Its functions rely on packaging DNA into a smaller volume.
-
2.
Euchromatin (open state, active) is the non-compact form of chromatin that allows transcription and replication, while heterochromatin (condensed, inactive) is the less active, more condensed form, which inhibits transcription and replication. The chromatin complex functions are directly or indirectly regulated by whtt or mhtt.
-
3.
DNA methylation is the process that contributes to increase the methylated chromatin level, inducing a more compact DNA configuration leading to a decreased transcriptional state. In health conditions, there is a balance between methylation and de-methylation [31,32].
-
4.
Histone deacetylation and acetylation balance. Histone is an alkaline protein, enriched in lysine and arginine amino acids. There are five major histone families. Apart from compacting DNA strands, histones are also associated with chromatin regulation. Histones are primary regulators of the chromatin structure, which is a dynamic process.
Mutant HTT induces an imbalance between histone deacetylation and acetylation (mediated by acetyl-coA).
Acetylation is regulated by histone acetyltransferase (HAT) enzyme and correlates with active gene transcription, producing an “open” chromatin.
Deacetylation is mediated by histone deacetylase (HDAC) enzyme and its activation is linked with transcriptional repression.
In HD there is a balance promoting deacetylation and formation of condensed inactive heterochromatin that represses the transcription of different genes [30,31,33,34].
There are many deacetylases; one of them is called Sirtuin (SIRT) that is normally blocked by the wthtt. In HD, mhtt induces its overexpression, and in turn a number of transcription factors are repressed or blocked.
There are 7 different SIRTs (SIRT1, 6, and 7 are primarily nuclear, while SIRT3, 4, and 5 are mitochondrial) that correspond to class III histone deacetylase. Among them, in HD SIRT1 modulates transcription factors related with brain-derived neurotrophic factor (BDNF), mitochondrial biogenesis, while SIRT 3 exerts its modulation on transcriptional factors involved in mitochondrial biogenesis and detoxification of reactive oxygen species (ROS) [35,36].
-
4.
Transcription factors represent a great number of proteins involved in the first step of DNA transcription.
Transcription factors involved in HD:
-
a.
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α). PGC-1α is the master regulator of genes involved in mitochondrial function, survival and biogenesis, along with energy metabolism. This protein can interact and regulate other protein activities such as transcription factor cAMP response element binding protein (CREB) which binds to DNA elements that contain a cAMP response element (CRE) sequence, such as in the PGC-1α gene promoter. In turn, mhtt interferes with CREB and another transcription complex, TFIID, leading to reduced activation of the PGC1-α gene, impacting on protein levels [37].
-
b.
Specificity Protein 1 (Sp1). Sp1 is a basal transcription factor highly regulated by post-translational modifications with a role in DNA damage response and apoptosis.
On the other hand, Sp1 modulates the expression of a number of essential oncogenes and tumor suppressors, as well as genes involved in essential cellular functions. It also plays a part in inflammation and genomic instability, as well as epigenetic silencing [38]. mhtt represses transcription of Sp1-dependent promoters.
-
c.
Repressor element-1 silencing transcription factor (REST)/neuron-restrictive silencer factor (NRSF). REST/NRSF is a transcriptional repressor factor which acts via epigenetic remodeling to silence targeted genes. The evidence suggests that REST/NRSF is a master transcription factor, which modulates neuron-specific genes not only at neurogenesis and neuronal differentiation, but also at postnatal period brain development, monitoring genes involved in synaptic plasticity and normal aging. It also plays a neuroprotective role by repressing genes that participate in oxidative stress and amyloid beta toxicity [39].
mhtt fails to interact with REST/NRSF in the cytoplasm, leading to increased REST/NRSF levels in the nucleus. Under these conditions, mhtt suppresses the transcription of BDNF and other RE1/NRSE regulated neuronal genes.
-
d.
SREBP Transcription factor. SREBP binds to SRE to regulate the transcription of genes involved in the cholesterol biosynthesis pathway. mhtt leads to a reduced expression of SREBP-dependent genes and decreases the biological effects of cholesterol biosynthesis.
-
e.
Brain-derived neurotrophic factor (BDNF). BDNF is probably one of the most relevant neurotrophic factors involved in HD, as it is involved in glutamatergic transmission, corticostriatal signaling, trafficking, astrocyte activation, and dopaminergic modulation.
whtt stimulates gene transcription from exon II promoter by binding to REST/NRSF, reducing the activity of the RE1/NRSE, allowing transcription. Moreover, mhtt affects BDNF gene transcription, impairing HAP1 function, and thus reducing BDNF vesicle delivery via the microtubules. As BDNF is released in an activity-dependent manner, reduced synaptic activity could lead to the loss of a feed-forward loop for BDNF release as well as its transcription [40].
The reduction of BDNF leads to an imbalance of two different signaling pathways, called TrkB/p75NTR complex. TrkB is involved in neuron survival, dendritic spine morphology and long-term potential (LTP) [29].
mhtt represses the BDNF-TrkB pathway, impairs synaptogenesis and neuronal survival; while mhtt action at BDNF-p75NRT interaction induces neuroinflammation, apoptosis and neurodegeneration [41].
-
f.
HSF1 Transcription factor. HSF1 has been reported as the major transcriptional regulator factor impaired in HD. This factor acts in the regulation of PGC-1α and p53 (antitumor transcription factor), as a major transcriptional regulator of the heat shock response. It is also involved in cell proliferation, inflammation, synapse formation, and energy metabolism, regulating mitochondrial activity [42].
3.1.1. Non-coding RNA
Non-coding RNAs (ncRNAs) are small fragments of RNA molecules that are transcribed from DNA but not translated to a protein. They represent 85%–90% of the genome. These ncRNAs play a significant role in:
-
a.
the differentiation of neuronal stem cells
-
b.
neuronal survival regulation and maturation
-
c.
neuronal homoeostasis along with neurite outgrowth
-
d.
neurotransmitter signaling and synaptic plasticity
-
e.
LTP and degradation of messenger RNA (mRNA) or translational silencing [30,31,43,44].
Immature ncRNAs inside the nucleus are modified by a system called Drosha that allows them to be exported to the cytoplasm. Once there, a Dicer complex (Dicer/TRBP/AGO2) cuts these single-stranded ncRNAs into small RNAs (<23 bases) or long RNAs (>200 bases).These non-coding RNAs induce mRNA degradation or transcription inhibition.
ncRNA constitutes a new therapeutic path able to interfere with mRNA leading cleavage and/or degradation, mediated by the synthetic production of antisense oligonucleotides (ASOs), ribozymes and interfering RNA (iRNA) to silence mhtt.
3.2. Huntingtin acts as a protein scaffold
As mentioned above, htt is a scaffold protein. This property is related to its capacity to bind to a HAP1 protein which then modulates other bindings with a microtubule protein complex, dynactin or kinesin, respectively. The dynactin complex promotes retrograde trafficking while kinesin promotes anterograde trafficking towards the synaptic area [45].
Htt controls organelle transport, in anterograde and retrograde directions, and by phosphorylation it modulates the capacity to bind microtubule-associated proteins in axons and dendrites within neurons. During mitosis, HTT is important for spindle pole assembly and it also regulates the kinesin 1-dependent trafficking of dynein/dynactin/NUMA/LGN to the cell cortex.
It mediates the dynein/dynactin/HAP1-dependent protein transport of to the pericentriolar material, including PCM1 protein, required for ciliogenesis.
Finally, N17 phosphorylation not only modulates the binding capacity of mhtt but also modulates the mhtt at N-terminal fragments to aggregate [[46], [47], [48]].
Recent findings show that Htt is associated and co-localized in neurons with the Anp32e phosphatase inhibitor.
Through co-precipitation experiments, Radrizzani et al. have been able to confirm the presence of the Htt protein in the synapse bound to proteins associated with fast vesicle transport in synaptic terminals: myosin-Va, spectrin alpha-II and Myosin-10/II-B. These results agree with a model in which motor proteins act as carriers using Htt as a scaffold protein, whereas vesicles move by means of microtubules or actin filaments in the cytoskeleton, as well as by post-synapse vesicle recycling involving Myosin-10.
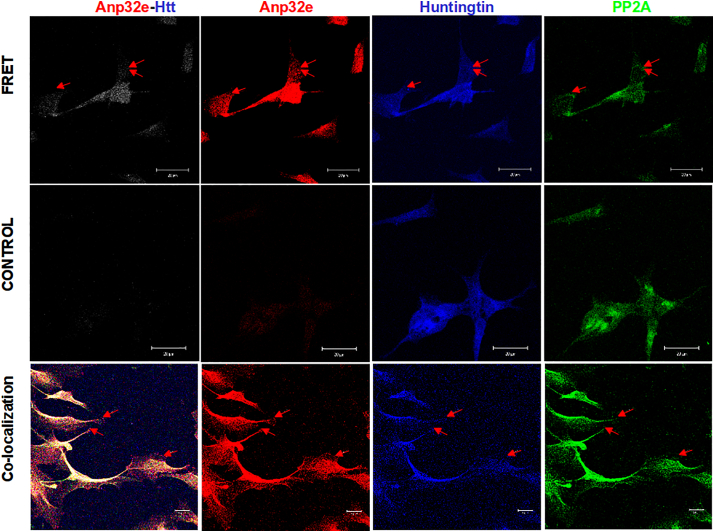
These results reinforce the proposal suggested by the Holzbaur group [57] that Htt is a scaffold protein that joins different motor proteins and might suggest a role in the transport of vesicles to their final targets. Phosphoprotein Anp32e appears to participate in signaling pathways involved in transport by means of the PP2A activity (M. Radrizzani et al., unpublished results) (see Fig. 5)
Fig. 5.
Colocalization between Huntingtin and Anp32e in SH-SY5Y cell cultures using fluorescence resonance energy transfer (FRET).
The image shows PP2A in green co-localized with Anp32e and Htt in the nucleus and cytoplasm of dopaminergic retinoic-acid differentiated SH-SY5Y cells after 10 days of exposure. The three proteins were bound with aptamers whose 5′ ends have FITC for PP2A, Cy3 for ANP32e, and Cy5 for Htt. The figure shows the images acquired with 543 nm excitation and reading above 700 nm in a confocal laser microscope. In this experiment, FRET occurs between Anp32e excited at 543 nm, and Htt receives the emission from Anp32e and emits fluorescence above 630 nm. Control cells were SH-SY5Y cells with Anp32e expression silenced with siRNA 72 h before the test. The third row shows the co-localization of all three proteins. The bar represents 20 nm.
3.3. Impairment of protein degradation systems: ubiquitin–proteasome system and autophagy
The cells possess different pathways in order to degrade abnormal proteins or organelles. Regarding proteins, the first step is played by chaperones, a number of proteins that contribute to fold the proteins in the correct helices, followed by ubiquitination, a process where the product to be degraded is bound for degradation inside the proteasome.
Whether or not proteasome is impaired in HD is still under discussion; in any case, the excess of unfolded proteins induces aggregation. These aggregates sequester a number of proteins, transcription factors, ncRNA, as well as wthtt.
Finally, the last stage is autophagy. In this case, the abnormal protein is included in a double membrane to conform an autophagosome that in turn fuses with the lysosome to initiate degradation.
When this system fails due to a deficiency in autophagosome conformation, fusion to the lysosome is impaired, and a number of misfolded proteins, ROS, and dysfunctional mitochondria remain in the cytosol, promoting cellular apoptosis.
Chaperones. Several chaperones participate in mhtt degradation. Heat shock proteins are a key regulator of proteostasis under both physiological and stress conditions [49]. HSP40, HSP70 and HSP90 (chaperones of 40, 70 and 90 kDa, respectively) promote proper protein folding and localization. HSP90 contributes to stabilize client proteins (whtt and mhtt) and inhibit their ubiquitylation, while HSP70 promotes proper folding and localization of proteins and function as well as a Chaperone-Dependent Ubiquitin Ligase CHIP dependent ubiquitylation and proteasomal degradation [50,51].
The Sigma 1 receptor (S1R) molecular chaperone is an evolutionarily conserved ligand-operated molecular chaperone, involved in neuromodulation and neuroplasticity. It is a transmembrane protein localized in an endoplasmic reticulum (ER) subdomain, the mitochondria-associated ER membrane (MAM). In specific conditions, as ER stress, S1R is activated by calcium dependent modulation. This activation attenuates the mitochondrial apoptotic pathway, with a suppressor effect on ROS and oxidative damage. Moreover, activation of S1P receptors modulates transport of BDNF, GDNF and increases proteosomal activity and mhtt degradation, leading to a protective effect in HD [[52], [53], [54]].
-
2.
Ubiquitin Proteasome System in HD. Co-chaperones, Parkin E3 ligase (ubiquitin ligase), CHIP and nucleotide exchange factor BAG3, have varied roles in assisting the proteostatic chaperone functions. In fact, mhtt misfolding can lead to subsequent aggregation and ubiquitylation. Smaller ubiquitylated aggregates may be cleared by the proteasome, whereas larger aggregates forming aggresomes or inclusion bodies will be degraded via autophagic routes [55].
-
3.
Autophagic process. Autophagy is an evolutionary process of great importance on proteins, aggregates and dysfunctional organelle clearance, besides providing energy and macromolecular precursors [62]. Physiological autophagy begins by initiation, nucleation, elongation and conformation of autophagosomes (double-membrane structures), which sequester portions of cytoplasm along with proteins or damaged cell organelles to be degraded.
In the next step, the autophagosome fuses with the lysosome (autophagolysosome/acidic proteases). The mhtt/HAP1 motor protein complex on autophagosomes disrupts the retrograde transport of autophagosomes necessary for degradation [[56], [57], [58]].
Aggregates of mhtt (oligomers) sequester proteins, chaperones, proteosome subunits, transcription factors and whtt [55,59].
The mammalian target of the rapamycin (mTOR) signaling pathway plays a critical role in regulating cell growth, proliferation, autophagy, and life span. Moreover, mTOR participates in the induction, process and finalization of autophagy. Mhtt induces a higher rate of autophagic flux, altering its protective role against mhtt cytotoxicity. mTOR phosphorylates autophagy-initiating kinase ULK1. Autophagy induction has been shown to relieve cellular toxicity caused by mhtt in the absence or failure of UPS activity.
The overexpression of HDAC6 (a cytosolic deacetylase) increases autophagy and promotes aggresomal clearance through tubulin deacetylation [55,60,61].
3.4. Mitochondrial dysfunction in HD
We have mentioned the PGC-1 alpha transcription factor, which is relevant for the mitochondrial biogenesis in HD, and which on the other hand is suppressed by mhtt.
Additionally, mhtt induces abnormalities inside the mitochondria, failures in complex II and IV of the respiratory chain, favoring the opening of transition pore with increased calcium concentration inside mitochondria, promoting the release of cytochrome C, and ROS. Moreover, N-terminal mhtt fragments inhibit the protein import machinery involved in the translocation of mitochondrial DNA or proteins from the cytosol to the mitochondria by blocking the TIM22–23 complex, and particularly at the TIM23 translocase of the inner membrane [[62], [63], [64], [65]].
Last but not least, mhtt inhibits mitophagy by sequestering mTOR, a negative regulator of autophagy.
Mitochondrial dynamics in HD. Mitochondria undergo dynamic cycles of fission and fusion. Both cycles are regulated by various GTPases. Fusion is mediated by mitofusin-1 (MFN1), mitofusin-2 (MFN2), and optic atrophy-1 (OPA1). On the other hand, fission is mediated by dynamin-related protein 1 (Drp1) [66,67]. Mhtt increases the expression of Drp1, inducing an imbalance in mitochondrial dynamics [[68], [69], [70]].
3.4.1. Inflammation and HD
A key role of neuroinflammation has been suggested in neurodegenerative disorders including HD. PET showing microglia activation with increased binding of radioligand in the striatum of the premanifest HD gene carriers contributes to support the inflammatory mechanism. The increased binding correlates with decreased raclopride binding in the striatum and enables to estimate the conversion by 5 years.
In HD, glia appear to be involved through different mechanisms: a) oligodendroglial myelination defect and failure, b) astrocyte activation that releases cytokines, activating the microglia and inducing proinflammatory and proapoptotic interleukin release, including activation of metalloproteins that induce blood brain barrier opening with migration of inflammatory blood cells into CNS [71,72].
miRNA and histone modifications affect glial and immune cells in HD. Microglia and peripheral cell mhtt content showed a decrease in migration response to chemotactic signals [[73], [74], [75]].
3.4.2. Sphingosine 1-phosphate (S1P) receptor
S1P functions in HD are many:
-
1.
Activation of Mitogen-Activated Protein Kinases (MAPK), a central signaling pathway that regulates a variety of stimulated cellular processes, including proliferation, differentiation, apoptosis and stress response [76].
-
2.
Promotion of mhtt phosphorylation.
-
3.
Increase in BDNF production and reduction in NMDA excitotoxicity.
-
4.
Anti-apoptotic effect.
-
5.
Reduction in mhtt aggregates in the striatum.
-
6.
Down-regulation of TNFα and p75 NTR [77].
3.5. Cell-to-cell transmission of aggregates
The prion hypothesis has been proposed in neurodegenerative disorders including HD. mhtt aggregates have shown a seeding capacity by tunneling, exocytosis, endocytosis [78].
Fluorescence resonance energy transfer (FRET) is a new technology able to quantify the cell-to-cell spreading capacity of mhtt aggregates. This capacity has been demonstrated in the cortex, the putamen and the caudate, and showed a survival correlation.
3.5.1. Phosphodiesterase 10 A
Phosphodiesterase 10 A enzyme appears as one of the most promising biomarkers in HD. This enzyme hydrolyzes cAMP and CGMP, acting as a signal transducer. The enzyme is highly expressed in the GABAergic medium spiny neurons and shows greatest affinity for cAMP [[79], [80], [81]].
PDE10A is involved in the direct and indirect nigrostriatal pathway affecting cortex stimulation. The radioligand 18F MNI-659 demonstrated to be a useful marker for disease progression with a marked annual binding loss at the caudate and striatum, and an estimated conversion of 10 years prior to symptom onset. A correlation between putaminal decrease, burden disease activity and UHDRS motor score has also been demonstrated. This binding reduction has been identified before volumetric changes and raclopride D2r binding reduction in the striatum.
Another PDE10A radioligand supported an early binding decrease in the striatum with a thalamic motor nucleus binding increase. These findings were able to predict disease conversion by 25 years [82,83]. The same radioligand was reported to be lower in insular and occipital areas.
To support the role of PDE10A, three different families with PDE10A gene mutations have been reported with a choreic phenotype and striatal abnormalities [[84], [85], [86]]. They have shown decreased binding not only at the striatum but also at the caudate, the thalamus, and some cortical areas.
In summary, PET striatal alterations in gene carriers have been identified. A recent meta-analysis supports early change in the putamen, caudate and striatum in premanifest individuals [82,83].
In a timeline, we can summarize that PDE10A PET is the earliest marker, on average 25 years before estimated symptom onset, followed by raclopride D2r binding, and finally PET for microglia activation.
3.6. Interplay between Mutant huntingtin and other aggregate-prone proteins
3.6.1. Tau protein
The interaction of mhtt and tau splicing factor SRSF6 may cause an imbalance between tau isoforms (4R 53R). mhtt decreases PP2B (calcineurin) levels, promoting tau hyperphosphorylation (p-tau) [87,88].
3.6.2. Α-synuclein
A number of early experimental and in vivo studies demonstrated a co-localization of a-synuclein and htt aggregates. In experimental models, the overexpression of a-synuclein increased mhtt aggregation, as well as interfered with mhtt autophagy [89,90].
3.7. Cancer and HD. A provocative concept
Epidemiological studies conducted in patients with trinucleotide repeat expansion showed a decreased incidence of cancer [91]. Two hypotheses have been suggested: one proposes the fact that CAG expansion may increase the production of ncRNA that promotes less cancer [92,93].
On the other hand, it increases the expression of the P53 tumor suppressor factor inducing programmed cell death [92,93].
Finally, experimental therapies are ongoing to demonstrate the antitumor capacity of some small inhibitor RNA produced by CAG expansions.
4. Conclusions
Many mechanisms are involved in the pathophysiology and development of HD. Dysfunctional transcription, dysfunctional mitochondria, abnormal protein degradation, abnormal trafficking, and dysregulated glutamatergic signaling are among the most important.
Disorders in the conformation and processing of mutant htt represent the core in HD molecular pathology. To date, a number of molecular pathways have been identified which either initiate or speed up the progression of the neurodegenerative process. This is important as regards developing fundamentally new approaches to treat this condition.
Funding
This research has not received any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Financial disclosures
Authors have nothing to disclose in relation with this paper.
Footnotes
Financial disclosures related to the manuscript: None.
References
- 1.Tabrizi S.J., Leavitt B.R., Landwehrmeyer G.B., Wild E.J., Saft C., Barker R.A., Blair N.F., Craufurd D., Priller J., Rickards H., Rosser A., Kordasiewicz H.B., Czech C., Swayze E.E., Norris D.A., Baumann T., Gerlach I., Schobel S.A., Paz E., Smith A.V., Bennett C.F., Lane R.M. Targeting huntingtin expression in patients with Huntington’s disease. N. Engl. J. Med. 2019;380:2307–2316. doi: 10.1056/NEJMoa1900907. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh R., Tabrizi S.J. Clinical features of Huntington’s disease. Adv. Exp. Med. Biol. 2018;1049:1–28. doi: 10.1007/978-3-319-71779-1_1. [DOI] [PubMed] [Google Scholar]
- 3.Quarrell O.W.J., Nance M.A., Nopoulos P., Reilmann R., Oosterloo M., Tabrizi S.J., Furby H., Saft C., Roos R.A.C., Squitieri F., Landwehrmeyer G.B., Burgunder J.M. Juvenile Huntington Disease Working Group of the European Huntington Disease Network. Defining pediatric Huntington disease: time to abandon the term juvenile Huntington disease? Mov. Disord. 2019;34:584–585. doi: 10.1002/mds.27640. [DOI] [PubMed] [Google Scholar]
- 4.Gatto E.M., Parisi V., Etcheverry J.L., Sanguinetti A., Cordi L., Binelli A., Persi G., Squitieri F. Juvenile Huntington disease in Argentina. Arq. Neuropsiquiatr. 2016;74:50–54. doi: 10.1590/0004-282X20150192. (PubMed PMID: 26602194) [DOI] [PubMed] [Google Scholar]
- 5.Fusilli C., Migliore S., Mazza T., Consoli F., De Luca A., Barbagallo G., Ciammola A., Gatto E.M., Cesarini M., Etcheverry J.L., Parisi V., Al-Oraimi M., Al-Harrasi S., Al-Salmi Q., Marano M., Vonsattel J.G., Sabatini U., Landwehrmeyer G.B., Squitieri F. Biological and clinical manifestations of juvenile Huntington’s disease: a retrospective analysis. Lancet Neurol. 2018;17:986–993. doi: 10.1016/S1474-4422(18)30294-1. [DOI] [PubMed] [Google Scholar]
- 6.Downing Nancy R., Lourens Spencer, De Soriano Isabella, Long Jeffrey D., Paulsen Jane S., the PREDICT-HD Investigators, Coordinators of the Huntington Study Group Phenotype characterization of HD intermediate alleles in PREDICT-HD. J Huntingtons Dis. 2016;5:357–368. doi: 10.3233/JHD-160185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cubo E., Ramos-Arroyo M.A., Martinez-Horta S., Martínez-Descalls A., Calvo S., Gil-Polo C. European HD Network. Clinical manifestations of intermediate allele carriers in Huntington disease. Neurology. 2016;87:571–578. doi: 10.1212/WNL.0000000000002944. [DOI] [PubMed] [Google Scholar]
- 8.McColgan P., Tabrizi S.J. Huntington’s disease: a clinical review. Eur. J. Neurol. 2018;25:24–34. doi: 10.1111/ene.13413. (Epub 2017 Sep 22) [DOI] [PubMed] [Google Scholar]
- 9.Kay C., Hayden M.R., Leavitt B.R. Epidemiology of Huntington disease. Handb. Clin. Neurol. 2017:31–46. doi: 10.1016/b978-0-12-801893-4.00003-1. [DOI] [PubMed] [Google Scholar]
- 10.Wild E.J., Tabrizi S.J. Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol. 2017 Oct;16(10):837–847. doi: 10.1016/S1474-4422(17)30280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Migliore S., Jankovic J., Squitieri F. Genetic counseling in Huntington’s disease: potential new challenges on horizon? Front Neurol. 2019;10(453):1–6. doi: 10.3389/fneur.2019.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cubo E., Martinez-Horta S.I., Santalo F.S., Descalls A.M., Calvo S., Gil-Polo C., Muñoz I., Llano K., Mariscal N., Diaz D., Gutierrez A., Aguado L., Ramos-Arroyo M.A. European HD Network. Clinical manifestations of homozygote allele carriers in Huntington disease. Neurology. 2019;92:e2101–e2108. doi: 10.1212/WNL.0000000000007147. [DOI] [PubMed] [Google Scholar]
- 13.Holmans P.A., Massey T.H., Jones L. Genetic modifiers of Mendelian disease: Huntington’s disease and the trinucleotide repeat disorders. Hum. Mol. Genet. 2017;26:R83–R90. doi: 10.1093/hmg/ddx261. [DOI] [PubMed] [Google Scholar]
- 14.Jain N., Chen-Plotkin A.S. Genetic modifiers in neurodegeneration. Curr Genet Med Rep. 2018;6:11–19. doi: 10.1007/s40142-018-0133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goold R., Flower M., Moss D.H., Medway C., Wood-Kaczmar A., Andre R., Farshim P., Bates G.P., Holmans P., Jones L., Tabrizi S.J. FAN1 modifies Huntington’s disease progression by stabilizing the expanded HTT CAG repeat. Hum. Mol. Genet. 2019;28:650–661. doi: 10.1093/hmg/ddy375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabrizi S.J., Scahill R.I., Owen G., Durr A., Leavitt B.R., Roos R.A., Borowsky B., Landwehrmeyer B., Frost C., Johnson H., Craufurd D., Reilmann R., Stout J.C., Langbehn D.R. TRACK-HD Investigators. Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington’s disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol. 2013;7:637–649. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- 17.Long J.D., Paulsen J.S. PREDICT-HD Investigators and Coordinators of the Huntington Study Group. Multivariate prediction of motor diagnosis in Huntington’s disease: 12 years of PREDICT-HD. Mov. Disord. 2015;12:1664–1672. doi: 10.1002/mds.26364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siebzehnrübl F.A., Raber K.A., Urbach Y.K., Schulze-Krebs A., Canneva F., Moceri S., Habermeyer J., Achoui D., Gupta B., Steindler D.A., Stephan M., Nguyen H.P., Bonin M., Riess O., Bauer A., Aigner L., Couillard-Despres S., Paucar M.A., Svenningsson P., Osmand A., Andreew A., Zabel C., Weiss A., Kuhn R., Moussaoui S., Blockx I., Van der Linden A., Cheong R.Y., Roybon L., Petersén Å., von Hörsten S. Early postnatal behavioral, cellular, and molecular changes in models of Huntington disease are reversible by HDAC inhibition. Proc Natl Acad Sci U S A. 2018;115:E8765–E8774. doi: 10.1073/pnas.1807962115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Hörsten Stephan, von Hörsten S., Schmitt I., Nguyen H.P., Holzmann C., Schmidt T., Walther T., Bader M., Pabst R., Kobbe P., Krotova J., Stiller D., Kask A., Vaarmann A., Rathke-Hartlieb S., Schulz J.B., Grasshoff U., Bauer I., Vieira-Saecker A.M., Paul M., Jones L., Lindenberg K.S., Landwehrmeyer B., Bauer A., Li X.J., Riess O. Transgenic rat model of Huntington’s disease. Hum. Mol. Genet. 2003;12:617–624. doi: 10.1093/hmg/ddg075. (PubMed PMID: 12620967) [DOI] [PubMed] [Google Scholar]
- 20.Bhide P.G., Day M., Sapp E., Schwarz C., Sheth A., Kim J., Young A.B., Penney J., Golden J., Aronin N., DiFiglia M. Expression of normal and mutant huntingtin in the developing brain. J. Neurosci. 1996;16:5523–5535. doi: 10.1523/JNEUROSCI.16-17-05523.1996. (PubMed PMID: 8757264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rangel-Barajas C., Rebec G.V. Overview of Huntington’s disease models: neuropathological, molecular, and behavioral differences. Curr Protoc Neurosci. 2018;83 doi: 10.1002/cpns.47. [DOI] [PubMed] [Google Scholar]
- 22.Haremaki T., Deglincerti A., Brivanlou A.H. Huntingtin is required for ciliogenesis and neurogenesis during early Xenopus development. Dev. Biol. 2015;408:305–315. doi: 10.1016/j.ydbio.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Świtońska K., Szlachcic W.J., Handschuh L., Wojciechowski P., Marczak Ł., Stelmaszczuk M., Figlerowicz M., Figiel M. Identification of altered developmental pathways in human juvenile HD iPSC with 71Q and 109Q using transcriptome profiling. Front. Cell. Neurosci. 2019;12:528. doi: 10.3389/fncel.2018.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.HD iPSC Consortium Developmental alterations in Huntington's disease neural cells and pharmacological rescue in cells and mice. Nat Neurosci. 2017;5:648–660. doi: 10.1038/nn.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caterino M., Squillaro T., Montesarchio D., Giordano A., Giancola C., Melone M.A.B. Huntingtin protein: a new option for fixing the Huntington’s disease countdown clock. Neuropharmacology. 2018;135:126–138. doi: 10.1016/j.neuropharm.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Cisbani G., Cicchetti F. An in vitro perspective on the molecular mechanisms underlying mutant huntingtin protein toxicity. Cell Death Dis. 2012;3 doi: 10.1038/cddis.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munoz-Sanjuan I., Bates G.P. The importance of integrating basic and clinical research toward the development of new therapies for Huntington disease. J. Clin. Invest. 2011;121:476–483. doi: 10.1172/JCI45364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franco-Iborra S., Plaza-Zabala A., Montpeyo M., Sebastian D., Vila M., Martinez-Vicente M. Mutant HTT (huntingtin) impairs mitophagy in a cellular model of Huntington disease. Autophagy. 2020:1–18. doi: 10.1080/15548627.2020.1728096. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saavedra A., García-Díaz Barriga G., Pérez-Navarro E., Alberch J. Huntington’s disease: novel therapeutic perspectives hanging in the balance. Expert Opin. Ther. Targets. 2018;22:385–399. doi: 10.1080/14728222.2018.1465930. [DOI] [PubMed] [Google Scholar]
- 30.Klein C.J., Benarroch E.E. Epigenetic regulation: basic concepts and relevance to neurologic disease. Neurology. 2014;82:1833–1840. doi: 10.1212/WNL.0000000000000440. [DOI] [PubMed] [Google Scholar]
- 31.Arenas F., Garcia-Ruiz C., Fernandez-Checa J.C. Intracellular cholesterol trafficking and impact in neurodegeneration. Front. Mol. Neurosci. 2017;10:382. doi: 10.3389/fnmol.2017.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francelle L., Lotz C., Outeiro T., Brouillet E., Merienne K. Contribution of neuroepigenetics to Huntington’s disease. Front. Hum. Neurosci. 2017;11:17. doi: 10.3389/fnhum.2017.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassi S., Tripathi T., Monziani A., Di Leva F., Biagioli M. Epigenetics of Huntington’s disease. Adv. Exp. Med. Biol. 2017;978:277–299. doi: 10.1007/978-3-319-53889-1_15. [DOI] [PubMed] [Google Scholar]
- 34.Valor L.M., Guiretti D. What’s wrong with epigenetics in Huntington’s disease? Neuropharmacology. 2014;80:103–114. doi: 10.1016/j.neuropharm.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 35.Neo S.H., Tang B.L. Sirtuins as modifiers of Huntington’s disease (HD) pathology. Prog. Mol. Biol. Transl. Sci. 2018;154:105–145. doi: 10.1016/bs.pmbts.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 36.Herskovits A.Z., Guarente L. Sirtuin deacetylases in neurodegenerative diseases of aging. Cell Res. 2013;23:746–758. doi: 10.1038/cr.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui L., Jeong H., Borovecki F., Parkhurst C.N., Tanese N., Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. (PubMed PMID: 17018277) [DOI] [PubMed] [Google Scholar]
- 38.Beishline K., Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015;282:224–258. doi: 10.1111/febs.13148. [DOI] [PubMed] [Google Scholar]
- 39.Hwang J.Y., Zukin R.S. REST, a master transcriptional regulator in neurodegenerative disease. Curr. Opin. Neurobiol. 2018;48:193–200. doi: 10.1016/j.conb.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyebji S., Hannan A.J. Synaptopathic mechanisms of neurodegeneration and dementia: insights from Huntington’s disease. Prog. Neurobiol. 2017;153:18–45. doi: 10.1016/j.pneurobio.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 41.Smith-Dijak A.I., Sepers M.D., Raymond L.A. Alterations in synaptic function and plasticity in Huntington disease. J. Neurochem. 2019;150:346–365. doi: 10.1111/jnc.14723. [DOI] [PubMed] [Google Scholar]
- 42.Intihar T.A., Martinez E.A., Gomez-Pastor R. Mitochondrial dysfunction in Huntington’s disease; interplay between HSF1, p53 and PGC-1α transcription factors. Front. Cell. Neurosci. 2019;13:103. doi: 10.3389/fncel.2019.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Padaro P.A., Bredy T.W. Emerging role of non-coding RNA in neural plasticity, cognitive function, and neuropsychiatric disorders. Front. Genet. 2012 Jul 13;3:132. doi: 10.3389/fgene.2012.00132. (eCollection 2012. PubMed PMID: 22811697; PubMed Central PMCID: PMC3395882) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan J.J., Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19051310. pii: E1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humbert S., Saudou F. Huntington’s disease: intracellular signaling pathways and neuronal death. J Soc Biol. 2005;199:247–251. doi: 10.1051/jbio:2005026. (Review. French. PubMed PMID: 16471265) [DOI] [PubMed] [Google Scholar]
- 46.Cariulo C., Azzollini L., Verani M., Martufi P., Boggio R., Chiki A., Deguire S.M., Cherubini M., Gines S., Marsh J.L., Conforti P., Cattaneo E., Santimone I., Squitieri F., Lashuel H.A., Petricca L., Caricasole A. Phosphorylation of huntingtin at residue T3 is decreased in Huntington’s disease and modulates mutant huntingtin protein conformation. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E10809–E10818. doi: 10.1073/pnas.1705372114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeGuire S.M., SM DeGuire, Ruggeri F.S., Fares M.B., Chiki A., Cendrowska U., Dietler G., Lashuel H.A. N-terminal Huntingtin (Htt) phosphorylation is a molecular switch regulating Htt aggregation, helical conformation, internalization, and nuclear targeting. J. Biol. Chem. 2018;293(2018):18540–18558. doi: 10.1074/jbc.RA118.004621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X.J., Saudou F., Humbert S. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27:2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schopf F.H., Biebl M.M., Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 50.Saudou F., Humbert S. The biology of Huntingtin. Neuron. 2016;89:910–926. doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 51.Bhat K.P., Yan S., Wang C.E., Li S., Li X.J. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc. Natl. Acad. Sci. U. S. A. 2014;111:5706–5711. doi: 10.1073/pnas.1402215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ionescu A., Gradus T., Altman T., Maimon R., Saraf Avraham N., Geva M., Hayden M., Perlson E. Targeting the sigma-1 receptor via pridopidine ameliorates central features of ALS pathology in a SOD1(G93A) model. Cell Death Dis. 2019;10:210. doi: 10.1038/s41419-019-1451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su T.P., Su T.C., Nakamura Y., Tsai S.Y. The sigma-1 receptor as a pluripotent modulator in living systems. Trends Pharmacol. Sci. 2016 Apr;37(4):262–278. doi: 10.1016/j.tips.2016.01.003. (Epub 2016 Feb 9. Review. PubMed PMID: 26869505; PubMed Central PMCID: PMC4811735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shacham T., Sharma N., Lederkremer G.Z. Protein misfolding and ER stress in Huntington’s disease. Front. Mol. Biosci. 2019;6:20. doi: 10.3389/fmolb.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harding R.J., Tong Y.F. Proteostasis in Huntington’s disease: disease mechanisms and therapeutic opportunities. Acta Pharmacol. Sin. 2018;39:754–769. doi: 10.1038/aps.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Z., Yang C., Iyaswamy A., Krishnamoorthi S., Sreenivasmurthy S.G., Liu J., Wang Z., Tong B.C., Song J., Lu J., Cheung K.H., Li M. Balancing mTOR signaling and autophagy in the treatment of Parkinson’s disease. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20030728. pii: E728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong Y.C., Holzbaur E.L. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci. 2014;34:1293–1305. doi: 10.1523/JNEUROSCI.1870-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin D.D., Ladha S., Ehrnhoefer D.E., Hayden M.R. Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci. 2015 Jan;38(1):26–35. doi: 10.1016/j.tins.2014.09.003. (Epub 2014 Oct 2. Review. PubMed PMID: 25282404) [DOI] [PubMed] [Google Scholar]
- 59.Labbadia J., Morimoto R.I. Huntington’s disease: underlying molecular mechanisms and emerging concepts. Trends Biochem. Sci. 2013;38:378–385. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abd-Elrahman K.S., Ferguson S.S.G. Modulation of mTOR and CREB pathways following mGluR5 blockade contribute to improved Huntington’s pathology in zQ175 mice. Mol Brain. 2019;12:35. doi: 10.1186/s13041-019-0456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Child N.D., Benarroch E.E. mTOR: its role in the nervous system and involvement in neurologic disease. Neurology. 2014;83:1562–1572. doi: 10.1212/WNL.0000000000000922. [DOI] [PubMed] [Google Scholar]
- 62.Carmo C., Naia L., Lopes C., Rego A.C. Mitochondrial dysfunction in Huntington’s disease. Adv. Exp. Med. Biol. 2018;1049:59–83. doi: 10.1007/978-3-319-71779-1_3. (Review. PubMed PMID: 29427098) [DOI] [PubMed] [Google Scholar]
- 63.Costa V., Scorrano L. Shaping the role of mitochondria in the pathogenesis of Huntington’s disease. EMBO J. 2012;31:1853–1864. doi: 10.1038/emboj.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franco-Iborra S., Vila M., Perier C. Mitochondrial quality control in neurodegenerative diseases: focus on Parkinson’s disease and Huntington’s disease. Front. Neurosci. 2018;12:342. doi: 10.3389/fnins.2018.00342. (eCollection 2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacPherson L., Tokatlidis K. Protein trafficking in the mitochondrial intermembrane space: mechanisms and links to human disease. Biochem. J. 2017;474:2533–2545. doi: 10.1042/BCJ20160627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 67.Cherubini M., Ginés S. Mitochondrial fragmentation in neuronal degeneration: toward an understanding of HD striatal susceptibility. Biochem. Biophys. Res. Commun. 2017;483:1063–1068. doi: 10.1016/j.bbrc.2016.08.042. [DOI] [PubMed] [Google Scholar]
- 68.Cha M.Y., Chen H., Chan D. Removal of the mitochondrial fission factor Mff exacerbates neuronal loss and neurological phenotypes in a Huntington’s disease mouse model. PLoS Curr. 2018;10 doi: 10.1371/currents.hd.a4e15b80c4915c828d39754942c6631f. pii:ecurrents.hd.a4e15b80c4915c828d39754942c6631f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kojer K., Hering T., Bazenet C., Weiss A., Herrmann F., Taanman J.W., Orth M. Huntingtin aggregates and mitochondrial pathology in skeletal muscle but not heart of late-stage R6/2 mice. J Huntingtons Dis. 2019;8:145–159. doi: 10.3233/JHD-180324. [DOI] [PubMed] [Google Scholar]
- 70.Roe A.J., Qi X. Drp1 phosphorylation by MAPK1 causes mitochondrial dysfunction in cell culture model of Huntington’s disease. Biochem. Biophys. Res. Commun. 2018;496:706–711. doi: 10.1016/j.bbrc.2018.01.114. (Epub 2018 Jan 31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Connolly C., Magnusson-Lind A., Lu G., Wagner P.K., Southwell A.L., Hayden M.R., Björkqvist M., Leavitt B.R. Enhanced immune response to MMP3 stimulation in microglia expressing mutant huntingtin. Neuroscience. 2016;325:74–88. doi: 10.1016/j.neuroscience.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 72.Bardile Costanza Ferrari, Garcia-Miralles Marta, Caron Nicholas S., Rayan Nirmala Arul, Langley Sarah R., Harmston Nathan, Rondelli Ana Maria, Teo Roy Tang Yi, Waltl Sabine, Anderson Lisa M., Bae Han-Gyu, Jung Sangyong, Williams Anna, Prabhakar Shyam, Petretto Enrico, Hayden Michael R., Pouladi Mahmoud A. Intrinsic mutant HTT-mediated defects in oligodendroglia cause myelination deficits and behavioral abnormalities in Huntington disease. PNAS. 2019;116:9622–9627. doi: 10.1073/pnas.1818042116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jimenez-Sanchez M., Licitra F., Underwood B.R., Rubinsztein D.C. Huntington’s disease: mechanisms of pathogenesis and therapeutic strategies. Cold Spring Harb Perspect Med. 2017;7 doi: 10.1101/cshperspect.a024240. pii: a024240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jimenez-Sanchez M., Lam W., Hannus M., Sönnichsen B., Imarisio S., Fleming A., Tarditi A., Menzies F., Dami T.E., Xu C., Gonzalez-Couto E., Lazzeroni G., Heitz F., Diamanti D., Massai L., Satagopam V.P., Marconi G., Caramelli C., Nencini A., Andreini M., Sardone G.L., Caradonna N.P., Porcari V., Scali C., Schneider R., Pollio G., O’Kane C.J., Caricasole A., Rubinsztein D.C. siRNA screen identifies QPCT as a druggable target for Huntington’s disease. Nat Chem Biol. 2015;11(5):347–354. doi: 10.1038/nchembio.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Niranjan R. Recent advances in the mechanisms of neuroinflammation and their roles in neurodegeneration. Neurochem. Int. 2018;120:13–20. doi: 10.1016/j.neuint.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 76.Plotnikov A., Zehorai E., Procaccia S., Seger R. The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Biophys. Acta. 2011;1813:1619–1633. doi: 10.1016/j.bbamcr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 77.DiPardo A., Maglione V., Di Pardo A., Maglione V. The S1P axis: new exciting route for treating Huntington’s disease. Trends Pharmacol Sci. 2018;39:468–480. doi: 10.1016/j.tips.2018.02.009. (2018) [DOI] [PubMed] [Google Scholar]
- 78.Jansen A.H., Batenburg K.L., Pecho-Vrieseling E., Reits E.A. Visualization of prion-like transfer in Huntington’s disease models. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:793–800. doi: 10.1016/j.bbadis.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 79.Nishi A., Kuroiwa M., Shuto T. Mechanisms for the modulation of dopamine d(1) receptor signaling in striatal neurons. Front. Neuroanat. 2011;5:43. doi: 10.3389/fnana.2011.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beaumont V., Zhong S., Lin H., Xu W., Bradaia A., Steidl E., Gleyzes M., Wadel K., Buisson B., Padovan-Neto F.E., Chakroborty S., Ward K.M., Harms J.F., Beltran J., Kwan M., Ghavami A., Häggkvist J., Tóth M., Halldin C., Varrone A., Schaab C., Dybowski J.N., Elschenbroich S., Lehtimäki K., Heikkinen T., Park L., Rosinski J., Mrzljak L., Lavery D., West A.R., Schmidt C.J., Zaleska M.M., Munoz-Sanjuan I. Phosphodiesterase 10A inhibition improves cortico-basal ganglia function in Huntington’s disease models. Neuron. 2016;92:1220–1237. doi: 10.1016/j.neuron.2016.10.064. [DOI] [PubMed] [Google Scholar]
- 81.Fazio P., Schain M., Mrzljak L., Amini N., Nag S., Al-Tawil N., Fitzer-Attas C.J., Bronzova J., Landwehrmeyer B., Sampaio C., Halldin C., Varrone A. Patterns of age related changes for phosphodiesterase type-10A in comparison with dopamine D(2/3) receptors and sub-cortical volumes in the human basal ganglia: a PET study with (18)F-MNI-659 and (11)C-raclopride with correction for partial volume effect. Neuroimage. 2017;152:330–339. doi: 10.1016/j.neuroimage.2017.02.047. [DOI] [PubMed] [Google Scholar]
- 82.Wood H. Neurodegenerative disease: changes in brain phosphodiesterase 10A levels in neurodegenerative basal ganglia disorders. Nat. Rev. Neurol. 2015;11:483. doi: 10.1038/nrneurol.2015.148. [DOI] [PubMed] [Google Scholar]
- 83.Boscutti G., A Rabiner E., Plisson C. PET radioligands for imaging of the PDE10A in human: current status. Neurosci. Lett. 2019;691:11–17. doi: 10.1016/j.neulet.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 84.Miyatake S., Koshimizu E., Shirai I., Kumada S., Nakata Y., Kamemaru A., Nakashima M., Mizuguchi T., Miyake N., Saitsu H., Matsumoto N. A familial case of PDE10A-associated childhood-onset chorea with bilateral striatal lesions. Mov. Disord. 2018;33:177–179. doi: 10.1002/mds.27219. [DOI] [PubMed] [Google Scholar]
- 85.Esposito S., Carecchio M., Tonduti D., Saletti V., Panteghini C., Chiapparini L., Zorzi G., Pantaleoni C., Garavaglia B., Krainc D., Lubbe S.J., Nardocci N., Mencacci N.E. A PDE10A de novo mutation causes childhood-onset chorea with diurnal fluctuations. Mov Disord. 2017;32:1646–1647. doi: 10.1002/mds.27175. [DOI] [PubMed] [Google Scholar]
- 86.Narayanan D.L., Deshpande D., Das Bhowmik A., Varma D.R., Dalal A. Familial choreoathetosis due to novel heterozygous mutation in PDE10A. Am. J. Med. Genet. A. 2018;176:146–150. doi: 10.1002/ajmg.a.38507. [DOI] [PubMed] [Google Scholar]
- 87.Coudert L., Nonaka T., Bernard E., Hasegawa M., Schaeffer L., Leblanc P. Phosphorylated and aggregated TDP-43 with seeding properties are induced upon mutant Huntingtin (mHtt) polyglutamine expression in human cellular models. Cell. Mol. Life Sci. 2019;76:2615–2632. doi: 10.1007/s00018-019-03059-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baskota S.U., Lopez O.L., Greenamyre J.T., Kofler J. Spectrum of tau pathologies in Huntington’s disease. Lab. Investig. 2019;99:1068–1077. doi: 10.1038/s41374-018-0166-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Monsellier E., Bousset L., Melki R. α-Synuclein and huntingtin exon 1 amyloid fibrils bind laterally to the cellular membrane. Sci Rep. 2016;6:19180. doi: 10.1038/srep19180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.St-Amour I., Turgeon A., Goupil C., Planel E., Hébert S.S. Co-occurrence of mixed proteinopathies in late-stage Huntington’s disease. Acta Neuropathol. 2018;135:249–265. doi: 10.1007/s00401-017-1786-7. [DOI] [PubMed] [Google Scholar]
- 91.Coarelli G., Diallo A., Thion M.S., Rinaldi D., Calvas F., Boukbiza O.L., Tataru A., Charles P., Tranchant C., Marelli C., Ewenczyk C., Tchikviladzé M., Monin M.L., Carlander B., Anheim M., Brice A., Mochel F., Tezenas du Montcel S., Humbert S., Durr A. Low cancer prevalence in polyglutamine expansion diseases. Neurology. 2017;88:1114–1119. doi: 10.1212/WNL.0000000000003725. [DOI] [PubMed] [Google Scholar]
- 92.Murmann A.E., Yu J., Opal P., Peter M.E. Trinucleotide repeat expansion diseases, RNAi, and cancer. Trends Cancer. 2018;4:684–700. doi: 10.1016/j.trecan.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Murmann A.E., Gao Q.Q., Putzbach W.E., Patel M., Bartom E.T., Law C.Y., Bridgeman B., Chen S., McMahon K.M., Thaxton C.S., Peter M.E. Small interfering RNAs based on huntingtin trinucleotide repeats are highly toxic to cancer cells. EMBO Rep. 2018;19 doi: 10.15252/embr.201745336. pii: e45336. [DOI] [PMC free article] [PubMed] [Google Scholar]