Abstract
Liver cancer is the third leading cause of cancer-associated mortality worldwide. By the time liver cancer is diagnosed, it is already in the advanced stage. Therefore, novel therapeutic strategies need to be identified to improve the prognosis of patients with liver cancer. In the present study, the profiles of GSE84402, GSE19665 and GSE121248 were used to screen differentially expressed genes (DEGs). Subsequently, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses for DEGs were conducted using the Database for Annotation, Visualization and Integrated Discovery. The protein-protein interaction network was established to screen the hub genes associated with liver cancer. Additionally, the expression levels of hub genes were validated using the Gene Expression Profiling Interactive Analysis and Oncomine databases. In addition, the prognostic value of hub genes in patients with liver cancer was analyzed using Kaplan-Meier Plotter. It was demonstrated that 132 and 246 genes were upregulated and downregulated, respectively, in patients with liver cancer. Among these DEGs, 10 hub genes with high connected node values were identified, which were AURKA, BIRC5, BUB1B, CCNA2, CCNB1, CCNB2, CDC20, CDK1, DLGAP5 and MAD2L1. CDK1 and CCNB1 had the most connection nodes and the highest score and were therefore, the most significantly expressed. In addition, it was demonstrated that high expression levels of CDK1 and CCNB1 were associated with poor overall survival time of patients with liver cancer. Dihydroartemisinin (DHA) is a Food and Drug Administration-approved drug, which is derived from the traditional Chinese medicine Artemisia annua Linn. DHA inhibits cell proliferation in numerous cancer types, including liver cancer. In our previous study, it was revealed that DHA inhibited the proliferation of HepG2215 cells. In the present study, it was further demonstrated that DHA reduced the expression levels of CDK1 and CCNB1 in liver cancer. Overall, CDK1 and CCNB1 were the potential therapeutic targets of liver cancer, and DHA reduced the expression levels of CDK1 and CCNB1, and inhibited the proliferation of liver cancer cells.
Keywords: liver cancer, bioinformatics analysis, dihydroartemisinin, CDK1, CCNB1
Introduction
Liver cancer is the third leading cause of cancer-associated mortality worldwide (1). Currently, it is treated with curative and palliative approaches. In clinical treatment, ablation (2), excision (3) and transplantation therapies (4) may be considered for patients with early-stage cancer. For patients with advanced cancer, interventional therapy (5) and targeted sorafenib therapy (2,6) are used as palliative treatment. However, current patients with liver cancer usually have a poor prognosis (3). This is caused by complex reasons, and poor diagnostic and prognostic assessments are the most noteworthy aspects (3). Therefore, understanding the pathogenesis of liver cancer is of great significance for its treatment.
In recent years, microarray technology and bioinformatics tools have been used to identify novel genes associated with the development, diagnosis and prognosis of tumors (7,8). Numerous cancer-related databases, such as Gene Expression Omnibus (GEO), Oncomine and Gene Expression Profiling Interactive Analysis (GEPIA), have emerged. The analysis of the expression and association with prognosis of cancer-related genes is helpful for the diagnosis and treatment of cancer. Previous studies have indicated that CDK1 and CCNB1 promote G2/M transformation (9) and serve a key role in regulating the cell cycle of mammalian cells (10). Therefore, CDK1 and CCNB1 are potential therapeutic targets of liver cancer, and the inhibition of their expression is crucial for the treatment of liver cancer.
Dihydroartemisinin (DHA) is the main extraction ingredient of artemisinin, which extracted from the traditional Chinese medicine of Artemisia annua Linn (11). In our previous study, it was identified that DHA inhibited the proliferation of HepG2215 cells (12). However, to the best of our knowledge, the mechanism by which DHA inhibits the proliferation of liver cancer cells remains unclear. The aim of the present study was to investigate the mechanism by which DHA inhibited the proliferation of liver cancer cells by inhibiting the expression of CDK1 and CCNB1.
Materials and methods
Acquisition of microarray data
The expression profile microarrays of three genes [GSE84402 (13), GSE19665 (14) and GSE121248 (13)] were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The samples were acquired on the same platform (GPL570). These profile microarrays included liver cancer and corresponding para-cancerous tissues. The array data of GSE84402 corresponded to 14 liver cancer tissues and 14 corresponding para-cancerous tissues, those of GSE19665 corresponded to 10 liver cancer tissues and 10 non-cancerous tissues, and those of GSE121248 corresponded to 70 liver cancer tissues and 37 non-cancerous tissues.
Identification of differentially expressed genes (DEGs)
The DEGs between tumor tissues and non-tumor tissues were identified using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) with the cut-off criteria of |log fold change|>1.0 and adjusted P-value (adjust-P) <0.05. These genes were identified using the Venn diagram webtool (bioinformatics.psb.ugent.be/webtools/Venn/).
Analysis of Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
GO analysis is often used for functional enrichment research in the three aspects of biological processes (BPs), molecular functions (MFs) and cellular component (CCs) (15). The pathways were analyzed using KEGG pathway enrichment (16). Subsequently, GO and KEGG analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID version 6.8) tools (https://david.ncifcrf.gov/). Visual analysis was performed on bioinformatics tools (http://www.bioinformatics.com.cn/) to display the bubble diagrams of DEGs. Adjust-P<0.01 and counts >10 were considered statistically significant.
Construction of the protein-protein interaction (PPI) network
Protein interaction network analysis was performed using the Search Tool for the Retrieval of Interacting Genes/Proteins (String version 11) (https://string-db.org/) database to evaluate protein interactions. Subsequently, Cytoscape version 3.6.0 software (https://cytoscape.org/) was used to visualize the PPI network. The connected nodes are important to maintain the stability of the whole network. MCODE plugin was used to screen the key subnetworks in PPI network. The degree of each protein node was calculated using CytoHubba, a plug-in for Cytoscape software (17). The first 10 genes which included AURKA, BIRC5, BUB1B, CCNA2, CCNB1, CCNB2, CDC20, CDK1, DLGAP5 and MAD2L1were identified as the hub genes with the screening criteria of degree >10.
Analysis of hub gene expression using GEPIA and the Oncomine database
GEPIA (http://gepia.cancer-pku.cn/) and Oncomine (https://www.oncomine.org/resource/login.html) are newly developed tools, which can analyze the gene expression data of tumor tissues and normal tissues. The cut-off values of some parameters were as follows: P-value, <0.05; fold change, ≥2; gene rank, top 10%; data type, mRNA. The gene names were entered according to these parameters and the expression of hub genes in liver cancer was visualized.
Immunohistochemical analysis using Human Protein Atlas (HPA)
The HPA version 20.1 website (https://www.proteinatlas.org/) contains the immunohistochemical expression data for ~20 of the more common cancers. The expression of nine hub genes in normal tissues and hepatocellular carcinoma (HCC) tissues was analyzed using HPA. However, the immunohistochemical image of BUB1B was not found.
Survival analysis of the hub genes
Kaplan-Meier Plotter Liver cancer RNA-seq (KM Plotter; http://kmplot.com/analysis/) is a survival analysis database, which is clinically used to evaluate the relationship between genes and overall survival with log-rank P-value and hazard ratios (HR) with 95% confidence intervals. Kaplan-Meier Plotter database divided patients into 2 groups based on the median expression level of each of the 10 genes. Log-rank test results with P<0.05 considered as statistically significant.
Cell line and drug treatment
HepG2215 and HepG2 cells were purchased from American Type Culture Collection. HepG2215 and HepG2 cells were identified by Shanghai Fuheng Biological Technology Co., Ltd. and Shanghai Biowing Applied Biotechnology Co., Ltd., respectively.
The cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in an atmosphere with 100% humidity and 5% CO2.
DHA was purchased from Tokyo Chemical Industry Co., Ltd. DHA was dissolved in DMSO (Sigma-Aldrich; Merck KGaA) and stored at −20°C. HepG2215 and HepG2 cells were treated with DHA (21.5 µM) for 24 h at 37°C in an atmosphere with 100% humidity and 5% CO2. Culture medium containing 0.1% DMSO was used as the control.
Assay of cell viability
HepG2 cells were seeded in 96-well plates (1×104 cells/well) and treated with DHA at different concentrations (5, 10, 20 and 40 µM) for 12, 24, 36 and 48 h at 37°C in an atmosphere with 5% CO2. A Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) was used to determine cell viability according to the manufacturer's protocol. Cells were mixed with CCK8 and incubated at 37°C in an atmosphere with 5% CO2 for 2 h. Subsequently, optical density was monitored at 450 nm with 650 nm as a reference wavelength by a Multiskan Spectrum Microplate Reader (Thermo Fisher Scientific, Inc.). The infected cells were observed under a phase contrast microscope (Motic AE31; Motic; magnification, ×100). Finally, the cell viability values were calculated as previously described (18). IC50 values were obtained from the cytotoxicity curves using Softmax Pro 5 Software (19).
Transcriptomic analysis
HepG2215 and HepG2 cells (1×107 cells/well) were seeded into 6-well plates. The total RNA of HepG2 and HepG2215 cells was isolated using TRIzol (cat. no. R1100; Beijing Solarbio Science and Technology Co. Ltd.). RNA integrity was detected with 1.2% agarose gel (cat. no. 111860; Biowest). Gel imaging was performed using a Bio-RAD (Bio-RAD GelDoc 2000; Bio-Rad Inc.) and analyzed by Personal Biotechnology Co., Ltd. The sequencing kit was NovaSeq 6000 Reagent kit v.1.5 (Illumina, Inc.). In brief, the mRNA was purified from total RNA using poly-T oligomeric magnetic beads. Subsequently, the RNA was broken into fragments of 300 bp in length by ion interruption. The first strand of cDNA was synthesized by using random primers and reverse transcriptase and a specific library was established when the second strand was synthesized. The sequencing type was PE150, with 150-base nucleotides and the direction of sequencing was double-ended sequencing. The library was amplified and were selected at 450 bp according to the fragment size. Subsequently, the total and effective concentrations of the libraries were detected using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). The libraries were mixed in a certain proportion and diluted to 2 nM. The single-chain libraries were formed by alkali denaturation. After the samples were extracted, purified and stored in the form of RNA, they were sequenced with next-generation Sequencing (NGS) based on the Illumina Hiseq (20) sequencing platform (Illumina, Inc.) by Personal Biotechnology Co., Ltd.
Reverse transcription-quantitative PCR (RT-qPCR) analysis
HepG2215 and HepG2 cells were treated with DHA (21.5 µM) for 24 h at 37°C in an atmosphere with 100% humidity and 5% CO2. Culture medium containing 0.1% DMSO was used as the control. Total RNA was extracted from the cells using the Ambion TRIzol reagent (Ambion; Thermo Fisher Scientific, Inc.). All steps were performed under RNase-free conditions. RNA and purity were assessed according to the ratio of A260/A280. Oligonucleotides (CDK1 forward, 5′-GGATGTGCTTATGCAGGATTCC-3′ and reverse, 5′-CATGTACTGACCAGGAGGGATAG-3′; CCNB1 forward, 5′-ATAAGGCGAAGATCAACATGGC-3′ and reverse, 5′-TTTGTTACCAATGTCCCCAAGAG-3′; and actin forward, 5′-CATGTACGTTGCTATCCAGGC-3′ and reverse, 5′-CTCCTTAATGTCACGCACGAT-3′) were reverse transcribed into cDNA according to the instruction of Prime Script TM RT reagent kit with gDNA Eraser (Perfect Real Time) (cat. no. RR047A; Takara Bio Inc.) and TB Green Premix Ex TaqTM II (Tli RNaseH Plus) (cat. no. RR820A; Takara Bio Inc.). The conditions used for reverse transcription were as follows: 37°C for 3 min, 85°C for 5 sec and 4°C for 13 min. Real time fluorescence RT-qPCR was performed on a Real-Time PCR system (Hangzhou Bioer Co. Ltd.) in reaction mixtures (25 µl) containing cDNA, primer pairs, and platinum SYBR Green QPCR Super Mix-UDG (Invitrogen; Thermo Fisher Scientific Inc.). The thermocycling conditions were as follows: Pre-denaturation at 95°C for 3 min followed by 39 cycles at 95°C for 10 sec and 58°C for 30 sec. The dissolution curve was from 65–95°C and the temperature rose 1°C every 20 sec. The relative quantitative values [2−ΔΔCq, Ct of the threshold cycle (21)] used in RT-qPCR were calculated for each sample. Actin was used as an endogenous control.
Western blot analysis
HepG2215 and HepG2 cells were seeded into 6-well plates (4×105 cells/well) and treated as aforementioned. After they were washed in PBS, the cells were directly lysed in SDS sample buffer (50 mM Tris-HCl pH 6.8, 1% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue). According to the instructions of Epizyme Protein Extraction kit (cat: PC201Plus; Shanghai Epizyme Biotech Co., Ltd.) proteins were added to 4X protein loading buffer (cat: P1016; Beijing Solarbio Science & Technology Co., Ltd.) and protein was determined by the BCA method (cat: PC0020; Beijing Solarbio Science & Technology Co., Ltd.). The amount of protein loaded was 10 µl/lane. Equal amounts of total protein were separated by 12% gel and transferred onto PVDF membranes. After blocking with 5% skimmed milk for 2.5 h at room temperature. The primary antibodies were added overnight at 4°C. The primary antibodies were rabbit anti-CDK1 polyclonal antibody (dilution, 1:1,000; cat. no. DF6024; Affinity Biosciences Ltd.), anti-CCNB1 polyclonal antibody (dilution, 1:500; cat. no. AF6168; Affinity Biosciences Ltd.), β-tubulin antibody (dilution, 1:1,000; cat. no. 2146; Cell Signaling Technology, Inc.) and rabbit anti-GAPDH antibody (dilution, 1:1,000; cat. no. 2118; Cell Signaling Technology, Inc.). The secondary antibody was goat anti-rabbit IgG-horse radish peroxidase (HRP) [dilution, 1:10,000; cat. no. abs20002; Absin (Shanghai) Biotechnology Co., Ltd.]. The secondary antibody was incubated at room temperature for 2 h. Finally, chemiluminescence and development were performed using general-purpose ECL luminescent substrate (cat. no. sb-wb012; Shanghai Shenger Biotechnology Co., Ltd.). The results were analyzed using Image-Pro Plus v6.0 software (Media Cybernetics, Inc.).
Statistical analysis
All statistical tests were performed using SPSS23.0 statistics software (IBM Corp.). All experiments were repeated at least 3 times and data were presented as means ± SD. For the comparison between two groups, unpaired t-test Student's t-test was used to determine the statistical significance. When more than two groups were compared, one-way ANOVA was used and then post-hoc Bonferroni test was used for the pairwise comparison. All in vitro experiments were repeated at least three times and at least three samples were taken at a time. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DEGs in liver cancer and construction of PPI network
A total of 1,220, 2,663 and 755 DEGs were identified in GSE84402, GSE19665 and GSE121248, respectively, according to GEO2R. Venn diagrams were used to examine the intersection among the DEGs, including 132 upregulated genes and 246 downregulated genes (Fig. 1A). Protein interaction network analysis was performed using the String database and visualized Cytoscape. The PPI network showed that there were 132 upregulated genes and 246 downregulated genes. Red and green rectangles indicate upregulated and downregulated genes, respectively (Fig. 1B).
Figure 1.
Identification of 10 hub genes. (A) Identification of the common DEGs from the GSE84402, GSE19665 and GSE121248 datasets. Venn diagrams of upregulated (left) and downregulated (right) DEGs based on the three Gene Expression Omnibus datasets. There were 132 upregulated genes and 246 downregulated genes identified in all three datasets. (B) PPI network and hub gene identification. A PPI network of 378 DEGs was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins database. The red and green boxes represent upregulated and downregulated genes, respectively. The cut-off criteria used were |log fold change|>1.0 and adjusted P-value <0.05. DEGs, differentially expressed genes; PPI, protein-protein interaction.
Analysis of hub genes
Subsequently, MCODE, a plug-in for Cytoscape, was used to screen the key subnetworks in PPI network. There were 376 edges and 103 nodes in the key subnetworks of PPI network. The greater the connection value of the node was, the higher the degree of network connection was, and the greater the correlation degree with disease was. The top 10 hub genes were identified according to the degree >10, and these were AURKA, BIRC5, BUB1B, CCNA2, CCNB1, CCNB2, CDC20, CDK1, DLGAP5 and MAD2L1. Among them, CDK1 and CCNB1, CCNA2 had the highest scores (Fig. 2; Table I). Normally, CDK1 and CCNB1 tend to form a complex, which is conducive to cell mitosis (22). CDK1 and CCNB1 had the most connection nodes and the highest score and were therefore, the most significantly expressed. Hence, the present study mainly studied CDK1 and CCNB1.
Figure 2.
Analysis of 10 hub genes. (A) The 10 hub genes in the protein-protein interaction network were screened using the Cytoscape (v3.6.0) plugin CytoHubba based on their connectivity degree. The red and blue boxes represent upregulated and downregulated genes, respectively. Red boxes represent genes that have high degree value. Orange boxes represent genes that have lower degree value compared with red. Yellow boxes represent genes that have lower degree value compared with orange. The screening criteria used were degree >10. (B) The 10 hub genes (AURKA, BIRC5, BUB1B, CCNA2, CCNB1, CCNB2, CDC20, CDK1, DLGAP5 and MAD2L1) were displayed as red (high degree value) to yellow (low degree value). The expression of CDK1 and CCNB1 was the most significant.
Table I.
Top 10 hub genes identified using CytoHubba.
| Rank | Name | Official full name | Score |
|---|---|---|---|
| 1 | CDK1 | Cyclin-dependent kinase 1 | 192 |
| 2 | CCNB1 | Cyclin B1 | 186 |
| 2 | CCNA2 | Cyclin A2 | 186 |
| 4 | AURKA | Aurora kinase A | 184 |
| 5 | MAD2L1 | Mitotic arrest deficient 2 like 1 | 182 |
| 5 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B | 182 |
| 5 | CCNB2 | Cyclin B2 | 182 |
| 8 | BIRC5 | Baculoviral inhibitor of apoptosis repeat containing | 180 |
| 8 | CDC20 | Cell division cycle 20 | 180 |
| 10 | DLGAP5 | DLG associated protein 5 | 176 |
GO and KEGG enrichment analysis of DEGs
The present study explored the influence of DEGs on the function and pathways of genes using GO and KEGG enrichment analysis using DAVID. The GO function analysis of these genes was conducted in three parts (BP, MF and CC). The results were considered to be statistically significant if count >10 and adjust-P<0.05. The results of the three parts of GO analysis are shown in Fig. 3. The bigger the dot, the more genes; the redder the color, the more significant the P-value. The main altered BP terms were ‘cell division’ (adjust-P=3.37×10−11) and ‘mitotic nuclear division’ (adjust-P=6.24×10−10). The main altered CC terms were ‘condensed chromosome kinetochore’ (adjust-P=2.33×10−7) and ‘chromosome, centromeric region’ (adjust-P=1.05×10−6). The main altered MF terms were ‘heme binding’ (adjust-P=2.31×10−4) and ‘iron ion binding’ (adjust-P=0.001189). The altered KEGG pathways were ‘Cell cycle’ (adjust-P=3.66×10−5), ‘p53 signaling pathway’ (adjust-P=3.21×10−4) and ‘Oocyte meiosis’ (adjust-P=0.007786; Fig. 3; Table II). Notably, these genes were mainly involved in cell cycle function and pathways.
Figure 3.
GO and KEGG pathway enrichment analysis of DEGs. (A) GO analysis results revealed that upregulated DEGs and downregulated DEGs were enriched in BPs. The bigger the dot, the more genes; the redder the color, the more significant the P-value. The main BP terms were ‘cell division’ and ‘mitotic nuclear division’. (B) GO analysis results demonstrated that upregulated DEGs and downregulated DEGs were enriched in CCs. The main altered CC terms were ‘condensed chromosome kinetochore’ and ‘chromosome, centromeric region’. (C) GO analysis results demonstrated that upregulated DEGs and downregulated DEGs were enriched in MFs. The main altered MF terms were ‘heme binding’ and ‘iron ion binding’. (D) Most significant KEGG enrichment pathways of the upregulated and downregulated DEGs. The altered KEGG pathways were ‘Cell cycle’, ‘p53 signaling pathway’ and ‘Oocyte meiosis’. Adjusted P-value <0.01 and counts >10 were considered statistically significant. BP, biological process; CC, cellular component; DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MF, molecular function.
Table II.
Analysis of Gene Ontology functions and Kyoto Encyclopedia of Genes and Genomes pathways.
| Category | Term | Enrichment | Count | FDR |
|---|---|---|---|---|
| BP | Cell division | 0.06641 | 35 | 3.37×10−11 |
| BP | Mitotic nuclear division | 0.05313 | 28 | 6.24×10−10 |
| BP | Sister chromatid cohesion | 0.03036 | 16 | 1.10×10−6 |
| BP | G1/S transition of mitotic cell cycle | 0.02277 | 12 | 0.001308 |
| BP | Oxidation-reduction process | 0.05503 | 29 | 0.003476 |
| BP | G2/M transition of mitotic cell cycle | 0.02277 | 12 | 0.009342 |
| CC | Condensed chromosome kinetochore | 0.02846 | 15 | 2.33×10−7 |
| CC | Chromosome, centromeric region | 0.02277 | 12 | 1.05×10−6 |
| CC | Extracellular region | 0.12144 | 64 | 1.45×10−6 |
| CC | Midbody | 0.03036 | 16 | 1.45×10−6 |
| CC | Organelle membrane | 0.02277 | 12 | 3.84×10−5 |
| CC | Kinetochore | 0.02087 | 11 | 1.19×10−4 |
| CC | Spindle | 0.02467 | 13 | 1.19×10−4 |
| CC | Extracellular space | 0.09298 | 49 | 5.03×10−4 |
| CC | Blood microparticle | 0.02467 | 13 | 9.58×10−4 |
| CC | Cytosol | 0.17457 | 92 | 0.002001 |
| MF | Heme binding | 0.02846 | 15 | 2.31×10−4 |
| MF | Iron ion binding | 0.02657 | 14 | 0.001189 |
| Pathway | Cell cycle | 0.03036 | 16 | 3.66×10−5 |
| Pathway | p53 signaling pathway | 0.02087 | 11 | 3.21×10−4 |
| Pathway | Oocyte meiosis | 0.02087 | 11 | 0.007786 |
BP, biological process; CC, cellular component; FDR, false discovery rate; MF, molecular function.
Expression levels of 10 genes in HCC
The present study further explored the expression levels of these genes in liver cancer using the GEPIA and Oncomine databases. The results of GEPIA database analysis revealed that AURKA, BIRC5, BUB1B, CCNA2, CCNB1, CCNB2, CDC20, CDK1, DLGAP5 and MAD2L1 (Fig. 4) were upregulated in liver cancer tissues compared with in normal tissues. T and N indicated liver cancer tissues and normal tissues, respectively.
Figure 4.
Expression levels of 10 hub genes based on GEPIA and Oncomine database analysis. (A) The 10 hub genes (AURKA, BIRC5, BUB1B, CCNA2, CCNB1, CCNB2, CDC20, CDK1, DLGAP5 and MAD2L1) were more highly expressed in liver cancer tissues compared with in normal tissues according to GEPIA. The red and blue boxes represent cancer and normal tissues, respectively. *P<0.05. (B) The 10 hub genes were more highly expressed in liver cancer tissues compared with in normal tissues according to Oncomine. The red box means up and blue box means down. GEPIA, Gene Expression Profiling Interactive Analysis; LIHC, liver hepatocellular carcinoma; TPM, transcripts per million.
In addition, the present study verified the mRNA expression levels of AURKA, BIRC5, BUB1B, CCNA2, CCNB1, CCNB2, CDC20, CDK1, DLGAP5 and MAD2L1 in different types of cancer using the Oncomine database. Similarly, the present study demonstrated that AURKA, BIRC5, BUB1B, CDK1 and MAD2L1 were upregulated in four datasets in liver cancer, whereas CCNA2, CCNB1, CCNB2, CDC20 and DLGAP5 were upregulated in three datasets of liver cancer (Fig. 4). These results demonstrated that the expression levels of the hub genes were upregulated in liver cancer.
Analysis of the protein expression levels of hub genes in liver cancer
Subsequently, the present study analyzed the protein expression levels of the 10 hub genes using the HPA database. Notably, in HPA database, the results showed that CDK1, CCNB1, AURKA, CCNA2, CDC20 and DLGAP5 proteins were not expressed in normal liver tissues, whereas low expression, and even medium (CDK1 and CCNB1) and high expression (AURKA) of these hub genes was observed in liver cancer tissues (Fig. 5). These results demonstrated that these genes except MAD2L1 were upregulated in liver cancer.
Figure 5.
Immunohistochemical analysis of the 10 hub genes using HPA. The immunohistochemical images of (A) AURKA, (B) BIRC5, (C) CCNA2, (D) CCNB1, (E) CCNB2, (F) CDC20, (G) CDK1, (H) DLGAP5 and (I) MAD2L1 in HCC and liver tissues from the HPA database. The results demonstrated that CDK1, CCNB1, AURKA, CCNA2, CDC20, DLGAP5CCNB1 and AURKA were upregulated in HCC. However, BIRC5, CCNB2 and MAD2L1 were not changed significantly in liver cancer. HCC, hepatocellular carcinoma; HPA, Human Protein Atlas.
Survival analysis of 10 genes
To illustrate the overall survival time of these genes in patients with liver cancer, KM plotter was used to draw the survival curves of patients with liver cancer. As shown in Fig. 6, AURKA [hazard ratio (HR)=1.9 (1.33–2.71), log-rank P=3.6×10−4], BIRC5 [HR=2.56 (1.79–3.66), log-rank P=8.6×10−8], BUB1B [HR=2.07 (1.45–2.97), log-rank P=4.7×10−5], CCNA2 [HR=2.76 (1.65–4.62), log-rank P=5.5×10−5], CCNB1 [HR=2.72 (1.74–4.26), log-rank P=5.1×10−6], CCNB2 [HR=2.2 (1.42–3.39), log-rank P=2.7×10−4], CDC20 [HR=2.8 (1.9–4.13), log-rank P=5.7×10−8], CDK1 [HR=2.2 (1.53–3.16), log-rank P=1.2×10−5], DLGAP5 [HR=2.83 (1.77–4.54), log-rank P=6.4×10−6] and MAD2L1 [HR=2.47 (1.65–3.7), log-rank P=5.3×10−6] were highly expressed and associated with poor overall survival time (Fig. 6). It was demonstrated that the hazard ratio and expression of these ten hub genes were generally higher in male patients than in female patients. However, there was little difference among ethnicities (Table SI). Furthermore, it was identified that the hazard ratios expression levels of the 10 hub genes were generally associated with the grade and stage of liver cancer (Table SI).
Figure 6.
Analysis of the prognostic value of the 10 hub genes in HCC using Kaplan-Meier Plotter. The 10 hub genes were represented by hazard ratios with a 95% confidence interval. AURKA [HR=1.9 (1.33–2.71), log-rank P=3.6×10−4], BIRC5 [HR=2.56 (1.79–3.66), log-rank P=8.6×10−8, BUB1B [HR=2.07 (1.45–2.97), log-rank P=4.7×10−5], CCNA2 [HR=2.76 (1.65–4.62), log-rank P=5.5×10−5], CCNB1 [HR=2.72 (1.74–4.26), log-rank P= 5.1×10−6], CCNB2 [HR=2.2 (1.42–3.39), log-rank P=2.7×10−4], CDC20 [HR=2.8 (1.9–4.13), log-rank P= 5.7×10−8], CDK1 [HR=2.2 (1.53–3.16), log-rank P=1.2×10−5], DLGAP5 [HR=2.83 (1.77–4.54), log-rank P=6.4×10−6] and MAD2L1 [HR=2.47 (1.65–3.7), log-rank P=5.3×10−6] High expression levels of AURKA, BIRC5, BUB1B, CCNA2, CCNB1, CCNB2, CDC20, CDK1, DLGAP5 and MAD2L1 were associated with poor overall survival. Log-rank P<0.05 was considered to indicate a statistically significant difference. HR, hazard ratio.
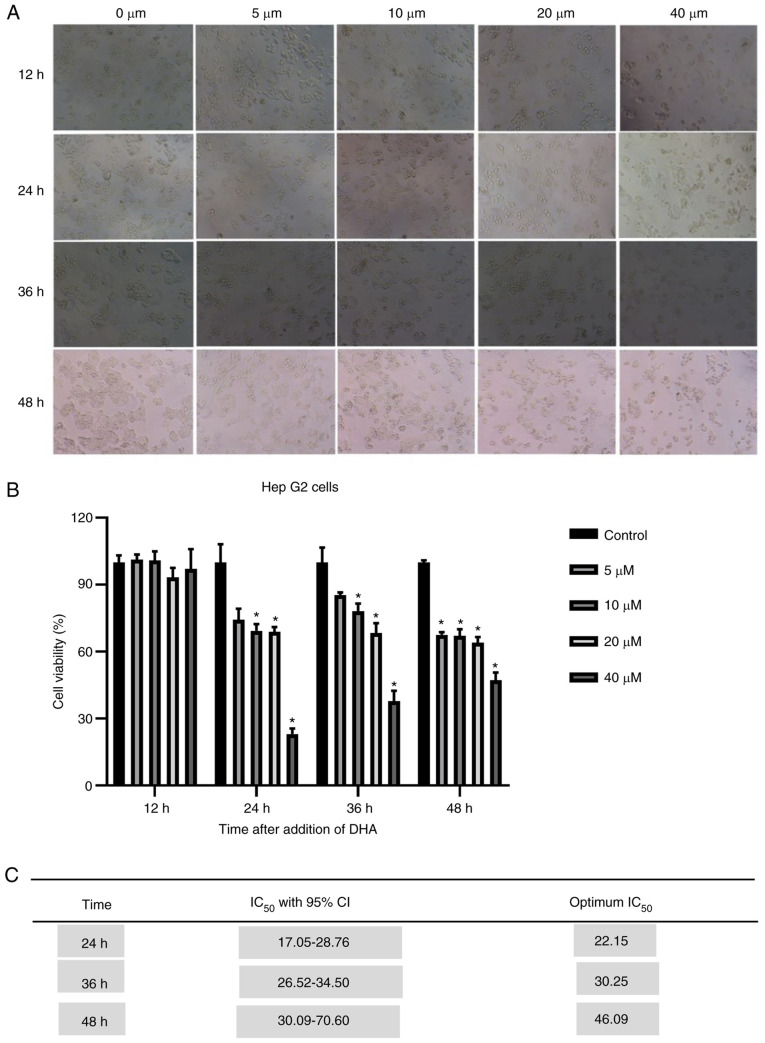
DHA inhibits the proliferation of HepG2 cells in vitro
Our previous study revealed that DHA was selectively cytotoxic for a number of cancer cell lines, including HepG2215 cells (12). HepG2 cells were treated with DHA (5, 10, 20 and 40 µM) for 12, 24, 36 and 48 h to test the anti-proliferative effect of DHA on the cells in vitro. The cell viability was measured by the CCK-8 assay. It was revealed that DHA cytotoxicity was dose- dependent. In addition, it was identified that the cancer cells viability decreased with the increase of concentration at 24, 36 and 48 h in the DHA group (Fig. 7A and B). Compared with that at other times, DHA had little inhibitory effect on liver cancer cells at 12 h. Therefore, 24 h was the optimal time, with an IC50 of 22.15 µM (Fig. 7C).
Figure 7.
DHA inhibits the proliferation of HepG2 cells. (A) Decreased cell numbers were detected at 12, 24, 36 and 48 h after treatment with different concentrations of DHA. All experiments were repeated at least three times. (B) A Cell Counting Kit-8 assay was used to detect the inhibitory effect of DHA on the proliferation of HepG2 cells. HepG2 cells were treated with DHA (mean ± SD; n=3). (C) IC50 (half maximal inhibitory concentration) is the semi-inhibitory concentration. The mortality rate of cells at 12 h was <50%, hence this column has not been included in the table. IC50 values of cells were measured at 24, 36 and 48 h. IC50 values were obtained from the cytotoxicity curves using the Soft Max Pro software. *P<0.05. 95% CI, 95% confidence interval; DHA, dihydroartemisinin.
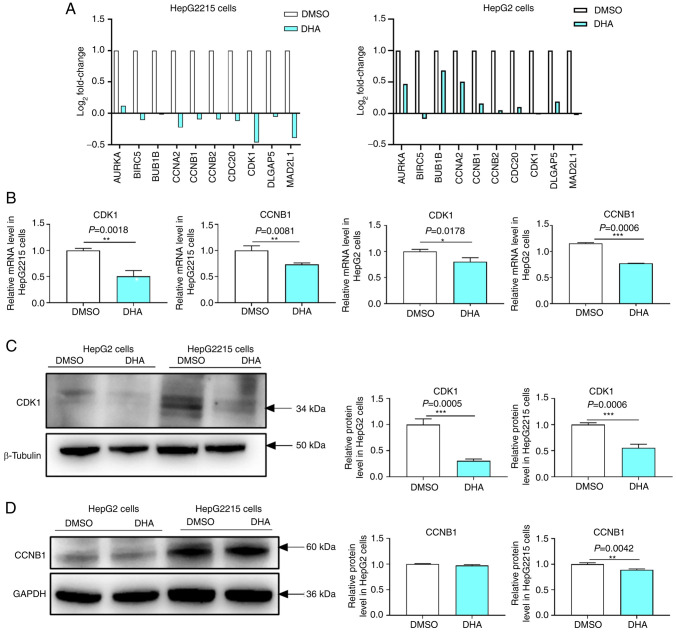
DHA reduces the mRNA and protein expression levels of CDK1 and CCNB1
CDK1 and CCNB1 were associated with the occurrence of liver cancer. Our previous study also indicated that DHA inhibited the proliferation of HepG2215 cells (12). Transcriptome analysis revealed that DHA reduced the mRNA expression levels of the 10 hub genes including CDK1 and CCNB1 in HepG2 and HepG2215 cells (Fig. 8A). Similarly, RT-qPCR results demonstrated that the mRNA expression levels of CDK1 (P=0.0018) and CCNB1 (P=0.0081) in HepG2215 cells were lower (Fig. 8B) in the DHA group compared with in the DMSO group. DHA also reduced the mRNA expression levels of CDK1 (P=0.0178) and CCNB1 (P=0.0006) in HepG2 cells (Fig. 8B). Western blotting demonstrated that DHA reduced the protein expression levels of CDK1 in HepG2 and HepG2215 cells (Fig. 8C). However, DHA did not reduce CCNB1 expression in HepG2 cells but reduced it in HepG2215 cells (Fig. 8D). These results suggested that DHA reduced CDK1 and CCNB1 expression, and inhibited cell proliferation in HepG2215 cells.
Figure 8.
Effects of DHA on CDK1 and CCNB1 in HepG2215 and HepG2 cells. (A) The transcriptome results demonstrated that DHA reduced the expression levels of CDK1 and CCNB1 in liver cancer cells. White represents the DMSO group and blue represents the DHA group. |log2 fold change|>1 and P<0.05 were considered to be statistically significant. (B) DHA reduced the mRNA expression levels of CDK1 and CCNB1 in HepG2 and HepG2215 cells. White represents the DMSO group and blue represents the DHA group. All experiments were repeated at least three times and at least three samples were taken at a time. P<0.05 was considered to indicate a statistically significant difference. (C) DHA reduced the protein expression levels of CDK1 in HepG2 and HepG2215 cells. White represents the DMSO group and blue represents the DHA group. All experiments were repeated at least three times and at least three samples were taken at a time. P<0.05 was considered to indicate a statistically significant difference. (D) DHA reduced the protein expression levels of CCNB1 in HepG2215 cells. White represents the DMSO group and blue represents the DHA group. All experiments were repeated at least three times and at least three samples were taken at a time. P<0.05 was considered to indicate a statistically significant difference. *P<0.05, **P<0.01, ***P<0.001. DHA, dihydroartemisinin.
Discussion
Gene-targeted therapy serves an important role in the treatment of liver cancer, and has attracted increasing attention. Therefore, bioinformatics experiments were performed to identify several potential target molecules for liver cancer. First, it was revealed through the analysis of DEGs of liver cancer that AURKA, BIRC5, BUB1B, CCNA2, CCNB1, CCNB2, CDC20, CDK1, DLGAP5 and MAD2L1 were upregulated in liver cancer and high expression of these genes was associated with poor prognosis. It was demonstrated through GO and KEGG enrichment analysis that these genes were mainly involved in cell cycle function and pathways. The CDK1, CCNB1, AURKA, CCNA2, CDC20 and DLGAP5 genes were not expressed in normal liver tissues, whereas low expression, and even moderate (CDK1 and CCNB1) and high (AURKA) expression of these hub genes emerged in liver cancer tissues in the HPA database. Secondly, it was revealed that CDK1 and CCNB1 were the most significantly expressed compared with other genes and were more closely related to liver cancer. Finally, the present results suggested that DHA reduced the expression levels of CDK1 and CCNB1 and inhibited the proliferation of liver cancer cells.
CDK1 is a member of the Ser/Thr protein kinase family. Previous studies have indicated that CDK1-CCNB1 regulate cell mitosis (23,24). CCNB1 is the main activator of CDK1, which promotes G2/M transformation together with CDK1 (9). A study has suggested that the dysfunction of the cell cycle leads to the generation of tumor stem cells, which is currently considered to be the cause of tumor formation (25). CDK1 is highly expressed in HCC (25), which promotes the growth of cancer and has a marked influence on the overall survival of patients (26). Studies have demonstrated that reduced CCNB1 expression inhibits the occurrence and development of HCC, and activated CCNB1 expression promotes the proliferation of human HCC cells (27,28). In addition, a previous study has revealed that CDK1 and CCNB1 promote the occurrence of rhabdomyosarcoma (29). Similarly, the present study also identified that CDK1 and CCNB1 were upregulated using GEO, GEPIA and Oncomine databases. High expression levels of CDK1 and CCNB1 were associated with poor overall survival. These results suggested that CDK1 and CCNB1 were potential biomarkers of liver cancer. Accumulated evidence has suggested that the upregulation of cyclin B1 and CDK1 contributes to cancer occurrence and progression (30,31). Therefore, it is important to understand the role of CDK1 and CCNB1 for the treatment of liver cancer. DHA is an antimalarial drug extracted from the traditional Chinese medicine of Artemisia annua Linn (11). Studies have demonstrated that DHA induces cell cycle arrest at different phases in various types of cancer (32), that DHA causes the cell cycle arrest mediated by forkhead box protein M1 (33) and induces cell cycle arrest at the G0/G1 phase in Lewis lung carcinoma cells (34), and that DHA also induces G2/M phase cell cycle arrest in NCI-H1975 human lung adenocarcinoma cells by reducing the protein expression levels of cyclin B1 and CDK1 (35). Similarly, in our previous study, it was identified that DHA inhibited the proliferation of HepG2215 cells (12). In the present study, it was demonstrated that DHA inhibited the proliferation of HepG2 cells, and that DHA reduced mRNA expression levels of CDK1 and CCNB1 in HepG2215 and HepG2 cells. Furthermore, DHA also reduced the protein expression levels of CDK1 in HepG2215 and HepG2 cells and reduced the protein expression levels of CCNB1 in HepG2215 cells. Consistently, certain studies have also found that DHA exerted its anticancer effect by changing the expression levels of cell cycle-related genes, such as CCNB1 and cyclinD1, inhibiting the growth of leukemia K562 (36) and human osteosarcoma cells (37). These results suggested that DHA inhibited the proliferation of HepG2215 and HepG2 cells by reducing the expression levels of CDK1 and CCNB1. However, the present study revealed that DHA had no significant effect on the protein expression levels of CCNB1 in HepG2 cells. This result may be due to the fact that the integration of hepatitis B virus (HBV) induced CCNB1 expression in HepG2215 cells compared with HepG2 cells (38). Similarly, a study has demonstrated that HBV X protein induces cell cycle progression (38). Sirtuin 4 (SIRT4) suppresses CyclinB1/Cdc2 in HCC, and the expression levels of SIRT4 in HBV-infected patients are lower than those in uninfected patients (38).
Furthermore, expression levels of other eight hub genes (AURKA, BIRC5, BUB1B, CCNA2, CCNB2, CDC20, DLGAP5 and MAD2L1) were upregulated in liver cancer and high expression of these was associated with poor prognosis of liver cancer. Similarly, studies have demonstrated that AURKA, BIRC5, BUB1B, CCNA2, CCNB2 and CDC20 are upregulated in liver cancer High expression of DLGAP5 is associated with poor overall survival of patients with lung cancer (39). A study also demonstrated that the expression levels of MAD2L1 in colorectal cancer tissues are higher than those in normal tissues (40). Collectively, this suggested that these 10 hub genes were associated with the prognosis of liver cancer and other cancer types, and they may also serve an important role in the development of numerous cancer types.
The present study demonstrated through bioinformatics methods that some hub genes (AURKA, BIRC5, BUB1B, CCNA2, CCNB1, CCNB2, CDC20, CDK1, DLGAP5 and MAD2L1) were upregulated in HCC. CDK1 and CCNB1 were the most significantly expressed compared with other genes. Furthermore, DHA reduced the expression levels of CDK1 and CCNB1 in HepG2215 cells. However, the present study may have some limitations. There were no available clinical patients with liver cancer. The specific mechanism by which hub genes are involved in liver cancer remains unclear. Therefore, further clinical studies are required to verify the reliability of the results to identify the genes most closely associated with the pathogenesis of liver cancer and to provide a novel treatment for liver cancer.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was financially supported by the National Natural Science Foundation of China (grant no. 81873112), Talent Engineering Training Funding Project of Hebei Province (grant no. A201902015), and The Project to Support Hundreds of Outstanding Innovative Talents from the Universities in Hebei Province (grant no. SLRC2019043).
Funding
The present study was financially supported by the National Natural Science Foundation of China (grant no. 81873112), Talent Engineering Training Funding Project of Hebei Province (grant no. A201902015), and The Project to Support Hundreds of Outstanding Innovative Talents from the Universities in Hebei Province (grant no. SLRC2019043).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the NCBI repository, SRA accession: PRJNA733334 (https://www.ncbi.nlm.nih.gov/sra/).
Authors' contributions
XS and LH designed the research. LH, SL, QP, YG, ZZ, JJ, YX and YL performed the experiments. LH and XS wrote the manuscript with contributions from all authors. XS and LH confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1:188314. doi: 10.1016/j.bbcan.2019.188314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu HZ, Zhou WJ, Wan YF, Ge K, Lu J, Jia CK. Downregulation of orosomucoid 2 acts as a prognostic factor associated with cancer-promoting pathways in liver cancer. World J Gastroenterol. 2020;26:804–817. doi: 10.3748/wjg.v26.i8.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu CY, Chen KF, Chen PJ. Treatment of Liver Cancer. Cold Spring Harb Perspect Med. 2015;5:a021535. doi: 10.1101/cshperspect.a021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su S, Huang XW. Advances in the study of cellular immunotherapy for liver cancer. Zhonghua Gan Zang Bing Za Zhi. 2020;28:461–465. doi: 10.3760/cma.j.cn501113-20200420-00199. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol. 2019;12:133. doi: 10.1186/s13045-019-0806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhuang L, Yang Z, Meng Z. Upregulation of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in tumor tissues predicted worse overall survival and disease-free survival in hepatocellular carcinoma patients. Biomed Res Int. 2018;30:7897346. doi: 10.1155/2018/7897346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song X, Du R, Gui H, Zhou M, Zhong W, Mao C, Ma J. Identification of potential hub genes related to the progression and prognosis of hepatocellular carcinoma through integrated bioinformatics analysis. Oncol Rep. 2020;43:133–146. doi: 10.3892/or.2019.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y, Sramkoski RM, Jacobberger JW. The kinetics of G2 and M transitions regulated by B cyclins. PLoS One. 2013;8:e80861. doi: 10.1371/journal.pone.0080861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou Y, Ruan S, Jin L, Chen Z, Han H, Zhang Y, Jian Z, Lin Y, Shi N, Jin H. CDK1, CCNB1, and CCNB2 are prognostic biomarkers and correlated with immune infiltration in hepatocellular carcinoma. Med Sci Monit. 2020;31:925289. doi: 10.12659/MSM.925289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Gao S, Zhu J, Zheng Y, Zhang H, Sun H. Dihydroartemisinin induces apoptosis and inhibits proliferation, migration, and invasion in epithelial ovarian cancer via inhibition of the hedgehog signaling pathway. Cancer Med. 2018;7:5704–5715. doi: 10.1002/cam4.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi X, Wang L, Ren L, Li J, Li S, Cui Q. Dihydroartemisinin, an antimalarial drug, induces absent in melanoma 2 inflammasome activation and autophagy in human hepatocellular carcinoma HepG2215 cells. Phytother Res. 2019;33:1413–1425. doi: 10.1002/ptr.6332. [DOI] [PubMed] [Google Scholar]
- 13.Ouyang G, Yi B, Pan G, Chen X. A robust twelve-gene signature for prognosis prediction of hepatocellular carcinoma. Cancer Cell Int. 2020;20:020–01294. doi: 10.1186/s12935-020-01294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yue C, Ren Y, Ge H, Liang C, Xu Y, Li G, Wu J. Comprehensive analysis of potential prognostic genes for the construction of a competing endogenous RNA regulatory network in hepatocellular carcinoma. Onco Targets Ther. 2019;12:561–576. doi: 10.2147/OTT.S188913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Sun T, Li S, Zhang Z, Jia J, Shan B. Protein anabolism is key to long-term survival in high-grade serous ovarian cancer. Transl Oncol. 2021;14:9. doi: 10.1016/j.tranon.2020.100885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen B, Sun D, Qin X, Gao XH. Screening and identification of potential biomarkers and therapeutic drugs in melanoma via integrated bioinformatics analysis. Invest New Drugs. 2021 Jan 26; doi: 10.1007/s10637-021-01072-y. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 17.Zhan Z, Chen Y, Duan Y, Li L, Mew K, Hu P, Ren H, Peng M. Identification of key genes, pathways and potential therapeutic agents for liver fibrosis using an integrated bioinformatics analysis. PeerJ. 2019;22:e6645. doi: 10.7717/peerj.6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X, Ye T, Liu C, Lu W, Lu M, Zhang J, Wang M, Cao P. Luteolin induced G2 phase cell cycle arrest and apoptosis on non-small cell lung cancer cells. Toxicol In Vitro. 2011;25:1385–1391. doi: 10.1016/j.tiv.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Mishra RK, Mishra V, Pandey A, Tiwari AK, Pandey H, Sharma S, Pandey AC, Dikshit A. Exploration of anti-Malassezia potential of Nyctanthes arbor-tristis L. and their application to combat the infection caused by Mala s1 a novel allergen. BMC Complement Altern Med. 2016;16:114. doi: 10.1186/s12906-016-1092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin W, Jia G, Sun H, Sun T, Hou D. Genome sequence of the fungus Pycnoporus sanguineus, which produces cinnabarinic acid and pH- and thermo-stable laccases. Gene. 2020;742:13. doi: 10.1016/j.gene.2020.144586. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Fang L, Du WW, Awan FM, Dong J, Yang BB. The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex suppressing cell invasion and tumorigenesis. Cancer Lett. 2019;459:216–226. doi: 10.1016/j.canlet.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 23.Alfonso-Pérez T, Hayward D, Holder J, Gruneberg U, Barr FA. MAD1-dependent recruitment of CDK1-CCNB1 to kinetochores promotes spindle checkpoint signaling. J Cell Biol. 2019;218:1108–1117. doi: 10.1083/jcb.201808015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu CX, Wang XQ, Chok SH, Man K, Tsang SHY, Chan ACY, Ma KW, Xia W, Cheung TT. Blocking CDK1/PDK1/β-Catenin signaling by CDK1 inhibitor RO3306 increased the efficacy of sorafenib treatment by targeting cancer stem cells in a preclinical model of hepatocellular carcinoma. Theranostics. 2018;8:3737–3750. doi: 10.7150/thno.25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Cho H, Shin HY, Chung JY, Kang ES, Lee EJ, Kim JH. Accumulation of cytoplasmic Cdk1 is associated with cancer growth and survival rate in epithelial ovarian cancer. Oncotarget. 2016;7:49481–49497. doi: 10.18632/oncotarget.10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chai N, Xie HH, Yin JP, Sa KD, Guo Y, Wang M, Liu J, Zhang XF, Zhang X, Yin H, et al. FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem Biophys Res Commun. 2018;500:924–929. doi: 10.1016/j.bbrc.2018.04.201. [DOI] [PubMed] [Google Scholar]
- 28.Meng Z, Wu J, Liu X, Zhou W, Ni M, Liu S, Guo S, Jia S, Zhang J. Identification of potential hub genes associated with the pathogenesis and prognosis of hepatocellular carcinoma via integrated bioinformatics analysis. J Int Med Res. 2020;48:300060520910019. doi: 10.1177/0300060520910019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Zhang L, Jiang J, Zhang Y, Wang X, Zhang Q, Wang Y, Liu C, Li F. CDK1 and CCNB1 as potential diagnostic markers of rhabdomyosarcoma: Validation following bioinformatics analysis. BMC Med Genomics. 2019;12:198. doi: 10.1186/s12920-019-0645-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Chen YL, Xie YT, Zheng LY, Han JY, Wang H, Tian XX, Fang WG. Association study of germline variants in CCNB1 and CDK1 with breast cancer susceptibility, progression, and survival among Chinese Han women. PLoS One. 2013;8:e84489. doi: 10.1371/journal.pone.0084489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomathinayagam R, Sowmyalakshmi S, Mardhatillah F, Kumar R, Akbarsha MA, Damodaran C. Anticancer mechanism of plumbagin, a natural compound, on non-small cell lung cancer cells. Anticancer Res. 2008;28:785–792. [PubMed] [Google Scholar]
- 32.Cheong DHJ, Tan DWS, Wong FWS, Tran T. Anti-malarial drug, artemisinin and its derivatives for the treatment of respiratory diseases. Pharmacol Res. 2020;158:13. doi: 10.1016/j.phrs.2020.104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin R, Zhang Z, Chen L, Zhou Y, Zou P, Feng C, Wang L, Liang G. Dihydroartemisinin (DHA) induces ferroptosis and causes cell cycle arrest in head and neck carcinoma cells. Cancer Lett. 2016;381:165–175. doi: 10.1016/j.canlet.2016.07.033. [DOI] [PubMed] [Google Scholar]
- 34.Zhang B, Zhang Z, Wang J, Yang B, Zhao Y, Rao Z, Gao J. Dihydroartemisinin sensitizes Lewis lung carcinoma cells to carboplatin therapy via p38 mitogen-activated protein kinase activation. Oncol Lett. 2018;15:7531–7536. doi: 10.3892/ol.2018.8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin H, Jiang AY, Wang H, Cao Y, Wu Y, Jiang XF. Dihydroartemisinin and gefitinib synergistically inhibit NSCLC cell growth and promote apoptosis via the Akt/mTOR/STAT3 pathway. Mol Med Rep. 2017;16:3475–3481. doi: 10.3892/mmr.2017.6989. [DOI] [PubMed] [Google Scholar]
- 36.Yao L, Xie H, Jin QY, Hu WL, Chen LJ. Analyzing anti-cancer action mechanisms of dihydroartemisinin using gene chip. Zhongguo Zhong Yao Za Zhi. 2008;33:1583–1586. (In Chinese) [PubMed] [Google Scholar]
- 37.Ji Y, Zhang YC, Pei LB, Shi LL, Yan JL, Ma XH. Anti-tumor effects of dihydroartemisinin on human osteosarcoma. Mol Cell Biochem. 2011;351:99–108. doi: 10.1007/s11010-011-0716-6. [DOI] [PubMed] [Google Scholar]
- 38.Huang FY, Wong DK, Seto WK, Mak LY, Cheung TT, Yuen MF. Tumor suppressive role of mitochondrial sirtuin 4 in induction of G2/M cell cycle arrest and apoptosis in hepatitis B virus-related hepatocellular carcinoma. Cell Death Discov. 2021;7:88. doi: 10.1038/s41420-021-00470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Chen Y, Feng H, Zhang B, Wang H. Prognostic and predictive value of HURP in non-small cell lung cancer. Oncol Rep. 2018;39:1682–1692. doi: 10.3892/or.2018.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding X, Duan H, Luo H. Identification of core gene expression signature and key pathways in colorectal cancer. Front Genet. 2020;11:45. doi: 10.3389/fgene.2020.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the NCBI repository, SRA accession: PRJNA733334 (https://www.ncbi.nlm.nih.gov/sra/).