Abstract
Long non-coding RNAs (lncRNAs) serve an important role in tumor progression, and their abnormal expression is associated with tumor development. The lncRNA narcolepsy candidate region 1 gene C (lnc-NLC1-C) is involved in numerous types of cancer, but its biological function in glioma remains unknown. In the present study, lnc-NLC1-C expression was detected using reverse transcription-quantitative (RT-q)PCR in U251, SHG44, U87MG and U118MG glioma cells. U87MG cells were transfected with lnc-NLC1-C overexpression or interference vectors. Cell proliferation was detected using a Cell Counting Kit-8 assay. Cell migration and invasion were examined using a Transwell assay, while apoptosis, cell cycle and reactive oxygen species production were evaluated using flow cytometry, and the expression levels of lnc-NLC1-C, microRNA (miR)-383 and peroxiredoxin 3 (PRDX-3) were measured using western blotting and RT-qPCR. Rescue experiments were performed to verify the function of the lnc-NLC1-C/miR-383/PRDX-3 axis. The highest expression levels of lnc-NLC1-C were identified in U87MG glioma cells. Overexpression of lnc-NLC1-C expression promoted cell proliferation, G1 phase blocking, migration and invasion, while inhibiting apoptosis and autophagy in U87MG cells. Mechanistically, miR-383 could bind to lnc-NLC1-C to regulate PRDX-3 expression and improve its oncogenic effect. Rescue experiments confirmed that the lnc-NLC1-C/miR-383/PRDX-3 axis was involved in the molecular mechanism of glioma progression. Therefore, lnc-NLC1-C may be a tumor promoter that affects multiple biological functions, such as migration, invasion and autophagy, in glioma cells.
Keywords: glioma, long non-coding RNA narcolepsy candidate region 1 gene C, microRNA-383, biological functions
Introduction
Neuroglioma is the most common primary central nervous system tumor, accounting for ~50% of intracranial tumors, with high morbidity and mortality rates (1). Surgical resection is currently the main treatment method. However, since tumor cells often infiltrate into normal brain tissue, and the boundary with normal brain tissue is not obvious, it is often difficult to completely remove the tumor during surgery (2,3). Therefore, it is extremely necessary to actively explore more effective treatment methods (3). With the development of medical technology, the mode of medical development has changed from symptomatic treatment to treatment of the cause. Therefore, studying the pathogenesis of glioma may help to identify more effective treatments.
Long non-coding RNAs (lncRNAs) serve an important role in cellular and biological processes (4), and their abnormal expression is associated with a variety of diseases, including colorectal cancer (5) and gastric cancer (6). LncRNAs, such as HOTAIR (7) and H19 (8), can act as tumor suppressors or oncogenes, and they may serve as competitive endogenous RNAs. Therefore, the interaction between lncRNA and microRNA (miRNA/miR) has gradually become a research hotspot (9–12). In a previous study, He et al (13) found that downregulation of miR-383 promotes glioma cell invasion, and identified that miR-383 functions as a tumor suppressor in glioma, targeting peroxiredoxin 3 (PRDX-3). However, the reason for the low miR-383 expression in glioma remains unknown.
Narcolepsy candidate region 1 gene C (NLC1-C), also known as RNA162 (LINC00162), PRED74, C21orf113 and NCRNA00162, is involved in tumor development and is expressed in specific tissues. NLC1-C reportedly targets miR-383, and the accumulation of lnc-NLC1-C in the nucleus can inhibit the transcriptional levels of miR-320a and miR-383 (14–19). Nevertheless, lnc-NLC1-C has rarely been studied in tumor cells, and its binding with miR-383 in glioma has not been reported.
The present study hypothesized that lnc-NLC1-C may regulate the proliferation, migration, invasion and apoptosis of glioma cells by regulating the expression levels of miR-383-5p, PRDX-3, autophagy-related protein 9 (ATG9), LC3II/I, Rab1 and P62. The aim of the study was to investigate the association between the lnc-NLC1-C/miR-383/PRDX-3 axis and the occurrence of glioma, and to investigate the possible underlying mechanism. The current study may provide a future basis for the clinical treatment of glioma.
Materials and methods
Cell lines
The human glioma U251, SHG44, U87MG (glioblastoma of unknown origin, HTB-14) and U118MG cell lines were purchased from the American Type Culture Collection. All cells were verified by STR profiling. U87MG cells were cultured in RPMI-1640 (cat. no. SH30809.01B; HyClone; Cytiva) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 5% CO2 and 37°C, and U251, SHG44 and U118MG cells were cultured in DMEM (cat. no. c11095500bt; Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS at 5% CO2 and 37°C.
Cell transfection
Lnc-NLC1-C-short hairpin (sh)RNA plasmids and pcDNA3.1-lnc-NLC1-C plasmids were purchased from Wuhan Hualianke Biotechnology Co., Ltd. The primer sequences were as follows: Lnc-NLC1-C-shRNA sense, 5′-GATCCGGAAGAGGCAGACACGGAAGGTTCAAGAGACCTTCCGTGTCTGCCTCTTCCTTTTTG-3′ and antisense, 5′-AATTCAAAAAGGAAGAGGCAGACACGGAAGGTCTCTTGAACCTTCCGTGTCTGCCTCTTCCG-3′; pcDNA3.1-lnc-NLC1-C forward, 5′-CGGAATTCCGTGAAGTGCTGACGGG-3′ and reverse, 5′-GCTCTAGAGCTTTTTTTTTTTTTTTTTT-3′. All primers were synthesized by Nanjing Genscript Biotechnology Co., Ltd. Lnc-NLC1-C-shRNA and pcDNA3.1 (ov)-lnc-NLC1-C plasmids (4 µg) were transfected into U87MG cells using the Lipofectamine® 2000 liposome transfection kit (Thermo Fisher Scientific, Inc.) at 37°C for 48 h before subsequent experiments. Negative controls (ov-NC and sh-NC; scramble sequence; Wuhan Hualianke Biotechnology Co., Ltd.) were established. The control mimic (5 nmol; cat. no. miR1N0000001-1-5) and control inhibitor (5 nmol; cat. no. miR2N0000001-1-5) were bought from Guangzhou RiboBio Co., Ltd. miR-383 mimics (5 nmol; cat. no. MC10353; 5′-AGAUCAGAAGGUGAUUGUGGCU-3′) and inhibitors (5 nmol; cat. no. MH10353; 5′-AGAUCAGAAGGUGAUUGUGGCU-3′) were bought from Thermo Fisher Scientific, Inc., and transfection was performed as aforementioned for the plasmids.
Cell proliferation
Cells in the logarithmic growth phase (100 µl) were seeded into a 96-well plate at 5×103 cells/ml and cultured overnight, after which the culture supernatant was discarded. The cells were divided into five groups (Control, ov-NC, ov-lnc-NLC1-C, sh-NC and sh-lnc-NLC1-C) according to the experiment. After 0, 24, 48 and 72 h of culture, 10 µl CCK-8 reagent (cat. no. PAB180031; Bioswamp; Wuhan Bienle Biotechnology Co., Ltd.) was added to each well, and the plates were incubated at 37°C for 4 h. The optical density was measured at 450 nm using a plate reader, and the cell proliferative activity was calculated.
Cell migration and invasion
U87MG cells (2×104) were cultured in serum-free medium and seeded into the upper chamber of a Transwell plate without coating (for migration) or coated with Matrigel® (for invasion; cat. no. 354230; BD Biosciences) at 37°C for 30 min. Subsequently, 750 µl culture medium with 10% FBS was added to the bottom chamber. After 48 and 72 h of culture at 37°C, cells in the upper chamber were removed, and those in the lower chamber were fixed with 4% paraformaldehyde at 4°C for 10 min and stained with 0.1% crystal violet at 4°C for 30 min. The numbers of migrating and invading cells were counted using an inverted fluorescence microscope (magnification, ×200; MIL LED; Leica Microsystems GmbH).
Reactive oxygen species (ROS) assay
ROS production was detected using an active oxygen detection kit (Beijing Solarbio Science & Technology Co., Ltd.). A DCFH-DA probe was diluted with a dimethyl sulfoxide solution at 1:1,000, resulting in a final concentration of 20 µmol/l. Cells were collected and suspended in diluted DCFH-DA at a density of 1×107 cells/ml and incubated at 37°C for 1 h. The cells were washed twice with PBS, and the ROS levels were measured using flow cytometry (FC500 MCL; Beckman Coulter, Inc.), and the data were analyzed with the CytExpert software (Beckman Coulter, Inc.; version 2.0)
Apoptosis and cell cycle analysis
To evaluate cell apoptosis, a single suspension of transfected U87MG cells was prepared using 0.25% trypsin. A total of 5×105 cells were centrifuged at 1,000 × g and 4°C for 5 min and washed twice with 1 ml precooled PBS. After 200 µl binding buffer was added to the cells, 10 µl Annexin V-fluorescein isothiocyanate and 10 µl propidium iodide (part of the AnnexinV-FITC/PI Apoptosis Detection kit; cat. no. 556547; BD Biosciences) were added, and the cells were incubated at 4°C in the dark for 30 min and subjected to flow cytometry (FC500 MCL; Beckman Coulter, Inc.). Data were analyzed with the CytExpert software (Beckman Coulter, Inc.; version 2.0).
To evaluate cell cycle progression, transfected U87MG cells were fixed for 24 h with 70% cold ethanol at 4°C. The fixed cells were washed twice with PBS and incubated with 10 µg/ml RNase A and 20 µg/ml propidium iodide (Sigma-Aldrich; Merck KGaA) at 37°C for 30 min. The cell cycle was analyzed using flow cytometry (FC500 MCL; Beckman Coulter, Inc.), and the results were analyzed using ModFit LT software (BD Biosciences).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cultured cells using TRIzol® (Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. RNA concentration was measured using a spectrophotometer (Thermo Fisher Scientific, Inc.) and total RNA (500 ng) was reverse transcribed into cDNA at a final volume of 10 µl using 5X reaction buffer (Takara Bio, Inc.), 10 mM dNTP Mix, oligodT primers and SuperScript II reverse transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.), and the temperature protocol is 42°C for 60 min, 70°C for 15 min and hold at 16°C. qPCR was performed using the SYBR Green reaction mix (Qiagen GmbH). The primer sequences were as follows: Lnc-NLC1-C forward (F), 5′-GCCAACACCCTACCCA-3′ and reverse (R), 5′-TGCCCATCTTCACCACA-3′; GAPDH F, 5′-CCACTCCTCCACCTTTG-3′ and R, 5′-CACCACCCTGTTGCTGT-3′; miR-383 F, 5′-GGGAGATCAGAAGGTGA-3′ and R, 5′-AACTGGTGTCGTGGAGTCGGC−3′; U6 F, 5′-CTCGCTTCGGCAGCACA-3′ and R, 5′-AACGCTTCACGAATTTGCGT-3′; PRDX-3 F, 5′-GCCTGGATAAATACACC-3′ and R, 5′-ACTGGGAGATCGTTGA-3′. GAPDH and U6 were used as internal controls for mRNA and miRNA, respectively. The reaction proceeded according to the manufacturer's protocol. The relative mRNA expression was calculated using the 2−∆∆Cq method (20).
Western blotting
U87MG cells were lysed using radioimmunoprecipitation assay buffer, and the total protein was extracted. A bicinchoninic acid assay was performed to determine the protein concentration, and the proteins were heated to the thermal denaturation temperature. SDS-PAGE (12%) was performed, and the proteins (20 µg/lane) were transferred to polyvinylidene fluoride membranes (EMD Millipore). The membranes were blocked at 37°C for 1 h using 5% skimmed milk powder and then incubated at 4°C overnight with rabbit antibodies against PRDX-3 (1:1,000; cat. no. PAB31341; Bioswamp; Wuhan Bienle Biotechnology Co., Ltd.), LC3II/I (1:3,000; cat. no. ab51520R; Abcam), ATG9 (1:1,000; cat. no. PAB35248; Bioswamp; Wuhan Bienle Biotechnology Co., Ltd.), Rab1 (1:1,000; cat. no. PAB32093; Bioswamp; Wuhan Bienle Biotechnology Co., Ltd.), p62 (1:1,000; ab455686; Abcam) and GAPDH (1:10,000; cat. no. PAB36269; Bioswamp; Wuhan Bienle Biotechnology Co., Ltd.). Thereafter, the membranes were incubated with secondary goat anti-rabbit IgG antibodies (1:10,000; cat. no. SAB43711; Bioswamp; Wuhan Bienle Biotechnology Co., Ltd.) at 37°C for 1 h. An enhanced chemiluminescence kit (Beijing Solarbio Science & Technology Co., Ltd.) was used for membrane development and images were processed using Image J software (version 1.8.0; National Institutes of Health).
Statistical analysis
Data were analyzed using SPSS 19.0 (IBM Corp.) statistical software and expressed as the mean ± standard deviation. Comparisons between two or multiple groups were performed using the unpaired t-test or one-way ANOVA followed by Tukey's post-hoc test, respectively. P<0.05 was considered to indicate a statistically significant difference. All experiments were repeated three times.
Results
Cell screening and lnc-NLC1-C expression in U87MG cells
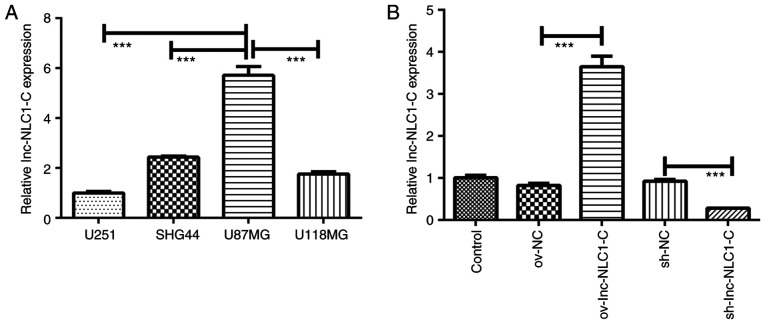
Compared with U251, SHG44 and U118MG glioma cells, lnc-NLC1-C expression was significantly increased in U87MG cells (P<0.001; Fig. 1A). Thus, U87MG cells were selected for subsequent experiments. Compared with the control, ov-NC and sh-NC showed no difference in lnc-NLC1-C expression (Fig. 1B). Compared with ov-NC, ov-lnc-NLC1-C significantly increased lnc-NLC1-C expression (P<0.001), while compared with sh-NC, sh-lnc-NLC1-C significantly decreased lnc-NLC1-C expression (P<0.001) (Fig. 1B).
Figure 1.
Lnc-NLC1-C expression in different glioma cell lines and construction of lnc-NLC1-C plasmids. (A) Lnc-NLC1-C expression in U251, SHG44, U118MG and U87MG glioma cells. (B) Lnc-NLC1-C expression in U87MG glioma cells transfected with ov-Lnc-NLC1-C and sh-lnc-NLC1-C. ***P<0.001. n=3. Lnc-NLC1-C, long non-coding RNA narcolepsy candidate region 1 gene C; ov, overexpression; sh, short hairpin RNA; NC, negative control.
Lnc-NLC1-C promotes the proliferation, migration and invasion of U87MG cells
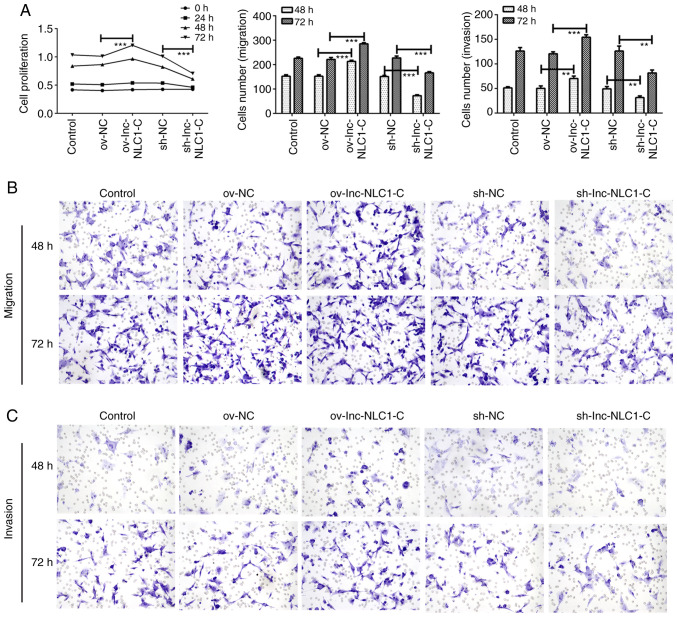
Compared with ov-NC, ov-lnc-NLC1-C significantly enhanced the proliferation (P<0.001; Fig. 2A) and significantly promoted the migration (Fig. 2B) and invasion (Fig. 2C) of U87MG cells at 48 and 72 h. Compared with sh-NC, sh-lnc-NLC1-C significantly decreased the proliferation (P<0.001; Fig. 2A) and inhibited the migration (Fig. 2B) and invasion (Fig. 2C) of U87MG cells at 48 and 72 h.
Figure 2.
Proliferation, migration and invasion of U87MG cells. (A) Statistical analysis of U87MG cell proliferation, migration and invasion at different time points. (B) U87MG cell migration and (C) invasion were detected using a Transwell assay at 48 and 72 h (magnification, ×200). **P<0.01; ***P<0.001. n=3. Lnc-NLC1-C, long non-coding RNA narcolepsy candidate region 1 gene C; ov, overexpression; sh, short hairpin RNA; NC, negative control.
Lnc-NLC1-C inhibits ROS generation and promotes cell cycle blocking in U87MG cells
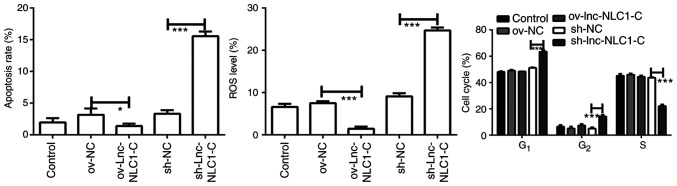
Compared with ov-NC, ov-lnc-NLC1-C significantly decreased the ROS production (P<0.001; Figs. 3 and S1B). Compared with sh-NC, sh-lnc-NLC1-C significantly promoted ROS generation and apoptosis of U87MG cells (P<0.001; Figs. 3 and S1A and B). Compared with the sh-NC group, U87MG cells in the G1 phase were significantly higher in the sh-lnc-NLC1-C group (P<0.001), whereas cells in the S phase were significantly decreased (P<0.001) (Figs. 3 and S1C). There was no significant effect on apoptosis or cell cycle following lnc-NLC1-C overexpression compared with the ov-NC group (P>0.05).
Figure 3.
Analysis of apoptosis, ROS level and cell cycle in U87MG cells. *P<0.05 and ***P<0.001. n=3. Lnc-NLC1-C, long non-coding RNA narcolepsy candidate region 1 gene C; ov, overexpression; sh, short hairpin RNA; NC, negative control; ROS, reactive oxygen species.
Lnc-NLC1-C inhibits autophagy in U87MG cells
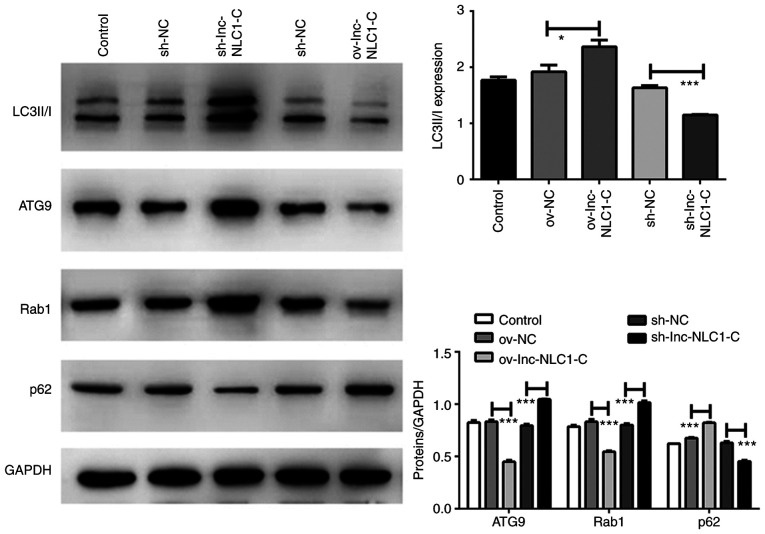
Compared with ov-NC, the protein expression levels of LC3II/I (P<0.05) and p62 (P<0.001) were significantly increased by ov-lnc-NLC1-C, whereas those of ATG9 and Rab1 were significantly decreased (P<0.001) (Fig. 4). Compared with sh-NC, the protein expression levels of LC3II/I and p62 were significantly decreased by sh-lnc-NLC1-C (P<0.001), whereas those of ATG9 and Rab1 were significantly increased (P<0.001) (Fig. 4).
Figure 4.
Expression levels of autophagy-associated proteins in U87MG cells subjected to lnc-NLC1-C overexpression or silencing. *P<0.05; ***P<0.001. n=3. Lnc-NLC1-C, long non-coding RNA narcolepsy candidate region 1 gene C; ov, overexpression; sh, short hairpin RNA; NC, negative control; ATG9, autophagy-related protein 9.
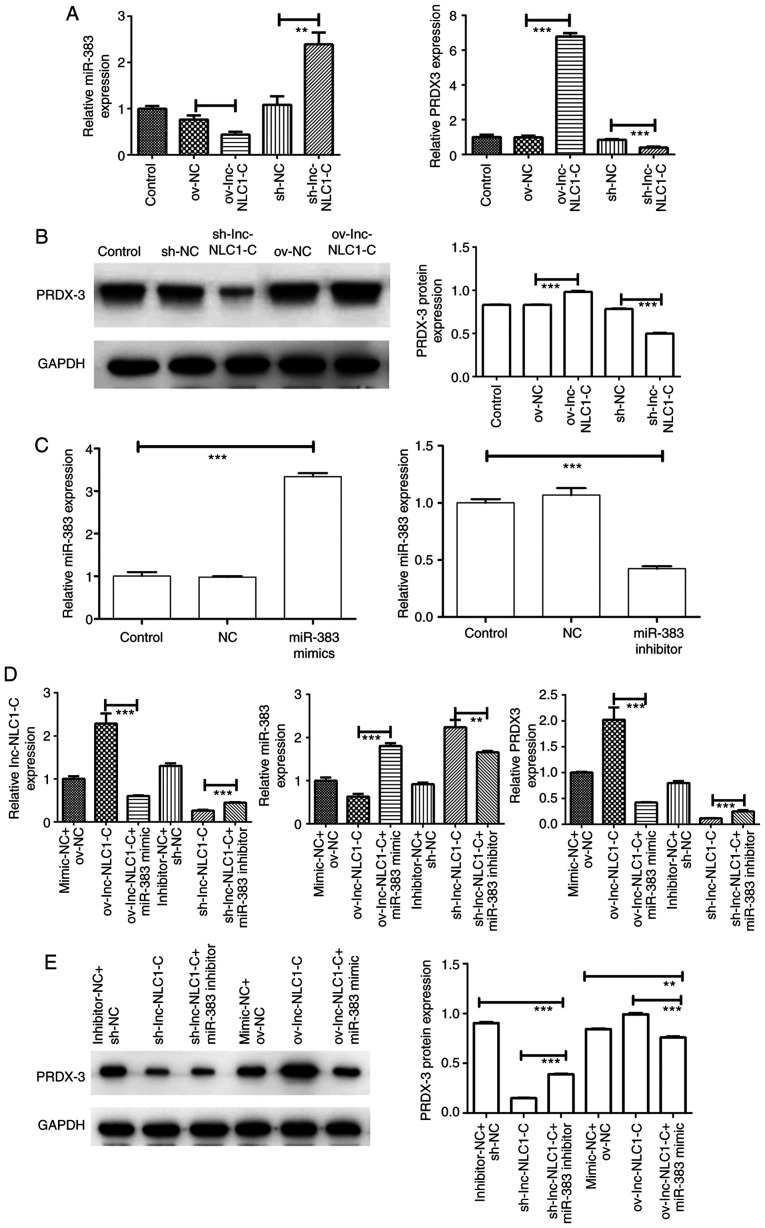
Expression of the lnc-NLC1-C/miR-383/PRDX-3 axis in U87MG cells
Compared with ov-NC, ov-lnc-NLC1-C significantly increased PRDX-3 expression (P<0.001) and significantly decreased miR-383 expression (P<0.01) (Fig. 5A and B). Compared with sh-NC, sh-lnc-NLC1-C significantly decreased PRDX-3 expression (P<0.001) and increased miR-383 expression (P<0.01) (Fig. 5A and B). As shown in Fig. 5C, miR-383 expression was significantly increased in the miR-383 mimic group (P<0.001) and was significantly decreased in the miR-383 inhibitor group compared with in the control group (P<0.001), indicating that the miR-383 mimic and inhibitor were successfully transfected. Compared with ov-lnc-NLC1-C, ov-lnc-NLC1-C + miR-383 mimic significantly decreased the expression levels of lnc-NLC1-C and PRDX-3, and significantly increased miR-383 expression (P<0.001; Fig. 5D and E). Compared with sh-lnc-NLC1-C, sh-lnc-NLC1-C + miR-383 inhibitor significantly increased the expression levels of lnc-NLC1-C and PRDX-3 (P<0.001), and decreased miR-383 expression (P<0.01) (Fig. 5D and E).
Figure 5.
Expression of the lnc-NLC1-C/miR-383/PRDX-3 axis in U87MG cells and rescue experiments. (A) miR-383 and PRDX-3 expression in U87MG cells was detected using RT-qPCR. (B) PRDX-3 protein expression in U87MG cells was detected using western blotting. (C) miR-383 expression in U87MG glioma cells transfected with miR-383 mimics and miR-383 inhibitors. (D) Expression levels of lnc-NLC1-C, miR-383 and PRDX-3 in rescue experiments were detected using RT-qPCR. (E) PRDX-3 protein expression in rescue experiments was detected using western blotting. **P<0.01; ***P<0.001. n=3. RT-qPCR, reverse transcription-quantitative PCR; lnc-NLC1-C, long non-coding RNA narcolepsy candidate region 1 gene C; ov, overexpression; sh, short hairpin RNA; NC, negative control; miR, microRNA; PRDX-3, peroxiredoxin 3.
Discussion
Carcinogenic factors affecting glioma include high-risk congenital genetic and environmental factors. Previous studies have shown that glioma accounts for ~80% of all malignant tumors of the central nervous system (21,22). Glioma has a high incidence, a high recurrence rate and a poor prognosis (23,24). Tumor heterogeneity and infection pose great challenges to the clinical treatment of glioma (25,26). In recent years, with the development of treatment for glioma, the mechanism of lncRNAs has attracted the attention of experts and scholars.
LncRNAs are non-coding RNAs composed of 200–100,000 nucleotides (27) that are closely associated with the occurrence and development of a number of tumors, such as gastric and colon cancer (28). A previous study has found that lncRNAs regulate gene expression, mainly by interacting with DNA, RNA and proteins (29). For example, lncRNAs activate or deactivate protein complexes by binding to chromosomal DNA (30). Currently known lncRNAs associated with glioma include HOTAIR, H19, CRNDE, MEG3 and ADAMTS9-AS2 (31–35). The present study revealed that lnc-NLC1-C expression was highest in U87MG cells compared with that in U251, SHG44 and U118MG cells. Lnc-NLC1-C overexpression promoted the proliferation, cell cycle blocking, migration and invasion of U87MG cells, and inhibited apoptosis and autophagy, indicating that lnc-NLC1-C may be a cancer-promoting factor in glioma.
NLC1-C is weakly expressed in cutaneous squamous cell carcinoma and highly expressed in the breasts, testis and parotid gland, thus participating in tumor development (19,36). Lnc-NLC1-C targets miR-383, and the accumulation of NLC1-C in the nucleus inhibits the transcription of miR-320a and miR-383, and promotes the proliferation of embryonic testicular cancer cells by binding to nucleolin (19). These results indicate that lnc-NLC1-C can target miR-383 and affect the expression of downstream genes and proteins. It has been previously demonstrated that miR-383 reverses mitochondrial ROS inhibition caused by PRDX-3 overexpression, promotes autophagy and oxidative stress in cancer cells, and enhances cancer cell apoptosis (37). Tian et al (38) showed that PRDX3 is a potential target gene of miR-383, and there is a targeting relationship between the two genes. miR-383 can inhibit the proliferation, migration and invasion of human hepatocellular carcinoma HepG2 cells by down-regulating PRDX3 expression (39). Wang et al (40) observed that miR-383 inhibited the proliferation of human medulloblastoma cells and promoted their apoptosis by downregulating the expression levels of PRDX3. Currently, to the best of our knowledge, there are no studies on the role of the PRDX3/miR-383 axis in glioma, and based on the fact that lnc-NLC1-C targets miR-383, the present study predicted that lnc-NLC1-C may affect the development of glioma through the miR-383/PRDX-3 axis. The present results revealed that lnc-NLC1-C downregulated miR-383 expression and upregulated PRDX-3 expression. miR-383 inhibition rescue experiments confirmed that miR-383 reversed the effect of lnc-NLC1-C and downregulated PRDX-3 expression. Li et al (41) observed that miR-383 acts as a regulator controlling cell proliferation of medulloblastoma via targeting PRDX3; although the studied diseases were different, they both belong to the neurological system, which indirectly confirmed the current results. In the present study, only one cell line was used for experimental manipulation, and there were no in vivo experiments, which are the main limitations of the study. Therefore, multiple cell lines and animal experiments should be used in follow-up experiments to further verify the conclusions of the present study. In conclusion, the lnc-NLC1-C/miR-383/PRDX-3 axis may serve an important role in glioma development.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZX and QC performed the experiments and confirmed the authenticity of all the raw data. XZ, ML and JL analyzed the data. ZX drafted the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lai NS, Wu DG, Fang XG, Lin YC, Chen SS, Li ZB, Xu SS. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer. 2015;112:1241–1246. doi: 10.1038/bjc.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabian C, Han M, Bjerkvig R, Niclou SP. Novel facets of glioma invasion. Int Rev Cell Mol Bio. 2021;360:33–64. doi: 10.1016/bs.ircmb.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Mansouri A, Mansouri S, Hachem LD, Klironomos G, Vogelbaum MA, Bernstein M, Zadeh G. The role of 5-aminolevulinic acid in enhancing surgery for high-grade glioma, its current boundaries, and future perspectives: A systematic review. Cancer. 2016;122:2469–2478. doi: 10.1002/cncr.30088. [DOI] [PubMed] [Google Scholar]
- 4.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20:5758. doi: 10.3390/ijms20225758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei GH, Wang X. LncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3850–3856. [PubMed] [Google Scholar]
- 7.Yu GJ, Sun Y, Zhang DW, Zhang P. Long non-coding RNA HOTAIR functions as a competitive endogenous RNA to regulate PRAF2 expression by sponging miR-326 in cutaneous squamous cell carcinoma. Cancer Cell Int. 2019;19:270. doi: 10.1186/s12935-019-0992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang K, Luo Z, Zhang Y, Zhang L, Wu L, Liu L, Yang J, Song X, Liu J. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. 2016;17:187–194. doi: 10.3233/CBM-160630. [DOI] [PubMed] [Google Scholar]
- 9.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA-lncRNA interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 10.Ballantyne MD, McDonald RA, Baker AH. LncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther. 2016;99:494–501. doi: 10.1002/cpt.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Zhou Y, Li H. LncRNA, miRNA and lncRNA-miRNA interaction in viral infection. Virus Res. 2018;257:25–32. doi: 10.1016/j.virusres.2018.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Wei L, Wang X, Lv L, Liu J, Xing H, Song Y, Xie M, Lei T, Zhang N, Yang M. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer. 2019;18:147. doi: 10.1186/s12943-019-1086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Z, Cen D, Luo X, Li D, Li P, Liang L, Meng Z. Downregulation of miR-383 promotes glioma cell invasion by targeting insulin-like growth factor 1 receptor. Med Oncol. 2013;30:557. doi: 10.1007/s12032-013-0557-0. [DOI] [PubMed] [Google Scholar]
- 14.Kawashima M, Tamiya G, Oka A, Hohjoh H, Juji T, Ebisawa T, Honda Y, Inoko H, Tokunaga K. Genomewide association analysis of human narcolepsy and a new resistance gene. Am J Hum Genet. 2006;79:252–263. doi: 10.1086/505539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mills JD, Kavanagh T, Kim WS, Chen BJ, Kawahara Y, Halliday GM, Janitz M. Unique transcriptome patterns of the white and grey matter corroborate structural and functional heterogeneity in the human frontal lobe. PLoS One. 2013;8:e78480. doi: 10.1371/journal.pone.0078480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zong L, Hattori N, Yasukawa Y, Kimura K, Mori A, Seto Y, Ushijima T. LINC00162 confers sensitivity to 5-Aza-2′-deoxycytidine via modulation of an RNA splicing protein, HNRNPH1. Oncogene. 2019;38:5281–5293. doi: 10.1038/s41388-019-0792-8. [DOI] [PubMed] [Google Scholar]
- 17.Lou X, Li J, Yu D, Wei YQ, Feng S, Sun JJ. Comprehensive analysis of five long noncoding RNAs expression as competing endogenous RNAs in regulating hepatoma carcinoma. Cancer Med. 2019;8:5735–5749. doi: 10.1002/cam4.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piipponen M, Nissinen L, Farshchian M, Riihila P, Kivisaari A, Kallajoki M, Peltonen J, Peltonen S, Kähäri VM. Long noncoding RNA PICSAR promotes growth of cutaneous squamous cell carcinoma by regulating ERK1/2 activity. J Invest Dermatol. 2016;136:1701–1710. doi: 10.1016/j.jid.2016.03.028. [DOI] [PubMed] [Google Scholar]
- 19.Lu M, Tian H, Cao YX, He X, Chen L, Song X, Ping P, Huang H, Sun F. Downregulation of miR-320a/383-sponge-like long non-coding RNA NLC1-C (narcolepsy candidate-region 1 genes) is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation. Cell Death Dis. 2015;6:e1960. doi: 10.1038/cddis.2015.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Morgan LL, Miller AB, Sasco A, Davis DL. Mobile phone radiation causes brain tumors and should be classified as a probable human carcinogen (2A) (review) Int J Oncol. 2015;46:1865–1871. doi: 10.3892/ijo.2015.2908. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Long H, Zhang M, Wang Y, Lu Q, Yuan H, Qu Q, Qu J. Glutathione S-transferase genes variants and glioma risk: A case-control and meta-analysis study. J Cancer. 2019;10:4679–4688. doi: 10.7150/jca.29398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng M, Liu L. MUC15 promotes growth and invasion of glioma cells by activating Raf/MEK/ERK pathway. Clin Exp Pharmacol Physiol. 2020;47:1041–1048. doi: 10.1111/1440-1681.13277. [DOI] [PubMed] [Google Scholar]
- 24.Hu L, Zhao T, Wang J, Wang R, Bu H, Zheng S, Li Y, Liu J, Wang S. Beauvericin inhibits the growth of U251 glioma cells and promotes apoptosis in vitro. J Biobased Materials Bioenergy. 2020;14:639–644. doi: 10.1166/jbmb.2020.2006. [DOI] [Google Scholar]
- 25.Hanaei S, Afshari K, Hirbod-Mobarakeh A, Mohajer B, Amir Dastmalchi D, Rezaei N. Therapeutic efficacy of specific immunotherapy for glioma: A systematic review and meta-analysis. Rev The Neurosci. 2018;29:443–461. doi: 10.1515/revneuro-2017-0057. [DOI] [PubMed] [Google Scholar]
- 26.Baumert BG, Hegi ME, van den Bent MJ, von Deimling A, Gorlia T, Hoang-Xuan K, Brandes AA, Kantor G, Taphoorn MJB, Hassel MB, et al. Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033–26033): A randomised, open-label, phase 3 intergroup study. Lancet Oncol. 2016;17:1521–1532. doi: 10.1016/S1470-2045(16)30313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jathar S, Kumar V, Srivastava J, Tripathi V. Technological developments in lncRNA biology. Adv Exp Med Biol. 2017;1008:283–323. doi: 10.1007/978-981-10-5203-3_10. [DOI] [PubMed] [Google Scholar]
- 28.Kumar MM, Goyal R. LncRNA as a therapeutic target for angiogenesis. Curr Top Med Chem. 2017;17:1750–1757. doi: 10.2174/1568026617666161116144744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 30.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren J, Zhang X, Cao J, Tian J, Luo J, Yu Y, Wang F, Li J, Zhao Q. Radiosynthesis of a novel antisense imaging probe targeting lncRNA HOTAIR in malignant glioma. Res Sq. doi: 10.1186/s12885-022-09170-7. doi: 10.21203/rs.3.rs-152169/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J, Qian J, Liu N, You Y. Long Non-Coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9:e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jing SY, Yang ZG, Cheng ZH, Fang JS, Li X. Changes in expression levels of long-chain non-coding RNA CRNDE in glioma and its association with clinical and pathological characteristics. J Clin Neurosurgery. 2019;16:11–16. [Google Scholar]
- 34.Wang D, Fu CW, Fan DQ. Participation of tumor suppressors long non-coding RNA MEG3, microRNA-377 and PTEN in glioma cell invasion and migration. Pathol Res Pract. 2019;215:152558. doi: 10.1016/j.prp.2019.152558. [DOI] [PubMed] [Google Scholar]
- 35.Yang JL, Ge HM, Yan ZJ, Yan H. Correlativity between low expression level of long non-coding RNA ADAMTS9-AS2 and poor prognosis in patients with gliomas. Chin J Clin Neurosurgery. 2019;24:278–278. [Google Scholar]
- 36.Luo Y, Morgan SL, Wang KC. PICSAR: Long noncoding RNA in cutaneous squamous cell carcinoma. J Invest Dermatol. 2016;136:1541–1542. doi: 10.1016/j.jid.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li KK, Pang JC, Lau KM, Zhou L, Mao Y, Wang Y, Poon WS, Ng HK. MiR-383 is downregulated in medulloblastoma and targets peroxiredoxin 3 (PRDX3) Brain Pathol. 2013;23:413–425. doi: 10.1111/bpa.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian S, Jiang H, Liu H, Cao X, Zhang D, Mou SD, Li Z. The effects of miR-383 targeting PRDX3 on the proliferation and invasion of HepG2 cells. J Gastroenterol Hepatol. 2020;29:1335–1340. [Google Scholar]
- 39.Ng HK. Proceedings of the 88th Annual Meeting of the American Association of Neuropathologists. AANP; Chicago, IL: 2012. Hsa-miR-383 and its Target Peroxiredoxin 3 (PRDX3) Have Major Roles Controlling Cell Growth in Medulloblastoma. [Google Scholar]
- 40.Wang SZ, Gao ML, Hou JH. Regulation of PRDX3 expression by miR383 and its effect on proliferation and apoptosis of human medulloblastoma. Labeled Immunoassays Clin Med. 2019;26:139–142. (In Chinese) [Google Scholar]
- 41.Li KK, Pang JC, Lau KM, Zhou L, Mao Y, Wang Y, Poon WS, Ng HK. miR-383 is downregulated in medulloblastoma and targets peroxiredoxin 3 (PRDX3) Brain Pathol. 2013;23:413–425. doi: 10.1111/bpa.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.