Abstract
The emergence of new SARS-CoV-2 variants and their rapid spread pose a threat to both human and animal health and may conceal unknown risks. This report describes an Italian human-to-cat outbreak of SARS-CoV-2 lineage B.1.1.7 (the Alpha variant) . On March 7th, 2021, approximately ten days after COVID-19 appeared in the family, the onset of respiratory signs in a cat by COVID-19-affected owners led to an in-depth diagnostic investigation, combining clinical and serological data with rt-qPCR-based virus detection and whole genome sequencing. The Alpha variant was confirmed first in the owners and a few days later in the cat that was then monitored weekly: the course was similar with one-week lag time in the cat. In addition, based on comparative analysis of genome sequences from our study and from 200 random Italian cases of Alpha variant, the familial cluster was confirmed. The temporal sequence along with the genomic data support a human-to-animal transmission. Such an event emphasizes the importance of studying the circulation and dynamics of SARS-CoV-2 variants in humans and animals to better understand and prevent potential spillover risks or unwarranted alerts involving our pet populations.
Keywords: Sars-CoV-2 alpha variant lineage B.1.1.7, Infection cluster, Human-to-animal transmission, One health approach, Cat
Highlights
-
•
Respiratory signs in a household cat ten days after the appearance of COVID-19 in the family led to a thorough investigation.
-
•
The Alpha variant (B.1.1.7) was first detected in owners and one week later in the cat which showed a similar course.
-
•
A familial cluster was confirmed based on a comparative sequence analysis of the study subjects and 200 Italian random cases.
-
•
The temporal sequence together with the genomic data support human-to-animal transmission.
-
•
The human epidemic, the easy crossing of species barriers and the abundance of pets suggest monitoring for spillover risks.
1. Introduction
On March 11th, 2020 the World Health Organization declared Global Pandemic the Coronavirus Disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 is supposed to have originated in bats [1], but the involvement of animals is not only limited to its origin. Susceptibility to SARS-CoV-2 infection has been demonstrated in a wide range of mammals under laboratory conditions, with cats and ferrets appearing to be the most permissive hosts [2]. A narrow number of animals worldwide, including dogs, cats, zoo tigers and lions, and farmed minks, have been infected with SARS-CoV-2, mostly through suspected human-to-animal transmission [3,4]. In the last year few cases of household cats infected by cohabiting COVID-19 positive owners have been reported in Spain [5], France [6], Italy [7], Belgium [8] and US [9,10]. High seroprevalence was shown when pets from COVID-19 positive households were tested [11]. This was not only likely but expected given the ability of coronaviruses to cross species barriers, the massive human outbreak and the abundance of the pets populations [12]. Cats appear to be particularly susceptible to the virus, and cat-to-cat transmission of SARS-CoV-2 has been demonstrated under experimental conditions [4]: this could be compatible with a role as reservoir and the development of new variants [12].
To date there is no evidence of transmission from cats to humans, but it is not difficult to imagine effective ways of exposure for both humans and cats [13]. Pets are often perceived as family members and cohabitation is increasingly close: owners allow animals to jump over the kitchen sink, sleep in the bed, lick faces or wounds [14]. On the other hand, cats can carry infections acquired in shelters from which they are adopted or in boarding catteries where they spend short periods.
In this respect, the frequent emergence of new SARS-CoV-2 variants and their rapid spread around the world may pose an even greater threat to both human and animal health and may conceal additional and unknown risks: this makes it increasingly important to develop the capacity to investigate in depth disease outbreaks where human and animal cases coexist.
The SARS-CoV-2 Alpha variant - Lineage B.1.1.7 (one of the four “variants of concern” along with Beta, Gamma and Delta), reported in autumn 2020, is characterized by several mutations, the most characteristic of which are the Spike 69–70 deletion and the Spike N501Y mutation: the former is linked to immune escape in immunocompromised patients and increased viral infectivity in vitro [15], while the latter involves enhanced binding affinity to human ACE2 receptor leading to substantially faster spread than pre-existing SARS-CoV-2 variants [16]. To date, infection with Alpha variant in cats has been reported in Texas (USA) [17] and the UK [18].
Here we describe an Italian human-to-cat outbreak: this is the first Italian case of a household cat detected as positive to Alpha variant and the infection with the same variant had been confirmed in his owners first. The cat showed overt respiratory signs one week after the clinical onset of COVID-19 disease of his owners.
2. Material and methods
2.1. Outbreak detection
The cat is a European Domestic Shorthair male, neutered, 8 years old, that lives without contact with other pets and with free access to the property garden and to the adjacent woods, in Novara province, Piedmont, Northern West Italy.
Cat's owners, a young girl and her father, started to show clinical signs referable to COVID-19 (asthenia, fever, rhinitis, loss of taste) on February 26th and March 2nd, 2021 respectively and on March 4th and 5th, owners' oropharyngeal swab samples were declared positive for the SARS-CoV-2 genome by rt-qPCR.
On March 7th, 2021, approximately ten days after COVID-19 appeared in the family, owners noticed an acute respiratory syndrome in their cat; and on March 10th, the cat underwent his first clinical examination and collection of blood sample and oropharyngeal and rectal swabs. After the confirmation of SARS-CoV-2 by rt-qPCR, the animal was subjected to a weekly monitoring program of both clinical and laboratory checks with the collection of swabs until complete remission and viral clearance.
2.2. RNA extraction and Sars-CoV-2 rt-qPCR detection and confirmation
Total RNA was extracted from oropharyngeal and rectal cat swabs collected on March 10th: 200 μl of swab medium (COPAN Diagnostics Inc., Italy) were submitted to a magnetic beads based method (MagMax Viral Isolation kit, Thermofisher, US) in an automated extractor (KingFisher, Thermofisher, US), according to the manufacturers' instructions. Ten μl of eluted RNA was then used for the RT-qPCR assay using the commercial kit TaqPath™ COVID-19 CE-IVD RT-PCR Kit (Thermofisher, US), amplified on QuantStudio™ 5 Real-Time PCR System (Thermofisher, US). TaqPath™ COVID-19 is based on a multiplex amplification of three different SARS-CoV-2 genomic regions [Orf1ab, N (Nucleocapsid) gene and S (Spike protein) gene], over the internal process control with a MS2 phage.
Deletion at position 69–70 of the spike (S) protein gene leads to a loss of amplification of TaqPath™ kit probe S, drawn in silico on the region containing the deletion.
To monitor virus clearance, cat swabs were collected and examined with the TaqPath™ COVID-19 CE-IVD RT-PCR Kit weekly. Oropharyngeal swabs collected from the owners on March 16th, about ten days after the first diagnosis, were also investigated with the same kit.
All extracted RNA from swab samples were submitted to a first step in genome characterization pipeline with SARS-CoV-2 Variants ELITe MGB® Kit (ElitecGroup, France), a reflex real time RT-PCR test for the detection and discrimination of the mutations E484K and N501Y of the S gene of SARS-CoV-2, based on melting curves analysis.
2.3. Serological analysis on cat
Two blood samples were collected from the cat, on March 10th, during the first clinical visit and 12 days after. Serum samples were promptly tested by ELISA method using the Eradikit COVID19 Multispecies (IN3 DIAGNOSTIC, Italy), a commercial dual antigen enzyme immunoassay in which the microtiter plates were coated with a recombinant SARS-CoV-2 antigen, accordingly to manufacturer's instruction. Reaction is considered positive if the sample to positive (S/P) ratio is ≥20% and negative if S/P ratio ˂ 20%.
2.4. Whole genome sequencing, lineage identification and clustering confirmation
Analyses were conducted in parallel with two different approaches.
In the first protocol two μl of total RNA obtained from cat swab was retrotranscribed using the QuantiTect Reverse Transcription kit (Qiagen, Germany), according to the manufacturers' instructions. Ten μl of cDNA were used for the Whole Genome Sequencing (WGS) of SARS-CoV-2 with the EasySeq™ RC-PCR SARS-CoV-2 (novel coronavirus) Whole Genome Sequencing kit (NimaGen, Netherlands), enabling the amplification of overlapping fragments round 300 bp to cover the entire viral genome. 10 pM libraries added with 3% of PhiX control were sequenced on an Illumina MiSeq platform with a 2 × 150 bp paired-end protocol. Raw data were analysed using a specific SARS-CoV-2 workflow on CLC Genomic Workbench (Qiagen, Germany). After quality check and adapter trimming, the sequences were mapped to the SARS-CoV-2 reference MN908947.3 with the identification of the nucleotide variants and amino acid changes and relative coverage. The consensus sequence was analysed on PANGO lineages [19] and Nextclade [20] software for lineage and clade identification.
In the second approach ten μl of cat and his owner total RNA were retrotranscribed using random hexamers, RNaseOUT™ Recombinant Ribonuclease Inhibitor and SuperScript™ II Reverse Transcriptase (all Life Technologies - Invitrogen), following manufacturers' instructions. The virus genome was then amplified in a two-step PCR protocol, 1st tiling in overlapping fragments of ~250 bp, and 2nd amplicon indexing. Libraries were sequenced on an Illumina NexSeq 500 platform, PE 150 bp mode. Analysis pipeline consisted in: reads filtering for adapters and low-quality positions, alignment to the reference by BWA-MEM (v 0.7.17) [21], variant calling by GATK (v 4.0.3.0) [[22], [23], [24]] and functional annotation by CorGAT [25]. Clustering was performed by randomly sampling from GISAID [26] 200 full-length accessions of Italian origin, belonging to the B.1.1.7 lineage. Multi-sequence alignment was performed by MAFFT v7.475 [27], with option –auto. Maximum-likelihood phylogenetic tree was generated by IQ-TREE [28] applying the ultrafast bootstrap option with 1000 replicates and was graphically reproduced by the Interactive Tree Of Life [29].
3. Results
The cat showed no clinical signs until March 7th when the owners noticed episodes of acute dyspnoea with air hunger and breathing rales, although regular major organic functions were preserved. The owners reported past recurrent episodes of rhinitis related to seasonal changes and regular vaccination against Feline Panleukopenia Virus, Feline Herpesvirus Type 1 and Feline Calicivirus. On March 8th and 9th, the respiratory effort subsided, even if rattle persisted. The clinical evaluations on March 10th, showed a subject in good body condition, about 5 kg in weight, senses on alert, with coat in tidy condition without soiling, normal temperature and regular mucous membranes. Auscultation of the respiratory tract revealed a continuous snoring respiratory noise, but pulmonary auscultation did not highlight respiratory noises, only a vesicular murmur partially covered by snoring noise. The cat was prescribed antibiotic therapy based on Amoxicillin and Clavulanic acid. During the clinical examination, blood sample, oropharyngeal and rectal swabs were collected and sent to the laboratory for Covid status testing.
SARS-CoV-2 RNA was detected on each cat's swabs firstly collected on March 10th. In particular, in the oropharyngeal swab Orf1ab and N genes gave a positive signal while no amplification signal was recorded for S gene; on rectal swab a weaker amplification signal occurred for N gene only. Drop out of S gene was the first indication that the cat would be positive for a SARS-CoV-2 variant.
In the following days, the owners reported the appearance of a bilateral, and intermittent nasal discharge. X-ray of the thoracic region showed a slightly decreased radio transparency and it could be assumed that the interstitium was involved; in addition, a radiopaque area in the apical lobe could be detected [Fig. 1].On March 17th, oropharyngeal swabs still showed positive reaction for N gene and Orf 1ab, while the rectal swab was negative. In the meanwhile, owners' swabs collected on March 16th confirmed similar profiles characterized by S gene dropout. A first characterization by reflex Real-Time PCR analysis resulted in the detection of the N501Y mutation in viral genomes extracted from cat as well as from owners' samples and no E484K mutation was observed. Viral genomic RNA was still evident in the oropharyngeal swabs up to four weeks after, when viral clearance was completed, while the rectal swab was always negative [Table 1].
Fig. 1.
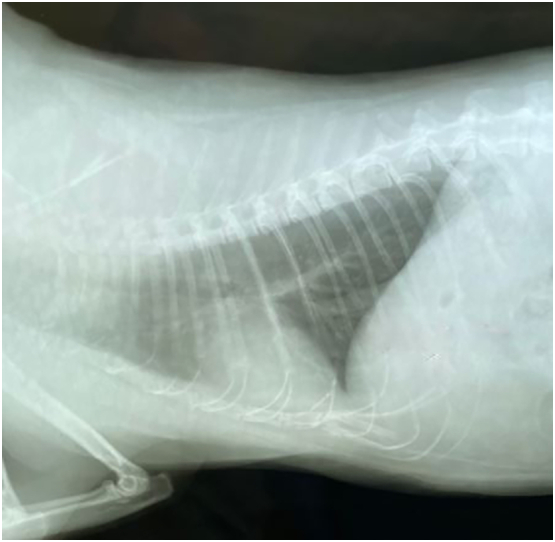
X-rays image on lateral view of cat thoracic region.
Table 1.
Sars-CoV-2 qRT-PCR detection in cat: gene amplification profiles during observation period.
| Week 0 (onset) |
Week 1 |
Week 2 |
Week 3 |
Week 4 |
||
|---|---|---|---|---|---|---|
| Ct | Ct | Ct | Ct | Ct | ||
| Oropharyngeal swab | N gene | 24,4 | 31,4 | 29,5 | 26,3 | – |
| Orf 1ab | 25,7 | 32,5 | 30,2 | 27,2 | – | |
| S gene | (dropout) | (dropout) | (dropout) | (dropout) | (dropout) | |
| Rectal swab | N gene | 33,3 | – | – | – | – |
| Orf 1ab | – | – | – | – | – | |
| S gene | (dropout) | (dropout) | (dropout) | dropout) | (dropout) | |
At symptoms onset, the specific antibody titre was 45 S/P percentage, while 12 days later it remained positive at 20 S/P, a bit lower due to lipemic interference occurred in the serum sample.
The RNA extracted from the cat swab collected first was submitted to WGS of SARS-CoV-2 by two different approaches. In the first experiment, the run produced 407.560 raw reads and the average coverage was 709.1×. with a read depth of 30× covering 92.4% of the full-length genome. The total mapped reads were 167.214. In the second experiment, the run produced 5.3 M, 4 M and 3.5 M raw reads for cat, father and daughter, respectively. The average coverage ranged between 13,166.7× and 20,308.2× with minimum of 92% of the virus reference genome covered at least 10×. Sequence analyses on PANGO lineages and Nextclade showed that both cat and owners SARS-CoV-2 genomes belong to the lineage B.1.1.7 and the phylogenetic clade 20I/501Y.V1.
The sequencing reads used in this study are available in Sequence Read Archive, under the accession PRJNA744045.
A closer look to two datasets of viral genome from cat showed that both sequencing approaches found 16 out of 17 VOC 202012/01/major variants defining B.1.1.7 lineage. The two datasets overlapped for most of the minor variants as well, with few discrepancies caused by sub-threshold coverage or differences in the methodologies used for sample preparation and analysis.
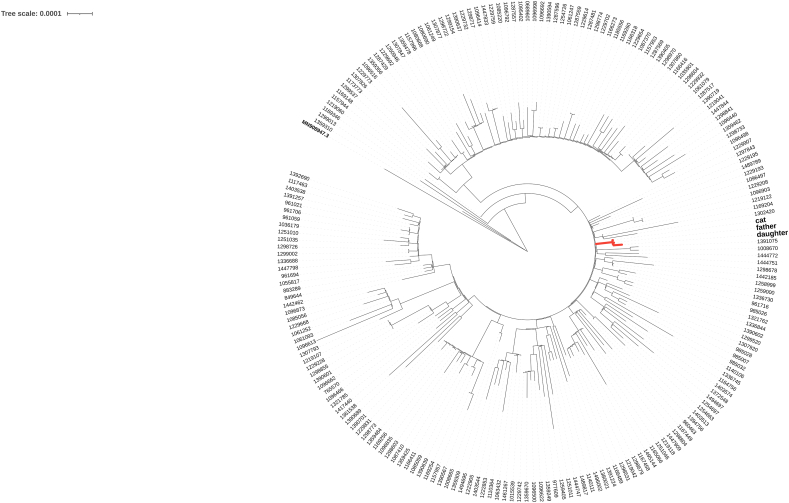
To identify the possibility of an owner-to-cat transmission event, data obtained from the second WGS experiment were used to compare virus variant patterns of the owners and their cat. Only variants with a MAF >0.7 in at least one sample were considered. In total 26 variants have been identified [Table 2]. Twenty-two were informative for all 3 samples, i.e. coverage greater than 10×. Twenty-one variants were concordant between both owners and cat. The unique discordant variant at the position 11,451 was found in the daughter, while cat and father were identical at the reference. The extremely high level of shared variants suggests infection from a common source and supports the hypothesis of the familial transmission. To further test this hypothesis, we created for each sample consensus sequences and compared them with a subset of 200 SARS-CoV-2 sequences of Italian origin belonging to the B.1.1.7 lineage deposited in GISAID [Fig. 2]. The three samples reported in this study formed a separate cluster giving a strong evidence for the familial transmission pattern. Given the timeline of the onset of the disease, with the daughter being the first to show symptoms, the father the second and the cat the third, we can infer that the virus was transmitted from the owners to the cat.
Table 2.
Functional annotation of variants identified in father, daughter, and cat. Variants with MAF >0.7 in at least one sample are shown. NA, not assigned allele frequencies due to low coverage (<10×). Coverage, number of high-quality reads with mapping quality >20.
| Position | Annotation | Allele frequency |
Coverage |
||||
|---|---|---|---|---|---|---|---|
| Cat | Father | Daughter | Cat | Father | Daughter | ||
| 241 | 5’UTR:nc.C241T,NA,NA; | 1 | 0.99 | 0.99 | 11,613 | 12,446 | 11,923 |
| 913 | orf1ab:c.648C > T,p.S216S,synonymous;nsp2:c.108C > T,p.S36S,synonymous; | 0.93 | 0.94 | 0.94 | 11,407 | 1783 | 519 |
| 3037 | orf1ab:c.2772C > T,p.F924F,synonymous;nsp3:c.318C > T,p.F106F,synonymous; | 0.99 | 0.99 | 0.99 | 1948 | 2097 | 1635 |
| 3267 | orf1ab:c.3002C > T,p.T1001I,missense;nsp3:c.548C > T,p.T183I,missense; | 0.92 | 0.93 | 0.72 | 2924 | 561 | 134 |
| 5388 | orf1ab:c.5123C > A,p.A1708D,missense;nsp3:c.2669C > A,p.A890D,missense; | 0.9 | 0.73 | 0.75 | 96 | 114 | 44 |
| 5986 | orf1ab:c.5721C > T,p.F1907F,synonymous;nsp3:c.3267C > T,p.F1089F,synonymous; | 0.8 | 0.81 | 0.6 | 225 | 289 | 163 |
| 11,288 | orf1ab:c.11023TCTGGTTTT > .........,p.SGF3675-,inframeDel;nsp6:c.316TCTGGTTTT > .........,p.SGF106-,inframeDel; | 0.94 | 0.98 | 0.93 | 3405 | 6662 | 2896 |
| 11,451 | orf1ab:c.11186A > G,p.Q3729R,missense;nsp6:c.479A > G,p.Q160R,missense; | 0 | NA | 1 | 853 | 6 | 69 |
| 14,408 | orf1ab:c.14144C > T,p.P4715L,missense;nsp12:c.968C > T,p.P323L,missense; | 1 | 1 | NA | 43 | 78 | 0 |
| 14,676 | orf1ab:c.14412C > T,p.P4804P,synonymous;nsp12:c.1236C > T,p.P412P,synonymous; | 0.95 | 1 | NA | 19 | 20 | 1 |
| 15,279 | orf1ab:c.15015C > T,p.H5005H,synonymous;nsp12:c.1839C > T,p.H613H,synonymous; | 0.94 | 0.92 | 0.94 | 81,871 | 10,882 | 1803 |
| 16,176 | orf1ab:c.15912 T > C,p.T5304T,synonymous;nsp12:c.2736 T > C,p.T912T,synonymous; | 0.81 | 0.66 | 0.8 | 270 | 362 | 201 |
| 21,765 | spike:c.203TACATG > ......,p.IHV68I,inframeDel; | 0.74 | 0.66 | 0.89 | 176 | 140 | 56 |
| 21,993 | spike:c.431ATT > …,p.YY144Y,inframeDel; | 0.91 | 0.94 | 1 | 350 | 191 | 72 |
| 23,063 | spike:c.1501A > T,p.N501Y,missense; | 0.92 | 0.98 | 0.94 | 59,839 | 92,889 | 73,230 |
| 23,271 | spike:c.1709C > A,p.A570D,missense; | 0.94 | 1 | NA | 235 | 187 | 6 |
| 23,403 | spike:c.1841A > G,p.D614G,missense; | 1 | 1 | 1 | 164 | 214 | 98 |
| 23,604 | spike:c.2042C > A,p.P681H,missense; | 0.66 | 0.78 | 0.87 | 2906 | 924 | 457 |
| 23,709 | spike:c.2147C > T,p.T716I,missense; | 0.73 | 0.63 | 0.78 | 995 | 1197 | 632 |
| 24,914 | spike:c.3352G > C,p.D1118H,missense; | 0.93 | 0.97 | 0.79 | 252 | 110 | 38 |
| 27,972 | orf8:c.79C > T,p.Q27*,stopGain; | 0.95 | 0.99 | 0.96 | 65,155 | 8235 | 5423 |
| 28,048 | orf8:c.155G > T,p.R52I,missense; | 0.95 | 0.99 | 0.94 | 32,546 | 4154 | 2774 |
| 28,273 | NA | 0.97 | 0.96 | 0.85 | 6187 | 3072 | 377 |
| 28,280 | geneN:c.7GAT > CTA,p.D3L,missense; | 0.97 | 0.97 | 0.88 | 6180 | 3047 | 366 |
| 28,881 | geneN:c.608GGG > AAC,p.RG203KR,missense; | 0.92 | 0.91 | 0.8 | 6842 | 11,609 | 7074 |
| 28,977 | geneN:c.704C > T,p.S235F,missense; | 0.97 | 0.99 | 0.92 | 21,515 | 8684 | 658 |
Fig. 2.
Phylogenetic analysis of SARS-CoV-2 genomes. A total of 204 viral genomes are displayed, including (i) father, daughter and cat genomes, (ii) 200 genomes from GISAID randomly selected among B.1.1.7 lineage of Italian origin; and (iii) the SARS-CoV-2 reference Wuhan Hu-1 genome. The father, daughter and cat genomes cluster together (node highlighted in red). GISAID sequences are labelled with EPI accession numbers. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1. Outbreak control measures
Based on clinical suspicion of COVID-19, the family members were placed in isolation; from the time of symptom development (February 26th). On March 7th as their cat was symptomatic, in accordance with the official Italian guidelines (issued by the Italian National Institute of Health) [30] on the management of pets potentially infected with Sars-CoV2, Veterinary Services prescribed isolation of the animal in a separate room. Because of the isolation regime the owners were under, on March 10th, to allow a clinical examination and the collection of blood sample and oropharyngeal and rectal swabs, the cat was taken to the nearby veterinary practice using a pet ambulance service and ensuring all precautionary safety measures to avoid exposure of the operators. After confirmation of SARS-CoV-2, the animal was kept in isolation and subjected to a weekly monitoring program of both clinical and laboratory checks until complete remission and viral clearance.
4. Discussion
By integrating anamnestic, clinical and analytical data it was possible to confirm the concomitant presence of Alpha variant infection and the sequence clustering in a cat and its owners and to understand the sequence of events. We were able to confirm the susceptibility to the infection to Alpha variant and the potential for subsequent respiratory disease in household cats. Together with the molecular virus detection, serological data and the clinical history of this outbreak, as shown in a situation where the Alpha variant was not involved [31], NGS analysis was helpful to provide evidence of human-to-animal transmission.
We performed SARS-CoV-2 variant tracking analysis at genomic level to gain the resolution required for the identification of transmission events. Based on comparative analysis of sequences from our study and additional 200 SARS-CoV-2 B.1.1.7 sequences of Italian origin from GISAID, we identified a specific cluster containing the three samples analysed in this study. To carry out this comparative analysis no previous reported Italian cats affected by this variant existed and were available in GISAID; however, the additional human sequences of Alpha variant used for comparison were randomly sampled to reduce any potential for selection bias.
The observation of sequence clustering combined with clinical data, i.e. cat having an acute respiratory syndrome one week after the onset of COVID-19 in the family and showing a similar evolution with about one-week lag time in respect to its owners, strongly support transmission inside the family cluster from human to cat.
A unique discordant variant was found in the daughter. The time course of onset of clinical signs and positivity suggest that the daughter, the first family member with COVID-19 symptoms, diffused virus to the rest of the family. The presence of the new variant in the daughter could be explained by the acquisition of new variants in the 18-day period that passed before the sample used for genomic analysis was collected. Alternatively, due to the presence of within-host virus diversity, i.e. intra-host variants, after transmission, variants may either disappear due to transmission bottleneck or occasionally become fixed and spread more widely [32] [33].
The evolution of the SARS-CoV-2 pandemic and the spread of oncoming variants may represent a new challenge to the Health Services of all countries: on one hand, our investigation strengthened the hypothesis, already reported in the literature [2], of human-to-cat transmission. On the other hand, it served to understand that the susceptibility of household cats to the SARS-CoV-2 variants should prompt infected owners to limit close contact with their pets. In such situations it has been suggested to handle cats only when wearing a mask, to wash hands with soap and water for at least 20 s before and after contact with cats, their food or litter, and to avoid kissing domestic cats or sharing food, towels or the bed with them [34].
In this outbreak, communication between owners, veterinary practitioners, veterinary officers, public health officers and diagnostic laboratories played a key role both in the early diagnosis of viral infection in the involved cat and in the appropriate health management of the case. Passive surveillance of pets living with COVID-19 positive patients should always be ensured, as close interactions between humans and their animals create opportunities for zoonotic transmission and spillover events (as it was shown in mink farms [35]). The presence of possible animal reservoirs like the cat underlines the importance of a detailed knowledge of the circulation of SARS-CoV-2 and its variants in humans and animals [2]. To date, despite the available evidence that B.1.1.7 lineage spreads easily and quickly, there is no evidence of its greater morbidity among pets: however the accumulating data from this and other studies [17,18] suggest that attention should be paid to possible spillover risks.
5. Conclusion
This report emphasizes the importance of the One Health approach when dealing with public health issues in the field of zoonotic disease transmission: this is particularly true in the case of the spread of SARS-CoV-2 variants and their potential zoonotic nature. It also confirms the usefulness of integrating epidemiological investigations and advanced diagnostic techniques to fully understand the origin and dynamics of outbreaks and to prevent unexpected modes of transmission or unwarranted alerts involving our pet populations.
Funding statement
This work was co-funded by the CRT Foundation - Welfare and Environment Grants 2020. Project title: Presenza di SARS-CoV-2 negli animali d'affezione in Piemonte: strumenti diagnostici ed. epidemiologici a supporto di una corretta gestione del rischio.
Ethical statement
Not relevant, as all the activities on animal, persons and their derivatives are carried out in the framework of established Public Health interventions: this report is based on management of clinical and laboratory data not produced for research purposes.
Authors' contributions
Roberto Zoccola and Maria Goria conceived the study, performed molecular diagnosis of COVID-19 on the cat, as well as molecular characterization of cat and owners Sars-CoV-2 genomes and wrote the article. Chiara Beltramo, Gabriele Magris, Simone Peletto, Pierluigi Acutis, Elena Bozzetta, Slobodanka Radovic carried out all Sars-CoV-2 NGS-WGS and corresponding data sets analysis. Maria Silvia Gennero was applied to serological examination of cat blood samples. Francesco Zappulla and Anna Maria Porzio were involved in Vet Public Health and cat clinical management respectively, collecting all cat samples and performing clinical examinations. Alessandro Dondo and Chiara Pasqualini dealt with management of samples by vet and human side respectively. Bartolomeo Griglio and Angelo Ferrari coordinated the communication between all the Public Health Authorities involved both on Human side and Vet side. Giuseppe Ru conceived the study, entered and managed the epidemiological data and wrote and critically revised the article. All the Authors internally contributed to discussion and paper revision.
Declaration of Competing Interest
None.
Acknowledgments
The Authors gratefully acknowledge Dr. Giovanni Camisasca, Local Health Unit of Novara, for having kindly provided rhino-pharyngeal swabs of cat's owners, making possible the comparative molecular and sequencing analysis.
References
- 1.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decaro N., Lorusso A. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet. Microbiol. 2020 May;244:108693. doi: 10.1016/j.vetmic.2020.108693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sit T.H.C., Brackman C.J., Ip S.M. Infection of dogs with SARS-CoV-2. Nature. 2020;586:776–778. doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi J., Wen Z., Zhong G. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segalés J., Puig M., Rodon J., Avila-Nieto C., Carrillo J., Cantero G. Detection of SARS-CoV-2 in a cat owned by a COVID-19-affected patient in Spain. Proc. Natl. Acad. Sci. U. S. A. 2020 Oct 6;117(40):24790–24793. doi: 10.1073/pnas.2010817117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sailleau C., Dumarest M., Vanhomwegen J., Delaplace M., Caro V., Kwasiborski A. First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound. Emerg. Dis. 2020 Nov;67(6):2324–2328. doi: 10.1111/tbed.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pagani G., Lai A., Bergna A., Rizzo A., Stranieri A., Giordano A. Human-to-cat SARS-CoV-2 transmission: case report and full-genome sequencing from an infected pet and its owner in northern Italy. Pathogens. 2021 Feb 23;10(2):252. doi: 10.3390/pathogens10020252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garigliany M., Van Laere A., Clercx C., Giet D., Escriou N., Huon C. SARS-CoV-2 natural transmission from human to cat, Belgium, march 2020. Emerg. Infect. Dis. 2020;26(12):3069–3071. doi: 10.3201/eid2612.202223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman A., Smith D., Ghai R., Wallace R.M., Torchetti M. First Reported Cases of SARS-CoV-2 Infection in Companion Animals - New York, March–April 2020-Morbidity and Mortality Weekly Report June 12, 2020 / Vol. 69 / No. 23 US Department of Health and Human Services/Centers for Disease Control and Prevention. https://www.cdc.gov/mmwr/volumes/69/wr/pdfs/mm6924-H.pdf [DOI] [PMC free article] [PubMed]
- 10.Hamer S.A., Pauvolid-Corrêa A., Zecca I.B., Davila E., Auckland L.D., Roundy C.M. Natural SARS-CoV-2 infections, including virus isolation, among serially tested cats and dogs in households with confirmed human COVID-19 cases in Texas, USA. bioRxiv. 2020 doi: 10.1101/2020.12.08.416339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritz M., Rosolen B., Krafft E., Becquart P., Elguero E., Vratskikh O. High prevalence of SARS-CoV-2 antibodies in pets from COVID-19+ households. One Health. 2021;11:100192. doi: 10.1016/j.onehlt.2020.100192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leroy E.M., Gouilh M.A., Brugère-Picoux J. The risk of SARS-CoV-2 transmission to pets and other wild and domestic animals strongly mandates a one-health strategy to control the COVID-19 pandemic. One Health. 2020;10 doi: 10.1016/j.onehlt.2020.100133. 100133. Published online 2020 Apr 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Totton S.C., Sargeant J.M., O’Connor A.M. How could we conclude cat-to-human transmission of SARS-CoV-2? Zoonoses Public Health. 2021;68(1):67–68. doi: 10.1111/zph.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Overgaauw P.A.M., Vinke C.M., Hagen M.A.E.V., Lipman L.J.A. A one health perspective on the human-companion animal relationship with emphasis on zoonotic aspects. Int. J. Environ. Res. Public Health. 2020;17(11):3789. doi: 10.3390/ijerph17113789. Published 2020 May 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemp S., Harvey W., Datir R., Collier D., Ferreira I., Carabelii A. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion ΔH69/V70. bioRxiv. 2020 doi: 10.1101/2020.12.14.422555. [DOI] [Google Scholar]
- 16.Davies N, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, et al. Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv and bioRxiv doi. 10.1101/2020.12.24.20248822. [DOI]
- 17.Gauntt J. Texas a&M research uncovers first known COVID-19 UK variant in animals. Texas A&M University College of Veterinary Medicine & Biomedical Sciences March. 2021;15 https://today.tamu.edu/2021/03/15/texas-am-research-uncovers-first-known-covid-19-uk-variant-in-animals/ Available from: [Google Scholar]
- 18.Ferasin, L, Fritz, M, Ferasin, H, Becquart P, Legros V, Leroy E.M. Myocarditis in naturally infected pets with the British variant of COVID-19 March 18, 2021. bioRxiv preprint doi doi. 10.1101/2021.03.18.435945. [DOI]
- 19.O'Toole A, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, et al. Pangolin: lineage assignment in an emerging pandemic as an epidemiological tool. Available from: github.com/cov-lineages/pangolin. [DOI] [PMC free article] [PubMed]
- 20.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Durbin R. BWA.Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mc Kenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo M., Banks E., Poplin R., Garimella K., Maguire J., Hartl C. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Auwera G.A., Carneiro M., Hartl C., Poplin R., del Angel G., Levy-Moonshine A. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiara M., Zambelli F., Tangaro M.A., Mandreoli P., Horner D.S., Pesole G. CorGAT: a tool for the functional annotation of SARS-CoV-2 genomes. Bioinformatics. 1 December 2020;36:22–23. doi: 10.1093/bioinformatics/btaa1047. 5522–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. DOI:10.1002/gch2.1018 PMCID: 31565258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letunic I., Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gruppo di lavoro ISS Sanità Pubblica Veterinaria e Sicurezza Alimentare COVID-19 . Roma: Istituto Superiore di Sanità; 2020. Animali da compagnia e SARS-CoV-2: cosa occorre sapere, come occorre comportarsi. Versione del 19 aprile 2020.https://www.iss.it/documents/20126/0/Rapporto+ISS+COVID-19+n.+16_2020+animali+%281%29.pdf/151c05b1-7f1e-4419-e929-57684b1931a1?t=1587536267422 (Rapporto ISS COVID-19, n. 16/2020). Italian. Avaible from. [Google Scholar]
- 31.Klaus J., Meli M.L., Willi B., Nadeau S., Beisel C., Stadler T. Detection and genome sequencing of SARS-CoV-2 in a domestic cat with respiratory signs in Switzerland. Viruses. 2021;13(3):496. doi: 10.3390/v13030496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lythgoe KA, Hall M, Ferretti L, de Cesare M, MacIntyre-Cockett G, Trebes A et al. SARS-CoV-2 within-host diversity and transmission, 2021/04/16 Science, 372 6539 doi. 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed]
- 33.Pfefferle S, Günther T, Kobbe R, Czech-Sioli M, Nörz D, Santer R et al. SARS Coronavirus-2 variant tracing within the first Coronavirus Disease 19 clusters in northern Germany. (2021). Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases, 27(1), 130.e5–130.e8. doi. 10.1016/j.cmi.2020.09.034. [DOI] [PMC free article] [PubMed]
- 34.Hosie M.J., Hofmann-Lehmann R., Hartmann K., Egberink H., Truyen U., Addie D.D. Anthropogenic infection of cats during the 2020 COVID-19 pandemic. Viruses. 2021;13(2):185. doi: 10.3390/v13020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oude Munnink B., Sikkema R., Nieuwenhuijse D., Molenaar R., Munger E., Molenkamp R. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371 doi: 10.1126/science.abe5901. 172–17. [DOI] [PMC free article] [PubMed] [Google Scholar]