The outbreak of COVID-19, which originated in Wuhan, China, was first noticed in December 2019. The genome of the causative agent, known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), had been sequenced and made publicly available a month later [1]. The virus rapidly spread to other countries in East Asia out of the Mainland China, and just a few days later, there were confirmed cases in the USA and Europe. As of July 2021, COVID-19 is still an ongoing pandemic and thousands of SARS-CoV-2 variants have been emerging and circulating globally.
The global health emergency imposed by the COVID-19 pandemic boosted research development that is now reflected in the more than 2 million worldwide SARS-CoV-2 genomes available through public databases.
The first detection of SARS-CoV-2 in the USA was in the state of Washington, on January 21, 2020, involving a patient who had returned after traveling to Wuhan. The virus was fully sequenced and revealed only three nucleotides and one amino acid distinct from the Wuhan reference genome [2]. A month later, a second SARS-CoV-2 infection was reported in Washington, but this episode was likely caused by a second introduction from Europe [3]. As of March 25, 2020, the state of Washington had already confirmed 2580 cases and 132 deaths [4].
In the issue of The Lancet Regional Health – Americas, Tordoff et al. [5] used 11,422 complete genomes of SARS-CoV-2 publicly available on the GISAID database (gisaid.org) to estimate the number of introductions and exportations of SARS-CoV-2 in the state of Washington during the first year of COVID-19 pandemics.
By using phylogenetic-based methods, they estimated that there were at least 287 distinct introductions in the state of Washington, most of them originating elsewhere in the United States. As expected, the lineages B.1 and B.1.1 belonging to the G clade, which is associated with increased transmissibility, accounted for the majority of the introductions that occurred mostly in March and April 2020.
Late in March 2020, when the number of cases in several states was on the rise, the Governor of Washington issued public health measures that severely restricted human mobility. The restrictions took effect and the number of cases and deaths decreased. Critically, despite the lack of air restrictions at the time, Tordoff et al. [5] also demonstrated that the number of virus introductions/exportations to and from the state dropped, reflecting the effectiveness of such policies in reducing the overall virus transmission.
Although non-pharmaceutical interventions were proven to be effective in decreasing the effective reproduction number (Rt) of SARS-CoV-2 [6], containment measures alone are not enough to tackle the pandemic. Openly shared and real-time information regarding the lineages circulating worldwide and close monitoring of the worrying variants are also critical to adequately respond to the COVID-19 health crisis.
COVID-19 is the first global emergency in which next-generation sequencing capacity has been available to public health since the very beginning. Netherlands and Australia are good examples where prompt efforts for sequencing the SARS-CoV-2 genome resulted in early decisions that helped to control local transmission.
In fact, to actually conduct public health measures, results must be obtained and communicated in a timely and efficient manner so that strategies can be rapidly defined. During the first half of the pandemic, phylogenetic and phylodynamic studies using SARS-CoV-2 genomic data were mostly dedicated to describing the epidemics and virus dispersion. Significant advances in diagnosis and vaccine development were also achieved thanks to the amount of data generated and openly shared in the first months of 2020. At that time, due to the apparently low evolutionary rate of SARS-CoV-2, little attention was paid to the risks for the emergence of variants with clinical and epidemiological consequences. But the picture has changed. As of June 2021, as a consequence of uncontrolled virus circulation, thousands of new mutations and at least four variants of concern (V.O.C) were described and it is very likely that other variants yet to be identified are also circulating.
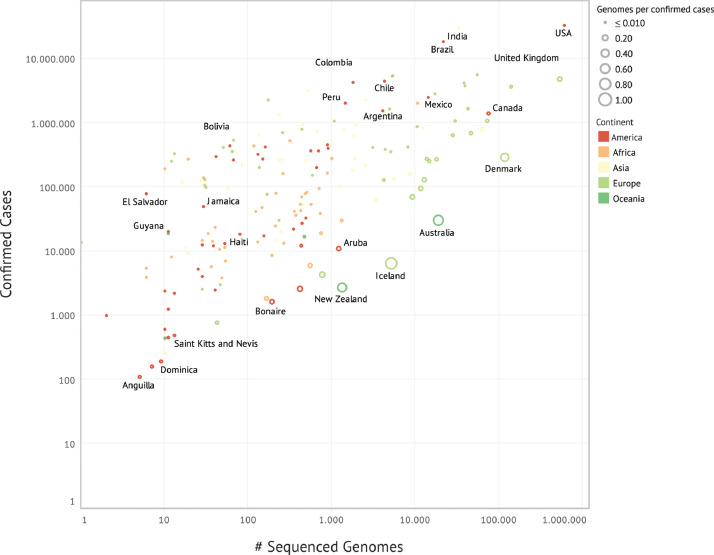
The number of SARS-CoV-2 sequences posted in the GISAID database has been substantially increasing every day. With few exceptions, such as Iceland and Australia, where the highest proportion of reported cases (~40–50% of all cases) was sequenced, most countries still face problems regarding genomic surveillance capacity, which is far from being sufficient in terms of number of genomes sequenced per total number of cases (Fig. 1). Public and private initiatives to implement and extend the sequencing capability on a global basis, particularly in developing countries, such as the Regional Network for COVID-19 surveillance (COVGEN) from the Pan American Health Organization (PAHO), GENOV, funded by the largest private medical diagnostic company in Brazil and the Africa CDC with the Pathogens Genomic Initiative, are helping to find blind spots of SARS-COV-2 genetic information in Latin America and Caribbean, and Africa [7], [8], [9].
Fig. 1.
The proportion of global SARS-CoV-2 cases sequenced and shared on the GISAID database until July 07, 2021. The size of each circle is proportional to the number of SARS-CoV-2 genomes sequenced per number of confirmed cases. Countries are colored by continent. For clarity, only some countries were labeled.
However, the sequencing capacity is not the only issue. Structured and efficient responses also depend on the ability of the researchers to translate the genomic data into insights for policy makers so that the results of this “open science” lead to pragmatic and rapid solutions. The COVID-19 pandemic exposed the gap between scientific research and policy makers. Therefore, it is mandatory that scientists and policy makers work closely to defeat this pandemic, otherwise the uncontrolled emergence of variants and vaccination lag in several low-income countries will prevent us from ending the COVID-19 pandemic.
Declaration of interests
The authors have nothing to disclosure.
Funding
CMR receives grant #2019/03859–9, São Paulo Research Foundation (FAPESP), and from The National Council for Scientific and Technological Development (CNPq) grant #402794/2020–6. FLM is granted by The National Council for Scientific and Technological Development (CNPq).
References
- 1.Wu F., Zhao S., Yu B., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holshue M.L., DeBolt C., Lindquist S. Washington State 2019-nCoV case investigation team. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford T., Greninger A.L., Roychoudhury P., et al. Cryptic transmission of SARS-CoV-2 in Washington state. Science. 2020;370:571–575. doi: 10.1126/science.abc0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Washington State Department of Health, “2019 novel coronavirus outbreak (COVID-19)” (2020); www.doh.wa.gov/Emergencies/Coronavirus.
- 5.Tordoff D.M., Greninger A.L., Roychoudhury P., et al. Phylogenetic estimates of SARS-CoV-2 introductions into Washington State. Lancet Reg Health Am. 2021 doi: 10.1016/j.lana.2021.100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inglesby T.V. Public health measures and the reproduction number of SARS-CoV-2. JAMA. 2020;323:2186–2187. doi: 10.1001/jama.2020.7878. [DOI] [PubMed] [Google Scholar]
- 7.COVID-19 Genomic Surveillance Regional Network. Available at: https://www.paho.org/en/topics/influenza/covid-19-genomic-surveillance-regional-network.
- 8.GENOV. Available at. https://medicinasa.com.br/dasa-banco-genomico/.
- 9.Africa Pathogen Genomics Initiative. Available at https://africacdc.org/news-item/us100-million-africa-pathogen-genomics-initiative-to-boost-disease-surveillance-and-emergency-response-capacity-in-africa/.