Abstract
SARS-CoV-2, the cause of the currently ongoing COVID-19 pandemic, encodes its own mRNA capping machinery. Insights into this capping system may provide new ideas for therapeutic interventions and drug discovery. In this work, we employ a previously developed Py-FLINT screening approach to study the inhibitory effects of compounds against the cap guanine N7-methyltransferase enzyme, which is involved in SARS-CoV-2 mRNA capping. We screened five commercially available libraries (7039 compounds in total) to identify 83 inhibitors with IC50 < 50 μM, which were further validated using RP HPLC and dot blot assays. Novel fluorescence anisotropy binding assays were developed to examine the targeted binding site. The inhibitor structures were analyzed for structure-activity relationships in order to define common structural patterns. Finally, the most potent inhibitors were tested for antiviral activity on SARS-CoV-2 in a cell based assay.
Keywords: nsp14, Inhibitor screening, COVID-19, Fluorescence assay
1. Introduction
In 2019, a novel β–coronavirus (β–CoV) emerged, causing coronavirus disease 19 (COVID-19) (Guo et al., 2020) and leading to more than one hundred million infections around the world as a serious threat to global health. Due to its high similarity to the SARS and MERS coronaviruses, it was named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). While multiple vaccines (i.e. Comirnaty® by BioNTech and Pfizer, mRNA-1273 by Moderna, as well as ChAdOx1 nCoV-19 by AstraZeneca) (Krammer, 2020) have been developed to prevent the spread of COVID-19, effective medical treatment is currently lacking. The U.S. Food and Drug Administration (FDA) recently approved Veklury (remdesivir) for the treatment of hospitalized COVID-19 patients (ClinicalTrials.gov: NCT04323761). Other repurposed drugs, such as favipiravir, lopinavir, and ritonavir (Abd El-Aziz and Stockand, 2020), are still subject to clinical trials. Several monoclonal antibody treatments (e.g., tocilizumab, ClinicalTrials.gov: NCT02735707) (Gordon et al., 2021) have also been approved for treatment of COVID-19 patients with mild or moderate symptoms at risk of progressing to severe disease and hospitalization.
Nevertheless, the ongoing pandemic necessitates the development of novel treatment for COVID-19 and possible future CoV infections. Compounds that target processes essential for virus multiplication and specifically inhibit crucial viral proteins may have potential as antiviral therapeutics. An example of a druggable target process is the 5’ end capping of newly synthesized viral mRNAs, crucial for transcript stability and protein expression in infected human cells.
In eukaryotes, mRNA is modified at its 5′ end via the addition of a 7-methylguanosine (m7G) cap, which protects transcripts from premature degradation, augments translation, and allows for discrimination between endogenous mRNAs and foreign RNA material (e.g. viral). The process of cap biosynthesis engages three enzymes, namely RNA 5′-triphosphatase (TPase), guanylyltransferase (GTase), and N7-methyltransferase (N7-MTase), resulting in cap 0 structure formation, typical for yeasts and plants (Shatkin and Manley, 2000). In the canonical (human) RNA capping pathway, TPase catalyzes RNA 5′ triphosphate cleavage to release RNA 5′ diphosphate and a free phosphate group. Thereafter, GTase transfers a GMP molecule from GTP onto the 5′ end of RNA. Finally, N7-MTase specifically methylates the guanine N7 position, utilizing S-adenosyl-L-methionine (SAM) as a methyl group donor. In higher eukaryotes, the mRNA cap is subjected to additional methylation at the 2′-O position of the first transcribed nucleotide (cap 1) under the action of 2′-O methyltransferase. Some mRNAs undergo a second 2′-O methylation at the second transcribed nucleotide, forming the so-called cap 2. The functional significance of such 2′-O-methylation is still under investigation, but it presumably protects mRNA from translational shutdown triggered via the innate immune response as a result of type I interferon signaling pathway. Due to the binding specificities of human IFIT proteins, the innate immune system can effectively distinguish and target triphosphate RNAs and cap 0-carrying mRNAs (which come from exogenous sources), but not endogenous cap 1 and cap 2 mRNAs (Daffis et al., 2010).
Viruses often harness their own mRNA capping pathways in order to augment the expression of viral proteins and escape the innate immune response of the host (Decroly et al., 2011a; Furuichi and Shatkin, 2000). These may include the canonical capping pathway or distinct non-canonical mechanisms, including m7GTP RNA capping way (alphaviruses) (Delang et al., 2016), GDP RNA capping (non-segmented negative-sense viruses e.g. Rabies virus) (Ogino et al., 2016) or “cap snatching” (influenza virus). (Plotch et al., 1981).
Coronaviruses (CoVs) are positive-stranded RNA viruses with some of the most complex and largest viral genomes. Severe acute respiratory syndrome coronavirus (SARS-CoV) expresses 16 non-structural proteins (nsps), which are responsible for essential processes and are no part of the viral capsid. The SARS-CoV-2 genome shares 85% identity with that of SARS-CoV (Wu et al., 2020), also encoding 16 non-structural proteins. In contrast to small RNA viruses, CoVs encode unique proteins to cap their mRNAs, presumably following the canonical or alphavirus-like capping pathway (Chen and Guo, 2016). While the nsp13 enzyme is responsible for TPase and RNA helicase function for both SARS-CoV and SARS-CoV-2 (Shu et al., 2020; Ivanov et al., 2004), there is still no experimental evidence of proteins with GTase activity engaged in RNA capping (Ramanathan et al., 2016). SARS-CoV mRNAs are N7-methylated by nsp14 (Chen et al., 2009). The nsp14 enzyme has also been reported to possess 3′–5′ exonuclease (ExoN) activity, thus acting as an RNA proofreading enzyme (Eckerle et al., 2010). The two catalytic domains of nsp14, ExoN and N7-MTase, are located at its N- and C-termini, respectively, and function independently. Nonetheless, the 62–527 amino acid sequence is required for both activities. The exonuclease activity of nsp14 is significantly enhanced upon binding with nsp10, while N7-MTase activity is not affected (Bouvet et al., 2010, 2012). The nascent cap 0 structure can be further methylated by SAM-dependent nsp16, which acts as a 2′-O-methyltransferase, requiring nsp10 as a cofactor for activation. The nsp16/nsp10 complex utilizes m7GpppA-RNA to synthesize the cap 1 structure (Decroly et al., 2011b; Krafcikova et al., 2020; Rosas-Lemus et al., 2020).
Differences in capping machinery and mechanisms between coronaviruses and humans create new opportunities for antiviral drug development. Viral N7-methyltransferases have already been highlighted by various groups as a potential targets for antiviral therapies(Coutard et al., 2014; Lim et al., 2015; Singh et al., 2016; Ferron et al., 2012), including ones for SARS-CoV (Tong, 2009). N7-MTase inhibition has been shown to suppress viral replication (Bouvet et al., 2010; Dong et al., 2008). Various small-molecule inhibitors of SARS-CoV nsp14 N7-MTase have already been discovered through different in vitro approaches, including large library screening (Aouadi et al., 2017) and rational drug design based on screening results or substrate structures (Ahmed-Belkacem et al., 2020).
Currently, the crystal structure of SARS-CoV-2 nsp14 N7-MTase domain remains unknown, impeding rational drug design (although the ExoN-nsp10 complex is already available – PDB code: 7DIY (Lin et al., 2021)). However, the N7-MTase active site is highly conserved among Coronaviridae (Becares et al., 2016), and the high amino acid identity with SARS-CoV nsp14 (>95%) enables homology modelling (Dong et al., 2020). This approach has been widely used for compound library virtual screening, including docking studies (Gurung, 2020; Liu et al., 2020; Martin and Cheng, 2020; Selvaraj et al., 2020).
Based on the structural comparison of SARS-CoV-2 nsp14 to its SARS-CoV homolog, Otava et al. carried out structure-guided design of substrate-based inhibitors, generating analogs of S-adenosyl-L-homocysteine (SAH) modified with aromatic groups at position 7 of the 7-deazaadenine derivative of SAH (Otava et al., 2021). Inhibitors have also been synthesized and tested in vitro via radioactivity-based assays using biotinylated RNA and 3H-SAM substrates (Devkota et al., 2021) as well as via surface plasmon resonance.
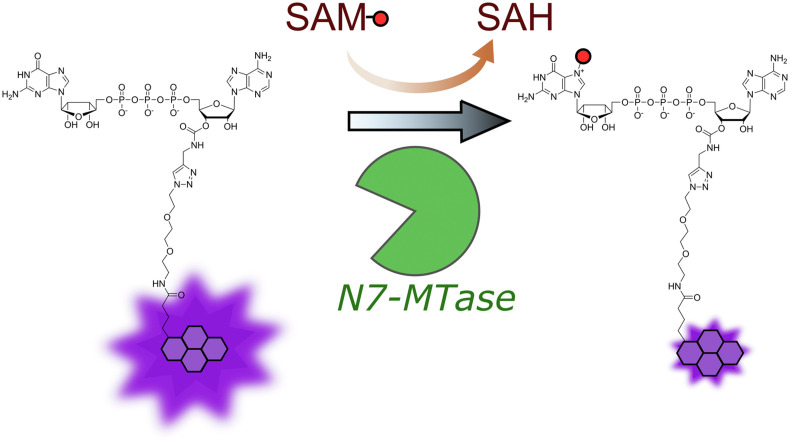
We recently developed a fluorescence-based assay for the high-throughput screening (HTS) of potential N7-methyltransferase inhibitors. The assay is based on a Gp3A analog labeled with pyrene at the 3′-O- position of adenosine that undergoes fluorescence intensity change (quenching) upon methylation (Py-FLINT probe; Fig. 1 ). We have applied the assay in studies on model N7-MTases, including Ecm1 from E. cuniculi, human RNMT-RAM, and Vaccinia capping enzyme (VCE) from Vaccinia virus (Kasprzyk et al., 2020). In the current work, we adapted the assay to explore the N7-MTase activity of nsp14 from SARS-CoV-2 in order to identify potent inhibitors of viral mRNA N7-methylation with antiviral drug potential. The method allows for the direct monitoring of nsp14 activity in real time and is suitable for the exploration of inhibitors of both the nucleotide- and SAM-binding sites of N7-MTase. We employed the method for HTS experiments of five commercially available compound libraries, namely LOPAC®1280, Machine Learning SARS Targeted Library (OTAVA Chemicals), SARS-CoV-2 nsp16 Targeted Library (OTAVA Chemicals), FDA-approved Drug Library, and part of the Flavonoids Compound Library (ChemFaces), containing a total of 7039 compounds. The most potent hits selected from the screening were further evaluated to determine their IC50, validate their inhibitory properties using RNA substrates, determine selectivity against the human N7-MTase RNMT-RAM, and performed structure-activity relationship (SAR) analysis. To study the mechanism of action (MOA), we developed fluorescence anisotropy (FA) binding assays using either fluorescently labeled Gp3A or SAH analogs. Finally, the antiviral activity of identified nsp14 inhibitors was studied using human β–coronavirus SARS-CoV-2-infected Huh 7 (human hepatocellular carcinoma) cells (Gurard-Levin et al., 2020). We identified three compounds that inhibited viral replication in the cell culture model.
Fig. 1.
Principle of the Py-FLINT N7-MTase assay (Kasprzyk et al., 2020).
The screening of large numbers of compounds together with SAR analysis for the identification of small-molecule inhibitors with high selectivity towards SARS-CoV-2 capping enzymes could benefit rational drug design. We hope that our results are considered in the development of new drugs and facilitate the introduction of COVID-19 therapeutics.
2. Materials and methods
Chemical synthesis of Gp3ApG and SAH-N6-6tFluo, nsp14 expression and purification, Py-FLINT assay optimization for nsp14, short RNA synthesis, and dot blot analysis were described in details in Supplementary Information. Py-FLINT and FA binding assays were performed using point fluorescence measurements in 96-well black, non-binding assay plates at 30 °C with microplate reader Biotek Synergy H1. For FA binding assays we used excitation (485 ± 20 nm) and emission (528 ± 20 nm) polarization filters.
2.1. Screening experiments and IC50 determination using Py–FLINT assay
For screening experiments each sample (well) contained 50 mM Tris-HCl, pH 7.5, a substrate (1 μM Py-FLINT probe), the SAM cosubstrate (20 μM), nsp14 enzyme (40 nM), an inhibitor (50 μM), and 5% of DMSO (if compound in library was dissolved in pure DMSO). The reaction components were preincubated for 15 min at 30 °C with mixing at 300 rpm. Before adding the enzyme, the plate was additionally incubated in a plate reader for 20 min at 30 °C with mixing at 300 rpm and point fluorescence registered every minute to obtain stable fluorescence signal. Immediately before fluorescence reading, 10 μL of an enzyme solution was added into each well to a total reaction volume of 150 μL. The reaction was monitored for 0.5–1 h. Initial rates were calculated by fitting a linear curve to the first 10 points (10 min).
IC50 determination experiments were performed analogously, but with different inhibitor concentrations (half-log dilutions logC inh <−2.5; 2> for nucleotide-like inhibitors or logC inh. <−3; 1.5>). If the inhibitor fluorescence interfered with the Py-FLINT probe emission, a control measurement of the inhibitor solution was performed under the same conditions and then the result was subtracted from the inhibition data. To determine IC50 parameters, a four-parameter dose-response equation was fitted as follows:
| (1) |
where A1 and A2 are the bottom and top asymptotes, respectively; C inh. the inhibitor concentration; p is the Hill coefficient, and V/V 0 is the ratio of the initial reaction rate with the inhibitor to that without the inhibitor.
2.2. FA binding assay
To determine dissociation constant values K D of two binding probes – Gp3A-3′-O-(5)FAM and SAH-N6-6tFluo direct binding experiment was performed. Each well contained a probe (10 nM), and nsp14 protein (0–4 μM) in 50 mM Tris-HCl pH 7.5 buffer in a total volume of 150 μL, together with FA point measurements. The components were preincubated for 15 min at 30 °C with mixing at 300 rpm. The FA signals were recorded for 60 min with 2.5 min time interval. For each timepoint the FA values were calculated using the following equation:
| (2) |
where is the parallel emission intensity, the perpendicular emission intensity, and G is a grating factor equal to 0.994. For each sample the final FA values were calculated as the mean FA values from datapoints between 50 and 60 min. To calculate the total emission intensity we used the following equation:
| (3) |
As the I values did not change more than 10% we did not corrected FA values on enhancement factor resulting from emission intensity changes. To determine K D the FA values were plotted as a function of protein concentration and the following binding curve was fitted:
| (4) |
where FA F is the fluorescence anisotropy of free probe, FA B is the fluorescence anisotropy of bound probe, C p is the nsp14 concentration, and L T is the total probe concentration. Competition experiments were carried out for 100 μM inhibitor concentration at constant probe (10 nM) and protein (2 μM) concentrations. The samples were prepared in 50 mM Tris-HCl pH 7.5 buffer in total volume of 150 μL.The FA signals were recorded for 60 min with 2.5 min time interval. For each timepoint the FA values were calculated using equation (3). For each sample the final FA values were calculated as the mean FA values from datapoints between 50 and 60 min.
2.3. Antiviral assay
The human hepatoma cell line Huh 7 (kindly provided by Ralf Bartenschlager, University of Heidelberg, Germany) was maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco cat no 41965-039) supplemented with 10% fetal bovine serum (FBS), 2% HEPES 1M (Gibco cat no 15630106), 1% non-essential amino acids (NEAA Gibco cat no 11140050) and 1% Penicillin-Streptomycin 10,000 U/mL (Gibco cat no 15140148) in a humidified 5% CO2 incubator at 37 °C. Assay medium, used for producing virus stocks and antiviral testing, is prepared by supplementing DMEM with 4% FBS, 2% HEPES 1M, 7.5% and 1% NEAA. In order to quantify antiviral activity on Huh7 cells we selected a SARS-CoV-2 virus strain that produces sufficient CPE on this cell line. For this we started from passage 6 of the SARS-CoV-2 strain BetaCov/Belgium/GHB-03021/2020 (EPI ISL 407976|2020-02-03), that has been described previously (BoudewijnsThibaut et al., 2021), and passaged this 3 additional times on Huh 7 cells while selecting those cultures that showed most CPE. This resulted in a virus stock (passage 9) that confers full CPE on Huh 7 (5.6 × 10^4 TCID50/mL) as well as on VeroE6 cells (1.8 × 10^7 TCID50/mL). For antiviral testing Huh 7 cells were seeded in 96-well plates (Corning® CellBIND® 96-well Microplate cat no 3300) at a density of 6000 cells per well in assay medium. After overnight growth, cells were treated with the indicated compound concentrations and infected with a MOI of 0.005 TCID50/cell (final volume 200 μL/well in assay medium). On day 4 p.i. differences in cell viability caused by virus-induced CPE or by compound-specific side effects were analyzed using MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt). For this an MTS/Phenazine methosulphate (PMS) stock solution (2 mg/mL MTS (Promega, Leiden, The Netherlands) and 46 μg/mL PMS (Sigma–Aldrich, Bornem, Belgium) in PBS at pH 6–6.5) was diluted 1/20 in MEM without phenol red (Gibco cat no 51200038). Medium was aspirated from wells of the test plates and 70 μL of MTS/PMS solution was added. After 0.5–1 h incubation at 37 °C absorbance was measured at 498 nm. Cytotoxic effects caused by compound treatment alone were monitored in parallel plates containing mock-infected cells. All virus-related work was conducted in the high-containment BSL3+ facilities of the KU Leuven Rega Institute (3CAPS) under licenses AMV 30112018 SBB 219 2018 0892 and AMV 23102017 SBB 219 2017 0589 according to institutional guidelines.
3. Results
3.1. Assay development
We first assessed whether the nsp14 N7-MTase could methylate small dinucleotide substrate Gp3A. To that end, Gp3A methylation in the presence of nsp14 was monitored via RP HPLC and mass spectrometry, which confirmed the formation of N7-methylated product m7Gp3A and SAH as a byproduct (Fig. S1). We then determined the steady-state parameters of SARS-CoV-2 nsp14-catalyzed Py-FLINT probe (Fig. 1) N7-methylation based on fluorescence changes. Since nsp14 does not require the nsp10 subunit for its N7-MTase activity (Bouvet et al., 2010; Devkota et al., 2021) all experiments were carried out with nsp14 alone. Different concentrations of the Py-FLINT probe (0–15 μM) were incubated with the SAM cosubstrate (20 μM) as well as nsp14 (40 nM) at 30 °C, and fluorescence intensities were measured over time with 1 min interval. We did not include SAH-degrading enzymes in the assay, since product inhibition is negligible in the initial phase of the reaction. The initial reaction rates were plotted as a function of Py-FLINT probe concentration (Fig. S2A). To calculate N7-methylation kinetic parameters (K M, V max, and k cat), Michaelis-Menten modeling was applied (Table S1). Based on the determined K M value (3.5 ± 1.1 μM), the optimal probe concentration was set at 1 μM (approximate to K M). The catalytic efficiency (k cat/K M) (Eisenthal et al., 2007) of nsp14-catalyzed N7-methylation was 0.025 s−1μM−1, which was almost two-fold higher than previously calculated for the RNMT-RAM enzyme (0.014 s−1μM−1) (Kasprzyk et al., 2020), indicating that the Py-FLINT probe is a slightly better substrate for SARS-CoV-2 than human N7-MTase.
To determine the optimal concentration of the SAM cosubstrate, we incubated the Py-FLINT probe (1 μM) with different SAM concentrations (0–200 μM) and nsp14 (40 nM) at 30 °C. The initial reaction rates were plotted as a function of SAM concentration, and the Michaelis-Menten equation was fitted to calculate K M value (5.2 ± 1.0 μM; Fig. S2B). The optimal SAM concentration was set to 20 μM based on the plateau position. Finally, the optimal conditions of nsp14 N7-MTase activity monitoring were established as follows: 1 μM Py-FLINT probe, 20 μM SAM, and 40 nM nsp14 (Table S2).
To validate the method for HTS experiments, we determined z-factor value by measuring fluorescence intensity changes of the positive and negative control samples (Fig. S3). The positive control samples (no inhibition) were prepared as a mixture of the Py-FLINT probe, SAM, and nsp14, while the negative control samples additionally contained 40 μM of sinefungin (universal MTase inhibitor). The calculated z-factor value of 0.79 implied that the assay met the requirement for HTS (z factor > 0.5) and could therefore be applied for compound library screening.
3.2. Screening experiments
We employed the optimized Py-FLINT method for the HTS screening of commercially available small-molecule compound libraries, Library of Pharmacologically Active Compounds (LOPAC®1280) and FDA-approved Drug Library, against nsp14. Both libraries have been well characterized in the literature, providing a wide range of different structures with inhibitory properties against various molecular targets. We screened two additional specialized libraries offered by OTAVA Chemicals, namely the Machine Learning SARS Targeted Library with compounds of predicted anti-SARS activity and the SARS-CoV-2 nsp16 Targeted Library, dedicated for studies on the SARS-CoV-2 methyltransferase nsp16 enzyme (2′-O-MTase). Throughout the course of the screening, we observed a repeating flavonoid backbone pattern present within inhibitor structures. Hence, we decided to examine part of the Flavonoids Compound library from ChemFaces. Altogether, we screened 7039 compounds against nsp14 N7-MTase activity.
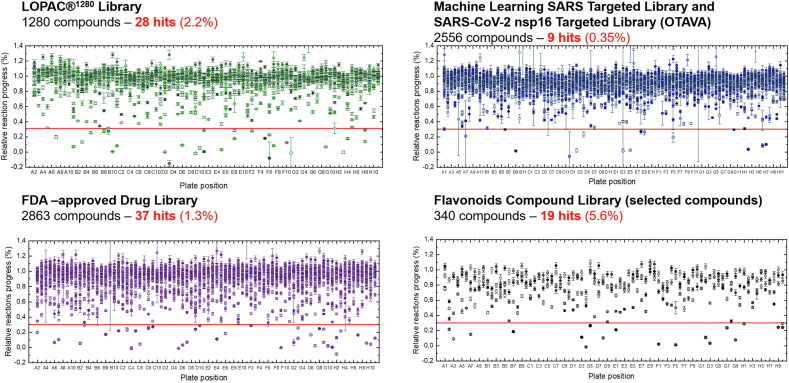
The screening experiments were performed under optimized conditions (1 μM Py-FLINT probe, 20 μM SAM, 40 nM nsp14, Tris-HCl pH 7.5 buffer, 30 °C) with 50 μM of library compound in a 96-well plate format. Relative reaction progress values were calculated as the ratio of the reaction initial rate in the presence of library compounds to the maximum initial rate without inhibitor. Among the tested compounds, 177 interfered with pyrene emission, including 36 from LOPAC®1280 (2.8%), 63 from OTAVA (2.5%), and 78 from FDA (2.7%). These were successfully characterized on separate plates. The cutoff for candidate selection was set at 30% of the relative reaction progress, leading to the.
Identification of 93 potential hits, including 28 from the LOPAC®1280 library, 9 from OTAVA libraries, 37 from FDA-approved Drug Library, and 19 out of 340 selected compounds from the Flavonoid Library (Fig. 2 ). When comparing the number of hits to the total number of compounds included in each library, the largest contribution was observed for the Flavonoid Library (5.6%) and the lowest for the two combined libraries from OTAVA Chemicals (0.35%; Fig. 2). The inhibitory properties of hit compound were subsequently explored.
Fig. 2.
Screening results. Relative reaction progress of nsp14–catalyzed N7-methylation of the Py-FLINT (Gp3A-3′-O-Py3) probe was determined in the presence of compounds from the LOPAC®1280, Machine Learning SARS Targeted (OTAVA), SARS-CoV-2 nsp16 Targeted (OTAVA), FDA-approved Drug, and Flavonoids Compound libraries. Compounds inhibiting the reaction progress by 70% or more were identified as potential hits. Data are presented as the mean ± SD of two independent experiments.
3.3. Py-FLINT inhibition assay
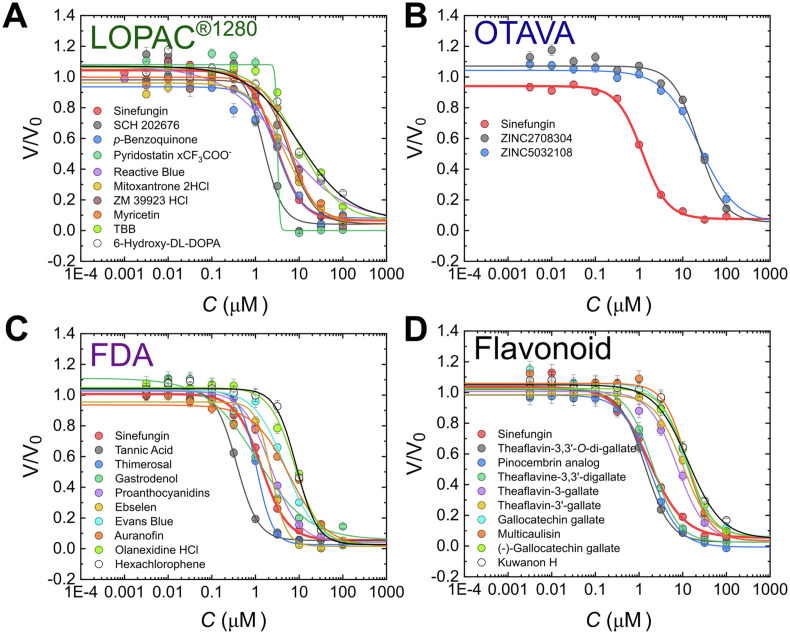
All hits were further evaluated to determine their IC50 values. To this end, Py-FLINT probe (1 μM), SAM cosubstrate (20 μM), and nsp14 (40 nM) were incubated with half-log dilutions of the hit compounds. The reaction progress was monitored based on fluorescence intensity read-outs at 378 nm (excitation at 345 nm) with 1-min time intervals. The initial rates (V) were calculated and then plotted as a function of the inhibitor concentration (C). To determine the IC50, a four-parameter dose-response curve (Hill equation with free coefficient p) was fitted to the obtained V(C) dependencies (Fig. 3 , S4–7, Table 1 ).
Fig. 3.
Inhibition curves for the most potent inhibitors of SARS-CoV-2 N7-MTase nsp14 selected from the A) LOPAC®1280 library; B) Machine Learning SARS Targeted Library and SARS-CoV-2 nsp16 Targeted Library (OTAVA Chemicals); C) FDA-approved Drug Library; and the D) Flavonoids Compound Library (selected compounds).
Table 1.
Biochemical characterization of the inhibitors identified in screening experiments.
| Compound[a] | IC50 (μM) Py-FLINT (nsp14) |
IC50 (μM) Py-FLINT (RNMT-RAM) |
EC50 (μM) Antiviral assay |
CC50 (μM) Toxicity assay |
CC50/EC50 Selectivity index |
|---|---|---|---|---|---|
| Tannic acid (FI1) | 0.37 ± 0.03 | 0.19 ± 0.04 | >100 | 68 | not active |
| Thimerosal (FI2) | 1.05 ± 0.08 | >100 | >100 | >100 | not active |
| Gastrodenol (FI3) | 1.08 ± 0.36 | 21.1 ± 2.1 | >100 | >100 | not active |
| Theaflavin-3,3′-di-O-gallate (fI1) | 1.36 ± 0.08 | 1.07 ± 0.17 | >100 | 34.8 ± 1.0 | not active |
| SCH 202676 HBr (LI1) | 1.50 ± 0.20 | 16.0 ± 3.3 | >100 | 14.0 ± 1.3 | not active |
| Sinefungin | 1.79 ± 0.07 | 0.22 ± 0.02 | >100 | >100 | not active |
| Pinocembrin 7-O-(3″-gallolyl-4″,6"-(S)-hexahydroxydiphenoyl)-beta-D-glucose (fI2) | 1.80 ± 0.22 | 1.73 ± 0.20 | >100 | n.d. | not active |
| Theaflavine-3,3′-digallate (fI3) | 2.06 ± 0.11 | 1.33 ± 0.47 | >100 | n.d. | not active |
| Proanthocyanidins (FI5) | 2.22 ± 0.15 | 1.00 ± 0.10 | >100 | 74.2 | not active |
| Ebselen (FI6) | 2.29 ± 0.20 | 23.8 ± 1.3 | >100 | n.d | not active |
| p-Benzoquinone (LI2) | 3.16 ± 0.72 | 30.6 ± 4.3 | >100 | 36.2 ± 0.7 | not active |
| Pyridostatin/CF3COO− (LI3) | 3.19 | 3.10 | 3.58 ± 0.16 | 59.3 ± 21.6 | 16.6 ± 6.1 |
| Reactive Blue 2 (LI4) | 4.12 ± 0.74 | 1.50 ± 0.28 | 16.3 ± 0.3 | 52.6 ± 10.2 | 3.23 ± 0.62 |
| Evans Blue (FI7) | 4.84 ± 0.32 | 0.33 ± 0.05 | 31.0 ± 0.9 | 35.6 ± 1.0 | 1.15 ± 0.04 |
| Mitoxantrone 2HCl (LI5) | 4.90 ± 0.42 | 3.92 ± 0.21 | >100 | <0.04 | not active |
| ZM 39923 HCl (LI7) | 6.0 ± 1.1 | 15.9 ± 3.3 | >100 | 12.2 ± 0.6 | not active |
| Myricetin (LI8) | 6.17 ± 0.54 | 3.57 ± 0.36 | >100 | 41.6 ± 7.1 | not active |
| Auranofin (FI9) | 5.99 ± 1.37 | >100 | >100 | n.d | not active |
| TBB (LI9) | 9.9 ± 2.0 | 38.1 ± 0.36 | >100 | 30.9 ± 1.6 | not active |
| Theaflavin-3-gallate (fI5) | 7.37 ± 0.57 | 2.66 ± 0.64 | >100 | n.d | not active |
| Olanexidine HCl (FI10) | 7.97 ± 0.50 | 26.5 ± 4.9 | >100 | n.d | not active |
| Hexachlorophene (FI11) | 8.65 ± 0.30 | 8.0 ± 3.2 | >100 | n.d | not active |
| Theaflavin-3′-gallate (fII1) | 10.00 ± 0.67 | 4.3 ± 1.8 | >100 | n.d | not active |
| 6-Hydroxy-DL-DOPA (LI10) | 10.0 ± 2.2 | 3.17 ± 0.44 | >100 | 30.0 ± 1.8 | not active |
| Gallocatechin gallate (fII2) | 11.2 ± 1.1 | 2.88 ± 0.92 | >100 | n.d | not active |
| Multicaulisin (fII3) | 12.45 ± 0.63 | 21.8 ± 1.9 | >100 | n.d | not active |
| (−)-Gallocatechin gallate (fII4) | 12.77 ± 0.56 | 2.74 ± 0.54 | >100 | 12 ± 12 | not active |
| Kuwanon H (fII5) | 13.3 ± 1.6 | 31.89 ± 0.32 | >100 | 18.1 ± 3.8 | not active |
| Cyanidin-3-O-glucoside chloride (fII6) | 15.3 ± 1.8 | 5.25 ± 0.58 | >100 | 59 ± 24 | not active |
| (−)-Catechin gallate (fII7) | 17.3 ± 1.4 | 1.87 ± 0.66 | >100 | 33.8 ± 7.9 | not active |
| (−)-Epigallocatechin gallate (fII8) | 17.44 ± 0.81 | 3.61 ± 0.70 | >100 | 23.2 ± 7.5 | not active |
| ZINC2708304 (OI1) | 22.8 ± 2.6 | >100 | >100 | >100 | not active |
| ZINC5032108 (OI2) | 24.3 ± 1.7 | >100 | >100 | 71.3 ± 5.9 | not active |
[a] Color: green – compounds from the LOPAC®1280 Library; blue – compounds from the OTAVA libraries; purple – compounds from the FDA-approved Drug Library; grey – compounds from the Flavonoids Compound Library; yellow – reference.
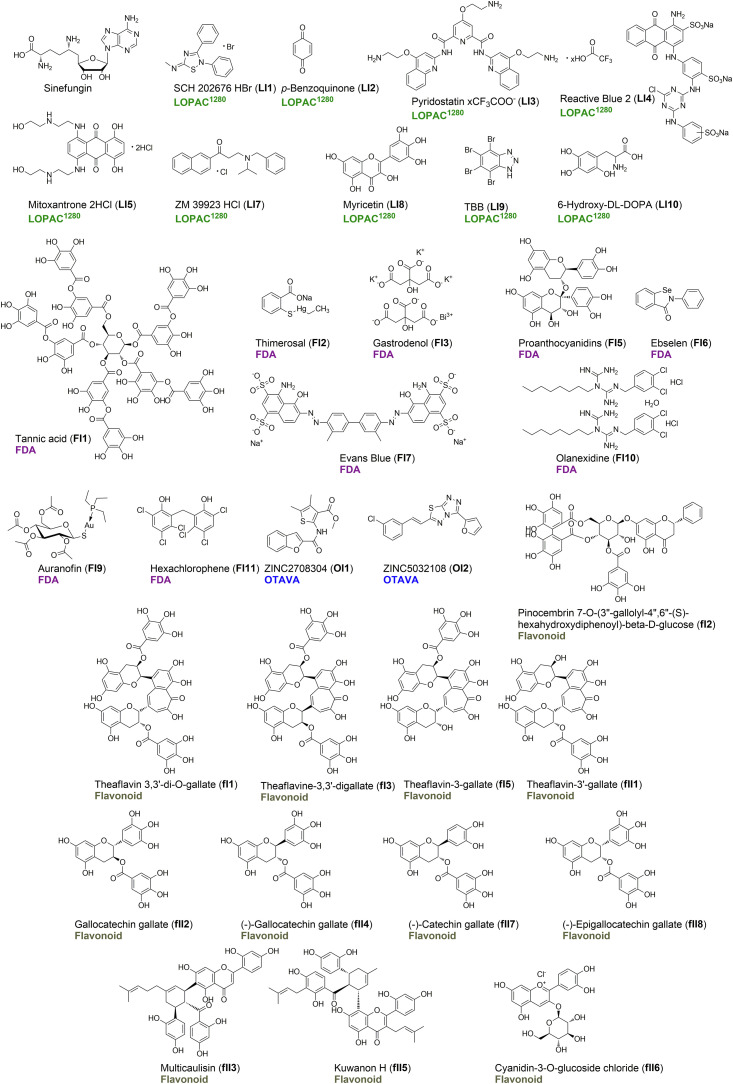
Based on the determined IC50 values for LOPAC®1280 hits, we divided the compounds into three sets. The first set (LI; Fig. 3, Fig. 4 , Table 1) contained the ten most potent inhibitors (IC50 < 10 μM), including SCH 202676 hydrobromide (LI1) with an IC50 of 1.50 ± 0.20 μM as the most potent compound, similar to sinefungin (IC50 1.79 ± 0.07 μM). The first set also included p-benzoquinone, (LI2), pyridostatin (LI3), Reactive Blue 2 (LI4), mitoxantrone (LI5), ruthenium red (LI6), ZM 39923 (LI7), myricetin (LI8), TBB (LI9), and 6-hydroxy-DL-DOPA (LI10). The second set (LII) consisted of 10 compounds with 10 μM < IC50 < 20 μM, and the third LIII included 8 compounds with IC50 > 20 μM (Fig. S4, Table S3). For pyridostatin (set I), we observed a very high Hill slope value p (18.1), which may suggests positive cooperativity in mechanism of action (Barcroft and Hill, 1910).
Fig. 4.
Structures of the 32 most potent inhibitors identified from the screening, including 9 from the LOPAC®1280 library, 2 from OTAVA library, 9 from the FDA-approved Drug Library, and 12 from the Flavonoids Compound Library. The IC50 values determined for these compounds are shown in Table 1.
The Machine Learning SARS Targeted and SARS-CoV-2 nsp16 Targeted libraries from OTAVA Chemicals were analyzed together and labeled as OTAVA. IC50 values were determined for 31 hits from this library. Among these, only 9 hits met the initially set criterion for inhibition level of at least 70%, and the other 22 yielded at least 50% inhibition (Fig. S5, Table S4). Overall, the identified inhibitors (most of them having ZINC and MCULE database structures) were weaker inhibitors compared to those from other libraries. ZINC2708304 (OI1), the most potent inhibitor, had an IC50 of 22.8 ± 2.6 μM (Fig. 3, Fig. 4, Table 1), one order of magnitude higher than that of sinefungin. Other identified compounds included ZINC5032108 (OI2) and ZINC118535 (OI3), which is the analog of ZINC2708304 (Fig. S5, Table S4). Despite their relatively high IC50 values, the small molecule character of OTAVA compounds facilitated SAR analysis for the identification of structural fragments of promise for nsp14-targeted drug design.
The third compound series that we characterized included 37 hits from the FDA-approved Drug Library. Based on the determined IC50 values, we divided compounds into three sets. The ten most potent inhibitors (FI set) had IC50 values lower than 10 μM and included i.a. tannic acid (FI1), thimerosal (FI2), and gastrodenol (FI3), proanthocyanidins (FI5), ebselen (FI6), Evans blue (FI7), auranofin (FI9), olanexidine HCl (FI10), and hexachlorophene (FI11 Fig. 3, Fig. 4, Table 1), which The first three compounds were even more potent than the sinefungin reference (Table 1). The second compound set (FII) included inhibitors with 10 μM < IC50 < 30 μM (10 compounds), while the third one (FIII) those with IC50 > 30 μM (16 compounds, Fig. S6, Table S5). Some of the hits, such as p-benzoquinone, mitoxantrone, morin, quercetin, auranofin, sanguinarine chloride, and candesartan cilexetil from LOPAC®1280 were also present in the FDA library, further confirming their inhibitory properties determined via Py-FLINT assays.
Among the identified nsp14 inhibitors, we found several compounds of plant origin, including flavonoid analogs, such as myricetin (LI8), morin (LII3), quercetin (LII4), luteolin (FIII13), proanthocyanidins (FI5), and proflavine (FIII3; Tables S3 and S5). Hence, we decided to test part of the Flavonoids Compound Library and identified 19 more hits for which the IC50 values were also determined (Fig. S7, Table S6). The identified inhibitors were structurally related, mostly including theaflavin or catechin analogs in the form of gallic acid esters (Fig. 4), and differed in the number of gallate groups and/or bond stereochemistry. Theaflavin-3,3′-di-O-gallate (fI1) with an IC50 of 1.36 ± 0.08 μM exhibited stronger inhibitory properties than the sinefungin reference. Other compounds with IC50 < 10 μM included a pinocembrin analog (fI2; pinocembrin 7-O-(3″-gallolyl-4″,6"-(S)-hexahydroxydiphenoyl)-beta-D-glucose), multicaulisin (fII3), kuwanon H (fII5), and cyanidin-3-O-glucoside chloride (fII6).
3.4. RP HPLC inhibition studies
In order to independently confirm whether the hits identified and characterized via Py-FLINT assay could inhibit nsp14 N7-MTase activity, we used RP HPLC to analyze the N7-methylation reaction of dinucleotide substrate Gp3A in the presence of the selected inhibitors. To that end, 20 μM Gp3A was incubated with 300 μM SAM, 500 nM nsp14, and 500 μM of the tested inhibitor for 3 h at 30 °C. The mixtures were analyzed via RP HPLC with UV detection at 254 nm (Fig. S8). The percentage of formed m7Gp3A was calculated as a ratio of the area under the m7Gp3A signal to the total area of m7Gp3A and Gp3A signals. Indeed, all of the tested compounds exhibited inhibitory activity against nsp14, as indicated by the decrease of m7Gp3A when compared to the reference reaction without inhibitor (Figs. S8A–C). However, some compounds (pyridostatin LI3, mitoxantrone LI5, and ruthenium red LI6) exhibited lower potency in the RP HPLC analysis when considering their IC50 value (Fig. S8D). Moreover, we observed that pyridostatin significantly affected the chromatographic properties of both Gp3A and m7Gp3A (Fig. S8A). We speculate that these compounds may interact with nucleotide substrates, influencing the IC50 values determined via Py-FLINT assays.
3.5. IVT RNA inhibition assay
To additionally confirm nsp14 N7-MTase inhibition, we carried out the reaction on longer (35 nt) in vitro transcribed RNA substrates capped with the Gp3ApG trinucleotide, which ensures high RNA capping efficiency (Sikorski et al., 2020). To analyze the reaction progress, we employed dot blot assays with product detection using the m7G-cap-specific antibody. To calculate the amount of N7-methylated product, a calibration curve was prepared using mixtures of Gp3ApG-RNA35 and m7Gp3ApG-RNA35 at different ratios. Unfortunately, many of the inhibitors, including SCH 202676, pyridostatin, tannic acid, or ebselen, interfered with RNA blotting and detection (Fig. S9). Hence, the IC50 values were determined only for selected non-interfering compounds sinefungin and p-benzoquinone (Fig. S10). We confirmed specific N7-methylation by nsp14 and observed that p-benzoquinone (IC50 1.93 ± 0.42 μM) was an even more potent inhibitor than sinefungin (21.0 ± 4.6 μM).
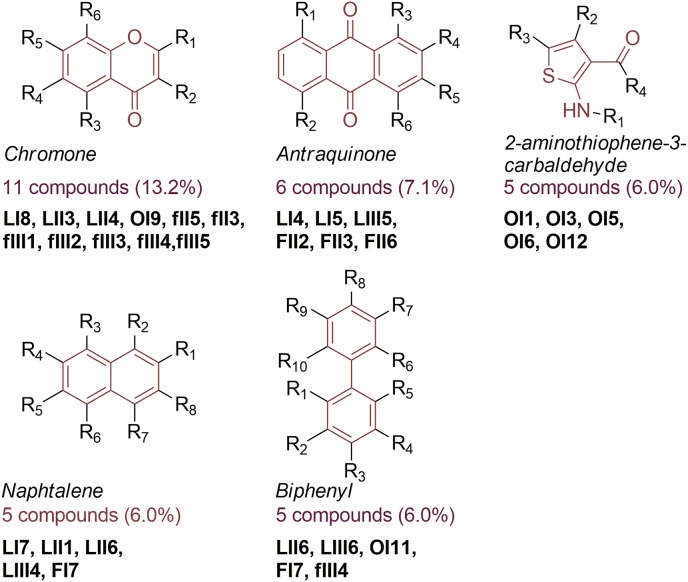
3.6. SAR analysis
We observed several frequently repeating structural patterns among the identified nsp14 inhibitors. In order to systematically review the structures and identify scaffolds potentially related to compound bioactivity, we used the SARvision|SM (Small Molecules) software (by Altoris, Inc., San Diego, CA). We selected nsp14 inhibitors with IC50 < 50 μM for the analysis, obtaining a total of 83 different structures. The most common pattern was chromone, found in 11 compounds, constituting 13% of the nsp14 inhibitors with an IC50 lower than 50 μM (Fig. 5 ). Usually the Chromone was present in flavonoid analogs, such as myricetin, morin, or multicaulisin. The other identified scaffolds are presented in Fig. 5 and Fig. S11. Nsp14 inhibitors found in the OTAVA library were weaker compared to inhibitors from other libraries, however their fragment-based drug character is of interest for SAR analysis. Hence, we separately analyzed scaffolds identified in the structures of OTAVA inhibitors (Fig. S12).
Fig. 5.
The most common scaffolds identified among compounds with inhibitory activity against SARS-CoV-2 nsp14 N7-MTase based on Py-FLINT assay results.
3.7. RNMT-RAM inhibition
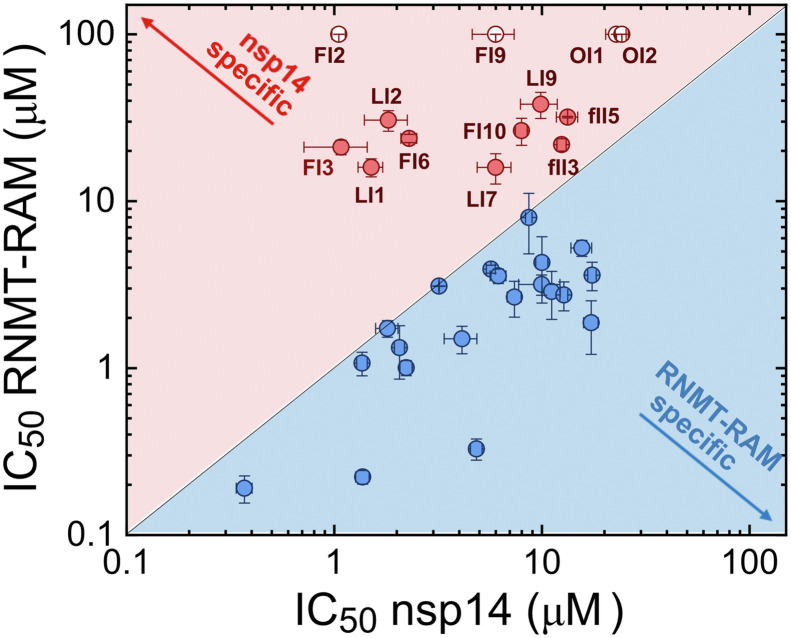
Finally, the selectivity of the 32 most potent inhibitors from all libraries was tested in inhibition experiments with human N7-MTase RNMT-RAM using the Py-FLINT assay under the previously optimized conditions (2 μM Py-FLINT probe, 20 μM SAM, and 20 nM RNMT-RAM) (Kasprzyk et al., 2020). For all compounds, the IC50 values were calculated and compared to the values determined for the viral nsp14 enzyme (Table 1; Fig. S13). The correlation between determined IC50 values is illustrated in Fig. 6 . Compounds with an IC50 for nsp14 lower than that for RNMT-RAM were considered more selective towards the viral enzyme and were marked in red. Four of the tested compounds, namely thimerosal, (FI2), auranofin (FI9), ZINC2708304 (OI1), and ZINC5032108 (OI2), exhibited no human N7-MTase inhibition under experimental conditions. Additionally, 9 compounds had a lower IC50 towards nsp14 than against RNMT-RAM.
Fig. 6.
Correlation of IC50 values determined for nsp14 and human N7-MTase RNMT-RAM using the Py-FLINT assay. Inhibitors more specific towards RNMT-RAM or nsp14 were marked in blue and red, respectively. Open circle data points indicate that the IC50 value was higher than 100 μM.
3.8. Mechanism of action (MOA)
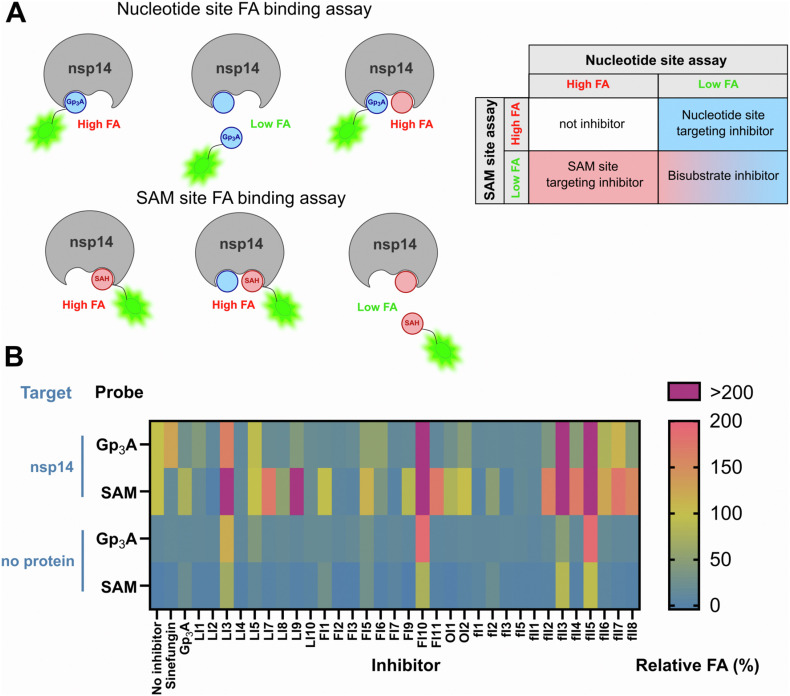
All validated compounds inhibited the N7-MTase activity of nsp14. However, none of the methods we used (Py-FLINT assay, RP HPLC, dot blot) revealed the area of inhibitor interaction within the nsp14 active site. The active site of RNA N7-methyltransferases includes two regions – one that binds nucleotide substrate subjected to N7-methylation and another that binds the cofactor (methyl donor) SAM. Both substrates are located close to each other with the cap N7-nucleophile oriented towards the S–CH3 bond in SAM (Fabrega et al., 2004). The lack of knowledge on the targeted region within nsp14 hinders the application of in silico methods, including docking studies. Moreover, the lack of information on the type of ligand-protein interactions complicates the rational design of more potent inhibitor structures. To address this, we developed two nsp14-fluorescence anisotropy (FA)-based competition binding assays using either Gp3A- or SAH-derived probes labeled with fluorescein. The structural information available for the SARS-CoV nsp14 complex with Gp3A and SAH (PDB code: 5C8S) guided the selection of optimal positions for probe labeling. To target the nucleotide-binding site, we used the previously synthesized Gp3A labeled with (5)-carboxyfluorescein via the 3′-O- position of adenosine (Gp3A-3′-O-(5)FAM) (Kasprzyk et al., 2020). The second probe, designed for the SAM-binding site, was synthesized in four steps, starting from 6-chloropurine riboside (Fig. S14; SAH-N6-6tFluo).
We then confirmed that the fluorescence anisotropy (FA) of the probe-nsp14 complexes is higher than that of free probes (Fig. S15). Using Gp3A and sinefungin, ligands targeting nucleotide- and SAM-binding sites, respectively, we validated the assay's functionality (Fig. 7 , columns 1–3). We used bovine serum albumin (BSA), a non-interacting protein, as a negative control. Both probes were also characterized for their binding affinity in FA-monitored saturation-binding experiments (Fig. S15B). The changes in FA were plotted as a function of nsp14 concentration, and saturation binding curves were fitted to determine the dissociation constant K D (Fig. S15C). The calculated K D values were 1.66 ± 0.31 μM for Gp3A-3′-O-(5)FAM and 3.16 ± 0.54 μM for SAH-N6-6tFluo.
Fig. 7.
A) The idea of two binding assays based on fluorescein-labeled Gp3A or SAH analog targeting two substrate binding sites of nsp14; B) Determination of inhibitor binding sites in nsp14 via FA-based competition assays. The upper two rows represent relative FA values for samples containing the indicated probe (targeting Gp3A- or SAM-binding site), nsp14, and inhibitor; bottom rows represent values for the corresponding samples without protein. For samples containing nsp14, an FA value lower than 100% indicated that the fluorescent probe was partially displaced from the binding site by the inhibitor. Data are presented as the mean values of two independent experiments. Numerical values are summarized in Table S7. Extended data including control samples with BSA are shown in Fig. S16.
To verify which binding site is targeted by nsp14 inhibitors identified in Py-FLINT assays, we carried out two independent competitive binding experiments. The fluorescent probe (Gp3A-3′-O-(5)FAM or SAH-N6-6tFluo) at 10 nM were incubated with 2 μM nsp14 and 100 μM of inhibitor, followed by FA measurement (Fig. S15A). To examine whether the inhibitor may non-specifically interact with the protein or substrate, we additionally ran control experiments monitoring the inhibitor-induced FA changes of both probes in the presence of BSA or in the absence of any protein. Relative FA values were calculated in reference to the FA of the probe/nsp14 complex (100%) and are presented in the form of a heatmap (Fig. 7; S16). FA values lower than 100% suggested that the fluorescent probe was partially displaced from the binding site by the inhibitor, whereas the lack of FA change indicated that the particular site is not targeted. Interestingly, some inhibitors significantly increased the FA value relative to the control sample, which may indicate that these target the protein at a different binding site (specifically or non-specifically), inducing conformational rigidity or targeting the probe itself. Most of the analyzed compounds behaved as either bisubstrate- or nucleotide binding site-targeting inhibitors. None of the.
Tested compounds targeted the SAM-binding site alone. Of the 32 nsp14 inhibitors, 12 exhibited low FA in the Gp3A site binding assay and a high FA in the SAM site binding assay, meaning that they were bound by nsp14 within the nucleotide binding pocket. These included ZM 39923 (LI7), TBB (LI9), tannic acid (FI1), auranofin (FI9), hexachlorophene (FI11), ZINC2708304 (OI1), ZINC5032108 (OI2), as well as flavonoids proanthocyanidins (FI5), gallocatechin gallate (fII2), (−)-gallocatechin gallate (fII4), cyanidin-3-O-glucoside chloride (fII6), and (−)-epigallocatechin gallate (fII8). Interestingly, catechin gallate analogs targeted the nucleotide-binding site, but additional expansion of the structure as in fI1, fI3–fII1 led to bisubstrate inhibition. Some compounds, including pyridostatin (LI3), olanexidine (FI10), multicaulisin (fII3), kuwanon H (fII5) and, to a lesser extent, mitoxantrone (LI5), displayed high FA values in both assays as well as in the control sample. Compounds with alkyl chains in their structures (olanexidine, multicausilin, and kuwanon H) interacted non-specifically with BSA and/or probe (Fig. S16). Pyridostatin and mitoxantrone contain flexible alkyl chains with amine groups, hence, they may electrostatically interact with the negatively charged probe. Such interactions may explain the relatively high FA values in the absence of protein.
3.9. Viral replication assay
The most potent nsp14 N7-MTase inhibitors identified from all tested libraries were further examined for their ability to inhibit SARS–CoV-2 replication in Huh 7 (human hepatocellular carcinoma) cells. To this end, Huh 7 cells were seeded in 96-well plates and treated with indicated compound concentrations, followed by infection after overnight incubation. A colorimetric MTS based staining for cell viability was performed at 4 days post infection to determine inhibition curves. Cytotoxic effects of the compounds were monitored in parallel plates containing mock-infected cells.
Three out of the 33 tested inhibitors induced suppression of viral replication, including pyridostatin (3.58 ± 0.16 μM), Reactive Blue 2 (16.3 ± 0.3 μM), and Evans Blue (31.0 ± 0.9 μM). To compare the potential efficacy and safety of the compounds we calculated the ratio of CC50/EC50, known as the selectivity index (SI). We found that pyridostatin, with an SI of 16.6 ± 6.1 and the lowest EC50 value, exhibited the greatest and selective antiviral activity. Interestingly the IC50's of this compound obtained in the biochemical assays correlate very well with the EC50 in the antiviral assay. The second most selective compound was Reactive Blue 2, with an SI of 3.23 ± 0.62. Evans Blue presented CC50 value similar to its EC50 (35.6 ± 1.0 μM), which is visible in its low SI value of 1.15 ± 0.04.
4. Discussion
Herein, we sought to apply our previously developed N7-MTase Py-FLINT assay (Kasprzyk et al., 2020) for the identification of inhibitors against the nsp14 N7-methyltansferase, which is involved in mRNA capping of SARS-CoV-2. We first confirmed that our assay could be used for monitoring nsp14 activity in real time. Thereafter, we optimized experimental conditions (probe, cosubstrate, and protein concentrations) and used the assay to screen five commercially available compound libraries. Setting the relative reaction progress cut-off at 30%, we identified 93 hits (among the 7039 compounds tested) with inhibitory properties against nsp14. Further evaluation of these compounds revealed tannic acid (FI1), thimerosal (FI2), gastrodenol (FI3), theaflavin-3,3′-O-digallate (fI1), and SCH 202676 hydrobromide (LI1) as the most potent nsp14 inhibitors with IC50 values from 4.8 to 1.2 times lower than that of sinefungin, a well-known methyltransferase inhibitor (Hausmann et al., 2005). We previously identified some of the inhibitors from the LOPAC®1280 library, including myricetin and Reactive Blue 2, as compounds targeting Ecm1 N7-MTase from the E. cuniculi parasite (Kasprzyk et al., 2020). Overall, the hit compounds from the two OTAVA libraries were weaker nsp14 inhibitors. The two most potent ones with IC50 ~20 μM were selected for further studies together with inhibitors from the other libraries.
The hits found via Py-FLINT assays were validated through HPLC with Gp3A as a substrate. All of the identified nsp14 compounds were confirmed as inhibitors of nsp14 activity. However, the RP HPLC analysis revealed pyridostatin (LI3), mitoxantone (LI5) and ruthenium red (LI6) as weaker inhibitors of nsp14 than indicated by Py-FLINT assay results. We speculate that ruthenium red, a complex of ruthenium ion and ammonia molecules, pyridostatin, and mitoxantrone, which has positively charged amine groups located at alkyne chains, electrostatically interacted with the Py-FLINT probe, influencing fluorescence intensity. Ruthenium red targets various proteins, which makes it non-selective (Vincent and Duncton, 2011) and was thus excluded from further studies.
For further hit validation, we used 35 nt RNA capped with Gp3ApG trinucleotide as a substrate. m7Gp3ApG-RNA35, the product of N7-methylation by nsp14, was detected using an m7G-cap-specific antibody via dot blot assay. Unfortunately, most inhibitors showed interferences with the antibody, in which case the reaction product could not be reliably quantified. Therefore, the IC50 values were determined for two selected compounds, namely sinefungin (reference) and p-benzoquinone, confirming their inhibitory properties against nsp14.
To identify the structural patterns that could enhance the potency of inhibition against nsp14 N7-MTase, we performed SAR analysis of all 83 inhibitors with IC50 < 50 μM using SARvision|SM software by Altoris Inc. As a result, we identified chromone in the structures of 11 nsp14 inhibitors. Chromone is found in various compounds of plant origin, such as alkaloids and flavonoids, which often possess anti-inflammatory and antiviral properties (AslamS.Henidi, 2021). Analogs of chromone have already been identified as inhibitors of the SARS-CoV-2 3C-like protease (3CLpro) (Jo et al., 2020). Herein, we demonstrated their ability to target another essential SARS-CoV-2 protein – the nsp14 N7-MTase. The flavonoid baicalein was previously identified as an nsp14 N7-MTase inhibitor via an in silico approach (Liu et al., 2020). However, we did not observe such activity in our Py-FLINT assays during Flavonoids compound library screening. An antraquinone motif was identified within 6 nsp14 inhibitors, including anthracyclines (i.e. doxorubicin), known for their anticancer activity. Three other scaffolds present in at least 5 compounds identified via SAR analysis were 2-aminothiophene-3-carbaldehyde, naphthalene, and biphenyl. Although compounds selected from OTAVA libraries were relatively weak nsp14 inhibitors (IC50 > 20 μM), we found them of interest for SAR analysis. Scaffold identification for these compounds revealed frequently repeating heterocycle fragments containing sulfur atoms, such as thiophene, thiazole, or thiadiazole. All of the identified scaffolds could potentially favor SARS-CoV-2 ns14 N7-MTase inhibition.
As a next step of inhibitor evaluation, we tested the selectivity of identified compounds towards human RNA N7-MTase (RNMT) in complex with RNMT-activating miniprotein (RAM) using Py-FLINT assay (Kasprzyk et al., 2020). Among the 33 tested nsp14 inhibitors, we identified 13 with IC50 values for nsp14 lower than for human N7-MTase, including thimerosal (FI2), auranofin (FI9), ZINC2708304 (OI1), and ZINC5032108 (OI2), which displayed no activity towards RNMT-RAM within the used concentration range (IC50 > 100 μM). Moreover, four compounds, namely SCH 202676 (LI1), p-benozoquinone (LI2), gastrodenol (FI3), and ebselen (FI6), had nsp14 IC50 values one order of magnitude lower compared to those against RNMT-RAM. Such a difference in inhibition between viral and host (human) protein suggested possible selectivity, allowing for lower cytotoxicity.
Enzymatic N7-methyltation is a two-substrate reaction based on nucleophilic substitution mechanism with SAM acting as a methyl group donor (cofactor) and the N7-guanine position in nucleotide as a methyl acceptor. The enzyme active site consists of two binding sites – one responsible for nucleotide/RNA binding and the other for SAM binding (Ma et al., 2015). Small-molecule inhibitors identified in the screen could interact with one of these sites, both, or neither (allosteric or unspecific inhibitors). To clarify the MOA of nsp14 inhibitors, we developed two fluorescence anisotropy (FA) binding assays aimed at nucleotide (Gp3A)- or SAM-binding sites, respectively. The 32 most potent inhibitors were examined in both FA assays. As a result, we defined whether the specific compound targets the nucleotide-binding site, SAM-binding site, is a bisubstrate inhibitor, or is another type of ligand. To verify the specificity of inhibitor-nsp14 interactions, we carried out control experiments for probe/BSA/inhibitor and probe/inhibitor mixtures. We identified 12 inhibitors that bound within the nucleotide site, while none targeted the SAM-binding region. The quality of the former may be beneficial for their selectivity as many other methyltransferases (RNA, DNA, histone, etc.) utilize SAM molecules for their catalytic activity. We observed that flavonoid-like nsp14 inhibitors with relatively small structures (e.g., proanthocyanidins) targeted the nucleotide-binding site. However, expansion of the structure (e.g., theaflavin gallates) extended the area of interactions to the SAM-binding site, making the compounds bisubstrate inhibitors. Increased FA values were observed for pyridostatin (LI3), olanexidine (FI10), multicaulisin (fII3), kuwanon H (fII5), and mitoxantrone (LI5) for both probes in the presence of nsp14, as well as in control samples containing either probe/BSA or probe alone. This result suggested interaction with the protein via a different mechanism or at a different binding site and/or the presence of non-specific interactions between inhibitors and probes. Interestingly, pyridostatin has been recently shown to stabilize G-quadruplex structures formed within the SARS-CoV-2 RNA genome, leading to a decrease of viral N protein levels (Zhao et al., 2021). Our results suggest that this compound may independently target other RNA structures as well, including 5’ caps. The remaining compounds had lower FA values in both assays, thereby act as bisubstrate inhibitors.
Finally, we characterized the antiviral properties of identified compounds in SARS-CoV-2 infected Huh 7 cells. Pyridostatin (G-quadruplex interacting molecule), Reactive Blue 2, and Evans Blue (dyes) exhibited EC50 values lower than 50 μM. The most selective compound with an EC50 of 3.58 ± 0.16 μM and an SI of 16.6 ± 6.1, was pyridostatin. This potency is in the same range as the EC50 of GS-441524 (the parent nucleoside of remdesivir) in this assay system. In binding assays, we observed that FA were higher for probe/nsp14/pyridostatin samples compared to probe/BSA/pyridostatin or probe/pyridostatin. This result suggested that pyridostatin may interact with both nucleotide probes and probe-nsp14 complexes. Hence, its antiviral mechanism may be more complex. Further studies are required to elucidate the mechanism of action. Although most of the compounds did not exhibit potent antiviral activity, which may be due insufficient cellular permeation, the current results provide a basis for the design of novel biologically active compounds targeting SARS-CoV-2 capping machinery. We hope that the current results will contribute to future rational drug design for COVID-19 treatment.
Author contributions
R.K., J.K. and J.J. designed the study; R.K. performed nucleotide synthesis, screening and inhibition experiments, binding assays development and application, and SAR analysis; T.S. carried out cloning, expression and purification of nsp14 protein; S.G. synthesized fluorescently labeled SAH analog; M.S. performed short RNA synthesis and dot-blot experiments; L.V., S.J., D.J. and J.N. carried out viral assay and cytotoxicity studies; R.K., J.K. and J.J. wrote the first draft of the manuscript. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Funding
This work was financially supported by the National Science Centre (grant number UMO-2020/01/0/ST4/00124) to J.J. and (grant number UMO-2019/32/T/ST4/00091) to R.K., and Foundation for Polish Science (grant number TEAM/2016-2/13 (POIR.04.04.00-00-20A2/16-00)) to J.J.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Jacek Jemielity.
June 25, 2021.
Acknowledgment
We thank Professor Victoria Cowling (University of Dundee) for the human N7-MTase RNMT-RAM, Professor Marcin Drag (Wroclaw University of Science and Technology) for sharing FDA-approved Drug Library, and Dr Karolina Drazkowska (University of Warsaw) for designing the cloning.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2021.105142.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abd El-Aziz T.M., Stockand J.D. Infect. Genet. Evol. 2020;83:104327. doi: 10.1016/j.meegid.2020.104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed-Belkacem R., Sutto-Ortiz P., Guiraud M., Canard B., Vasseur J.J., Decroly E., Debart F. Eur. J. Med. Chem. 2020;201:112557. doi: 10.1016/j.ejmech.2020.112557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouadi W., Eydoux C., Coutard B., Martin B., Debart F., Vasseur J.J., Contreras J.M., Morice C., Quérat G., Jung M.L., Canard B., Guillemot J.C., Decroly E. Antivir. Res. 2017;144:330–339. doi: 10.1016/j.antiviral.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam S A.M., Henidi H.A., B.-J. M. N. Zia-Ul-Haq M., Alothman S.I., Henidi H.A. Alternative Medicine Interventions for COVID-19. Springer; 2021. pp. 111–152. ch. 4. [Google Scholar]
- Barcroft J., Hill A.V. J Physiol. 1910;39:411–428. doi: 10.1113/jphysiol.1910.sp001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becares M., Pascual-Iglesias A., Nogales A., Sola I., Enjuanes L., Zuñiga S. J. Virol. 2016;90:5399–5414. doi: 10.1128/JVI.03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudewijns R., Thibaut H.J., Kaptein S.J.F., Li R., Vergote V.O.P.V., Seldeslachts L., De Keyzer C., Bervoets L., Sharma S., Van Weyenbergh J., Liesenborghs L., Ma J., Jansen S., Van Looveren D., Vercruysse T., Jochmans D., Wang X., Martens E., Roose K., De Vlieger D., Schepens B., Van Buyten T., Jacobs S., Liu Y., Martí-Carreras J., Vanmechelen B., Schramm G., Van Laere K., Saelens X., Callewaert N., Opdenakker G., Maes P., Weynand B., Cawthorne C., Velde G.V., Wang Z., Neyts J., Dallmeier K. bioRxiv. 2021 doi: 10.1101/2020.04.23.056838. [DOI] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Guo D. Virol. Sin. 2016;31:3–11. doi: 10.1007/s12250-016-3726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Decroly E., Li C., Sharff A., Lescar J., Bricogne G., Barral K. Antivir. Res. 2014;106:61–70. doi: 10.1016/j.antiviral.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H., Thiel V., Sen G.C., Fensterl V., Klimstra W.B., Pierson T.C., Buller R.M., Gale M., Shi P.Y., Diamond M.S. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Ferron F., Lescar J., Canard B. Nat. Rev. Microbiol. 2011;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delang L., Li C., Tas A., Quérat G., Albulescu I.C., De Burghgraeve T., Guerrero N.A., Gigante A., Piorkowski G., Decroly E., Jochmans D., Canard B., Snijder E.J., Pérez-Pérez M.J., van Hemert M.J., Coutard B., Leyssen P., Neyts J. Sci. Rep. 2016;6:31819. doi: 10.1038/srep31819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota K., Schapira M., Perveen S., Yazdi A.K., Li F., Chau I., Ghiabi P., Hajian T., Loppnau P., Bolotokova A., Satchell K.J.F., Wang K., Li D., Liu J., Smil D., Luo M., Jin J., Fish P.V., Brown P.J., Vedadi M. bioRxiv. 2021 [Google Scholar]
- Dong H., Zhang B., Shi P.Y. Antivir. Res. 2008;80:1–10. doi: 10.1016/j.antiviral.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Sun J., Mao Z., Wang L., Lu Y.L., Li J. J. Med. Virol. 2020 doi: 10.1002/jmv.25768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., Scherbakova S., Graham R.L., Baric R.S., Stockwell T.B., Spiro D.J., Denison M.R. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenthal R., Danson M.J., Hough D.W. Trends Biotechnol. 2007;25:247–249. doi: 10.1016/j.tibtech.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Fabrega C., Hausmann S., Shen V., Shuman S., Lima C.D. Mol Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- Ferron F., Decroly E., Selisko B., Canard B. Antivir. Res. 2012;96:21–31. doi: 10.1016/j.antiviral.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y., Shatkin A.J. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A.C., Mouncey P.R., Al-Beidh F., Rowan K.M., Nichol A.D., Arabi Y.M., Annane D., Beane A., van Bentum-Puijk W., Berry L.R., Bhimani Z., Bonten M.J.M., Bradbury C.A., Brunkhorst F.M., Buzgau A., Cheng A.C., Detry M.A., Duffy E.J., Estcourt L.J., Fitzgerald M., Goossens H., Haniffa R., Higgins A.M., Hills T.E., Horvat C.M., Lamontagne F., Lawler P.R., Leavis H.L., Linstrum K.M., Litton E., Lorenzi E., Marshall J.C., Mayr F.B., McAuley D.F., McGlothlin A., McGuinness S.P., McVerry B.J., Montgomery S.K., Morpeth S.C., Murthy S., Orr K., Parke R.L., Parker J.C., Patanwala A.E., Pettilä V., Rademaker E., Santos M.S., Saunders C.T., Seymour C.W., Shankar-Hari M., Sligl W.I., Turgeon A.F., Turner A.M., van de Veerdonk F.L., Zarychanski R., Green C., Lewis R.J., Angus D.C., McArthur C.J., Berry S., Webb S.A., Derde L.P.G., Investigators R.-C. N. Engl. J. Med. 2021 [Google Scholar]
- Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurard-Levin Z.A., Liu C., Jekle A., Jaisinghani R., Ren S., Vandyck K., Jochmans D., Leyssen P., Neyts J., Blatt L.M., Beigelman L., Symons J.A., Raboisson P., Scholle M.D., Deval J. Antivir. Res. 2020;182:104924. doi: 10.1016/j.antiviral.2020.104924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung A.B. Gene Rep. 2020;21:100860. doi: 10.1016/j.genrep.2020.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S., Zheng S., Fabrega C., Schneller S.W., Lima C.D., Shuman S. J. Biol. Chem. 2005;280:20404–20412. doi: 10.1074/jbc.M501073200. [DOI] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.S. J. Enzym. Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzyk R., Fido M., Mamot A., Wanat P., Smietanski M., Kopcial M., Cowling V.H., Kowalska J., Jemielity J. Chem. Eur J. 2020 doi: 10.1002/chem.202001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafcikova P., Silhan J., Nencka R., Boura E. Nat. Commun. 2020;11:3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Lim S.P., Noble C.G., Shi P.Y. Antivir. Res. 2015;119:57–67. doi: 10.1016/j.antiviral.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Lin S., Chen H., Chen Z., Yang F., Ye F., Zheng Y., Yang J., Lin X., Sun H., Wang L., Wen A., Dong H., Xiao Q., Deng D., Cao Y., Lu G. Nucleic Acids Res. 2021;49:5382–5392. doi: 10.1093/nar/gkab320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhu X., Lu Y., Zhang X., Jia X., Yang T. J Pharm Anal. 2020 doi: 10.1016/j.jpha.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W.R., Cheng F. J. Proteome Res. 2020;19:4670–4677. doi: 10.1021/acs.jproteome.0c00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino M., Ito N., Sugiyama M., Ogino T. Viruses. 2016;8 doi: 10.3390/v8050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otava T., Šála M., Li F., Fanfrlík J., Devkota K., Pakarian P. ChemRxiv. Preprint. 2021 doi: 10.26434/chemrxiv.14075408.v1. [DOI] [PubMed] [Google Scholar]
- Plotch S.J., Bouloy M., Ulmanen I., Krug R.M. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- Ramanathan A., Robb G.B., Chan S.H. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Brunzelle J., Satchell K.J.F. Sci. Signal. 2020:13. doi: 10.1126/scisignal.abe1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj C., Dinesh D.C., Panwar U., Abhirami R., Boura E., Singh S.K. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1778535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin A.J., Manley J.L. Nat. Struct. Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- Shu T., Huang M., Wu D., Ren Y., Zhang X., Han Y., Mu J., Wang R., Qiu Y., Zhang D.Y., Zhou X. Virol. Sin. 2020;35:321–329. doi: 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski P.J., Warminski M., Kubacka D., Ratajczak T., Nowis D., Kowalska J., Jemielity J. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Kumar M., Mansuri R., Sahoo G.C., Deep A. J. Pharm. BioAllied Sci. 2016;8:188–194. doi: 10.4103/0975-7406.171682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T.R. Infect. Disord. - Drug Targets. 2009;9:223–245. doi: 10.2174/187152609787847659. [DOI] [PubMed] [Google Scholar]
- Vincent F., Duncton M.A. Curr. Top. Med. Chem. 2011;11:2216–2226. doi: 10.2174/156802611796904861. [DOI] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Qin G., Niu J., Wang Z., Wang C., Ren J., Qu X. Angew Chem. Int. Ed. Engl. 2021;60:432–438. doi: 10.1002/anie.202011419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.