Abstract
Wilms' tumor (WT) is a major type of kidney cancer in children; however, the therapeutic measures for control of tumor metastasis, recurrence and death for this type of cancer remain unsatisfactory. The present study aimed to verify the expression of T-cell factor 3 (TCF3) in WT, and to explore its role in regulating the viability, migration and apoptosis of kidney tumor cells. Tumor tissues were collected from 10 patients with WT, and adjacent tissues were collected as normal controls. The expression levels of TCF3 were detected in WT tissues and adjacent tissues by reverse transcription-quantitative PCR (RT-qPCR), western blotting and immunohistochemistry. In addition, TCF3 expression was silenced in G401 kidney tumor cells via small interfering RNA transfection. Cell viability, cell cycle progression and cell apoptosis were assessed using the MTT assay and flow cytometry; the migration and invasion of kidney tumor cells were examined using Transwell and wound-healing assays; and the expression levels of Wnt signaling pathway-related genes (Wnt1, β-catenin and c-myc) were detected by RT-qPCR and western blotting. The results revealed that the expression levels of TCF3 were high in WT tissues from patients. Silencing TCF3 expression in G401 kidney tumor cells in vitro significantly inhibited cell viability and migration, and promoted cell apoptosis. Moreover, silencing TCF3 expression in G401 cells inhibited the expression levels of Wnt signaling pathway-related genes. Overall, these data indicated that TCF3 may be involved in WT development through regulation of Wnt signaling pathways. The findings of the present study provide a novel potential marker for the treatment and prognostic evaluation of WT.
Keywords: Wilms' tumor, T-cell factor 3, knockdown, viability, migration, apoptosis
Introduction
Wilms' tumor (WT) is the most common renal malignancy in children, ranking second among all primary abdominal malignancies in European children and fifth among all childhood malignancies in terms of incidence; notably, it accounts for 95% of pediatric renal tumors (1,2). According to statistics, WT accounts for 8–10% of all pediatric tumors worldwide (3). Although a comprehensive treatment approach has been developed for WT and refined for patients with an overall good prognosis, the prognosis is poor for patients with tumors that develop embryonic phenotypes after preoperative chemotherapy, and 13% of patients with WT relapse within 2 years of diagnosis (1,4). In addition, malignant metastasis can occur in late-stage disease, and the mortality rate of WT is still 5% (5). Therefore, exploring the molecular mechanism underlying WT cell proliferation, metastasis and invasion to identify a method to intervene in tumor development and improve the treatment approach is important.
T-cell factor (TCF) 3 is an important member of the TCF/lymphoid enhancer-binding factor (LEF) family, which includes TCF1, TCF3, TCF4 and LEF1 (6,7). It was previously reported that loss of the TCF3 gene could lead to the death of early embryos (8). Furthermore, TCF3 is expressed in some poorly differentiated malignant tumors (9); however, the role and mechanism of TCF3 in malignant tumors have been less well studied. Kehl et al (10) demonstrated that TCF3 is a potential master regulator in blastemal WT, and that TCF3 and other regulators serve a central role in drug-resistant chemotherapeutic blastemal WT. The Wnt/β-catenin signaling pathway is a highly conserved key signaling pathway in mammalian cells, which is closely related to the occurrence and development of renal carcinoma (11,12). TCF3 is one of the effector molecules of the classical Wnt signaling pathway (13), which regulates cell proliferation and activation, and is closely related to tumor occurrence and development (14). Previous studies have shown that TCF3 plays an important role in the development of human tumors, such as renal carcinoma (15), embryonal carcinoma (9) and breast cancer (16). However, few studies have examined the role and mechanism of TCF3 in WT, and the relationship between the Wnt signaling pathway and WT is not clear.
The present study preliminarily measured TCF3 expression in WT, and then silenced TCF3 expression in G401 kidney tumor cells in vitro via small interfering RNA (siRNA)-mediated knockdown. Subsequently, the present study assessed the effect of TCF3 knockdown on the viability, apoptosis, migration and invasion of G401 cells in vitro, and investigated the possible molecular mechanisms. Furthermore, to illuminate its regulatory mechanism in kidney tumor cells in vitro, TCF3 in the regulation of the Wnt pathway was studied. The aim of the present study was to provide a theoretical basis for development of a drug targeting TCF3 for WT.
Materials and methods
WT samples
WT tissues (n=10) and adjacent tissues (n=10) were obtained from five girls and five boys (age range, 1–8 years) with resection surgery between June 2018 and September 2020 from Kunming Children's Hospital (Kunming, China). The mean distance between WT and adjacent tissue was ~2 cm. Written informed consent for the use of tissue samples in the present study was obtained from the parents/guardians of the patients. The present study was approved by the Medical Ethics Committee of Kunming Children's Hospital (approval no. 2020-03-105-K01). None of the patients received any medication, radiotherapy or chemotherapy before the operation. The tissue samples were quickly separated from the surgical specimen, rinsed with sterile saline and fixed with 4% paraformaldehyde (cat. no. P1110; Beijing Solarbio Science & Technology Co., Ltd.) for 48 h at room temperature. Unfixed tissue samples were frozen in liquid nitrogen in a cryopreservation tube and then stored in a −80°C freezer for reverse transcription-quantitative (RT-q)PCR and western blotting.
Hematoxylin & eosin (HE) staining
The WT and adjacent tissue samples were dehydrated, embedded in paraffin, sliced (5 µm), dewaxed with xylene, washed with a series of ethanol solutions and rinsed with 1X phosphate-buffered saline (PBS). For HE staining (cat. no. G1120; Beijing Solarbio Science & Technology Co., Ltd.), the tissue sections were stained with hematoxylin solution for 10 min at room temperature, washed with running water, differentiated with 0.5% hydrochloric acid ethanol solution for 3–10 sec, stained with eosin solution for 2 min, and then sealed after dehydration and clearing. Sections were observed under a light microscope (Nikon Corporation).
Immunohistochemistry (IHC)
The sections (5 µm) were fixed, paraffin embedded, dewaxed and washed as aforementioned and subjected to antigen retrieval with 0.01 M citric acid buffer (pH 6.0) for 15 min at 100°C and 80 kPa, then cooled at room temperature. Subsequently, the cells were blocked with 5% goat serum (cat. no. SL038; Beijing Solarbio Science & Technology Co., Ltd.) for 15 min at room temperature. The sections were incubated with anti-TCF3 antibody (1:100 dilution; cat. no. 67140-1-Ig; Wuhan Sanying Biotechnology) at 4°C overnight. The slices were washed with PBS-0.05% Tween-20 (PBST) and incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (1:200 dilution; cat. no. SA00001-1; Wuhan Sanying Biotechnology) for 2 h at room temperature. Staining of the sections was detected using an Elivision™ super HRP IHC kit (cat. no. KIT-9922; MXB Biotechnologies) and the sections were observed under a light microscope (Nikon Corporation). The mean optical density (MOD) of each section was measured using ImageJ2 software (National Institutes of Health).
Cell culture and cell transfection
The G401 kidney tumor cell line was purchased from the Cell Bank of Kunming, Type Culture Collection of The Chinese Academy of Sciences. Cells were authenticated using STR profiling. The cells were cultured in McCoy's 5A (modified) medium (cat. no. 16600082; Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (cat. no. S9020; Beijing Solarbio Science & Technology Co., Ltd.) at 37°C in a 5% CO2 cell incubator. The G401 cells were seeded into a 6-well plate at 3×105 cells/well. When the cells were well adhered and had reached ~70% confluence, they were transfected with 30 pmol TCF3 siRNA (5′-AAGCAACAAAACATACACT-3′) or negative control (NC) siRNA (5′-TTCTCCGAACGTGTCACGT-3′) synthesized by the Invitrogen company using Lipofectamine® 2000 (cat. no. 11668019; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The cells were divided into three groups for subsequent experiments: The control group, which did not undergo transfection and the cells grew normally; the NC group, which was transfected with NC siRNA; and the TCF3 siRNA group, which was transfected with TCF3 siRNA. After transfection for 48 h, the cells were collected for subsequent experiments.
RT-qPCR
Total RNA was extracted from tissues and from cells 48 h post-transfection using an RNA Simple kit (Tiangen Biotech, Co., Ltd.), and the RNA concentration and purity were determined using a micro nucleic acid quantitative analyzer. Subsequently, RNA was reverse transcribed into cDNA using the PrimeScript RT Reagent kit (cat. no. RR036Q; Takara Biotechnology Co., Ltd.) according to the manufacturer's instructions, and cDNA was amplified via qPCR using TB Green Premix Ex Taq II (cat. no. RR420A; Takara Biotechnology Co., Ltd.) as follows: 95°C for 5 min; followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec. The primers were synthesized by Shanghai Sangon Biotech Co., Ltd., and the sequences were as follows: TCF3, forward 5′-AGGAGAAGGAGGACGAGGAG-3′ and reverse, 5′-AAAGGCCTCGTTGATGTCAC-3′; Wnt1 forward, 5′-CGGCGTTTATCTTCGCTATC-3′ and reverse, 5′-GCCTCGTTGTGAAGGTT-3′; β-catenin forward, 5′-GAAACGGCTTTCAGTTGAGC-3′ and reverse, 5′-CTGGCCATATCCACCAGAGT-3′; c-myc forward, 5′-AAAGGCCCCCAAGGTAGTTA-3′ and reverse, 5′-GCACAAGAGTTCCGTAGCTG-3′; and β-actin forward, 5′-GCTCTTTTCCAGCCTTCCTT-3′ and reverse, 5′-GAGCCAGAGCAGTGATCTCC-3′. β-actin served as the internal control for RT-qPCR. Relative expression levels were calculated using the 2−ΔΔCq method (17).
Western blotting
After transfection for 48 h, total protein was extracted from tissue and cells using RIPA lysis buffer (cat. no. P0013B; Beyotime Institute of Biotechnology), and the total protein concentration was determined using a BCA kit (cat. no. P0012; Beyotime Institute of Biotechnology). Each protein sample (30 µg) was separated on a 10% gel using SDS-PAGE and then transferred onto a PVDF membrane (cat. no. FFP36; Beyotime Institute of Biotechnology). The membranes were blocked with 5% non-fat milk in 1X PBST (cat. no. P0216; Beyotime Institute of Biotechnology) for 2 h at 4°C and incubated with primary antibodies against TCF3 (1:1,000; cat. no. 67140-1-Ig), Wnt1 (1:2,000 dilution; cat. no. 27935-1-AP), β-catenin (1:1,000 dilution; cat. no. 51067-2-AP), c-myc (1:1,000 dilution; cat. no. 10828-1-AP) and β-actin (1:1,000 dilution; cat. no. 66009-1-Ig) (all from Wuhan Sanying Biotechnology) overnight at 4°C. After washing with 1X PBST (cat. no. P1031; Beijing Solarbio Science & Technology Co., Ltd.), the membranes were probed with goat anti-rabbit IgG-HRP secondary antibodies (1:5,000 dilution; cat. no. SA00001-2) or goat anti-mouse IgG-HRP secondary antibodies (1:5,000 dilution; cat. no. SA00001-1) (both from Wuhan Sanying Biotechnology) for 1 h at room temperature. Subsequently, protein bands were observed using an enhanced chemiluminescence kit (cat. no. P0018S; Beyotime Institute of Biotechnology) on a Bio-Rad Gel Imaging Analysis system (GelDoc XR+ IMAGELAB; Bio-Rad Laboratories, Inc.). β-actin served as an internal control. ImageJ 2× software was used to semi-quantify the protein bands.
MTT assay
After transfection for 48 h, MTT solution (cat. no. M1020; Beijing Solarbio Science & Technology Co., Ltd.) was added to each well and cultured with the cells (5×104) for 4 h at 37°C. The culture solution was discarded and then 150 µl DMSO was added to each well. The culture plate was placed on an oscillator and shaken for 10 min at room temperature to fully dissolve the purple crystals. The absorbance values of each well were measured at 570 nm using a microplate reader (Bio-Rad Laboratories, Inc.) and the cells optical density (OD) value was recorded.
Flow cytometry
After transfection for 48 h, an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) detection kit (cat. no. C1062L; Beyotime Institute of Biotechnology) was used to analyze cell apoptosis. After transfection, the cells (1×106) were harvested with 0.25% trypsin, incubated with 10 µl Annexin V-FITC at 4°C for 30 min and then incubated with 5 µl PI solution for 15 min at 4°C in the dark. The stained cells were detected using BD FACSCanto II flow cytometry (BD Biosciences) with CellQuest software (version 5.1; BD Biosciences). Early apoptotic cells were Annexin V-FITC-positive and PI-negative, and late apoptotic cells were Annexin V-FITC-positive and PI-positive. The cell apoptosis rate is presented as the percentage of cells in the early and late apoptotic phases.
A cell cycle analysis kit (cat. no. C1052; Beyotime Institute of Biotechnology) was used to analyze cell cycle progression according to the manufacturer's instructions. After transfection for 48 h, the cells (1×106) were harvested with 0.25% trypsin. Subsequently, the cells were fixed with 1 ml precooled 70% ethanol at 4°C for 2 h. Subsequently, 0.5 ml PI staining solution was added to each cell sample, and the cells were incubated at 37°C for 30 min in the dark. Red fluorescence was detected via BD FACSCanto II flow cytometry (BD Biosciences) with CellQuest software (version 5.1; BD Biosciences) at an excitation wavelength of 488 nm.
Cell invasion assay
The cell invasion assay was performed using Transwell chambers (8 µm; cat. no. 3422; Corning, Inc.) coated with Matrigel (cat. no. 356234; Corning, Inc.) at 37°C for 30 min. Briefly, 300 µl of a 5×105 cells/ml suspension (McCoy's 5A medium without FBS) was added into the upper chamber and 500 µl medium (with 10% FBS) was added to the lower chamber. After transfected for 48 h in the Transwell chambers, the invaded cells were fixed in 90% alcohol at room temperature for 10 min, stained with 0.1% crystal violet at room temperature for 30 min and observed under a light microscope (Olympus Corporation).
Cell migration assay
Cell migration was detected using a cell wound-healing assay. When cell confluence reached 90%, 1-ml sterile pipette tips were used to draw a straight vertical line across each plate. The plates were washed with PBS three times to remove the suspended cells, and 2 ml medium with 10% FBS (cat. no. S9020; Beijing Solarbio Science & Technology Co., Ltd.) was added. Subsequently, images of the cells were captured and assessed under a light microscope, and cells were then transfected with TCF3 siRNA or NC siRNA. After transfection for 48 h, images of the cells were captured and assessed under a light microscope, the migration distance was measured using ImageJ (version 2) software, and the migration ratio of the cells in each group was calculated.
Statistical analysis
Each experiment was repeated three times independently. Data are expressed as the mean ± standard error of the mean. Using GraphPad Prism 5.0 software (GraphPad Software Inc.), differences between two groups were compared using Student's t-test (paired), and differences among three or more groups were compared using one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
TCF3 expression in WT tissues and adjacent tissues
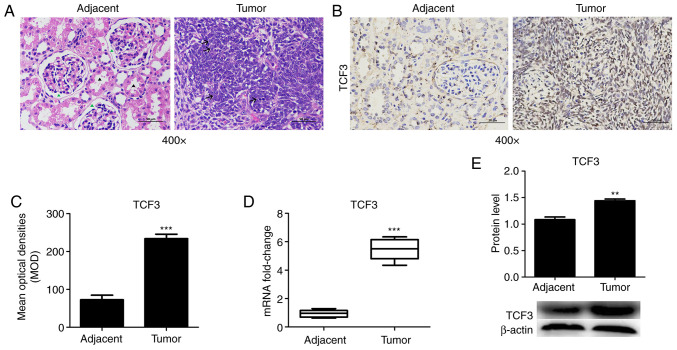
To measure the expression levels of TCF3 in patients with WT, WT and adjacent tissues were assessed via IHC, RT-qPCR and western blotting. HE staining was used to detect histopathological tissue morphology. The HE staining results revealed that the glomerulus (green arrowhead) and kidney tubules (black arrowhead) were intact in the adjacent tissue group, whereas the tumor cells in the WT tissues were diffuse or patchy, cells appear short fusiform or elliptical, partially deviated nucleoli and pathological mitoses (arrow; Fig. 1A). TCF3 protein was mainly localized in the nuclei of the kidney tumor cells (Fig. 1B). TCF3 protein expression was detected via IHC, and the results revealed that TCF3 expression was much higher in WT tissues compared with that in adjacent tissues (Fig. 1B). The MOD of TCF3 expression was significantly increased in WT tissues compared with that in adjacent tissues (Fig. 1C). Furthermore, RT-qPCR (Fig. 1D) and western blotting (Fig. 1E) revealed that the mRNA and protein expression levels of TCF3 were significantly increased in WT tissues compared with those in adjacent tissues. The results demonstrated that changes in TCF3 were consistent at the protein and mRNA levels (Fig. 1D and E). These results suggested that patients with WT may exhibit high TCF3 expression.
Figure 1.
TCF3 expression in WT tissues. WT tissues and adjacent tissues were collected to detect the expression levels of TCF3. (A) Hematoxylin and eosin staining of WT and adjacent tissues (scale bar, 50 µm). The glomerulus (green arrowhead) and kidney tubules (black arrow) were intact in the adjacent tissue group. Tumor cells in the WT tissue were diffuse or patchy and cells appeared short fusiform or elliptical with partially deviated nucleoli (arrow). (B) TCF3 expression in WT and adjacent tissues determined by immunohistochemistry (scale bar, 50 µm). (C) MOD of TCF3 was semi-quantified using ImageJ2 software. (D) Reverse transcription-quantitative PCR and (E) western blotting were performed to detect the mRNA and protein expression levels of TCF3. All experiments were repeated at least three times. Data are presented as the mean ± standard error of the mean and were analyzed via Student's t-test. **P<0.01 and ***P<0.001 compared with the adjacent tissue group. MOD, mean optical density; TCF3, T-cell factor 3; WT, Wilms' tumor.
Effect of TCF3 silencing on the viability, cell cycle progression and apoptosis of G401 kidney tumor cells
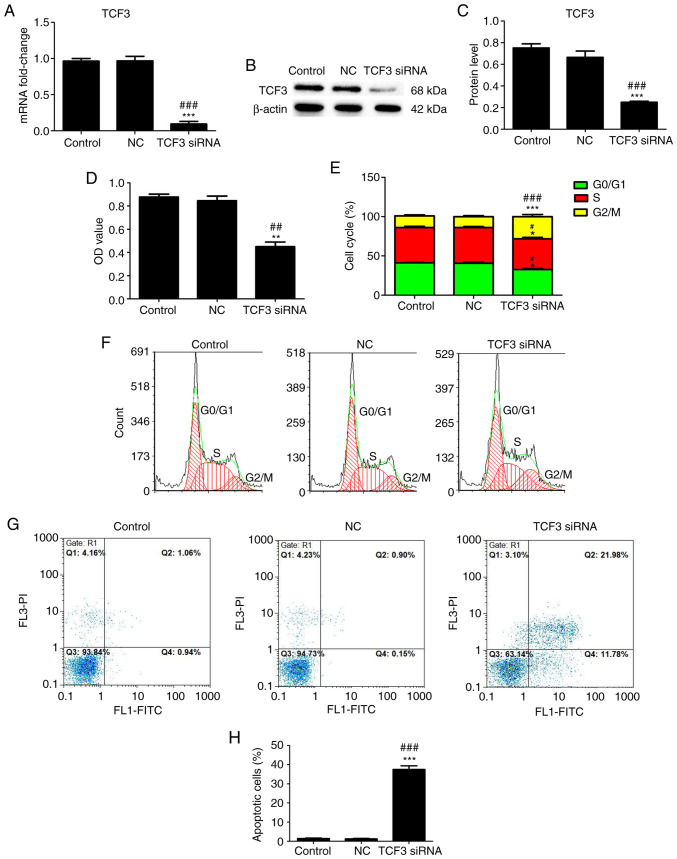
To investigate the effect of TCF3 silencing on G401 kidney tumor cell viability, cell cycle progression and apoptosis, the cells were transfected with TCF3 siRNA or NC siRNA, RT-qPCR and western blotting were performed to detect the transfection efficiency and expression levels of TCF3, MTT assays were performed to assess cell viability, and flow cytometry was performed to assess cell cycle distribution and apoptosis. RT-qPCR (Fig. 2A) and western blotting (Fig. 2B and C) revealed that the mRNA and protein expression levels of TCF3 were significantly decreased in the TCF3 siRNA group compared with those in the control and NC groups. As shown in Fig. 2D, the OD value in the TCF3 siRNA group was lower than that in the control and NC groups. The cell cycle analysis results showed an increase in G2/M-phase cells, which was accompanied by a decrease in G0/G1- and G2/M-phase cells, in the TCF3 siRNA group compared with the control and NC groups (Fig. 2B and C). The apoptosis assay results revealed that the percentage of apoptotic cells was higher in the TCF3 siRNA group than that in the control and NC groups (Fig. 2D and E). These results indicated that silencing TCF3 expression could inhibit the viability and promote the apoptosis of G401 kidney tumor cells.
Figure 2.
Effect of TCF3 silencing on the viability, cell cycle progression and apoptosis of G401 kidney tumor cells. Reverse transcription-quantitative PCR (A) and western blotting (B) were performed to detect the mRNA and protein expression levels of TCF3 in all groups. (C) Semi-quantification of western blotting was performed using ImageJ2 software. (D) MTT assays were performed to detect viability. (E) Cell cycle distribution was determined. The effect of TCF3 silencing on the (F) cell cycle progression and (G) apoptosis of G401 kidney tumor cells were examined via flow cytometry. (H) Percentage of apoptotic cells was quantified. Control group, untransfected cells; NC group, cells transfected with NC siRNA; TCF3 siRNA group, cells transfected with TCF3 siRNA. All experiments were repeated at least three times. Data are presented as the mean ± standard error of the mean and were analyzed via one-way ANOVA with Tukey's post hoc test. *P<0.05, **P<0.01 and ***P<0.001 compared with the control group; #P<0.05, ##P<0.01 and ###P<0.001 compared with the NC group. FITC, fluorescein isothiocyanate; NC, negative control; OD, optical density; PI, propidium iodide; siRNA, small interfering RNA; TCF3, T-cell factor 3.
Effect of TCF3 silencing on the migration and invasion of G401 kidney tumor cells
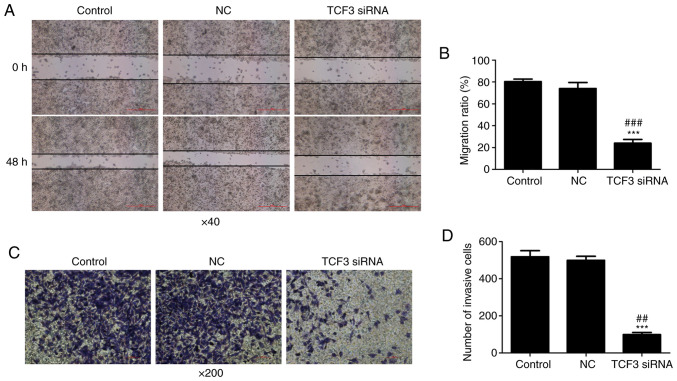
To investigate the effect of TCF3 silencing on the migration and invasion of G401 kidney tumor cells, wound-healing and Transwell assays were performed. The results of the wound-healing assay demonstrated that the cell migration ratio was decreased in the TCF3 siRNA group compared with that in the control and NC groups (Fig. 3A and B). The results of Transwell assays revealed that the number of invasive cells in the TCF3 siRNA group was significantly lower than that in the control and NC groups (Fig. 3C); quantification of cells stained with crystal violet is shown in Fig. 3D. These results suggested that TCF3 may promote the migration and invasion of G401 kidney tumor cells.
Figure 3.
Effect of TCF3 silencing on G401 kidney tumor cell migration and invasion. (A) Cell migration was detected using a wounding-healing assay (scale bar, 500 µm). (B) Migration ratio was calculated. (C) Cell invasion was detected using a Transwell assay (scale bar, 100 µm). (D) Cells were stained with crystal violet and number of invasive cells was assessed. Control group, untransfected cells; NC group, cells transfected with NC siRNA; TCF3 siRNA group, cells transfected with TCF3 siRNA. All experiments were repeated at least three times. Data are presented as the mean ± standard error of the mean and were analyzed via one-way ANOVA with Tukey's post hoc test. ***P<0.001 compared with the control group; ##P<0.01 and ###P<0.001 compared with the NC group. NC, negative control; siRNA, small interfering RNA; TCF3, T-cell factor 3.
Effect of TCF3 silencing on the mRNA and protein expression levels of Wnt pathway-related genes in G401 kidney tumor cells
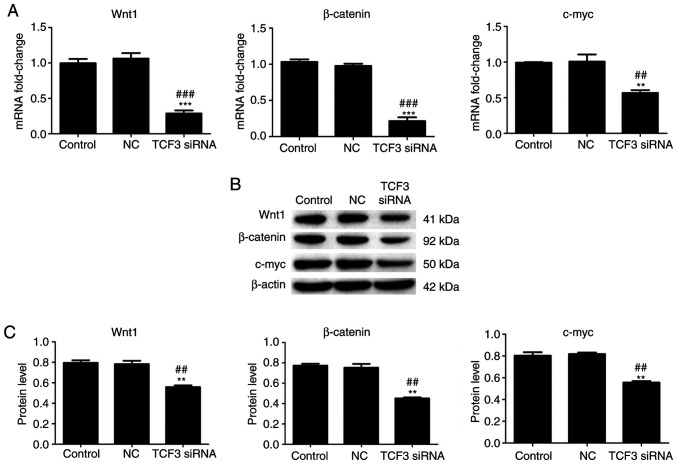
A previous study reported that Wnt pathway activation is associated with TCF3 (18). Therefore, to further investigate the effect of TCF3 silencing on genes involved in the Wnt signaling pathway, the mRNA and protein expression levels of Wnt1, β-catenin and c-myc in the Wnt signaling pathway were detected using RT-qPCR and western blotting. The results of RT-qPCR demonstrated that the mRNA expression levels of Wnt1, β-catenin and c-myc were significantly lower in the TCF3 siRNA group compared with those in the control and NC groups (Fig. 4A). The results of western blotting regarding changes in the protein expression levels of Wnt1, β-catenin and c-myc were consistent with the changes detected in the mRNA expression levels. Compared with those in the control and NC groups, the protein expression levels of Wnt1, β-catenin and c-myc were significantly decreased in the TCF3 siRNA group (Fig. 4B and C). These results indicated that TCF3 silencing may inactivate the Wnt signaling pathway in G401 kidney tumor cells.
Figure 4.
Effect of TCF3 silencing on the expression of Wnt signaling pathway-related genes in G401 kidney tumor cells. (A) Reverse transcription-quantitative PCR was performed to detect the mRNA expression levels of Wnt1, β-catenin and c-myc in all groups. (B) Western blotting was performed to detect the protein expression levels of Wnt1, β-catenin and c-myc in all groups. (C) Semi-quantification of western blotting was performed using ImageJ2 software. Control group, untransfected cells; NC group, cells transfected with NC siRNA; TCF3 siRNA group, cells transfected with TCF3 siRNA. All experiments were repeated at least three times. Data are presented as the mean ± standard error of the mean and were analyzed via one-way ANOVA with Tukey's post hoc test. **P<0.01 and ***P<0.001 compared with the control group; ##P<0.01 and ###P<0.001 compared with the NC group. NC, negative control; siRNA, small interfering RNA; TCF3, T-cell factor 3.
Discussion
In the present study, the expression of TCF3 in WT tissue was higher than that in normal adjacent tissue, and the results in vitro and in vivo suggested that high TCF3 expression may promote G401 cell viability and WT development. The effects of TCF3 silencing on kidney tumor cell viability, apoptosis, migration and invasion were investigated via MTT assay, flow cytometry, and cell migration and invasion assays. The results revealed that the viability, migration and invasion of G401 kidney tumor cells were significantly decreased after TCF3 was silenced. In addition, when TCF3 was silenced, the percentage of apoptotic G401 cells was significantly increased, the proportion of G0/G1 phase cells was decreased and the proportion of G2/M phase cells was significantly increased. These results indicated that TCF3 silencing may inhibit G401 kidney tumor cell viability, migration and invasion, and promote apoptosis.
TCF3 is a common transcription factor that regulates tumor cell proliferation, migration and invasion (19). The present study revealed that TCF3 regulated the viability, migration and invasion of kidney tumor cells. TCF3 is also one of the final effector molecules of the highly conserved classical Wnt signaling pathway in mammalian cells, which is closely related to tumor occurrence and development (20). TCF3 has dual regulatory functions: TCF3 binds to the Groucho/TLE family of transcription inhibitors to inhibit gene transcription in the absence of Wnt signaling (21,22); however, TCF3 binds to β-catenin to promote gene transcription in the presence of Wnt signaling (22). In addition, TCF3 has different regulatory roles for different Wnt target genes (23). Overexpression of TCF3 has been reported to stimulate division of human hair follicle bulge epidermal progenitor cells; TCF3 detaches from the corresponding gene promoter when the Wnt pathway is activated and target gene transcription is activated to enhance the division activity of pluripotent stem cells (18). The present study revealed that TCF3 silencing inactivated the Wnt pathway in kidney tumor cells. TCF3 has also been shown to promote the ability of breast cancer cells to initiate the formation of tumor tissue and maintain the growth of cancer cells (16). Conversely, the prognosis of patients with colon cancer was reported to be worse in patients with low TCF3 expression, and the absence of TCF3 was shown to cause colon cancer cells to proliferate faster and metastasize more easily (24). Thus, TCF3 serves an important role during the course and development of cancer, and it may have different roles in various tumor types. In the present study, TCF3 was revealed to have a positive role in the development of WT. Although TCF3 has been shown to be a potential master regulator in blastemal WT, the effect of TCF3 expression on kidney tumor cells was unclear in the present study, and exploring WT pathogenesis and finding novel targets for diagnosis and treatment is still of great significance (25).
The cell cycle-related gene c-myc is a target gene of the Wnt signaling pathway (26,27). The Wnt1/β-catenin signaling pathway is known to be related to tumor development (28–30). Genes related to the Wnt signaling pathway, such as Wnt1 and β-catenin, have an important role in tumor cell proliferation, dedifferentiation and metastasis (31,32). The present study hypothesized that TCF3 might affect the viability of kidney tumor cells by regulating the Wnt1/β-catenin signaling pathway and its target gene c-myc. To investigate this hypothesis, the expression levels of TCF3, Wnt1, β-catenin and c-myc were detected using RT-qPCR and western blotting. The results revealed that in G401 kidney tumor cells, TCF3 silencing significantly inhibited the mRNA and protein expression levels of TCF3, Wnt1, β-catenin and c-myc. The downregulation of Wnt signaling pathway-related genes after TCF3 knockdown in G401 cells suggested that TCF3 may serve a role in transcriptional activation of Wnt signaling in kidney tumor cells. One possible mechanism is that a large amount of dissociated β-catenin enters the nucleus when the Wnt pathway is activated, at which point TCF3 binds to β-catenin, forming a transcriptional activator that regulates Wnt target genes (33). Thus, TCF3 silencing could inactivate the Wnt pathway and inhibit the Wnt pathway target gene c-myc. These results indicated that TCF3 silencing may inhibit the malignant biological behavior of kidney tumor cells, and that TCF3 siRNA may suppress the viability of G401 cells by inhibiting the expression of Wnt signaling pathway-related genes.
To the best of our knowledge, the present study is the first to demonstrate the important role of TCF3 in the biological function of kidney tumor cells in vitro. TCF3 silencing was shown to suppress the viability, migration and invasion, and promote the apoptosis of G401 kidney tumor cells in vitro by regulating the expression of Wnt signaling-related genes at the cellular level. The present study provided novel insight into the molecular mechanism underlying the effects of TCF3 on kidney tumor cells and may indicate a path towards a novel treatment for WT. In subsequent in-depth studies, animal experiments should be conducted to confirm that TCF3 silencing can contribute to inhibition of kidney tumor development in vivo.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Kunming Health Science and Technology Talent project [grant no. 2020-SW (reserve)-115], the Health Research project of Kunming Health Committee (grant no. 2020-06-02-001) and the Scientific Research project of Yunnan Education Department (grant no. 2021J0286).
Funding
The present study was supported by the Kunming Health Science and Technology Talent project [grant no. 2020-SW (reserve)-115], the Health Research project of Kunming Health Committee (grant no. 2020-06-02-001) and the Scientific Research project of Yunnan Education Department (grant no. 2021J0286).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
NZ and ZY conceived and designed the study. NZ, BY, JM, HJ, LL, HT and FJ performed the experiments. NZ, JM and HJ processed data. NZ and BY wrote the manuscript. NZ and ZY reviewed and edited the manuscript. NZ and ZY confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All human tissue samples were collected by the Kunming Children's Hospital and the present study was approved by the Medical Ethics Committee of Kunming Children's Hospital (approval no. 2020-03-105-K01). Written informed consent was obtained from the parents/guardians of the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pastore G, Znaor A, Spreafico F, Graf N, Pritchard-Jones K, Steliarova-Foucher E. Malignant renal tumours incidence and survival in European children (1978–1997): Report from the automated childhood cancer information system project. Eur J Cancer. 2006;42:2103–2114. doi: 10.1016/j.ejca.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 2.Davidoff AM. Wilms tumor. Adv Pediatr. 2012;59:247–267. doi: 10.1016/j.yapd.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brok J, Lopez-Yurda M, Tinteren HV, Treger TD, Furtwängler R, Graf N, Bergeron C, van den Heuvel-Eibrink MM, Pritchard-Jones K, Olsen ØE, et al. Relapse of Wilms' tumour and detection methods: A retrospective analysis of the 2001 Renal Tumour Study Group-International Society of Paediatric Oncology Wilms' tumour protocol database. Lancet Oncol. 2018;19:1072–1081. doi: 10.1016/S1470-2045(18)30293-6. [DOI] [PubMed] [Google Scholar]
- 4.Kaste SC, Dome JS, Babyn PS, Graf NM, Grundy P, Godzinski J, Levitt GA, Jenkinson H. Wilms tumour: Prognostic factors, staging, therapy and late effects. Pediatr Radiol. 2008;38:2–17. doi: 10.1007/s00247-007-0687-7. [DOI] [PubMed] [Google Scholar]
- 5.Lee JS, Padilla B, DuBois SG, Oates A, Boscardin J, Goldsby RE. Second malignant neoplasms among children, adolescents and young adults with Wilms tumor. Pediatric Blood Cancer. 2015;62:1259–1264. doi: 10.1002/pbc.25484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected] Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 7.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merrill BJ, Pasolli HA, Polak L, Rendl M, García-García MJ, Anderson KV, Fuchs E. Tcf3: A transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- 9.Lin G, Zhao L, Yin F, Lan R, Li L, Zhang X, Zhang H, Yang B. TCF3 inhibits F9 embryonal carcinoma growth by the down-regulation of Oct4. Oncol Rep. 2011;26:893–899. doi: 10.3892/or.2011.1376. [DOI] [PubMed] [Google Scholar]
- 10.Kehl T, Schneider L, Kattler K, Stöckel D, Wegert J, Gerstner N, Ludwig N, Distler U, Tenzer S, Gessler M, et al. The role of TCF3 as potential master regulator in blastemal Wilms tumors. Int J Cancer. 2019;144:1432–1443. doi: 10.1002/ijc.31834. [DOI] [PubMed] [Google Scholar]
- 11.Huang H, He X. Wnt/beta-catenin signaling: New (and old) players and new insights. Curr Opin Cell Biol. 2008;20:119–125. doi: 10.1016/j.ceb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, Biechele TL, Gingras AC, Zheng N, Maccoss MJ, et al. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007;316:1043–1046. doi: 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- 13.Huelsken J, Behrens J. The Wnt signalling pathway. J Cell Sci. 2002;115((Pt 21)):3977–3978. doi: 10.1242/jcs.00089. [DOI] [PubMed] [Google Scholar]
- 14.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: Diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 15.Uematsu K, He B, You L, Xu Z, McCormick F, Jablons DM. Activation of the Wnt pathway in non small cell lung cancer: Evidence of dishevelled overexpression. Oncogene. 2003;22:7218–7221. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- 16.Slyper M, Shahar A, Bar-Ziv A, Granit RZ, Hamburger T, Maly B, Peretz T, Ben-Porath I. Control of breast cancer growth and initiation by the stem cell-associated transcription factor TCF3. Cancer Res. 2012;72:5613–5624. doi: 10.1158/0008-5472.CAN-12-0119. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Yi F, Pereira L, Hoffman JA, Shy BR, Yuen CM, Liu DR, Merrill BJ. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat Cell Biol. 2011;13:762–770. doi: 10.1038/ncb2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah M, Rennoll SA, Raup-Konsavage WM, Yochum GS. A dynamic exchange of TCF3 and TCF4 transcription factors controls MYC expression in colorectal cancer cells. Cell Cycle. 2015;14:323–332. doi: 10.4161/15384101.2014.980643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 22.Stamos JL, Weis WI. The β-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 24.Zhao H, Zhao C, Li H, Zhang D, Liu G. E2A attenuates tumor-initiating capacity of colorectal cancer cells via the Wnt/beta-catenin pathway. J Exp Clin Cancer Res. 2019;38:276. doi: 10.1186/s13046-019-1261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan CC, To KF, Yuen HL, Shing Chiang AK, Ling SC, Li CH, Cheuk DK, Li CK, Shing MM. A 20-year prospective study of Wilms tumor and other kidney tumors: A report from Hong Kong pediatric hematology and oncology study group. J Pediatr Hematol Oncol. 2014;36:445–450. doi: 10.1097/MPH.0000000000000018. [DOI] [PubMed] [Google Scholar]
- 26.Pajic A, Spitkovsky D, Christoph B, Kempkes B, Schuhmacher M, Staege MS, Brielmeier M, Ellwart J, Kohlhuber F, Bornkamm GW, et al. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int J Cancer. 2000;87:787–793. doi: 10.1002/1097-0215(20000915)87:6<787::AID-IJC4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 27.Doose G, Haake A, Bernhart SH, López C, Duggimpudi S, Wojciech F, Bergmann AK, Borkhardt A, Burkhardt B, Claviez A, et al. MINCR is a MYC-induced lncRNA able to modulate MYC's transcriptional network in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 2015;112:E5261–E5270. doi: 10.1073/pnas.1505753112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trowbridge JJ, Xenocostas A, Moon RT, Bhatia M. Glycogen synthase kinase-3 is an in vivo regulator of hematopoietic stem cell repopulation. Nat Med. 2006;12:89–98. doi: 10.1038/nm1339. [DOI] [PubMed] [Google Scholar]
- 29.Ying Y, Tao Q. Epigenetic disruption of the WNT/beta-catenin signaling pathway in human cancers. Epigenetics. 2009;4:307–312. doi: 10.4161/epi.4.5.9371. [DOI] [PubMed] [Google Scholar]
- 30.Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol. 2003;129:199–221. doi: 10.1007/s00432-003-0431-0. [DOI] [PubMed] [Google Scholar]
- 31.Nusse R, Brown A, Papkoff J, Scambler P, Shackleford G, McMahon A, Moon R, Varmus H. A new nomenclature for int-1 and related genes: The Wnt gene family. Cell. 1991;64:231. doi: 10.1016/0092-8674(91)90633-A. [DOI] [PubMed] [Google Scholar]
- 32.Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 33.Chodaparambil JV, Pate KT, Hepler MR, Tsai BP, Muthurajan UM, Luger K, Waterman ML, Weis WI. Molecular functions of the TLE tetramerization domain in Wnt target gene repression. EMBO J. 2014;33:719–731. doi: 10.1002/embj.201387188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.