Abstract
Osteosarcoma is a quickly developing, malignant cancer of the bone, which is associated with a bad prognosis. In osteosarcoma, hypoxia promotes the malignant phenotype, which results in a cascade of immunosuppressive processes, poor prognosis, and a high risk of metastasis. Nonetheless, additional methodologies for the study of hyperoxia in the tumor microenvironment also need more analysis. We obtained 88 children patients with osteosarcoma from the Therapeutically Applicable Research to Generate Effective Treatment (TARGET) database and 53 children patients with RNA sequence and clinicopathological data from the Gene Expression Omnibus (GEO). We developed a four-gene signature related to hypoxia to reflect the immune microenvironment in osteosarcoma that predicts survival. A high-risk score indicated a poor prognosis and immunosuppressive microenvironment. The presence of the four-gene signature related to hypoxia was correlated with clinical and molecular features and was an important prognostic predictor for pediatric osteosarcoma patients. In summary, we established and validated a four-gene signature related to hypoxia to forecast recovery and presented an independent prognostic predictor representing overall immune response strength within the osteosarcoma microenvironment.
1. Introduction
Osteosarcomas are primary malignant tumors of the bone that are defined by the malignant cells' production of osteoid or immature bone. Osteosarcomas are rare; in the United States, around 750 to 900 cases are identified each year, with 400 cases occurring in children and adolescents under the age of 20 [1, 2]. It is most likely to develop in children and teenagers [3]. Osteosarcoma is particularly malignant, and it grows across the body and metastasizes to the lungs [4, 5]. The 5-year rate of patients with localized osteosarcoma is 80%, while that of patients with metastatic osteosarcoma is 15-30% [6]. For those with metastatic osteosarcoma, the survival rate is very low [7]. The advances in surgery, chemotherapy, and radiation therapy are highly significant since they can decrease the occurrence of lung metastasis and increase the long-term survival in patients with osteosarcoma [8]. The poor prognosis is due to difficulty in early detection, high incidence of metastasis, and relapse. The process by which the tumor grows was not completely elucidated.
Hypoxia is a characteristic that happens when the intake of oxygen is too inadequate to support the growth rate of cancerous tumor cells [9]. Current reports have shown the crucial effect of hypoxia on cell proliferation, tumor growth, and cell differentiation [10]. Bone is one of the most vulnerable organs to hypoxia and has been shown to play a major role in metastasis, weak prognosis, and radiation tolerance of osteosarcoma [11, 12]. Nonetheless, the regulatory structure remains uncertain. The study focuses on the tumor microenvironment and immune cells that are active in tumor progression. Studies have shown that the hypoxic microenvironment can encourage the mobilization of innate immune cells in tumors and reduce adaptive immunity in the tumor microenvironment [13, 14]. As a result of the limited literature on the relationship between hypoxia and immunity, new therapeutic techniques are required.
Approximately one-third of ongoing osteosarcoma clinical studies (as reported by ClinicalTrials.gov) use targeted medications that target all parts of cellular functions. It is obvious that targeted therapy plays a critical role in the treatment of osteosarcoma. In an SJSA-1 osteosarcoma xenograft model, Chessari et al. discovered a potent isoindolinone inhibitor of the MDM2-p53 interaction, which demonstrated critical anticancer efficacy [15]. Trametinib and trastuzumab, two MEK inhibitors, were investigated in a phase I/II clinical study and a phase II clinical study for osteosarcoma or recurrent osteosarcoma, respectively [16]. However, there is still a disconnect between what we know about osteosarcoma biology and how it affects patients. In this research, we aimed at hypoxia-related genes in osteosarcoma using the TARGET and GEO databases to create a predictive model for predicting its immune microenvironment in children with overall survival (OS). This study may play a major role in aiding medical professionals in making critical clinical choices.
2. Materials and Methods
2.1. Data Selection and Analysis
A total of 88 osteosarcoma samples with RNA-Seq data and clinical details were retrieved from the TARGET (https://ocg.cancer.gov/programs/target) database. The GSE21257 collection containing 53 samples was downloaded from GEO (https://www.ncbi.nlm.gov/geo) as a validation set. The clinical features of the samples are provided in Table S1.
2.2. The Integration of Immune Cell Types
CIBERSORT, a method developed by Newman et al. (https://cibersort.stanford.edu/), is a process for estimating the relative abundances of types of cells in a mixed population of cells [17]. We calculated how proportionally immune cell types were distributed across the low and high hypoxia risk groups. The score given to each immune cell class in each study is equivalent to 1.
2.3. The Development of a Hypoxia Risk Model
The genes related to hypoxia statistically relevant in univariate analysis were then shown to be significant in multivariate analysis which produced the risk score formula [18], which is risk score = β1X1 + β2X2 + ⋯+βnXn, where X1, X2, ⋯, Xn were the level of the considered predictors and the “β” parameters were respective regression coefficients.
2.4. Survival Analysis
Overall survival between the high and low hypoxia risk groups was evaluated by a log-rank test in R software (version 3.8.2). Univariate analysis was carried to identify prognosis predictors, and multivariate analysis was performed to assess if the score for risk is predictive of survival. We gathered data to determine the model's capability to assess the outcomes of patients.
2.5. GSEA
The gene set enrichment study was conducted to decide whether there was a substantial variation in the genes that are expressed between the high and low-risk classes of the MSigDB (c5.bp.v7.0.symbols.gmt; h.all.v7.0.cymbols.gmt) [19]. The gene set permutations were tested 1,000 times to illustrate its capacity to function consistently. The phenotype label was used to forecast adverse events.
2.6. PPI Network
The STRING database was used in building a PPI network. Cytoscape was also used to measure the node degree of the PPI [20], which is the number of relationships to filter the key genes.
3. Results
3.1. Application of a Hypoxia Risk Signature to Assess Prognosis of Children with Osteosarcoma
The genes involved in hypoxic responses were derived through the GSEA study (hallmark; hypoxia), which was performed under conditions with hypoxia. To better explain the molecular interactions that arise while a stressed cell is deoxygenated, we performed PPI studies utilizing the STRING database (http://string-db.org). Figure S1 charts the top 150 genes which have the highest degrees involved in the reaction to hypoxia.
To assess the prognosis for osteosarcoma patients based on an analysis of gene expression, by using univariate and multivariate regression, we conducted the list of top 100 genes in the TARGET training set. The researchers observed that 19 genes related to hypoxia were correlated with children's OS (Figure 1(a)). Four hypoxia-related genes were selected to create a predictive model. These genes were EFNA1, P4HA1, STC2, and MAFF (Figure 1(b)). The risk score formula was constructed as
| (1) |
Figure 1.
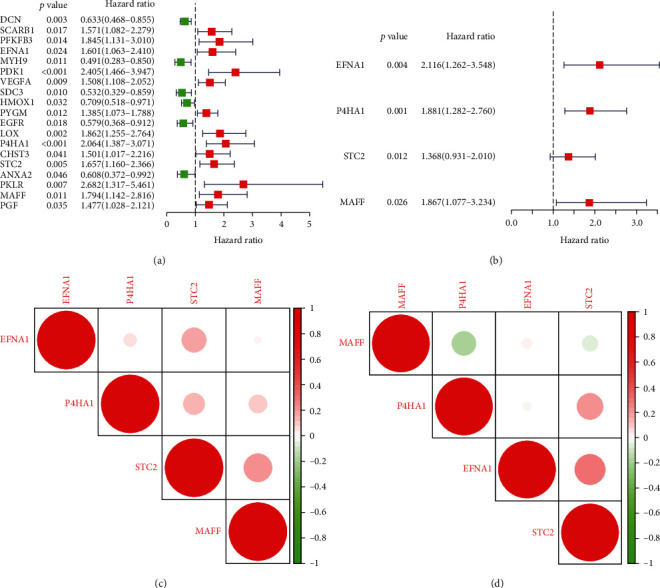
Characterizing hypoxia risk signatures in osteosarcoma to assess mortality and clinical outcomes: (a, b) development of a hypoxia susceptibility signature to assess children with osteosarcoma prognosis through univariate and multivariate Cox analyses; (c, d) Spearman relation study of a four-gene signature in TARGET and GEO databases.
All four genes were shown to be linked to each other in association with other genes (Figures 1(c) and 1(d)).
3.2. The Predictive Consequences of a Hypoxia-Induced Signature in Osteosarcoma Patients
Due to the effect on tumor cells under hypoxia, we also investigated whether the hypoxia signature has prognostic significance. Using the scores, we determined the rank and then grouped the patients into high- and low-risk categories depending on the median ranking (Figure 2(b)). Figure 2(a) reveals that the expressions of EFNA1, P4HA1, STC2, and MAFF increased as risk scores increased in the TARGET and GEO datasets, which means that children with elevated risk appear to establish a hypoxic microenvironment. Our studies suggest that the people who are in the high-risk group had a consistently higher risk of death (Figures 2(c) and 2(d)). In addition, Kaplan-Meier research was conducted to determine the predictive importance of the hypoxia-related signature in children with osteosarcoma. The figure depicts that a high risk of hypoxia was correlated with poor overall survival in the TARGET cohort, which is confirmed by GEO results.
Figure 2.
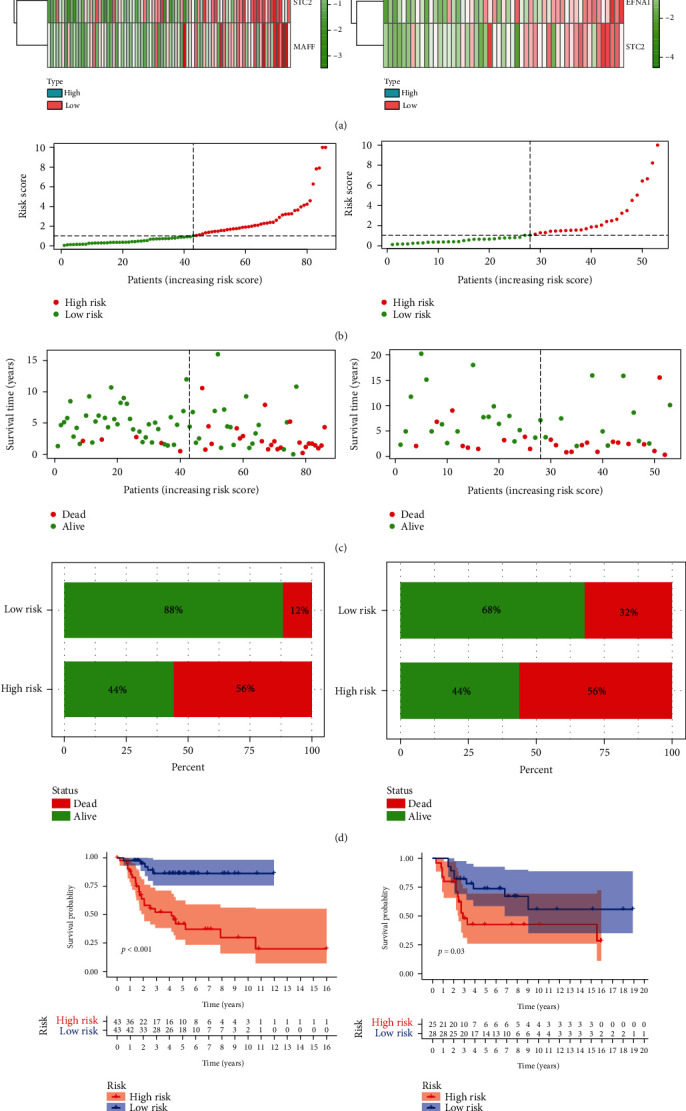
Prognostic importance of the four-gene signature in osteosarcomas: (a) a heat map displaying four-gene expression patterns in two groups from TARGET and GEO databases; (b, c) the patient status distributed in the two risk groups; (d) the death ratios for the two groups; (e) the survival curves for patients depending on risk.
3.3. Gene Expression in reaction to Hypoxia Is Associated with Clinicopathological Features in Osteosarcoma in Children
In order to explore the biological functions of hypoxia in tumorigenesis and development, we examined the relationships between four genes linked to hypoxia and certain characteristics of children with osteosarcoma, including if they had metastasized. Gene expression values and metastasis status were compared (Figure 3(a)), showing that gene P4HA1 increased in the metastasis group (Figure 3(b)).
Figure 3.
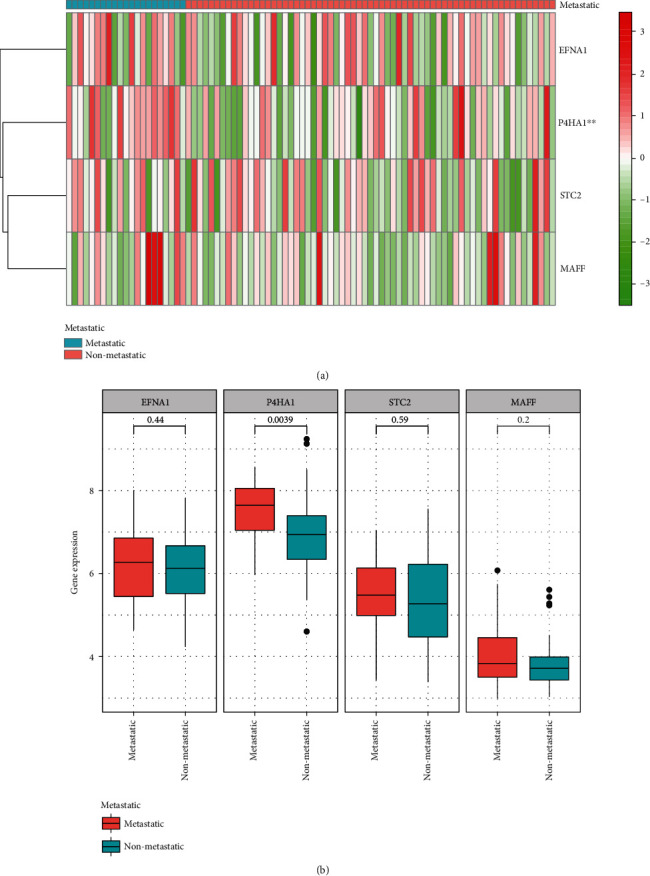
Hypoxia-associated gene is associated with clinicopathological characteristics of osteosarcoma in children: (a) a heat map displaying hypoxia gene expression profiles in TARGET database of metastatic and nonmetastatic patients; (b) the expression levels of four genes in osteosarcoma patients.
3.4. The Hypoxia Risk Signature Is an Important Instrument for Prognosis Evaluation
The risk signature was modeled using the ROC (receiver operating characteristic) curve method on survival analysis from the TARGET and GEO datasets. AUC was 0.779 at one year, 0.783 at three years, and 0.801 at five years, suggesting a strong predictive value (Figure 4(a)). This research was tested using GEO criteria (Figure 4(b)).
Figure 4.
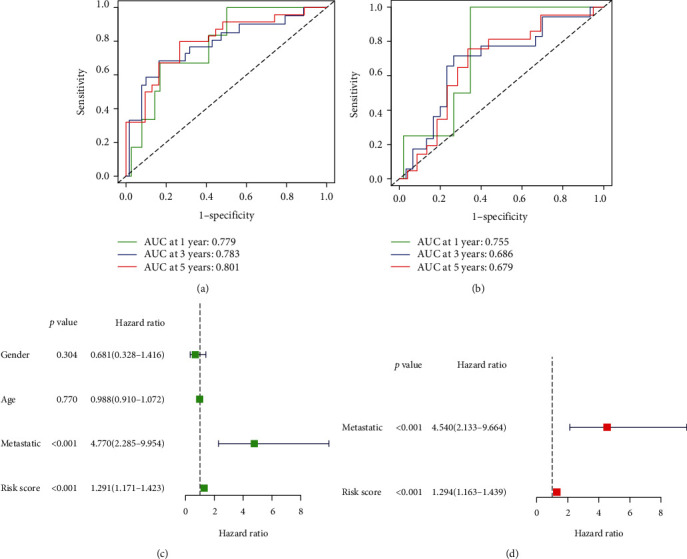
Prognostic importance of the 4-gene signature in osteosarcomas: (a, b) the predictive strength of the four-gene signature was measured with Kaplan-Meier survival curves for contrast; (c, d) univariate and multivariate Cox studies assessing the independent prognostic significance of the four-gene signature in terms of overall survival in pediatric osteosarcoma patients in the TARGET dataset.
Univariate and multivariate Cox survival analyses were done to assess the prognostic importance of a hypoxia signature on the survival of patients with osteosarcoma. The univariate study found that a high risk for hypoxia was linked to low survival (Figure 4(c)). Other factors such as metastatic status were also related to poor survival. Multivariate analysis found that a high risk of hypoxia for children with osteosarcoma was substantially correlated with a significantly lower overall survival of children with osteosarcoma.
3.5. Gene Set Enrichment Analysis
To further validate that these elevated risk groups had altered signaling mechanisms, we evaluated the GSEA comparing the high vs. low hypoxia groups. Gene sets were elevated in the high-risk group in the TARGET database, such as hedgehog signaling pathway, nitrogen metabolism, and abc transporters (Figure 5, Table S2).
Figure 5.
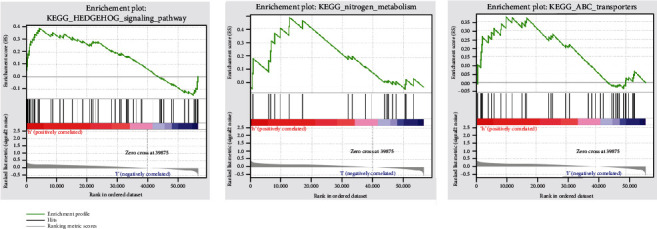
GSEA shows enrichment between the two risk categories.
3.6. Immune Landscape in Children with Osteosarcoma between High and Low Hypoxia Risk
Accumulating evidence shows that tumor microenvironments can protect tumors from immune defenses by promoting immune escape and inhibiting immune effector cells. Here, we studied whether a hypoxic hypoxia signature of an immune microenvironment could be evaluated.
CIBERSORT was used to identify variations in the immune penetration of 22 different immune cell forms between low- and high-risk children with osteosarcoma. In Figure 6(a), the outcomes of patients in the TARGET database and GEO dataset are summarized. Patients with elevated levels of hypoxia risk have a significantly higher number of immune suppression and immunosuppressive cells (Figure 6(b)). The microenvironment can be influenced by the immunosuppressive cells.
Figure 6.
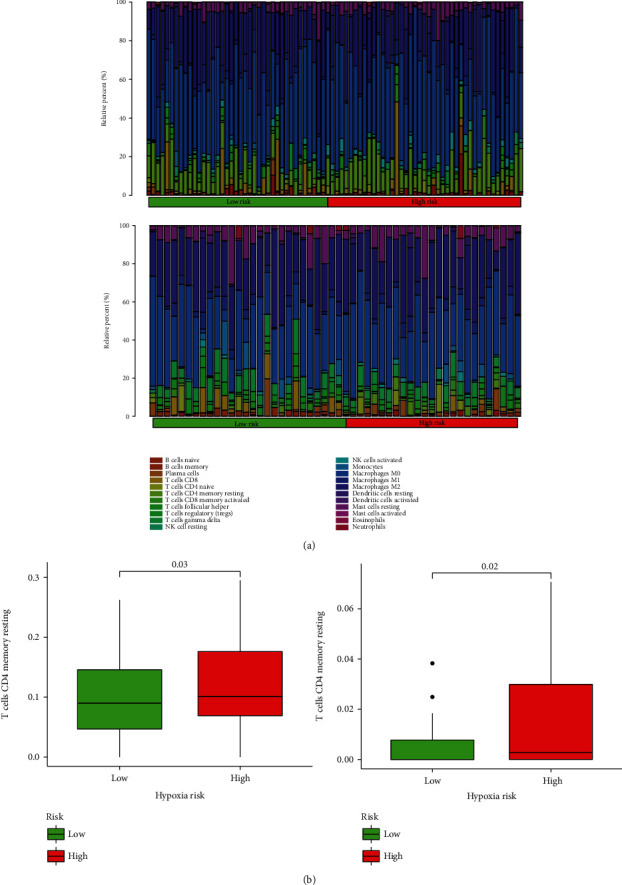
The immune landscape of children with osteosarcoma in two groups: (a) the relative frequency of different immune cells between patients in two groups with osteosarcoma; (b) box plots indicating different immune cells in children with osteosarcoma between two groups.
3.7. An Elevated Hypoxia Risk Suggests a Suppressive Immunologic Environment
The cancer-immunity cycle helps explain how cancer and immunotherapy function. The research examines how cancer cells are destroyed by the immune system: cancer cell antagonism, cancer antigen introduction, priming and activating, trafficking of T cells to tumors, intratumoral T cell aggregation, cancer cell recognition by T cells, and cancer cell death [21]. The study analyzes the expression of various genes in the high- and low-risk groups. Gene details are downloaded from the TTI database [22] (http://biocc.hrbmu.edu.cn/TIP/index.jsp). As shown in Figures 7(a) and 7(b), immune genes that lead to a reduced chance of contracting cancer were more highly expressed in the high-risk group, suggesting low activity of these mechanisms in these patients.
Figure 7.
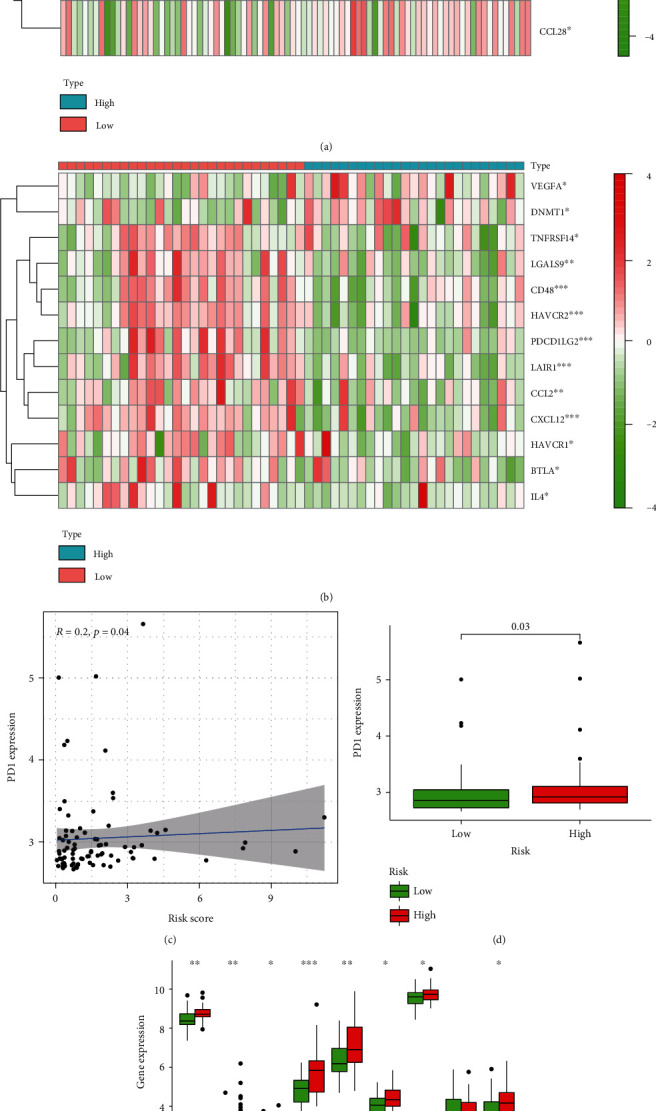
High-risk score suggests a microenvironment with immunosuppression: (a) a heat map of the gene profiles participating in cancer/immune system cycle in two groups in TARGET and GEO databases; (b) association between PD1 expression and risk score; (c) correlation between PD1 and risk score; (d) PD1 level in two groups; (e) tumor immune cytokine expression in two groups.
Focused on previous studies showing that immune checkpoint molecules can be upregulated in reaction to hypoxia [23], we conducted an analysis of the expression level of molecules in the low- and high-risk groups in the TARGET dataset. We found that PD1, which is thought to be upregulated in hypoxic environments, has high expression in the high-risk group (Figures 7(c) and 7(d)). Certain cytokines were elevated in high-risk groups attributed to hypoxia (Figure 7(e)).
4. Discussion
The treatment of osteosarcoma and the prognosis have not improved significantly over the past three decades. In recent decades, high-throughput sequencing has become an essential instrument in biomedical science and is used for prognosis forecasting, cancer recurrence monitoring, and clinical stratification since it will be important to apply to osteosarcoma and identify possible key targets for therapy.
Malignant tumors often exhibit dysregulation in cellular oxygen metabolism. It was shown in various studies that hypoxia played a role in the development of this cancer [24]. Beyond the role of oxygen supply, the precise role of hypoxia in osteosarcoma remains uncertain. In this study, we explored the role of four genes (EFNA1, P4HA1, STC2, and MAFF) profiled in a previous study on osteosarcoma. EFNA1 is a transmembrane protein, and EFNA11 expression is upregulated in a number of tumor cells, including gastric cancer, colorectal cancer, and hepatocellular cancer [25]. Large levels of an enzyme linked with cancer survivability have a weak prognosis for cancer patients. P4HA1 is an essential enzyme in hemoglobin synthesis [26]. Previously, it has been shown that P4HA1 stabilizes hypoxia-inducible factor 1α (HIF1α) by modulating the levels of its glycolytic substrates, α-ketoglutarate, and succinate, thus mediating cellular transformation of cancer cells [27]. The STC2 protein encodes a secreted homodimer and is expressed in a broad range of tissues. The protein has autocrine and/or paracrine roles [28]. Studies have demonstrated that STC2 is oncogenic in several forms of cancer. The aberrant STC2 expression has significant consequences for the prediction, incidence, metastasis, and prognosis of different cancers, including various cancers of the liver, colon, and prostate [29]. MAFF is a part of the essential leucine zipper (bZIP) transcription factor Maf family and plays a key role in numerous physiological and pathological processes, including signal transduction, hematopoiesis, central nervous system activity, and tumorigenesis [30, 31]. With the four-gene signatures, it was seen that this was an independent factor that affects the prognosis of osteosarcoma and that the model had a stronger effect in forecasting osteosarcoma.
Our GSEA revealed that hedgehog signaling pathway, nitrogen metabolism, and abc transporters were enriched in the high-risk group in the TARGET database. Onishi et al. reported that hypoxia can activate the hedgehog signaling pathway in a ligand-independent manner by upregulation of Smo transcription in pancreatic cancer [32]. Kodama et al. found that accumulation in glutamine nitrogen metabolism can contribute to the malignant progression of tumors [33]. abc transporters also have been considered for their contribution to cancer cell biology recently, and they may play a potential role in cancer development [34].
Many studies have been conducted investigating the processes by which hypoxia controls the immune response in tumor cells [35, 36]. The lack of oxygen will negatively affect the differentiation and work of immune cells and their subsequent cytokine development [37]. The immune system can detect and eliminate tumor cells [38, 39]. Nonetheless, the tumor microenvironment could interact with this immunotherapy reaction by blocking various checkpoints through which the therapy operates [40]. Regulation of NK cells and tumor antigen-specific T cells is a key step that must take place for an antitumor immune response [41]. Hypoxia prevents T and NK cell development and activation in animal models [42]. Our analysis showed that children with osteosarcoma at high risk of experiencing hypoxia had an elevated number of CD4 cells, suggesting an immunosuppressive disorder.
Cytokines have the capacity to control immune responses. Immune cell exhaustion is one of the main factors supporting tumor progression [43]. The hypoxic microenvironment of tumors is associated with VEGF and stimulates tumor development [44]. CCL28, a crucial mediator between oncogenic β-catenin signaling and the stomach tumor microenvironment, is shown to have an integral part of cancer development by either facilitating cancer cell proliferation or metastasis or by forming the tumor immune microenvironment [45]. In our research, the immune system is weakened in the high hypoxia community, contributing to greater suppression of the immune system. Immune checkpoint proteins safeguard against cancer and support tumor immunosuppression. Tumors defend themselves using immunotherapy modalities and clinical goals (e.g., CTLA-4, PD-L1, and PD1) [46, 47]. In our research, immunosuppressive cytokines and PD1 have high expressions in participants with higher risk of developing hypoxia.
This is the first analysis to construct and evaluate a risk model related to hypoxia based on four genes and then use it as a prognostic factor for osteosarcoma patients. Our results indicate that hypoxia triggers immune cell death in osteosarcoma and can contribute to better management of the condition.
Acknowledgments
We would like to thank the Core Facility of the First Affiliated Hospital of Nanjing Medical University for its help in the experiment.
Data Availability
The generated and analyzed datasets of the current research are available freely in the TARGET database (https://ocg.cancer.gov/programs/target) and GEO database (https://www.ncbi.nlm.nih.gov/gds).
Conflicts of Interest
The authors declare no competing interests.
Authors' Contributions
Data curation and writing (original draft) were performed by Feng Jiang and Xiao-Lin Miao; methodology was performed by Chu-Yan Wu; software was taken care of by Feng Yan; verification was performed by Yan Mao; visualization was performed by Xiao-Tian Zhang; and writing (review and editing) was performed by Guo-Ping Zhou. Feng Jiang and Xiao-Lin Miao contributed equally to this work.
Supplementary Materials
Table S1: sample clinical features.
Table S2: summary of GSEA results.
Figure S1: list of the top 150 hypoxia-associated genes. The size and color of the genes in the word cloud reflect how closely related they are to one another.
References
- 1.Jackson T. M., Bittman M., Granowetter L. Pediatric malignant bone tumors: a review and update on current challenges, and emerging drug targets. Current Problems in Pediatric and Adolescent Health Care. 2016;46(7):213–228. doi: 10.1016/j.cppeds.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Meyer J. S., Mackenzie W. Malignant bone tumors and limb-salvage surgery in children. Pediatric Radiology. 2004;34(8):606–613. doi: 10.1007/s00247-004-1226-4. [DOI] [PubMed] [Google Scholar]
- 3.Czarnecka A. M., Synoradzki K., Firlej W., et al. Molecular biology of osteosarcoma. Cancers. 2020;12(8):p. 2130. doi: 10.3390/cancers12082130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Setty B. A., Jin Y., Houghton P. J., Yeager N. D., Gross T. G., Nelin L. D. Hypoxic proliferation of osteosarcoma cells depends on arginase II. Cellular Physiology and Biochemistry. 2016;39(2):802–813. doi: 10.1159/000447790. [DOI] [PubMed] [Google Scholar]
- 5.Zhang D., Cui G., Sun C., et al. Hypoxia promotes osteosarcoma cell proliferation and migration through enhancing platelet-derived growth factor-BB/platelet-derived growth factor receptor-β axis. Biochemical and Biophysical Research Communications. 2019;512(2):360–366. doi: 10.1016/j.bbrc.2019.03.040. [DOI] [PubMed] [Google Scholar]
- 6.Anderson M. E. Update on survival in osteosarcoma. The Orthopedic Clinics of North America. 2016;47(1):283–292. doi: 10.1016/j.ocl.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Harrison D. J., Geller D. S., Gill J. D., Lewis V. O., Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Review of Anticancer Therapy. 2018;18(1):39–50. doi: 10.1080/14737140.2018.1413939. [DOI] [PubMed] [Google Scholar]
- 8.Isakoff M. S., Bielack S. S., Meltzer P., Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. Journal of Clinical Oncology. 2015;33(27):3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Multhoff G., Vaupel P. Hypoxia compromises anti-cancer immune responses. Advances in Experimental Medicine and Biology. 2020;1232:131–143. doi: 10.1007/978-3-030-34461-0_18. [DOI] [PubMed] [Google Scholar]
- 10.Xiong Q., Liu B., Ding M., Zhou J., Yang C., Chen Y. Hypoxia and cancer related pathology. Cancer Letters. 2020;486:1–7. doi: 10.1016/j.canlet.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Gola C., Iussich S., Noury S., et al. Clinical significance and in vitro cellular regulation of hypoxia mimicry on HIF-1α and downstream genes in canine appendicular osteosarcoma. Veterinary Journal. 2020;264 doi: 10.1016/j.tvjl.2020.105538. [DOI] [PubMed] [Google Scholar]
- 12.Prudowsky Z. D., Yustein J. T. Recent insights into therapy resistance in osteosarcoma. Cancers. 2021;13(1):p. 83. doi: 10.3390/cancers13010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar A., Deep G. Hypoxia in tumor microenvironment regulates exosome biogenesis: molecular mechanisms and translational opportunities. Cancer Letters. 2020;479:23–30. doi: 10.1016/j.canlet.2020.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Jing X., Yang F., Shao C., et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Molecular Cancer. 2019;18(1):p. 157. doi: 10.1186/s12943-019-1089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chessari G., Hardcastle I. R., Ahn J. S., et al. Structure-based design of potent and orally active isoindolinone inhibitors of MDM2-p53 protein-protein interaction. Journal of Medicinal Chemistry. 2021;64(7):4071–4088. doi: 10.1021/acs.jmedchem.0c02188. [DOI] [PubMed] [Google Scholar]
- 16.Qadeer Z. A., Valle-Garcia D., Hasson D., et al. ATRX in-frame fusion neuroblastoma is sensitive to EZH2 inhibition via modulation of neuronal gene signatures. Cancer Cell. 2019;36(5):512–527.e9. doi: 10.1016/j.ccell.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman A. M., Liu C. L., Green M. R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nature Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X., Piantadosi S., Le-Rademacher J., Mandrekar S. J. Statistical considerations for subgroup analyses. Journal of Thoracic Oncology. 2021;16(3):375–380. doi: 10.1016/j.jtho.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A., Tamayo P., Mootha V. K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D. S., Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39(1):1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 22.Xu L., Deng C., Pang B., et al. TIP: a web server for resolving tumor immunophenotype profiling. Cancer Research. 2018;78(23):6575–6580. doi: 10.1158/0008-5472.CAN-18-0689. [DOI] [PubMed] [Google Scholar]
- 23.Ohta A. Oxygen-dependent regulation of immune checkpoint mechanisms. International Immunology. 2018;30(8):335–343. doi: 10.1093/intimm/dxy038. [DOI] [PubMed] [Google Scholar]
- 24.Wigerup C., Pahlman S., Bexell D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacology & Therapeutics. 2016;164:152–169. doi: 10.1016/j.pharmthera.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Hao Y., Li G. Role of EFNA1 in tumorigenesis and prospects for cancer therapy. Biomedicine & Pharmacotherapy. 2020;130 doi: 10.1016/j.biopha.2020.110567. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X., Liu S., Yang X., Wang W., Shao W., Ji T. P4HA1 as an unfavorable prognostic marker promotes cell migration and invasion of glioblastoma via inducing EMT process under hypoxia microenvironment. American Journal of Cancer Research. 2021;11(2):590–617. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q., Yin Y., Zhao H., et al. P4HA1 regulates human colorectal cancer cells through HIF1alpha-mediated Wnt signaling. Oncology Letters. 2020;21 doi: 10.3892/ol.2020.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J., Yang Z., Huang S., Li D. BIRC7 and STC2 expression are associated with tumorigenesis and poor outcome in extrahepatic cholangiocarcinoma. Technology in Cancer Research & Treatment. 2020;19 doi: 10.1177/1533033820971676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q., Zhou X., Fang Z., Pan Z. Effect of STC2 gene silencing on colorectal cancer cells. Molecular Medicine Reports. 2019;20(2):977–984. doi: 10.3892/mmr.2019.10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X., Zhang Y., Wan X., et al. Responsive expression of MafF to β-amyloid-induced oxidative stress. Disease Markers. 2020;2020:14. doi: 10.1155/2020/8861358.8861358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen S., Yang C., Liu X., et al. RBFOX1 regulates the permeability of the blood-tumor barrier via the LINC00673/MAFF pathway. Molecular Therapy - Oncolytics. 2020;17:138–152. doi: 10.1016/j.omto.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onishi H., Kai M., Odate S., et al. Hypoxia activates the hedgehog signaling pathway in a ligand-independent manner by upregulation of Smo transcription in pancreatic cancer. Cancer Science. 2011;102(6):1144–1150. doi: 10.1111/j.1349-7006.2011.01912.x. [DOI] [PubMed] [Google Scholar]
- 33.Kodama M., Oshikawa K., Shimizu H., et al. A shift in glutamine nitrogen metabolism contributes to the malignant progression of cancer. Nature Communications. 2020;11(1):p. 1320. doi: 10.1038/s41467-020-15136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fletcher J. I., Haber M., Henderson M. J., Norris M. D. ABC transporters in cancer: more than just drug efflux pumps. Nature Reviews. Cancer. 2010;10(2):147–156. doi: 10.1038/nrc2789. [DOI] [PubMed] [Google Scholar]
- 35.Jarosz-Biej M., Smolarczyk R., Cichon T., Kulach N. Tumor microenvironment as a "game changer" in cancer radiotherapy. International Journal of Molecular Sciences. 2019;20(13):p. 3212. doi: 10.3390/ijms20133212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noman M. Z., Hasmim M., Messai Y., et al. Hypoxia: a key player in antitumor immune response. A review in the theme: cellular responses to hypoxia. American Journal of Physiology. Cell Physiology. 2015;309(9):C569–C579. doi: 10.1152/ajpcell.00207.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ceribelli A., De Santis M., Isailovic N., Gershwin M. E., Selmi C. The immune response and the pathogenesis of idiopathic inflammatory myositis: a critical review. Clinical Reviews in Allergy and Immunology. 2017;52(1):58–70. doi: 10.1007/s12016-016-8527-x. [DOI] [PubMed] [Google Scholar]
- 38.Bauer S., Groh V., Wu J., et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285(5428):727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 39.Senovilla L., Vitale I., Martins I., et al. An immunosurveillance mechanism controls cancer cell ploidy. Science. 2012;337(6102):1678–1684. doi: 10.1126/science.1224922. [DOI] [PubMed] [Google Scholar]
- 40.Koukourakis M. I., Giatromanolaki A. Tumor microenvironment, immune response and post-radiotherapy tumor clearance. Clinical & Translational Oncology. 2020;22(12):2196–2205. doi: 10.1007/s12094-020-02378-8. [DOI] [PubMed] [Google Scholar]
- 41.Chung L., Maestas D. J., Housseau F., Elisseeff J. H. Key players in the immune response to biomaterial scaffolds for regenerative medicine. Advanced Drug Delivery Reviews. 2017;114:184–192. doi: 10.1016/j.addr.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 42.Spetz J., Presser A. G., Sarosiek K. A. T cells and regulated cell death: kill or be killed. International Review of Cell and Molecular Biology. 2019;342:27–71. doi: 10.1016/bs.ircmb.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang S., Xu T., Xiong M., et al. Role of immune response, inflammation, and tumor immune response-related cytokines/chemokines in melanoma progression. Journal of Investigative Dermatology. 2019;139(11):2352–2358.e3. doi: 10.1016/j.jid.2019.03.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanotelli M. R., Reinhart-King C. A. Mechanical forces in tumor angiogenesis. Advances in Experimental Medicine and Biology. 2018;1092:91–112. doi: 10.1007/978-3-319-95294-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji L., Qian W., Gui L., et al. Blockade of β-catenin–induced CCL28 suppresses gastric cancer progression via inhibition of Treg cell infiltration. Cancer Research. 2020;80(10):2004–2016. doi: 10.1158/0008-5472.CAN-19-3074. [DOI] [PubMed] [Google Scholar]
- 46.Dalvin L. A., Shields C. L., Orloff M., Sato T., Shields J. A. Checkpoint inhibitor immune therapy: systemic indications and ophthalmic side effects. Retina. 2018;38(6):1063–1078. doi: 10.1097/IAE.0000000000002181. [DOI] [PubMed] [Google Scholar]
- 47.Alsaab H. O., Sau S., Alzhrani R., et al. PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: mechanism, combinations, and clinical outcome. Frontiers in Pharmacology. 2017;8:p. 561. doi: 10.3389/fphar.2017.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: sample clinical features.
Table S2: summary of GSEA results.
Figure S1: list of the top 150 hypoxia-associated genes. The size and color of the genes in the word cloud reflect how closely related they are to one another.
Data Availability Statement
The generated and analyzed datasets of the current research are available freely in the TARGET database (https://ocg.cancer.gov/programs/target) and GEO database (https://www.ncbi.nlm.nih.gov/gds).


