Abstract
Currently, there are over 230 different COVID-19 vaccines under development around the world. At least three decades of scientific development in RNA biology, immunology, structural biology, genetic engineering, chemical modification, and nanoparticle technologies allowed the accelerated development of fully synthetic messenger RNA (mRNA)-based vaccines within less than a year since the first report of a SARS-CoV-2 infection. mRNA-based vaccines have been shown to elicit broadly protective immune responses, with the added advantage of being amenable to rapid and flexible manufacturing processes. This review recapitulates current advances in engineering the first two SARS-CoV-2-spike-encoding nucleoside-modified mRNA vaccines, highlighting the strategies followed to potentiate their effectiveness and safety, thus facilitating an agile response to the current COVID-19 pandemic.
Keywords: SARS-CoV-2; Spike protein; MRNA; Nucleoside-modified; LNP, vaccines
1. Introduction
Since naked in vitro transcribed mRNA molecules were expressed in vivo after direct injection into mouse muscle, mRNA has been investigated extensively as a preventive and therapeutic modality [1], [2], [3].
Messenger RNA (mRNA) vaccines are designed to direct cells to express virtually any desired protein inside the host cells and tissues that can have a therapeutic or preventive benefit, potentially addressing a broad spectrum of diseases [3], [4].
Following this approach, mRNA is synthesized in a cell-free system and manufactured within standardized and controlled conditions, allowing a comparatively fast design and a relatively inexpensive and straightforward large-scale production. The complete mRNA code, for example, for the Moderna Therapeutics vaccine, was compiled in two days, and the materials for the first clinical trials were manufactured and delivered within 45 days. On March 16, 2020, the first trial participants were vaccinated, just 66 days after the SARS-CoV-2 genome was made public on January 10, 2020 [5].
Delivery of mRNA is safer than whole viral particles, as the former is a non-infectious transient carrier of information. Furthermore, recombination among single-stranded RNA species is rarely possible, and cytosolic mRNA cannot be integrated into the host genome [6]. Moreover, mRNA exhibits self-adjuvating properties as a result of its capacity to bind to pattern-recognition receptors (PRRs) like Toll-like receptor 7 (TLR7), promoting cellular immunity [7], and at the same time provides the technological basis to deliver in a single molecule, open reading frames encoding a wide variety of antigens, modulators, and cell-signaling factors [8].
mRNA therapeutics combine safety with fine dose control and the potential for multiple administrations with a reduced risk of pre-existing or anti-vector immunity [9]. For example, upon intravenous injection into mice of a single dose of lipid nanoparticle (LNP)-encapsulated nucleoside-modified mRNAs encoding the heavy and light chains of the anti-HIV-1 neutralizing antibody VRC01, Pardi and colleagues observed high levels of functional antibody in the serum, protecting the humanized mice from HIV-1 infection. [9], [10]. Thran and colleagues employed an unmodified-mRNA–LNP complex to protect from otherwise lethal challenges with rabies virus, botulinum toxin, and a B cell lymphoma through a system expressing three monoclonal antibodies [11]. Furthermore, nucleoside-modified mRNA-LNP influenza vaccines induced humoral immune responses in the recipients, with a safety profile comparable to an influenza vaccine using inactivated influenza virus [12], [13]. The above are just a few representative examples illustrating the potential of mRNA vaccines to provide successful prophylactic approaches.
Eleven months after discovering the SARS-CoV-2 virus, it has been confirmed that a mRNA vaccine for coronavirus disease 2019 (COVID-19) is effective and safely tolerated [14]. More than three decades of scientific advances into RNA biology, chemical modifications, lipid-based delivery systems, and nanotechnology have led to fast progress in the development of mRNA-based vaccines against infectious diseases and their move into clinical trials [3], [4], [9], [15], [16].
Herein, we overview molecular aspects of engineering the two lipids nanoparticle-encapsulated, nucleoside-modified mRNAs vaccines for COVID-19, highlighting the main differences between Moderna's mRNA-1273 vaccine and the Pfizer/BioNTech (BNT162b2) vaccine in terms of the strategies followed to optimize their effectiveness and safety.
2. Relevant molecular features of the SARS-CoV2 spike protein
The SARS-CoV-2 RNA genome is introduced in the host cell using a highly glycosylated homotrimeric S protein (spike glycoprotein) to achieve fusion with target cell membranes [17], [18].
The transmembrane SARS-CoV S-protein spike trimer possesses an N-terminal receptor-binding S1 subunit and a C-terminal S2 subunit. The S1 subunit is subdivided into the N-terminal domain (NTD), followed by the receptor-binding domain (RBD) and two structurally conserved subdomains (SD1 and SD2) [19]. Binding to the host receptor via the RBD in S1 is followed by proteolytic cleavage of the spike by host proteases into the membrane-associated S2 part and the distal S1 part, which remain associated through non-covalent interactions [20], [21]. S1binds the angiotensin-converting enzyme 2 (ACE2) on the host cell surface, while S2 mediates membrane fusion [21], [22]. Proteolytic cleavage of the S protein by furin or other cellular proteases like TMPRSS2 [21] at the S1/S2 site is essential for the infection, as it separates two functions of spike [23]. For example, the furin cleavage site has been shown to be essential for efficient viral entry into human lung cells, especially in terms of cell-cell fusion, to form a syncytium to facilitate viral spread from one cell to another [24].
The aforementioned structural insights reveal the S protein as a crucial target for vaccine development, therapeutic antibody generation, and clinical diagnosis of COVID-19 [25], [26]. Specifically, a vaccine targeting the S protein could prevent the spread of SARS-CoV-2 by impeding its initial activation through blockage of S protein binding to the host receptor ACE2 [17], [25], [26], mainly if the antibody titer against S protein is high enough to prevent the virus from being engulfed into endosomes or undergoing fusion at the host cell surface [25], [27].
3. Key features for the mRNA-based vaccines engineering
mRNA vaccines constitute a subtype of nucleic acid vaccines, which is divided into two categories: self-amplifying RNA (encoding both an antigen and an RNA replicase that amplifies the recombinant mRNA) and non-replicating mRNA (encoding only the antigen) [28], which has been the model approved and employed in the current vaccines against COVID-19.
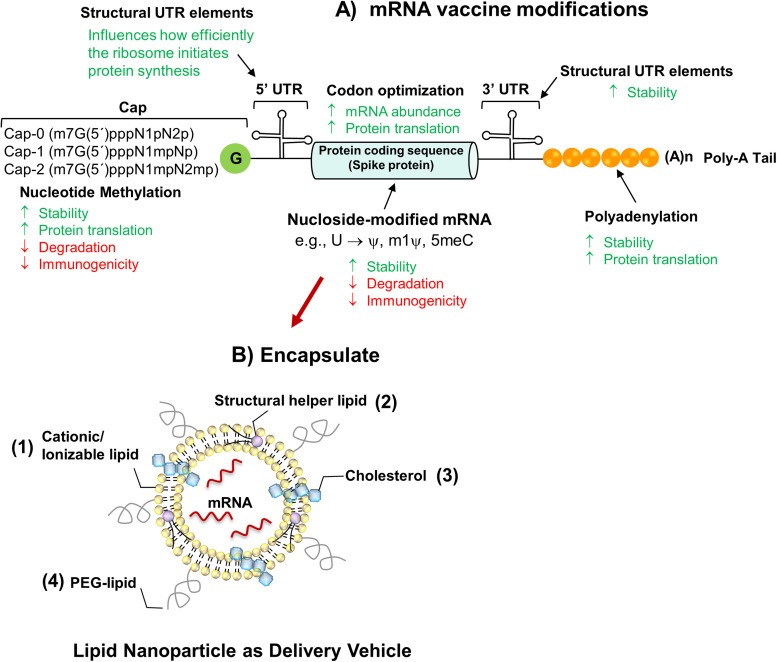
The basic structure of non-replicating mRNA closely resembles “mature” eukaryotic mRNA. Usually, it consists of an in vitro-transcribed mRNA (IVT-mRNA) is composed of the antigen-encoding open Reading frame (ORF) flanked by the 5′ and 3′ untranslated regions (UTRs), a 7-methyl guanosine 5′cap structure incorporated to the first nucleotide of the transcript, and a 3′poly(A) tail [9] ( Fig. 1A). Recent remarkable progress has been achieved within this approach, in terms of 1) optimization of the half-life of mRNA, 2) magnitude and duration of protein expression, 3) modifications of the 5′ and 3′ UTRs to fine-tuning of the immunogenicity, 4) optimization of the length of the poly-A tail, 5) incorporation of modified nucleosides in the coding sequence, and 6) capping strategies [9], [29], [30]. The above improvements have made possible the adaptation of this strategy to many mRNA-based vaccines, such as the current COVID-19 mRNA vaccines.
Fig. 1.
Design of the nucleoside-modified SARS-CoV-2 mRNA-LNP vaccines. A) Design of the nucleoside-modified SARS-CoV-2 mRNA-LNP vaccines. A) The critical structures of mRNA are the 5ʹ cap (e.g., the 7–methylguanosine cap), the 5ʹ and 3ʹ untranslated regions (UTRs), sequence encoding the full-length S protein and the poly(A) tail. mRNA cap is incorporated either in one step during transcription in the presence of CAP analogs (e.g., Clean-Cap) or in two steps, after IVT-mRNA production, by enzymatic capping reaction. Replacement of native nucleosides in in-vitro-transcribed mRNA with chemically modified versions reduces immunogenicity and increases translation efficiency. mRNA-1273 and BNT162b2 are nucleoside-modified transcripts with substitution of uridines for N1-methyl pseudouridine is (1mψ). Each of these structural elements of mRNA can be optimized and modified to modulate the stability, translation capacity, and immune-stimulatory profile of mRNA. B) Schematic depiction of mRNA vaccine encapsulated into LNP formulations for improved in vivo mRNA delivery, which are typically composed of (1) an ionizable or cationic lipid [e.g., SM-102 (mRNA-1273) and ALC-0315 (BNT162b2)], bearing tertiary or quaternary amines to encapsulate the polyanionic mRNA; (2) a helper lipid [LNPs of Moderna and BioNTech contain the same helper lipid 1,2-distearoyl-snglycero-3-phosphocholine (DSPC)] that resembles the lipids in the cell membrane; (3) cholesterol to stabilize the lipid bilayer of the LNP; and (4) a polyethylene glycol (PEG)-lipid [(2-[(polyethylene glycol)− 2000]-N,N-ditetradecylacetamide (PEG2000-DMA) in BNT162b2 or 1,2-dimyristoyl-rac-glycero3-methoxypolyethylene glycol-2000 (PEG2000-DMG) in mRNA-1273] to lend the nanoparticle a hydrating layer, improve colloidal stability, and reduce protein absorption.
3.1. mRNA capping
Eukaryotic mRNAs, as well as RNA viral genomes, have a 7-methylguanosine (m7G) cap at the 5′ end of the mRNA sequence (m7GpppN structure), attached to the first RNA nucleotide through a 5′,5′-triphosphate bridge (ppp) during mRNA in vitro transcription [31], [32].
The cap structure plays essential functions in mRNA translation by recruiting translation initiation factors, and different 5′ caps can be incorporated into mRNAs [30] (Fig. 1A). For example, CAP0 protects endogenous mRNA from nuclease attack and is also involved in nuclear export and translation initiation. Both CAP1 and CAP2 are two 5′ caps that contain additional methyl groups on the second or third ribonucleotide and are less immunogenic than CAP0 [32].
Cap analogs such as anti-reverse-Cap-analog [CAP-0 (N7MeGpppN) structure] or Clean Cap [CAP-1 (N7MeGpppN2′-OMe) structure] [33] are the two main approaches used to produce capped mRNA in in-vitro transcription (IVT) [30]. Uncapped (5′ppp or 5′pp) or inadequately capped mRNAs can be recognized by PRRs such as Retinoic acid-Inducible Gene-I-like (RIG-I-like) receptor [34]. However, treatment of the uncapped IVT-mRNAs with phosphatases might avoid recognition by RIG-I, where the latter recognizes the 5′ triphosphate of uncapped mRNA, thus avoiding mRNA translation [30], [34].
3.2. Untranslated Regions
The 5′- and 3′-UTR elements flanking the coding sequence influence the stability and translation of mRNA. Optimization of the 5′-UTR of mRNA (Fig. 1A), whose secondary structures are recognized by cell-specific RNA binding proteins, can maximize the translational yield of mRNAs useful for therapeutics and vaccines [35]. The use of α-globin or β-globin UTRs from Xenopus laevis or human has been a standard approach in mRNA vaccine design due to their high stability [36]. However, recently, Orlandini von Niessen and colleagues, through the Systematic Evolution of Ligands by Exponential enrichment (SELEX) method, developed a cell culture-based systematic selection process to identify novel 3′ UTR motifs that could induce approximately threefold higher protein production via IVT compared to the classical human β-globin 3′-UTR used in effective mRNA vaccines [13]. Therefore, these regulatory sequences can be derived from viral or eukaryotic genes and modified through the systematic enrichment of naturally occurring RNA sequences to improve the protein synthesis efficiency of mRNA in vitro and in vivo and to significantly increase the half-life of therapeutic mRNAs [13], [37].
3.3. Optimizing translation of IVT-mRNA
The antigen-coding sequence can be modified at specific locations and/or codon-optimized to improve the translation. IVT-mRNA can also be engineered to direct the product carrying the antigen to the desired compartment or to keep it in a soluble form as well as to be optimized to improve antigenicity or immunogenicity, either through point mutations, deletion of some segment, or deletion of putative glycosylation sites [29], [38], [39].
In terms of codon usage, replacing rare codons with frequently used synonymous codons can be recognized by abundant cognate tRNA in the cytosol is a recurrent strategy to increase the protein expression level of mRNAs [9], [40]. ORF sequence can also be modified to obtain comparable ratios for every codon found naturally in genes encoding highly expressed proteins in human cells [30], and also G•C content optimization has been shown to increase steady-state mRNA levels in-vitro and protein expression in-vivo [41], [42]. For example, Acuitas Therapeutics has used LNPs to deliver erythropoietin (EPO)-encoding mRNA rich in GC codons into pigs, demonstrating that it can elicit EPO-related responses with no associated immunogenicity [41].
3.4. Poly-A tail
One of the last steps of mRNA biogenesis is poly-A tail synthesis, an essential element for efficient translation. According to earlier studies, the addition of long poly-A sequences (~250 units in length) is preferable for enhancing mRNA stability. However, poly-A sequences of 120 units provide more stable IVT-mRNAs [43] and more efficient translation than shorter tails in human monocyte-derived DCs [37], while in human primary T cells, a poly-A tail longer than 300 nucleotides is more conducive to efficient translation [44].
3.5. Purification of IVT-mRNA
After transcription, the sample contains the antigen-encoding mRNA and short transcripts derived from abortive cycling during transcription initiation or incomplete transcripts derived from premature termination during the elongation process and other reaction components such as triphosphate nucleotides, salts, and enzymes [45], [46]. A high-performance liquid chromatography (HPLC) step allows the separation of the intended mRNA from shorter and longer transcripts, yielding a pure single mRNA product [46], [47]. Thus, implementing this purification by chromatography within a Good Manufacturing Practice (GMP) during the production process of mRNA increases the activity of mRNA preparations several-fold in terms of protein expression in vivo [47]. However, although usually, HPLC purification is amenable to small-scale processes, it can be suboptimal for large-scale manufacturing [15]. To address this issue, Baiersdorfer and colleagues described a simple, inexpensive, and highly scalable method for mRNA purification via the adsorption of double-stranded RNA contaminants employing polysaccharide cellulose with a performance comparable to HPLC to remove double-stranded RNA (dsRNA) contaminants from IVT mRNA. Therefore, such a method can be an alternative to HPLC purification at laboratory and industrial scales of therapeutic mRNAs [48].
3.6. Chemical modifications in RNA nucleosides
Protection of antigen-coding IVT mRNAs by chemical modifications, encapsulation, or other means against degradation by extracellular RNases are present in the skin and muscle tissues is critical [29]. Karikó and colleagues have not only shown that nucleoside-modified mRNA is translated more efficiently than unmodified mRNA in-vitro [49], particularly in primary DCs, and in-vivo in mice [50] but also that the incorporation of modified nucleotides in the mRNA sequence, such as methylated nucleosides or pseudouridine, strongly reduces the immune-modulatory capacity of endogenous mRNA [51].
Cytidines can also be replaced with numerous chemical modifications, including 5-methylcytidine (m5C); uridines can be converted into 5-methyluridine (m5U), 2-thiouridine (s2U), 5-methoxyuridine (5moU), pseudouridine (ψ) and N1-methylpseudouridine (m1ψ), while adenosines can be replaced by N1-methyladenosine (m1A) and N6-methyladenosine (m6A) [9], [52].
Post-transcriptional modifications naturally occurring in the mRNA nucleotides sequence prevent its immune detection, which, in contrast, is commonly triggered by pathological or invading mRNA. Recognition from innate immune sensors can be avoided by incorporating chemically modified nucleosides, such as ψ [53] and m1ψ [50] (present in transfer and ribosomal RNAs) to prevent activation of TLR7, TLR8, and other innate immune sensors [54], hence reducing type I interferon production [51].
Moreover, the highest levels of protein production and immunogenicity are observed when nucleoside-modified mRNA is also purified by HPLC or fast protein liquid chromatography (FPLC), where aberrant double-stranded transcripts are removed [46], [55]. For example, Pardi and colleagues demonstrated that a single intradermal injection of LNP-formulated mRNA encoding Zika virus prM-E, modified with 1-methylpseudouridine and FPLC purification, elicited protective immune responses in mice and rhesus macaques with as little as 50 μg (0.02 mg kg−1) of the vaccine in macaques [56].
Although these modifications can enhance the efficacy of mRNA-based vaccines, their inappropriate incorporation can negatively affect the translation of transcripts [57].
3.7. Lipid-nanoparticles formulations used for mRNA vaccines delivery
Thanks to their nano-size and physicochemical characteristics, biocompatible rationally engineered materials can protect drug cargos from degradation and confer controllable biodistribution and intracellular localization and release [6], [58].
Lipid nanoparticles (LNPs) containing ionizable lipids represent the most advanced nucleic acid carrier enabling package, protection, and efficient in vivo delivery of mRNA, are currently being deployed as delivery vectors for mRNA vaccines in clinical trials [29], [59]. For example, it has been demonstrated that lipid nanoparticles protect the mRNA from enzymatic attack and enhance cell uptake and expression by up to 1000-fold compared to naked mRNA when administered to animal models [60], [61].
Ionizable lipids, previously optimized for the delivery of small interfering RNAs (siRNAs), have a neutral to mild cationic charge under physiological pH conditions, thereby reducing nonspecific lipid-protein interactions and facilitating the nucleic acid release in the cytosol. This property gives those advantages over lipids with a permanent charge of reduced toxicity and prolonged blood circulation lifetime [8], [58]. The other lipidic species are considered "helper lipids" because they may affect the structural arrangement of complexes LNP-mRNAs (Fig. 1B) to improve their stability or to facilitate their intracellular uptake and the cytosolic entry [16], [62].
Therefore, typical LNP formulations consist of an RNA-lipid complex, lipids that provide structural rigidity, and a lipidized polymer coating able to modify the particles' surface properties [4], [62] (Fig. 1B). As an example, cationic liposomes composed of the cationic lipid 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), together with the helper lipid 1,2-dioleoyl-snglycero-3-phosphoethanolamine (DOPE) in combination with mRNA, have been developed as mRNA-based vaccines [4], [63], [64].
Kranz et al. demonstrated that mRNA and DOTMA/DOPE lipid formulations, designed to change its surface charge from positive to negative gradually, protected the mRNA from the action of extracellular ribonucleases, allowing it to accumulate in the spleen efficiently and to be subsequently released into DCs after systemic administration, thus inducing an antigen-specific immune response [65].
mRNA uptake mechanisms seem, however, to be influenced by cell type, and the physicochemical properties of the mRNA–carrier complex have also shown to influence cellular mRNA delivery and organ distribution [66], [67].
Lipid systems formulations for mRNA vaccines commonly contain ionizable cationic lipids, phospholipids, cholesterol, and lipid-anchored polyethylene glycol (PEG) [8], [59] (Fig. 1B). Dlin-MC3-DMA, also known as MC3, is an ionizable amino lipid developed for siRNA [68] that has recently been employed in developing of LNPs for the delivery of mRNA vaccines [61].
Phospholipids are known to facilitate the membrane formation and disruption processes that facilitate endosomal escape. The ionizable lipids being neutral at physiological pH avoid any cationic charge in circulation, becoming protonated in the endosome at pH ~6.5 to prompt endosomal release [61]. The ionizable lipid complexes with the mRNA form a core structure while helper lipids (such as a phospholipid, cholesterol, and/or a PEG-lipid) envelop the lipid–mRNA complex. In turn, the PEG-lipid protects the nanoparticle shell [69], [70] (Fig. 1B). Thus, cholesterol functions as a stabilizing element in LNPs and plays a crucial role in the transfection of cells [71]. Lipid-anchored PEGs preferentially deposit on the LNP surface, stabilizing it sterically, also reducing nonspecific binding to proteins [8], [70]. Moreover, a higher PEG content can increase the blood circulation time of LNPs by reducing a specific absorption to plasma proteins while reducing cellular uptake and interaction with the endosomal membrane, thus enhancing mRNA delivery efficiency [72], [73].
4. Current nucleoside-modified SARS-CoV-2 mRNA-LNP vaccines
4.1. Moderna/US NIAID (mRNA-1273) COVID-19 vaccine
Boston-based Moderna Therapeutics partnered with the National Institute of Allergy and Infectious Diseases (NIAID) to produce the first potential mRNA vaccine candidate against SARS-CoV-2, which entered clinical trials 66 days after identifying the genome virus sequence [5].
The mRNA-1273 vaccine candidate is a lipid nanoparticle–encapsulated, nucleoside-modified mRNA–based vaccine that harbors only the elements required for transient expression of the S-2P antigen (Fig. 1A), consisting of the SARS-CoV-2 spike (S) glycoprotein with a transmembrane anchor and an intact furin S1–S2 cleavage site [5]. Two consecutive proline substitutions stabilize S-2P at amino acid positions 986 and 987 of the prefusion spike conformation at the top of the central helix in the S2 subunit [5].
Moderna’s platform applies bioinformatic, biochemical, and biological screening approaches, most of which have been invented internally, and they aim to optimize the amount of protein produced per mRNA. Specifically, Moderna has identified proprietary sequences for the 5’-UTR region to increase the likelihood that a ribosome bound to the 5’-end of the mRNA transcript will find the desired start codon and reliably initiate translation of the coding region [74] ( Table 1).
Table 1.
The main differences, limitations, and optimizations suggested for Moderna's mRNA-1273 Vaccine and the Pfizer/BioNTech (BNT162b2) vaccine.
| Feature | mRNA-1273 | BNT162b2 | Limitations | Optimizations needed |
|---|---|---|---|---|
| Size (nucleotides) | 4004 | 4284 | The assembled contig includes the full coding region but could lack some sequence from the ends of the Moderna vaccine RNA[75]. | |
| Cap | Cap 1 structure[76] | Cap1 structure | None | |
| 5’-UTR | Not revealed GC-rich tract CCCCGGCGCC was included, just upstream the Kozak consensus[77]. |
A fragment of 35-nt from 5’-UTR of the highly expressed human gene α-globin (HBA1) was incorporated. | The GC-rich tract and the secondary structure this propitiates in mRNA-1273 may reduce translation initiation efficiency and overall protein output[78]. | Potential optimization of the speed of the 5'-UTR to load ribosomes onto the mRNA. |
| Kozak sequence | GCCACCAUG | GCCACCAUG | None | Suppression of secondary structures spanning the start codon for efficient translation initiation[77]. |
| Coding sequence | GAG codon in the spike protein gene replaced all GAA codons[77]. | 14 GAA codons unchanged[77] | a) ψ wobbles more in base-pairing than U and can hybridize with A and G and, to a lesser extent, with C and U[79]. b) Excessive use of CGG (is not an optimal codon) in Moderna's mRNA-1273 vaccine[77]. |
Replacement of all or most synonymous codons according to usage in highly expressed genes, either ubiquitously (e.g., CGC instead of CGG), or in muscle tissues[77]. |
| Stop codon | Three different stop codons are used (ψGAψAAψAG)[77]. | Two consecutive UGA stop codons are used (ψGAψGA)[77]. | a) Uracil replacement by ψ increases the rate of misreading of stop codons by a near-cognate tRNAs[79]. b) Decrease in the number of immunogenic proteins[77]. c) Production of larger proteins with an unknown destination and with potentially deleterious effects[77]. |
a) UAA is the more efficient stop codon[80]. b) The optimal stop signal should be UAAA instead of UGAU/UAGU/UAAU in both mRNA vaccines[77]. |
| 3’-UTR | The 110-nt 3’-UTR of human β-globin gene (HBA1) placed between the last stop codon and the Poly(A) tail[77]. | The 3'-UTR comprises the human AES/TLE5 gene segment (136-nt), inserted 6-nt downstream the second stop codon, and the human mitochondrial 12S rRNA (mtRNR1) segment (139-nt), positioned downstream of the first[77]. | Decrease the number of destabilizing elements of the mRNA (e.g., reducing the number of predicted miRNAs binding sites introducing point mutations). | |
| Poly-A tail | Not revealed | A30(GCATATGACT)A70 | Increase and sustain protein expression. | |
| Dosage | 100 µg | 30 μg | The Pfizer/BioNTech vaccine likely produces about 3.3 times as many Spike proteins as the Moderna vaccine[77]. | Decrease the amount of vaccine mRNA (μg) by increasing the mRNA translation efficiency. |
Moderna implemented the incorporation of a mΨ modified nucleotide into the mRNA-1273 and removed dsRNA fragments during the mRNA production process, which strongly reduces the innate immune signaling in response to mRNA through decreased activation of TLR signaling and cytosolic RNA sensors [81].
This preparation employs four lipid components to encapsulate the mRNA and form the nanoparticle complex, although the ratios at which these elements are combined have not yet been disclosed. The aforementioned lipid components are SM-102 [Heptadecan-9-yl 8-((2-hydroxyethyl)(8-(nonyloxy)− 8-oxooctyl)amino)octanoate)]; PEG2000-DMG (1,2- dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000); cholesterol; and DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine). The preparation also contains tromethamine, tromethamine hydrochloride, acetic acid, sodium acetate, and sucrose [82], [83].
The mRNA-1273-LNP follows the prototype MC3 LNP but replaces MC3 with Lipid H (SM-102) [84]. The ionizable lipids that have replaced MC3 for mRNA delivery allow more significant endosomolytic activity by inducing more branching into the alkyl tails, as Lipid H and Lipid 5 display three alkyl tails, instead of two in MC3. Presumably, this enhances cone-shaped morphology; thus, the cross-section of the lipid tails is larger than that of its head group, leading to greater endosomal release [61].
The Phase 3 trial of mRNA-1273 was published in the New England Journal of Medicine at the end of December 2021, confirming the preparation's 94.1% efficacy and safety profile [85] ( Table 2).
Table 2.
Comparison of the first two mRNA-based COVID-19 vaccine candidates.
| Developers | Vaccine | Vaccine platform | Coronavirus target | Type of Candidate Vaccine | Emergency use authorization | Dosage, schedule, and route of administration | Confirmed efficacy | Clinical trial registry number |
|---|---|---|---|---|---|---|---|---|
| Moderna/NIAID | mRNA-1273 | mRNA-based therapeutics | SARS-CoV-2 Spike protein | LNP-encapsulated nucleoside-modified mRNA | US (Dec 18, 2020), Canada (Dec 23, 2020), Israel (January 4, 2021), EMA (Jan 6, 2021) | Two intramuscular injections (100 μg per dose), 28 days apart | 94.1% (measured starting from 14 days after the second dose) | NCT04470427 |
| BioNTech/Pfizer | BNT162b2 | mRNA-based therapeutics | SARS-CoV-2 Spike protein | LNP-encapsulated nucleoside-modified mRNA | UK (Dec 2, 2020), Canada (Dec 9, 2020), US (Dec 11, 2020), EMA (Dic 21, 2021), other countriesa | Two intramuscular doses, 21 days apart (30 μg per dose) | 95% (measured starting from seven days after the second dose)b | NCT04368728 |
US, The United States; EMA, European Medicines Agency; UK, The United Kingdom
December 31, 2020 (Argentina, Ecuador, Chile, Panama, Mexico, Costa Rica, Kuwait, Singapure, Switzerland, South Arabia)
Differences in efficacy (between 94.5% and 95%) are small compared to the potential variables between the studies.
4.2. Pfizer/BioNTech/Fosun Pharma (mRNA-BNT162b2/Comirnaty) COVID‑19 vaccine
BNT162b2 is the second candidate mRNA platform, developed by Pfizer in collaboration with German-based Biopharmaceutical New Technologies (BioNTech) and Shanghai-based Fosun Pharma [86]. This vaccine is also based on a nucleoside-modified mRNA molecule that encodes the stabilized prefusion form of the SARS-CoV-2 Spike protein encapsulated in a LNP to enhance its uptake by host immune cells [87].
The Pfizer–BioNTech is undergoing the different stages of vaccine production at its three-plant network [88], [89]. The first stage involves cloning the SARS-CoV segment spike protein gene into DNA plasmid vectors, then their propagation into Escherichia coli. After four days of growth, bacterial cultures are lysed, and the DNA plasmid is purified over a week and a half [88].
The second stage is being conducted at Andover, Massachusetts, in the United States and Germany [89]. The DNA is utilized as a template to build the desired mRNA strands. A plasmid DNA template for in-vitro transcription contains at least 1) a bacteriophage promoter, 2) an ORF, 3) a poly[d(A/T)] sequence transcribed into poly(A), and 4) a unique restriction site for linearization of the plasmid to ensure defined termination of transcription [2]. A chromatographic purification step removes mainly short or double-stranded RNA molecules and yields a pure single mRNA product. This purification step within the production process of mRNA can increase their expression several-fold in vivo [47]. Once transcribed, the mRNA is transported in different packages on a scale of 5–10 million doses for the next stage, the third is conducted at plants in Kalamazoo, Michigan, in the United States and Puurs in Belgium. There, the mRNA-containing lipid nanoparticles are constructed and sterilized [90].
BioNTech began developing its SARS-CoV-2 vaccine with four mRNA-encoded immunogens, two of which were nucleoside modified [61]. BNT162b1 consists of a sequence (approximately 1 kb) encoding the receptor-binding domain of the spike protein, modified by a foldon trimerization domain to increase immunogenicity by the multivalent display [61]. The RNA sequence of the vaccine candidate BNT162b2 is 4284 nucleotides in length, with a molecular weight of approximately 1388 kDa [91]. It consists of a modified 5′ CAP1 structure (m7G+ −5′-ppp-5′-Am); a 5’ UTR region (ΨCΨΨCΨGGΨCCCCACAGACΨCAGAGAGAACCCGCC) derived from the 5’-UTR of the highly expressed human gene α-globin, as well as an optimized downstream Kozak consensus GCCACCAUG instead of ACCAUG (where AUG is the start codon); a codon-optimized ORF encoding the full-length spike protein of SARS-CoV-2 (nucleotides 55–3879), including the signal peptide (nucleotides 55–102) and two proline substitutions designated as "2 P" (K986P and V987P) [91]. The latter causes the spike to adopt a prefusion-stabilized conformation reducing its membrane fusion ability, resulting in increased expression and stimulation of neutralizing antibodies [14], [92]. Towards the 3' end, it has a 3' UTR (nucleotides 3880–4174), which is a combination of the AES (Amino-terminal enhancer of split) gene sequence and mtRNR1 (mitochondrial 12S ribosomal RNA), selected to increase protein expression and mRNA stability [13], [91]. These sequences are followed by a 30-mer poly(A) tail, a 10-nucleotide linker sequence (GCAUAUGACU), and 70 other adenosine residues (nucleotides 4175–4284) to increase and prolongate the protein expression [91] (Table 1).
During the review process of the current manuscript, a paper by Xuhua Xia [77] appeared, critically analyzing both mRNA-based COVID-19 vaccines in terms of design optimization. In Table 1, we summarize crucial design aspects highlighted by Xuhua Xia.
The incorporation of chemically, naturally occurring, modified nucleosides, including but not limited to pseudouridine and 1–methylpseudouridine, prevents activation of TLR7, TLR8, and other innate immune sensors, thus reducing IFN-I signaling [9], [49], [51].
Consequently, in the Pfizer–BioNTech COVID‑19 vaccine, uridine residues were replaced by 1-methyl-3′-pseudouridine modifications [91] (Fig. 1A).
The lipid nanoparticle delivery system in the trials of BioNTech is composed of the cationic lipid ALC-0315 (licensed from Acuitas Therapeutics) combined with the phospholipid 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, and a PEG–lipid [86] (Fig. 1B).
BNT162b2 possesses the ability to mimic the process by which natural SARS-CoV-2 viral infection occurs and confers protection against COVID-19 by the transient expression of the full-length spike antigen, once expressed on the surface of the host cells, to induce neutralizing antibody generation and cellular immune responses against it [93].
The vaccine candidate BNT162b2 was chosen as the most promising in terms of better safety profile among three others with similar technology developed by BioNTech [94], [95]. Ninety to one hundred percent of vaccine efficacy was observed across subgroups defined by age, sex, race, ethnicity, baseline body-mass index, and the presence of coexisting conditions [93].
All participants received two doses, 21 days apart, of either BNT162b2 or placebo, delivered into the deltoid muscle [93] (Table 2). A regimen of two-dose BNT162b2 conferred 95% protection against COVID-19 in subjects 16 years of age or older, showing a safety profile observed for other viral vaccines for about two months [93].
5. Production of the nucleoside-modified SARS-CoV-2 mRNA-LNP vaccines
Development of an mRNA vaccine is faster than inactivated vaccines, attenuated live vaccines, and subunit vaccines [96], and speed, the feasibility of manufacturing scale-up, and global access are three criteria that should be considered [97].
Globally, Pfizer/BioNTech expects to produce about 2 billion doses in 2021 (compared with the more than 50 million doses of vaccines generated during 2020 [98]. However, at the end of March of this year, they raised to 2.5 billion doses [99]. Instead, Moderna has announced that it would boost the production of its vaccines, stating that it would produce no less than 800 million doses in 2021 (compared with the 20 million doses generated by the end of 2020) and potentially triple its production in 2022 due to improvements in vaccine manufacturing methods [100].
Interestingly, Pascolo [101] has recently calculated that to produce one billion doses of prophylactic vaccines (to vaccinate ~10% of the population, 500 million people twice), Moderna will need to generate 100 kg of IVT mRNA (100 μg per dose) while BioNTech will need to produce 30 kg of IVT mRNA (30 μg per dose). Thus, the necessary volumes of in vitro transcription reactions will be at least 20,000 and 6000 L, respectively, assuming to reach a concentration of at least 5 mg/mL of mRNA at the end of the process [101].
Following practices like these, mRNA vaccines can be easily and quickly adapted to produce new vaccines against future epidemics.
6. Administration route
The in vivo local delivery of the lipid-based mRNA vaccines (subcutaneous, intramuscular, intradermal, and intranodal administration) is used to initiate the stimulatory reaction in small areas and to eliciting locally strong and long-lasting immune responses [28]. For example, it has been demonstrated that subcutaneous administration of the PEGylated LNPs facilitated the uptake by the DCs located in the lymph nodes and allowed the rapid release of mRNA vaccines [102]. Moreover, the administration route can affect the extent and quality of immune responses independently of the administered dose [103]. Subcutaneous and intramuscular administrations have been the two most frequently used injection routes for mRNA vaccination because they are less invasive and therefore do not require much training for their implementation [4], [8]. However, the administration route and vaccine formulation also determine how the immune response is modulated and when the peak of antigen expression is reached [4], [104].
Some recent studies highlight the importance of mucosal immune responses against SARS-CoV-2 infection [105], [106]. Nevertheless, although intranasal and oral vaccinations are theoretically the most straightforward solution to elicit mucosal immunity, there are no commercial vaccines that use the pulmonary delivery route, given the additional requirement for a suitable and the resulting technical challenges in the formulation. Therefore, almost all COVID-19 vaccine candidates that have undergone clinical trials are given by injection, although they may not induce specific mucosal immunity [107].
7. Immune response evoked by both nucleoside-modified SARS-CoV-2 mRNA-LNP vaccines
After cellular internalization of LNPs, mRNA can be recognized by endosomal or cytosolic PRRs, specific for GU-rich single-stranded RNA (ssRNA), (which can be sensed by TLR-7 and −8) [108] and also by dsRNA (sensed by TLR-3) [64], in terms of the structure that the mRNA molecules could adopt. mRNA can also bind to cytosolic RNA sensors RIG-1[109] and MDA5 [64]. Upon RNA sensing, PRRs can lead to activation of the IFN-I pathway characterized by upregulation of multiple genes, including those encoding proinflammatory cytokines such as tumor necrosis factor-α (TNF-α), IL-6, and IL-12, leading to antigen-presenting cell (APC) activation [104], [110]. Antigen presentation on B and T cells ultimately leads to the production of the typical immunoglobulin M (IgM) and immunoglobulin G (IgG) antibodies, where IgM antibodies have been shown to last up to week 12. In contrast, IgG antibodies can provide prolonged protection [97], [111].
DCs represent an attractive target by mRNA vaccines, both in vivo and ex vivo, because DCs can internalize naked mRNA through various endocytic pathways [112], [113] and may present whole antigens to B cells to trigger an antibody response [114].
Exogenously delivered mRNA uses the host cell translation machinery to produce the S protein of the virus. However, antigen expression is not the final aim for assessing the effectiveness of a vaccine. The capability to induce cellular immunity (T cell activation) may better reflect whether an immune response directed against the S protein will be protective [115], being a requirement to eradicate the intracellular SARS-CoV-2 reservoir. Nevertheless, an indiscriminate immune activation can also induce mRNA degradation and reduce antigen expression [116].
It has been shown that the immune responses elicited by the mRNA-1273 vaccine increased with time and dose of the vaccine and that highly-neutralizing antibody responses were also elicited in a dose-dependent fashion in the vaccinated group [97]. However, both the mRNA-1273 and BNT162b2 vaccines induced in the inoculated participants the production of similarly high dose-dependent neutralizing antibody titers against SARS-CoV-2 [117]. Moreover, in humans, both preparations elicited S-specific CD4 + T cell responses targeting the S1 (including RBD) and S2 regions of the S glycoprotein [81]. In terms of the induction of CD8 + T cell responses, results indicate that BNT162b2 outperforms mRNA-1273. Ninety-one percent of the study participants vaccinated with BNT162b2 mounted significant S-specific CD8 + T cell responses [118], compared to low or undetectable levels in the clinical evaluation of mRNA-1273 [5], [81].
Concerning this, vaccine strategies that induce strong cellular responses in addition to humoral immunity in an adequate balance present a significant advantage in the current outbreak [117], [119].
8. Concluding remarks
While established conventional approaches have allowed the development of many currently available vaccines, mRNA-based vaccines have benefited significantly through breakthroughs in novel technologies based on at least three decades of scientific development, allowing them to reach the market of mass vaccination for the first time at relatively high speed [20].
On the other hand, whereas the majority of early work in mRNA vaccines focused on cancer therapeutics, several reports have demonstrated the potency and versatility of mRNA to protect against a wide variety of viral agents, including influenza virus [120], [121], Ebola virus [121] Zika virus [56], and now SARS-CoV-2 virus [5], [93].
The Pfizer–BioNTech COVID‑19 and Moderna COVID-19 vaccines are two mRNA vaccine platforms that have shown good tolerability, inducing antigen-specific T and B cell immune responses with minimal differences in immunostimulatory profiles [5], [14], [85], [93], [122]. However, many experimental approaches are needed to prove that the optimizations suggested and implemented by both developers are the most appropriate. Only until then, we could do an entirely rational critical analysis to improve the design of both vaccines, leading to their more optimized development in the future. In this regard, further research is needed to determine how different human populations respond to the mRNA vaccine components and how these nucleoside-modified mRNA molecules elicit long-lasting and robust cellular and humoral immune responses against SARS-CoV-2 in humans, in terms of differences between individuals [14], [107].
The inherent structural instability of the spike protein has been an essential aspect in the design of vaccines because the loss of its native conformation may lead to the induction of antibodies with lower neutralizing activity [20], [119]. Furthermore, new strategies and optimizations are required to decrease the doses employed by these mRNA vaccines and elicit more robust and extended memory responses with just one immunization [123]. Efforts to develop thermostable formulations more suitable for wide distribution and long-term storage, while preserving biological activity, have been gaining interest, specifically freeze-dried mRNA [124], lyophilized mRNA[125] as well as protamine-complexed mRNA formulations [126], [127].
In order to meet global demand, mRNA vaccines can be designed and produced massively at relatively high speed, even though this relies on the synthesis of RNA and the procurement of the required supplies, like capping compounds, triphosphate nucleotides, cholesterol, and other components for the lipid nanoparticles, as well as large numbers of vials glass vials, syringes, dry ice, and cold packs for distribution.
Although several of the current COVID-19 vaccine platforms elicit neutralizing antibodies against the S protein of SARS-CoV-2, these may exhibit different protection grades among different population groups such as children, pregnant women, immunocompromised populations, and immunosenescent age groups ≥ 65 years [117], [128], an aspect that deserves to be evaluated in the medium and long term.
Pfizer-BioNTech and Moderna Therapeutics plan to update their vaccines and develop booster doses to improve their efficacy against any future variants [129]. More extensive relevant human clinical experience will give us a more comprehensive insight into mRNA vaccine approaches.
CRediT authorship contribution statement
Javier T. Granados-Riveron: Conceptualization, Writing – review & editing. Guillermo Aquino-Jarquin: Conceptualization, Writing – original draft, Funding acquisition.
Conflict of interest statement
None
Acknowledgments
The APC was funded by The Mexican Council of Science and Technology (CONACyT), grant number SEP-CONACyT-CB-286421.
References
- 1.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247(4949 Pt 1):1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 2.Schlake T., Thess A., Fotin-Mleczek M., Kallen K.J. Developing mRNA-vaccine technologies. RNA Biol. 2012;9(11):1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA vaccines for infectious diseases. Front Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27(4):710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., 2nd, Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nanomedicine and the COVID-19 vaccines, Nat Nanotechnol 15(12) (2020) 963. [DOI] [PMC free article] [PubMed]
- 7.Fotin-Mleczek M., Duchardt K.M., Lorenz C., Pfeiffer R., Ojkic-Zrna S., Probst J., Kallen K.J. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J. Immunother. 2011;34(1):1–15. doi: 10.1097/CJI.0b013e3181f7dbe8. [DOI] [PubMed] [Google Scholar]
- 8.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7(5):319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Disco. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pardi N., Secreto A.J., Shan X., Debonera F., Glover J., Yi Y., Muramatsu H., Ni H., Mui B.L., Tam Y.K., Shaheen F., Collman R.G., Kariko K., Danet-Desnoyers G.A., Madden T.D., Hope M.J., Weissman D. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thran M., Mukherjee J., Ponisch M., Fiedler K., Thess A., Mui B.L., Hope M.J., Tam Y.K., Horscroft N., Heidenreich R., Fotin-Mleczek M., Shoemaker C.B., Schlake T. mRNA mediates passive vaccination against infectious agents, toxins, and tumors. EMBO Mol. Med. 2017;9(10):1434–1447. doi: 10.15252/emmm.201707678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman R.A., Fuhr R., Smolenov I., Mick Ribeiro A., Panther L., Watson M., Senn J.J., Smith M., Almarsson, Pujar H.S., Laska M.E., Thompson J., Zaks T., Ciaramella G. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37(25):3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- 13.Orlandini von Niessen A.G., Poleganov M.A., Rechner C., Plaschke A., Kranz L.M., Fesser S., Diken M., Lower M., Vallazza B., Beissert T., Bukur V., Kuhn A.N., Tureci O., Sahin U. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3’ UTRs identified by cellular library screening. Mol. Ther. 2019;27(4):824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.Y., Tureci O., Tompkins K.R., Lyke K.E., Raabe V., Dormitzer P.R., Jansen K.U., Sahin U., Gruber W.C. Safety and Immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383(25):2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pardi N., Hogan M.J., Weissman D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020;65:14–20. doi: 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Cheng X., Lee R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016;99(Pt A):129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 17.Xia X. Domains and functions of spike protein in Sars-Cov-2 in the context of vaccine design. Viruses. 2021;13(1):109. doi: 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M.C., Kopp M., Li D., Parks R., Hsu A.L., Borgnia M.J., Haynes B.F., Acharya P. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27(10):925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinz F.X., Stiasny K. Wien Klin Wochenschr; 2021. Profiles of current COVID-19 vaccines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;183(6):1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ord M., Faustova I., Loog M. The sequence at spike S1/S2 site enables cleavage by furin and phospho-regulation in SARS-CoV2 but not in SARS-CoV1 or MERS-CoV. Sci. Rep. 2020;10(1):16944. doi: 10.1038/s41598-020-74101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78(4):779–784. doi: 10.1016/j.molcel.2020.04.022. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., Acton O.J., Jaconi S., Guarino B., Minola A., Zatta F., Sprugasci N., Bassi J., Peter A., De Marco A., Nix J.C., Mele F., Jovic S., Rodriguez B.F., Gupta S.V., Jin F., Piumatti G., Lo Presti G., Pellanda A.F., Biggiogero M., Tarkowski M., Pizzuto M.S., Cameroni E., Havenar-Daughton C., Smithey M., Hong D., Lepori V., Albanese E., Ceschi A., Bernasconi E., Elzi L., Ferrari P., Garzoni C., Riva A., Snell G., Sallusto F., Fink K., Virgin H.W., Lanzavecchia A., Corti D., Veesler D. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183(4):1024–1042. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F., Kream R.M., Stefano G.B. An evidence based perspective on mRNA-SARS-CoV-2 vaccine development. Med Sci. Monit. 2020;26 doi: 10.12659/MSM.924700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., Zatta F., De Marco A., Guarino B., Bianchi S., Lauron E.J., Tucker H., Zhou J., Peter A., Havenar-Daughton C., Wojcechowskyj J.A., Case J.B., Chen R.E., Kaiser H., Montiel-Ruiz M., Meury M., Czudnochowski N., Spreafico R., Dillen J., Ng C., Sprugasci N., Culap K., Benigni F., Abdelnabi R., Foo S.C., Schmid M.A., Cameroni E., Riva A., Gabrieli A., Galli M., Pizzuto M.S., Neyts J., Diamond M.S., Virgin H.W., Snell G., Corti D., Fink K., Veesler D. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370(6519):950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Zhang Z., Luo J., Han X., Wei Y., Wei X. mRNA vaccine: a potential therapeutic strategy. Mol. Cancer. 2021;20(1):33. doi: 10.1186/s12943-021-01311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alameh M.G., Weissman D., Pardi N. Messenger RNA-based vaccines against infectious diseases. Curr. Top. Microbiol Immunol. 2020 doi: 10.1007/82_2020_202. [DOI] [PubMed] [Google Scholar]
- 30.Linares-Fernandez S., Lacroix C., Exposito J.Y., Verrier B. Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol. Med. 2020;26(3):311–323. doi: 10.1016/j.molmed.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Xu S., Yang K., Li R., Zhang L. mRNA vaccine era-mechanisms, drug platform and clinical prospection. Int. J. Mol. Sci. 2020;21(18) doi: 10.3390/ijms21186582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramanathan A., Robb G.B., Chan S.H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44(16):7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaidyanathan S., Azizian K.T., Haque A., Henderson J.M., Hendel A., Shore S., Antony J.S., Hogrefe R.I., Kormann M.S.D., Porteus M.H., McCaffrey A.P. Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC Purification. Mol. Ther. Nucleic Acids. 2018;12:530–542. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M., Endres S., Hartmann G. 5’-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 35.Trepotec Z., Aneja M.K., Geiger J., Hasenpusch G., Plank C., Rudolph C. Maximizing the translational yield of mRNA therapeutics by minimizing 5’-UTRs. Tissue Eng. Part A. 2019;25(1–2):69–79. doi: 10.1089/ten.TEA.2017.0485. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z., Day N., Trifillis P., Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell Biol. 1999;19(7):4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holtkamp S., Kreiter S., Selmi A., Simon P., Koslowski M., Huber C., Tureci O., Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108(13):4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 38.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., Ciaramella G., Diamond M.S. Modified mRNA vaccines protect against zika virus infection. Cell. 2017;168(6):1114–1125. doi: 10.1016/j.cell.2017.02.017. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maruggi G., Chiarot E., Giovani C., Buccato S., Bonacci S., Frigimelica E., Margarit I., Geall A., Bensi G., Maione D. Immunogenicity and protective efficacy induced by self-amplifying mRNA vaccines encoding bacterial antigens. Vaccine. 2017;35(2):361–368. doi: 10.1016/j.vaccine.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 40.Gustafsson C., Govindarajan S., Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22(7):346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 41.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23(9):1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kudla G., Lipinski L., Caffin F., Helwak A., Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4(6):180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lima S.A., Chipman L.B., Nicholson A.L., Chen Y.H., Yee B.A., Yeo G.W., Coller J., Pasquinelli A.E. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 2017;24(12):1057–1063. doi: 10.1038/nsmb.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grier A.E., Burleigh S., Sahni J., Clough C.A., Cardot V., Choe D.C., Krutein M.C., Rawlings D.J., Jensen M.C., Scharenberg A.M., Jacoby K. pEVL: a linear plasmid for generating mRNA IVT templates with extended encoded poly(A) sequences. Mol. Ther. Nucleic Acids. 2016;5:306. doi: 10.1038/mtna.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong P., Martin C.T. Mechanism of instability in abortive cycling by T7 RNA polymerase. J. Biol. Chem. 2006;281(33):23533–23544. doi: 10.1074/jbc.M604023200. [DOI] [PubMed] [Google Scholar]
- 46.Kariko K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39(21):142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Probst J., Weide B., Scheel B., Pichler B.J., Hoerr I., Rammensee H.G., Pascolo S. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007;14(15):1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 48.Baiersdorfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Kariko K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kariko K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16(11):1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 51.Kariko K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23(2):165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Weng Y., Li C., Yang T., Hu B., Zhang M., Guo S., Xiao H., Liang X.J., Huang Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020;40 doi: 10.1016/j.biotechadv.2020.107534. [DOI] [PubMed] [Google Scholar]
- 53.Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Kariko K. Nucleoside modifications in RNA limit activation of 2’-5’-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39(21):9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maruggi G., Zhang C., Li J., Ulmer J.B., Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther. 2019;27(4):757–772. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R., Wagner W., Granados A., Greenhouse J., Walker M., Willis E., Yu J.S., McGee C.E., Sempowski G.D., Mui B.L., Tam Y.K., Huang Y.J., Vanlandingham D., Holmes V.M., Balachandran H., Sahu S., Lifton M., Higgs S., Hensley S.E., Madden T.D., Hope M.J., Kariko K., Santra S., Graham B.S., Lewis M.G., Pierson T.C., Haynes B.F., Weissman D. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543(7644):248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaefer M., Kapoor U., Jantsch M.F. Understanding RNA modifications: the promises and technological bottlenecks of the ‘epitranscriptome’. Open Biol. 2017;7(5) doi: 10.1098/rsob.170077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rietwyk S., Peer D. Next-generation lipids in RNA interference therapeutics. ACS Nano. 2017;11(8):7572–7586. doi: 10.1021/acsnano.7b04734. [DOI] [PubMed] [Google Scholar]
- 59.Eygeris Y., Patel S., Jozic A., Sahay G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 2020;20(6):4543–4549. doi: 10.1021/acs.nanolett.0c01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E., LaBranche C.C., Montefiori D.C., Lobby J.L., Saunders K.O., Liao H.X., Korber B.T., Sutherland L.L., Scearce R.M., Hraber P.T., Tombacz I., Muramatsu H., Ni H., Balikov D.A., Li C., Mui B.L., Tam Y.K., Krammer F., Kariko K., Polacino P., Eisenlohr L.C., Madden T.D., Hope M.J., Lewis M.G., Lee K.K., Hu S.L., Hensley S.E., Cancro M.P., Haynes B.F., Weissman D. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215(6):1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buschmann M.D., Carrasco M.J., Alishetty S., Paige M., Alameh M.G., Weissman D. Nanomaterial delivery systems for mRNA vaccines. Vaccin. (Basel) 2021;9(1) doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maugeri M., Nawaz M., Papadimitriou A., Angerfors A., Camponeschi A., Na M., Holtta M., Skantze P., Johansson S., Sundqvist M., Lindquist J., Kjellman T., Martensson I.L., Jin T., Sunnerhagen P., Ostman S., Lindfors L., Valadi H. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019;10(1):4333. doi: 10.1038/s41467-019-12275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Midoux P., Pichon C. Lipid-based mRNA vaccine delivery systems. Expert Rev. Vaccin. 2015;14(2):221–234. doi: 10.1586/14760584.2015.986104. [DOI] [PubMed] [Google Scholar]
- 64.De Beuckelaer A., Grooten J., De S. Koker, Type I Interferons Modulate CD8(+) T Cell Immunity to mRNA Vaccines. Trends Mol. Med. 2017;23(3):216–226. doi: 10.1016/j.molmed.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 65.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., Grunwitz C., Vormehr M., Husemann Y., Selmi A., Kuhn A.N., Buck J., Derhovanessian E., Rae R., Attig S., Diekmann J., Jabulowsky R.A., Heesch S., Hassel J., Langguth P., Grabbe S., Huber C., Tureci O., Sahin U. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534(7607):396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 66.Veiga N., Goldsmith M., Granot Y., Rosenblum D., Dammes N., Kedmi R., Ramishetti S., Peer D. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 2018;9(1):4493. doi: 10.1038/s41467-018-06936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uchida S., Perche F., Pichon C., Cabral H. Nanomedicine-based approaches for mRNA delivery. Mol. Pharm. 2020;17(10):3654–3684. doi: 10.1021/acs.molpharmaceut.0c00618. [DOI] [PubMed] [Google Scholar]
- 68.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., Rajeev K.G., Hafez I.M., Akinc A., Maier M.A., Tracy M.A., Cullis P.R., Madden T.D., Manoharan M., Hope M.J. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int Ed. Engl. 2012;51(34):8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cullis P.R., Hope M.J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 2017;25(7):1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ponti F., Campolungo M., Melchiori C., Bono N., Candiani G. Cationic lipids for gene delivery: many players, one goal. Chem. Phys. Lipids. 2021;235 doi: 10.1016/j.chemphyslip.2020.105032. [DOI] [PubMed] [Google Scholar]
- 71.Allen T.M., Cullis P.R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 2013;65(1):36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 72.Li S.D., Huang L. Stealth nanoparticles: high density but sheddable PEG is a key for tumor targeting. J. Control Release. 2010;145(3):178–181. doi: 10.1016/j.jconrel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buyens K., De Smedt S.C., Braeckmans K., Demeester J., Peeters L., van Grunsven L.A., de Mollerat du Jeu X., Sawant R., Torchilin V., Farkasova K., Ogris M., Sanders N.N. Liposome based systems for systemic siRNA delivery: stability in blood sets the requirements for optimal carrier design. J. Control Release. 2012;158(3):362–370. doi: 10.1016/j.jconrel.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 74.The U.S. Securities and Exchange Commission, Moderna, Inc. 10-K (annual report). Available online: https://www.sec.gov/Archives/edgar/data/1682852/000168285221000006/mrna-20201231.htm. (Retrieved on 1 July 2021).
- 75.D.E. Jeong, M. McCoy, K. Artiles, O. Ilbay, A. Fire, K. Nadeau, H. Park, B. Betts, S. Boyd, R. Hoh, et al., Assemblies of Putative SARS-CoV2 Spike Encoding mRNA Sequences for Vaccines BNT-162b2 and mRNA-1273. Available online: https://virological.org/t/assemblies-of-putative-sars-cov2-spike-encoding-mrna-sequences-for-vaccines-bnt-162b2-andmrna-1273/663 (Accessed 14 July 2021).
- 76.Vaccines and Related Biological Products Advisory Committee December 17, Available online: 〈https://www.fda.gov/media/144452/download〉 (Accessed 16 July 2021).
- 77.Xia X. Detailed dissection and critical evaluation of the Pfizer/BioNTech and Moderna mRNA vaccines. Vaccines. 2021;9(734) doi: 10.3390/vaccines9070734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mauger D.M., Cabral B.J., Presnyak V., Su S.V., Reid D.W., Goodman B., Link K., Khatwani N., Reynders J., Moore M.J., McFadyen I.J. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. USA. 2019;116(48):24075–24083. doi: 10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adachi H., De Zoysa M.D., Yu Y.T. Post-transcriptional pseudouridylation in mRNA as well as in some major types of noncoding RNAs. Biochim Biophys. Acta Gene Regul. Mech. 2019;1862(3):230–239. doi: 10.1016/j.bbagrm.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCaughan K.K., Brown C.M., Dalphin M.E., Berry M.J., Tate W.P. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. USA. 1995;92(12):5431–5435. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. The dawn of mRNA vaccines: the COVID-19 case. J. Control Release. 2021;333:511–520. doi: 10.1016/j.jconrel.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moderna COVID-19 Vaccine EUA Letter of Authorization. Available online: 〈https://www.fda.gov/media/144636/download〉. (Retrieved on 14 December 2020).
- 83.European Medicines Agency, EMA Recommends First COVID-19 Vaccine for Authorisation in the EU. Available online: https://www.ema.europa.eu/en/news/ema-recommends-first-covid-19-vaccine-authorisation-eu, 2020. (Retrieved on 20 December 2020).
- 84.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., Mihai C., Lynn A., McFadyen I., Moore M.J., Senn J.J., Stanton M.G., Almarsson O., Ciaramella G., Brito L.A. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O’Dell S., Schmidt S.D., Corbett K.S., Swanson P.A., 2nd, Padilla M., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Martinez D.R., Baric R., Buchanan W., Luke C.J., Phadke V.K., Rostad C.A., Ledgerwood J.E., Graham B.S., Beigel J.H. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383(25):2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pfizer-BioNTech COVID-19 Vaccine FDA EUA Letter of Authorization (25 February 2021). Available online: 〈https://www.fda.gov/media/144412/download〉. (Retrieved on 14 March 2021).
- 87.Lamb Y.N. BNT162b2 mRNA COVID-19 vaccine: first approval. Drugs. 2021;81(4):495–501. doi: 10.1007/s40265-021-01480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gray B. Pfizer's Chesterfield workforce playing a key role in coronavirus vaccine development. St. Louis Post-Dispatch (Retrieved 24 April 2021).
- 89.Hughes M. (27 February 2020)."Andover's piece of the vaccine: Pfizer". The Eagle-Tribune. (Retrieved 17 December 2020).
- 90.Johnson CY (17 November 2020). "A vial, a vaccine and hopes for slowing a pandemic — how a shot comes to be". The Washington Post. (Retrieved 17 December 2020).
- 91.World Health Organization (September 2020). "Messenger RNA encoding the full-length SARS-CoV-2 spike glycoprotein" (DOC). WHO MedNet. (Retrieved 25 February 2021).
- 92.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., Kong W.P., Andres E.L., Kettenbach A.N., Denison M.R., Chappell J.D., Graham B.S., Ward A.B., McLellan J.S. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114(35):E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr., Hammitt L.L., Tureci O., Nell H., Schaefer A., Unal S., Tresnan D.B., Mather S., Dormitzer P.R., Sahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gaebler C., Nussenzweig M.C. All eyes on a hurdle race for a SARS-CoV-2 vaccine. Nature. 2020;586(7830):501–502. doi: 10.1038/d41586-020-02926-w. [DOI] [PubMed] [Google Scholar]
- 95.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.Y., Tureci O., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Sahin U., Jansen K.U. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586(7830):589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 96.Abbasi J. COVID-19 and mRNA Vaccines-First Large Test for a New Approach. JAMA. 2020;324(12):1125–1127. doi: 10.1001/jama.2020.16866. [DOI] [PubMed] [Google Scholar]
- 97.Rawat K., Kumari P., Saha L. COVID-19 vaccine: a recent update in pipeline vaccines, their design and development strategies. Eur. J. Pharm. 2021;892 doi: 10.1016/j.ejphar.2020.173751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Herper M. Covid-19 vaccine from Pfizer and BioNTech is strongly effective, early data from large trial indicate. STAT. https://www.statnews.com/2020/11/09/covid-19-vaccine-from-pfizer-and-biontech-is-strongly-effective-early-data-from-large-trial-indicate/. (Retrieved 27 January 2020).
- 99.Naomi Kresge, (30 March 2021), BioNTech Raises Covid Vaccine Target to 2.5 Billion Doses, www.bloomberg.com, Retrieved 6 May 2021.
- 100.Loftus, Peter (29 April 2021). "Moderna to Boost Covid-19 Vaccine Production to Meet Rising Global Demand". Wall Street Journal. ISSN 0099–9660. Retrieved 6 May 2021.
- 101.Pascolo S. Synthetic messenger RNA-Based vaccines: from scorn to hype. Viruses. 2021;13(2) doi: 10.3390/v13020270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Y., Zhang L., Xu Z., Miao L., Huang L. mRNA vaccine with antigen-specific checkpoint blockade induces an enhanced immune response against established melanoma. Mol. Ther. 2018;26(2):420–434. doi: 10.1016/j.ymthe.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mohanan D., Slutter B., Henriksen-Lacey M., Jiskoot W., Bouwstra J.A., Perrie Y., Kundig T.M., Gander B., Johansen P. Administration routes affect the quality of immune responses: a cross-sectional evaluation of particulate antigen-delivery systems. J. Control Release. 2010;147(3):342–349. doi: 10.1016/j.jconrel.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 104.Moon C. Fighting COVID-19 exhausts T cells. Nat. Rev. Immunol. 2020;20(5):277. doi: 10.1038/s41577-020-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hassan A.O., Kafai N.M., Dmitriev I.P., Fox J.M., Smith B.K., Harvey I.B., Chen R.E., Winkler E.S., Wessel A.W., Case J.B., Kashentseva E., McCune B.T., Bailey A.L., Zhao H., VanBlargan L.A., Dai Y.N., Ma M., Adams L.J., Shrihari S., Danis J.E., Gralinski L.E., Hou Y.J., Schafer A., Kim A.S., Keeler S.P., Weiskopf D., Baric R.S., Holtzman M.J., Fremont D.H., Curiel D.T., Diamond M.S. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183(1):169–184. doi: 10.1016/j.cell.2020.08.026. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M., Pu A., Christie-Holmes N., Gervais C., Ceccarelli D., Samavarchi-Tehrani P., Guvenc F., Budylowski P., Li A., Paterson A., Yue F.Y., Marin L.M., Caldwell L., Wrana J.L., Colwill K., Sicheri F., Mubareka S., Gray-Owen S.D., Drews S.J., Siqueira W.L., Barrios-Rodiles M., Ostrowski M., Rini J.M., Durocher Y., McGeer A.J., Gommerman J.L., Gingras A.C. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5(52) doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang J., Peng Y., Xu H., Cui Z., Williams R.O., 3rd The COVID-19 vaccine race: challenges and opportunities in vaccine formulation. AAPS PharmSciTech. 2020;21(6):225. doi: 10.1208/s12249-020-01744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 109.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Devoldere J., Dewitte H., De Smedt S.C., Remaut K. Evading innate immunity in nonviral mRNA delivery: don’t shoot the messenger. Drug Disco Today. 2016;21(1):11–25. doi: 10.1016/j.drudis.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 111.Li G., Chen X., Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 2003;349(5):508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 112.Lorenz C., Fotin-Mleczek M., Roth G., Becker C., Dam T.C., Verdurmen W.P., Brock R., Probst J., Schlake T. Protein expression from exogenous mRNA: uptake by receptor-mediated endocytosis and trafficking via the lysosomal pathway. RNA Biol. 2011;8(4):627–636. doi: 10.4161/rna.8.4.15394. [DOI] [PubMed] [Google Scholar]
- 113.Diken M., Kreiter S., Selmi A., Britten C.M., Huber C., Tureci O., Sahin U. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 2011;18(7):702–708. doi: 10.1038/gt.2011.17. [DOI] [PubMed] [Google Scholar]
- 114.Selmi A., Vascotto F., Kautz-Neu K., Tureci O., Sahin U., von Stebut E., Diken M., Kreiter S. Uptake of synthetic naked RNA by skin-resident dendritic cells via macropinocytosis allows antigen expression and induction of T-cell responses in mice. Cancer Immunol. Immunother. 2016;65(9):1075–1083. doi: 10.1007/s00262-016-1869-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Karlsson A.C., Humbert M., Buggert M. The known unknowns of T cell immunity to COVID-19. Sci. Immunol. 2020;5(53) doi: 10.1126/sciimmunol.abe8063. [DOI] [PubMed] [Google Scholar]
- 116.Landesman-Milo D., Peer D. Toxicity profiling of several common RNAi-based nanomedicines: a comparative study. Drug Deliv. Transl. Res. 2014;4(1):96–103. doi: 10.1007/s13346-013-0158-7. [DOI] [PubMed] [Google Scholar]
- 117.Kyriakidis N.C., Lopez-Cortes A., Gonzalez E.V., Grimaldos A.B., Prado E.O. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccin. 2021;6(1):28. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Walsh E.E., Frenck R., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.Y., Tureci O., Thompkins K.R., Lyke K.E., Raabe V., Dormitzer P.R., Jansen K.U., Sahin U., Gruber W.C. RNA-based COVID-19 vaccine BNT162b2 selected for a pivotal efficacy study. medRxiv. 2020 doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sariol A., Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53(2):248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Scorza F.B., Pardi N. New kids on the block: RNA-based influenza virus vaccines. Vaccin. (Basel) 2018;6(2) doi: 10.3390/vaccines6020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chahal J.S., Khan O.F., Cooper C.L., McPartlan J.S., Tsosie J.K., Tilley L.D., Sidik S.M., Lourido S., Langer R., Bavari S., Ploegh H.L., Anderson D.G. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. USA. 2016;113(29):E4133–E4142. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., Nagata B.M., Andersen H., Martinez D.R., Noe A.T., Douek N., Donaldson M.M., Nji N.N., Alvarado G.S., Edwards D.K., Flebbe D.R., Lamb E., Doria-Rose N.A., Lin B.C., Louder M.K., O’Dell S., Schmidt S.D., Phung E., Chang L.A., Yap C., Todd J.M., Pessaint L., Van Ry A., Browne S., Greenhouse J., Putman-Taylor T., Strasbaugh A., Campbell T.A., Cook A., Dodson A., Steingrebe K., Shi W., Zhang Y., Abiona O.M., Wang L., Pegu A., Yang E.S., Leung K., Zhou T., Teng I.T., Widge A., Gordon I., Novik L., Gillespie R.A., Loomis R.J., Moliva J.I., Stewart-Jones G., Himansu S., Kong W.P., Nason M.C., Morabito K.M., Ruckwardt T.J., Ledgerwood J.E., Gaudinski M.R., Kwong P.D., Mascola J.R., Carfi A., Lewis M.G., Baric R.S., McDermott A., Moore I.N., Sullivan N.J., Roederer M., Seder R.A., Graham B.S. Vol. 383. 2020. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates; pp. 1544–1555. (N Engl J Med). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jeyanathan M., Afkhami S., Smaill F., Miller M.S., Lichty B.D., Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020;20(10):615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D., Fotin-Mleczek M., Hoerr I., Clemens R., von Sonnenburg F. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390(10101):1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 125.Jones K.L., Drane D., Gowans E.J. Long-term storage of DNA-free RNA for use in vaccine studies. Biotechniques. 2007;43(5):675–681. doi: 10.2144/000112593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alfagih I.M., Aldosari B., AlQuadeib B., Almurshedi A., Alfagih M.M. Nanoparticles as adjuvants and nanodelivery systems for mRNA-based vaccines. Pharmaceutics. 2020;13(1) doi: 10.3390/pharmaceutics13010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stitz L., Vogel A., Schnee M., Voss D., Rauch S., Mutzke T., Ketterer T., Kramps T., Petsch B. A thermostable messenger RNA based vaccine against rabies. PLoS Negl. Trop. Dis. 2017;11(12) doi: 10.1371/journal.pntd.0006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., Kato Y., Crotty E.G., Rawlings S.A., Mateus J., Tse L.P.V., Frazier A., Baric R., Peters B., Greenbaum J., Ollmann Saphire E., Smith D.M., Sette A., Crotty S. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and Associations with age and disease severity. Cell. 2020;183(4):996–1012. doi: 10.1016/j.cell.2020.09.038. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Deva Priya S.A., Kavitha S., Venugopal P., Sriram D.K., George M. Can mRNA vaccines turn the tables during the COVID-19 pandemic? Current status and challenges. Clin. Drug Invest. 2021 doi: 10.1007/s40261-021-01022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]