Abstract
We introduce a target capture next-generation sequencing methodology, the ONETest Coronaviruses Plus, to sequence the SARS-CoV-2 genome and select loci of other respiratory viruses. We applied the ONETest on 70 respiratory samples (collected in Florida, USA between May and July, 2020), in which SARS-CoV-2 had been detected by a PCR assay. For 48 of the samples, we also applied the ARTIC protocol. Of the 70 ONETest libraries, 45 (64%) had a (near-)complete sequence (>29,000 bases and >90% covered by >9 reads). Of the 48 ARTIC libraries, 25 (52%) had a (near-)complete sequence. In 19 out of 25 (76%) samples in which both the ONETest and ARTIC yielded (near-)complete sequences, the lineages assigned were identical. As a target capture approach, the ONETest is less prone to loss of sequence coverage than amplicon approaches, and thus can provide complete genomic information more often to track and monitor SARS-CoV-2 variants.
Keywords: COVID-19, Genome sequencing, Target hybridization, Respiratory viruses
1. Introduction
SARS-CoV-2 genome sequencing is widely achieved using the amplicon next-generation sequencing (NGS) ARTIC methodology (Tyson et al., 2020). Because of its ease of use and low cost of sequencing, ARTIC has become the method of choice among many laboratories. Notwithstanding its advantages, the ARTIC PCR primer set needs to be maintained and updated due to amplicon dropouts (Tyson et al., 2020), which may be caused by primer interactions (Itokawa et al., 2020) or mutations at primer binding sites (Kim et al., 2021). Without continual upkeep, amplicon sequencing may yield incomplete SARS-CoV-2 genome sequences and therefore create a loss of valuable genetic information. This could weaken our vigilance towards SARS-CoV-2 mutations, which may impact our diagnostic, therapeutic, and vaccination efforts (Chen et al., 2021), and SARS-CoV-2 lineages, especially variants of concern such as B.1.1.7 and B.1.135 that may enhance the virus’ transmissibility or lethality (Iacobucci, 2021; Tegally et al., 2021; Volz et al., 2021).
Alternatively, SARS-CoV-2 genome sequencing can be accomplished using probe-based liquid-phase hybridization followed by NGS (Charre et al., 2020; Kim et al., 2021; Nasir et al., 2020). A major appeal of target capture NGS methodologies is its capacity to enrich samples for a practically limitless repertoire of genetic loci without needing to constantly update the primers and to deal with multiplexing issues encountered with amplicon-based approaches. Indeed, virome target capture NGS methodologies have been developed (e.g., Briese et al., 2015; Chalkias et al., 2018). Another advantage is that target capture NGS approaches perform better than amplicon NGS approaches in degraded samples (e.g., archived FFPE samples [Zakrzewski et al., 2019]). A validated target capture NGS solution with end-to-end automation for concurrent detection and sequence characterization of SARS-CoV-2 and other common respiratory pathogens can be a powerful tool for genomic surveillance of respiratory infectious disease in the post COVID-19 era and can play a crucial role in timely generation and dissemination of genomic data.
The ONETestTM is a pre-commercial target capture NGS platform developed by Fusion Genomics Corp. (Burnaby, BC, Canada). The platform offers a sequencer-agnostic end-to-end NGS workflow that includes library preparation, probe-based liquid phase hybridization, and bioinformatics analysis (see the workflow in Fig. 1 ). The ONETestTM Coronaviruses Plus (http://www.fusiongenomics.com/onetestplatform/coronavirusesplus/), based on the ONETestTM platform, has been demonstrated to enrich samples for select genetic loci of various respiratory viruses (e.g., influenza A viruses) in a separate study (in preparation). Furthermore, the ONETestTM EnviroScreen, also based on the ONETestTM platform, has been shown to detect diverse subtypes of avian influenza viruses in wetland sediments (Himsworth et al., 2020; Kuchinski et al., 2020).
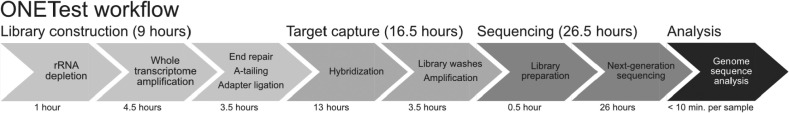
Fig. 1.
The major steps of the ONETest protocol. The ONETest workflow has 4 stages: (1) library construction, (2) target capture, (3) sequencing, and (4) bioinformatics analysis. Input to the ONETest protocol is extracted nucleic acids, or specifically total RNA in this study. Library construction and target capture, which respectively took 9 hours and 16.5 hours, were performed using proprietary kits from Fusion Genomics Corp. Sequencing of the libraries was conducted using an Illumina NextSeq 500 instrument (2 × 150 nt) in this study, and took 26.5 hours. Finally, SARS-CoV-2 genome sequences were reconstructed using the ONETest pipeline described in Materials and Methods, which took less than 10 minutes to run per library. In total, the ONETest workflow in this study took 52 hours.
To capture the full-length genome of SARS-CoV-2, we have expanded the probe design of the ONETest Coronaviruses Plus. Here, using the updated ONETest, we sequenced the SARS-CoV-2 genomes in 70 retrospectively selected samples, which were initially tested at the University of Florida (UF) Health Shands Hospital Clinical Laboratory during the COVID-19 pandemic in 2020. We also processed a subset of them (n = 48) using the ARTIC protocol for Illumina sequencing. These data allowed us to demonstrate the ability of the ONETest to determine the genome sequence of SARS-CoV-2 from respiratory samples.
2. Materials and methods
2.1. Ethics
Approval for this study was obtained from the University of Florida Institutional Review Board (IRB202001328).
2.2. Sample collection
We retrospectively selected 70 samples in which SARS-CoV-2 had been detected by a PCR assay. Nasopharyngeal (NP) swabs (n = 61) and endotracheal aspirates (n = 9) were collected from patients, who had respiratory illness and were suspected to have COVID-19, at UF Health Shands Hospital in May (n = 31) and in July (n = 39), 2020. Among the patients, 30 (43%) were male and 40 (57%) were female. The mean age of the patients (±standard deviation) was 46.1 (±19.8) years (range, 5 to 102 years; interquartile range, 27.8 to 54.0 years). Three of the patients had 2 separate samples collected 7 to 12 days apart; one patient had 4 samples, of which 2 samples were collected in May (1 NP swab and 1 endotracheal aspirate on the same day) and 2 samples were collected in July that were duplicate samples. The samples were initially tested for SARS-CoV-2 using a FDA Emergency Use Authorization qualitative PCR assay (GeneFinder™ COVID-19 Plus RealAmp Kit from OSANG Healthcare Co. Ltd., South Korea), which targets the RdRp, N, and E genes. We retrieved from storage the Ct values from the OSANG PCR assay for 50 out of the 70 samples (71%), but we were unable to obtain the Ct values for the other 20 samples due to hard drive failure on one of the PCR instruments. We retrospectively selected 70 samples in which SARS-CoV-2 had been detected by the PCR assay.
2.3. RNA extraction
Nucleic acids were isolated from 200 μL of the samples and eluted in 100 µL, of which 10 µL was tested for SARS-CoV-2 by the ELlTe InGenius® platform (ELITechGroup, Puteaux, France) using the GeneFinder™ COVID-19 Plus RealAmp Kit, as per the manufacturer's instructions. The remaining 90 µL of de-identified RNA extracts were then shipped to Fusion Genomics Corp. (Burnaby, BC, Canada). Each RNA extract was treated with DNAse (MilliporeSigma Canada, Ontario) and partitioned into 2 aliquots. One aliquot of 11 μL of RNA extract was processed using the ONETest protocol, and the other aliquot of 2 μL of RNA extract was processed using the ARTIC protocol. A higher input volume was allocated for the ONETest because the ONETest protocol involves depletion of human and bacterial ribosomal RNA, whereas the ARTIC protocol does not. Hence, in this study, we ensured that the ONETest had adequate input material for successful library construction following rRNA depletion.
2.4. ONETest: probe design
We expanded the ONETest probe set (QuantumProbesTM; http://www.fusiongenomics.com/onetestplatform/), which originally targets non-SARS-CoV-2 respiratory pathogens, to capture the entire SARS-CoV-2 genome based on the Wuhan-Hu-1 reference sequence (NC_045512.2). Additionally, we designed probes to capture the nucleotide variants frequently observed in SARS-CoV-2 genomes (>1%; retrieved from NCBI GenBank in July, 2020) and to cover the GC-poor regions (<35% GC) of the virus’ genome.
2.5. ONETest: library preparation, target capture, and NGS
Next, we processed 11 μL of total RNA extract from each sample using the ONETest protocol (Fig. 1). Target-enriched Illumina-compatible libraries were prepared from total RNA using the ONETest kit from Fusion Genomics Corp. (Burnaby, BC, Canada). In brief, total RNA was subject to removal of human and bacterial (Gram positive and Gram negative) rRNA using targeted rRNA probes and enzymatic digestion. Depleted RNA was then reverse transcribed using adapted random primers, resulting in fragmented cDNA. Whole transcriptome amplification was then performed, and resulting cDNA was ligated with Illumina-compatible indexed adapters, according to the manufacturer's instructions. The indexed libraries were mixed with Illumina adapter-specific blocking reagents, and target-specific biotin-labeled QuantumProbes (Fusion Genomics, Burnaby, BC, Canada) in a hybridization solution. Hybridization was performed overnight at 50°C. The target-probe duplexes were then captured by using streptavidin coated magnetic beads and non-specific fragments were iteratively removed by washing off with increasingly stringent wash buffers. Enriched libraries were universally re-amplified for 20 cycles using Illumina adapter-specific primers. Normalization and pooling of the enriched libraries were based on quantification using the Quant-iT HS dsDNA kit (Thermo Fisher Scientific, ON, Canada). Molar quantification of the pooled library was performed using NEB Library Quant Kit (New England Biolabs, Whitby, ON, Canada). The pooled library was sequenced as 2 × 150 nt reads on an Illumina NextSeq 500 instrument (Illumina Canada, Vancouver, BC, Canada), as per the manufacturer's instructions. The entire ONETest workflow, as performed manually in this study, took approximately 52 hours (Fig. 1).
2.6. ONETest: NGS data analysis
Reads from the ONETest libraries were analyzed using an in-house bioinformatics pipeline. The pipeline preprocesses raw NGS reads using a custom C/C++ program (removing adapter sequences, trimming off poor-quality bases of <Q30, and filtering out reads of <50 nt and reads with low complexity of normalized trimer entropy of <60, poor mean base quality of <Q27, or percent G of >40%). Reads were discarded that mapped to the human genome sequence (GRCh38.p13, release 35) using bowtie2 v2.4.2 (Langmead and Salzberg, 2012). Then, it aligned the remaining reads to the SARS-CoV-2 Wuhan-Hu-1 reference sequence (MN996528.1) using bowtie2 (with the settings “–very-sensitive-local –score-min G,100,9”), marking duplicate reads using samtools v1.11 (Li et al., 2009). Finally, the pipeline performed comparative assembly to reconstruct consensus SARS-CoV-2 genome sequences using bcftools v1.11 and in-house scripts. Nucleotides were called at positions that were covered by >9 reads (excluding duplicate reads); otherwise, they were masked as Ns. Discounting poor-quality bases of <Q15 and excluding duplicate reads, nucleotide variants were filtered out unless (1) their quality score was ≥Q15, (2) they were supported by >1 forward aligned read and >1 reverse aligned read, and (3) they were supported by >25% of the reads; a maximum depth of 300,000 was allowed during pileup. Indels were normalized after calling. For a position to be considered as a starting point for any indel, it was checked whether >9 and ≥80% of the reads support any indel starting at that position. If the aforementioned filters were passed for a position, candidate indels were filtered out unless they were supported by (1) ≥50% of the reads, and (2) >1 forward aligned read and >1 reverse aligned read. The pipeline was implemented in C/C++ and Python using a combination of in-house software and third-party tools, including Biopython v1.78 (Cock et al., 2009), bedtools v2.29.2 (Quinlan and Hall, 2010), pybedtools v0.8.1 (Dale et al., 2011), samtools/bcftools/htslib v1.11 (Li et al., 2009), pysam v0.16.0.1, pandas v1.1.3, and Snakemake v5.26.1 (Koster and Rahmann, 2012).
2.7. ARTIC protocol
We processed 2 μL of RNA extract from each sample using the ARTIC Illumina protocol (https://www.protocols.io/view/covid-19-artic-v3-illumina-library-construction-an-bibtkann). This protocol utilizes 2 pools of ARTIC V3 primer pairs to amplify 98 ∼400 nt partially overlapping regions that tile the entire SARS-CoV-2 genome (https://github.com/artic-network/artic-ncov2019/blob/master/primer_schemes/nCoV-2019/V3/), which were ordered from Sigma-Aldrich (Oakville, ON, Canada). Libraries were constructed using TruSeq Nano from Illumina Inc. (Illumina Canada, Vancouver, BC, Canada), as per the manufacturer's instructions. Libraries were normalized, pooled together, and sequenced as 2 × 150 nt reads on an Illumina NextSeq 500 instrument (Illumina Canada, Vancouver, BC, Canada). Reads from these libraries were analyzed using a bioinformatics pipeline (v1.3.0; https://github.com/connor-lab/ncov2019-artic-nf) that automates the ARTIC data analysis protocol for Illumina reads (https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html), which utilizes bwa mem (Li, 2013), samtools (Li et al., 2009), and iVar (Grubaugh et al., 2019).
2.8. Sub-sampling analysis of the ONETest libraries
We sequenced the ONETest libraries at 2.66 million 2 × 150 nt reads on average, nearly 4 times as deep as that of the ARTIC libraries (0.63 million 2 × 150 nt reads on average). To assess whether the observed differences in genome coverage between the ONETest and ARTIC libraries might have resulted from deeper sequencing of the ONETest libraries, we conducted a sub-sampling analysis in which we compared down-sampled ONETest libraries with the full ARTIC libraries. Using seqtk v1.3 (https://github/com/lh3/seqtk), we randomly down-sampled (without replacement) the 2 × 150 nt reads of each ONETest library so that the resulting library had the same number of reads as the matched ARTIC library; each ONETest library was sub-sampled 3 times in this manner to generate 3 simulated replicates of the library. Then, we analyzed those sub-sampled reads to determine which positions were poorly covered across the SARS-CoV-2 genome in the simulated ONETest libraries.
2.9. Depth of sequence coverage analysis
Using bedtools, we generated depth of sequence coverage profiles for the full ONETest libraries and the sub-sampled ONETest libraries based on bowtie2 read alignments and the ARTIC libraries based on the bwa mem read alignments. For the ONETest libraries, we excluded duplicate reads, but for the ARTIC libraries, we included duplicate reads. Visualization of the depth of coverage profiles was done in R using ggplot2 (Wickham, 2016).
2.10. Lineage analysis
We identified the lineages of SARS-CoV-2 in the samples based on the ONETest and ARTIC consensus sequences using pangolin v3.0.3 (https://github.com/cov-lineages/pangolin). This tool assigns SARS-CoV-2 lineages according to a dynamic nomenclature system (Rambaut et al., 2020).
2.11. Data availability
The complete or near-complete consensus SARS-CoV-2 genome sequences from the ONETest libraries are available via GISAID (accessions: EPI_ISL_2648013 to EPI_ISL_2648057). All de-identified FastQ files (with human reads removed) of the ONETest and ARTIC libraries are available via the NCBI Short Read Archive (BioProject ID: PRJNA741220).
3. Results
3.1. ONETest yields complete or near-complete SARS-CoV-2 genome more often than ARTIC
The ONETest libraries of the 70 samples had a total of ∼186 million paired-end reads, and each of the libraries had ∼2.66 million paired-end reads on average (range, ∼0.45 to ∼6.14 million) (Table S1). This per-sample amount of sequencing is comparable to that used in a study (Kim et al., 2021) evaluating another target capture product (7.4 million 1 × 100 nt filtered reads per sample). Of the 70 ONETest libraries, 45 (64%) had a complete or near-complete SARS-CoV-2 genome sequence that was >29,000 nucleotides (nt) long and had >90% well covered bases (>9x depth). After sub-sampling, the ONETest libraries had a complete or near-complete genome sequence for 39 (56%) of the samples (this percent was identical for all the 3 sets of sub-samples). Additionally, we processed 48 (69%) of the 70 samples using ARTIC. The ARTIC libraries had a total of ∼30 million paired-end reads, and each of the libraries had ∼0.63 million paired-end reads on average (range, ∼0.20 to ∼2.1 million) (Table S1). This amount of sequencing was comparable to that in the ARTIC experiments performed by other groups (Figure S1). Of the 48 ARTIC libraries, 25 (52%) had a complete or near-complete SARS-CoV-2 genome sequence.
When considering the 48 samples for which both ONETest and ARTIC libraries were made, the mean percent poorly covered positions in the ONETest sequences was 23% (range, 0% to 100%), whereas that in the ARTIC sequences was 25% (range, 3% to 99%) (Table S1). For 31 (71%) of the samples, there was sufficient sequence information in both the ONETest and ARTIC libraries so that lineage could be assigned to both the ONETest and ARTIC sequences using pangolin (see below), regardless of whether or not the genome sequences were complete or near-complete. We focused on these lineage-assigned matched ONETest and ARTIC library pairs to compare the genome sequences from the 2 methodologies.
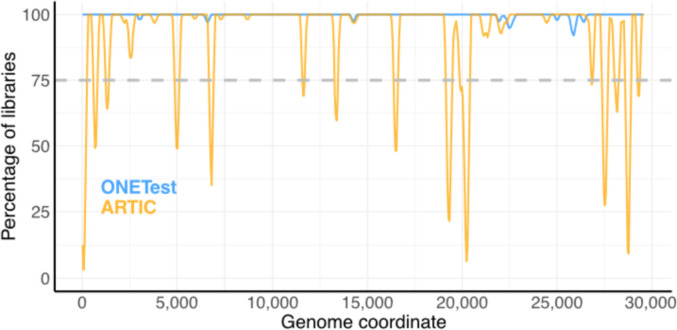
In the matched ONETest and ARTIC library pairs, there were fewer poorly covered positions (<10x depth) across the SARS-CoV-2 genome in the ONETest libraries than in the ARTIC libraries (Fig. 2 ; Figure S2). Some of this difference may be explained by the fact that the ONETest libraries were sequenced deeper than the ARTIC libraries (almost 4 times deeper on average). However, a sub-sampling analysis indicated that even at similar sequencing depths, the ONETest libraries yielded better sequence coverage than the ARTIC libraries (Figure S3).
Fig. 2.
Aggregate summary of sequence coverage over the SARS-CoV-2 genome in the ONETest and ARTIC libraries from the samples examined in this study. Here, we considered only the 31 samples for which lineage could be assigned to both its ONETest and ARTIC sequences using pangolin. For each position in the SARS-CoV-2 reference sequence targeted by the ARTIC PCR primers (MN996528.1: 30 to 29,866), we computed the percentage of samples in which its depth of sequence coverage was >9 (excluding duplicates for the ONETest libraries and including duplicates for the ARTIC libraries). This percentage was averaged across the positions of each 200 nt partially overlapping window across the genome (skip size of 50 nt). Poorly covered regions in the SARS-CoV-2 genome appear as troughs below the dashed line.
3.2. Regions with poorer sequence coverage in the ARTIC libraries than the ONETest libraries
While there were several regions of the SARS-CoV-2 genome in the ARTIC libraries that had poor sequence coverage compared to the ONETest libraries, we closely examined one region that had particularly poor sequence coverage in the ARTIC libraries (Fig. 2). We observed that depth of coverage was generally poor in the ∼19,900-20,500 region of the SARS-CoV-2 genome in the ARTIC libraries (Fig. 2). This region is targeted by the ARTIC primer pairs 66_LEFT/66_RIGHT (pool 2, MN908947.3: 19,844-20,255) and 67_LEFT/67_RIGHT (pool 1, MN908947.3: 20,172-20,572). In contrast, the ∼19,900-20,500 region was well covered overall in the ONETest libraries (Fig. 2). For example, depth of coverage across the SARS-CoV-2 genome in the ARTIC library of sample 27 was high (mean, 2,592x), except in that region amplified by the 2 primer pairs (visualized using IGV (Robinson et al., 2011) in Figure S4); on the other hand, the ONETest library of sample 27 had high depth of coverage across the entire genome of the virus (mean, 1,237x), even in the region targeted by those 2 problematic ARTIC PCR primer pairs (Figure S4).
3.3. Negative correlation between the percent of well covered positions in the SARS-CoV-2 genome sequences from the ONETest and ARTIC libraries and the Ct values from a PCR test
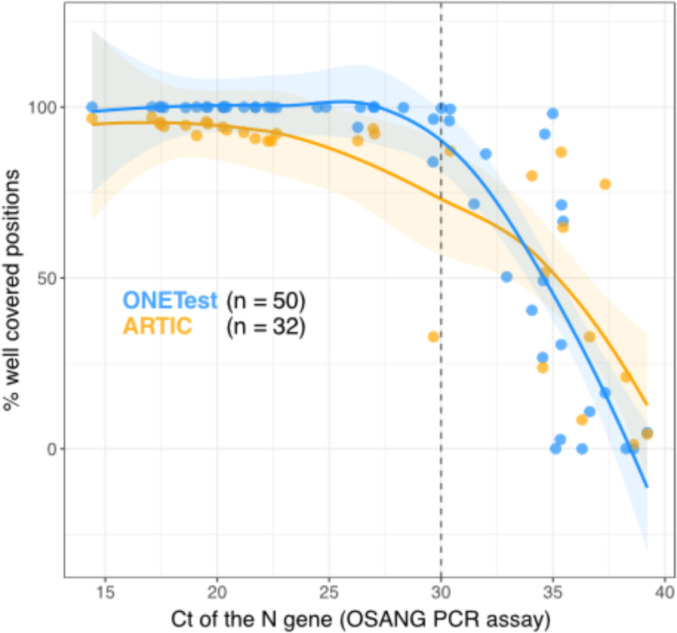
In some ONETest and ARTIC libraries, incomplete SARS-CoV-2 genome sequences might have arisen from low-titer samples. To test this, we examined the relationship between the percent of well covered positions in the SARS-CoV-2 genome in the ONETest and ARTIC libraries and the Ct values obtained using the OSANG PCR assay. Because the N gene was the only gene that was detected by the PCR assay in the 50 samples for which Ct values were available, we analyzed the Ct values of only the N gene (nevertheless, within each sample, the Ct values of the 3 genes were highly similar; see Table S1). The percent of well covered positions in the SARS-CoV-2 genome was negatively correlated in the ONETest and ARTIC libraries (Fig. 3 ). Recovery of the SARS-CoV-2 genome sequence was poor at a Ct value of ∼30 or higher in the ONETest and ARTIC libraries (Fig. 3).
Fig. 3.
Relationship between the percent of well covered positions in the SARS-CoV-2 genome sequences from the ONETest and ARTIC libraries and the Ct values of the SARS-CoV-2 N gene. Well covered positions in the SARS-CoV-2 genome were supported by at least 10 reads. The Ct values were obtained using the OSANG PCR assay. Ct values were available for only 50 of the 70 samples analyzed in this study (for 50 out of the 70 ONETest libraries, and for 32 out of the 48 ARTIC libraries). Lines of best fit and 95% confidence intervals around the lines were estimated using local regression in R.
3.4. ONETest and ARTIC determined SARS-CoV-2 genome sequences with concordant lineage assignments
For 31 samples, the consensus sequences from both the ONETest and ARTIC libraries could be assigned to a SARS-CoV-2 lineage using pangolin. In 24 (77%) of these samples, the lineage assignment was identical for the ONETest and ARTIC libraries (e.g., in sample 50, both the ONETest and ARTIC sequences were assigned to B.1.509). In the other 7 samples, the lineage assignment was nevertheless in the same major lineage (e.g., in sample 46, both the ONETest and ARTIC sequences were assigned to the B.1 lineage rather than the A.1 lineage). These differences in lineage assignment likely stemmed from differences in sequence coverage between the ONETest and ARTIC libraries. In the 7 samples, the mean difference in percent poorly covered positions between the ARTIC and ONETest sequences was 6.6%.
3.5. SARS-CoV-2 lineages detected in the ONETest libraries
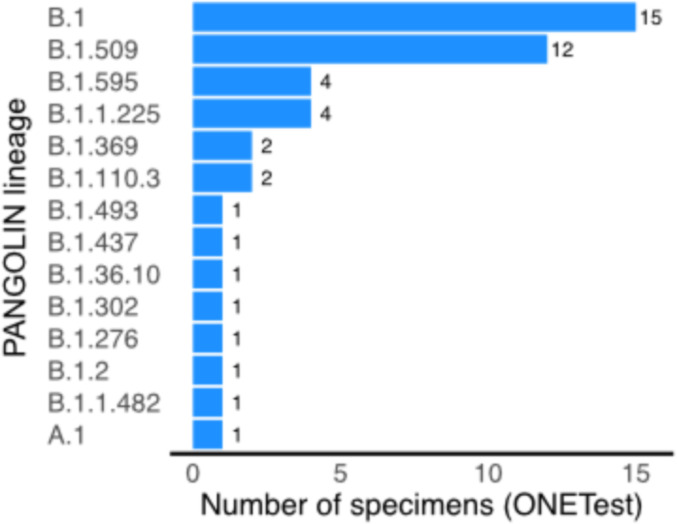
Of the 70 samples sequenced in this study using the ONETest, 45 had a complete or near-complete SARS-CoV-2 genome sequence. We found 14 genetically distinct SARS-CoV-2 lineages (as assigned by pangolin) to the ONETest sequences of the samples (Fig. 4 ).
Fig. 4.
SARS-CoV-2 lineages identified in the samples examined in this study using the ONETest. Lineage was assigned to the complete or near-complete SARS-CoV-2 genome sequences from the ONETest libraries of 37 samples.
4. Discussion
Vaccines against SARS-CoV-2 are presently being administered around the globe, but we have yet to see how effectively the vaccines will protect our populations from the new variants of concerns. Having multiple technologies in our SARS-CoV-2 genome sequencing toolbox should help to heighten our vigilance towards new SARS-CoV-2 variants that may escape our vaccines. Here, we propose the ONETest target capture NGS methodology to sequence the SARS-CoV-2 genome to aid in efforts to track SARS-CoV-2 variants.
Using the ONETest and ARTIC, we sequenced SARS-CoV-2 genomes from archived samples in which SARS-CoV-2 had been detected by a FDA EUA qualitative PCR assay. Our data demonstrate that the ONETest can yield complete SARS-CoV-2 genome sequences more often than ARTIC (64% vs 52%). The ability of the ONETest and the ARTIC to recover complete or near-complete SARS-CoV-2 genomes begins to decline at a similar Ct value (∼30 as per a PCR assay), indicating that the partial genome sequences from some of the ONETest and ARTIC libraries were likely due to low viral titre. While relatively shallow sequencing of the ARTIC libraries may account for some of the other poorly covered regions, a sub-sampling analysis indicates that the ONETest produces complete genome sequences more often than ARTIC even at about one fourth the amount of sequencing on average. Nonetheless, there are consistently poorly covered regions in the SARS-CoV-2 genome across the ARTIC libraries. In particular, the ∼19,900-20,500 SARS-CoV-2 genome region targeted by 2 ARTIC PCR primer pairs (e.g., sample 27) is poorly covered in many ARTIC libraries, even though other genomic regions in the same libraries are well covered. As shown by an analysis of the SARS-CoV-2 genome sequences deposited in GISAID (Cotten et al., 2021), many publicly available sequences contain problematic regions (i.e., contiguous stretches of 200 Ns) around the 20,000th nucleotide position. Many of the genome sequences were produced using an amplicon NGS methodology, in particular ARTIC. Furthermore, by comparing the lineage assignments of the ONETest and ARTIC sequences, which are generally concordant, we show that the ONETest can provide quality genome sequences to study the evolution and epidemiology of SARS-CoV-2.
Target capture NGS methodologies, such as the ONETest, can detect mutations that impact the performance of amplicon NGS methodologies, such as ARTIC. Kim et al., (2021) showed a case in which target capture NGS detected a large 382 nt deletion in the ORF8 gene of SARS-CoV-2 that ablated sequence coverage in 4 contiguous genes (ORF3a, E, M, and ORF6) in the ARTIC library due to PCR amplification failure. Although we did not encounter such a dramatic case in this study, we anticipate that as we sequence more samples using the ONETest, the ONETest will detect large deletions in the SARS-CoV-2 genome that could severely reduce sequence coverage when using amplicon NGS methodologies. This advantage of target capture NGS approaches is important as new SARS-CoV-2 genetic mutations of unpredictable nature continue to emerge.
The ONETest was performed manually in this study. Library preparation (library construction plus target capture) took a total of 25.5 hours (Fig. 1), taking extracted RNA as the input. At the time of this writing, Fusion Genomics Corp. is developing and testing a fully automated ONETest workflow. In the automated ONETest workflow, the target capture step is reduced to 8.5 hours from 16.5 hours, thereby shortening the library preparation time from 25.5 hours to 17.5 hours. This reduced time is still longer than the library preparation time of the ARTIC workflow of 9 hours (5 hours of library construction plus 4 hours of target-specific multiplex PCR amplification), but the hands-on time of both the ONETest and ARTIC, when automated, will be the same. Moreover, the ONETest, when automated, allows for flexible sample batching. When automated using a robotic liquid handling platform, 24 to 96 ONETest libraries can be processed in a single run. Alternatively, when automated using a “lab on a chip” technology, 1 to 4 ONETest libraries can be built in a single run providing a simplified solution for low throughput labs wishing to perform this assay. With automation, the complexity of the ONETest workflow, as compared to a PCR amplicon-based workflow, should no longer be a barrier for laboratories with access to the appropriate equipment.
Our data show the ability of the ONETest to determine the genome sequences of SARS-CoV-2 in respiratory samples. Importantly, our data indicate that the ONETest is less prone to loss of sequence coverage that may be caused by poor or failed target binding (e.g., the amplicon dropouts in the ARTIC libraries shown here and in studies by other groups), which can ultimately result in inaccurate SARS-CoV-2 genotyping and lineage identification. The added value of the ONETest to characterize multiple respiratory pathogens, although not assessed in this study, should help us to better understand the epidemiology of respiratory pathogens in the post COVID-19 era.
Acknowledgments
We thank Dr. Mohammad A. Qadir (Fusion Genomics Corp.) for providing guidance throughout this study and constructive feedback on this manuscript and Greg Stazyk (Fusion Genomics Corp.) for setting up the computing infrastructure that enabled this study. We are grateful to Compute Canada and Simon Fraser University for providing the computing resources that facilitated this study. Also, we gratefully acknowledge the support of the staff from the University of Florida Health Shands Hospital Laboratory.
Funding
This study was funded by Fusion Genomics Corporation and supported in part by the Department of Pathology, Immunology and Laboratory Medicine, University of Florida (Gainesville, FL, USA ).
Declaration of competing interests
S. H. Z., S. M. A., H. D., B. S. K., M. H. L., and J. K. are current or former employees and/or shareholders of Fusion Genomics Corp. H. J. H. and K. H. R. do not have competing interests to declare.
Authors’ contributions
SHZ: Conceptualization, Methodology, Software, Formal analysis, Investigation, Data curation, Visualization, Writing – original draft preparation, Writing – review and editing.
SMA: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Project administration, Writing – review and editing.
HD: Formal analysis, Methodology, Software, Data curation, Investigation, Writing – review and editing.
BSK: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Project administration, Writing – review and editing.
MHL: Methodology, Software, Formal analysis, Investigation, Writing – review and editing.
JK: Methodology, Formal analysis, Data curation, Investigation.
HJH: Methodology, Data curation, Resources.
KHR: Conceptualization, Methodology, Investigation, Data curation, Resources, Writing – review and editing.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.diagmicrobio.2021.115508.
Appendix. Supplementary materials
References
- Briese T, Kapoor A, Mishra N, Jain K, Kumar A, Jabado OJ, et al. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. mBio. 2015;6 doi: 10.1128/mbio.01491-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkias S, Gorham JM, Mazaika E, Parfenov M, Dang X, DePalma S, et al. ViroFind: a novel target-enrichment deep-sequencing platform reveals a complex JC virus population in the brain of PML patients. PLoS One. 2018;13 doi: 10.1371/journal.pone.0186945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charre C, Ginevra C, Sabatier M, Regue H, Destras G, Brun S, et al. Evaluation of NGS-based approaches for SARS-CoV-2 whole genome characterisation. Virus Evol. 2020;6 doi: 10.1093/ve/veaa075. veaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AT, Altschuler K, Zhan SH, Chan YA, Deverman BE. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. Elife. 2021;10 doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M, Bugembe DL, Kaleebu P, Phan MVT. Alternate primers for whole-genome SARS-CoV-2 sequencing. Virus Evolution. 2021 doi: 10.1093/ve/veab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale RK, Pedersen BS, Quinlan AR. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–3424. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, De Jesus JG, Main BJ, et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20(8) doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himsworth CG, Duan J, Prystajecky N, Coombe M, Baticados W, Jassem AN, et al. Targeted resequencing of wetland sediment as a tool for avian influenza virus surveillance. J Wildl Dis. 2020;56:397–408. doi: 10.7589/2019-05-135. [DOI] [PubMed] [Google Scholar]
- Iacobucci G. Covid-19: new UK variant may be linked to increased death rate, early data indicate. BMJ. 2021;372:n230. doi: 10.1136/bmj.n230. [DOI] [PubMed] [Google Scholar]
- Itokawa K, Sekizuka T, Hashino M, Tanaka R, Kuroda M. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS One. 2020;15 doi: 10.1371/journal.pone.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Deveson IW, Pang CNI, Yeang M, Naing Z, Adikari T, et al. Respiratory viral co-infections among SARS-CoV-2 cases confirmed by virome capture sequencing. Sci Rep. 2021;11 doi: 10.1038/s41598-021-83642-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster J, Rahmann S. Snakemake–a scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- Kuchinski K, Duan J, Coombe M, Himsworth C, Hsiao W, Prystajecky N. Recovering influenza genomes from wild bird habitats for better avian flu surveillance. Int J Infect Dis. 2020;101:371–372. doi: 10.1016/j.ijid.2020.09.977. [DOI] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bioGN] 2013.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. Vol. 25. 2009. The sequence alignment/map format and SAMtools; pp. 2078–2079. (Bioinformatics). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir JA, Kozak RA, Aftanas P, Raphenya AR, Smith KM, Maguire F, et al. A comparison of whole genome sequencing of SARS-CoV-2 using amplicon-based sequencing, random hexamers, and bait capture. Viruses. 2020;12 doi: 10.3390/v12080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Holmes EC, Á O'Toole, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature. 2021 doi: 10.1038/s41586-021-03402-9. [DOI] [Google Scholar]
- Tyson JR, James P, Stoddart D, Sparks N, Wickenhagen A, Hall G, et al. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv. 2020 doi: 10.1101/2020.09.04.283077. [DOI] [Google Scholar]
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2nd ed. Springer-Verlag; New York: 2016. https://ggplot2.tidyverse.org. Accessed August 9, 2021. [Google Scholar]
- Zakrzewski F, Gieldon L, Rump A, Seifert M, Grützmann K, Krüger A, et al. Targeted capture-based NGS is superior to multiplex PCR-based NGS for hereditary BRCA1 and BRCA2 gene analysis in FFPE tumor samples. BMC Cancer. 2019;19:396. doi: 10.1186/s12885-019-5584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete or near-complete consensus SARS-CoV-2 genome sequences from the ONETest libraries are available via GISAID (accessions: EPI_ISL_2648013 to EPI_ISL_2648057). All de-identified FastQ files (with human reads removed) of the ONETest and ARTIC libraries are available via the NCBI Short Read Archive (BioProject ID: PRJNA741220).