Abstract
The ongoing COVID-19 outbreak, caused by SARS-CoV-2, has posed a massive threat to global public health, especially to people with underlying health conditions. Type 2 diabetes (T2D) is lethal comorbidity of COVID-19. However, its pathogenetic link remains unclear. This research aims to determine the genetic factors and processes contributing to the synergistic severity of SARS-CoV-2 infection among T2D patients through bioinformatics approaches. We analyzed two sets of transcriptomic data of SARS-CoV-2 infection obtained from lung epithelium cells and PBMCs, and two sets of T2D data from pancreatic islet cells and PBMCs to identify the associated differentially expressed genes (DEGs) followed by their functional enrichment analyses in terms of protein-protein interaction (PPI) to detect hub-proteins and associated comorbidities, transcription factors (TFs), microRNAs (miRNAs) as well as the potential drug candidates. In PPI analysis, four potential hub-proteins (i.e., BIRC3, C3, MME, and IL1B) were identified among 25 DEGs shared between the disease pair. Enrichment analyses using the mutually overlapped DEGs revealed the most prevalent GO and cell signalling pathways, including TNF signalling, cytokine-cytokine receptor interaction, and IL-17 signalling, which are related to cytokine activities. Furthermore, as significant TFs, we identified IRF1, KLF11, FOSL1, and CREB3L1 while miRNAs including miR-1-3p, 34a-5p, 16–5p, 155–5p, 20a-5p, and let-7b-5p were found to be noteworthy. The findings illustrated the significant association between COVID-19 and T2D at the molecular level. These genetic determinants can further be explored for their specific roles in disease progression and therapeutic intervention, while significant pathways can also be studied as molecular checkpoints. Finally, the identified drug candidates may be evaluated for their potency to minimize the severity of COVID-19 patients with pre-existing T2D.
Keywords: COVID-19, SARS-CoV-2, Type 2 diabetes, Differentially expressed genes, Protein–protein interactions, Drug molecules
1. Introduction
After the outbreak of SARS-CoV [1] and MERS-CoV [2] in 2002 and 2012, respectively, a novel coronavirus SARS-CoV-2 has emerged in Wuhan city of Hubei province of China at the end of 2019 with its massive infectious threat [3,4]. This virus causes respiratory tract infection with clinical syndromes including fever, cough, sore throat, pneumonia, and in some severe cases, acute respiratory distress syndrome (ARDS), sepsis and septic shock, multiorgan failure, such as acute kidney injury and cardiac arrest [5,6]. The virus SARS-CoV-2 has been identified as the causative pathogen of the outbreak of coronavirus disease (COVID-19) by deep sequencing and etiological investigations [7]. The World Health Organization (WHO) has already declared COVID-19 as a pandemic. As of June 29, 2021, 181,007,816 cases have been confirmed as SARS-CoV-2 infected in more than 218 countries and regions with 3,927,222 deaths according to WHO [8].
Comorbid diseases, such as diabetes, hypertension, and heart diseases have been reported as risk factors for COVID-19 patients [6,9]. In Ref. [6], it was reported that, out of 41, 20 SARS-CoV-2 infected patients had comorbidities including diabetes (20%), hypertension (15%), and cardiovascular disease (15%). In Ref. [9], 191 COVID-19 patients’ records from two hospitals of Wuhan, China were investigated. Among them, 91 (48%) patients had comorbidity, of which 58 (30%) patients had hypertension, 36 (19%) had diabetes, and 15 (8%) were suffering from coronary heart disease. Moreover, several recent studies reported that type II diabetes (T2D) increases the risk of infection, severity, and mortality of COVID-19 patients [10,11]. However, there is a lack of sufficient information regarding genetic interactions of T2D on COVID-19.
Viruses are obligate intracellular pathogens and they cannot replicate without host cellular factors during infection [12]. Consequently, virus-host protein-protein interaction (PPI), might be an effective way toward elucidating the mechanism of viral infection [13]. Recently, integrative network-based approaches showed better effectiveness to identify disease-associated biomarkers and therapeutic targets [14] of various diseases, including respiratory system diseases [15], neurodegenerative diseases [[16], [17], [18]], cardiomyopathy [19], and viral infections, such as SARS-CoV, HIV [20] and Zika [21]. The network-based strategies can potentially be used to identify effective repurposable drugs [[22], [23], [24]] as well as the combination of drugs [25] for various human diseases as well. Recently, genetic investigations into the gene expression data offered a better understanding of the molecular mechanism of the SARS-CoV-2 infection and its genetic association with various complications [[26], [27], [28], [29], [30]].
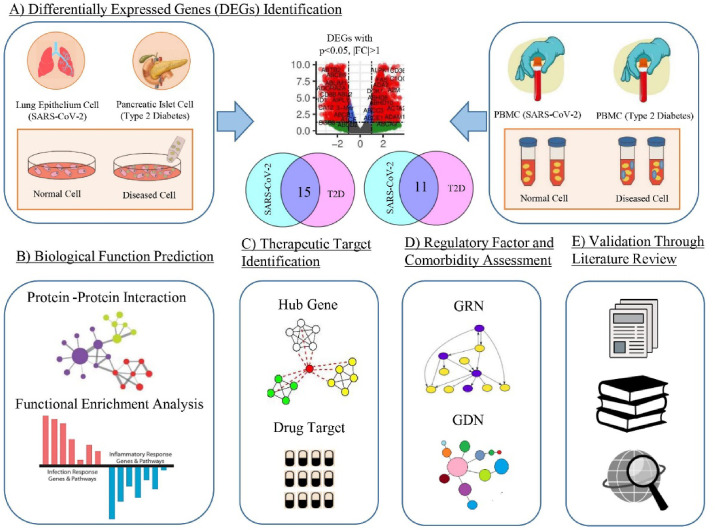
In this study, we investigated the genetic interaction of SARS-CoV-2 with T2D using this network-based strategy that incorporates studies regarding gene expression profiling, protein-protein interaction, gene ontologies, molecular pathways, and regulatory analysis. The findings may lead to determining the significant therapeutic target to fight against the ongoing pandemic due to SARS-CoV-2 infection. The analytical approach adopted in this research work is illustrated in Fig. 1 .
Fig. 1.
Schematic view of the systematic pipeline used in this study. (A) At first, transcriptomic analyses of two RNA-Seq data, one NanoString and one microarray data and cross-comparison identified 15 common DEGs between SARS-CoV-2 infection and T2D for lung epithelium and pancreatic islet cells respectively, and 11 common DEGs for peripheral blood mononuclear cells (PBMCs). (B) Biological functions of these 26 (25 distinct) DEGs were assessed by PPI analysis and functional enrichment analysis using GO and cell signalling pathway databases. (C) Therapeutic targets were identified by obtaining hub genes and putative drug candidates. (D) Regulatory elements and possible comorbidities were determined. (E) All the gained results were validated through an extensive literature review.
2. Materials and methods
2.1. Data
To reveal the genetic association of SARS-CoV-2 with T2D, we collected the RNA-Seq and gene expression microarray data from National Center for Biotechnology Information Gene Expression Omnibus (NCBI-GEO) and EBI array express. In this study, we analyzed the datasets with accession numbers GSE147507 and E-MTAB-8871 for SARS-CoV-2 while the datasets with accession numbers GSE9006 and E-MTAB-5060 for T2D. The GSE147507 is an RNA-Seq data obtained by transcriptional profiling of SARS-CoV-2 infected and mock-treated cells from humans and ferrets using Illumina NextSeq 500 platform [31]. Herein, the human lung epithelium (NHBE) cell replicates from 3 SARS-CoV-2 and 3 mock-treated samples were collected. The E-MTAB-8871 was obtained from human peripheral blood mononuclear cells (PBMCs) of 10 healthy individuals and 23 samples from 3 SARS-CoV-2 infected patients using NanoString profiling [32]. The T2D dataset (GSE9006) was obtained by gene expression microarray using Affymetrix HG-U133 array of PBMCs collected from 24 healthy individuals and 12 T2D patients [33]. The E-MTAB-5060 was produced by RNA-sequencing of human pancreatic islet cells from 4 T2D patients and 3 healthy individuals using Illumina TruSeq protocol [34]. Since the complications and severity of COVID-19 usually vary drastically, we compiled two sets of experimental data to ensure the validity of our results. Firstly, we analyzed the differentially expressed genes (DEGs) for SARS-CoV-2 infection and T2D with data obtained from lung epithelial and pancreatic islet cells, respectively. Secondly, we compared the DEGs for the same disease-pair with data collected from human PBMCs.
2.2. Identification of significant dysregulated genes
The RNA-Seq is a next generation sequencing technology to measure gene expression with a high level of accuracy and mitigates many limitations of microarrays [35]. Using this high-throughput sequencing technology and global transcriptome analyses, we compared the gene expression profiles of SARS-CoV-2 and T2D. To identify DEGs associated with the respective diseases, we used an R Bioconductor package DESeq2 [36] which is explicitly designed for RNA-Seq data. This package uses the Student unpaired -test for the identification of DEGs.
As the GSE9006 dataset is a microarray data set, we analyzed it with the R Bioconductor package Limma [37] to obtain the dysregulated genes. Since various errors can be introduced in preparing and analyzing microarray data of different platforms and experimental systems, the gene expression data in each sample (case or control) were normalized using the -score transform which is defined as
| (1) |
where and are the mean and standard deviation of the expression value of -th gene estimated over all the samples, respectively. After obtaining the DEGs for each disease condition, we selected the significant genes by setting the threshold level of the absolute value of base-2 log Fold Change 1 and adjusted -value (FDR) . The shared DEGs by the SARS-CoV-2 and T2D were then obtained through a cross-comparative analysis.
2.3. Profiling associated comorbidities
To assess the possible health implications linked to COVID-19, we anticipated the comorbidities associated with the shared DEGs via Enrichr web-platform [38] using DisGeNET database [39]. This database is a publicly available repository of genes that are associated with human diseases and the latest version (v7.0) includes information regarding the association of 21,671 genes with 30,170 disorders (https://disgenet.org). In this analysis, we only considered diseases or complications with gene enrichment and adjusted -value. Likewise, we ran the shared DEGs using the same database via Metascape server [40], where statistical significance was set to adjusted p-value. For visualization of the gene-disease association (GDA), a bipartite network was created and designed with Cytoscape v3.8 software [41]. It is an open source software to produce high resolution interactomes and complex networks.
2.4. Functional enrichment analysis
Functional enrichment analysis, also referred to as gene set enrichment analysis (GSEA), identifies a group of significantly enriched genes engaging in the course of biological action or residing at the central chromosomal location by integrating all prior knowledge [42]. Such a group of genes can be associated with a biological function by means of gene ontology (GO) that has 3 separate classes including biological process (BP), molecular function (MF), and cellular component (CC) [43]. Similarly, pathway analysis also plays an important role to reveal the molecular or biological function that underlies the development of complex diseases and it also helps us to understand how to intervene therapeutically in disease processes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database constitutes metabolic pathways representing the network of molecules interconnected for a general function [44]. Besides KEGG, Wikipathways [45], and Reactome [46] pathway databases are also widely used as annotation sources for analyzing signalling pathways. We performed the GSEA using the online platform Enrichr by integrating the pathway and GO term annotations for all the shared DEGs between the SARS-CoV-2 and T2D to have a better understanding of the metabolic pathways active in SARS-CoV-2. Enrichr integrates available gene-set libraries to provide enrichment outcomes for a gene list of interest. For statistical significance, the adjusted -value was considered as less than 0.05 for the significance assessment of the obtained enrichment results.
2.5. Protein-protein interaction analysis
Proteins usually interact with each other to form molecular machines that perform cellular functions. Such protein-protein interactions (PPIs) can be mapped to discern the functional and structural knowledge of cellular protein networks [47]. We incorporated the web-based visualization software NetworkAnalyst v3.0 to construct the PPI network of the common DEGs. This web-tool interprets the gene expression data from PPI network perspective [48]. Herein, we employed the STRING database with a confidence score of 600 for network construction. This database comprises published and estimated interactions that include both functional and physical contacts [49]. In this case, we only considered an experimentally validated interaction dataset. To determine potential hubs within the PPI network, we then applied three different methods, i.e., degree, maximal clique centrality (MCC), and betweenness using cytoHubba plugin [50] in Cytoscape v3.8. Next, we compared the results and identified the common nodes as the most potential hubs. Finally, the obtained networks were customized in Cytoscape v3.8.
2.6. Probing the regulatory networks
Transcription factors (TFs) control the gene expression level by switching it on and off at the transcription level. Thus, TFs and their interactions with genes are essential for their functional implications [51]. We utilized the NetworkAnalyst platform to investigate the interactions of TFs with the shared DEGs. For this purpose, we selected the Encyclopedia of DNA Elements (ENCODE) database in NetworkAnalyst that uses peak intensity signal 500 and the predicted regulatory potential score 1. The ENCODE project provides genomic and transcriptomic annotations to characterize the human genomic elements [52]. Likewise, microRNAs (miRNAs) are non-coding RNAs that affect gene expression via post-transcriptional regulation [51]. To generate the gene-miRNA interactome, we provided the shared DEGs into the same platform utilizing the TarBase 8.0 database, which provides an experimentally validated collection of curated miRNA-gene interaction data as well as function-related data derived from various present-day empirical methods [53]. Both gene-miRNA and gene-TF networks were initially filtered with the degree centrality. Next, the obtained networks were analyzed with degree method via CytoHubba plugin to determine the key regulatory components and their interacting gene partners.
2.7. Identification of drug candidates
One of the primary objectives of this line of research focuses on pinpointing the potential drug molecules. We searched the Drug Signatures Database (DSigDB) based on the shared DEGs between SARS-CoV-2 and T2D using the Enrichr web-platform. The DSigDB is a collection of 22,527 gene sets related to the drug and small molecules considering the dysregulation in gene expression due to drug/compounds [54]. The significant drug molecules were filtered through manual curation based on adjusted -value.
3. Results
3.1. Shared differentially expressed genes
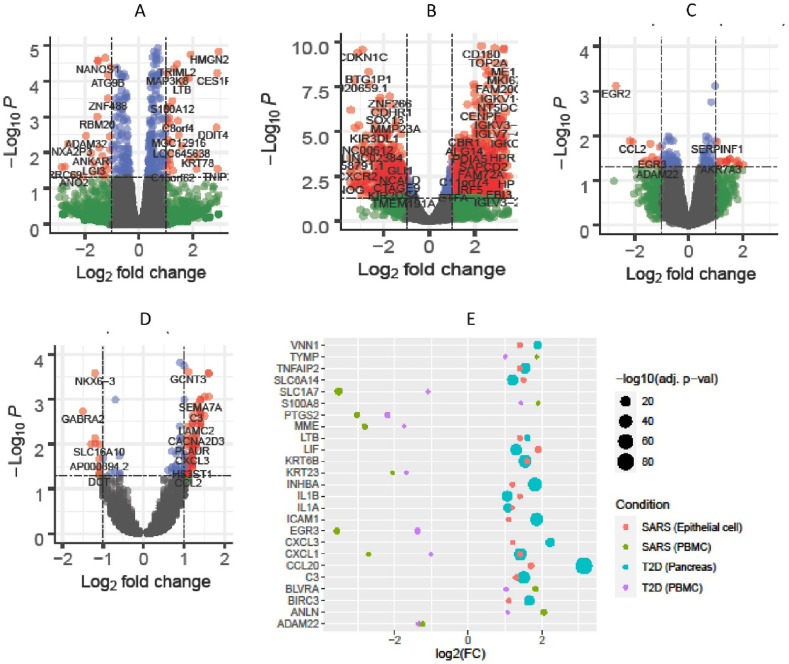
We first compared the gene expression levels of lung epithelium cells from SARS-CoV-2 patients (GSE147507) and pancreatic islet cells from T2D individuals (E-MTAB-5060) with the corresponding cells from healthy controls. Similarly, we also compared the expression profiling of PBMCs from SARS-CoV-2 (E-MTAB-8871) and T2D (GSE9006) patients with normal individuals. By filtering DEGs for each disease condition, we found 156 and 78 significant DEGs in the first experiment for SARS-CoV-2 and T2D, respectively. In the second experiment, we found 1289 and 73 significant DEGs in PBMCs for the same disease-pair. The four volcano plots in Fig. 2 presented the expression pattern of the genes in both experiments.
Fig. 2.
Differential gene expression and common DEGs. Volcano plots depict the genes expression in A) SARS-CoV-2 infected lung epithelium cells, B) SARS-CoV-2 infected PBMCs, C) PBMCs of T2D patients, and D) T2D diseased pancreatic islet cells (red dots indicate significant DEGs), while bubble plot shows (E) the expression pattern of 25 common DEGs between SARS-CoV-2 and T2D in lung epithelium cells and pancreatic islet cells, respectively as well as in PBMCs.
The cross-comparative analysis revealed 15 common DEGs between SARS-CoV-2 and T2D for lung epithelium and pancreatic islet cells, respectively. On the other hand, for PBMCs, we found 11 common DEGs between the disease pair. By combining these two sets of DEGs, we found a total of 25 unique shared DEGs. Negative 10-base logarithmic transformed FDR adjusted -value of these 25 common DEGs are shown in Fig. 2-E. Notably, all the common genes were strikingly up-regulated for the lung epithelium and pancreatic islet cell in SARS-CoV-2 and T2D, respectively, whereas CXCL1 was irregularly expressed in both scenarios.
3.2. Significant gene ontologies and signalling pathways
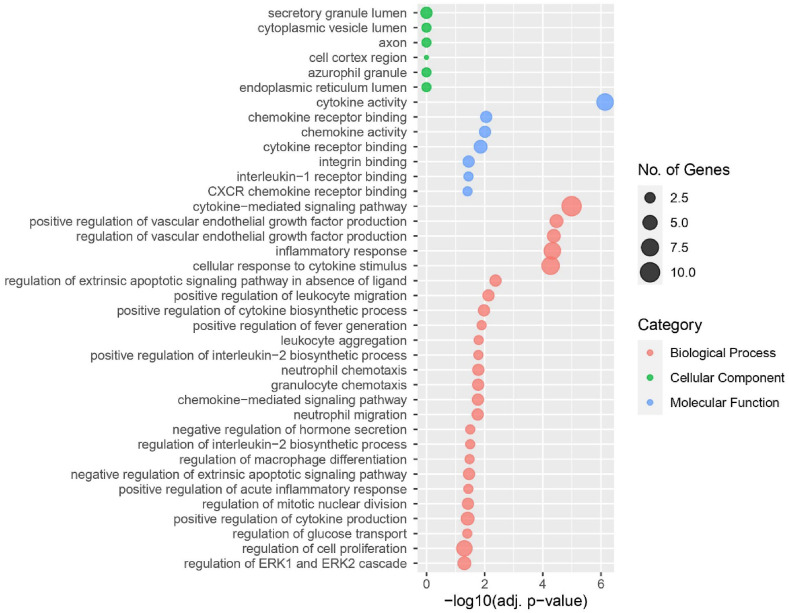
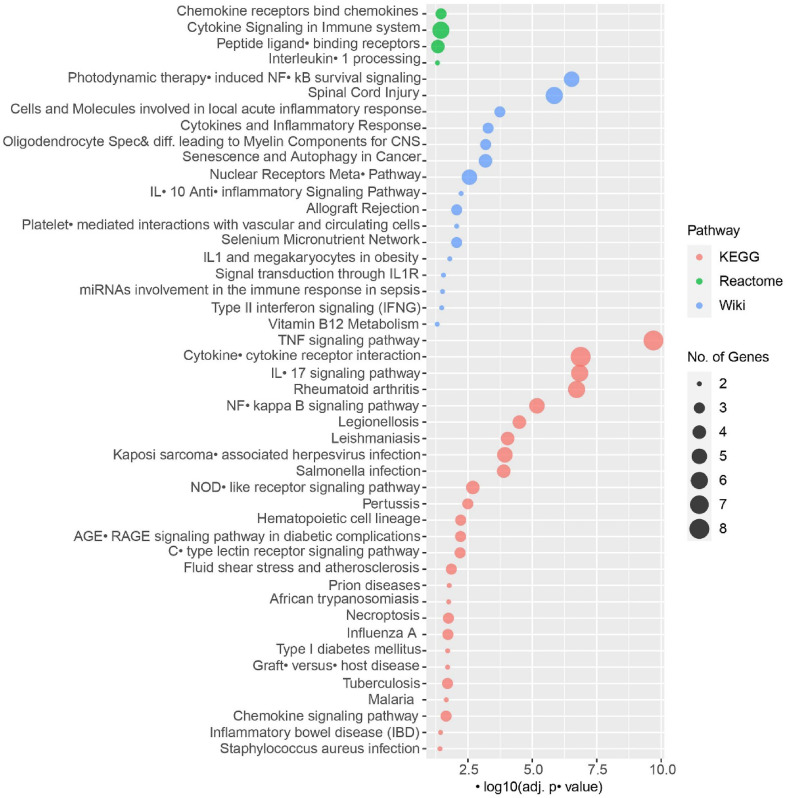
After identifying the overlapping DEGs for the disease pair, we performed extensive GO and signalling pathway analyses accessing relevant curated databases. The adjusted -value filtration found a total of 38 GO terms (Fig. 4) and 46 molecular pathways (Fig. 5) to be significantly enriched. Curation found cytokine activity and cytokine-mediated signalling pathway to be predominant. Similarly, signalling pathway analyses exhibited TNF signalling pathway and cytokine-cytokine receptor interaction having noteworthy enrichment.
Fig. 4.
Gene ontology analysis revealed significant GO terms associated with SARS-CoV-2 and T2D. The biological process, cellular component, and molecular function datasets were considered for this analysis.
Fig. 5.
Significant signalling pathways associated with SARS-CoV-2 and T2D. The human KEGG (2019), reactome (2016), and human WikiPathways (2019) datasets were considered for this analysis.
3.3. Genetic relationship of COVID-19 with other diseases
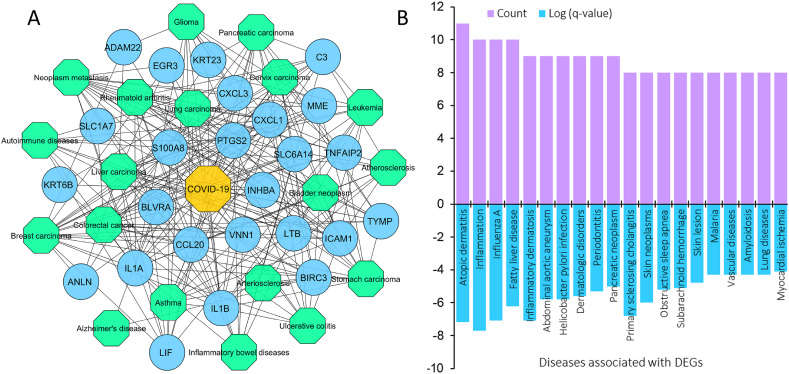
By analyzing the GDA, we identified 19 highly significant (-value ) diseases associated with up to 18 shared DEGs. Fig. 3 shows the different cancers and diseases related to DEGs identified in COVID-19. Most of the comorbidities were different forms of cancers. The highest number of DEGs (18) were found to be associated with breast cancer and its metastasis while at least 10 DEGs were affiliated with ulcerative colitis, cervix carcinoma, bladder neoplasm, atherosclerosis, autoimmune diseases, and inflammatory bowel disease (IBD). Other diseases include rheumatoid arthritis (13), asthma (12), Alzheimer's disease (11), and arteriosclerosis (11). Additionally, the Metascape server predicted the top 20 diseases that are associated with 8–11 genes as shown in Fig. 3-B. These DEGs and diseases could provide the directionality of the COVID-19 progression and its long-term health impact.
Fig. 3.
Gene-disease association network. In this figure, (A) the bipartite network includes circular nodes (blue) representing the shared DEGs and hexagonal nodes indicating COVID-19 (yellow) and different diseases (green), and (B) the bar graph indicating the top 20 diseases associated with DEGs in terms of expression and number of DEGs involved as predicted by the Metascape server.
3.4. Hub-proteins identified in the PPI network
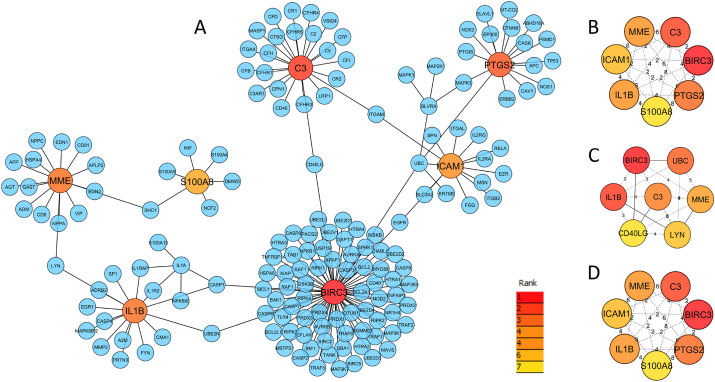
We constructed the PPI network using all the distinct DEGs shared between the disease pair through NetworkAnalyst. The resulted PPI network comprises 171 nodes and 176 edges as shown in Fig. 6 -A. Each node in the network represents a protein and an edge indicates the functional interaction between two proteins. We employed three methods for topological analysis where each method identified the top 7 hub-nodes within the PPI network (Fig. 6B–D). Interestingly, four out of 7 hub-proteins were anticipated by all methods. As validated by multiple methods and exhibited a minimum of eight interconnections, we recognized those four hub-nodes as potential hub-proteins, i.e., BIRC3, C3, MME, and IL1B.
Fig. 6.
The protein-protein interaction network and hub-proteins. This network was constructed with the DEGs shared by SARS-CoV-2 and T2D using STRING database (confidence cut off 600). The network depicting (A) a total of 171 proteins including 11 shared DEGs in which 7 hubs were indicated as predicted by the degree method. Additionally, three smaller networks are depicting hub-proteins anticipated by (B) degree, (C) betweenness, and (D) maximal clique centrality (MCC) methods. For all methods, the top seven hub-proteins are indicated by the color ranging from red (higher) to yellow (lower).
3.5. Regulatory networks revealed highly active TFs and miRNAs
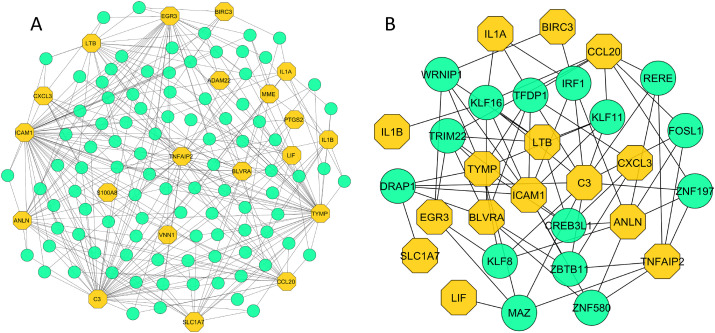
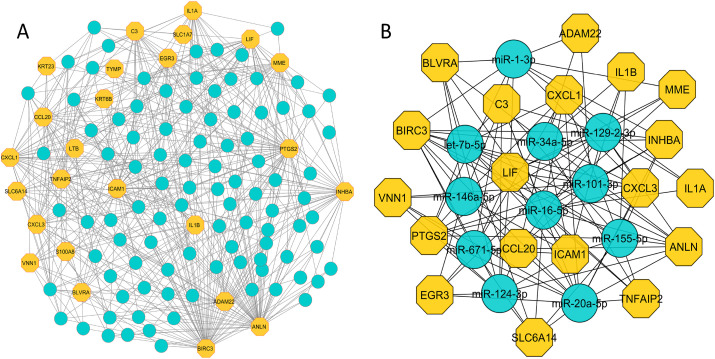
The TF-gene interactome obtained from NetworkAnalyst contains a total of 116 nodes with 341 interactions, where 20 nodes were from our common DEGs. The network comprises circular and hexagonal nodes representing TFs and common DEGs, respectively. Out of total 116 nodes, 111 nodes showed at least 3 TF-gene interactions. Notably, ICAM1, C3 and TYMP evidenced 49, 45 and 42 TF-gene interactions, respectively. The high number of interactions between the TFs and common DEGs are shown in Fig. 7 -A. Further network analysis with degree method revealed 30 most significant nodes including 15 TFs (IRF1, RERE, FOSL1, KLF8, MAZ, ZNF580, DRAP1, TRIM22, KLF16, WRNIP1, TFDP1, KLF11, CREB3L1, ZBTB11, and ZNF197) and their partner DEGs (Fig. 7-B). Similarly, the constructed gene-miRNA interaction network identified a total of 535 interactions among 121 nodes where 96 nodes represent miRNAs as shown in Fig. 8 -A. The gene ANLN exhibited maximum 59 interactions whereas BIRC3, PTGS2, INHBA, LIF, and ICAM1 were involved with 30 connections and/or more. All the nodes regulated at least three neighbours suggesting high level of miRNA association with the shared DEGs. Finally, topological study of the network identified 11 most important miRNAs (miR-1-3p, 34a-5p, 129-2-3p, 146a-5p, 16–5p, 101–3p, 671–5p, 155–5p, 124–3p, 20a-5p, and let-7b-5p) interacting with 19 DEGs (Fig. 8-B).
Fig. 7.
TF-gene interaction network. The network was constructed using the shared DEGs and filtered with degree centrality. It shows (A) the interactions of 96 TFs with 20 DEGs, and (B) the 30 most significant nodes of the gene-TF network, which included 15 TFs and their interactions with 15 DEGs.The hexagonal (yellow) and circular (green) shaped nodes in the network indicate DEGs and TFs, respectively.
Fig. 8.
Gene-miRNA interaction network. The network was constructed using the shared DEGs and filtered with degree centrality. It shows (A) the interactions of 96 miRNAs with 25 DEGs, and (B) the 30 most significant nodes of the gene-miRNA network that includes 11 miRNAs and their interactions with 19 DEGs.The hexagonal (yellow) and circular (blue) shaped nodes in the network indicate DEGs and miRNAs, respectively.
3.6. Candidate drug identification
Based on the shared DEGs between SARS-CoV-2 and T2D, we identified some potential drug molecules. We obtained 438 significant drugs/compounds to be associated with the DEGs that resulted from manual curation by considering FDR adjusted -value. Among them, estradiol and tetradioxin were the top 2 molecules based on the number of DEGs involved. Again, phencyclidine and profenamine molecules were found to be the most significant ones. The top 20 significant drug molecules along with the associated DEGs are provided in Table 1 .
Table 1.
Top 20 significant drug candidates identified for shared DEGs between SARS-CoV-2 and T2D.
| Drug/small molecule | -value | Adj. -value | Associated genes |
|---|---|---|---|
| Phencyclidine | 1.16E-15 | 4.68E-12 | IL1A, CCL20, IL1B, TNFAIP2, LIF, CXCL1, CXCL3, PTGS2, ICAM1, TYMP, BIRC3 |
| Profenamine | 1.94E-14 | 3.91E-11 | IL1B, LIF, CXCL1, INHBA, CXCL3, PTGS2, ICAM1, BIRC3 |
| 8-Azaguanine | 5.99E-14 | 8.04E-11 | IL1A, CCL20, IL1B, LIF, CXCL1, INHBA, CXCL3, PTGS2, KRT6B, ICAM1, BIRC3 |
| Nickel chloride | 6.02E-13 | 6.06E-10 | IL1A, CCL20, IL1B, TNFAIP2, CXCL1, INHBA, LTB, CXCL3, PTGS2, ICAM1, BIRC3 |
| Chloropyramine | 2.62E-12 | 2.11E-09 | LIF, CXCL1, INHBA, PTGS2, ICAM1, BIRC3 |
| Pizotifen | 2.36E-11 | 1.58E-08 | CXCL1, INHBA, CXCL3, PTGS2, ICAM1, BIRC3 |
| MS-275 | 4.76E-11 | 2.74E-08 | C3, IL1A, CCL20, IL1B, TNFAIP2, CXCL1, PTGS2, KRT6B, S100A8, BIRC3 |
| 0297417-0002B | 1.53E-10 | 7.74E-08 | LIF, CXCL1, LTB, PTGS2, KRT6B, ICAM1, BIRC3 |
| Silica | 1.89E-10 | 8.45E-08 | ANLN, IL1A, EGR3, CCL20, IL1B, TNFAIP2, CXCL1, LTB, CXCL3, PTGS2, ICAM1, BIRC3 |
| Gemcitabine | 2.93E-10 | 1.07E-07 | IL1A, CCL20, IL1B, CXCL1, CXCL3, ICAM1, TYMP, BIRC3 |
| Peptidoglycan | 3.313E-10 | 1.11E-07 | IL1A, CCL20, IL1B, PTGS2, ICAM1 |
| 1-Nitropyrene | 2.83E-10 | 1.14E-07 | IL1A, CCL20, IL1B, CXCL1, LTB, CXCL3, ICAM1, BIRC3 |
| Estradiol | 8.55E-10 | 2.45E-07 | EGR3, MME, CCL20, SLC6A14, ADAM22, LIF, KRT23, INHBA, CXCL3, PTGS2, ICAM1, C3, ANLN, IL1A, VNN1, IL1B, LTB, KRT6B, S100A8, BIRC3 |
| Nickel sulphate | 8.04E-10 | 2.49E-07 | IL1A, CCL20, IL1B, CXCL1, INHBA, CXCL3, PTGS2, ICAM1, TYMP, BIRC3 |
| 3-Nitrofluoranthene | 9.33E-10 | 2.50E-07 | CCL20, CXCL1, LTB, CXCL3 |
| Dexamethasone | 1.12E-09 | 2.67E-07 | IL1B, SLC6A14, LIF, LTB, PTGS2, S100A8, ICAM1, TYMP, BIRC3 |
| Niclosamide | 1.10E-09 | 2.77E-07 | CCL20, CXCL1, INHBA, CXCL3, PTGS2, ICAM1, BIRC3 |
| MG-132 | 1.35E-09 | 3.02E-07 | IL1A, IL1B, CXCL1, PTGS2, ICAM1, TYMP, BIRC3 |
| Simvastatin | 2.24E-09 | 4.75E-07 | IL1A, IL1B, LIF, LTB, PTGS2, S100A8, ICAM1, BIRC3 |
| Thioridazine | 3.22E-09 | 6.49E-07 | LIF, INHBA, LTB, CXCL3, PTGS2, ICAM1, BIRC3 |
4. Discussion
This research work aimed to uncover the obscure genetic interactions between SARS-CoV-2 and T2D by maneuvering a series of bioinformatics approaches. To our best knowledge, this is the first endeavor to divulge the genomic and transcriptomic connections shared by the disease pair. In this study, we first performed the gene expression analysis of the SARS-CoV-2 and T2D in lung epithelium and pancreatic islet cells, respectively, as well as in peripheral blood using four different datasets. Considering the expression patterns, we identified 25 unique DEGs shared by the disease pair that manifested their genetic proximity. The subsequent course of actions in this study revolved around these shared DEGs employing functional enrichments in terms of GO, molecular pathways, protein-protein interactome, regulatory network, and drug target identification.
Functional enrichment analysis using the overlapping DEGs derived significant GO terms associated with diseases under consideration. Markedly, inflammation related signalling pathways, including cytokine activity, cytokine-mediated signalling pathway, chemokine activity, and cellular response to cytokine stimulus were found to be prevalent. Evidently, the comprehensive activity of these pathways may contribute to the cytokine storm, thus, playing a crucial role in the progression of COVID-19 [55]. A recent study revealed chronic inflammation as a prominent feature of T2D that may lead to abnormal blood clot formation, which explains the severity of T2D upon SARS-CoV-2 infection [56]. Another biological process is the regulation of cell proliferation, which is plausible since cell cycle regulation actively occurs in T2D patients to counteract insulin resistance by increasing insulin-secreting -cells despite the lack of success [57]. Therefore, the DEGs associated with this GO term may directly or indirectly involve in the regulation of -cell proliferation and hence, can be potential therapeutic targets [58].
In our GDA analysis, we found that most of the shared DEGs (18) were associated with breast carcinoma and cancer metastasis. The ICAM1 is crucial for infectious disease in viral replication modulation and has been evidenced to have key involvement in lung disease of severe COVID-19 patients [59]. Again, genetic and biological evidence suggest that ICAM1 has a vital role in the development of diabetes [60]. The dysregulation of ANLN (anillin actin binding protein) gene is associated with pulmonary carcinogenesis and it can be characterized as a prognostic biomarker and therapeutic candidate for lung cancer [61]. A recent study identified ANLN as a potential biomarker for bladder urothelial carcinoma [62]. Besides, 10 DEGs from this study were found to be linked to bladder cancers. A recent review highlighted the higher risk of COVID-19 and/or disease severity in individuals with breast cancer [63]. On the other hand, the least associated diseases were atherosclerosis, IBD, bladder and cervix cancers, and ulcerative colitis, which involved with 10 shared DEGs each. In particular, the intense inflammations due to IL-6, TNF-, and IL-1 can lead to aggravated atherosclerosis which in turn contribute to developing cardiovascular diseases [64]. Although no association was found between IBD and increased susceptibility towards SARS-CoV-2, a recent study suggested gut inflammation may contribute to viral entry by up-regulating ACE2 expression [65]. Other notable diseases were lung carcinoma, pancreatic carcinoma, rheumatoid arthritis, and asthma, which were also evident in some previous studies [[66], [67], [68], [69]]. Therefore, the associated DEGs could be of interest for risk management and genetic manipulation for therapeutic purposes.
Pathway analysis focusing on the common DEGs was carried out and identified similar pathways for COVID-19 and T2D. Amongst all, TNF signalling pathway, cytokine-cytokine receptor interaction, IL-17 signalling pathway, and photodynamic therapy-induced NF–B survival signalling were the top annotated pathways. Ouyang et al. found TNF-signalling pathway to be significantly enriched in severe COVID-19 conditions [70]. Recently, Akash et al. reviewed the direct involvement of TNF- with the pathogenesis of T2D. Activation of TNF- reduces the expression of glucose transporter type 4 (GLUT4) which is related to insulin metabolism. Besides, TNF- expression coincides with the inhibition of insulin receptor through serine phosphorylation of insulin receptor substrate-1 (IRS-1) [71]. Likewise, IL-17 pathway is involved in cytokine storm that regulates normal immune response [72]. Intriguingly, IL-17 also promotes insulin resistance by inducing angiotensin II type 1 receptor (AT1R). This lead to the production of nitric oxide (NO) in diabetic nephropathy resulting in complications in T2D patients [73]. Therefore, the increased disease severity of COVID-19 in T2D patients is not surprising. Now that we know the genetics of this complex interplay, blocking the involved DEGs could be an effective strategy in alleviating disease severity by regulating these signalling pathways. For instance, there are some clinical trials of antibodies against IL-17 for chronic inflammatory diseases such as psoriasis and arthritis [74]. These approaches can also be adopted to ameliorate the hyper-inflammation in SARS-CoV-2-infected T2D. Moreover, AT1R antagonists or vitamin D or in combinations are suggested to reduce the COVID-19 complications [75].
The PPI network analysis using the shared DEGs promisingly identified BIRC3, C3, MME, and IL1B as the potential hub-proteins, the proteins exhibiting a high degree of interactions and identified in multiple networks analyses. Notably, BIRC3 (Baculoviral IAP repeat containing 3) is connected with Marginal Zone B-Cell Lymphoma and is reported as NK cell immune checkpoint in tumor [76]. Likewise, C3 (Complement C3) is a principal element of the protective immune system against pathogens [77]. A recent study noticed hyper-activation of C3 in COVID-19 and suggested its suppression as a therapeutic strategy [78]. Importantly, C3 is suggested as a potential therapeutic target for diabetic nephropathy in a recent study [79]. Another hub-protein MME (Membrane metalloendopeptidase) in cerebrospinal fluid (CSF) was found to be involved in Alzheimer's disease progression [80]. Interestingly, in this study, we also identified Alzheimer's disease as the comorbidity of COVID-19. This protein directly employs neutrophil recruitment and consequently intensifies the immunopathology of COVID-19 [81]. The IL1B (Interleukin 1 beta) belongs to the interleukin 1 cytokine family and plays a vital role in arbitrating the inflammatory response. It is also involved in osteoarthritis pathogenesis [82], which was also evident in our comorbidity analysis. Moreover, 1L1B like pro-inflammatory cytokine level upraises in the bronchial alveolar lavage fluid of serious COVID-19 patients [83]. Consequently, the inflammatory response promoted by IL1B induces lung damage in COVID-19 [84]. Additionally, PTGS2 (Prostaglandin-endoperoxide synthase 2), exclusively detected by MCC method, is demonstrated to be a prognostic biomarker for gastric cancer [85]. Moreover, PTGS2 is lately reported as a key target molecule of the Qing-Fei-Pai-Du decoction (QFPDD), the traditional Chinese medicine that was successfully applied to treat COVID-19 [86]. Therefore, the identified hub-proteins could be considered as potential biomarkers for COVID-19. Further investigation could be conducted to confirm their biological involvement to highlight them as drug targets.
Furthermore, the regulatory network analyses elucidated notably high occurrence of TF-gene and gene-miRNA interactions. The ICAM1 (Intercellular adhesion molecule 1) gene was found to be involved with the highest number (49) of TF-gene interactions. In this study, we found interferon regulatory factor 1 (IRF1) as a potential TF. A previous study showed IRF1 directly regulates gene expression in response to herpes simplex virus (HSV) and MERS-CoV infections [87], probably by promoting IRF3 activation [88]. It is also responsible for the upregulation of interleukin-1 (IL1B) as we evidenced in our gene-TF network [89]. Also, krüppel-like factors (KLFs) are TFs that regulate various pathways and rendered metabolic abnormalities if altered. Notably, phenotypic expression related to KLF-11 includes decreased circulating insulin [90]. We identified KLF8, KLF11 and KLF16 as highly active TFs. Besides, we detected TF protein CREB3L1 which is in line with a previous study [91]. This TF is associated with ER stress [92] and PCSK1 gene, which was found to be linked with diabetes mellitus [93]. Another TF protein FOSL1 interacts with CCL20 gene, which was found upregulated in this study. The elevated level of CCL20 is implicated in cytokine storm of COVID-19, obesity, and insulin resistance [94,95]. Also, the CCL20 is regulated by the NF–B subunits in pancreatic -cells [95]. Importantly, FOSL1 is reported to negatively regulate the type I interferon (IFN–I) response in host against malaria and viral infections and is identified as a potential drug target for controlling malaria and other diseases [96].
We also detected 11 potential miRNAs that might have shared pathogenetic relationship between COVID-19 and diabetes. For example, miR-1-3p and 20a-5p identified in this study reported to be involved in viral respiratory diseases and can be used to design anti-viral drugs [97]. Another miRNA miR-34a-5p increases high glucose-mediated apoptosis in cardiomyocytes by reducing anti-apoptotic BCL2 protein [98]. Thus, the upregulation of miR-34a-5p in the diabetic condition is most likely to increase apoptosis [99]. Interestingly, the elevated level of miRNA-34a was also observed in patients who experienced acute myocardial infarction (MI) leading to heart failure [99]. Further, some miRNAs associated with lung diseases (e.g., let-7b-5p) and asthma (e.g., miR-155–5p, miR-16–5p) were highly prevalent in COVID-19 [100]. Furthermore, miR-16 and miR-155 modulate inflammatory responses by targeting the MCL1 and NFKBIA hub-proteins [100,101]. Therefore, the association of identified TFs and miRNAs with COVID-19 pathogenesis and diabetes could reveal other potential drug targets with further study.
Moreover, taking the common DEGs into consideration with the DSigDB database, several drug/molecule candidates were proposed. As expected, the top interacted component estradiol was found to have a positive protective effect on COVID-19 patients in several experimental investigations that suggests its prospect as a therapeutic candidate against SARS-CoV-2 [102]. Again, dexamethasone which had association with 9 shared DEGs, was found effective in COVID-19 patients who require oxygen supplement and/or mechanical ventilation [103]. Another drug candidate tetradioxin (not shown in Table 1) showed interactions with 18 shared DEGs. Previously, dioxin showed immune suppression activity through the inhibition of CD4+ T-cell differentiation into Th1, Th2 and Th17 effector cells [104]. Strikingly, activation of Th17 is mainly responsible for IL-17 release, the prominent cause of cytokine storm in COVID-19 [105,106]. Therefore, the identified drug candidates can further be assessed for their potency in alleviating the COVID-19 complication. Altogether, the obtained results demand clinical expeditions that may improve our understanding of the genetic impact of T2D on SARS-CoV-2 as well as to propose putative therapeutic targets.
5. Conclusions
To conclude, the pathophysiological outcome of SARS-CoV-2 infection significantly differs in different people based on their underlying health conditions. In this study, we explored the molecular pathogenesis of COVID-19 in patients with pre-existing T2D hoping to find the genetic determinants that cause higher infection risk and disease severity. In doing so, we observed that both diseases exhibited similarity in gene expression profile to some extent. We also identified several key proteins that could be the source of potential biomarkers in early disease prognosis. In addition, the identified regulatory molecules such as TFs and miRNAs may highlight the disease-modifying factors involved in the COVID-19 progression. These regulatory molecules along with the signalling pathways can be targeted for the development of novel COVID-19 therapeutics. Furthermore, the identified potential drug candidates need experimental validation for their therapeutic evaluation. Thus, we believe that this study will shed light on the understanding of the intertwined pathobiology of the complexity of SARS-CoV-2 infected T2D patients.
Contributions
All authors contributed to the manuscript. M.B.I., U.N.C., and M.A.M. conceived and designed the study. M.B.I. and U.N.C. analyzed the data and wrote the R programming code for the development of the pipeline. M.B.I., U.N.C., Z.N. and M.A.M. conducted all other bioinformatic analyses and wrote the manuscript. S.U. and M.B.A. provided insightful guidance and feedback for the study. All authors read and approved the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Drosten C., Günther S., Preiser W., Van Der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. others. [DOI] [PubMed] [Google Scholar]
- 2.Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China, Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Organization W.H. World Health Organization; 2020. Clinical Management of Severe Acute Respiratory Infection (SARI) when COVID-19 Disease Is Suspected: Interim Guidance, 13 March 2020. others. [Google Scholar]
- 6.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microb. Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO coronavirus (COVID-19) dashboard. https://covid19.who.int/ World Health Organization. (n.d.
- 9.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection?, the Lancet. Respir. Med. 2020;8 doi: 10.1016/S2213-2600(20)30116-8. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roncon L., Zuin M., Rigatelli G., Zuliani G. Diabetic patients with COVID-19 infection are at higher risk of ICU admission and poor short-term outcome. J. Clin. Virol. 2020 doi: 10.1016/j.jcv.2020.104354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.-Y. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C., Ma Y., Zhao J., Nussinov R., Zhang Y.-C., Cheng F., Zhang Z.-K. Computational network biology: Data, model, and applications. Phys. Rep. 2019;846:1–66. [Google Scholar]
- 14.Barabási A.-L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease, Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rana H.K., Akhtar M.R., Islam M.B., Ahmed M.B., Lio P., Quinn J.M., Huq F., Moni M.A. Genetic effects of welding fumes on the development of respiratory system diseases. Comput. Biol. Med. 2019;108:142–149. doi: 10.1016/j.compbiomed.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Chowdhury U.N., Islam M.B., Ahmad S., Moni M.A. Network-based identification of genetic factors in ageing, lifestyle and type 2 diabetes that influence to the progression of alzheimer's disease. Informatics in Medicine Unlocked. 2020 [Google Scholar]
- 17.Rahman M.R., Islam T., Zaman T., Shahjaman M., Karim M.R., Huq F., Quinn J.M., Holsinger R.D., Gov E., Moni M.A. Identification of molecular signatures and pathways to identify novel therapeutic targets in alzheimer's disease: insights from a systems biomedicine perspective. Genomics. 2020;112:1290–1299. doi: 10.1016/j.ygeno.2019.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Moni M.A., Rana H.K., Islam M.B., Ahmed M.B., Xu H., Hasan M.A.M., Lei Y., Quinn J.M. A computational approach to identify blood cell-expressed Parkinson's disease biomarkers that are coordinately expressed in brain tissue, Comput. Biol. Med. 2019;113 doi: 10.1016/j.compbiomed.2019.103385. [DOI] [PubMed] [Google Scholar]
- 19.Haidar M.N., Islam M.B., Chowdhury U.N., Rahman M.R., Huq F., Quinn J.M., Moni M.A. Network-based computational approach to identify genetic links between cardiomyopathy and its risk factors. IET Syst. Biol. 2020;14:75–84. doi: 10.1049/iet-syb.2019.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moni M.A., Liò P. Network-based analysis of comorbidities risk during an infection: SARS and HIV case studies. BMC Bioinf. 2014;15:333. doi: 10.1186/1471-2105-15-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moni M.A., Lio’ P. Genetic profiling and comorbidities of zika infection. J. Infect. Dis. 2017;216:703–712. doi: 10.1093/infdis/jix327. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery. 2020;6:1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng X., Zhu S., Lu W., Liu Z., Huang J., Zhou Y., Fang J., Huang Y., Guo H., Li L. Target identification among known drugs by deep learning from heterogeneous networks. Chem. Sci. 2020;11:1775–1797. doi: 10.1039/c9sc04336e. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng X., Zhu S., Hou Y., Zhang P., Li L., Li J., Huang L.F., Lewis S.J., Nussinov R., Cheng F. Network-based prediction of drug–target interactions using an arbitrary-order proximity embedded deep forest. Bioinformatics. 2020;36(9):2805–2812. doi: 10.1093/bioinformatics/btaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng F., Kovács I.A., Barabási A.-L. Network-based prediction of drug combinations. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-09186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nain Z., Rana H.K., Liò P., Islam S.M.S., Summers M.A., Moni M.A. Pathogenetic profiling of COVID-19 and SARS-like viruses. Briefings Bioinf. 2020;22(2):1175–1196. doi: 10.1093/bib/bbaa173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taz T.A., Ahmed K., Paul B.K., Kawsar M., Aktar N., Mahmud S., Moni M.A. Network-based identification genetic effect of SARS-CoV-2 infections to idiopathic pulmonary fibrosis (IPF) patients. Briefings Bioinf. 2020;22(2):1254–1266. doi: 10.1093/bib/bbaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taz T.A., Ahmed K., Paul B.K., Al-Zahrani F.A., Mahmud S.H., Moni M.A. Identification of biomarkers and pathways for the SARS-CoV-2 infections that make complexities in pulmonary arterial hypertension patients. Briefings Bioinf. 2021;22(2):1451–1465. doi: 10.1093/bib/bbab026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satu M.S., Khan M.I., Rahman M.R., Howlader K.C., Roy S., Roy S.S., Quinn J.M., Moni M.A. Diseasome and comorbidities complexities of SARS-CoV-2 infection with common malignant diseases. Briefings Bioinf. 2021;22(2):1415–1429. doi: 10.1093/bib/bbab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nashiry A., Sumi S.S., Islam S., Quinn J.M., Moni M.A. Bioinformatics and system biology approach to identify the influences of COVID-19 on cardiovascular and hypertensive comorbidities. Briefings Bioinf. 2021;22(2):1387–1401. doi: 10.1093/bib/bbaa426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanco-Melo D., Nilsson-Payant B., Liu W.-C., Møller R., Panis M., Sachs D., Albrecht R. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. BioRxiv. 2020 others. Preprint. [Google Scholar]
- 32.Ong E.Z., Chan Y.F.Z., Leong W.Y., Lee N.M.Y., Kalimuddin S., Mohideen S.M.H., Chan K.S., Tan A.T., Bertoletti A., Ooi E.E. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020;27(6):879–882. doi: 10.1016/j.chom.2020.03.021. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaizer E.C., Glaser C.L., Chaussabel D., Banchereau J., Pascual V., White P.C. Gene expression in peripheral blood mononuclear cells from children with diabetes. J. Clin. Endocrinol. Metab. 2007;92:3705–3711. doi: 10.1210/jc.2007-0979. [DOI] [PubMed] [Google Scholar]
- 34.Segerstolpe Å, Palasantza A., Eliasson P., Andersson E.-M., Andréasson A.-C., Sun X., Picelli S., Sabirsh A., Clausen M., Bjursell M.K. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabol. 2016;24:593–607. doi: 10.1016/j.cmet.2016.08.020. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z., Gerstein M., Snyder M. RNA-seq: a revolutionary tool for transcriptomics, Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update, Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piñero J., Bravo À., Queralt-Rosinach N., Gutiérrez-Sacristán A., Deu-Pons J., Centeno E., Sanz F., Furlong L.I., J. García-García DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016;45(D1):D833–D839. doi: 10.1093/nar/gkw943. gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1–10. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles, Proc. Natl. Acad. Sci. Unit. States Am. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pico A.R., Kelder T., Van Iersel M.P., Hanspers K., Conklin B.R., Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060184. e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B. The reactome pathway knowledgebase, Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Las Rivas J., Fontanillo C. Protein–protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou G., Soufan O., Ewald J., Hancock R.E., Basu N., Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis, Nucleic Acids Res. 2019;47:W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016;45(D1):D362–D368. doi: 10.1093/nar/gkw937. others. gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chin C.-H., Chen S.-H., Wu H.-H., Ho C.-W., Ko M.-T., Lin C.-Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8:1–7. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 52.Maher B. ENCODE: the human encyclopaedia, Nature News. 2012;489:46. doi: 10.1038/489046a. [DOI] [PubMed] [Google Scholar]
- 53.Karagkouni D., Paraskevopoulou M.D., Chatzopoulos S., Vlachos I.S., Tastsoglou S., Kanellos I., Papadimitriou D., Kavakiotis I., Maniou S., Skoufos G. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA–gene interactions, Nucleic Acids Res. 2018;46:D239–D245. doi: 10.1093/nar/gkx1141. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoo M., Shin J., Kim J., Ryall K.A., Lee K., Lee S., Jeon M., Kang J., Tan A.C. DSigDB: drug signatures database for gene set analysis. Bioinformatics. 2015;31:3069–3071. doi: 10.1093/bioinformatics/btv313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mangalmurti N., Hunter C.A. Cytokine storms: understanding COVID-19. Immunity. 2020;53(1):19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Randeria S.N., Thomson G.J., Nell T.A., Roberts T., Pretorius E. Inflammatory cytokines in type 2 diabetes mellitus as facilitators of hypercoagulation and abnormal clot formation. Cardiovasc. Diabetol. 2019;18:72. doi: 10.1186/s12933-019-0870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keller M.P., Choi Y., Wang P., Davis D.B., Rabaglia M.E., Oler A.T., Stapleton D.S., Argmann C., Schueler K.L., Edwards S. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 2008;18:706–716. doi: 10.1101/gr.074914.107. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang W.-J., Peng Y.-C., Yang K.-M. Cellular signaling pathways regulating β-cell proliferation as a promising therapeutic target in the treatment of diabetes. Experimental and Therapeutic Medicine. 2018;16:3275–3285. doi: 10.3892/etm.2018.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pouya F., Saber Z.I., Kerachian M.A. Molecular aspects of co-morbidities in COVID-19 infection. Archives of Bone and Joint Surgery. 2020;8:226. doi: 10.22038/abjs.2020.47828.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu H.F., Ma J., Gu K.T., Brismar K. Association of intercellular adhesion molecule 1 (ICAM1) with diabetes and diabetic nephropathy. Front. Endocrinol. 2013;3:179. doi: 10.3389/fendo.2012.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki C., Daigo Y., Ishikawa N., Kato T., Hayama S., Ito T., Tsuchiya E., Nakamura Y. ANLN plays a critical role in human lung carcinogenesis through the activation of RHOA and by involvement in the phosphoinositide 3-kinase/AKT pathway. Canc. Res. 2005;65:11314–11325. doi: 10.1158/0008-5472.CAN-05-1507. [DOI] [PubMed] [Google Scholar]
- 62.Zeng S., Yu X., Ma C., Song R., Zhang Z., Zi X., Chen X., Wang Y., Yu Y., Zhao J. Transcriptome sequencing identifies ANLN as a promising prognostic biomarker in bladder urothelial carcinoma. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-02990-9. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Derosa L., Melenotte C., Griscelli F., Gachot B., Marabelle A., Kroemer G., Zitvogel L. The immuno-oncological challenge of COVID-19. Nat. Can. (Que.) 2020:1–19. doi: 10.1038/s43018-020-00122-3. [DOI] [PubMed] [Google Scholar]
- 64.Vinciguerra M., Romiti S., Fattouch K., De Bellis A., Greco E. Atherosclerosis as pathogenetic substrate for sars-Cov2 cytokine storm. J. Clin. Med. 2020;9:2095. doi: 10.3390/jcm9072095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Popa I.V., Diculescu M., Mihai C., Cijevschi-Prelipcean C., Burlacu A. COVID-19 and inflammatory bowel diseases: risk assessment, shared molecular pathways and therapeutic challenges. Gastroenterol. Res. Pract. 2020;2020 doi: 10.1155/2020/1918035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calles A., Aparicio M.I., Alva M., Bringas M., Gutierrez N., Soto J., Arregui M., Tirado V.C., Álvarez E.L., del Monte-Millán M. Outcomes of COVID-19 in patients with lung cancer treated in a tertiary hospital in madrid. Frontiers in Oncology. 2020;10:1777. doi: 10.3389/fonc.2020.01777. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chai P., Yu J., Ge S., Jia R., Fan X. Genetic alteration, RNA expression, and DNA methylation profiling of coronavirus disease 2019 (COVID-19) receptor ACE2 in malignancies: a pan-cancer analysis, J. Hematol. Oncol. 2020;13:1–5. doi: 10.1186/s13045-020-00883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roongta R., Ghosh A. Managing rheumatoid arthritis during COVID-19. Clin. Rheumatol. 2020:1–8. doi: 10.1007/s10067-020-05358-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peters M.C., Sajuthi S., Deford P., Christenson S., Rios C.L., Montgomery M.T., Woodruff P.G., Mauger D.T., Erzurum S.C., Johansson M.W. COVID-19–related genes in sputum cells in asthma. Relationship to demographic features and corticosteroids. Am. J. Respir. Crit. Care Med. 2020;202:83–90. doi: 10.1164/rccm.202003-0821OC. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ouyang Y., Yin J., Wang W., Shi H., Shi Y., Xu B., Qiao L., Feng Y., Pang L., Wei F. Down-regulated gene expression spectrum and immune responses changed during the disease progression in COVID-19 patients. Clin. Infect. Dis. 2020;71(16):2052–2060. doi: 10.1093/cid/ciaa462. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akash M.S.H., Rehman K., Liaqat A. Tumor necrosis factor-alpha: role in development of insulin resistance and pathogenesis of type 2 diabetes mellitus. J. Cell. Biochem. 2018;119:105–110. doi: 10.1002/jcb.26174. [DOI] [PubMed] [Google Scholar]
- 72.Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor fedratinib. J. Microbiol. Immunol. Infect. 2020;53(3):368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abouzeid S., Sherif N. Role of alteration in treg/Th17 cells' balance in nephropathic patients with type 2 diabetes mellitus. Electron. Physician. 2015;7:1613. doi: 10.19082/1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lavoz C., Rayego-Mateos S., Orejudo M., L. Opazo-Ríos. Marchant V., Marquez-Exposito L., Tejera-Muñoz A., Navarro-González J.F., Droguett A., Ortiz A. Could IL-17A be a novel therapeutic target in diabetic nephropathy? J. Clin. Med. 2020;9:272. doi: 10.3390/jcm9010272. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rafiullah M. Can a combination of AT1R antagonist and vitamin d treat the lung complication of COVID-19? Am. J. Med. Sci. 2020;360(4):338–341. doi: 10.1016/j.amjms.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ivagnès A., Messaoudene M., Stoll G., Routy B., Fluckiger A., Yamazaki T., Iribarren K., Duong C.P., Fend L., Caignard A. TNFR2/BIRC3-TRAF1 signaling pathway as a novel NK cell immune checkpoint in cancer. OncoImmunology. 2018;7 doi: 10.1080/2162402X.2017.1386826. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Delanghe J.R., Speeckaert R., Speeckaert M.M. Complement C3 and its polymorphism: biological and clinical consequences. Pathology. 2014;46:1–10. doi: 10.1097/PAT.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 78.Gao T., Hu M., Zhang X., Li H., Zhu L., Liu H., Dong Q., Zhang Z., Wang Z., Hu Y. Highly pathogenic coronavirus n protein aggravates lung injury by MASP-2-mediated complement over-activation. MedRxiv. 2020 others. Preprint. [Google Scholar]
- 79.Tang S., Wang X., Deng T., Ge H., Xiao X. Identification of C3 as a therapeutic target for diabetic nephropathy by bioinformatics analysis. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-70540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grimmer T., Goldhardt O., Yakushev I., Ortner M., Sorg C., Diehl-Schmid J., Förstl H., Kurz A., Perneczky R., Miners S. Associations of neprilysin activity in CSF with biomarkers for alzheimer's disease. Neurodegener. Dis. 2019;19:43–50. doi: 10.1159/000500811. [DOI] [PubMed] [Google Scholar]
- 81.Didangelos A. COVID-19 hyperinflammation: what about neutrophils? mSphere. 2020;5 doi: 10.1128/mSphere.00367-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu H., Zhang M., Li W., Zhu S., Zhang D. Stachydrine attenuates IL-1 -induced inflammatory response in osteoarthritis chondrocytes through the NF-. Chem. Biol. Interact. 2020 doi: 10.1016/j.cbi.2020.109136. [DOI] [PubMed] [Google Scholar]
- 83.Crisci C.D., Ardusso L.R., Mossuz A., Müller L. A precision medicine approach to SARS-CoV-2 pandemic management. Current Treatment Options in Allergy. 2020:1–19. doi: 10.1007/s40521-020-00258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perricone C., Triggianese P., Bartoloni E., Cafaro G., Bonifacio A.F., Bursi R., Perricone R., Gerli R. The anti-viral facet of anti-rheumatic drugs: Lessons from COVID-19, Journal of Autoimmunity. 2020 doi: 10.1016/j.jaut.2020.102468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoo H.J., Kim T.J., Kim D.J., Kim W. Role of COX2 as a biomarker for estimating survival of patients with clinical stage i gastric cancer. Anticancer Res. 2020;40:341–347. doi: 10.21873/anticanres.13958. [DOI] [PubMed] [Google Scholar]
- 86.Zhao J., Tian S., Lu D., Yang J., Zeng H., Zhang F., Tu D., Ge G., Zheng Y., Shi T. Systems pharmacological study illustrates the immune regulation, anti-infection, anti-inflammation, and multi-organ protection mechanism of qing-fei-pai-du decoction in the treatment of COVID-19. Phytomedicine. 2020 doi: 10.1016/j.phymed.2020.153315. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Irving A.T., Zhang Q., Kong P.-S., Luko K., Rozario P., Wen M., Zhu F., Zhou P., Ng J.H., Sobota R.M. Interferon regulatory factors IRF1 and IRF7 directly regulate gene expression in bats in response to viral infection. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108345. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang J., Li H., Xue B., Deng R., Huang X., Xu Y., Chen S., Tian R., Wang X., Xun Z. IRF1 promotes the innate immune response to viral infection by enhancing the activation of IRF3. J. Virol. 2020;94 doi: 10.1128/JVI.01231-20. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Masuda T., Iwamoto S., Mikuriya S., Tozaki-Saitoh H., Tamura T., Tsuda M., Inoue K. Transcription factor IRF1 is responsible for IRF8-mediated IL-1β expression in reactive microglia. J. Pharmacol. Sci. 2015;128:216–220. doi: 10.1016/j.jphs.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 90.Pollak N.M., Hoffman M., Goldberg I.J., Drosatos K. Krüppel-like factors: Crippling and uncrippling metabolic pathways, JACC: Basic to Translational Science. 2018;3:132–156. doi: 10.1016/j.jacbts.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sajuthi S.P., DeFord P., Jackson N.D., Montgomery M.T., Everman J.L., Rios C.L., Pruesse E., Nolin J.D., Plender E.G., Wechsler M.E., et al. Type 2 and interferon inflammation strongly regulate SARS-CoV-2 related gene expression in the airway epithelium. Nat. Commun. 2020;11.1:1–18. doi: 10.1038/s41467-020-18781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Greenwood M., Greenwood M.P., Paton J.F., Murphy D. Transcription factor CREB3L1 regulates endoplasmic reticulum stress response genes in the osmotically challenged rat hypothalamus. PloS One. 2015;10 doi: 10.1371/journal.pone.0124956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greenwood M., Paterson A., Rahman P.A., Gillard B.T., Langley S., Iwasaki Y., Murphy D., Greenwood M.P. Transcription factor Creb3l1 regulates the synthesis of prohormone convertase enzyme PC1/3 in endocrine cells. J. Neuroendocrinol. 2020;32 doi: 10.1111/jne.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system, Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burke S.J., Karlstad M.D., Regal K.M., Sparer T.E., Lu D., Elks C.M., Grant R.W., Stephens J.M., Burk D.H., Collier J.J. CCL20 is elevated during obesity and differentially regulated by NF-κ b subunits in pancreatic β-cells. Biochimica Et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2015;1849:637–652. doi: 10.1016/j.bbagrm.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cai B., Wu J., Yu X., Su X., Wang R.-F. FOSL1 inhibits type i interferon responses to malaria and viral infections by blocking TBK1 and TRAF3/TRIF interactions. mBio. 2017;8 doi: 10.1128/mBio.02161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sardar R., Satish D., Gupta D. Identification of novel SARS-CoV-2 drug targets by host microRNAs and transcription factors co-regulatory interaction network analysis. Front. Genet. 2020;11:1105. doi: 10.3389/fgene.2020.571274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao F., Li B., Wei Y., Zhou B., Wang H., Chen M., Gan X., Wang Z., Xiong S. MicroRNA-34a regulates high glucose-induced apoptosis in H9c2 cardiomyocytes. J. Huazhong Univ. Sci. Technol. - Med. Sci. 2013;33:834–839. doi: 10.1007/s11596-013-1207-7. [DOI] [PubMed] [Google Scholar]
- 99.Hathaway Q.A., Pinti M.V., Durr A.J., Waris S., Shepherd D.L., Hollander J.M. Regulating microRNA expression: at the heart of diabetes mellitus and the mitochondrion. Am. J. Physiol. Heart Circ. Physiol. 2018;314:H293–H310. doi: 10.1152/ajpheart.00520.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nain Z., Rana H.K., Liò P., Islam S.M.S., Summers M.A., Moni M.A. Pathogenetic profiling of COVID-19 and SARS-like viruses. Briefings Bioinf. 2021;22:1175–1196. doi: 10.1093/bib/bbaa173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leon-Icaza S.A., Zeng M., Rosas-Taraco A.G. microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA. 2019;1:1–7. doi: 10.1186/s41544-018-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seeland U., Coluzzi F., Simmaco M., Mura C., Bourne P.E., Heiland M., Preissner R., Preissner S., et al. Evidence for treatment with estradiol for women with SARS-CoV-2 infection. BMC Med. 2020;18.1:1–9. doi: 10.1186/s12916-020-01851-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., et al. Dexamethasone in hospitalized patients with covid-19-preliminary report. N. Engl. J. Med. 2020;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marshall N.B., Kerkvliet N.I. Dioxin and immune regulation: emerging role of aryl hydrocarbon receptor in the generation of regulatory t cells. Ann. N. Y. Acad. Sci. 2010;1183:25. doi: 10.1111/j.1749-6632.2009.05125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Megna M., Napolitano M., Fabbrocini G. May IL-17 have a role in COVID-19 infection? Med. Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shibabaw T. Inflammatory cytokine: IL-17A signaling pathway in patients present with COVID-19 and current treatment strategy. J. Inflamm. Res. 2020;13:673. doi: 10.2147/JIR.S278335. [DOI] [PMC free article] [PubMed] [Google Scholar]