Abstract
Breast cancer is one of the most common malignant tumors in women worldwide. Circular RNA (circRNA) is a class of structurally stable non-coding RNA with a covalently closed circular structure. In recent years, with the development of high-throughput RNA sequencing, many circRNAs have been discovered and have proven to be clinically significant in the development and progression of breast cancer. Importantly, several regulators of circRNA biogenesis have been discovered. Here, we systematically summarize recent progress regarding the network of regulation governing the biogenesis, degradation, and distribution of circRNAs, and we comprehensively analyze the functions, mechanisms, and clinical significance of circRNA in breast cancer.
Keywords: circRNA, breast cancer, biogenesis, degradation, distribution, oncogenic, tumor suppressive, biomarker
Introduction
Breast cancer is the most common female malignancy worldwide. According to authoritative cancer statistics in 2021 (1), breast cancer has become the most prevalent of all cancers. An abundance of epidemiological studies has led to the identification of a variety of risk factors for developing breast cancer, including age, family history, early menarche, late pregnancy and menopause, high estrogen level, and excessive dietary fat intake (2).
Breast cancer can be generally divided into four molecular subtypes according to the status of the expression of the joint hormone receptors (estrogen receptors [ER] and progesterone receptors [PR]) and of the human epidermal growth factor receptor 2 (HER2). These subtypes include Luminal A, in which hormone receptors are expressed but HER2 is not (ER+PR+HER2−); Luminal B, in which hormone receptors and HER2 are expressed (ER+PR+HER2+); HER2 overexpression, in which hormone receptors are absent but HER2 is expressed (ER-PR-HER2+); and triple negative breast cancer (TNBC), in which all of the noted receptors are absent (ER-PR-HER2−). Endocrine and targeted therapies can be effective for ER+ or HER2+ breast cancer patients. These treatments include the use of tamoxifen to block the effects of estrogen and trastuzumab to target HER2 receptors on breast cancer cells (3, 4). However, such treatments are ineffective for TNBC, and chemotherapy is regarded as the main systematic treatment for TNBC. Because the efficacy of chemotherapy is unsatisfactory, there exists a desperate need for effective therapies for TNBC (5). Therefore, it is important to explore new treatment regimens to improve current therapeutic strategies, especially for TNBC patients.
One potentially underappreciated class of biological molecules that may yield effective therapeutic targets is circular RNAs (circRNA). CircRNAs generally form single-stranded covalently closed circular structures by the joining of the 3′ and 5′ ends (6). Two key pathways leading to circRNA formation have been proposed in recent authoritative articles. One mechanism involves the backsplicing of exons during transcriptional activities; this splicing is facilitated by base pairing of reverse repeat elements, such as Alu elements, located in flanking introns or by RNA binding proteins (RBP). In another key mechanism, circRNAs can be generated from lariat precursors formed in exon-skipping events, as well as from intronic lariat precursors escaping from debranching (7–9). Approximately 80% of circRNAs come from the backsplicing of exons from precursor mRNA or lncRNA (10), but the first circRNA molecules to be discovered, viroids, were found more than 40 years ago to be produced independent of a backsplicing mechanism (11). No matter the mechanism of formation, circRNAs tend to have the following characteristics: (1) CircRNAs are widely and abundantly present in tissues and bodily fluids, and some circRNAs accumulate to a higher level than their respective linear counterparts because of their stable covalently closed structure (9). (2) Many circRNAs are evolutionarily conserved in eukaryotes, in part because they exert critical biological functions (8). (3) Specific circRNAs are typically expressed in a tissue- or cell-specific manner (8, 12).
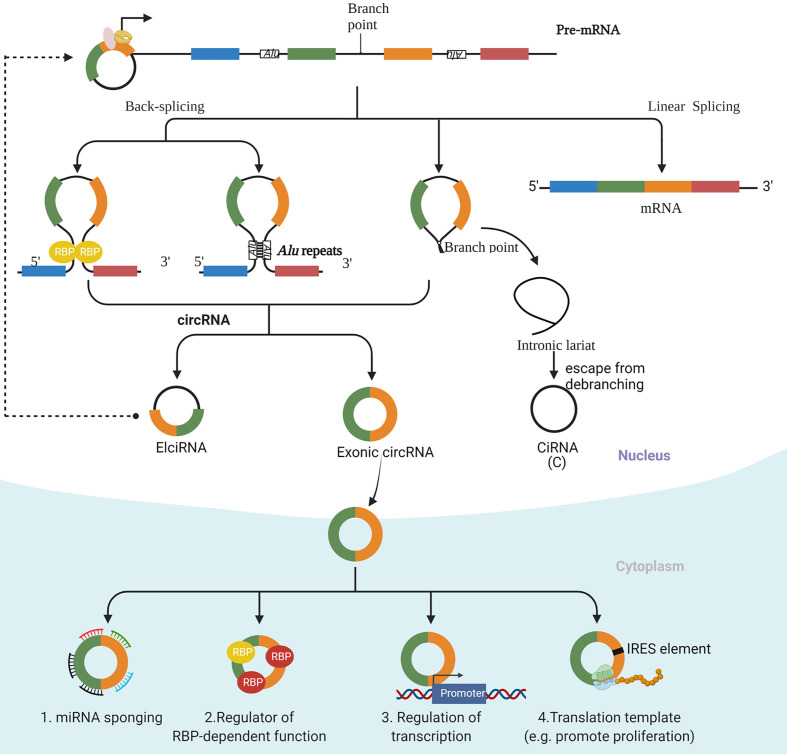
Improvements in RNA sequencing technology have led to the identification of several types of circRNAs, including exonic circRNAs, which consist of only exon(s), exon-intron circRNAs (EIciRNA), which consist of both exon(s) and intron(s), and intronic circRNAs (CiRNA), which consist of only intron(s) ( Figure 1 ). Cytoplasmic exonic circRNAs tend to have much longer intracellular half-lives compared with their linear counterparts (half-lives of circRNAs average approximately 48 h as opposed to roughly 10 h for many linear RNA molecules). A generally higher intracellular level of circRNAs than their linear counterparts results from this stability, and the stability can be explained at least partly by the resistance of circular RNA molecules to digestion by exonucleases (13, 14).
Figure 1.
The biogenesis and function of circRNAs. There are three main types of circRNAs, including exonic circRNAs, which consist of only exon(s), exon-intron circRNAs (ElciRNA), which consist of both exon(s) and intron(s), and intronic circRNAs (CiRNA), which consist of only intron(s). An exon-intron circRNA can regulate the expression of its parental gene through binding to its promoter. The mechanism of regulation by exonic circRNAs include serving as a decoy for binding to microRNAs (miRNAs); interacting with specific binding elements on RBPs and acting as a sponge to regulate the functions of RBPs and to affect the activities of associated proteins; regulating transcription; and acting as templates to encode proteins.
Other characteristics that are specific to the size and sequence of particular circRNAs have also been discovered among exonic circRNAs. Jeck et al. have reported that the length of a given exon appears to influence circularization, a factor that is especially evident for those circles that consist of a single exon (15). Memczak et al. has shown that exonic circRNAs described to date generally involve the GT-AG pair of canonical splice sites (16). Moreover, the flanking intron sites that are involved in the backsplicing process generally have relatively long intron sequences, but some exceptions to the lengths of these sequences exist (9). Finally, the sequence of the circRNA can impact cellular localization and activity, as EIciRNA and CiRNA are mainly located in the nucleus and regulate the transcription of specific genes (17, 18).
The most well-established biological function of circRNA involves its serving as a decoy for binding to microRNAs (miRNAs). This activity of circRNA thus modulates the effects of miRNA effects on their target transcripts. In addition, circRNAs can interact with specific binding elements on RBPs and can act as a sponge to indirectly regulate the functions of RBPs and to affect the activities of associated proteins. Finally, circRNAs can recruit specific proteins to certain loci, such as promoter regions, so as to facilitate the transcription of their own host genes (8, 10). Interestingly, under certain circumstances, some circRNAs with internal ribosome entry site (IRES) elements and AUG sites have been reported to be translated into specific peptides, which points to their underestimated cap-independent translational potency (19) ( Figure 1 ).
In recent years, regulators of circRNA biogenesis, degradation, and distribution have been identified with increasing rapidity. In addition, various types of circRNA have been found to be abnormally expressed in breast cancer, and their aberrant expression correlates with the occurrence, development, and prognosis of breast cancer; thus, circRNAs may be an important therapeutic target for breast cancer. In this review, we summarize the biogenesis, degradation, and distribution of circRNAs and the mechanisms of action of their regulators. We focus on cutting-edge discoveries concerning functions of circRNAs as well as associated mechanisms and their clinical significance in breast cancer.
The Regulators of circRNA Biogenesis
RNA Splicing Factor Regulates the Cyclization and Biogenesis of circRNA
CircRNAs are generally formed by backsplicing of mRNA from canonical splice sites (19). Because RNA splicing factors are central to this process, these factors are important to the regulation of circRNA biosynthesis. For example, Ashwal-Fluss et al. discovered that circRNA exon cyclization and canonical pre-mRNA splicing are mutually exclusive and that this competition is a tissue-specific and conservative mechanism of regulation of circRNA in animals (20). They found that a circular RNA related to the muscleblind gene (circMbl) and its flanking introns contain conserved binding sites for the MBL protein. Therefore, the level of MBL present strongly affects circMbl biosynthesis, and the authors confirmed that this effect depends on specific binding sites for MBL (20). Similarly, Pagliarini et al. found that SAM68 can bind to the distal intron inverted repeat Alu sequence of the spinal muscular atrophy gene (SMN), thereby supporting circRNA biosynthesis in vivo and in vitro (21). Splicing factor proline/glutamine-rich (SFPQ) is specifically enriched in introns flanking a type of Distal-Alu-Long-Intron (DALI) circRNA that is characterized by distal inverted Alu elements and long flanking introns, and depletion of SFPQ has been shown to significantly repress DALI circRNA production (22).
The importance of circRNA and its regulation have been demonstrated for several cancer types. Li et al. for example, found a global downregulation of circRNA levels in hepatocellular carcinoma. They further demonstrated that inhibition of the RNA splicing factor nudix hydrolase 21 (NUDT21) in this cancer type can promote the formation of circRNA and the production of UGUA sequences. Notably, UGUA sequences are crucial for circRNA formation, and thus this system establishes a positive feedback acceleration of circRNA production (23). In another carcinoma, oral squamous cell carcinoma, Zhao et al. found that circUHRF1 could bind to and inhibit miR-526b-5p and thus up-regulate the expression of c-MYC. Overexpression of c-MYC then facilitates the transcription of TGF-β1 and epithelial splicing regulatory protein 1 (ESRP1). Interestingly, ESRP1 itself targets flanking introns of circUHRF1, thereby accelerating circUHRF1 cyclization and biosynthesis (24). In gliomas, Liu et al. found that splicing factor serine and arginine-rich splicing factor 10 (SRSF10) can bind to Alu sequences of DNA surrounding a circular RNA related to ataxin-a (circATXN1) to regulate biosynthesis of this circRNA. Circ-ATXN1 levels were, therefore, decreased significantly upon knocking down of SRSF10 (25).
RNA Binding Proteins Regulate the Biogenesis of circRNA
Several other proteins with RNA binding activity have been shown to affect circRNA levels. For example, the immune factor NF90/NF110 contains double-stranded RNA binding domains and can bind to circRNA in the cytoplasm. In addition, however, the production of circRNA has been shown to increase upon association of nuclear NF90/NF110 with intronic RNA pairs flanking circRNA-forming exons. During viral infection, NF90/NF110 translocates from the nucleus to the cytoplasm, leading to a decrease in circRNA formation (26).
These regulatory abilities have, importantly, been shown to be important mediators in several developmental processes. In lung cancer, trinucleotide repeat-containing gene 6A (TNRC6A) can bind to introns near the exons that form circRNA to regulate production of circ0006916 (27). In induced pluripotent stem cell (iPSC) cells, the RNA binding protein fused in sarcoma (FU) can bind to introns near circular RNA reverse splicing sites to regulate circRNA biosynthesis (28). In prostate cancer, P53 can regulate activation of RNA binding motif protein 25 (RBM25), thus affecting the binding of RBM25 to circAMOTL1L. This binding is important for the induction of the biosynthesis of circAMOTL1L (29), because RBM25 mainly binds to the splicing complex, which in turn regulates selective splicing in combination with poly-G or exon splicing enhancer 5′- CGGGCA-3′ sequences (30, 31).
In TNBC, transcription factor E2F1 binds to the promoter of the gene that encodes septin-9 (SEPT9) and promotes the transcription of SEPT9 and the biogenesis of circSEPT9. Another transcription factor, EIF4A3, also regulates the biogenesis of circSETP9 by binding to the upstream and downstream sequences of the circSEPT9 exon (32). In hepatocellular carcinoma, RBM3 binds to SCD-circRNA 2 and regulates its biosynthesis, though the actual mechanisms underlying this regulation are not yet clear (33).
m6A Modification Regulates circRNA Biogenesis and Stability
Cis factors are also responsible for the control of biogenesis and stability of circRNAs. A recent study found that backsplicing occurs mostly at sites enriched in N-6-methyladenosine (m6A). For example, in male germ cells, approximately half of the circRNA molecules contain an m6A-modified initiation codon (34). Similarly, in macrophages of patients with acute coronary syndrome, knockdown of the N6-adenosine-methyltransferase METTL3 can down-regulate the m6A modification of hsa_circ_0029589 and promote biogenesis of this circRNA (35). Specifically relating to cancer, in hepatocellular carcinoma, m6A modification has been found to increase the cellular levels of circ_SORE by enhancing its stability (36).
Di Timoteo et al. found that m6A modification regulates the accumulation of circ-ZNF609 by regulating the back-splicing of circ-ZNF609, and they found direct correlations among a requirement for the presence of the METTL3 methyltransferase, binding of YTHDC1, and the back-splicing of the m6A modified exon (37), suggesting that methylation-mediated regulation might be a general phenomenon in circRNAs. Zhou et al. found that the m6A reader proteins YTHDF1, YTHDF2, and YTHDC1 bind to m6A-modified circRNA and that binding of YTHDF2 in particular reduces the stability of the circRNA (38). Thus, it can be concluded that m6A modification sites can function together with accessory proteins, including m6A writers, erasers, and readers, to control circRNA biogenesis.
The Regulation of circRNA Degradation
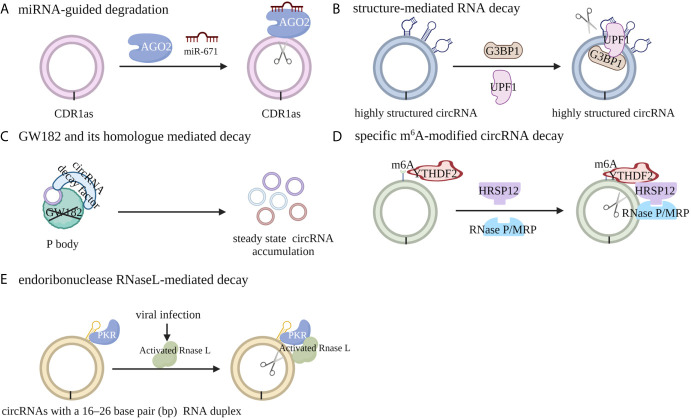
The levels of circRNA can be controlled at both the synthesis and degradation levels. It has been universally acknowledged that the degradation of most mRNAs initiates with poly(A)-tail shortening at the 3′ end, whereas some mRNAs undergo interior cleavage by endonucleases. Because of the lack of a 5′ 7-methylguanosine cap or a 3′ poly (A) tail, the cleavage of circRNAs, on the other hand, are generally dependent on endonucleases, which initiate degradation internally (39, 40). Cleavage of circRNA CDR1 by endonuclease AGO2 has been shown to be assisted by miR-671. This discovery served as the first evidence that some circRNAs can be degraded by endonucleases in a sequence-dependent manner (41).
In general, the pathways leading to circRNA degradation can be divided into five categories: miRNA-guided degradation, structure-mediated RNA decay (SRD), decay mediated by GW182 and its human homolog, specific m6A-modified circRNA decay and endoribonuclease RNaseL-mediated decay. Fischer et al. reported that the degradation of some highly structured circRNAs can be regulated by UPF1 and G3BP1, both of which recognize and unwind the overall structures of circRNAs (42). The absence of GW182, which is a key component of P body and RNAi complexes, can lead to the accumulation of endogenous circRNA and increase the steady state of cytoplasmic circRNAs, whereas the absence of other factors in the P body or RNAi complex has no similar effect (43). Further study indicated that the MID domain of GW182 protein may mediate the interaction between circRNAs and circRNA decay factors; the absence of TNRC6A, TRNC6B, or TRNC6C, which are the human homologs of GW182, in HEK293 cells, results in the same accumulation of steady-state circRNAs in human cells, indicating a conserved role of P-body and RNAi-mediated degradation of circRNA (43). Park et al. reported that m6A-modified circRNA can recruit the m6A reader protein YTHDF2 as well as the adaptor protein HRSP12, and HRSP12 can serve as the bridge to connect YTHDF2 with the endoribonuclease RNase P/MRP, thus enabling downregulation of m6A modified circRNA by RNase P/MRP (44). Liu et al. discovered that RNase L, a widely expressed cytoplasmic endoribonuclease, can be activated upon viral infection through an undefined mechanism and degrade the circRNAs with 16 to 26 bp RNA duplex (45) ( Figure 2 ).
Figure 2.
Mechanisms of circRNA degradation. (A) CircRNA CDR1 can be cleaved by endonuclease AGO2 with the assistance of miR-671 in a miRNA target sequence-dependent manner. (B) The degradation of highly structured circRNAs can be regulated by UPF1 and G3BP1, both of which recognize and unwind the overall structures of circRNAs. (C) m6A-modified circRNA can recruit the m6A reader protein YTHDF2 as well as the adaptor protein HRSP12, and HRSP12 can serve as a bridge to connect YTHDF2 with the endoribonuclease RNase P/MRP, thus enabling degradation of m6A modified circRNA by RNase P/MRP. (D) The absence of GW182 or its human homologue TRNC6A/B/C, the key component of P body and RNAi complexes, can lead to the accumulation of endogenous circRNA and increase the steady state of cytoplasmic circRNAs. (E) Upon viral infection, RNase L can be activated and can degrade circRNAs with RNA duplexes containing 16 to 26 bp through an undefined mechanism.
Regulators of circRNA Distribution
Once produced, the majority of circRNAs are transported to the cytoplasm where they exhibit further biological functions, whereas some intronic circRNAs and exon-intron circRNAs are thought to reside in the nucleus (17, 18). Since circRNAs lack the classical RNA transport sequence, the nuclear transport mechanism of circRNA remains unclear. As existing studies have indicated that the location of circRNAs determines their functions, it is important to continue to explore the potential mechanisms of circRNA nucleus export.
In order to explore circRNA nuclear transporting mechanisms, Li et al. knocked down 26 reported RNA transport-related proteins in Drosophila. Their results showed that one circRNA, with a length of 1120 bp, remained in the nucleus, whereas another circRNA, with a length of 490 bp, mainly localized to the cytoplasm after deletion of Hel25E. Both of these circRNAs mainly localize in the cytoplasm in wild-type Drosophila. The localizations of 12 other endogenous circRNAs that reside in the cytoplasm in wild-type Drosophila were further studied in Hel25E-knockout flies, and it was discovered that circRNAs with lengths of more than 811 bp mainly accumulated in the nucleus, whereas circRNA lengths of less than 702 bp resided in the cytoplasm in cells of the knockout Drosophila. An experiment exploring the human Hel25E homologs UAP56 (DDX39B) and URH49 (DDX39A), which share more than 90% similarity with Drosophila Hel25E, found that human UAP56 mainly regulates the nuclear transport of long circRNA (more than 1200 bp), whereas URH49 mainly regulates the nuclear export of short circRNA (less than 400 bp). Therefore, the nuclear accumulation of circRNA results mainly from defects of nuclear transport, and the length of circRNA is an important determinant that determines nuclear export (46, 47).
In addition, a very recent study found that m6A modification can also influence circRNA nuclear export. It was reported that circNSUN2 could bind to the m6A reader YTHDC1, which interacted with the m6A binding motif at the GAACU m6A within the splice junction of circNSUN2 (48). In studies of mRNA splicing, YTHDC1 has been shown to bind to the RNA splicing factors SRSF1, SRSF3, SRSF7, and SRSF10 to regulate mRNA splicing (49). YTHDC1 can also bind to SRSF3 and the classical nuclear receptor NXF1, thereby regulating the metabolism and transport of m6A modified mRNAs (50), suggesting that circRNA may also share the nuclear transporting mechanisms with mRNA.
CircRNA Profiles in Breast Cancer
By using high-throughput RNA sequencing and microarray analysis, profiles of circRNA expression in breast cancer have been comprehensively analyzed. The RNA sequencing or microarray data of circRNA expression profiles in samples related to breast cancer are listed in Table 1 . Unlike the studies in gastric cancer and hepatocellular carcinoma that found more down-regulated circRNAs than up-regulated ones (30, 66), in breast cancer, more circRNAs have been found to be up-regulated. These results indicated that the regulation of circRNA levels is tissue specific and that circRNAs likely regulate different process in different systems.
Table 1.
RNA sequencing and microarray analyses of differentially expressed circRNAs in breast cancer.
| Samples | Detection method | Differentially expressed circRNAs | References |
|---|---|---|---|
| 4 pairs of TNBC and adjacent noncancerous tissues | RNA sequencing | 47 were up-regulated and 307 were down-regulated (FC≥2, P< 0.05) | (32) |
| 3 pairs of breast cancer and corresponding adjacent non-cancerous tissues | RNA sequencing | 85 were upregulated and 67 were downregulated.(FDR ≤ 0.001, FC≥2) | (51) |
| 8 patients’ specimens (TNBC, N = 4; luminal A, N = 4) and 3 normal mammary gland tissues (NMGT) | RNA sequencing | 140 upregulated and 95 downregulated circRNA were identified, including 215 and 73 circRNAs for the TNBC and LA subtypes, respectively (FC>1.5, P<0.05) | (52) |
| 4 TNBC and 3 NMGT | RNA sequencing | 122 upregulated and 93 downregulated circRNA were identified (FC>1.5, P<0.05) | (52) |
| 4 LA and 3 NMGT | RNA sequencing | 55 upregulated and 18 downregulated circRNA were identified (FC>1.5, P<0.05) | (52) |
| 3 pairs of breast cancer tissues with or without metastasis | RNA sequencing | A total of 51 circRNAs were differentially expressed (FC>1.5, P< 0.05) | (53) |
| 4 pairs of TNBC tissues and adjacent non-cancerous tissues | RNA sequencing | 47 were upregulated, whereas 307 were downregulated (FC>2.0, P< 0.05) | (54) |
| 4 pairs of breast cancer lesions and matched adjacent normal-appearing tissues | microarray | 715 circRNAs were upregulated and 440 circRNAs were downregulated (FC≥2, P<0.05) | (55) |
| 3 pairs of TNBC tissues and matched normal mammalian tissues | microarray | 173 circRNAs were up-regulated, and 77 circRNAs were down-regulated (FC≥1.5, P<0.05, FDR< 0.05) | (56) |
| 4 pairs of breast cancer tissues and adjacent noncancerous tissues | microarray | A total of 2,587 circRNAs with 1.5 fold of upregulation or downregulation (FC>1.5, P<0.05) | (57) |
| 4 pairs of breast cancer tissues and adjacent normal tissues | microarray | 15 circRNAs upregulated and 16 circRNAs downregulated (FC>4.0, P<0.05) | (58) |
| 5 pairs of breast tumor tissues and matched non-tumor tissues | microarray | A total of 716 circRNAs that were differently expressed (FC>1.5, P<0.05) | (59) |
| 3 pairs of breast cancer tissues and their adjacent non-tumor tissues | microarray | 1953 circRNAs were upregulated and 1700 circRNAs were downregulated (FC>2.0 and P< 0.05) | (60) |
| 3 pairs of TNBC and adjacent normal tissues | RNA sequencing | 1307 circRNAs were up-regulated and 3726 circRNAs were down-regulated (FC≥2, P< 0.05) | (61) |
| 5 pairs of breast cancer tissues and corresponding nontumorous tissues | RNA sequencing | 30 were up-regulated and 19 were down-regulated (FC>2, P< 0.001) | (62) |
| 2 pairs of BCLM tissues and primary tumor tissues | microarray | 559 were upregulated and 661 were downregulated (FC>2, P< 0.05) | (63) |
| 6 pairs of breast cancer/adjacent tissues | microarray | 2,375 were upregulated, whereas 1,995 were downregulated (FC>2.0 or <0.5, P<0 .05) | (64) |
| 3 pairs of breast cancer tissues and paired noncancerous tissues | microarray | 292 were upregulated and 228 were downregulated (FC≥2, P< 0.05) | (65) |
BC, breast cancer; LA, luminal A; TNBC, triple-negative breast cancer; NMGT, normal mammary gland tissues; BCLM, breast cancer liver metastases; FC, fold change; FDR, false discovery rate.
The Diverse Roles of circRNAs in Breast Cancer Progression and Associated Mechanisms
Mechanisms Leading to Oncogenic Functions of circRNAs in Breast Cancer
Most reported circRNAs have been proposed to function by acting as molecular sponges that bind to and inhibit the functions of miRNA molecules (67, 68). As shown in the RNA sequencing or microarray data, many circRNAs are dysregulated in breast cancer. Among them, the up-regulated circRNAs mainly function as oncogenic factors, whereas the circRNAs down-regulated in breast cancer generally play tumor suppressive roles. The circRNAs that play oncogenic roles in breast cancer are listed in Table 2 . Here, we discuss several mechanisms through which circRNAs might influence the development or progression of breast cancer.
Table 2.
CircRNAs that play oncogenic roles in breast cancer.
| CircRNA name | Circbase ID | Length | Gene Name | Distribution | Phenotype | Target | Downstream genes/ pathways |
Reference |
|---|---|---|---|---|---|---|---|---|
| circSEPT9 | hsa_circ_ 0005320 | 645 | SEPT6 | cytoplasm | promote cell proliferation and migration in vitro | miR-637 | LIF/STAT3 | (32) |
| circular RNA CDR1as | hsa_circ_0001946 | 1485 | CDR1 | \ | increase chemosensitivity of 5-FU-resistant BC cells | miR-7 | CCNE1 | (69) |
| hsa_circ_0006528 | hsa_circ_0006528 | 496 | PRELID2 | \ | promote adriamycin resistance | miR-7-5p | Raf1 | (70) |
| hsa_circRNA_0006528 | hsa_circ_0006528 | 496 | PRELID2 | \ | promote cell proliferation, invasion, and migration, inhibit apoptosis | miR-7-5p | Raf1-MAPK/ERK | (71) |
| hsa_circRNA_0000518 | hsa_circ_0000518 | 150 | RPPH1 | \ | and promote cell proliferation, migration, invasion, inhibit cell cycle arrest, apoptosis, in vitro, and promote tumor growth in vivo | miR-326 | FGFR1 | (72) |
| circIQCH | hsa_circ_0104345 | 864 | IQCH | cytoplasm | promote cell proliferation, migration in vitro, and promote tumor growth, and metastasis in vivo | miR-145 | DNMT3A | (73) |
| circZNF609 | hsa_circ_0000615 | 874 | ZNF609 | cytoplasm | promote cell proliferation, invasion and migration in vitro, and promote tumor growth in vivo | miR-145-5p | p70S6K1 | (74) |
| hsa_circ_0136666 | hsa_circ_0136666 | 477 | PRKDC | \ | promote cell proliferation | miR-1299 | CDK6 | (75) |
| circ_0006528 | hsa_circ_0006528 | 496 | PRELID2 | \ | promote proliferation, migration, invasion, autophagy, and inhibit cell apoptosis of PTX-resistant breast cancer cells | miR-1299 | CDK8 | (76) |
| circ_0001667 | hsa_circ_0001667 | 456 | HEATR2 | \ | promote cell proliferation, invasion, and migration in vitro | miR-125a-5p | TAZ | (77) |
| hsa_circ_0052112 | hsa_circ_0052112 | 209 | ZNF83 | cytoplasm | promote cell migration and invasion in vitro | miR-125a-5p | ZNF83 | (78) |
| circHMCU | hsa_circ_0000247 | 792 | MCU | cytoplasm | promote cell proliferation, migration, and invasion in vitro, and promote tumor growth and lung metastasis in vivo | let-7 | MCY, HMGA2, and CCND1 | (79) |
| circ-ABCB10 | \ | \ | ABCB10 | \ | decrease PTX sensitivity and apoptosis, promote cell invasion and autophagy of PTX-resistant BC cells | let-7a-5p | DUSP7 | (80) |
| circPLK1 | hsa_circ_0038632 | 1708 | PLK1 | cytoplasm | promote cell proliferation, invasion in vitro, and tumor occurrence and metastasis in vivo | miR-296-5p | PLK1 | (81) |
| hsa_circ_0000515 | hsa_circ_0000515 | 229 | RPPH1 | cytoplasm | promote cell proliferation, invasion, angiogenesis, and enhance inflammatory response in vitro, promote tumor growth in vivo | miR-296-5p | CXCL10 | (82) |
| hsa_circ_0011946 | hsa_circ_0011946 | 782 | SCMH1 | \ | promote cell migration and invasion | miR-26a/b | RFC3 | (51) |
| circWWC3 | hsa_circ_0089866 and hsa_circ_0001910 | 1068 and 825 | WWC3 | cytoplasm | promote cell proliferation, migration, and invasion | miR-26b-3p and miR-660-3p | ZEB1 | (83) |
| circ-DNMT1 | hsa_circ_0049224 | 155 | DNMT1 | cytoplasm | promote cell proliferation | \ | interact with p53 and AUF1 |
(84) |
| FECR1 | \ | 571 | FLI1 | both cytoplasm and nucleus | promote cell invasion in vitro | \ | binds to the FLI1 promoter in cis and recruits TET1, binds to and downregulates DNMT1 in trans | (85) |
| circular HER2 | hsa_circ_0007766 | 676 | ERBB2 | cytoplasm | promote cell proliferation, invasion, and tumorigenesis in vitro and in vivo | encoded a novel protein, HER2-103 | promote homo/hetero dimerization of EGFR/HER3, sustain AKT phosphorylation | (86) |
| circIFI30 | hsa_circ_0005571 | 351 | IFI30 | cytoplasm | promote cell proliferation, migration, invasion, inhibit apoptosis in vitro, and promote tumorigenesis and metastasis in vivo | miR-520b-3p | CD44 | (87) |
| circAGFG1 | hsa_circ_0058514 | 527 | AGFG1 | cytoplasm | promote cell proliferation, mobility, and invasion in vitro, promote tumorigenesis and metastasis in vivo | miR-195-5p | CCNE1 | (54) |
| circKIF4A | hsa_circ_0007255 | 355 | KIF4A | cytoplasm | promote cell proliferation, migration in vitro, and promote tumor growth and metastasis in vivo | miR-375 | KIF4A | (88) |
| circEPSTI1 | hsa_circ_0000479 | 375 | EPSTI1 | cytoplasm | promote cell proliferation and inhibit apoptosis | miR-4753 and miR-6809 | BCL11A | (56) |
| circGNB1 | hsa_circ_0009362 | 152 | GNB1 | cytoplasm | promote cell proliferation, migration in vitro, and tumor growth in vivo | miR-141-5p | IGF1R | (89) |
| circ-ABCB10 | hsa_circ_0008717 | 724 | ABCB10 | \ | promote cell proliferation and inhibit apoptosis in vitro | miR-1271 | \ | (57) |
| hsa_circ_0001982 | hsa_circ_0001982 | 899 | RNF111 | \ | promote cell proliferation, invasion, and inhibit apoptosis | miR-143 | \ | (59) |
| circANKS1B | hsa_circ_0007294 | 459 | ANKS1B | cytoplasm | promote cell invasion and migration in vitro, promote metastasis in vivo | miR-148a-3p and miR-152-3p | USF1-TGF-β1/SMAD | (61) |
| hsa_circ_0004771 | hsa_circ_0004771 | 203 | NRIP1 | \ | promote tumor growth and inhibit apoptosis | miR-653 | ZEB2 | (62) |
| circGFRA1 | hsa_circ_0005239 | 427 | GFRA1 | cytoplasm | promote cell proliferation and inhibit apoptosis in vitro | miR-34a | GFRA1 | (90) |
| circRNA_069718 | hsa_circ_0069718 | 590 | DCUN1D4 | \ | promote cell proliferation, invasion in vitro | \ | reduce the expression of Wnt/β-catenin pathway-related genes | (91) |
| circular RNA ciRS-7 | hsa_circ_0001946 | 1485 | antisense strand of CDR1 | cytoplasm | promote cell migration and invasion in vitro | miR-1299 | MMPs | (92) |
| circRNA-CER | hsa_circ_0023404 | 180 | RNF121 | \ | promote cell proliferation and migration in vitro | miR−136 | MMP13 | (93) |
| circ-RNF111 | hsa_circ_0001982 | 899 | RNF111 | \ | promote cell proliferation, invasion, glycolysis, and PTX resistance in PTX-resistant BC cells in vitro, and enhanced PTX sensitivity in vivo | miR-140-5p | E2F3 | (94) |
| circACAP2 | \ | \ | \ | cytoplasm | promote cell proliferation and motility | miR-29a/b-3p | COL5A1 | (95) |
| circVAPA | hsa_circ_0006990 | 338 | VAPA | cytoplasm | promote cell proliferation, migration, invasion | miR-130a-5p | \ | (96) |
| hsa_circ_002178 | \ | \ | \ | cytoplasm | promote cell proliferation, invasion, and migration | miR-1258 | KDM7A | (97) |
| hsa-circ-0083373, hsa-circ-0083374, hsa-circ-0083375 | hsa_circ_0083373, hsa_circ_0083374, hsa_circ_0083375 | 3868,4045,5469 | DLC1 | \ | promote pathogenesis and development of breast cancer | hsa-miR-511 | \ | (98) |
| circRNA_100876 | \ | \ | \ | \ | promote cell proliferation and invasion | miR-4753 and miR-6809 | BCL11A | (99) |
| circ_0103552 | hsa_circ_0103552 | 920 | UBR1 | \ | promote cell proliferation, migration, invasion, and inhibit apoptosis | miR-1236 | \ | (100) |
| circMMP11 | hsa_circ_0062558 | 906 | MMP11 | cytoplasm | promote cell proliferation and migration in vitro | miR-1204 | MMP11 | (101) |
| circ-UBE2D2 | \ | \ | UBE2D2 | \ | promote cell proliferation, migration, and invasion in vitro | miR-1236 and miR-1287 | \ | (102) |
| circ_UBE2D2 | hsa_circ_0005728 | 280 | UBE2D2 | cytoplasm | promote tamoxifen resistance | miR-200a-3p | \ | (103) |
| circUBE2D2 | hsa_circ_0005728 | 280 | UBE2D2 | cytoplasm | promote cell proliferation, migration, and invasion, and doxorubicin resistance in vitro, promote tumor growth in vivo | miR-512-3p | CDCA3 | (104) |
| hsa_circ_0091074 | hsa_circ_0091074 | 373 | TCONS_00016926 | \ | promote cell proliferation and invasion | miR−1297 | TAZ/TEAD4 | (105) |
| circ_0000043 | hsa_circ_0000043 | 438 | PUM1 | \ | promote cell proliferation, migration, invasion, and EMT | miR-136 | SMAD3 | (106) |
| hsa_circ_0003645 | hsa_circ_0003645 | 356 | C16orf62 | cytoplasm | promote cell proliferation and inhibit cell apoptosis in vitro and in vivo | miR-139-3p | HMGB1 | (107) |
| circCDYL | hsa_circ_0008285 | 667 | CDYL | cytoplasm | promote the malignant progression in vitro and in vivo | miR-1275 | ATG7 and ULK1 | (108) |
| circ_0000520 | hsa_circ_0000520 | 123 | RPPH1 | cytoplasm | promote cell proliferation, migration, and invasion, but inhibit cell cycle arrest and apoptosis | miR-1296 | SP1 | (109) |
| circular RNA KIF4A | hsa_circ_0007255 | 355 | KIF4A | cytoplasm | promote cell migration, invasion, and inhibit apoptosis | miR-152 | ZEB | (110) |
| circABCC4 | hsa_circ_0030586 | 1192 | ABCC4 | \ | promote cell proliferation, migration, invasion, and inhibit apoptosis | miR-154-5p | promote NF-κB and WNT/β-catenin pathways | (111) |
| circDENND4C | \ | \ | DENND4C | \ | increase glycolysis, migration, and invasion under hypoxia | miR-200b and miR-200c | MMP2, MMP9 | (112) |
| circDENND4C | \ | \ | \ | \ | promote cell migration and invasion under hypoxia in vitro, promote tumor growth in vivo | \ | \ | (113) |
| circHIPK3 | hsa_circ_0000284 | 1099 | HIPK3 | cytoplasm | promote cell proliferation, migration, and invasion in vitro and tumor growth in vivo | miR-193a | HMGB1/PI3K/AKT | (114) |
| hsa_circ_001783 | \ | 34460 | EBLN3- ZCCHC7 | cytoplasm | promote cell proliferation and invasion | miR-200c-3p | \ | (115) |
| circABCB10 | hsa_circ_ 0008717 | 724 | ABCB10 | \ | promote cell proliferation, glycolysis, and increase IR resistance | miR-223-3p | PFN2 | (116) |
| circular RNA PVT1 | hsa_circ_0001821 | 410 | PVT1 | cytoplasm | promote tumor growth in vivo | miR-204-5p | upregulate E-cadherin, downregulate N-cadherin, Vimentin, Slug, and Twist | (117) |
| circRAD18 | hsa_circ_0002453 | 756 | RAD18 | cytoplasm | promote cell proliferation and migration, inhibit cell apoptosis in vitro, promote tumor growth in vivo | miR-208a and miR-3164 | IGF1 and FGF2 | (118) |
| circRAD18 | hsa_circ_0002453 | 756 | RAD18 | cytoplasm | promote cell proliferation, migration, invasion, EMT, and inhibit cell apoptosis | miR-613 | HK2 | (119) |
| hsa_circ_0131242 | hsa_circ_0131242 | 12400 | MAP3K4 | \ | promote cell proliferation and migration in vitro | miR-2682 | \ | (120) |
| *hsa_circ_0072995 | hsa_circ_0072995 | 435 | ARHGEF28 | both nucleus and cytoplasm | promote cell migration and invasion | miR-30c-2-3p | ARHGEF28 | (121) |
| circ-TFF1 | hsa_circ_0061825 | 492 | TFF1 | cytoplasm | promote cell proliferation, migration, invasion, EMT in vitro, and tumor growth in vivo | miR-326 | TFF1 | (122) |
| circ_0000291 | hsa_circ_0000291 | 311 | CD44 | \ | promote cell proliferation, migration, and invasion | miR-326 | ETS1 | (123) |
| circular RNA 0007255 | hsa_circ_0007255 | 355 | KIF4A | cytoplasm | promote oxygen consumption, colony formation, and cell mobility in vitro, inhibit tumor growth in vivo | miR-335-5p | SIX2 | (124) |
| circEIF3M | hsa_circ_0003119 | 361 | EIF3M | cytoplasm | promote cell proliferation, migration, and invasion | miR-33a | cyclin D1 | (125) |
| hsa_circRNA_002178 | hsa_circ_0000519 | 98 | RPPH1 | cytoplasm | promote cell proliferation, energy metabolism, and angiogenesis | miR-328-3p | COL1A1 | (126) |
| circ-PGAP3 | hsa_circ_0106800 | 537 | PGAP3 | cytoplasm | promote cell proliferation and invasion | miR-330-3p | Myc | (127) |
| circIRAK3 | hsa_circ_0005505 | 754 | IRAK3 | cytoplasm | promote cell migration and invasion in vitro, promote metastasis and in vivo | miR-3607 | FOXC1 | (128) |
| hsa_circ_0008039 | hsa_circ_0008039 | 462 | PRKAR1B | cytoplasm | promote cell proliferation and migration in vitro | miR-432-5p | E2F3 | (129) |
| hsa_circ_0008039 | hsa_circ_0008039 | 462 | PRKAR1B | \ | promote cell proliferation, migration, and invasion in vitro, and promote tumor growth in vivo | miR-515-5p | CBX4 | (130) |
| circular RNA circ-ZEB1 | \ | \ | \ | \ | promote cell proliferation and inhibit apoptosis | miR-448 | eEF2K | (131) |
| circular RNA-100219 | hsa_circ_0004619 | 377 | FAF1 | \ | promote cell proliferation and migration in vitro | miR-485-3p | NTRK3 | (132) |
| circMYO9B | hsa_circ_0000907 | 898 | MYO9B | \ | promote cell proliferation, migration, and invasion in vitro, and promote tumor growth in vivo | miR-4316 | FOXP4 | (133) |
| circRNF20 | hsa_circ_0087784 | 499 | RNF20 | cytoplasm | promote cell proliferation, inhibit cell apoptosis in vitro, promote tumor growth in vivo | miR-487a | HIF1α-HK2 | (134) |
| hsa_circ_0007534 | hsa_circ_0007534 | 400 | DDX42 | \ | promote cell proliferation, invasion, and inhibit apoptosis in vitro | miR-593 | MUC19 | (135) |
| circ_DCAF6 | \ | \ | \ | cytoplasm | promote cell proliferation and stemness | miR-616-3p | GLI1 | (136) |
| circ_0005230 | hsa_circ_0005230 | 3958 | DNM3OS | \ | promote cell invasion, migration in vitro, and promote cell growth in vivo | miR-618 | CBX8 | (137) |
| circZFR | hsa_circ_0072088 | 693 | ZFR | \ | promote cell proliferation, migration, invasion, glycolysis, and inhibit cell apoptosis in vitro, promote tumor growth in vivo | miR-578 | HIF1A | (138) |
| circ-UBAP2 | hsa_circ_0001846 | 747 | UBAP2 | cytoplasm | promote cell proliferation, migrate, and inhibit cell apoptosis in vitro, and promote tumor growth and metastasis in vivo | miR-66 | MTA1 | (139) |
| circFBXL5 | hsa_circ_0125597 | 912 | FBXL5 | cytoplasm | promote cell proliferation and migration in vitro, promote tumor growth and metastasis in vivo | miR-660 | SRSF6 | (140) |
| circTP63 | \ | 295 | TP63 | cytoplasm | promote cell proliferation, invasion, migration in vitro, promote tumor growth in vivo | miR-873-3p | FOXM1 | (141) |
| hsa_circRPPH1_015 | hsa_circ_0000517 | 88 | RPPH1 | cytoplasm | promote cell proliferation and aggressiveness in vitro, promote tumor growth in vivo | miR-326 | ELK1 | (142) |
| #circANKRD12 | hsa_circ_0046841 or \ | 286 or 925 | ANKRD12 | cytoplasm | promote cell proliferation, invasion, migration, and alter cell metabolism | \ | CCND1 | (143) |
| circCNOT2 | \ | \ | \ | \ | increase cell proliferation | \ | \ | (144) |
| circAMOTL1 | \ | \ | \ | \ | promote cell proliferation and invasion, and inhibit cell apoptosis when exposed to PAX in vitro | \ | AKT | (145) |
| circ_103809 | \ | \ | \ | \ | promote cell proliferation | \ | PI3K/AKT signaling | (146) |
| hsa_circ_0008673 | hsa_circ_0008673 | 689 | BRCA1 | \ | promote cell proliferation and migration | \ | \ | (147) |
| hsa_circ_001569 | \ | \ | \ | \ | promote cell proliferation, migration, invasion, and inhibit cell apoptosis in vitro | \ | PI3K/AKT signaling | (148) |
| circBACH2 | hsa_circ_0001627 | 2995 | BACH2 | cytoplasm | promote cell proliferation, invasion, and migration in vitro, promote tumor growth and metastasis in vivo | miR-186-5p/miR-548c-3p | CXCR4 | (149) |
| circPGR | \ | \ | PGR | cytoplasm | promote ER-positive breast cancer cell proliferation, invasion, and migration in vitro, promote tumor growth in vivo | miR-301a-5p | Cell cycle genes | (150) |
EMT, epithelial mesenchymal transition; PAX, paclitaxel; *The circRNA can simultaneously regulate an miRNA and genes, but the relationship between miRNA and genes is not clear. #Two circular isoforms of 286 bp (hsa_circ_0046841) and 925 bp (not found in the circbase) lengths share the same backsplice junction of ANKRD12, and the study did not distinguish these two circular isoforms.
Acting as Competing Endogenous RNAs (ceRNAs)
A major mechanism that explains impacts of circRNA levels on cancer development involves the interaction of these RNA molecules with miRNA molecules. This binding event, called “sponging,” can inhibit the interactions of these miRNA molecules with other cellular molecules, frequently leading to inhibition of the miRNA activity. Thus, circRNAs can serve as ceRNAs. The downstream effect of the competition depends on the ultimate target of the miRNA itself.
Multiple circRNAs can target the same miRNA and thus assist malignant progression of breast cancer. For example, hsa_circ_0006528 and circCDR1as can target CCNE1 and RAF1, respectively, through sponging miR-7. In this way, both of these circRNA molecules promote the proliferation, invasion, migration, and chemoresistance of breast cancer cells (69, 70). Similarly, hsa_circ_0000291, hsa_circRNA_000518, and circ_TFF1 can target ETS1, FGFR1, and TFF1, respectively, through sponging miR-326, to accelerate the proliferation, colony formation, invasion, and migration of breast cancer cells; these activities also serve to reduce cell apoptosis (71, 72, 151). CircZNF609 and circIQCH can target DNMT3A and p70S6K1 by sponging miR-145 to promote the malignant phenotype of breast cancer cells (73, 74). Hsa_circ_0136666 and hsa_circ_0006528 can target CDK6 and CDK8 by sponging miR-1299, respectively, to regulate cell cycle progression, proliferation, apoptosis, invasion, migration, and drug resistance of breast cancer cells (75, 76). Hsa_circ_0052112 and hsa_circ_0001667 mainly regulates ZNF36 and FOXO3a by sponging target miR-125a-5p, respectively, to regulate the invasion and migration of breast cancer cells (77, 78). Circ-ABCB10 and circHMCU can sponge let-7 then regulate the expression of MCY, HMGA2, CCND1, and DUSP7, respectively, to promote the proliferation, invasion, migration, and chemoresistance of breast cancer cells (79, 80). CircPLK1 and hsa_circ_0000515 can target miR-296-5p to regulate the expression of PLK1 and CXCL10, respectively, to promote the growth, proliferation, migration and inflammatory response of breast cancer cells (81, 82). And surprisingly, hsa_circ_0011946 can interact with miR-26a/b to regulate RFC3, to promote the invasion and migration of breast cancer cells, whereas circWWC3 can interact with miR-26b-3p and miR-660-3p to regulate the expression of ZEB1, and thus promote the proliferation, invasion and migration of breast cancer cells (51, 83).
It is important to note the potential for ceRNA-acting circRNA molecules to improve patient outcomes in addition to their roles in cancer progression. For example, TV-circRGPD6 can interact with miR-26b. This interaction serves to regulate the expression of YAF2 and thus leads to a better prognosis (152).
Protein Decoy or Scaffolding Functions
In some cases, circRNAs may modulate the progression of breast cancer through direct or indirect interactions with RBPs. These interactions may alter the functions of the RBPs either through competition, with the circRNA serving as a decoy, or through the recruitment of other interacting factors, with the circRNA serving a scaffolding role. For example, in breast cancer cells with wild-type p53, circ-CCNB1 can interact with both p53 and H2AX, and Bclaf1 is free to bind to Bcl2. In p53-mutant breast cancer cells, on the other hand, H2AX is not able to interact with p53, and circ-CCNB1 is thus free to form a complex with H2AX and Bclaf1 and to thus to slow the development of p53 mutation-induced breast cancer progression (153). Similarly, circ-Dnmt1 can interact with p53 and AUF1 to facilitate their nuclear translocation, and this nuclear translocation of p53 induces autophagy. The nuclear translocation of AUF1 can increase mRNA stability and protein expression of Dnmt1, whereas the nuclear translocation of Dnmt1 further inhibits p53 transcription (84). In addition, CircSKA3 can interact with Tks5 and integrin-β1 to promote the invadopodium formation, thereby promoting breast cancer invasion (154).
Transcriptional Regulation
In at least one important case, circRNA has been shown to impact cellular biological functions via a direct impact on transcription in breast cancer. Specifically, circRNA FECR1 (a circRNA consisting of exons 2, 3, and 4 of the Friend leukemia integration 1 (FLI1) gene) has been shown to bind to the promoter of the gene encoding DNA methyltransferase 1 (DNMT1) and to down-regulate DNMT1 transcription. Similarly, circRNA FECR1 has been shown to recruit the methylcytosine dioxygenase TET1 to the promoter of FLI1 and thus to induce demethylation at the FLI1 promoter (85).
Encoding of Functional Peptides
One circRNA that is related to the growth factor receptor HER2 (circHER2) encodes a 103- amino acid peptide known HER2-103. This peptide promotes the binding of EGFR with HER2 and increases EGFR kinase activity. This activity has been shown to correlate with increased proliferation and invasion of breast cancer cells and to enhance sensitivity to pertuzumab (86).
Mechanisms Leading to Tumor Suppressive Functions of circRNAs in Breast Cancer
Consistent with the smaller number of circRNAs that are down-regulated in breast cancer, the number of known tumor suppressive circRNAs is much fewer than the oncogenic circRNAs in breast cancer. Similar to the mechanisms leading to enhancement of tumor development, these circRNAs may act as ceRNAs, protein decoys or scaffolds as well as encode functional peptides to suppress breast cancer progression. The known tumor suppressive circRNAs are listed in Table 3 .
Table 3.
CircRNAs with tumor suppressive functions in breast cancer.
| CircRNA name | CircBase ID | Length | Gene | Distribution | Phenotype | Target | Downstream genes/ pathways |
Reference |
|---|---|---|---|---|---|---|---|---|
| TV-circRGPD6 | \ | \ | \ | \ | inhibit tumor-initiating properties in vitro, inhibit tumor growth and metastasis in vivo | miR-26b | YAF2 | (152) |
| circ-Ccnb1 | hsa_circ_0072758 | 342 | CCNB1 | nucleus | inhibit cell proliferation, increase cell apoptosis in vitro, inhibit tumor growth and extend mouse viability in vivo | \ | interact with H2AX, p53, and BCLAF1 | (153) |
| circular RNA 000554 | hsa_circ_0000376 | 48782 | PRH1-PRR4 | cytoplasm | inhibit cell invasion and migration in vitro, inhibit tumor growth in vivo | miR-182 | ZFP36 | (155) |
| circRNA_0025202 | hsa_circ_0025202 | 495 | GAPDH | cytoplasm | inhibit cell proliferation, migration, increase cell apoptosis and sensitivity to tamoxifen in HR(+) BC cells in vitro | miR-182-5p | FOXO3a | (156) |
| circular RNA-0001283 | hsa_circ_0001283 | 1400 | WDR48 | cytoplasm | inhibit cell proliferation, invasion, and promote cell apoptosis | miR-187 | HIPK3 | (157) |
| circTADA2A-E6 and circTADA2A-E5/E6 | hsa_circ_0006220 hsa_circ_0043278 | 158 and 250 | TADA2A | cytoplasm | inhibit cell proliferation, migration, invasion in vitro | miR-203a-3p | SOCS | (52) |
| circASS1 | hsa_circ_0089105 | 241 | ASS1 | both nucleus and cytoplasm(majority) | inhibit cell invasion and migration in vitro | miR-4443 | ASS1 | (158) |
| circKDM4C | hsa_circ_0001839 | 292 | KDM4C | cytoplasm | inhibit cell proliferation, invasion, migration, doxorubicin resistance, promote cell apoptosis in vitro, inhibit tumor growth and metastasis, in vivo | miR-548p | PBLD | (53) |
| circBMPR2 | hsa_circ_0003218 | 342 | BMPR2 | cytoplasm | inhibit cell proliferation, migration, and invasion and tamoxifen resistance | miR-553 | USP4 | (159) |
| circular RNA 0001073 | hsa_circ_0001073 | 473 | ACVR2A | both nucleus and cytoplasm | inhibit proliferation, migration, invasion, and promote apoptosis in vitro, inhibit tumor growth in vivo | \ | HuR | (160) |
| circ-Foxo3 | \ | \ | FOXO3 | \ | inhibit tumor growth and extend mouse lifespan in vivo | \ | increase Foxo3 protein level but decreasep53 level | (161) |
| circular RNA−MTO1 | hsa_circ_0007874 | 318 | MTO1 | both nucleus and cytoplasm | inhibit cell proliferation, promote monastrol-induced cell cytotoxicity and reverse monastrol resistance in vitro | \ | TRAF4/Eg5 | (162) |
| circFBXW7 | hsa_circ_0001451 | 1227 | FBXW7 | cytoplasm | inhibit cell proliferation, migration in vitro and inhibit tumor growth and lung metastasis in vivo | miR-197-3p and FBXW7-185aa | FBXW7 | (163) |
| circRNA-000911 | \ | \ | \ | \ | inhibit cell proliferation, migration, and invasion, and promote cell apoptosis | miR−449a | NOTCH1/NF-κB | (59) |
| circEHMT1 | \ | \ | EHMT1 | \ | inhibit cell migration and invasion in vitro and inhibit lung metastasis in vivo | miR-1233-3p | KLF4/MMP2 | (164) |
| circNFIC | hsa_circ_0002018 | 311 | NFIC | cytoplasm | inhibit cell proliferation and migration in vitro, inhibit cell growth and lung metastasis in vivo | miR-658 | UPK1A | (165) |
| circ_0000442 | hsa_circ_0000442 | 7273 | MED13L | \ | inhibit cell proliferation in vitro and inhibit tumor growth in vivo | miR-148b-3p | PTEN/PI3K/AKT | (166) |
| circCDYL | hsa_circ_0008285 | 667 | CDYL | cytoplasm | inhibit cell proliferation, migration, invasion, and promote cell apoptosis | miR-190a-3p | TP53INP1 | (167) |
| circ-ITCH | \ | \ | ITCH | \ | inhibit cell proliferation, migration, and invasion in vitro, and inhibit tumor growth and metastasis in vivo | miR-214 and miR-17 | ITCH | (168) |
| Circular RNA BARD1 | hsa_circ_0001098 | 1204 | BARD1 | \ | inhibit cell proliferation and promote cell apoptosis in vitro, inhibit tumor growth and metastasis in vivo | miR-3942-3p | BARD1 | (169) |
| circAHNAK1 | hsa_circ_0000320 | 384 | AHNAK1 | cytoplasm | inhibit cell proliferation, migration, and invasion in vitro, inhibit tumor growth and metastasis in vivo | miR-421 | RASA1 | (170) |
| hsa_circ_0072309 | hsa_ circ_0072309 | 580 | LIFR | cytoplasm | inhibit cell proliferation, migration, and invasion in vitro, promote tumor growth in vivo | miR-492 | \ | (171) |
| circRNA_103809 | \ | \ | ZFR | \ | inhibit cell proliferation, invasion, and migration, EMT in vitro | miR-532-3p | \ | (172) |
| circDDX17 | hsa_circ_0002211 | 927 | DDX17 | cytoplasm | inhibit cell proliferation and promote cell apoptosis | miR-605 | CDK1 and p21 | (173) |
| hsa_circ_0068033 | hsa_circ_0068033 | 900 | NAALADL2 | cytoplasm | inhibit cell proliferation, invasion, migration, and promote apoptosis in vitro, inhibit tumor growth in vivo | miR-659 | \ | (174) |
| hsa_circ_0001785 | hsa_circ_0001785 | 467 | ELP3 | cytoplasm | inhibit cell proliferation, migration, and invasion in vitro, inhibit tumor growth in vivo | miR-942 | SOCS3 | (175) |
| circSMARCA5 | hsa_circ_0001445 | 269 | SMARCA5 | nucleus | increase cisplatin sensitivity in vitro and in vivo | \ | SMARCA5 | (176) |
| circSCYL2 | hsa_circ_0006258 | 508 | SCYL2 | \ | inhibit cell migration and invasion | \ | EMT | (177) |
| circ-LARP4 | \ | \ | LARP4 | \ | increase the doxorubicin sensitivity in vitro | \ | \ | (178) |
| circular RNA VRK1 | hsa_circ_0141206 | 852 | VRK1 | \ | inhibit cell proliferation, promote cell apoptosis in vitro | \ | \ | (179) |
| circular RNA VRK1 | hsa_circ_0141206 | 852 | VRK1 | cytoplasm | inhibit cell proliferation, promote cell apoptosis, and inhibit self-renewal capacity in vitro | \ | \ | (180) |
| circUSP42 | hsa_circ_0007823 | 451 | USP42 | \ | inhibit cell invasion and migration | miR-4443 | ASS1 | (181) |
ceRNAs
Both CircRNAs 000554 and circRNA_0025202 can sponge miR-182 to regulate the expression of ZNF36 and FOXO3a, respectively, and thus inhibit cellular proliferation, invasion, migration, and sensitivity to chemotherapy agents (155, 156). Similarly, circRNA-0001283 functions to sponge miR-187 to suppress HIPK3 expression, thus inhibiting the proliferation and invasion and promoting the apoptosis of breast cancer cells (157). CircTADA2A-E6 mainly regulates suppressor of cytokine signaling (SOCS) expression by sponging miR-203a-3p, and this sponging effect correlates with inhibition of breast cancer proliferation, invasion, and migration (52). CircASS1 mainly inhibits breast cancer invasion and migration by sponging miR-4443; it has also been found that the expression of circASS1 is inversely related with that of its parental gene arginosuccinate synthase 1 (ASS1), which indicates that the splicing of ASS1 mRNA and circASS1 may compete with each other (158). CircKDM4C mainly regulates the expression of phenazine biosynthesis like protein domain containing (PBLD) by sponging miR-548p, thereby inhibiting breast cancer growth, metastasis, and drug resistance (53). CircBMPR2 regulates ubiquitin specific peptidase 4 (USP4) expression by sponging miR-553, to inhibit breast cancer proliferation, invasion, migration, and tamoxifen resistance (159).
Protein Decoys or Scaffolds
Some tumor suppressive circRNAs function through interacting with proteins. For example, circ-Ccnb1 interacts with p53, inhibits tumor growth and increase the survival time of mice (153). Yi et al. reported the direct interaction between circRNA 0001073 and the RNA binding protein HuR (160). Experiments with the apoptosis-related protein receptor-interacting protein (RIP) revealed that the tumor suppressive circRNA circ-FOXO3, which is enriched in normal cells, can interact with MDM2, p53, and FOXO3, facilitating the formation of the MDM2-P53 complex while inhibiting MDM2-FOXO3 binding. This results in promotion of p53 ubiquitination and inhibition of FOXO3 degradation and thus the induction of apoptosis of breast cancer cells (161). Circular RNA-MTO1 can block the interaction between TRAF4 and Eg5, and suppress the translation of Eg5, to inhibit breast cancer growth and reverse the resistance of breast cancer cells to monosterol (162).
Encoding of Functional Peptides
circFBXW7 not only can interact with miR-197-3p, but also encodes a 185-amino-acid peptide that regulates the expression of FBXW7 and the degradation of c-Myc, thus inhibiting proliferation and migration of ovarian cancer cells (163).
Analysis of CircRNAs in the Diagnosis and Prognosis of Breast Cancer
Although circRNAs in plasma are not as stable as those within cells, circRNAs are still considered important diagnostic and prognostic biomarkers for cancer. CircRNAs that are particularly important diagnostic or prognostic biomarkers in breast cancer are listed in Table 4 .
Table 4.
CircRNAs with diagnostic and prognostic value in breast cancer.
| CircRNA name | Circbase ID | Clinicopathological Association | Tumor type (Sample number) | Reference |
|---|---|---|---|---|
| circSEPT9 | hsa_circ_0005320 | TNM stage, T stage, N stage, OS, diagnosis | TNBC (cohort2: n=80; cohort1: n=60) | (32) |
| circZNF609 | hsa_circ_0000615 | lymph node metastasis, TNM stage, OS | (n=143) | (74) |
| circHMCU | hsa_circ_0000247 | lymph node metastasis, T stage, N stage, histological grade, OS | (n=267) | (79) |
| circPLK1 | hsa_circ_0038632 | tumor size, lymph node metastasis, TNM stage, DFS, OS | TNBC (n=240) | (81) |
| hsa_circ_0000515 | hsa_circ_0000515 | OS | (n=340) | (82) |
| circWWC3 | hsa_circ_0089866 and hsa_circ_0001910 | clinical stage, OS | (n=156) | (83) |
| circRGPD6 | \ | DFS, OS | (n=165) | (152) |
| circular HER2 | hsa_circ_0007766 | OS | TNBC (n=59) | (86) |
| circRNA_0025202 | hsa_circ_0025202 | lymph node metastasis, histological grade | HR positive breast cancer (n=230) | (156) |
| circular RNA 0001073 | hsa_circ_0001073 | tumor size, distant metastasis, TNM stage, RFS, diagnosis | (n=132) | (160) |
| circFBXW7 | hsa_circ_0001451 | tumor size, lymph node metastasis, DFS, OS | TNBC (n=473) | (163) |
| circular RNA VRK1 | hsa_circ_0141206 | tumor size, TNM stage, T stage, N stage, histological grade, OS, diagnosis | (n=350) | (179) |
| *circIFI30 | hsa_circ_0005571 | clinical stage, histological grade, OS, diagnosis | TNBC (n=78/n=38) | (87) |
| circAGFG1 | hsa_circ_0058514 | TNM stage, T stage, N stage, OS, diagnosis | TNBC (n=40) | (54) |
| hsa_circ_006054, hsa_circ_100219, and hsa_circ_406697 | \ | diagnosis | (n=51) | (55) |
| hsa_circ_0001785 | hsa_circ_0001785 | distant metastasis, TNM stage, histological grade, diagnosis | (n=57) | (182) |
| circKIF4A | hsa_circ_0007255 | tumor size, lymph node metastasis, TNM stage, DFS, OS | TNBC (n=240) | (88) |
| circEPSTI1 | hsa_circ_0000479 | tumor size, lymph node metastasis, TNM stage, DFS, OS | TNBC (n=240) | (56) |
| circGNB1 | hsa_circ_0009362 | tumor size, TNM stage, DFS, OS | TNBC (n=222) | (89) |
| circANKS1B | hsa_circ_0007294 | lymph node metastasis, clinical stage, OS | TNBC (n=165) | (61) |
| hsa_circ_0006220 | hsa_circ_0006220 | lymph node metastasis, pathological type, diagnosis | (n=50) | (65) |
| circGFRA1 | hsa_circ_0005239 | tumor size, lymph node metastasis, TNM stage, DFS, OS | TNBC (n=222) | (90) |
| circRNA_069718 | hsa_circ_0069718 | lymph node metastasis, TNM stage, OS | TNBC (n=35) | (91) |
| hsa_circ_002178 | \ | tumor size, lymph node metastasis, OS | (n=83) | (97) |
| circRNA_100876 | \ | OS | (n=50) | (99) |
| circMMP11 | hsa_circ_0062558 | lymph node, metastasis, TNM stage | (n=113) | (101) |
| circUBE2D2 | \ | tumor size, lymph node metastasis, TNM stage, OS, PFS | (n=80) | (102) |
| circUBE2D2 | hsa_circ_0005728 | lymph node metastasis, TNM stage, OS | TNBC (n=66) | (104) |
| circCDYL | \ | DFS, OS | (n=113) | (108) |
| circ_0000520 | hsa_circ_0000520 | lymph node metastasis, TNM stage, OS | (n=60) | (109) |
| circHIPK3 | hsa_circ_0000284 | lymph node metastasis, TNM stage, OS | (n=50) | (114) |
| hsa_circ_001783 | \ | tumor size, metastasis, TNM stage, DFS | (n=136) | (115) |
| circular RNA PVT1 | hsa_circ_0001821 | TNM stage, OS | (n=99) | (117) |
| circRAD18 | hsa_circ_0002453 | tumor size, TNM stage, T stage, OS | TNBC (n=126) | (118) |
| hsa_circ_0131242 | hsa_circ_0131242 | tumor size, TNM stage, OS | TNBC (n=86) | (120) |
| circ_0000291 | hsa_circ_0000291 | tumor size, lymph node metastasis | (n=37) | (123) |
| hsa_circRNA_002178 | hsa_circ_0000519 | OS | (n=70) | (126) |
| circ-PGAP3 | hsa_circ_0106800 | tumor size, lymph node metastasis, TNM stage, DFS,OS | TNBC (n=86) | (127) |
| circIRAK3 | hsa_circ_0005505 | RFS | (n=122) | (128) |
| circMYO9B | hsa_circ_0000907 | tumor size, lymph node metastasis, TNM stage, OS | (n=21) | (133) |
| circRNF20 | hsa_circ_0087784 | tumor size, lymph node metastasis, OS | (n=50) | (134) |
| circZFR | hsa_circ_0072088 | TNM stage, OS | (n=70) | (138) |
| circ-UBAP2 | hsa_circ_0001846 | tumor size, lymph node metastasis, distant metastasis, TNM stage, OS | TNBC (n=78) | (139) |
| circFBXL5 | hsa_circ_0125597 | OS | (n=150) | (140) |
| circCNOT2 | \ | PFS | (n=84) | (144) |
| hsa_circ_001569 | \ | lymph node metastasis, clinical stage, OS | (n=75) | (148) |
| circBACH2 | hsa_circ_0001627 | T stage, N stage, TNM stage | TNBC (n=38) | (149) |
| circNFIC | hsa_circ_0002018 | lymph node metastasis, OS | (n=145) | (165) |
| circAHNAK1 | hsa_circ_0000320 | TNM stage, T stage, N stage, DFS, OS | TNBC (n=136) | (170) |
| hsa_circ_0072309 | hsa_circ_0072309 | tumor size, lymph node metastasis, TNM stage, OS | (n=32) | (171) |
| circRNA_103809 | \ | distant metastasis, TNM stage, OS, HER2 status | (n=65) | (172) |
| circDDX17 | hsa_circ_0002211 | lymph node metastasis, TNM stage | \ | (173) |
| hsa_circ_0068033 | hsa_circ_0068033 | tumor size, lymph node metastasis, diagnosis | (n=36) | (174) |
| circ-LARP4 | \ | tumor size, TNM stage, T stage, N stage, DFS, OS | (n=283) | (178) |
| circUSP42 | hsa_circ_0007823 | lymph node metastasis, TNM stage, DFS, OS | TNBC (n=30) | (181) |
TNM, tumor node metastasis; DFS, disease free survival; PFS, progression free survival; OS, overall survival; *n = 78 for the study of relationships between circIFI30 expression and clinical stage, histological grade and OS, while n = 38 for the study of the diagnostic value of circIFI30.
As an example of this role, the analyses of several circRNAs can distinguish breast cancer tissues from noncancerous tissues. These distinctions have been shown to occur with favorable statistics, including having area under the curve (AUC) values greater than 0.7. These diagnostic circRNAs include the circular RNA VRK1 (AUC: 0.720, n = 350) (179), circIFI30 (AUC: 0.733, n = 38) (87), circSEPT9 (AUC: 0.711, n = 60) (32) and circAGFG1 (AUC: 0.767, n = 40) (54). In addition, the combination of hsa_circ_006054, hsa_circ_100219, and hsa_circ_406697 has proven useful in diagnosis (AUC: 0.82, n = 51) (55). Although these circRNAs have been shown to be useful as tissue markers, to date, only hsa_circ_0001785 in the plasma has been shown to serve as potentially non-invasive biomarker for breast cancer (AUC: 0.771, n = 20; AUC: 0.784, n = 57) (182).
In addition to diagnostic potential, several circRNAs are prognostic biomarkers for breast cancer. For example, the expressions of circPLK1, circKIF4A, circEPSTI1, circGNB1, or circGFRA1 are associated with the tumor size and TNM stage of TNBC, whereas circPLK1, circKIF4A, circEPSTI1, or circGFRA1 are also associated with the lymph node metastasis of TNBC (56, 81, 88, 89). Similarly, high expression of circPLK1, circKIF4A, circEPSTI1, circGNB1, or circGFRA1 is associated with worse disease-free survival and overall survival in TNBC (56, 81, 88, 89).
Prospectives
Despite the rapid progress of our understanding of circRNA biogenesis and degradation over the last several decades, many important issues remain unresolved. For example, what signals regulate circRNA-specific expression? What are the intracellular or extracellular signals that activate the mechanisms leading to degradation of circRNAs? Why have only a few blood-localized circRNAs been identified as useful diagnostic markers?
Yet another important question involves potential roles of circRNAs in the communication between the tumor cells and the tumor microenvironment? Although many breast cancer-associated dysregulated circRNAs have been discovered and functionally characterized, most related studies have focused only on the roles of circRNAs on the proliferation, migration, invasion, apoptosis, and chemoresistance of tumor cells. However, tumor heterogeneity is a great challenge for therapeutic management, so growing attention has recently been paid to the molecules that mediate communication between tumor cells and cells of the tumor microenvironment, including cancer-associated fibroblasts, tumor-associated macrophages and T cells. Accordingly, further studies regarding the function of circRNA in mediating the dialogs that occur within or between tumor, stromal and immune cells have great future prospects.
Recently, with the development of ribosome nascent-chain complex (RNC) sequencing and ribo-sequencing, several circRNAs with coding potential have been discovered; however, only a few circRNAs with coding potential have been definitively identified, and most of these studies have indicated that the circRNA-encoded peptides, but not the circRNAs themselves, are the functional unit. Given the broad prospects of peptide therapeutics in the pharmaceutical industry, studies on tumor suppressive circRNA-encoded peptides may provide new directions for the development of drugs for breast cancer.
In addition, as noted in this study, different circRNAs can sponge the same miRNA, but these interactions tend to regulate the expression of different targets. These circRNAs, miRNAs, and targets form regulatory networks and thus promote or inhibit the progression of breast cancer. Therefore, the study of the circRNAs, miRNAs, and target networks in different systems is an important direction for future research.
Another important question involves mechanisms leading to the intracellular localizations of circRNAs. By analyzing the distribution of the tumor suppressive and oncogenic circRNAs in breast cancer ( Table 2 , Table 3 ), we noticed that most of the circRNAs with known function are mainly distributed in the cytoplasm. Five circRNAs, with lengths of 241, 318, 435, 473, and 571 bp, are distributed in both the cytoplasm and nucleus, and two circRNAs, of 269 and 342 bp, are mainly distributed in the nucleus. Similarly, at least 22 cytoplasm-localized circRNAs are less than 400 bp, and this phenomenon cannot be explained by transport mechanisms that solely involve the length of the circRNA, indicating that regulation of the localization of circRNAs is more complex than is currently appreciated.
Conclusion
CircRNAs form a large class of regulators of breast cancer progression. Clinical studies have also indicated that circRNAs are potential biomarkers for breast cancer diagnosis, prognosis, and therapy. However, the regulation network of circRNAs in breast cancer is still incompletely defined, and this uncertainty impedes the clinical exploitation of circRNAs. Future studies on the precise mechanisms of the key circRNA regulators and clinical relevance to breast cancer will further promote the clinical use of circRNAs and/or their relevant products in the management of breast cancer.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author Contributions
JX wrote the manuscript text and prepared the tables. XC revised the manuscript and prepared the figures. CL and XJ provided advice and revised the manuscript. YuS, YaS, FT, and ML critically reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The study was financially supported by the National Natural Science Foundation of China (81902651), Jiangsu provincial key research and development program (BE2019621), Research Innovation Program for Graduates of Jiangsu Province (JX10413758, JX10413759).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
The reviewer [ZW] declared a shared affiliation, with no collaboration, with the authors to the handling editor at the time of review.
Acknowledgments
We thank BioRender for providing the template for our figures.
Abbreviations
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; BC, breast cancer; LA, luminal A; TNBC, triple-negative breast cancer; NMGT, normal mammary gland tissues; BCLM, breast cancer liver metastases; IPSC, induced pluripotent stem cells; HR, hormonal receptor; TNM, topography, lymph node, metastasis; OS, overall survival; DFS, disease free survival; RFS, recurrence-free survival; BCSC, breast cancer stem cell; EMT, epithelial–mesenchymal transition; PTX, paclitaxel; AUC, area under curve; DALI, distal-Alu-long-intron; SRD, structure-mediated RNA decay; RBP, RNA binding proteins; IRES, internal ribosome entry site; AUC, area under curve.
References
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71:7–33. 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2. Sun Y, Zhao Z, Yang Z, Xu F, Lu H, Zhu Z, et al. Risk Factors and Preventions of Breast Cancer. Int J Biol Sci (2017) 13:1387–97. 10.7150/ijbs.21635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eroles P, Bosch A, Alejandro Pérez-Fidalgo J, Lluch A. Molecular Biology in Breast Cancer: Intrinsic Subtypes and Signaling Pathways. Cancer Treat Rev (2011) 38:698–707. 10.1016/j.ctrv.2011.11.005 [DOI] [PubMed] [Google Scholar]
- 4. Howlader N, Cronin KA, Kurian AW, Andridge R. Differences in Breast Cancer Survival by Molecular Subtypes in the United States. Cancer Epidem Biomar (2018) 27:619–26. 10.1158/1055-9965.EPI-17-0627 [DOI] [PubMed] [Google Scholar]
- 5. Yin L, Duan J, Bian X, Yu S. Triple-Negative Breast Cancer Molecular Subtyping and Treatment Progress. Breast Cancer Res (2020) 22:61. 10.1186/s13058-020-01296-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Okholm T, Nielsen MM, Hamilton MP, Christensen LL, Vang S, Hedegaard J, et al. Circular RNA Expression is Abundant and Correlated to Aggressiveness in Early-Stage Bladder Cancer. NPJ Genom Med (2017) 2:36. 10.1038/s41525-017-0038-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat Rev Genet (2019) 20:675–91. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 8. Jeck WR, Sharpless NE. Detecting and Characterizing Circular RNAs. Nat Biotechnol (2014) 32:453–61. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim TB, Lavenniah A, Foo RS. Circles in the Heart and Cardiovascular System. Cardiovasc Res (2020) 116:269–78. 10.1093/cvr/cvz227 [DOI] [PubMed] [Google Scholar]
- 10. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA Profiling Reveals an Abundant Circhipk3 That Regulates Cell Growth by Sponging Multiple miRNAs. Nat Commun (2016) 7:11215. 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-Like Structures. P Natl Acad Sci USA (1976) 73:3852–6. 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patop IL, Wüst S, Kadener S. Past, Present, and Future of circRNAs. EMBO J (2019) 38:e100836. 10.15252/embj.2018100836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO, Preiss T. Circular RNAs are the Predominant Transcript Isoform From Hundreds of Human Genes in Diverse Cell Types. PloS One (2012) 7:e30733. 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Naeli P, Pourhanifeh MH, Karimzadeh MR, Shabaninejad Z, Movahedpour A, Tarrahimofrad H, et al. Circular RNAs and Gastrointestinal Cancers: Epigenetic Regulators With a Prognostic and Therapeutic Role. Crit Rev Oncol Hematol (2020) 145:102854. 10.1016/j.critrevonc.2019.102854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are Abundant, Conserved, and Associated With ALU Repeats. RNA (2013) 19:141–57. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a Large Class of Animal RNAs With Regulatory Potency. Nature (2013) 495:333–8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 17. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-Intron Circular RNAs Regulate Transcription in the Nucleus. Nat Struct Mol Biol (2015) 22:256–64. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 18. Lasda E, Parker R. Circular RNAs: Diversity of Form and Function. RNA (Cambridge) (2014) 20:1829–42. 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep-UK (2015) 5:16435. 10.1038/srep16435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA Biogenesis Competes With Pre-mRNA Splicing. Mol Cell (2014) 56:55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 21. Pagliarini V, Jolly A, Bielli P, Di Rosa V, de la Grange P, Sette C. Sam68 Binds Alu-Rich Introns in SMN and Promotes pre-mRNA Circularization. Nucleic Acids Res (2020) 48:633–45. 10.1093/nar/gkz1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stagsted LVW, O’Leary ET, Ebbesen KK, Hansen TB. The RNA-Binding Protein SFPQ Preserves Long-Intron Splicing and Regulates circRNA Biogenesis in Mammals. ELIFE (2021) 10:e63088. 10.7554/eLife.63088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li X, Ding J, Wang X, Cheng Z, Zhu Q. NUDT21 Regulates circRNA Cyclization and ceRNA Crosstalk in Hepatocellular Carcinoma. Oncogene (2020) 39:891–904. 10.1038/s41388-019-1030-0 [DOI] [PubMed] [Google Scholar]
- 24. Zhao W, Cui Y, Liu L, Qi X, Liu J, Ma S, et al. Splicing Factor Derived Circular RNA Circuhrf1 Accelerates Oral Squamous Cell Carcinoma Tumorigenesis via Feedback Loop. Cell Death Differ (2020) 27:919–33. 10.1038/s41418-019-0423-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu X, Shen S, Zhu L, Su R, Zheng J, Ruan X, et al. SRSF10 Inhibits Biogenesis of Circ-ATXN1 to Regulate Glioma Angiogenesis via miR-526b-3p/MMP2 Pathway. J Exp Clin Canc Res (2020) 39:121. 10.1186/s13046-020-01625-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li X, Liu C, Xue W, Zhang Y, Jiang S, Yin Q, et al. Coordinated circRNA Biogenesis and Function With NF90/NF110 in Viral Infection. Mol Cell (2017) 67:214–27. 10.1016/j.molcel.2017.05.023 [DOI] [PubMed] [Google Scholar]
- 27. Dai X, Zhang N, Cheng Y, Yang T, Chen Y, Liu Z, et al. RNA-Binding Protein Trinucleotide Repeat-Containing 6A Regulates the Formation of Circular RNA Circ0006916, With Important Functions in Lung Cancer Cells. Carcinogene (New York) (2018) 39:981–92. 10.1093/carcin/bgy061 [DOI] [PubMed] [Google Scholar]
- 28. Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, et al. FUS Affects Circular RNA Expression in Murine Embryonic Stem Cell-Derived Motor Neurons. Nat Commun (2017) 8:14741. 10.1038/ncomms14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang Z, Qu CB, Zhang Y, Zhang WF, Wang DD, Gao CC, et al. Dysregulation of P53-RBM25-Mediated Circamotl1l Biogenesis Contributes to Prostate Cancer Progression Through the Circamotl1l-miR-193a-5p-Pcdha Pathway. Oncogene (2019) 38:2516–32. 10.1038/s41388-018-0602-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carlson SM, Soulette CM, Yang Z, Elias JE, Brooks AN, Gozani O. RBM25 is a Global Splicing Factor Promoting Inclusion of Alternatively Spliced Exons and Is Itself Regulated by Lysine Mono-Methylation. J Biol Chem (2017) 292:13381–90. 10.1074/jbc.M117.784371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou A, Ou AC, Cho A, Jr., Benz EJ, Huang S. Novel Splicing Factor RBM25 Modulates Bcl-X Pre-mRNA 5 ‘ Splice Site Selection. Mol Cell Biol (2008) 28:5924–36. 10.1128/MCB.00560-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R, et al. The circRNA Circsept9 Mediated by E2F1 and EIF4A3 Facilitates the Carcinogenesis and Development of Triple-Negative Breast Cancer. Mol Cancer (2020) 19:73. 10.1186/s12943-020-01183-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dong W, Dai Z, Liu F, Guo X, Ge C, Ding J, et al. The RNA-Binding Protein RBM3 Promotes Cell Proliferation in Hepatocellular Carcinoma by Regulating Circular RNA SCD-circRNA 2 Production. EBIOMEDICINE (2019) 45:155–67. 10.1016/j.ebiom.2019.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang C, Xie Y, Yu T, Liu N, Wang Z, Woolsey RJ, et al. M6a-Dependent Biogenesis of Circular RNAs in Male Germ Cells. Cell Res (2020) 30:211–28. 10.1038/s41422-020-0279-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo M, Yan R, Ji Q, Yao H, Sun M, Duan L, et al. IFN Regulatory Factor-1 Induced Macrophage Pyroptosis by Modulating M6a Modification of Circ_0029589 in Patients With Acute Coronary Syndrome. Int Immunopharmacol (2020) 86:106800. 10.1016/j.intimp.2020.106800 [DOI] [PubMed] [Google Scholar]
- 36. Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N 6-Methyladenosine-Modified CircRNA-SORE Sustains Sorafenib Resistance in Hepatocellular Carcinoma by Regulating β-Catenin Signaling. Mol Cancer (2020) 19:163. 10.1186/s12943-020-01281-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Di Timoteo G, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F, et al. Modulation of circRNA Metabolism by M6a Modification. Cell Rep (Cambridge) (2020) 31:107641. 10.1016/j.celrep.2020.107641 [DOI] [PubMed] [Google Scholar]
- 38. Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, et al. Genome-Wide Maps of M6a circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns That Are Distinct From mRNAs. Cell Rep (Cambridge) (2017) 20:2262–76. 10.1016/j.celrep.2017.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schoenberg DR, Maquat LE. Regulation of Cytoplasmic mRNA Decay. Nat Rev Genet (2012) 13:246–59. 10.1038/nrg3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guo Y, Wei X, Peng Y. Structure-Mediated Degradation of CircRNAs. Trends Cell Biol (2020) 30:501–3. 10.1016/j.tcb.2020.04.001 [DOI] [PubMed] [Google Scholar]
- 41. Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. miRNA-Dependent Gene Silencing Involving Ago2-Mediated Cleavage of a Circular Antisense RNA. EMBO J (2011) 30:4414–22. 10.1038/emboj.2011.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fischer JW, Busa VF, Shao Y, Leung AKL. Structure-Mediated RNA Decay by UPF1 and G3BP1. Mol Cell (2020) 78:70–84. 10.1016/j.molcel.2020.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jia R, Xiao M, Li Z, Shan G, Huang C. Defining an Evolutionarily Conserved Role of GW182 in Circular RNA Degradation. Cell Discovery (2019) 5:1–4. 10.1038/s41421-019-0113-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic Cleavage of M 6 A-Containing RNAs by RNase P/MRP Complex. Mol Cell (2019) 74:494. 10.1016/j.molcel.2019.02.034 [DOI] [PubMed] [Google Scholar]
- 45. Liu C, Li X, Nan F, Jiang S, Gao X, Guo S, et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell (Cambridge) (2019) 177:865–80. 10.1016/j.cell.2019.03.046 [DOI] [PubMed] [Google Scholar]
- 46. Li Z, Kearse MG, Huang C. The Nuclear Export of Circular RNAs Is Primarily Defined by Their Length. RNA Biol (2019) 16:1–4. 10.1080/15476286.2018.1557498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang C, Liang D, Tatomer DC, Wilusz JE. A Length-Dependent Evolutionarily Conserved Pathway Controls Nuclear Export of Circular RNAs. Gene Dev (2018) 32:639–44. 10.1101/gad.314856.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen R, Chen X, Xia L, Zhang J, Pan Z, Ma X, et al. N6-Methyladenosine Modification of Circnsun2 Facilitates Cytoplasmic Export and Stabilizes HMGA2 to Promote Colorectal Liver Metastasis. Nat Commun (2019) 10:4695. 10.1038/s41467-019-12651-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiao W, Adhikari S, Dahal U. Nuclear M(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell (2016) 61:507–19. 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 50. Roundtree IA, Luo G, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 Mediates Nuclear Export of N6-Methyladenosine Methylated mRNAs. ELIFE (2017) 6:e31311. 10.7554/eLife.31311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou J, Zhang W, Peng F, Sun J, He Z, Wu S. Downregulation of Hsa_Circ_0011946 Suppresses the Migration and Invasion of the Breast Cancer Cell Line MCF-7 by Targeting RFC3. Cancer Manag Res (2018) 10:535–44. 10.2147/CMAR.S155923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu J, Shao C, Wang X, Zhao X, Chen J, Ouyang Y, et al. Circtada2as Suppress Breast Cancer Progression and Metastasis via Targeting miR-203a-3p/SOCS3 Axis. Cell Death Dis (2019) 10:175. 10.1038/s41419-019-1382-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liang Y, Song X, Li Y, Su P, Han D, Ma T, et al. Circkdm4c Suppresses Tumor Progression and Attenuates Doxorubicin Resistance by Regulating miR-548p/PBLD Axis in Breast Cancer. Oncogene (2019) 38:6850–66. 10.1038/s41388-019-0926-z [DOI] [PubMed] [Google Scholar]
- 54. Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA Circagfg1 Acts as a Sponge of miR-195-5p to Promote Triple-Negative Breast Cancer Progression Through Regulating CCNE1 Expression. Mol Cancer (2019) 18:4. 10.1186/s12943-018-0933-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55. Lü L, Sun J, Shi P, Kong W, Xu K, He B, et al. Identification of Circular RNAs as a Promising New Class of Diagnostic Biomarkers for Human Breast Cancer. Oncotarget (2017) 8:44096–107. 10.18632/oncotarget.17307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, et al. Circepsti1 as a Prognostic Marker and Mediator of Triple-Negative Breast Cancer Progression. Theranostics (2018) 8:4003–15. 10.7150/thno.24106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liang H, Zhang X, Liu B, Jia G, Li W. Circular RNA Circ-ABCB10 Promotes Breast Cancer Proliferation and Progression Through Sponging miR-1271. Am J Cancer Res (2017) 7:1566–76. [PMC free article] [PubMed] [Google Scholar]
- 58. Tang Y, Zhao P, Zou T, Duan J, Zhi R, Yang S, et al. Circular RNA Hsa_Circ_0001982 Promotes Breast Cancer Cell Carcinogenesis Through Decreasing miR-143. DNA Cell Biol (2017) 36:901–8. 10.1089/dna.2017.3862 [DOI] [PubMed] [Google Scholar]
- 59. Wang H, Xiao Y, Wu L, Ma D. Comprehensive Circular RNA Profiling Reveals the Regulatory Role of the circRNA-000911/miR-449a Pathway in Breast Carcinogenesis. Int J Oncol (2018) 52:743–54. 10.3892/ijo.2018.4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yuan C, Zhou L, Zhang L, Yin K, Peng J, Sha R, et al. Identification and Integrated Analysis of Key Differentially Expressed Circular RNAs in ER-Positive Subtype Breast Cancer. Epigenomics-UK (2018) 11:297–321. 10.2217/epi-2018-0147 [DOI] [PubMed] [Google Scholar]
- 61. Zeng K, He B, Yang BB, Xu T, Chen X, Xu M, et al. The Pro-Metastasis Effect of Circanks1b in Breast Cancer. Mol Cancer (2018) 17:160. 10.1186/s12943-018-0914-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xie R, Tang J, Zhu X, Jiang H. Silencing of Hsa_Circ_0004771 Inhibits Proliferation and Induces Apoptosis in Breast Cancer Through Activation of miR-653 by Targeting ZEB2 Signaling Pathway. Biosci Rep (2019) 39:BSR20181919. 10.1042/BSR20181919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lin X, Hong S, Chen J, Chen W, Wu Z. The Potential Targets for Metastases: a Study on Altered Circular RNA Profile in Breast Cancer Liver Metastases. Epigenomics-UK (2019) 11:1237–50. 10.2217/epi-2019-0099 [DOI] [PubMed] [Google Scholar]
- 64. Li Z, Chen Z, Hu G, Zhang Y, Feng Y, Jiang Y, et al. Profiling and Integrated Analysis of Differentially Expressed circRNAs as Novel Biomarkers for Breast Cancer. J Cell Physiol (2020) 235:7945–59. 10.1002/jcp.29449 [DOI] [PubMed] [Google Scholar]
- 65. Liu C, Chen M, Shi Y. Downregulation of Hsa_Circ_0006220 and its Correlation With Clinicopathological Factors in Human Breast Cancer. Gland Surg (2021) 10:816–25. 10.21037/gs-21-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: A Rising Star in Gastric Cancer. Cell Mol Life Sci (2020) 77:1661–80. 10.1007/s00018-019-03345-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA Profiling Reveals an Abundant Circhipk3 That Regulates Cell Growth by Sponging Multiple miRNAs. Nat Commun (2016) 7:11215. 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495:384–8. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 69. Yang W, Gu J, Wang X, Wang Y, Feng M, Zhou D, et al. Inhibition of Circular RNA CDR1as Increases Chemosensitivity of 5-FU-Resistant BC Cells Through Up-Regulating miR-7. J Cell Mol Med (2019) 23:3166–77. 10.1111/jcmm.14171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gao D, Zhang X, Liu B, Meng D, Fang K, Guo Z, et al. Screening Circular RNA Related to Chemotherapeutic Resistance in Breast Cancer. Epigenomics-UK (2017) 9:1175–88. 10.2217/epi-2017-0055 [DOI] [PubMed] [Google Scholar]
- 71. Gao D, Qi X, Zhang X, Fang K, Guo Z, Li L. Hsa_circRNA_0006528 as a Competing Endogenous RNA Promotes Human Breast Cancer Progression by Sponging miR-7-5p and Activating the MAPK/ERK Signaling Pathway. Mol Carcinogen (2019) 58:554–64. 10.1002/mc.22950 [DOI] [PubMed] [Google Scholar]
- 72. Jiang J, Lin H, Shi S, Hong Y, Bai X, Cao X. Hsa_circRNA_0000518 Facilitates Breast Cancer Development via Regulation of themiR-326/FGFR1axis. Thorac Cancer (2020) 11:3181–92. 10.1111/1759-7714.13641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li Y, Jiang B, He Z, Zhu H, He R, Fan S, et al. circIQCH Sponges miR-145 to Promote Breast Cancer Progression by Upregulating DNMT3A Expression. Aging (Albany NY.) (2020) 12:15532–45. 10.18632/aging.103746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang S, Xue X, Wang R, Li X, Li Q, Wang Y, et al. CircZNF609 Promotes Breast Cancer Cell Growth, Migration, and Invasion by Elevating P70s6k1 via Sponging miR-145-5p. Cancer Manag Res (2018) 10:3881–90. 10.2147/CMAR.S174778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu L, Tian Q, Liu J, Zhou Y, Yong H. Upregulation of Hsa_Circ_0136666 Contributes to Breast Cancer Progression by Sponging miR-1299 and Targeting CDK6. J Cell Biochem (2019) 120:12684–93. 10.1002/jcb.28536 [DOI] [PubMed] [Google Scholar]
- 76. Liu G, Zhang Z, Song Q, Guo Y, Bao P, Shui H. Circ_0006528 Contributes to Paclitaxel Resistance of Breast Cancer Cells by Regulating miR-1299/CDK8 Axis. Oncotargets Ther (2020) 13:9497–511. 10.2147/OTT.S252886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Geng Z, Wang W, Chen H, Mao J, Li Z, Zhou J. Circ_0001667 Promotes Breast Cancer Cell Proliferation and Survival via Hippo Signal Pathway by Regulating TAZ. Cell Biosci (2019) 9:104. 10.1186/s13578-019-0359-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang H, Jiang L, Hou J, Zhong S, Zhou S, Zhu L, et al. Circular RNA Hsa_Circ_0052112 Promotes Cell Migration and Invasion by Acting as Sponge for miR-125a-5p in Breast Cancer. BioMed Pharmacother (2018) 107:1342–53. 10.1016/j.biopha.2018.08.030 [DOI] [PubMed] [Google Scholar]
- 79. Song X, Liang Y, Sang Y, Li Y, Zhang H, Chen B, et al. circHMCU Promotes Proliferation and Metastasis of Breast Cancer by Sponging the Let-7 Family. Mol Ther-Nucl Acids (2020) 20:518–33. 10.1016/j.omtn.2020.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang W, Gong P, Yang Y, Yang C, Yang B, Ren L. Circ-ABCB10 Contributes to Paclitaxel Resistance in Breast Cancer Through Let-7a-5p/DUSP7 Axis. Cancer Manag Res (2020) 12:2327–37. 10.2147/CMAR.S238513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kong Y, Yang L, Wei W, Lyu N, Zou Y, Gao G, et al. CircPLK1 Sponges miR-296-5p to Facilitate Triple-Negative Breast Cancer Progression. Epigenomics-UK (2019) 11:1163–76. 10.2217/epi-2019-0093 [DOI] [PubMed] [Google Scholar]
- 82. Cai F, Fu W, Tang L, Tang J, Sun J, Fu G, et al. Hsa_circ_0000515 Is a Novel Circular RNA Implicated in the Development of Breast Cancer Through Its Regulation of the microRNA-296-5p/CXCL10 Axis. FEBS J (2021) 288:861–83. 10.1111/febs.15373 [DOI] [PubMed] [Google Scholar]
- 83. Meng L, Liu S, Liu F, Sang M, Ju Y, Fan X, et al. ZEB1-Mediated Transcriptional Upregulation of Circwwc3 Promotes Breast Cancer Progression Through Activating Ras Signaling Pathway. Mol Ther-Nucl Acids (2020) 22:124–37. 10.1016/j.omtn.2020.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, et al. A Circular RNA Circ-DNMT1 Enhances Breast Cancer Progression by Activating Autophagy. Oncogene (2018) 37:5829–42. 10.1038/s41388-018-0369-y [DOI] [PubMed] [Google Scholar]
- 85. Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, et al. A Novel FLI1 Exonic Circular RNA Promotes Metastasis in Breast Cancer by Coordinately Regulating TET1 and DNMT1. Genome Biol (2018) 19:218. 10.1186/s13059-018-1594-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li J, Ma M, Yang X, Zhang M, Luo J, Zhou H, et al. Circular HER2 RNA Positive Triple Negative Breast Cancer Is Sensitive to Pertuzumab. Mol Cancer (2020) 19:1–142. 10.1186/s12943-020-01259-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xing L, Yang R, Wang X, Zheng X, Yang X, Zhang L, et al. The circRNA Circifi30 Promotes Progression of Triple-Negative Breast Cancer and Correlates With Prognosis. Aging (Albany NY.) (2020) 12:10983–1003. 10.18632/aging.103311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tang H, Huang X, Wang J, Yang L, Kong Y, Gao G, et al. Circkif4a Acts as a Prognostic Factor and Mediator to Regulate the Progression of Triple-Negative Breast Cancer. Mol Cancer (2019) 18:23. 10.1186/s12943-019-0946-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu P, Zou Y, Li X, Yang A, Ye F, Zhang J, et al. Circgnb1 Facilitates Triple-Negative Breast Cancer Progression by Regulating miR-141-5p-IGF1R Axis. Front Genet (2020) 11:193. 10.3389/fgene.2020.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, et al. Circgfra1 and GFRA1 Act as ceRNAs in Triple Negative Breast Cancer by Regulating miR-34a. J Exp Clin Canc Res (2017) 36:145. 10.1186/s13046-017-0614-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang J, Xu H, Xing X, Liang Z, Xia Z, Zhao Y. CircRNA_069718 Promotes Cell Proliferation and Invasion in Triple-Negative Breast Cancer by Activating Wnt/β-Catenin Pathway. Eur Rev Med Pharmaco (2019) 23:5315–22. 10.26355/eurrev_201906_18198 [DOI] [PubMed] [Google Scholar]
- 92. Sang M, Meng L, Liu S, Ding P, Chang S, Ju Y, et al. Circular RNA ciRS-7 Maintains Metastatic Phenotypes as a ceRNA of miR-1299 to Target MMPs. Mol Cancer Res (2018) 16:1665–75. 10.1158/1541-7786.MCR-18-0284 [DOI] [PubMed] [Google Scholar]
- 93. Qu Y, Dou P, Hu M, Xu J, Xia W, Sun H. circRNA−CER Mediates Malignant Progression of Breast Cancer Through Targeting the Mir−136/MMP13 Axis. Mol Med Rep (2019) 19:3314–20. 10.3892/mmr.2019.9965 [DOI] [PubMed] [Google Scholar]
- 94. Zang H, Li Y, Zhang X, Huang G. Circ-RNF111 Contributes to Paclitaxel Resistance in Breast Cancer by Elevating E2F3 Expression via miR-140-5p. Thorac Cancer (2020) 11:1891–903. 10.1111/1759-7714.13475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhao B, Song X, Guan H. CircACAP2 Promotes Breast Cancer Proliferation and Metastasis by Targeting miR-29a/B-3p-COL5A1 Axis. Life Sci (2020) 244:117179. 10.1016/j.lfs.2019.117179 [DOI] [PubMed] [Google Scholar]
- 96. Zhou S, Chen W, Yang S, Li J, Zhang J, Zhang H, et al. Circular RNA circVAPA Regulates Breast Cancer Cell Migration and Invasion via Sponging miR-130a-5p. Epigenomics-UK (2020) 12:303–17. 10.2217/epi-2019-0124 [DOI] [PubMed] [Google Scholar]
- 97. Li W, Yang X, Shi C, Zhou Z. Hsa_circ_002178 Promotes the Growth and Migration of Breast Cancer Cells and Maintains Cancer Stem-Like Cell Properties Through Regulating miR-1258/KDM7A Axis. Cell Transplant (2020) 29:963689720960174. 10.1177/0963689720960174 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98. Ma X, Liu C, Gao C, Li J, Zhuang J, Liu L, et al. circRNA-Associated ceRNA Network Construction Reveals the circRNAs Involved in the Progression and Prognosis of Breast Cancer. J Cell Physiol (2020) 235:3973–83. 10.1002/jcp.29291 [DOI] [PubMed] [Google Scholar]
- 99. Yang C, Zhang F, He J, Wang S. CircRNA_100876 Promote Proliferation and Metastasis of Breast Cancer Cells Through Adsorbing microRNA-361-3p in a Sponge Form. Eur Rev Med Pharmacol Sci (2019) 23:6962–70. 10.26355/eurrev_201908_18736 [DOI] [PubMed] [Google Scholar]
- 100. Yang L, Song C, Chen Y, Jing G, Sun J. Circular RNA Circ_0103552 Forecasts Dismal Prognosis and Promotes Breast Cancer Cell Proliferation and Invasion by Sponging miR-1236. J Cell Biochem (2019) 120:15553–60. 10.1002/jcb.28822 [DOI] [PubMed] [Google Scholar]
- 101. Li Z, Chen Z, Feng Y, Hu G, Jiang Y. CircMMP11 Acts as a ce-circRNA in Breast Cancer Progression by Regulating miR-1204. Am J Transl Res (2020) 12:2585–99. [PMC free article] [PubMed] [Google Scholar]
- 102. Wang Y, Li J, Du C, Zhang L, Zhang Y, Zhang J, et al. Upregulated Circular RNA Circ-UBE2D2 Predicts Poor Prognosis and Promotes Breast Cancer Progression by Sponging miR-1236 and miR-1287. Transl Oncol (2019) 12:1305–13. 10.1016/j.tranon.2019.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hu K, Liu X, Li Y, Li Q, Xu Y, Zeng W, et al. Exosomes Mediated Transfer of Circ_UBE2D2 Enhances the Resistance of Breast Cancer to Tamoxifen by Binding to MiR-200a-3p. Med Sci Monit (2020) 26:e922253. 10.12659/MSM.922253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. ou D, Ren X, Han M, Xu X, Ge X, Gu Y, et al. CircUBE2D2 (Hsa_Circ_0005728) Promotes Cell Proliferation, Metastasis and Chemoresistance in Triple-Negative Breast Cancer by Regulating miR-512-3p/CDCA3 Axis. Cancer Cell Int (2020) 20:454. 10.1186/s12935-020-01547-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hu J, Ji C, Hua K, Wang X, Deng X, Li J, et al. Hsa_circ_0091074 Regulates TAZ Expression via microRNA-1297 in Triple Negative Breast Cancer Cells. Int J Oncol (2020) 56:1314–26. 10.3892/ijo.2020.5000 [DOI] [PubMed] [Google Scholar]
- 106. Leng X, Huang G, Ding J, Ma F. Circ_0000043 Promotes Breast Cancer Cell Proliferation, Migration, Invasion and Epithelial-Mesenchymal Transition via the miR-136/Smad3 Axis. Biochem Cell Biol (2021) 99:277–85. 10.1139/bcb-2020-0219 [DOI] [PubMed] [Google Scholar]
- 107. Zhang J, Ke S, Zheng W, Zhu Z, Wu Y. Hsa_circ_0003645 Promotes Breast Cancer Progression by Regulating miR-139-3p/HMGB1 Axis. Oncotargets Ther (2020) 13:10361–72. 10.2147/OTT.S265796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, et al. Autophagy-Associated circRNA circCDYL Augments Autophagy and Promotes Breast Cancer Progression. Mol Cancer (2020) 19:65. 10.1186/s12943-020-01152-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zang H, Li Y, Zhang X, Huang G. Blocking Circ_0000520 Suppressed Breast Cancer Cell Growth, Migration and Invasion Partially via miR-1296/SP1 Axis Both In Vitro and In Vivo . Cancer Manag Res (2020) 12:7783–95. 10.2147/CMAR.S251666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jin Y, Yang L, Li X, Liu F. Circular RNA KIF4A Promotes Cell Migration, Invasion and Inhibits Apoptosis Through miR-152/ZEB1 Axis in Breast Cancer. Diagn Pathol (2020) 15:55. 10.1186/s13000-020-00963-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Jiang J, Cheng X. Circular RNA Circabcc4 Acts as a ceRNA of miR-154-5p to Improve Cell Viability, Migration and Invasion of Breast Cancer Cells In Vitro . Cell Cycle (Georgetown Tex) (2020) 19:2653–61. 10.1080/15384101.2020.1815147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ren S, Liu J, Feng Y, Li Z, He L, Li L, et al. Knockdown of Circdennd4c Inhibits Glycolysis, Migration and Invasion by Up-Regulating miR-200b/C in Breast Cancer Under Hypoxia. J Exp Clin Canc Res (2019) 38:388. 10.1186/s13046-019-1398-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Liang G, Liu Z, Tan L, Su AN, Jiang WG, Gong C. Hif1α-Associated Circdennd4c Promotes Proliferation of Breast Cancer Cells in Hypoxic Environment. Anticancer Res (2017) 37:4337–43. 10.21873/anticanres.11827 [DOI] [PubMed] [Google Scholar]
- 114. Chen ZG, Zhao HJ, Lin L, Liu JB, Bai JZ, Wang GS. Circular RNA CirCHIPK3 Promotes Cell Proliferation and Invasion of Breast Cancer by Sponging miR-193a/HMGB1/PI3K/AKT Axis. Thorac Cancer (2020) 11:2660–71. 10.1111/1759-7714.13603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, et al. Circular RNA Hsa_Circ_001783 Regulates Breast Cancer Progression via Sponging miR-200c-3p. Cell Death Dis (2019) 10:55. 10.1038/s41419-018-1287-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhao Y, Zhong R, Deng C, Zhou Z. Circle RNA Circabcb10 Modulates PFN2 to Promote Breast Cancer Progression, as Well as Aggravate Radioresistance Through Facilitating Glycolytic Metabolism Via miR-223-3p. Cancer Biother Radio (2020). Online ahead of print. 10.1089/cbr.2019.3389 [DOI] [PubMed] [Google Scholar]
- 117. Bian Q. Circular RNA PVT1 Promotes the Invasion and Epithelial-Mesenchymal Transition of Breast Cancer Cells Through Serving as a Competing Endogenous RNA for miR-204-5p. Oncotargets Ther (2020) 12:11817–26. 10.2147/OTT.S180850 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 118. Zou Y, Zheng S, Xiao W, Xie X, Yang A, Gao G, et al. Circrad18 Sponges miR-208a/3164 to Promote Triple-Negative Breast Cancer Progression Through Regulating IGF1 and FGF2 Expression. Cacinogene (New York) (2019) 40:1469–79. 10.1093/carcin/bgz071 [DOI] [PubMed] [Google Scholar]
- 119. Zang H, Li Y, Zhang X, Huang G. Knockdown of Circrad18 Mitigates Breast Cancer Progression Through the Regulation of miR-613/HK2 Axis. Cancer Manag Res (2020) 12:3661–72. 10.2147/CMAR.S243300 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120. Li Y, Shi P, Zheng T, Ying Z, Jiang D. Circular RNA Hsa_Circ_0131242 Promotes Triple-Negative Breast Cancer Progression by Sponging hsa-miR-2682. Oncotargets Ther (2020) 13:4791–8. 10.2147/OTT.S246957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhang HD, Jiang LH, Hou JC, Zhou SY, Zhong SL, Zhu LP, et al. Circular RNA Hsa-Circ-0072995 Promotes Breast Cancer Cell Migration and Invasion Through Sponge for miR-30c-2-3p. Epigenomics-UK (2018) 10:1229–42. 10.2217/epi-2018-0002 [DOI] [PubMed] [Google Scholar]
- 122. Pan G, Mao A, Liu J, Lu J, Ding J, Liu W. Circular RNA Hsa_Circ_0061825 (Circ-TFF1) Contributes to Breast Cancer Progression Through Targeting miR-326/TFF1 Signalling. Cell Proliferat (2020) 53:e12720. 10.1111/cpr.12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Min J, Pan X, Lv G. The circRNA Circ_0000291 Acts as a Sponge of microRNA 326 to Regulate E26 Transformation-Specific Sequence-1 Expression and Promote Breast Cancer Progression. Pathol Int (2020) 70:953–64. 10.1111/pin.13011 [DOI] [PubMed] [Google Scholar]
- 124. Jia Q, Ye L, Xu S, Xiao H, Xu S, Shi Z, et al. Circular RNA 0007255 Regulates the Progression of Breast Cancer Through miR-335-5p/SIX2 Axis. Thorac Cancer (2020) 11:619–30. 10.1111/1759-7714.13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Li X, Ren Z, Yao Y, Bao J, Yu Q. The Circular RNA Circeif3m Promotes Breast Cancer Progression by Promoting Cyclin D1 Expression. Aging (Albany NY.) (2020) 12:14775–90. 10.18632/aging.103539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Liu T, Ye P, Ye Y, Lu S, Han B. Circular RNA Hsa_circRNA_002178 Silencing Retards Breast Cancer Progression via microRNA-328-3p-Mediated Inhibition of COL1A1. J Cell Mol Med (2020) 24:2189–201. 10.1111/jcmm.14875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. He D, Yang X, Kuang W, Huang G, Liu X, Zhang Y. The Novel Circular RNA Circ-PGAP3 Promotes the Proliferation and Invasion of Triple Negative Breast Cancer by Regulating the miR-330-3p/Myc Axis. Oncotargets Ther (2020) 13:10149–59. 10.2147/OTT.S274574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J, et al. CircIRAK3 Sponges miR-3607 to Facilitate Breast Cancer Metastasis. Cancer Lett (2018) 430:179–92. 10.1016/j.canlet.2018.05.033 [DOI] [PubMed] [Google Scholar]
- 129. Liu Y, Lu C, Zhou Y, Zhang Z, Sun L. Circular RNA Hsa_Circ_0008039 Promotes Breast Cancer Cell Proliferation and Migration by Regulating miR-432-5p/E2F3 Axis. Biochem Bioph Res Co (2018) 502:358–63. 10.1016/j.bbrc.2018.05.166 [DOI] [PubMed] [Google Scholar]
- 130. Huang F, Dang J, Zhang S, Cheng Z. Circular RNA Hsa_Circ_0008039 Promotes Proliferation, Migration and Invasion of Breast Cancer Cells Through Upregulating CBX4 via Sponging miR-515-5p. Eur Rev Med Pharmacol Sci (2020) 24:1887–98. 10.26355/eurrev_202002_20367 [DOI] [PubMed] [Google Scholar]
- 131. Pei X, Zhang Y, Wang X, Xue B, Sun M, Li H. Circular RNA Circ-ZEB1 Acts as an Oncogene in Triple Negative Breast Cancer via Sponging miR-448. Int J Biochem Cell B (2020) 126:105798. 10.1016/j.biocel.2020.105798 [DOI] [PubMed] [Google Scholar]
- 132. Yao Y, Zhao Z, Han X, Liu S, Nie C, Fang H. Circular RNA-100219 Promotes Breast Cancer Progression by Binding to microRNA-485-3p. J BUON (2019) 24:501. [PubMed] [Google Scholar]
- 133. Wang N, Gu Y, Li L, Wang F, Lv P, Xiong Y, et al. Circular RNA Circmyo9b Facilitates Breast Cancer Cell Proliferation and Invasiveness via Upregulating FOXP4 Expression by Sponging miR-4316. Arch Biochem Biophys (2018) 653:63–70. 10.1016/j.abb.2018.04.017 [DOI] [PubMed] [Google Scholar]
- 134. Cao L, Wang M, Dong Y, Xu B, Chen J, Ding Y, et al. Circular RNA Circrnf20 Promotes Breast Cancer Tumorigenesis and Warburg Effect Through miR-487a/HIF-1α/Hk2. Cell Death Dis (2020) 11:145. 10.1038/s41419-020-2336-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Song L, Xiao Y. Downregulation of Hsa_Circ_0007534 Suppresses Breast Cancer Cell Proliferation and Invasion by Targeting miR-593/MUC19 Signal Pathway. Biochem Bioph Res Co (2018) 503:2603–10. 10.1016/j.bbrc.2018.08.007 [DOI] [PubMed] [Google Scholar]
- 136. Ye G, Pan R, Zhu L, Zhou D. Circ_DCAF6 Potentiates Cell Stemness and Growth in Breast Cancer Through GLI1-Hedgehog Pathway. Exp Mol Pathol (2020) 116:104492. 10.1016/j.yexmp.2020.104492 [DOI] [PubMed] [Google Scholar]
- 137. Xu Y, Yao Y, Leng K, Ji D, Qu L, Liu Y, et al. Increased Expression of Circular RNA Circ_0005230 Indicates Dismal Prognosis in Breast Cancer and Regulates Cell Proliferation and Invasion via miR-618/CBX8 Signal Pathway. Cell Physiol Biochem (2018) 51:1710–22. 10.1159/000495675 [DOI] [PubMed] [Google Scholar]
- 138. Chen Z, Wang F, Xiong Y, Wang N, Gu Y, Qiu X. CircZFR Functions as a Sponge of miR-578 to Promote Breast Cancer Progression by Regulating HIF1A Expression. Cancer Cell Int (2020) 20:1–400. 10.1186/s12935-020-01492-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wang S, Li Q, Wang Y, Li X, Wang R, Kang Y, et al. Upregulation of Circ-UBAP2 Predicts Poor Prognosis and Promotes Triple-Negative Breast Cancer Progression Through the miR-661/MTA1 Pathway. Biochem Bioph Res Co (2018) 505:996–1002. 10.1016/j.bbrc.2018.10.026 [DOI] [PubMed] [Google Scholar]
- 140. Zhou H, Tang G, Zhao M, Xie L, Xie Y, Zhang Z, et al. Circfbxl5 Promotes Breast Cancer Progression by Sponging miR-660. J Cell Mol Med (2020) 24:356–61. 10.1111/jcmm.14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Deng Y, Xia J, Xu Y. Circular RNA Circtp63 Enhances Estrogen Receptor-Positive Breast Cancer Progression and Malignant Behaviors Through the miR-873-3p/FOXM1 Axis. Anti-Cancer Drug (2020) 32:44–52. 10.1097/CAD.0000000000001010 [DOI] [PubMed] [Google Scholar]
- 142. Zhao C, Li L, Li Z, Xu J, Yang Q, Shi P, et al. A Novel Circular RNA Hsa_Circrpph1_015 Exerts an Oncogenic Role in Breast Cancer by Impairing miRNA-326-Mediated ELK1 Inhibition. Front Oncol (2020) 10:906. 10.3389/fonc.2020.00906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Karedath T, Ahmed I, Al Ameri W, Al-Dasim FM, Andrews SS, Samuel S, et al. Silencing of ANKRD12 circRNA Induces Molecular and Functional Changes Associated With Invasive Phenotypes. BMC Cancer (2019) 19:565. 10.1186/s12885-019-5723-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Smid M, Wilting SM, Uhr K, Aacute FG, Iacute NR, Aacute G, et al. The Circular RNome of Primary Breast Cancer. Genome Res (2019) 29:356–66. 10.1101/gr.238121.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Ma J, Fang L, Yang Q, Hibberd S, Du WW, Wu N, et al. Posttranscriptional Regulation of AKT by Circular RNA Angiomotin- Like 1 Mediates Chemoresistance Against Paclitaxel in Breast Cancer Cells. Aging (Albany NY.) (2019) 11:11369–81. 10.18632/aging.102535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Qiu X, Wang Q, Song H, Shao D, Xue J. Circ_103809 Promotes Breast Cancer Progression by Regulating the PI3K/AKT Signaling Pathway. Oncol Lett (2020) 19:3725–30. 10.3892/ol.2020.11507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Hu Y, Song Q, Zhao J, Ruan J, He F, Yang X, et al. Identification of Plasma Hsa_Circ_0008673 Expression as a Potential Biomarker and Tumor Regulator of Breast Cancer. J Clin Lab Anal (2020) 34:e23393. 10.1002/jcla.23393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Xu JH, Wang Y, Xu D. Hsa-Circ-001569 is an Unfavorable Prognostic Factor and Promotes Cell Proliferation and Metastasis by Modulating PI3K-AKT Pathway in Breast Cancer. Cancer Biomark (2019) 25:193–201. 10.3233/CBM-182293 [DOI] [PubMed] [Google Scholar]
- 149. Wang X, Xue B, Zhang Y, Guo G, Duan X, Dou D. Up-Regulated Circbach2 Contributes to Cell Proliferation, Invasion, and Migration of Triple-Negative Breast Cancer. Cell Death Dis (2021) 12:412. 10.1038/s41419-021-03684-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Wang L, Yi J, Lu L, Zhang Y, Wang L, Hu G, et al. Estrogen-Induced circRNA, circPGR, Functions as a ceRNA to Promote Estrogen Receptor-Positive Breast Cancer Cell Growth by Regulating Cell Cycle-Related Genes. Theranostics (2021) 11:1732–52. 10.7150/thno.45302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Alink A, Charest I. Clinically Relevant Autistic Traits Predict Greater Reliance on Detail for Image Recognition. Sci Rep-UK (2020) 10:14239. 10.1038/s41598-020-70953-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lin X, Chen W, Wei F, Xie X. TV-Circrgpd6 Nanoparticle Suppresses Breast Cancer Stem Cell-Mediated Metastasis via the miR-26b/YAF2 Axis. Mol Ther (2021) 29:244–62. 10.1016/j.ymthe.2020.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Fang L, Du WW, Lyu J, Dong J, Zhang C, Yang W, et al. Enhanced Breast Cancer Progression by Mutant P53 Is Inhibited by the Circular RNA Circ-Ccnb1. Cell Death Differ (2018) 25:2195–208. 10.1038/s41418-018-0115-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. ?A3B2 twb 0.24w?>Du WW, Yang W, Li X, Fang L, Wu N, Li F, et al. The Circular RNA Circska3 Binds Integrin β1 to Induce Invadopodium Formation Enhancing Breast Cancer Invasion. Mol Ther (2020) 28:1287–98. 10.1016/j.ymthe.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Mao Y, Lv M, Cao W, Liu X, Cui J, Wang Y, et al. Circular RNA 000554 Represses Epithelial-Mesenchymal Transition in Breast Cancer by Regulating microRNA-182/ZFP36 Axis. FASEB J (2020) 34:11405–20. 10.1096/fj.201903047R [DOI] [PubMed] [Google Scholar]
- 156. Sang Y, Chen B, Song X, Li Y, Liang Y, Han D, et al. circRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the miR-182-5p/FOXO3a Axis in Breast Cancer. Mol Ther (2019) 27:1638–52. 10.1016/j.ymthe.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Hu Y, Guo F, Zhu H, Tan X, Zhu X, Liu X, et al. Circular RNA-0001283 Suppresses Breast Cancer Proliferation and Invasion via MiR-187/HIPK3 Axis. Med Sci Monit (2020) 26:e921502. 10.12659/MSM.921502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Hou J, Xu Z, Zhong S, Zhang H, Jiang L, Chen X, et al. Circular RNA Circass1 is Downregulated in Breast Cancer Cells MDA-MB-231 and Suppressed Invasion and Migration. Epigenomics-UK (2019) 11:199–213. 10.2217/epi-2017-0167 [DOI] [PubMed] [Google Scholar]
- 159. Liang Y, Song X, Li Y, Ma T, Su P, Guo R, et al. Targeting the Circbmpr2/miR-553/USP4 Axis as a Potent Therapeutic Approach for Breast Cancer. Mol Ther-Nucl Acids (2019) 17:347–61. 10.1016/j.omtn.2019.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Yi Z, Li Y, Wu Y, Zeng B, Li H, Ren G, et al. Circular RNA 0001073 Attenuates Malignant Biological Behaviours in Breast Cancer Cell and Is Delivered by Nanoparticles to Inhibit Mice Tumour Growth. Oncotargets Ther (2020) 13:6157–69. 10.2147/OTT.S248822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of Tumor Apoptosis Through a Circular RNA Enhancing Foxo3 Activity. Cell Death Differ (2017) 24:357–70. 10.1038/cdd.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Liu Y, Dong Y, Zhao L, Su L, Luo J. Circular RNA−MTO1 Suppresses Breast Cancer Cell Viability and Reverses Monastrol Resistance Through Regulating the TRAF4/Eg5 Axis. Int J Oncol (2018) 53:1752–62. 10.3892/ijo.2018.4485 [DOI] [PubMed] [Google Scholar]
- 163. Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou X, et al. Circfbxw7 Inhibits Malignant Progression by Sponging miR-197-3p and Encoding a 185-Aa Protein in Triple-Negative Breast Cancer. Mol Ther-Nucl Acids (2019) 18:88–98. 10.1016/j.omtn.2019.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Lu M, Wu Y, Zeng B, Sun J, Li Y, Luo J, et al. CircEHMT1 Inhibits Metastatic Potential of Breast Cancer Cells by Modulating miR-1233-3p/KLF4/MMP2 Axis. Biochem Bioph Res Co (2020) 526:306–13. 10.1016/j.bbrc.2020.03.084 [DOI] [PubMed] [Google Scholar]
- 165. Xu G, Ye D, Zhao Q, He R, Ma W, Li Y, et al. circNFIC Suppresses Breast Cancer Progression by Sponging miR-658. J Cancer (2020) 11:4222–9. 10.7150/jca.38830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Zhang X, Mao L. Circular RNA Circ_0000442 Acts as a Sponge of MiR-148b-3p to Suppress Breast Cancer via PTEN/PI3K/Akt Signaling Pathway. Gene (2021) 766:145113. 10.1016/j.gene.2020.145113 [DOI] [PubMed] [Google Scholar]
- 167. Wang S, Liu F, Ma H, Cui X, Yang S, Qin R. circCDYL Acts as a Tumor Suppressor in Triple Negative Breast Cancer by Sponging miR-190a-3p and Upregulating Tp53inp1. Clin Breast Cancer (2020) 20:422–30. 10.1016/j.clbc.2020.04.006 [DOI] [PubMed] [Google Scholar]
- 168. Wang ST, Liu LB, Li XM, Wang YF, Xie PJ, Li Q, et al. Circ-ITCH Regulates Triple-Negative Breast Cancer Progression Through the Wnt/β-Catenin Pathway. Neoplasma (2019) 66:232–9. 10.4149/neo_2018_180710N460 [DOI] [PubMed] [Google Scholar]
- 169. Zhao J, Zou H, Han C, Ma J, Zhao J, Tang J. Circlular RNA BARD1 (Hsa_circ_0001098) Overexpression in Breast Cancer Cells With TCDD Treatment Could Promote Cell Apoptosis via miR-3942/BARD1 Axis. Cell Cycle (2018) 17:2731–44. 10.1080/15384101.2018.1556058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Xiao W, Zheng S, Zou Y, Yang A, Xie X, Tang H, et al. CircAHNAK1 Inhibits Proliferation and Metastasis of Triple-Negative Breast Cancer by Modulating miR-421 and RASA1. Aging (Albany NY.) (2019) 11:12043–56. 10.18632/aging.102539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Yan L, Zheng M, Wang H. Circular RNA Hsa_Circ_0072309 Inhibits Proliferation and Invasion of Breast Cancer Cells via Targeting miR-492. Cancer Manag Res (2019) 11:1033–41. 10.2147/CMAR.S186857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Liu M, Luo C, Dong J, Quo J, Luo Q, Ye C, et al. CircRNA_103809 Suppresses the Proliferation and Metastasis of Breast Cancer Cells by Sponging MicroRNA-532-3p (miR-532-3p). Front Genet (2020) 11:485. 10.3389/fgene.2020.00485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Peng HH, Wen YG. CircDDX17 Acts as a Competing Endogenous RNA for miR-605 in Breast Cancer Progression. Eur Rev Med Pharmaco (2020) 24:6794–801. 10.26355/eurrev_202006_21668 [DOI] [PubMed] [Google Scholar]
- 174. Yuan P, Lei L, Dong S, Liu D. Circular RNA HsaCirc_0068033 Acts as a Diagnostic Biomarker and Suppresses the Progression of Breast Cancer Through Sponging miR-659. Oncotargets Ther (2020) 13:1921–9. 10.2147/OTT.S223542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Li Z, Zheng J, Lin W, Weng J, Hong W, Zou J, et al. Circular RNA Hsa_Circ_0001785 Inhibits the Proliferation, Migration and Invasion of Breast Cancer Cells In Vitro and In Vivo by Sponging miR-942 to Upregulate SOCS3. Cell Cycle (2020) 19:2811–25. 10.1080/15384101.2020.1824717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen W, et al. CircRNA Inhibits DNA Damage Repair by Interacting With Host Gene. Mol Cancer (2020) 19:1–128. 10.1186/s12943-020-01246-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Yuan C, Luo X, Zhan X, Zeng H, Duan S. EMT Related Circular RNA Expression Profiles Identify Circscyl2 as a Novel Molecule in Breast Tumor Metastasis. Int J Mol Med (2020) 45:1697–710. 10.3892/ijmm.2020.4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Zhang X, Su X, Guo Z, Jiang X, Li X. Circular RNA La-Related RNA-Binding Protein 4 Correlates With Reduced Tumor Stage, as Well as Better Prognosis, and Promotes Chemosensitivity to Doxorubicin in Breast Cancer. J Clin Lab Anal (2020) 34:e23272. 10.1002/jcla.23272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Li Y, Li H. Circular RNA VRK1 Correlates With Favourable Prognosis, Inhibits Cell Proliferation But Promotes Apoptosis in Breast Cancer. J Clin Lab Anal (2020) 34:e22980. 10.1002/jcla.22980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Yan N, Xu H, Zhang J, Xu L, Zhang Y, Zhang L, et al. Circular RNA Profile Indicates Circular RNA VRK1 is Negatively Related With Breast Cancer Stem Cells. Oncotarget (2017) 8:95704–18. 10.18632/oncotarget.21183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Yu J, Shen W, Xu J, Gong B, Gao B, Zhu J. Circusp42 is Downregulated in Triple-Negative Breast Cancer and Associated With Poor Prognosis. Technol Cancer Res T (2020) 19:1079218475. 10.1177/1533033820950827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Yin W, Yan M, Fang X, Guo J, Xiong W, Zhang R. Circulating Circular RNA Hsa_Circ_0001785 Acts as a Diagnostic Biomarker for Breast Cancer Detection. Clin Chim Acta (2018) 487:363–8. 10.1016/j.cca.2017.10.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.