Abstract
Background
Testing a hypothesis for ‘factors-outcome effect’ is a common quest, but standard statistical regression analysis tools are rendered ineffective by data contaminated with too many noisy variables. Expert Systems (ES) can provide an alternative methodology in analysing data to identify variables with the highest correlation to the outcome. By applying their effective machine learning (ML) abilities, significant research time and costs can be saved. The study aims to systematically review the applications of ES in urological research and their methodological models for effective multi-variate analysis. Their domains, development and validity will be identified.
Methods
The PRISMA methodology was applied to formulate an effective method for data gathering and analysis. This study search included seven most relevant information sources: WEB OF SCIENCE, EMBASE, BIOSIS CITATION INDEX, SCOPUS, PUBMED, Google Scholar and MEDLINE. Eligible articles were included if they applied one of the known ML models for a clear urological research question involving multivariate analysis. Only articles with pertinent research methods in ES models were included. The analysed data included the system model, applications, input/output variables, target user, validation, and outcomes. Both ML models and the variable analysis were comparatively reported for each system.
Results
The search identified n = 1087 articles from all databases and n = 712 were eligible for examination against inclusion criteria. A total of 168 systems were finally included and systematically analysed demonstrating a recent increase in uptake of ES in academic urology in particular artificial neural networks with 31 systems. Most of the systems were applied in urological oncology (prostate cancer = 15, bladder cancer = 13) where diagnostic, prognostic and survival predictor markers were investigated. Due to the heterogeneity of models and their statistical tests, a meta-analysis was not feasible.
Conclusion
ES utility offers an effective ML potential and their applications in research have demonstrated a valid model for multi-variate analysis. The complexity of their development can challenge their uptake in urological clinics whilst the limitation of the statistical tools in this domain has created a gap for further research studies. Integration of computer scientists in academic units has promoted the use of ES in clinical urological research.
Introduction
In the 1950’s J McCarthy in Stanford University and A Turing in Cambridge University proposed the concept of machine simulation of human learning and intelligence [1, 2]. Being keen mathematicians, they advanced the basic mathematical logic into programming languages enabling machines to perform more complex functions. E Shortliffe advanced those systems to develop MYCIN, which successfully simulated the reasoning of a human microbiologist in diagnosing and treating patients with microbial infection [3]. Their model introduced Expert Systems (ES) to the scientific literature and a ten year review by Liao et al. demonstrated their wide prevalence in the industrial fields with immense applications including health care [4]. In contrast to Liao’s review, other studies questioned their real time implementation in health care and suggested a lack of their uptake and integration in the health care systems [5]. This is despite evidence from systematic reviews demonstrating the positive impact of computer aid systems on patients’ outcome and health care [6, 7].
This study aimed to systematically review published ES in urological health care with a primary aim to demonstrate their availability, progression, testing and applications. The secondary aim was to evaluate their development life cycle against standards suggested by O’Keefe and Benbasat in their review articles on ES development [8, 9]. The later would evaluate the gap between their development and implementation in health care.
Methods
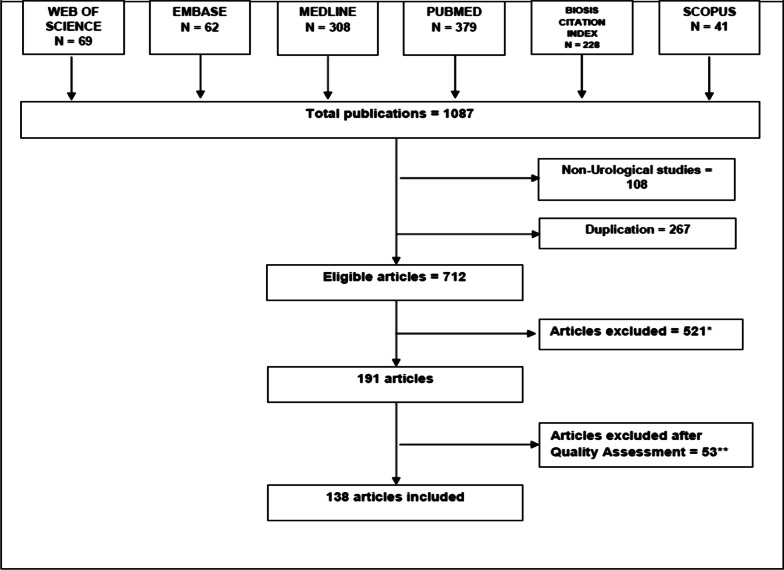
The study methodology followed the recommendations outlined in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (Fig. 1). No ethical approval was required because the type of the study waives this requirement.
Fig. 1.
PRISMA flow chart for the systematic review of articles included in the review of expert systems in urology
Search
Information sources including WEB OF SCIENCE, EMBASE, BIOSIS CITATION INDEX, SCOPUS, PUBMED, Google Scholar and MEDLINE were searched using key words in (Table 1). Articles published between 1960 and 2016 were considered and examined against the inclusion criteria. While tailoring the conducted search for each literature database, the key words were combined by “OR” in each domain, then domains were combined by “AND”.
Table 1.
Keywords used for literature search
| #1 |
TOPIC: ("expert system*") OR TOPIC: ("decision support") OR TOPIC: ("artificial intelligence") OR TOPIC: ("rule based") OR TOPIC: ("knowledge base* system*") OR TOPIC: ("neural network") OR TOPIC: ("fuzzy") DocType = All document types; Language = All languages; |
|---|---|
| #2 |
TOPIC: (urology) DocType = All document types; Language = All languages; |
| #3 |
#1 AND #2 DocType = All document types; Language = All languages; |
Eligibility criteria
For the primary aim, data search was conducted to yield the collected results then analyse them according to pre-planned eligibility criteria based on the system model, year of production, type and outcome of its validation, functional domain application, variables for input and output, target user and domain. This selection criteria were designed with an objective to identify expert system studies and demonstrate their prevalence, testing, and applications in clinical urology. Only articles and studies written in English were included.
Further qualitative analysis was required to meet the study secondary aim. For this, further data was gathered on credibility (user perception on the system), evaluation (system usability), validation (building the right system) and verification (building the system right) then compare against the standards reported in [8, 9].
Data filtering
The resultant reference list of each included article was checked to identify a potentially eligible item that had not been retrieved by the initial search. All retrieved articles were collated in a final reference list on a management software (Endnote, X8), then duplicate studies were removed from the list.
Upon including more than one hundred articles, the rest of the eligible articles were meticulously compared to the ones included, then excluded based on demonstrating clear similarity. This was applied to avoid expanding the size of the data without adding to the study analysis.
Results
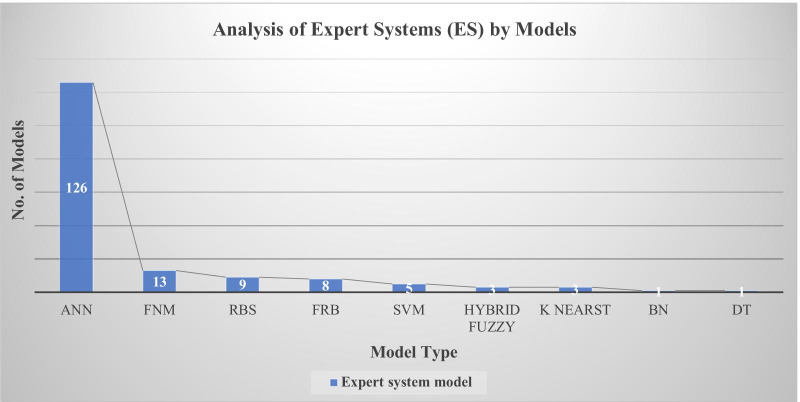
ANN was the commonest model to be applied in Urological ES (Fig. 2). The rest of the models demonstrated diversity which is consistent with other published industrial systems [4].
Fig. 2.
Analysis of Expert Systems (ES) by models (n = 169). ANN was the most common but other systems were applied on different domain as fuzzy neural model (FNM), rule-based system (RBS), fuzzy rule based (FRB), support vector machine (SVT), Bayesian network (BN) and decision trees (DT)
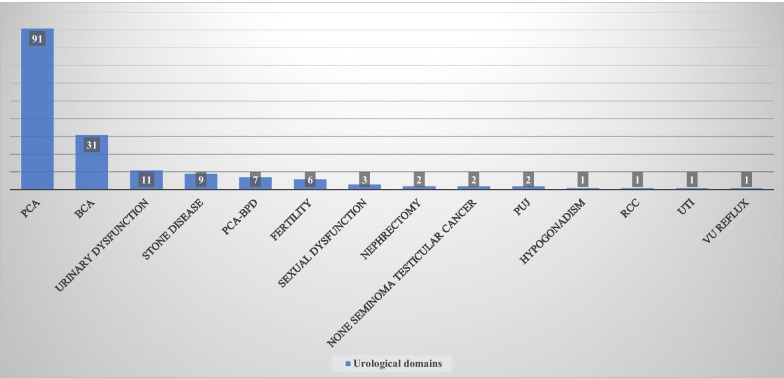
Prostate cancer was the commonest domain for urological ES with most of the system focusing on cancer diagnosis. These systems were applied to various domains (Fig. 3), and they were further stratified and analysed according to their core functional application as outlines in the methodology.
Fig. 3.
Urological domains (n = 168) applied by Expert Systems (ES). Prostate cancer (CaP) was the commonest domain followed by bladder cancer (Bca) then other diseases as benign prostatic disease (BPD), pelvi ureteric junction obstruction (PUJ), urinary tract infection (UTI), renal cell cancer (RCC), vesico ureteric reflux (VU reflux)
Quantitative analysis
Decision support systems
The main objective of ES in this domain was to facilitate the clinical decision making by identifying key elements from patients clinical and laboratory examinations then refine a theoretical diagnostic or treatment strategy [10]. They can guide the expert to find the right answer [11] or take over the decision making to support the none expert as [12] or even replace both to interact with the patient directly [13].
They have supported various aspects of urological decision making such as diagnosis, investigations analysis, radiotherapy dose calculation, the delivery of behavioural treatment and therapeutic dialogues.
Domains
Urinary dysfunction (U Dys) was the commonest domain to be covered in the decision support system application (n = 9), which could be further categorised into U Dys diagnostic, investigation analysis and therapeutic systems. They have demonstrated a range of methodologies, validation, and target users (Table 2) applicable to Decision support systems in Urological domain. For instance, Keles et al. [14] designed an ES to support junior nurses in diagnosing urinary elimination dysfunction in a selected group of patients while [15, 16] systems were able to support any medical user to diagnose urinary incontinence with an accuracy reaching higher than 90%. The target user of most of these systems were predominantly medical health care workers including both experts and none experts, with exception of [13, 17] which can be directly used by patients to receive an assessment of their urinary elimination dysfunction followed by a tailored treatment plan.
Table 2.
Decision support systems in urological domain
| Article | Mdl | Dom | Subdomain | Variables | Output | Knowledge acquisition | Validation method | Target user |
|---|---|---|---|---|---|---|---|---|
| [18] | RBR | U Dys | Incontinence in long-term care facilities | Disease related questions | Recommendations | Experts | Comparison to blinded experts and pilot RCT | Non-expert nurses |
| [15] | RBR | U Dys | U incont treatment | Incontinence symptoms | Behavioural treatment | Agency guidelines | RCT (60) reliability and validity by experts | Patients |
| [19] | RBR | U Dys | U incont treatment | 19 evaluation questionnaires | Individualised health information | An expert and patients’ feedback | No validation | Patients |
| [20] | RBR | U Dys | U incont | MH, incontinence symptoms, previous incidents and medication history | U incont treatment | Multiple experts, patients record and literature | Evaluation by experts, 95 retrospective data | Non-experts |
| [16] | RBR | U Dys | Ward management of micturition | LUTS, Urinary tract infection Anatomical obstruction, Multiple causality and sensory impairment | Diagnosis and risk of fall | Multiple experts | Se 0.95, Sp 0.72, Likert scale Cronbach α 0.9 | Urology ward nurses |
| [21] | FRB | U Dys | U dyn interpretation | U dyn variables | Detrusor and sphincter dysfunction | Not mentioned | Improve User Ac by 10% | Experts |
| [22] | ANN | U Dys | Uroflow interpretation | Value of slopes, frequency and value of maximums, ration of amplitude and total voiding time | Healthy or pathologic Uroflow | Patients data from U dyn | 78 test cases ROC 0.7 Ac 79% | Experts |
| [23] | SVM | U Dys | Diagnosis | Age, examination, Uroflow, U dyn | Healthy or pathologic Uroflow | Patients data | Ac 84%, Se 93%, Sp 33% | Experts |
| [17] | FNM | U Dys | Diagnosis | 46 defining Characteristics from NANDA-I | Diagnosis of U Dys | Multiple experts weighted the variables and literature review | kappa vs experts (0.92–0.42), Se 0.95, Sp 0.92 | Experts and non-experts |
| [14] | FNM | CaP-BPD | Diagnosis of BPE and CaP | Clinical and pathological variables | CaP, BPE medical, BPE surgery | Patients data | 10 folds CV AUC 0.86, se 100%, sp 98% | Non-experts |
| [24] | FRB | CaP-BPD | AP CP CaP BPE | LUTS, quality of life, fever, haematuria, haemospermia, painful ejaculation, fever, perineal pain, bone pain, pyuria, age, DRE | Diagnosis and treatment of prostatic disease | Multiple experts interviews, patients records and literature | Ac 0.76, Se 0.79, Sp 0.75, retrospective data (n = 105) | Residents, patients, medical students |
| [12] | FRB | CaP-BPD | AP CP CaP BPE | LUTS, quality of life, fever, haematuria, haemospermia, painful ejaculation, fever, perineal pain, bone pain, pyuria, age, DRE | Diagnosis and treatment | WEKA* to extract rules then experts to modify | 200 test cases Ac 0.93, Se 0.97, Sp 0.99, | Residents, patients, medical students |
| [25] | RBR | CaP | Diagnosis before 1st biopsy | Age, race, FH, DRE, PSA, PSAD, PSAV, TRUS findings | Cancer and benign | Not mentioned |
25 test cases Se 100% Sp 33% PPV 62%, NPV 100% |
Experts |
| [13] | F-CBR | CaP | Radiotherapy dose for CaP | Gl, PSA, Distribution Volume Histogram | Radiotherapy dose | 72 patients’ cases | Comparison to experts, Ac 85% | Experts |
| [26] | F-ONT | BPD | Diagnosis and treatment of BPE | LUTS, DRE | Watchful waiting, medical, surgery | Multiple experts weighted the variables | 44 prospective cases, agreement kappa = 0.89 | Experts and non-experts |
| [27] | RBR | S Dys | Diagnosis and treatment | Set of descriptors | Therapeutic dialogue | Not mentioned | 10 Patients' evaluations | Couples |
| [28] | RBR | S Dys | Male S dys diagnosis | 22 parameters from history and examination | ED diagnosis | GA rule extraction from 30 cases |
Se (73–94%), Sp (78–96%) Ac (89%) vs Residents |
Un specified |
| [29] | FRB | S Dys | Male S dys diagnosis and treatment | MH, non-coital erection, diabetes mellitus, coronary artery, neuropathies, sexual history, psychosocial history, depression, smoking, alcohol, examination, hormonal evaluation, cholesterol | Diagnosis and treatment of ED | Multiple experts’ interviews, Pearson analysis on variables from patients' data and literature | 70 test cases vs experts and non-experts (Ac79%) | Non-experts |
| [30] | FNM | UTI | UTI treatment | Clinical data on UTI | Antibiotics course | Patients data and guidelines | Ac 86.8%, 38 random cases | Experts and non-experts |
| [31] | ANN | VUR | Decision support for intervention | Age, gender, number of UTIs prior to VUR diagnosis, UTI, of complete ureteral duplication noted on Ultrasound, the presence of bowel or bladder dysfunction | UTI or not | 255 cases, 96 cases | AUC 0.76 | Experts |
| [32] | ANN | Nlt | ESWL dose calculation | Age, stone size, stone burden, number of sittings | Number and power of shock | 196 cases, 80 cases | coefficient of correlation 0.9 | Experts |
A total of 21 Expert Systems included supporting the decision making in Urological domains. Rule based reasoning was the most common model and urinary dysfunction was the commonest domain
Prostate diseases were represented in 6 systems while 3 of them modelled by [10, 12, 20] for diagnosing both benign and malignant prostatic disease, namely cancer prostate (CaP).
All systems in this domain were diagnosis support system with exception of [19] which also provided treatment for benign prostatic hyperplasia (BPH) and [11] calculated the required radiotherapy dose for treating CaP.
Sexual dysfunctions were modelled in 3 systems where [21] diagnosed male sexual dysfunction with an accuracy of 89%, while [22] added a therapeutic model for the same disease with an overall accuracy of 79%. Sexpert by [23] was the third system in this category developed in 1988 and in fact the oldest ES to be identified from our search in all urological domains. Interestingly this RB system was designed to interact directly with couples suffering from sexual dysfunction where the system responds to their query with a tailored therapeutic dialogue for treating their problem.
Urinary tract infection (UTI) was diagnosed and treated by one of the hybrid fuzzy systems FNM developed by [24] with an accuracy of 86.8%.
Diagnosis prediction
In this domain, ES quantifying the probability of a clinical diagnosis with a defined margin of error. They simulate a second expert opinion and it has been suggested that their use could eliminate unnecessary invasive investigation as the application of ANN by [26] could reduce up to 68% of repeated TRUS biopsies to diagnose CaP.
Domains
Prostate cancer was the main domain for this application with 19 systems out of 20. Most of them were designed to predict organ confinement before radical surgical excision of the prostate (Tables 3, 4). The target population were patients with clinically localised CaP and their accuracy reached high estimates as in [28], where the system was able to predict 98% of the low risk group for lymph node involvement using preoperative available date (PSA, clinical stage and Gleason score).
Table 3.
Diagnosis prediction application of Expert Systems (ES) in Urology
| Art | Mdl | Dom | Subdomain | Variables | Output | System training | Validation | Statistical outcome |
|---|---|---|---|---|---|---|---|---|
| [33] | ANN | CaP | Pre-biopsy diagnosis with TRUS variables | Age, PSA, number of biopsies, clinical diagnosis, PSAD, TRUS variables | Cancer or benign | N = 442 from single centre database |
ROC AUC NPV, PPV ½ CV |
NPV 97%, PPV 82% better than LR |
| [34] | ANN | CaP | Diagnosis PSA 2.5–4 | Age, tPSA, creatinine phospho kinase, prostatic acid phosphatase | Cancer or benign | Multicentre data 522 (PSA 2.5–4) |
ROC AUC CV 152 cases |
AUC 0.74 |
| [35] | ANN | CaP | Diagnosis PSA 4–10 | Age, tPSA, %fPSA, TPV, DRE | Risk of cancer | 656 data from Finnish trial | ROC, Sp, Se LOO | Se 79%, Sp 57%, Ac 62%, PPV 35, NPV 90 |
| [36] | ANN | CaP | Diagnosis PSA 2–20 | Age, tPSA, %fPSA, TPV, DRE | Risk of Cancer | 1188 multi centre | ROC, Sp, Se, 1/10 CV | Sp 90%, Se 64% |
| [36] | ANN | CaP | Diagnosis in trial patients with PSA 4–10 | Age, tPSA, %fPSA, TPV, DRE | Risk of Cancer | 1188 multi centre | ROC, Sp, Se, 204 trial data PSA 4–10 | Se 95%, Sp 23.3%, CI 17.4%–30.2%, P < 0.0002 |
| [37] | ANN | CaP | Diagnosis | fPSA, TZD, PSAV, %f PSA, TZV, t PSA, and PSAD | Cancer or benign | PSA 2.5–4, 272 patients, multicentre data |
ROC, AUC ¼ CV |
AUC 0.88 |
| [37] | ANN | CaP | Diagnosis | TZD, % f PSA, PSAD and TPV | Cancer or benign | PSA 4–10, 974 patients, multicentre data |
ROC, AUC ¼ CV |
AUC 0.91 |
| [38] | ANN | CaP | Diagnosis after initial negative biopsy PSA 4–10 | t PSA, %f PSA, TPV, TZV, PSAD, TZD | Cancer or benign | 820 patients with PSA 4–10 European cancer detection studies |
ROC AUC 1/3 CV |
AUC 0.83 |
| [39] | ANN | CaP | Diagnosis of BPE and CaP | Age, ethnicity, FH, IPSS, t PSA, %f PSA, DRE | Risk of cancer | Multicentre 354 patients, multicentre | ROC vs LR, 144 test set 40% CV |
AUC ANN 0.8, LR 0.5 |
| [40] | FRB | CaP | Early diagnosis | Age, t PSA, TPV | Risk of cancer | Experts aided in developing 77 fuzzy rules | Not published | None |
| [41] | ANN | CaP | Diagnosis PSA 2–10 | Age, tPSA, %fPSA, TPV, TZV, PSAD, TZD = ANNA 1 | Cancer and benign | 228 data one centre | ROC, 30% CV | AUC 0.78 |
| [41] | ANN | CaP | Diagnosis PSA 2–10 | ANNA 1 + presumed circle area ratio and DRE | Cancer and benign | 228 data one centre |
ROC 30% CV |
AUC 0.79, Sp 45%, Se 90% |
| [42] | ANN | CaP | Diagnosis | Age, tPSA, TPV, PSAD, DRE, and TRUS findings | Cancer and benign | 3814 prostate cancer screening data | ROC AUC 1/3 CV, 2 centres prospective data | AUC: 0.74, 0.76, and 0.75 prospective 0.73, 0.74 |
| [43] | ANN | CaP | Diagnosis | Age, DRE, PSA, PSAD, TZV, TZD = ANNA | Cancer and benign | TRUS, single centre 684 data |
ROC AUC 1/4 CV |
AUC 0.74 |
| [43] | ANN | CaP | Diagnosis | ANNA + TRUS findings | Cancer and benign | TRUS, single centre 684 data |
ROC AUC 1/4 CV |
AUC 0.86 |
| [44] | FNM | CaP | Diagnosis, PSA < 20 | Age, PSA, %f PSA | Cancer and benign | 1030 patients’ data, one centre | ROC, Sp, Se, 1/4 CV | AUC 0.8, Sp 52%, Se 90% |
| [45] | ANN | CaP | Prostate cancer early diagnosis PSA 4–10 | Age, tPSA, %fPSA, TPV, DRE | Cancer or benign | 606 multicentre group (PSA 4–10) | ROC AUC, 1/10 CV | AUC 0.83, AUC 0.74 in Finish group |
| [45] | ANN | CaP | Prostate cancer early diagnosis PSA 4–10 | Age, tPSA, %fPSA, TPV, DRE | Cancer or benign | 656 Finnish cancer survey group (PSA 4–10) | ROC AUC, 1/10 CV | AUC 0.77 |
| [46] | ANN | CaP | Diagnosis | Age, DRE, t PSA and f PSA | Cancer and benign | 1509 with PSA < 20, Single centre | ROC AUC, 1/5 CV | AUC 0.74 |
| [46] | ANN | CaP | Diagnosis | Age, DRE, t PSA, f PSA, TPV and TRUS findings | Cancer and benign | 1509 with PSA < 20, Single centre | ROC AUC, 1/5 CV | AUC 0.75 |
| [47] | ANN | CaP | Diagnosis with -2 Pro PSA | Age, TPV, tPSA, %fPSA, p2 PSA, %p2 PSA (-2 proPSA) | Cancer and benign | PSA 1–30, 586 one centre | ROC, Sp, Se LOO 586 | AUC 0.85, Sp 62%, Se 90% |
| [48] | ANN | CaP | Diagnosis pre-biopsy | Age, DRE, tPSA, PSAD, TZD, TRUS findings | Benign and malignant | 600 patients with suspected CaP | ROC AUC, 477 random | AUC 0.77 |
| [48] | SVM | CaP | Diagnosis pre-biopsy | Age, DRE, tPSA, PSAD, TZD, TRUS findings | Benign and malignant | 600 patients with suspected CaP | ROC AUC, 477 random | AUC 0.85 |
| [49] | ANN | CaP | Diagnosis PSA 2–20 | Age, tPSA, %f PSA, DRE, TPV | Cancer and benign | Testing Prostataclass | ROC AUC, 165 patients one centre | AUC (PSA 2–10) 63–69%, (PSA 10–20) 57–88% |
| [50] | ANN | CaP | Diagnosis | Age, tPSA, %f PSA | Prognosis: cancer or not | 121 Patients data from one centre | ROC AUC, 30% CV 29 patients | AUC 0.92 |
| [51] | ANN | CaP | Diagnosis of clinically significant cancer | Age, DRE, PSA, PRV, TRUS, Biopsy cores | Disease clinical significance | 3025 multicentre data | Accuracy estimation | Ac 57% |
| [52] | ANN | CaP | Diagnosis of cancer | Age, DRE, PSA, %fPSA, and TPV | Cancer and benign | 204 PSA between 4 -10 | ROC AUC | AUC 0.72 |
| [53] | ANN | CaP | PHI index and TPV in diagnosis | Age, TPV, %fPSA, tPSA, PHI, %P2PSA | Cancer and benign |
220 cases PSA < 10 |
ROC AUC | AUC 0.81 |
| [53] | ANN | CaP | PHI index and TPV in diagnosis | Age, %fPSA, tPSA, PHI, %P2PSA | Cancer and benign |
221 cases PSA < 10 |
ROC AUC | AUC 0.77 |
| [54] | FRB | CaP | Diagnosis | Age, PSA, TPV | Cancer and benign | 78 TRUS cases from Urology clinic | None | None |
| [55] | ANN | Fert | Sperm count | Age, duration of infertility, FSH, LH, TT and PRL, testicular volume | Presence of spermatozoa | 303 patient’s data | ROC AUC then kappa stats of LR, test set 73 random | Se 68%, Sp 87.5%, PPV 73.9%, NPV 84% |
| [56] | ANN | Fert | Endocrinopathy with low sperm count | Testis volume, total sperm count, | Endocrinopathy | 1035 Data from 2 centres | ROC AUC | AUC 0.95 |
| [57] | ANN | Fert | Microdissection testicular sperm extraction |
Age, FSH level, cryptorchidism and Klinefelter Syndrome |
Sperm retrieval | 1026 data, one centre | ROC AUC | Se 67% Sp 49.5% PPV, 63.9% NPV 52% Ac 60.8% |
| [58] | ANN | U Dys | Interpretation of U dyn and symptoms | Neurological and physical symptoms, flowmetry, cystometry, U dyn | Areflexia, hyper-reflexive, effort incontinence | 400 U Dyn data | 80 patients, 1/5 CV, | Accuracy 85% |
| [59] | ANN | U Dys | Interpretation of U dyn and symptoms | Neurological and physical symptoms, flowmetry, cystometry, U dynamics | Healthy or ill | 300 patients with LUT disease | ROC, Ac, 1/5 CV | Accuracy 89% |
| [60] | ANN | U Dys | Bladder outlet obstruction | values of the average flow rate, Qmax, PVR and TPV | Obstructed, non-obstructed, and equivocal | N = 457 cases from single centre | Accuracy estimation 157 cases | Ac 60% (testing) 75% (training) |
| [61] | ANN | BPD | IPSS interpretation | IPSS subdomain scores | Obstructed, non-obstructed, and equivocal | N = 460 from single centre | Accuracy estimation 157 cases | Ac 73% |
A total of 37 systems identified in this application of Expert Systems in Urology with evident prevalence of ANN as the model and CaP to be the dominant domain
Table 4.
Disease stage prediction
| Art | Mdl | Dom | Subdomain | Variables | Output | System training | Statistical outcome | Validation set |
|---|---|---|---|---|---|---|---|---|
| [62] | ANN | CaP | staging of localised disease | Age, race, DRE, tPSA, size of tumour on ultrasound, Gl, bilaterality of cancer and number of positive cores and perineural infiltration | Margin, seminal vesicle and lymph node positivity | 1200, patients’ data from multicentre | AUC 0.77, 0.79, 0.8 | 20% CV |
| [63] | FSS | CaP | Localised disease staging | Age, PSA, PSAD, DRE, TRUS, Gl, CT, bone scan, chest x-ray, MRI | Localised or advanced | 16 Cases | Se 92%, Sp 84%, Ac 82% | 43 cases RRP |
| [64] | ANN | CaP | Lymph node staging in CaP post RPP | Age, Gl, clinical stage | Lymph node spread | 736 data from one centre clinically localised CaP | Se 64%, Sp 81.5%, PPV 14%, NPV 98% | 1840 and 316 cases from 2 centres |
| [65] | ANN | CaP | Prostate cancer staging post RRP | Age, tPSA, Gl, clinical stage | Lymph node spread or organ confinement | 5744 data from one centre clinically localised CaP | AUC 77%, 88% for LN | 25% CV random |
| [66] | ANN | CaP | Stage prediction post RRP | Age, histological variables from biopsy | CaP stage | 97 cases with non-organ confined | Prediction accuracy ranged from 82 to 90% | |
| [66] | ANN | CaP | Stage prediction post RRP | Age, histological variables from biopsy, tPSA and TPV | CaP stage | 77 cases with non-organ confined and extracapsular spread | Prediction accuracy ranged from 82 to 90% | |
| [67] | ANN | CaP | Prostate cancer staging post RRP PSA 2–10 | tPSA, TNM, Gl (ANNA1) | localised disease | 124 data from 2 centres Clinically localised CaP | AUC 0.82 | 20% (n = 36 patients) |
| [67] | ANN | CaP | Prostate cancer staging post RRP PSA 2–10 | tPSA, TNM, Gl, maximum tumour length (ANNA2) | localised disease | 124 data from 2 centres Clinically localised CaP | AUC 0.88 | 20% (n = 36 patients) |
| [67] | ANN | CaP | Prostate cancer staging post RRP PSA 2–10 | tPSA, TNM, Gl, maximum tumour length, PSAD (ANNA3) | localised disease |
124 data 2 centres Clinically localised CaP |
Ac 83.3%, Se 85%, Sp 83%, PPV 73%, NPV 90% AUC 0.9 | 20% (n = 36 patients) |
| [67] | ANN | CaP | Prostate cancer staging post RRP PSA 2–10 | tPSA, TNM, Gl, maximum tumour length PSAD, age (ANNA4) | localised disease |
124 data 2 centres Clinically localised CaP |
AUC 0.87 | 20% 36 patients |
| [68] | ANN | CaP | Prostate cancer staging post RRP | tPSA, TPV, TZV, PSAD, TZ, Gl | Pathological stage t2-4 | 201 cases from multinational European cancer data base (PSA 10 or less) | AUC 0.87 | 61 prospective set |
| [69] | ANN | CaP | diagnosis of skeletal metastasis | Age, tPSA | skeletal Mets | 111 retrospective cases in one centre | AUC 0.88, Se 87.5%, Sp 83.3% | Bootstrap CV |
| [70] | ANN | CaP | Stage prediction post RRP | DRE, % of cancer, sum of tumour length, % cancer length and maximum cancer core length | advanced cancer (> pT3a) | 300 randomly selected from retrospective data | AUC 0.71, Se 63%, Sp 81%, Ac78% | 232 random selected set |
| [70] | SVM | CaP | Stage prediction post RRP | DRE, % of cancer, sum of tumour length, % cancer length and maximum cancer core length | advanced caner (> pT3a) | 300 randomly selected from retrospective data | AUC 0.81, Se 67%, Sp 79%, Ac77% | 232 random selected set |
| [71] | ANN | CaP | Define precise stage | PSA, clinical stage, pathological stage, Gl (other added for different set: erection, IPSS, TRUS size, MRI stage | margin, seminal vesicle and lymph node positivity | From 7500 patients’ data from BAUS database and remodelled with external data of 85 patients | AUC 0.38–0.67, concordance index for variables | 10 folds CV |
| [71] | BN | CaP | Define precise stage | PSA, clinical stage, pathological stage, Gl (other added for different set: erection, IPSS, TRUS size, MRI stage | margin, seminal vesicle and lymph node positivity | From 7500 patients’ data from BAUS database and remodelled with external data of 85 patients | AUC 0.01–0.67 concordance index for variables | 10 folds CV |
| [71] | kNN | CaP | Define precise stage | PSA, clinical stage, pathological stage, Gl (other added for different set: erection, IPSS, TRUS size, MRI stage | margin, seminal vesicle and lymph node positivity | From 7500 patients’ data from BAUS database and remodelled with external data of 85 patients | AUC 0.33–0.6 concordance index for variables | 10 folds CV |
| [71] | RBF | CaP | Define precise stage | PSA, clinical stage, pathological stage, Gl (other added for different set: erection, IPSS, TRUS size, MRI stage | margin, seminal vesicle and lymph node positivity | From 7500 patients’ data from BAUS database and remodelled with external data of 85 patients | AUC 0.45–0.5 concordance index for variables | 10 folds CV |
| [71] | SVM | CaP | Define precise stage | PSA, clinical stage, pathological stage, Gl (other added for different set: erection, IPSS, TRUS size, MRI stage | margin, seminal vesicle and lymph node positivity | From 7500 patients’ data from BAUS database and remodelled with external data of 85 patients | AUC 0.5 concordance index for variables | 10 folds CV |
| [72] | ANN | CaP | Staging post RRP | Age, tPSA, n Positive cores, involvement per core, % of positive core | Organ confinement and metastasis | 870 multicentre data | Ac 60% | 120 cases, Accuracy estimation |
| [73] | FNM | CaP | Cancer staging of organ confinement | Age, PSA, Primary Gleason Pattern, secondary Gleason pattern, clinical stage | Organ confinement and metastasis | 399 cases from research network database | AUC 0.8, FNM outperformed ANN, FCM, LR | ROC AUC vs other models |
| [74] | ANN | Nsc | staging | vascular, lymphatic, tunical invasion, percentage of embryonal carcinoma, yolk sac carcinoma, teratoma and seminoma | Stage one or two | 93 cancer specimen, single centre | Prediction accuracy 79.6 to 87.1%, | 10 folds CV |
This table demonstrated Expert Systems predicting urological diagnosis from variable clinical and radiological date. Artificial neural networks (ANN) diagnosing localised prostate cancer (CaP) before surgery were the most common systems in this application
Chiu et al. [29] modelled a system with clinical variables for patients undergoing nuclear bone scintigraphy for predicting skeletal metastasis. The system was able to predict metastatic disease in the test group with Se 87.5%, Sp 83.3%.
None seminoma testicular cancer was the other domain in this application with the system [27] able to predict the cancer disease stage (Table 4) with accuracy reaching 87%.
Treatment outcome prediction
In this application, ES combined disease and patient related factors to estimate the success of a specific treatment or intervention. As in [30, 38, 64, 69] where the system predicted the outcome of extra corporeal shock wave (ESWL) for treating kidney stones and [74, 75] providing an estimation of cancer recurrence after radical surgical treatment of prostate cancer.
Domains
Prostate cancer was also common domain in this application (n = 23). Potter [74, 75] described 4 models developed by data acquired from patients with clinically localised CaP and had radical prostatectomy with curative intent. The variables included clinical and histological findings of the surgical specimen and they were able to predict up to 81% who did not have evidence biochemical failure (rising PSA) in their follow up. Hamid et al. [76] and Gomha [77] models were not restricted to the clinically localised CaP cohort and their study population included patients at different disease stages and on any treatment pathway. Their models included 2 experimental histological markers (tumour suppressor gene p53 and the proto-oncogene bcl-2) in their input variables and the estimated predictive accuracy of the patient response to treatment were reaching 68% and 80% (p < 0.00001) respectively.
Nephrolithiasis treatment was expressed by 6 other systems applying the treatment outcome prediction concept. Cummings et al. targeted this group in his ANN [78] where he trained his network with patients’ data treated at the emergency service of 3 centres with ureteric stones, to identify patients failing conservative management and requiring further intervention. When tested on a different set of 55 cases, the system correctly predicted 100% of the patients who passed the stone spontaneously with an overall accuracy of 76%.
Extra corporeal shockwave lithotripsy (ESWL) is one of the favourable interventions in the nephrolithiasis treatment domain. The stone here receives strong external shock waves, which can subsequently reduce it into small fragment and eliminate the need for direct instrumentation of the renal tract. Their reported success rate can only provide a generalised prediction of outcome to the individual case and ANN was capable of providing an alternative multivariate analytical tool in the 4 models developed by [30, 38, 64, 69]. They estimated high accuracy of their models (Table 5), as in [64], the system predicted 97% of the patients who were confirmed to be stone free following ESWL for treating ureteric stone.
Table 5.
Treatment outcome prediction
| Art | Mdl | Dom | Subdomain | Variables | Output | System training | Validation methods | Statistical outcome |
|---|---|---|---|---|---|---|---|---|
| [79] | ANN | CaP | Outcome of RRP | Age, stage, bone scan, grade, PSA, treatment, bcl-2, p54 | No response, response then relapse, response and no relapse | cohort of CaP single centre 21 patients |
ROC, Sp, Se 20 patients randomly selected |
Ac 85% (60% without markers), K, 0.65; Cl, P < 0.00001 |
| [80] | ANN | CaP | BCF post RRP | Age Pathologic findings and GENN1 | Disease progression | Gl 5–7, T1B-2C, Single centre 136 |
ROC, Sp, Se Test set of 35 (20%) |
AUC 0.71, Ac 74%, Se 82%, Sp 61%, |
| [80] | ANN | CaP | BCF post RRP | DNA polyploidy and quantitative nuclear grade | Disease progression | Gl 5–7, T1B-2C, Single centre 136 | ROC, Sp, Se | AUC 0.74, Ac 80%, Se 75%, Sp 85% |
| [80] | ANN | CaP | BCF post RRP | Pathologic findings, age, DNA polyploidy and quantitative nuclear grade | Disease progression | Gl 5–7, T1B-2C, Single centre 136 | Test set of 35 (20%) | AUC 0.73, Ac 78%, Se 84%, Sp 72% |
| [81] | ANN | CaP | BCF post RRP | Age, PSA, Gl and stage | BCF post RRP all | 140 cases post RRP, one centre |
ROC, Sp, Se 35 (20%) for validity |
AUC 0.81, Se 74%, Sp 78%, PPV 71%, NPV 81%, |
| [82] | Fkn | CaP | Outcome of RRP | TM, Gl, PSA, P53, bcl-2, treatment method |
No response No progression after treatment, Relapse |
41 men with CaP | LOO and compare predictive accuracy of ANN, Fkn | Predictive accuracy ranged from 61–88% |
| [68] | ANN | CaP | Outcome of RRP | tPSA, TZV, PSAd, Gl | Local or advanced disease | 200 cases from multinational European cancer data base |
AUC ROC 60 prospective set |
AUC 0.91, Se 95%, Sp 64%, |
| [83] | ANN | CaP | Outcome of RRP, margin positive | tPSA, clinical stage, Gl (ANNA1) | Positive surgical margins | 218 post RRP and pelvic lymph adenectomy in one centre |
ROC AUC 48 cases 1/4 CV |
AUC 0.7 |
| [83] | ANN | CaP | Outcome of RRP, margin positive | tPSA, clinical stage, Gl, pMRI findings (ANNA2) | Positive surgical margins | 218 post RRRP and pelvic lymph adenectomy in one centre |
ROC AUC 48 cases 1/4 CV |
AUC 0.87 |
| [83] | ANN | CaP | Outcome of RRP, margin positive |
tPSA, clinical stage, Gl, pMRI findings, % of cancer in biopsy, PSAd ANNA3 |
Positive surgical margins | 218 post RRP and pelvic lymph adenopathy in one centre |
ROC AUC 48 cases 1/4 CV |
AUC 0.87 |
| [83] | ANN | CaP | Outcome of RRP, margin positive | tPSA, clinical stage, Gl, % of cancer in biopsy ANNA4 | Positive surgical margins | 218 post RRP and pelvic lymph adenopathy in one centre |
ROC AUC 48 cases 1/4 CV |
AUC 0.71 |
| [84] | ANN | CaP | Outcome of RRP, margin and LN | tPSA, clinical TNM Gl ANNA1 | Positive surgical margins, LN involvement | 41 post RRP and pelvic lymph adenopathy in one centre |
ROC AUC 160 cases randomly selected |
AUC 0.86 for positive margin, 0.88 for LN + ve |
| [84] | ANN | CaP | Outcome of RRP, margin and LN | tPSA, clinical TNM Gl, pMRI findings ANNA2 | Positive surgical margins, LN involvement | 41 post RRP and pelvic lymph adenopathy in one centre |
ROC AUC 160 cases randomly selected |
AUC 0.9 for positive margin, 0.89 for LN + ve |
| [84] | ANN | CaP | Outcome of RRP, margin and LN |
tPSA, clinical stage, Gl, pMRI findings, age ANNA3 |
Positive surgical margins, LN involvement | 41 post RRP and pelvic lymph adenopathy in one centre |
ROC AUC 160 cases randomly selected |
AUC 0.9 for positive margin, 0.9 for LN + ve |
| [85] | FRB | CaP | Outcome of RRP | Clinical stage, Gl, tPSA | Cancer stage (confined, capsule, vesicle and LN) | 116 rules developed from nomograms |
ROC Se, Sp 190 patients post RRP in one centre |
AUC 0.76 (95% CI 0.7–0.8), Se 85%, Sp 61%) |
| [86] | ANN | CaP | Outcome of RRP, margin positive | TNM stage, age, Gl, tPSA | Capsule penetration | 650 retrospective data for RRP at one centre | PPV, NPV 98 cases for testing and 1/2 CV | PPV 100%, NPV 95% |
| [86] | ANN | CaP | Outcome of RRP, margin positive | TNM stage, age, Gl, tPSA MLP | Capsule penetration | 650 retrospective data for RRP at one centre | PPV, NPV 98 cases for testing and 1/2 CV | PPV 97%, NPV 95% |
| [86] | ANN | CaP | Outcome of RRP, margin positive | TNM stage, age, Gl, tPSA, Partial RNN (recurrent neural network) | Capsule penetration | 650 retrospective data for RRP at one centre | PPV, NPV 98 cases for testing and 1/2 CV | PPV 97%, NPV 95% |
| [86] | ANN | CaP | Outcome of RRP, margin positive | TNM stage, age, Gl, tPSA, RBF-MLP | Capsule penetration | 650 retrospective data for RRP at one centre | PPV, NPV 98 cases for testing and 1/2 CV | PPV 97%, NPV 94% |
| [87] | FRB | CaP | Outcome of RPP | Clinical stage, Gl, tPSA | Capsule penetration | Genetic algorithm on 331 patients post RRP in one centre | 48 patients post RRP in one centre ROC | AUC 0.82 (95% CI 0.5–0.8) |
| [88] | ANN | CaP | Outcome of LAP RRP, BCF | Clinical and pathologic parameters, tPSA, margin status, TNM and Gl | BCF | 1575 patients at one centre post lap RRPP |
ROC AUC LOO |
AUC 0.75, Se 90%, Sp 35 |
| [32] | FNM | CaP | Outcome post RRP | Age, FH, DRE, tPSA, Gl, MR findings | tPSA at 6 months | 19 one centre post RRP | Correlation coefficient = 0.99 | 3 Cases |
| [89] | ANN | CaP | Outcome post RRP | Age, tPSA, staging, perineural infiltration, Gl, months of FU | BCF | 1400 multicentre data | Se 85% Sp74%, PPV 77% | 400 data |
| [90] | ANN | CaP | Outcome post RRP, organ confined | Gleason score, preoperative PSA and clinical stage, | Organ confined | 468 cases for training | NPV 83% | 47 cases 30% CV |
| [91] | ANN | CaP | Outcome of RRPP | PSA, BMI, DRE, TRUS, Gl score or grade | Capsule penetration | 225 patients’ data post RRP from 3 centres | 74 patients randomly selected ROC | AUC 0.79 LR 0.74 (P = 0.016) Partin AUC 0.7 |
| [78] | ANN | Nlt | Stone regrowth after ESWL | Anatomy, position, stone analysis, urine analysis, previous stone, medical treatment | Stone recurrence | single centre data base, 65 cases | ROC, Sp, Se33 cases | Se 91%, Sp 92%, AUC 0.96 |
| [92] | ANN | Nlt | Stone clearance with conservative treatment | Age, gender, duration, creatinine, nausea, vomiting, fever | Clearance or intervention | multi centre, Ureteric stone 125 cases | 55 cases ROC, Sp, Se | AC 76% Predict 100% of stones passed |
| [75] | ANN | Nlt | lower pole stone ESWL | Gender, BMI, radiology, stone size and composition, urine analysis, 24 h urine, serum ca and creatinine | Clearance or intervention | 321 patients with lower pole stone | 211 random set ROC, Sp, Se, vs LR | AUC 0.97 Se 95%, Sp 92%, |
| [76] | ANN | Nlt | Stone clearance with ESWL | Age, gender, body habitus, serum electrolytes, 24 h urine, radiological findings | Stone free | 60 patients, one centre | Correlation co-efficient 22 cases | 0.75 |
| [77] | ANN | Nlt | Stone clearance with ESWL | Age, gender, anatomy, location, side, number, length, width, new or recurrent, stent | Stone clearance | Ureteric stone ESWL, One centre 688 cases | 296 cases ROC, Sp, Se | Ac 78%, Se78%, Sp 75%, PPV 97% |
| [93] | ANN | Nlt | Outcome of conservative stone disease treatment | Age, gender, BMI, fever, previous treatments and stones, duration of the symptoms, dimension and position of the stone | Spontaneous expulsion or intervention | 402 patients from one centre |
50 patient, 1/4 cross validation ROC Se, Sp |
Se 95%, Sp 63% |
| [93] | SVM | Nlt | Outcome of conservative stone disease treatment | Age, gender, BMI, fever, previous treatments and stones, duration of the symptoms, dimension and position of the stone | Spontaneous expulsion or intervention | 402 patients from one centre |
50 patient, 1/4 cross validation ROC Se, Sp |
Se 85%, Sp 87% |
| [94] | ANN | Nlt | ESWL outcome prediction | The patients’ characteristics, stone location, burden, shape dimension, pre-ESWL procedure and cost of admission | unexpected post-ESWL visits | 1026 patients received ESWL at one centre` | AUC 0.66 | 506 patients |
| [95] | ANN | PUJ | Outcome of PUJ repair | Demographic, clinical and radiological findings | Sonographic outcome of pyeloplasty | Single centre unilateral paediatric pyeloplasty n = 100 |
16 cases (16%) ROC, Sp, Se |
Ac 100%, Se 100%, Sp 100% |
| [96] | ANN | PUJ | Outcome of PUJ conservative treatment | Age, gender, renal pelvis diameter, laterality, separated renal function on DMSA, urine culture and infections | Observation or surgery | 37 infants with PUJ obstruction | Prediction accuracy16 patients for validation | 75% prediction accuracy |
| [97] | ANN | Neph | Post lap partial nephrectomy hospital stay | Age, co-morbidities, tumour size and extension | Hospital stay less than 2 days | 334 one centre |
5 institutes 77, 19 prospective ROC |
AUC 0.6, 0.5 |
| [97] | ANN | Neph | Post lap nephrectomy hospital stay | Age, co-morbidities, tumour size and extension | Hospital stay less than 2 days | 392 One centre |
5 institutes 127, 29 prospective ROC |
AUC 0.7, 0.7 |
| [98]Z | ANN | Bca | Pathological stage after surgery | Age, gender, tumour (size, number, grade, invasion, lymph vascular invasion, stage), lymph nodes | Prognosis and advanced stage | 183 patients, one centre post cystectomy |
ROC and compare with LR 1/3 cross validation |
MANN AUC 0.86, Se 88%, Sp 77%, PPV 93%, NPV 63%, Ac 85% |
| [98] | ANN | Bca | Pathological stage after surgery | Age, gender, tumour (size, number, grade, invasion, lymph vascular invasion, stage), lymph nodes | Prognosis and advanced stage | 183 patients, one centre post cystectomy |
ROC and compare with LR 1/3 cross validation |
SANN AUC 0.85, Se 84%, Sp 71%, PPV 91%, NPV 67%, Ac 83% |
| [99] | ANN | VUR | outcome of endo repair of VU reflux | Age, gender, implant type, implant volume, number of treatments, side, endo findings, type of cystography | Ultrasound finding | Single centre data base, paediatric VU reflux 174 data |
87 cases for validation ROC, Sp, Se |
Se 71.4%, Sp 81.6%, PPV 58.8%, NPV 88.6% and success rate 78.9%, |
Is one of the common applications of urological expert system. They predicted treatment outcome of radical nephrectomy, radical cystectomy, radical prostatectomy, vesico ureteric reflux endoscopic repair, pelvi-ureteric junction obstruction conservative management, nephrolithiasis conservative management and extracorporeal shockwave treatment. The commonest domain was predicting negative surgical margins post radical prostatectomy
Paediatric pelvi-ureteric junction obstruction is primarily treated conservatively unless there is any evidence of renal function compromise, recurring infection or worsening radiological findings. For the failing group, pyeloplasty is the second line of treatment and [81] developed an ANN to estimate the success rate of this procedure for each individual case by predicting the post-operative degree of hydronephrosis with a reported 100% accuracy in the small tested sample.
Vesico ureteric reflux or reflux uropathy is another paediatric disease, characterised by back flow of urine from the bladder into the ureter through incompetent Vesico ureteric functional valve. Treatment is primarily conservative as it can be a self-limiting disease or surgery to reimplantation the ureters or endoscopic injection of bulking agent at the ureteric orifices [80]. The study authors trained a neural network using 261 cases whom have received endoscopic injection and the system predicted 94% of the patients who did not benefit from the treatment [80].
Laparoscopic partial and radical nephrectomy were the domain of the [82], which was developed by multi institutional case data (age, co-morbidities, tumour size, and extension) of patients having laparoscopic partial or radical nephrectomy. The system was able to predict the length of their postoperative hospital stay with an accuracy of 72%.
Bladder cancer can be treated with complete bladder excision and [79] developed systems to predict the cure rate with an accuracy of 83%.
Recurrence and survival prediction
The ES in this domain aimed to provide individualised risk analysis tools estimating the disease specific mortality and recognising the group whom may benefit from more aggressive or adjuvant treatment.
Domains
Bladder cancer survival and recurrence prediction following radical cystectomy (RC) with curative intention was the commonest domain in this application (24 out of 26 total systems). The lymph nodal involvement is highly predictive of the recurrence and these patients are considered for adjuvant or neoadjuvant systemic chemotherapy. The node free cohort will include high-risk patients who were not identified by the conventional linear stratification system. Catto et al. developed a FNM system to identify this high risk group in the nodal free cohort by predicting the disease recurrence rate (Se 81%, Sp 85%) and their survival with a median error of 8.15 months [92]. The high-risk group identified by this model can benefit from systemic treatment post cystectomy to improve their disease related morbidity and mortality [95, 96]. The 5 years survival post cystectomy was the output of 2 other ANN with a high prediction efficacy of 77% and 90% respectively (Table 6) [97, 99].
Table 6.
Recurrence and progression prediction
| Art | Mdl | Dom | Subdomain | Variables | Output | Knowledge acquisition | Validation | Statistical outcome |
|---|---|---|---|---|---|---|---|---|
| [83, 100] | ANN | Bca | Recurrence | Age, gender, smoking, tumour stage and grade, CIS, number, cytology, other mucosal biopsy | Recurrence or no | N = 432 patients’ data, multicentre |
Radom set of 200 ROC AUC |
Se 76%, Sp 55%, Ac 72% |
| [101] | ANN | Bca | Tumour progression recurrence | Tumour stage and grade, size, number, gender, eGFR | Stage progression | 105 Ta/T1 TCC multicentre | Compare to 4 clinicians McNemar test | 80% accuracy |
| [101] | ANN | Bca | 12 months cancer specific survival | Tumour stage and grade, size, number, gender, eGFR, smoking, cis, dysplasia tumour site, architecture, c-erbB2 (oncogene), p53 (tumour suppressor gene) | 6 months recurrence 12 months survival | 56 Ta/T1 (6 months recurrence), 40 T2-T4 (12 months survival) | Compare to 4 clinicians McNemar test | Accuracy to predict recurrence (75%) and to predict survival (82%) |
| [102] | ANN | Bca | Progression of non-invasive TCC | Age, gender, tumour (grade, stage, number and architecture) and mean nuclear volume | Tumour progression and recurrence | 68 patients’ specimen from one centre |
22 Random test set ROC, Sp, Se |
Recurrence: Se 33%, Sp 40%, PPV 40%, NPV 33% Progression: Se 100%, Sp 67%, PPV 40%, NPV 100% |
| [103] | FNM | Bca | Recurrence classifier | Age, gender, grade, smoking, previous cancer, p53, hMLH1, hMSH2 | Recurrence or not | 109 patients from one centre with TCC | 10% cross validation ROC, LR | AUC 0.98, Se 90%, Sp 80%, PPV 92%, NPV 74%, Ac 88% |
| [103] | FNM | Bca | Survival predictor | Age, gender, grade, smoking, previous cancer, p53, hMLH1, hMSH2 | Survival in months | 109 patients from one centre with TCC |
10% cross validation Root mean square |
RMS = 4.8 |
| [103] | ANN | Bca | Recurrence classifier | Age, gender, grade, smoking, previous cancer, p53, hMLH1, hMSH2 | Recurrence or not | 109 patients from one centre with TCC |
ROC, LR 10% cross validation |
AUC 0.91, Se 94%, Sp 96%, PPV 99%, NPV 84%, Ac 95% |
| [103] | ANN | Bca | Survival predictor | Age, gender, grade, smoking, previous cancer, p53, hMLH1, hMSH2 | Survival in months | 109 patients from one centre with bladder | 10% cross validation RMS | RMS = 11.7 |
| [104] | ANN | Bca | Survival predictor | Age, stage, Grade, smoking, previous cancer | Risk of relapse | 109 patients with primary TCC | Difference in RMS 1/4 CV ROC AUC | Se 90%, Sp 89%, PPV 98, NPV, 64%, Ac 90%, RMS 8.8 |
| [104] | ANN | Bca | Recurrence predictor | Stage, Grade, age, smoking, previous cancer, p53, hMLH1, hMLH2 | Time to relapse | 109 patients with primary TCC | Difference in RMS 1/4 CV ROC AUC | Se 94, Sp 96%, NPV 99%,PPV 84%, Ac 95%, RMS 7.6 |
| [104] | FNM | Bca | Survival predictor | Stage, Grade, age, smoking, previous cancer | Risk of relapse | 109 patients with primary TCC | Difference in RMS 1/4 CV ROC AUC | Se 92%, Sp 90%, PPV 98% NPV 72%, Ac 92%, RMS 8.5 |
| [104] | FNM | Bca | Recurrence predictor | Stage, Grade, age, smoking, previous cancer, p53, hMLH1, hMLH2 | Time to relapse | 109 patients with primary TCC | Difference in RMS 1/4 CV ROC AUC | Se 90% Sp 80%, NPV 92%,PPV 74%, Ac 88%, RMS 7.3 |
| [105] | FNM | Bca | Recurrence (classifier) | Age, gender, grade, smoking, previous cancer, p53, methylation index (% of loci on chromosomes), RARB methylation (gene locus) | Recurrence or not | 117 patients with 1ry TCC or UCC from one centre | 10% cross validation ROC, LR | AUC 0.98, Se 88–100%, Sp 94–100%, Ac 100% |
| [105] | FNM | Bca | Survival predictor | Age, gender, grade, smoking, previous cancer, p53, methylation index (% of loci on chromosomes), RARB methylation | Survival in months | 117 patients with 1ry TCC or UCC from one centre |
10% CV Kaplan Maier for survival |
Average error = 5 months |
| [105] | ANN | Bca | Recurrence (classifier) | Age, gender, grade, smoking, previous cancer, p53, methylation index (% of loci on chromosomes), RARB methylation | Recurrence or not | 117 patients with 1ry TCC or UCC from one centre | 10% cross validation ROC, LR | Ac 89–90%, Se 81–87%, Sp 95–100% |
| [105] | ANN | Bca | Survival predictor | Age, gender, grade, smoking, previous cancer, p53, methylation index (% of loci on chromosomes), RARB methylation | Survival in months | 117 patients with 1ry TCC or UCC from one centre |
10% CV Kaplan Maier for survival |
Average error = 9 months |
| [106] | ANN | Bca | Recurrence | Age, sex, previous recurrence, response to adjuvant therapy, number of lesions, adjuvant therapy | Recurrence or no | 403 patients |
1/3 CV 123 patients ROC AUC |
AUC 0.87,Se 79%, Sp 98% |
| [107] | ANN | Bca | 5 Years survival cystectomy | Age, gender, tumour stage, grade, ln, vascular in, perineural in, prostatic invasion, CaP | Survival at 5 years | 369 patients | ROC, Cox proportional hazard 1/3 CV | Se 63%, Sp 86%, PPV 76%, NPV 77% |
| [108] | FNM | Bca | Recurrence classifier | Gender, pathological stage, grade, CIS, lymph vascular invasion | Recurrence or not | 609 patients from multiple centres |
ROC, LR 10% CV |
Se 93%, Sp 68% |
| [108] | FNM | Bca | Survival predictor | Gender, pathological stage, grade, CIS, lymph vascular invasion | Survival in months | 172 multicentre data |
ROC, LR 10% CV |
Kaplan–Meier survival plots, median error of 8.15 months |
| [109] | ANN | Bca | Survival post cystectomy | Age, gender, bilhariziasis, histology, grade, lymph nodes, lymph vascular, type of diversion | Patient survival | 871 patients’ data post cystectomy |
30% CV ROC vs LR |
AUC 0.86, Se 79%, Sp 81% |
| [110] | ANN | Bca | bladder cancer 5 years survival | Age, gender, histology grade, tumour stage, positive LN, removed LN | 5 years survival | cystectomy data base, single centre 106 patients | Prediction error percent 11 and 29 patients | prediction error rate, > 90% efficiency |
| [111] | ANN | Bca | Recurrence and survival | Age, gender, tumour stage, grade, CIS, ln, lymph vascular invasion | 5 years recurrence and cancer specific death | cystectomy data base, multicentre 2111 | ROC, Kaplan Maier for survival, Cox Proportional Hazard | Se 59%, Sp 77%, PPV 67%, NPV 70% (30% cross validation) |
| [112] | ANN | Bca | Survival post cystectomy | Age, gender, albumin, surgical approach, tumour stage, follow up period, type of diversion | 5 years survival | 117 patients with post cystectomy from one centre |
10 Folds CV ROC, Se, Sp Ac |
Ac 72–80% RELM and ELM had best performance |
| [113] | ANN | Bca | Recurrence of G3 pTa after TURBT | Age, sex, previous histopathological data, previous recurrence rate response to previous BCG adjuvant therapy, number of lesions, size of lesions presence of inflammatory reaction and adjuvant therapy | Recurrence or No | 143 patients with G3 pTa at one centre |
AUC, Se, Se 1/3 cv 43 cases |
AUC 0.81, Se 82%, Sp 96% |
| [114] | ANN | RCC | RCC survival 36 months | Age, gender, BMI, performance status, histopathology, time interval between primary tumour and detection of Mets, type of systemic therapy, number and sites of Met | Recurrence within 36 months | 175 single centre |
30% CV ROC sensitivity analysis |
AUC 0.95 (95% CI 0.87–0.98) |
| [115] | ANN | Nsc | Disease recurrence in five years | (32 variables) age, tumour type, grade, invasion, Mets, ln, treatment, FBC, kidney function | Recurrence within five years | 202 multicentre cases |
1/4 CV ROC, Sensitivity analysis |
AUC 0.87 |
| [116] | FNM | CaP | Prognosis and survival | Age, BMI, PSA, DRE, Gl, clinical stage and treatment methods | Disease specific survival | 100 cases single centre | Sensitivity analysis mean square error, | MSE = 0.068907 (1/10 CV) |
| [117] | ANN | Nlt | Recurrence of Upper tract stones | Age, sex, history of previous calculi, radiologic type, location and composition of previous calculi, 24-h urine assay urine culture, treatment | Recurrence of Upper tract stones | 168 cases, single centre | PPV estimation 68 cases | PPV 100% |
The majority of the Expert Systems in this application were artificial neural network predicting recurrence and survival following bladder cancer treatment. Other systems were applied in non-seminomatous testicular cancer, prostate cancer, renal cell carcinoma and recurrence of upper renal tract stones
Renal cell cancer is primarily treated with partial or radical nephrectomy for clinically localised disease with systemic therapy for the metastatic disease. There is still a degree of uncertainty in stratifying individual disease risk in order to predict the indication and outcome of systemic therapy in the group with distant metastasis. Vukicevic et al. [98] attempted to clarify this uncertainty by training a neural network with patients’ data who had nephrectomy (partial or radical) and received systemic therapy. The mature model predicted the patients who survived the disease at 3 years with an overall accuracy of 95% (CI 0.878–0.987).
None seminoma testicular cancer 5 years recurrence was the domain of [118] ANN. The system was trained with multicentre data and in its testing phase and predicted 100% of the patients who did not suffer from disease recurrence at 5 years with an overall predictive accuracy of 94% (AUC = 87%).
Predicting research variables
In academia, testing a hypothesis for ‘factors-outcome effect’ is a popular quest and the standard statistical regression analysis tools may not be effective for data contaminated by irrelevant variables [119]. AI can provide an alternative methodology in the analysis to identify variables with high correlation to the outcome by applying machine learning as in ANN. The area under the curve (AUC) is estimated for the system predictive accuracy applying all researched variables. Those research variables can be given random values or randomised then the AUC is re estimated for comparison with the original [120]. Only variables that decreases the AUC are considered significant and the wider the discrepancy of the AUC the more significant they are (Table 7).
Table 7.
Research variable prediction
| Art | Mdl | DOM | Subdomain | Variables | Output | System training | Validation | Statistics | Research outcome |
|---|---|---|---|---|---|---|---|---|---|
| [121] | ANN | BPE/CaP | Analysis of variables of quality of life questionnaire | Questionnaire suggested by medical and allied professional | High- or low-quality group | Single centre recruitment with BPE or CaP, 63 cases | ROC, Linear quadratic and logistic regression | Ac 90%, Se 94%, Sp 85%, PPV 89%, NPV 92% | Identify relevant variables |
| [78] | ANN | Nlt | Stone recurrence after ESWL | Anatomy, position, stone analysis, urine analysis, previous stone, medical treatment | Stone recurrence | 65 patients post ESWL from single centre |
33 test set ROC AUC vs LR |
AUC 0.96, Se 91%, S 91% | Stone recurrence, fragments not risk factor |
| [122] | ANN | CaP | Biochemical failure post RRP | TNM, tPSA, Gleason, pathology stage |
BCF at 3 years Yes or no |
564 patients’ data post RRRP with Gl 7, single centre | ROC, Kaplan Meier and Cox Proportional Hazards Model | AUC 75%, NPV 84 | Gleason 7 is inversely correlated to disease free survival and direct to BCF |
| [122] | ANN | CaP | Biochemical failure post RRP | TNM, tPSA, Gleason, pathology stage | BCF post RRRP | 564 patients’ data post RRRP with clinically localised CaP Gl7, single centre | ROC, Kaplan Meier for survival and Cox Proportional Hazards | AUC 81%, NPV 93% | |
| [75] | ANN | Nlt | lower pole stone ESWL | Gender, BMI, radiology, stone size, composition, urine analysis, 24 h urine, serum ca and creatinine | Clearance or intervention | 321 patients with lower pole stone |
211 random set ROC, Sp, Se, vs LR |
AUC 0.97, Se 95%, Sp 92% | BMI, normal urinary transport and infundibular width of 5 mm or more and the infundibular ureteropelvic angle is 45° or more are correlated with stone clearance |
| [103] | FNM | Bca | Recurrence classifier | Age, gender, grade, smoking, previous cancer, p53, hMLH1, hMSH2 | Recurrence or not | 109 patients from one centre with bladder TCC | tenfold CV ROC, LR | AUC 0.98, Se 90%, Sp 80%, PPV 92%, NPV 74%, Ac 88% | p value calculated to compare all models, the effect of combining HK p53 with other variables |
| [103] | FNM | Bca | Survival predictor | Age, gender, grade, smoking, previous cancer, p53, hMLH1, hMSH2 | Survival in months | 109 patients from one centre with bladder TCC |
tenfold CV Root mean square |
RMS = 4.8 | |
| [103] | ANN | Bca | Recurrence classifier | Age, gender, grade, smoking, previous cancer, p53, hMLH1, hMSH2 | Recurrence or not | 109 patients from one centre with bladder TCC |
ROC, LR 10% cross validation |
AUC 0.91, Se 94%, Sp 96%, PPV 99%, NPV 84%, Ac 95% | |
| [103] | ANN | Bca | Survival predictor | Age, gender, grade, smoking, previous cancer, p53, hMLH1, hMSH2 | Survival in months | 109 patients from one centre with bladder TCC | 10% cross validation RMS | RMS = 11.7 | |
| [123] | ANN | Bca | diagnosis | Urine levels of nuclear matrix protein-22, monocyte chemoattractant protein-1 and urinary intercellular adhesion molecule-2 | Cancer and benign | 253 Data from one cystoscopy clinic | ROC, Sp, Se | Se 100%, Sp 75.7%, PPV 32.9%, NPV 100%, | The three factors improve diagnosis |
| [124] | ANN | BPE | Significant LUT symptoms in BPE | Age, PSA, Qmax, TZV, TPV, Oss, ISS, PVR | Progression or no | 397 patient with mild LUTS from 4 centres |
1/3 CV ROC, Sp, Se, Then sensitivity analysis |
Ac 79%, Se 82%, Sp 77%, PPV 78%, NPV 81% | PSA, Oss, TZV are correlated to disease progression |
| [125] | ANN | Hgon | Diagnosis of hypogonadism, | Age, ED, depression score, sexual health score, testosterone level | Risk of hypogonadism | 148 one centre | 70 test cases | Depression most significant, p < 0.0019 | |
| [126] | ANN | BPE/CaP | Diagnosis of BPE and CaP | Age, tPSA, %f PSA, TPV, MIC-1, Hk11, MIF | Cancer and benign | Single centre 371 patients | LOO | AUC 0.91, Se 90%, Sp 80% | Positive if all makers added together |
| [127] | ANN | Bca | Survival and recurrence predictor | 22 different genes variables | Risk and time to relapse | 67 bladder neoplasms and 8 normal bladder specimens |
Difference RMS 10 folds CV ROC AUC |
RMS 5.2 Ac 100% |
500 genes where reduced to 22 genes for creating the network, thus significant |
| [127] | FNM | Bca | Survival and recurrence predictor | 66 rules from 11 gene variables | Risk and time to relapse | 67 bladder neoplasms and 8 normal bladder specimens |
Difference RMS 10 folds CV ROC AUC |
RMS 2.2 Ac 100% |
500 genes where reduced to 22 genes for creating the network, thus significant |
| [105] | FNM | Bca | Recurrence (classifier) | Age, gender, grade, smoking, previous cancer, p53, methylation index (% of loci on chromosomes), RARB methylation | Recurrence or not | 117 patients with 1ry TCC or UCC from one centre | 10% cross validation ROC, LR | AUC 0.98, Se 88–100%, Sp 94–100%, Ac 100% | p value calculated to compare all models, the effect of combining HK p53 with other variables |
| [105] | FNM | Bca | Survival predictor | Age, gender, grade, smoking, previous cancer, p53, methylation index (% of loci on chromosomes), RARB methylation | Survival in months | 117 patients with 1ry TCC or UCC from one centre |
10% CV Kaplan Maier for survival |
Average error = 5 months | Interrogate different markers to suggest a predicative combination |
| [105] | ANN | Bca | Recurrence (classifier) | Age, gender, grade, smoking, previous cancer, p53, methylation index (% of loci on chromosomes), RARB methylation | Recurrence or not | 117 patients with 1ry TCC or UCC from one centre | 10% cross validation ROC, LR | Ac 89–90%, Se 81–87%, Sp 95–100% | |
| [105] | ANN | Bca | Survival predictor | Age, gender, grade, smoking, previous cancer, p53, methylation index (% of loci on chromosomes), RARB methylation | Survival in months | 117 patients with 1ry TCC or UCC from one centre |
10% CV Kaplan Maier for survival |
Average error = 9 months | p for comparison ANN and FNM calculated |
| [128] | ANN | CaP | Diagnosis of cancer in PSA 1–4 4–10 | Age, tPSA, %fPSA, TPV, DRE, -5pro PSA, -7, pro PSA | Risk of cancer | 2 centre PSA 1–10 and TRUS 6–12 cores, 898 patients | ROC, Spearman correlation co efficient LOO | AUC 84% | Pro PSA improved detection rate in 1–4 and improved %fPSA performance in 4–10 group |
| [129] | ANN | CaP | Early CaP diagnosis | Age, tPSA, %fPSA, hK11, hK11/tPSA, hK11/%tPSA | Cancer or benign | 357 with histologically proven cancer or BPE | ROC Se, Sp test set 206 with histologically proven cancer or BPE | AUC 0.84, Se 90%, Sp 52% | Sensitivity analysis of these variables to demonstrate their impact on AUC |
| [130] | ANN | CaP | Early CaP diagnosis | Age, tPSA, %f PSA, TPV, DRE (PSA done by five different assays) | Risk of cancer | 585 patients with suspected cancer PSA 0.49–27 |
ROC AUC 25% random set 195 patients and LOO |
AUC 0.91 (mean value) | Authors suggests developing PSA assay specific ANN to optimise function |
| [131] | ANN | CaP | Prostate cancer early diagnosis | Age, BMI, tPSA, fPSA, TPV, PSAD, smoking, systolic-diastolic pressure, pulse, Gl | Cancer or benign | 300 patients’ data with suspected cancer from one centre |
10- folds CV ROC Se, Sp |
Ac 79%, Se 81%, Sp 78% | |
| [131] | SVM | CaP | Prostate cancer early diagnosis | Age, BMI, tPSA, fPSA, TPV, PSAD, smoking, systolic-diastolic pressure, pulse, Gl | Cancer or benign | 300 patients’ data with suspected cancer from one centre |
10- folds CV ROC Se, Sp |
Ac 81%, Se 84%, Sp 75% | Smoking is a significant classifier but not BMI |
| [132] | ANN | CaP | Diagnosis | Age, tPSA, %f PSA, DRE, TPV | Risk of cancer | PSA2-20 393proscpective data |
ROC AUC LOO |
AUC 0.75, Se 90%, Sp 37% | Demonstrate the impact of different data cohorts on ANN performance |
| [133] |
FNM ANN |
Bca | Gene micro array to predict UCC progression | 200 genes reduced from 2800 by Pearson correlation | Cancer progression to muscle invasive or metastatic | 66 tumours from 34 patients in one centre |
COX multivariate analysis 10 folds CV |
11 new gene signatures | 200 gene micro array reduced to 11 gene signatures |
| [134] | ANN | U Dyn | Urodynamic interpretation | Age, BMI, menopause, sexual activity, UTI, number of vaginal deliveries, surgery, | U Dyn diagnosis | 802 data from single centre POP with symptoms and UDS performed | ROC and compare to multi linear regression CV 20% | AUC 80% (Average) | ANN cannot replace Urodynamic |
| [135] | ANN | Fert | Seminal profile from questionnaire about life habits and health status | Age, BMI, demographic, medical history facts, smoking, alcohol, life style and cloth | Seminal profile | 100 volunteers one centre study | ROC AUC Se, 10 Folds cross validation | Se 73–94%, Sp 25–45%, PPV 79–92%, NPV 7.4–54% | Comparison of different AI classifiers with same variables |
| [135] | SVM | Fert | Seminal profile from questionnaire about life habits and health status | Age, BMI, demographic, medical history facts, smoking, alcohol, life style and cloth | Seminal profile | 100 volunteers one centre study | ROC AUC Se, tenfold CV | Se 74_99%, Sp 12–21%, PPV 75–91%, NPV 4–86% | |
| [135] | DT | Fert | Seminal profile from questionnaire about life habits and health status | Age, BMI, demographic, medical history facts, smoking, alcohol, life style and cloth | Seminal profile | 100 volunteers one centre study | ROC AUC Se, tenfold CV | Se 72–96%, Sp 12–41%, PPV 77–90%, NPV 4–48% | |
| [136] | ANN | Fert | Seminal profile from questionnaire about life habits and health status | Age, season, childhood disease, surgery, trauma, smoking, alcohol, hours sitting ANNA1 | Sperm concentration | 100 volunteers one centre study |
ROC AUC Se, Sp 10 Folds CV |
Se 95%, Sp 50%, PPV 93%, NPV 60% | |
| [136] | ANN | Fert | Seminal profile from questionnaire about life habits and health status | Age, BMI, marital status, vaccines, siblings, allergy, baths, hours of sleep ANNA2 | Sperm motility | 100 volunteers one centre study | ROC AUC Se, Sp | Se 89%, Sp 44%, PPV 89%, NPV 44% | |
| [137] | ANN | CaP | Statistical evaluation of PSA INDEX | Age, TPV, DRE, tPSA, %fPSA | Risk of Cancer | 1362 from multiple centres with suspected CaP and PSA 1.6–8.0 | ROC AUC and comparison to other markers | AUC 0.7—0.74 | |
| [137] | ANN | CaP | Statistical evaluation of PSA INDEX | Age, TPV, DRE, tPSA, %fPSA, %p2PSA | Risk of Cancer | 1362 from multiple centres with suspected CaP and PSA 1.6–8.0 | ROC AUC and comparison to other markers | AUC 0.73—0.79 | |
| [137] | ANN | CaP | Statistical evaluation of PSA INDEX | Age, TPV, DRE, tPSA, %fPSA, %fPSA prostate health index (p2PSA / fPSA X square root tPSA) | Risk of Cancer | 1362 from multiple centres with suspected CaP and PSA 1.6–8.0 | ROC AUC and comparison to other markers | AUC 0.73- 0.8 | Prostate Health index improved ANN performance |
| [112] | ANN | Bca | Survival post cystectomy | Age, gender, albumin, surgical approach, tumour stage, follow up period, type of diversion | 5 years survival | 117 patients with post cystectomy from one centre | ROC, Se, Sp Ac, 10 Folds cross validation | Ac 72–80% |
Comparison of 7 different machine learning RELM and ELM had best performance |
| [138] | ANN | CaP | + ve lymph nodes to the total number of lymph nodes in predicting BCF | Age, tPSA, Clinical stage, Gl, seminal vesicle invasion, number of positive lymph nodes and laterality of lymph node involvement | BCF | 124 cases with lymph node dissection | hazard ration for each variable | LND, Gl, and stage were identified as independent prognostic | LND is more prognostic than their number |
| [139] | BN | BPE | Correlation between symptoms, decision and outcome of surgery | Age, Qmax, PVR, PSA, TPV, TZV, BOO on UDS, and IPSS scores (stratified) | surgical decision-BN model, the outcome of surgery | 1108 cases from one centre | ROC AUC and correlation coefficient | AUC 0.8 TZV (R = 0.396, P < 0.001), treating physician (R = 0.340, P < 0.001) and BOO on UDS (R = 0.300, P < 0.001) |
TPV, physician, BOO on UDS, and the IPSS item of intermittency were factors that directly influenced Decision-making in physicians treating patients with LUTS/BPE |
| [140] | ANN | CaP | Progression biomarkers | Gene microarray | Cancer progression and DSS | 192 tissue histology results | MSE for each variable, then Kaplan Meyer and Pearson’s × 2-tests | 10 gene microarrays identified by ANN | Ki67 and DLX2, appear to predict CaP specific survival and metastasis |
| [141] | ANN | VUR | Renal ultrasound to predict voiding cystourethrogram (VCUG) | Renal ultrasound findings | abnormal VCUG | 2259 cases post UTI and had VCUG | ROC AUC | Se 64.2%, Sp 59.6%, PPV 61.6%, NPV 62.2%, AUC 0.6852 | Renal ultrasound is a poor screening test for VCUG-identified abnormalities |
In this application, the system modifies their machine learning ability to identify the significant variables from the data in terms of their correlation to a specified outcome. This can save time, effort and cost specially when applied on gene microarrays
Domains
Prostate cancer was a common domain in this application with a total of 15 systems analysing predictive factors for diagnosis of cancer, response to treatment and quality of life with prostatic disease. One of the hot topics in Urological cancer is discovering alternative CaP diagnostic markers since serum PSA is not sensitive for distinguishing benign from malignant disease. Stephan et al. investigated the diagnostic value of three markers in this domain: Macrophage inhibitory cytokine-1, macrophage inhibitory factor and human kallikrein 11 [108]. These were used as variables (nodes) in ANN models and compared their accuracy to the linear regression of %fPSA. They have reported that only the ANN model including all three variables was more accurate (AUC 91%, Se 90%, Sp 80%) than all other models proving his hypothesis that they are only relevant as when combined.
Similarly, another study estimated the predictive values of serum PSA precursors (-5, -7 proPSA) in diagnosing prostate cancer using and comparing the accuracy to %fPSA [107]. The -5, -7 pro PSA were only significant in the cohort with PSA between 4 and10 µg/l and did not improve the predictive accuracy when added to the %fPSA. The same author tested this hypothesis on another free PSA precursor (-2 proPSA) by developing ANN with the %p2PSA (-2 ProPSA: fPSA) among other disease variables, which have improved the system accuracy (AUC 85% from 75%) [120].
Three systems evaluated the presence of bcl-2 and p53 (tumor suppressor genes) as a predictive variable for response to prostate cancer treatment [76, 77]. Their combination was reported to be significant (Ac 85%, p < 0.00001) in [77] but [76] found that only bcl-2 is relevant in the other two models (accuracy 63–68%).
Bladder cancer diagnosis and disease progression was the second most common domain with 13 systems. Kolasa et al. [110] have modeled an ANN with three novel urine markers: urine levels of nuclear matrix protein-22, monocyte chemoattractant protein-1 and urinary intercellular adhesion molecule-1, to predict the diagnosis of bladder cancer and it succeeded in predicting all cancer free patients when the three variables were used as a group. Catto.et al. [119] developed two AI models (ANN & FNM) performing microarray analysis on genes associated with bladder cancer progression. Their models narrowed down these genes from 200 to 11 progression-associated genes out of 200 ([OR] 0.70; 95% [CI] 0.56–0.87), which were found to be more accurate than the regression analysis when compared to the specimen immunohistology results.
Kolasa et al. [110] model predicting the pre-histology diagnosis of malignancy based on urine level of novel tumour markers. Their ANN was found to be more accurate (Se 100%, Sp 75.7%) than haematuria diagnosed on urine dipstick (Se 92.6%, Sp 51.8%) and atypical urine cytology (Se 66.7%, Sp 81%).
ESWL of renal stones was the research domain of [30, 69], where they aimed at identifying significant variables correlated to the treatment outcome (stone free) and developing a predictive model. Chiu et al. [69] model did not recognise residual fragments following ESWL as a significant risk for triggering further stone growth and [30] identified these factor: positive BMI, infundibular width (IW) 5 mm, infundibular ureteropelvic angle 45% or more (IUPA), to be all predictive of lower pole stone breaking and clearance.
Benign prostatic hyperplasia was modelled in a system [114] to link the disease specific clinical and radiological factors with the disease progression in patients with mild disease (IPSS < 7) and not receiving any treatment. His ANN identified: obstructive symptoms (Oss), PSA of more than 1.5 ng/ml and transitional zone volume of more than 25 cm3, to be correlated to disease progression and can accurately predict 78% of the cohort who will need further treatment.
Urinary dysfunction diagnosis accuracy by clinical symptoms was compared to urodynamic findings in female patients with pelvic organ prolapse by [115] and both the linear regression and ANN models could not establish relation between the symptoms and urodynamic based diagnosis hence dismissing the hypothesis of only relying on clinical symptoms to reach an accurate diagnosis and replace the need for urodynamics study.
Hypogonadism (Hgon) was represented in [133] system where the diagnosis was made based on patient’s age, erectile dysfunction and depression with AUC of 70% (p < 0.01).
Image analysis
This one of the advancing applications of AI in medicine where the system either analyse the variables in the reported medical images as data input or identifies these variables through a separate image analyser without the need for expert to report the scan or images. The first category was included among other systems mentioned above as in the diagnosis prediction domain where [47] included different variables from TRUS in the system input to predict CaP diagnosis. In this domain, we focused on the other group where the images are presented to the machine in the form raw data translated by the image analyser and the system will then apply their machine learning to identify the cause effect pattern (Table 8).
Table 8.
Image analysis
| Art | Mdl | Dom | Subdomain | Variables | Output | System training | Validation | Statistical outcome |
|---|---|---|---|---|---|---|---|---|
| [142] | ANN | CaP | Radiotherapy dose planning | Patient prostate contour points (anterior, posterior and 5 lateral) | Anterior, posterior and lateral beam | 12–68 patients record of radiotherapy treatment planning | Average asymmetry of ANN and acceptance by dosimetrists Small field prostate (n = 133) and for large field prostate (n = 64) | Average asymmetry of ANN 0.20% and acceptance by dosimetrists was 96% (small field prostate) and 88% for large field prostate |
| [143] | ANN | CaP | Diagnosis of localised disease from TRUS | Pixel distribution and grey levels of the TRUS images | Benign, malignant with Gleason grading | 53 images of benign and malignant sample images from 5 patients | Compare to histology results of 500 pictures from 61 patients post RRP for localised disease in one centre | Sp 99%, Se 83%, true positive for isoechoic is 97% |
| [144] |
ANN LDA |
CaP | progression post RPP | Prostate volume, PSA, Pathology morphometric variables LDA | Progression or no | Progression t2n0 post RRP, 228 patients from one centre |
ROC, Sp, Se, LOO 39 cases |
Ac 70%, Se 55%, Sp 85% |
| [144] | ANN LVQ | CaP | progression post RPP | Prostate volume, PSA, Pathology morphometric variables | Progression or no | Progression t2n0 post RRP, 228 patients from one centre |
ROC, Sp, Se, LOO 39 cases |
Ac 90%, Se 95%, Sp 85%, |
| [144] | ANN LVQPAK | CaP | progression post RPP | Prostate volume, PSA, Pathology morphometric variables | Progression or no | Progression t2n0 post RRP, 228 patients from one centre |
ROC, Sp, Se, LOO 39 cases |
Ac 83%, Se 85%, Sp 80% |
| [144] | ANN MLFF-bp | CaP | progression post RPP | Prostate volume, PSA, Pathology morphometric variables | Progression or no | Progression t2n0 post RRP, 228 patients from one centre |
ROC, Sp, Se, LOO 39 cases |
Ac 76%, Se 73%, Sp 80% |
| [145] | kNN | CaP | TRUS cancer image analysis | Image pixels segmented by tissue descriptor (spatial grey level dependence) | Predict cancer | Images of 202 patients with suspected CaP at one centre | 87 randomly selected patients Comparison to other classifiers and ROC | AUC 0.6 |
| [146] | ANN | CaP | TRUS Image segmentation | Pixel’s colour values from TRUS images | TRUS image segmentation | 212 CaP TRUS data | Overlap measure (compared to expert segmented boundary) on 10 random images | 81% mean overlap measurement |
| [147] | ANN | CaP | MRI cancer diagnosis | 256 MRSI spectra (resonance intensities at given PPM) | Cancer or benign | 5308 voxels of 18 patients with CaP in a retrospective study |
15% validation ROC Se, Sp |
AUC 0.95, Se 50%, Sp 99% |
| [147] | ANN | CaP | MRI cancer diagnosis | 256 MRSI spectra (resonance intensities at given PPM), peripheral and transition zone, periurethral and outside region | Cancer or benign | 5308 voxels of 18 patients with CaP in a retrospective study |
15% CV validation ROC Se, Sp |
AUC 0.97, Se 62%, Sp 99% |
| [148] | SVM | CaP | Diagnosis of cancer from pMRI images | Image segmentation then clustering voxels | Cancer or benign | 16 pMRI images with CaP | Correlation coefficients of voxel parameters | Mean accuracy of 84% |
| [149] | ANN | Bca | Image histology analysis | Image histology analysis (measurements of the segmentation of nuclei and other features) | Benign and malignant | 141 randomly chosen cell images (30%) |
329 cell images (70%) ROC, Sp, Se |
Sp 100%, Se 82%, PPV 96%, NPV 80%, Ac 88% |
| [150] | FCM | Bca | Diagnosis of tumour |
Bladder wall segmentation and tumour region extraction To detect bladder abnormalities, four volume-based morphological features: bent rate, shape index, wall thickness, and bent rate difference between the inner and outer surfaces |
Bladder neoplasm | 16 Bladder tumour MRI images | Overlap Ratio (OR) | OR 86.3% |
| [151] | ANN | Bca | Transitional cell cytology analysis | Cytology image analysis and pixel variations as variables | Cancer or benign | 16 cytology images | comparison to experts, × 2 test | 75% concordance with the experts |
| [152] | ANN | Nlt | Spectroscopy stone analysis | Absorbance infra-red spectrum of 91 wave lengths | Stone composition | 160 and 57 stone mixtures | Predictive accuracy, root mean square error on 36 independent stone samples | Overall good predictive value |
Expert Systems in this application analysed images from histology and radiological scans to learn patterns that are correlated to a specific diagnosis. They have proven to be effective in this domain and they facilitated diagnosis of cancer and even delivering radiotherapy dosage
Domains
Prostate cancer image analysis was modelled in 10 systems to enhance diagnostic accuracy as in [126] and disease progression prediction as in [128]. The first system represented each TRUS image pixel as one variable or neuron in a pulse coupled neural network and trained their system with 212 prostate cancer images to segment prostate gland boundary with an average overlap accuracy (overlap measure = difference between PCNN boundary and the expert) of 81% for ten images [126].
The other 4 systems analysed histological images of a cohort of patients post RP with clinically localised CaP to predict the disease progression. The histological images were given coloured coding and analysed by the system that used variables as % of epithelial cell and glandular Lumina to identify the high risk group for disease recurrence with an accuracy reaching 90% [128].
LUT disease urine cytology images were analysed by 2 models in [123], which identified all patients with benign disease with an overall accuracy of 97%.
Nephrolithiasis stone biochemistry analysis can be achieved through an expert analysis of infrared spectroscopy which was simulated by [124] where the infrared spectra wavelength numbers were modelled as input variables and the system prediction accuracy of the expert analysed stone specimen had a root square mean error of 3.471.
Qualitative analysis
The same articles were considered for the qualitative analysis against the four stages (validation, verification, evaluations and credibility) reported in Okeefe industrial survey [8] and Benbasat article [9]. The completion of the four stages examined in this qualitative analysis was demonstrated by none of the included systems. There is a possibility that some of these missing stages has been performed but not published in the scientific literature.
Validation was performed by almost all the systems (166 out 169) with varying degree of study strength, bias, and limitations (Table 9). Most of the data driven systems (ANN, SVM, BN, kNN and FNM) were validated by the ROC and AUC by having a training and validation set or cross validation or applying the leave one out technique. Samli et al. enhanced the validity of their system by estimating the kappa statistics with the ROC [134].
Table 9.
Qualitative assessment of urological Expert Systems
| Art | Mdl | Validation methods | Credibility | Evaluation | Validation | Verification | Strength and bias |
|---|---|---|---|---|---|---|---|
| [27] | RBR | Patients' evaluation | No | Yes | Yes | No | Only qualitative evaluation |
| [18] | RBR | Blinded comparison against 4 experts with independent experts rating and 3 centres RCT pilot trial | Yes | Yes | Yes | No | Consideration of system evaluation with real time testing but small number |
| [21] | FRB | Improve practitioner accuracy | No | No | No | No | Insufficient info on development and validation |
| [15] | RBR | RCT reliability and validity by experts’ reviews | Yes | Yes | Yes | No | Small number in the study and short duration of follow up |
| [95] | ANN | ROC, Sp, Se | No | No | Yes | No | Small number for validation |
| [63] | FSS | ROC, Sp, Se | No | No | Yes | No | 2 methods for validation, compared to experts and data |
| [143] | ANN | Compare to histology results | No | No | Yes | No | No comparison to human to demonstrate usability, no p value or CI |
| [103] | FNM | ROC, LR, RMS | No | No | Yes | No | p value calculated to compare all models |
| [103] | ANN | ROC, LR, RMS | No | No | Yes | No | p value calculated to compare all models, the effect of combining HK p53 with other variables |
| [102] | ANN | ROC, Sp, Se | No | No | Yes | No | No p value |
| [76] | ANN | Correlation co-efficient | No | No | Yes | No | Correlation co-efficient between expert and system? Kappa more accurate |
| [40] | FRB | Not published | No | No | No | No | Not validated |
| [68] | ANN | AUC ROC | No | No | Yes | No | p value calculated vs LR |
| [19] | RBR | Feedback from patients with no control group | No | Yes | No | No | No validation but user (patient evaluation) |
| [29] | FRB | Comparison to experts and non-experts | No | No | Yes | No | Expert as gold standard |
| [25] | RBR |
PPV 62%, NPV 100% Se 100% Sp 33% |
No | No | Yes | No | Small number, low specificity |
| [55] | ANN | ROC AUC then compare with LR, kappa stats | No | No | Yes | No | Multimodal of validation |
| [99] | ANN | ROC, Sp, Se | No | No | Yes | No | Not long term follows up |
| [43] | ANN | ROC (0.74 and 0.86) | No | No | Yes | No | TRUS finding from expert panel, human as gold standard |
| [105] | FNM | ROC, LR | No | No | Yes | No | p value calculated to compare all models |
| [105] | ANN | Kaplan Maier for survival | No | No | Yes | No | p for comparison ANN and FNM calculated |
| [145] | kNN | Comparison to other classifiers and ROC | No | Yes | Yes | No | Evaluated the usability of the product and was found to have less than significant effect |
| [129] | ANN | ROC Se, Sp | No | No | Yes | No | Sensitivity analysis of input variables |
| [22] | ANN | ROC 0.7, accuracy 79% | No | No | Yes | No | Compare to experts without accounting for human error |
| [85] | FRB | ROC Se, Sp | No | No | Yes | No | No user evaluation |
| [24] | FRB | Ac 0.76, Se 0.79, Sp 0.75 | No | No | Yes | No | Expert as gold standard |
| [109] | ANN | ROC Compare to LR | No | No | Yes | No | CI calculated |
| [12] | FRB | Ac 0.93, Se 0.97, Sp 0.99 | No | No | Yes | No | Expert as gold standard |
| [110] | ANN | Prediction error percent | No | No | Yes | No | Experimental results |
| [48] | SVM | ROC AUC | No | No | Yes | No | P value calculated to compare all models |
| [146] | ANN | Overlap measure (segmented by experts) | No | No | Yes | No | Expert as gold standard |
| [23] | ANN | Ac 0.84, Se 0.93, Sp 0.33 | No | No | Yes | No | Experts verified data no account for human error |
| [30] | FNM | Accuracy 86.8% | No | No | Yes | No | Guidelines as gold standard |
| [20] | RBR | Evaluation by experts, 95 retrospective | No | No | Yes | No | Expert as gold standard, qualitative evaluation |
| [26] |
HYB FUZZY ONT |
Kappa vs experts, k = 0.89 | No | No | Yes | No | Kappa limitation prospective, randomisation, |
| [16] | RBR | Se 0.95, Sp 0.72, Bayesian analysis S&S, usability of system by Likert scale (Cronbach’s alpha 0.9) | Yes | Yes | Yes | No | Full system evaluation but nurse as gold standard, no attempts to eliminate error |
| [91] | ANN | ROC AUC compare with Partin nomogram and LR | No | No | Yes | No | No correlation with user |
| [17] | FNM | Kappa vs experts, Se 0.95, Sp 0.92 | No | No | Yes | No | Human expert as gold standard and no qualitative evaluation (weight of error) |
| [60] | ANN | Ac 60% (testing) 75% (training) | No | No | Yes | No | Compare to gold standard, Urodynamic |
| [117] | ANN | PPV 100% | No | No | Yes | No | No calculation of NPP and overall accuracy |
| [32] | FNM | Correlation coefficient = 0.99 | No | No | Yes | No | Small number of cases for validation |
| [150] | FCM | OR 86.3% | No | No | Yes | No | Comparison with experts as gold standard than mapping to histology |
| [141] | ANN | ROC, Se 64.2%, Sp 59.6%, PPV 61.6%, NPV 62.2%, AUC 0.6852 | No | No | Yes | No | Similar to urodynamic as research tool |
| [54] | FRB | None | No | No | Yes | No | No validation |
All systems’ development was qualitatively assessed against the common industrial steps in the development pathway described by Okeefe and Benbasat. With exception of the system validation, the rest of the cycle was defective with no explanation. The validation had variable degree of strength with common application of the receiver operator characteristic for estimating the area under the curve for data driven systems
Evaluation was only performed by a small fraction of these systems (n = 6). Their evaluation was aiming at the user or the expert but rarely both. There is no evidence to support that these were performed at early stages to determine the substantiality of the system to the user.
System credibility and verification were never performed. It would be implied that the verification was performed to an extent but not reported as it is a technical part of the development.
‘System development limitation and bias evaluation’ demonstrated an overall acceptable validation methodology with valid statistical analysis. However, a few observed limitations (Table 9) were reported with the common encounter being the consideration human opinion as a gold standard (n = 9). For instance, the gold standard in diagnosing prostate cancer is tissue biopsy confirmation. The interpretation of the expert clinical diagnosis as the gold standard reference can lead to statistical errors and invalidate the study.
Discussion
Expert Systems are widely available in Urological domains, with a large range of models, applications, domains, and target users including patients, students, non-experts, experts, and researchers. The number of published systems has risen over the years but with a consistent lack of publications reporting their real time testing or healthcare implementation (Fig. 4).
Fig. 4.
Expert System (ES) analysis by year of publication showing an upward trend and increase in number of publications. Systems were included according to the keywords for expert system models and applied in urological domains
There is an increasing interest in analysing this gap which is reflected from the scope of AI historic review articles which aimed to only familiarise the readers with ES existence and application [33, 125]. In fact, the majority had a relatively narrow scope on the evolution and application of one ES models (artificial neural network) in prostate cancer diagnosis. Recently, similar to our research, there has been more interest in AI validation, and lack of uptake despite the faith in their ability. Therefore, in this study we quantified ES progression and applications in Urology while examining their developmental life cycle.
It was evident that CaP was the commonest domain in almost all applications contributing with more than two thirds of the systems (91 systems in total). Different aspects of this domain have been simulated by these systems to include diagnosis, therapeutics, predictions of disease progression or treatment outcome, researching variables and medical images analysis. Most of these systems were simulating urologist cognitive function with little guidance on their benefits and how they can be implemented to improve cancer decision making.
In industry, this is usually performed before the system development by evaluating the system usability from the user perspective. This part has lacked or not been acknowledged in the published studies and is possibly a core reason for the lack of their integration in urological health care. Furthermore, none of these systems has been a subject to live testing in a well-designed study to prove its efficacy over standard tools or in the clinical context to prove its validity to justify their complex structure to AI novice health care professionals. The qualitative analysis demonstrated that validation is the only stage of the development cycle to be applied by most of the systems and there is a lack of system evaluation, credibility, and verification. The evaluation can be subdivided into usability (usually by average user), utility and system quality (by experts) [9]. Despite the crucial stage of ES development, there has been a lack of attention in the published articles to integrate it into the development life cycle. This can mean the whole system can fail and also challenge its uptake [8].
An example can be drawn from this review where the majority of the systems focused on CaP diagnosis and treatment. Their implementation would be challenged by the standard decision-making tools of the cancer multidisciplinary team and the ethical concerns of relying on ANN in making such life changing and expensive decision. The utility analysis of those ES would have been essential for tailoring their development for real time applications where they can be more substantial to the user. One example is lack of community-based systems for the initial referral of suspected cancer patients and follow up of stable disease, where NICE have identified a need for such decision support models [152, 153].
There was a wide diversity of modelling in Urological ES with ANN being the most common model in this review. These would bypass the need for direct learning from experts and the exhaustive process of knowledge acquisition, which is a core requirement for knowledge-based systems to attest the whole system progress [55]. However, their analytical hidden layer of nodes “black box phenomenon” has been a subject for wide criticism and rejection from clinicians due to lack of transparency and understanding of its function.
Stephan et al. suggested a statistical solution to identify the variables significance by performing sensitivity analysis [154]. This estimates the variation of the AUC with introduction or elimination of each variable. This can only reflect the significance of each variable but does not explain how the cases are being solved nor quantify this to the user in a standard statistical value. This can be useful in research as they can identify significant variables in a large set data and has been successfully applied in the field of academic urology as in [119] where the system successfully identified the relevant gene signature for bladder cancer progression which saved time and cost of microarray analysis of all suspected genes.
Holzinger et al. emphasised on the importance of the explicability of the AI model specially in medicine which is a clear challenge for machine learning due to their complex reasoning [155]. Their study attempted to simplify the explanation by classifying the systems into post-hoc or ante-hoc. In post-hoc, explanations were provided for a specific decision as in model agnostic framework where the black box reasoning can be explained through transparent approximations of the mathematical models and variable [156, 157]. Those are reproduced on demand for a specific problem rather than the whole system which can shed more light on the system function. It is not certain if those can be easily interpreted by the AI novice clinician, but it has provided more explicit models for tackling the black box phenomenon.
Knowledge based systems can be explained by ante hoc models where the whole system reasoning can be represented. Those systems rely on expert knowledge in their development and face the bottle neck phenomenon in their applications. Furthermore, they are not always successful in identifying and mapping multilinear mathematical rules and machine learning is mandatory or at least more efficient [155]. Bologna and Hayashi et al. suggested that machine learning is more successful in complex problem solving with inverse relation between the machine performance, and it is built-in transparency [158].
Another common aspect lacking in these articles was the coupling of their system development methodology with the medical device registration requirements. This is essential as ES often function as standalone software with no human supervision to their calculation. This categorises the system as a medical device with mandatory perquisite to register with the relevant authorities as Medicines & Healthcare products Regulatory Agency in the UK [5].
Cabitza et al. compared AI validation to other medical interventions as drugs and emphasised on considering the “software as a medical device” [159]. Unlike other devices or drugs, AI models in healthcare are unique in being more dynamic which should be reflected in their validation cycle. They also quoted the known term “techno-vigilance” to learn from other medical device validation pathways. They recommended different outlook to validation where it is broken down to statistical (efficacy), relational (usability), pragmatic (effectiveness) and ecological (cost-effectiveness) with available standards for those steps (ISO 5725, ISO 9241 and ISO 14155). The latter is viewed as a novel standard for evaluating the cost benefits of applying specific AI model in healthcare which would require longitudinal modelling of health economics [159]. This was evidently lacking in articles that were included in our review and in fact most of the studies were non-randomised and retrospective.
Similarly, Nagendran et al. systematically analysed studies that compare AI performance to experts in classifying medical imaging into diseased and non-diseased, they concluded that AI performance was non-inferior to human experts with potential for out-performing [160]. Their 10 years review identified from literature 2 randomised clinical trials and 9 prospective non-randomised trials extracted from a total of 10 and 81 studies, respectively. Their review assessed the risk of bias using PROBAST (prediction model risk of bias assessment tool) criteria for non-randomised studies. The tool is designed for identifying the risk of bias by analysing four domains (participant, predictors, outcome, and analysis) [161], which is applicable to systematic review analysing prediction model with a target outcome.
In our study, as there was no unified outcome for the included prediction tools, the scope was on the role of validation rather than the outcome. Therefore, those tools assessing the risk of bias were not utilised due to the wide gaps in the tool checklist between the included articles. Such study design and data heterogeneities were also evident in Nagendran et al. and similar to our study, data synthesis was not possible. This will pose a challenge reinforcing the application of AI models in healthcare due to lack of level 1 evidence which is mandatory in healthcare for accepting a novel intervention.
Finally, the quality of the data analysis was beyond the scope of our systematic review despite being essential for developing quality AI systems. Cabitza et al. examined this gap and focused on the data governance [161]. There has been very limited evidence on data quality appraisal and standards with call for further research and allocation of more resources specially in healthcare where the data are notoriously limited with errors or discordance.
The potential application of AI in urology with focus on its future application has been recently discussed by Eminaga et al. [162]. They have shown an increasing interest in urology research, but with a challenged mechanistic update due to the model complexity and lack of end user understanding of its design and function. Furthermore, they identified discrepancy between AI engineering and clinical application which reflects some lack of communication between both disciplines.
This can be either a consequence or a cause for lack of clinical utility testing, which increases the need for research in this domain to be incorporated in the software development [163]. In fact, it has been recommended to perform the utility test before developing the system to tailor its application [164, 165]. Despite having different methodology to our systematic review, the recommendations were similar with strong emphasis on the lack of utility testing and its impact on AI uptake in healthcare [166–168].
Conclusion
ES have been advancing in Urology with demonstrated versatility and efficacy. They have suffered from lack of formality in their development, testing and methodology for registration, which has limited their uptake. Future research is recommended in identifying criteria for successful functional domain applications, knowledge engineering and integrating the system development with the registration requirement for their future implementation in the health care systems.
Acknowledgements
Not applicable.
Abbreviations
- Ac
Accuracy
- AI
Artificial intelligence
- ANN
Artificial neural networks
- AP
Acute prostatitis
- Bca
Bladder cancer
- BC
Backward chaining
- BCF
Biochemical failure
- BCG
Bacille Calmette–Guérin
- BP
Back propagation neural network
- BPD
Benign prostatic disease
- BPH
Benign prostatic hyperplasia
- CAD
Computer aided diagnosis
- CBR
Case based reasoning
- CP
Chronic prostatitis
- CV
Cross validation
- Dom
Domain
- DRE
Digital rectal exam
- ED
Erectile dysfunction
- ES
Expert Systems
- FC
Forward chaining
- Fert
Fertility
- FH
Family history
- FLS
Fuzzy logic systems
- F-ONT
Fuzzy ontology
- FNM
Fuzzy neural modelling
- FRB
Fuzzy rule-based systems
- FSH
Follicular stimulating hormone level
- GA
Genetic algorithm
- Gl
Gleason score
- Hgon
Hypogonadism
- Hk11
Human kallikrein 11
- Incont
Incontinence
- IS
Information systems
- ISS
Irritative symptoms
- IT
Information technology
- IUPA
Infundibular ureteropelvic angle
- IW
Infundibular width
- KA
Knowledge acquisition
- KMSP
Kaplan Meir Survival Plot
- KE
Knowledge engineer
- Lap
Laparoscopy
- LH
Luteinising hormone level
- LOO
Leave one out
- LUT
Lower urinary tract
- LVQ
Learning vector quanitizer
- MIC-1
Macrophage inhibitory cytokine-1
- MIF
Macrophage inhibitory factor
- MH
Medical history
- ML
Machine learning
- MHRA
Medicines and Healthcare products Regulatory Agency
- Mdl
Model
- Nep
Nephrectomy
- Nlt
Nephrolithiasis
- NICE
National Institute for Health and Care Excellence
- Nomo
Nomogram
- NPV
Negative predictive value
- Nsc
None seminoma testicular cancer
- Oss
Obstructive symptoms
- Pop
Pelvic organ prolapse
- Pca
Prostate cancer
- PPV
Positive predictive value
- PRL
Prolactin level
- PSAd
PSA density
- PSAv
PSA velocity
- PVR
Post void residual
- Qmax
Maximum flow rate
- RA
Requirement analysis
- RBR
Rule based reasoning
- RC
Radical cystectomy
- RCC
Renal cell carcinoma
- Recur
Recurrence
- Res
Response
- ROC
Receiver operating characteristic
- RP
Radical prostatectomy
- Sc
Single centre
- Se
Sensitivity
- SPC
Stable prostate cancer
- Sp
Specificity
- tPSA
Total PSA
- TPV
Total prostatic volume
- TRUS
Trans rectal ultrasound scan
- TT
Total Testosterone
- TZD
Transitional zone PSA density
- TZV
Transitional zone volume
- U Dyn
Urodynamic study
- U Dys
Urinary dysfunction
- UTI
Urinary tract infection
- V&V
Verification and validation
- VU rflx
Vesico-ureteric reflux
- %fPSA
Percentage free/total PSA
- %p2PSA
Percentage p2PSA/fPSA
- p2PSA
-2 ProPSA
- U incont
Urinary incontinence
Authors' contributions
All listed authors have read and approved the final manuscript. All listed authors contributed sufficiently to take responsibility for the whole content of the manuscript following the criteria in ICJME guidelines of authorship rights and responsibilities. HS for conceptualisation, literature review, data curation, formal analysis, methodology and original writing, review, and editing. DS and JNL for supervision, writing review and editing. AA for field investigation, validation, draft review and editing. All authors read and approved the final manuscript.
Funding
No sources of funding or any form of financial support of disclose.
Availability of data and material
For data and supporting materials access, please contact authors for data requests.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
No competing interests or exclusive licenses used in preparing this manuscript. The authors indicated no potential conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hesham Salem, Email: hesham.salem1@nhs.net.
Daniele Soria, Email: d.soria@westminster.ac.uk.
Jonathan N. Lund, Email: jon.lund@nottingham.ac.uk
Amir Awwad, Email: amir.awwad@lhsc.on.ca.
References
- 1.McCarthy J, Minsky ML, Shannon CE. A proposal for the Dartmouth summer research project on artificial intelligence—August 31, 1955. Ai Mag. 2006;27(4):12–14. [Google Scholar]
- 2.Turing A. Computing machinery and intelligence. In: Epstein R, Roberts G, Beber G, editors. Parsing the turing test. Netherlands: Springer; 2009. pp. 23–65. [Google Scholar]
- 3.Shortliffe EH, et al. computer as a consultant for selection of antimicrobial therapy for patients with bacteremia. Clin Res. 1975;23(3):A385–A385. [Google Scholar]
- 4.Jackson P. Introduction to expert systems. Boston: Addison-Wesley; 1999. [Google Scholar]
- 5.Liao SH. Expert system methodologies and applications—a decade review from 1995 to 2004. Expert Syst Appl. 2005;28(1):93–103. doi: 10.1016/j.eswa.2004.08.003. [DOI] [Google Scholar]
- 6.Ammenwerth E, et al. Clinical decision support systems: need for evidence, need for evaluation. Artif Intell Med. 2013;59(1):1–3. doi: 10.1016/j.artmed.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Garg AX, et al. Effects of computerized clinical decision support systems on practitioner performance and patient outcomes—a systematic review. J Am Med Assoc: JAMA. 2005;293(10):1223–1238. doi: 10.1001/jama.293.10.1223. [DOI] [PubMed] [Google Scholar]
- 8.Kawamoto K, et al. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ. 2005;330(7494):765. doi: 10.1136/bmj.38398.500764.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okeefe RM, Oleary DE. Expert system verification and validation—a survey and tutorial. Artif Intell Rev. 1993;7(1):3–42. doi: 10.1007/BF00849196. [DOI] [Google Scholar]
- 10.Benbasat I, Dhaliwal JS. A framework for the validation of knowledge acquisition. Knowl Acquis. 1989;1(2):215–233. doi: 10.1016/S1042-8143(89)80020-2. [DOI] [Google Scholar]
- 11.Pandey B, Mishra RB. Knowledge and intelligent computing system in medicine. Comput Biol Med. 2009;39(3):215–230. doi: 10.1016/j.compbiomed.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Koutsojannis C, et al. Using machine learning techniques to improve the behaviour of a medical decision support system for prostate diseases. In: 2009 9th international conference on intelligent systems design and applications. 2009. p. 341–6.
- 13.Petrovic S, Mishra N, Sundar S. A novel case based reasoning approach to radiotherapy planning. Expert Syst Appl. 2011;38(9):10759–10769. doi: 10.1016/j.eswa.2011.01.109. [DOI] [Google Scholar]
- 14.Keles A, et al. Neuro-fuzzy classification of prostate cancer using NEFCLASS-J. Comput Biol Med. 2007;37(11):1617–1628. doi: 10.1016/j.compbiomed.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Gorman R. Expert system for management of urinary incontinence in women. In: Proceedings of the annual symposium on computer application in medical care. 1995. p. 527–31. [PMC free article] [PubMed]
- 16.Hao ATH, et al. Nursing process decision support system for urology ward. Int J Med Inform. 2013;82(7):604–612. doi: 10.1016/j.ijmedinf.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Lopes M, et al. Fuzzy cognitive map in differential diagnosis of alterations in urinary elimination: a nursing approach. Int J Med Inform. 2013;82(3):201–208. doi: 10.1016/j.ijmedinf.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrucci K, et al. Evaluation of UNIS: urological nursing information systems. In: Proceedings of the annual symposium on computer application [sic] in medical care. Symposium on computer applications in medical care. 1991. [PMC free article] [PubMed]
- 19.Boyington AR, et al. Development of a computer-based system for continence health promotion. Nurs Outlook. 2004;52(5):241–247. doi: 10.1016/j.outlook.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Koutsojannis C, Lithari C, Hatzilygeroudis I. Managing urinary incontinence through hand-held real-time decision support aid. Comput Methods Programs Biomed. 2012;107(1):84–89. doi: 10.1016/j.cmpb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Sucevic D, Ilic I. Uncertain knowledge processing in urology diagnostic problems based Expert System. In: 6th Mediterranean electrotechnical conference, proceedings vols 1 and 2. 1991. p. 741–3.
- 22.Altunay S, et al. A new approach to urinary system dynamics problems: evaluation and classification of uroflowmeter signals using artificial neural networks. Expert Syst Appl. 2009;36(3):4891–4895. doi: 10.1016/j.eswa.2008.05.051. [DOI] [Google Scholar]
- 23.Gil D, Johnsson M. Using support vector machines in diagnoses of urological dysfunctions. Expert Syst Appl. 2010;37(6):4713–4718. doi: 10.1016/j.eswa.2009.12.055. [DOI] [Google Scholar]
- 24.Koutsojannis C, Tsimara M, Nabil E. HIROFILOS: a medical expert system for prostate diseases. In: Zaharim A, Mastorakis N, Gonos I, editors. Proceedings of the 7th Wseas international conference on computational intelligence, man-machine systems and cybernetics. 2008. 254–259.
- 25.Pereira M, Schaefer M, Marques JB. Remote expert system of support the prostate cancer diagnosis. In: Conference proceedings of the annual international conference of the IEEE engineering in medicine and biology society. IEEE engineering in medicine and biology society. Conference, vol 5. 2004. p. 3412–5. [DOI] [PubMed]
- 26.Torshizi AD, et al. A hybrid fuzzy-ontology based intelligent system to determine level of severity and treatment recommendation for Benign Prostatic Hyperplasia. Comput Methods Programs Biomed. 2014;113(1):301–313. doi: 10.1016/j.cmpb.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 27.Binik YM, et al. Intelligent computer-based assessment and psychotherapy - an expert system for sexual dysfunction. J Nerv Ment Dis. 1988;176(7):387–400. doi: 10.1097/00005053-198807000-00001. [DOI] [PubMed] [Google Scholar]
- 28.Beligiannis G, et al. A GA driven intelligent system for medical diagnosis. In: Knowledge-based intelligent information and engineering systems, Pt 1, proceedings, vol 4251. 2006. p. 968–75.
- 29.Koutsojannis C, Hatzilygeroudis L. FESMI: a fuzzy expert system for diagnosis and treatment of male impotence. In: Knowledge-based intelligent information and engineering systems, Pt 2, proceedings, vol 3214. 2004. p. 1106–13.
- 30.Papageorgiou EI. Fuzzy cognitive map software tool for treatment management of uncomplicated urinary tract infection. Comput Methods Programs Biomed. 2012;105(3):233–245. doi: 10.1016/j.cmpb.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Arlen AM, et al. Computer model predicting breakthrough febrile urinary tract infection in children with primary vesicoureteral reflux. J Pediatr Urol. 2016;5:288 e1. doi: 10.1016/j.jpurol.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Goyal NK, et al. Prediction of biochemical failure in localized carcinoma of prostate after radical prostatectomy by neuro-fuzzy. Indian J Urol. 2007;23(1):14–17. doi: 10.4103/0970-1591.30258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronco AL, Fernandez R. Improving ultrasonographic diagnosis of prostate cancer with neural networks. Ultrasound Med Biol. 1999;25(5):729–733. doi: 10.1016/S0301-5629(99)00011-3. [DOI] [PubMed] [Google Scholar]
- 34.Babaian RJ, et al. Performance of a neural network in detecting prostate cancer in the prostate-specific antigen reflex range of 2.5 to 4.0 ng/ml. Urology. 2000;56(6):1000–1006. doi: 10.1016/S0090-4295(00)00830-X. [DOI] [PubMed] [Google Scholar]
- 35.Finne P, et al. Predicting the outcome of prostate biopsy in screen-positive men by a multilayer perceptron network. Urology. 2000;56(3):418–422. doi: 10.1016/S0090-4295(00)00672-5. [DOI] [PubMed] [Google Scholar]
- 36.Stephan C, et al. Multicenter evaluation of an artificial neural network to increase the prostate cancer detection rate and reduce unnecessary biopsies. Clin Chem. 2002;48(8):1279–1287. doi: 10.1093/clinchem/48.8.1279. [DOI] [PubMed] [Google Scholar]
- 37.Djavan B, et al. Novel artificial neural network for early detection of prostate cancer. J Clin Oncol. 2002;20(4):921–929. doi: 10.1200/JCO.2002.20.4.921. [DOI] [PubMed] [Google Scholar]
- 38.Remzi M, et al. An artificial neural network to predict the outcome of repeat prostate biopsies. Urology. 2003;62(3):456–460. doi: 10.1016/S0090-4295(03)00409-6. [DOI] [PubMed] [Google Scholar]
- 39.Kalra P, et al. A neurocomputational model for prostate carcinoma detection. Cancer. 2003;98(9):1849–1854. doi: 10.1002/cncr.11748. [DOI] [PubMed] [Google Scholar]
- 40.Saritas I, Allahverdi N, Sert IU. A fuzzy expert system design for diagnosis of prostate cancer. In Proceedings of the 4th international conference conference on Computer systems and technologies: e-Learning (CompSysTech '03). Association for Computing Machinery, New York, NY, USA, 345–351.
- 41.Matsui Y, et al. The use of artificial neural network analysis to improve the predictive accuracy of prostate biopsy in the Japanese population. Jpn J Clin Oncol. 2004;34(10):602–607. doi: 10.1093/jjco/hyh112. [DOI] [PubMed] [Google Scholar]
- 42.Porter CR, et al. Model to predict prostate biopsy outcome in large screening population with independent validation in referral setting. Urology. 2005;65(5):937–941. doi: 10.1016/j.urology.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 43.Lee HJ, et al. Role of transrectal ultrasonography in the prediction of prostate cancer—artificial neural network analysis. J Ultrasound Med. 2006;25(7):815–821. doi: 10.7863/jum.2006.25.7.815. [DOI] [PubMed] [Google Scholar]
- 44.Benecchi L. Neuro-fuzzy system for prostate cancer diagnosis. Urology. 2006;68(2):357–361. doi: 10.1016/j.urology.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 45.Stephan C, et al. Networks for prostate biopsy indication in two different patient Populations comparison of two different artificial neural. Urology. 2007;70(3):596–601. doi: 10.1016/j.urology.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Kawakami S, et al. Development, validation, and head-to-head comparison of logistic regression-based nomograms and artificial neural network models predicting prostate cancer on initial extended biopsy. Eur Urol. 2008;54(3):601–611. doi: 10.1016/j.eururo.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 47.Stephan C, et al. A -2 proPSA-based artificial neural network significantly improves differentiation between prostate cancer and benign prostatic diseases. Prostate. 2009;69(2):198–207. doi: 10.1002/pros.20872. [DOI] [PubMed] [Google Scholar]
- 48.Lee HJ, et al. Image-based clinical decision support for transrectal ultrasound in the diagnosis of prostate cancer: comparison of multiple logistic regression, artificial neural network, and support vector machine. Eur Radiol. 2010;20(6):1476–1484. doi: 10.1007/s00330-009-1686-x. [DOI] [PubMed] [Google Scholar]
- 49.Meijer RP, et al. The value of an artificial neural network in the decision-making for prostate biopsies. World J Urol. 2009;27(5):593–598. doi: 10.1007/s00345-009-0444-7. [DOI] [PubMed] [Google Scholar]
- 50.Saritas I, Ozkan IA, Sert IU. Prognosis of prostate cancer by artificial neural networks. Expert Syst Appl. 2010;37(9):6646–6650. doi: 10.1016/j.eswa.2010.03.056. [DOI] [Google Scholar]
- 51.Lawrentschuk N, et al. Predicting prostate biopsy outcome: artificial neural networks and polychotomous regression are equivalent models. Int Urol Nephrol. 2011;43(1):23–30. doi: 10.1007/s11255-010-9750-7. [DOI] [PubMed] [Google Scholar]
- 52.Ecke TH, et al. Outcome prediction for prostate cancer detection rate with artificial neural network (ANN) in daily routine. Urol Oncol Semin Orig Investig. 2012;30(2):139–144. doi: 10.1016/j.urolonc.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 53.Filella X, et al. The influence of prostate volume in prostate health index performance in patients with total PSA lower than 10 μg/L. Clin Chim Acta. 2014;436:303–307. doi: 10.1016/j.cca.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 54.Yuksel S, et al. Application of soft sets to diagnose the prostate cancer risk. J Inequalities Appl. 2013;2013::229. doi: 10.1186/1029-242X-2013-229. [DOI] [Google Scholar]
- 55.Samli MM, Dogan I. An artificial neural network for predicting the presence of spermatozoa in the testes of men with nonobstructive azoospermia. J Urol. 2004;171(6, Part 1):2354–2357. doi: 10.1097/01.ju.0000125272.03182.c3. [DOI] [PubMed] [Google Scholar]
- 56.Powell CR, et al. Computational models for detection of endocrinopathy in subfertile males. Int J Impot Res. 2007;20(1):79–84. doi: 10.1038/sj.ijir.3901593. [DOI] [PubMed] [Google Scholar]
- 57.Ramasamy R, et al. A comparison of models for predicting sperm retrieval before microdissection testicular sperm extraction in men with nonobstructive azoospermia. J Urol. 2013;189(2):638–642. doi: 10.1016/j.juro.2012.09.038. [DOI] [PubMed] [Google Scholar]
- 58.Paya AS, et al. Development of an artificial neural network for helping to diagnose diseases in urology. In Proceedings of the 1st international conference on Bio inspired models of network, information and computing systems (BIONETICS '06). Association for Computing Machinery, New York, NY, USA, 9–es.
- 59.Gil D, et al. Application of artificial neural networks in the diagnosis of urological dysfunctions. Expert Syst Appl. 2009;36(3):5754–5760. doi: 10.1016/j.eswa.2008.06.065. [DOI] [Google Scholar]
- 60.Wadie BS, et al. Application of artificial neural network in prediction of bladder outlet obstruction: a model based on objective, noninvasive parameters. Urology. 2006;68(6):1211–4. doi: 10.1016/j.urology.2006.08.1079. [DOI] [PubMed] [Google Scholar]
- 61.Wadie BS, Badawi AM, Ghoneim MA. The relationship of the international prostate symptom score and objective parameters for diagnosing bladder outlet obstruction. Part II: the potential usefulness of artificial neural networks. J Urol. 2001;165(1):35–37. doi: 10.1097/00005392-200101000-00009. [DOI] [PubMed] [Google Scholar]
- 62.Tewari A, Narayan P. Novel staging tool for localized prostate cancer: a pilot study using genetic adaptive neural networks. J Urol. 1998;160(2):430–436. doi: 10.1016/S0022-5347(01)62916-1. [DOI] [PubMed] [Google Scholar]
- 63.Chang PL, et al. Evaluation of a decision-support system for preoperative staging of prostate cancer. Med Decis Making. 1999;19(4):419–427. doi: 10.1177/0272989X9901900410. [DOI] [PubMed] [Google Scholar]
- 64.Batuello JT, et al. Artificial neural network model for the assessment of lymph node spread in patients with clinically localized prostate cancer. Urology. 2001;57(3):481–485. doi: 10.1016/S0090-4295(00)01039-6. [DOI] [PubMed] [Google Scholar]
- 65.Han M, et al. Evaluation of artificial neural networks for the prediction of pathologic stage in prostate carcinoma. Cancer. 2001;91(8):1661–1666. doi: 10.1002/1097-0142(20010415)91:8+<1661::AID-CNCR1180>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 66.Mattfeldt T, et al. Prediction of postoperative prostatic cancer stage on the basis of systematic biopsies using two types of artificial neural networks. Eur Urol. 2001;39(5):530–536. doi: 10.1159/000052499. [DOI] [PubMed] [Google Scholar]
- 67.Matsui Y, et al. Artificial neural network analysis for predicting pathological stage of clinically localized prostate cancer in the Japanese population. Jpn J Clin Oncol. 2002;32(12):530–535. doi: 10.1093/jjco/hyf114. [DOI] [PubMed] [Google Scholar]
- 68.Zlotta AR, et al. An artificial neural network for prostate cancer staging when serum prostate specific antigen is 10 NG./ML. or less. J Urol. 2003;169(5):1724–1728. doi: 10.1097/01.ju.0000062548.28015.f6. [DOI] [PubMed] [Google Scholar]
- 69.Chiu JS, et al. Artificial neural network to predict skeletal metastasis in patients with prostate cancer. J Med Syst. 2009;33(2):91–100. doi: 10.1007/s10916-008-9168-2. [DOI] [PubMed] [Google Scholar]
- 70.Kim SY, et al. Pre-operative prediction of advanced prostatic cancer using clinical decision support systems: accuracy comparison between support vector machine and artificial neural network. Korean J Radiol. 2011;12(5):588–594. doi: 10.3348/kjr.2011.12.5.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Regnier-Coudert O, et al. Machine learning for improved pathological staging of prostate cancer: a performance comparison on a range of classifiers. Artif Intell Med. 2012;55(1):25–35. doi: 10.1016/j.artmed.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 72.Veltri RW, et al. Comparison of logistic regression and neural net modeling for prediction of prostate cancer pathologic stage. Clin Chem. 2002;48(10):1828–1834. doi: 10.1093/clinchem/48.10.1828. [DOI] [PubMed] [Google Scholar]
- 73.Cosma G, et al. Prediction of pathological stage in patients with prostate cancer: a neuro-fuzzy model. PLoS ONE. 2016;11(6):e0155856. doi: 10.1371/journal.pone.0155856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moul JW, et al. Neural-network analysis of quantitative histological factors to predict pathological stage in clinical stage-I nonseminomatous testicular cancer. J Urol. 1995;153(5):1674–1677. doi: 10.1016/S0022-5347(01)67502-5. [DOI] [PubMed] [Google Scholar]
- 75.Poulakis V, et al. Prediction of clearance of inferior caliceal calculi with extracorporeal shock wave lithotripsy. Using an artificial neural network analysis. Urol A. 2002;41(6):583–595. doi: 10.1007/s00120-002-0194-2. [DOI] [PubMed] [Google Scholar]
- 76.Hamid A, et al. Artificial neural networks in predicting optimum renal stone fragmentation by extracorporeal shock wave lithotripsy: a preliminary study. BJU Int. 2003;91(9):821–824. doi: 10.1046/j.1464-410X.2003.04230.x. [DOI] [PubMed] [Google Scholar]
- 77.Gomha MA, et al. Can we improve the prediction of stone-free status after extracorporeal shock wave lithotripsy for ureteral stones? A neural network or a statistical model? J Urol. 2004;172(1):175–179. doi: 10.1097/01.ju.0000128646.20349.27. [DOI] [PubMed] [Google Scholar]
- 78.Michaels EK, et al. Use of a neural network to predict stone growth after shock wave lithotripsy. Urology. 1998;51(2):335–338. doi: 10.1016/S0090-4295(97)00611-0. [DOI] [PubMed] [Google Scholar]
- 79.Naguib RNG, et al. Neural network analysis of combined conventional and experimental prognostic markers in prostate cancer: a pilot study. Br J Cancer. 1998;78(2):246–250. doi: 10.1038/bjc.1998.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Potter SR, et al. Genetically engineered neural networks for predicting prostate cancer progression after radical prostatectomy. Urology. 1999;54(5):791–795. doi: 10.1016/S0090-4295(99)00328-3. [DOI] [PubMed] [Google Scholar]
- 81.Porter C, et al. Artificial neural network model to predict biochemical failure after radical prostatectomy. Mol Urol. 2001;5(4):159–162. doi: 10.1089/10915360152745830. [DOI] [PubMed] [Google Scholar]
- 82.Seker H, et al. A fuzzy logic based-method for prognostic decision making in breast and prostate cancers. IEEE Trans Inf Technol Biomed. 2003;7(2):114–122. doi: 10.1109/TITB.2003.811876. [DOI] [PubMed] [Google Scholar]
- 83.Poulakis V, et al. Preoperative neural network using combined magnetic resonance imaging variables, prostate-specific antigen, and Gleason score to predict positive surgical margins. Urology. 2004;64(3):516–521. doi: 10.1016/j.urology.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 84.Poulakis V, et al. Preoperative neural network using combined magnetic resonance imaging variables, prostate specific antigen and Gleason score to predict prostate cancer stage. J Urol. 2004;172(4):1306–1310. doi: 10.1097/01.ju.0000139881.04126.b6. [DOI] [PubMed] [Google Scholar]
- 85.de Paula Castanho MJ, et al. Fuzzy expert system: an example in prostate cancer. Appl Math Comput. 2008;202(1):78–85. [Google Scholar]
- 86.Botoca C, et al. Prediction of prostate capsule penetration using neural networks. In: Proceedings of the 8th Wseas international conference on computational intelligence, man-machine systems and cybernetics (Cimmacs '09). 2009. p. 108–11.
- 87.Castanho MJP, et al. Fuzzy expert system for predicting pathological stage of prostate cancer. Expert Syst Appl. 2013;40(2):466–470. doi: 10.1016/j.eswa.2012.07.046. [DOI] [Google Scholar]
- 88.Hu XH, et al. Risk prediction models for biochemical recurrence after radical prostatectomy using prostate-specific antigen and Gleason score. Asian J Androl. 2014;16(6):897–901. doi: 10.4103/1008-682X.129940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tewari A, et al. Genetic adaptive neural network to predict biochemical failure after radical prostatectomy: a multi-institutional study. Mol Urol. 2001;5(4):163–169. doi: 10.1089/10915360152745849. [DOI] [PubMed] [Google Scholar]
- 90.Borque A, et al. The use of neural networks and logistic regression analysis for predicting pathological stage in men undergoing radical prostatectomy: a population based study. J Urol. 2001;166(5):1672–1678. doi: 10.1016/S0022-5347(05)65651-0. [DOI] [PubMed] [Google Scholar]
- 91.Tsao CW, et al. Artificial neural network for predicting pathological stage of clinically localized prostate cancer in a Taiwanese population. J Chin Med Assoc. 2014;77(10):513–518. doi: 10.1016/j.jcma.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 92.Cummings JM, et al. Prediction of spontaneous ureteral calculous passage by an artificial neural network. J Urol. 2000;164(2):326–328. doi: 10.1016/S0022-5347(05)67351-X. [DOI] [PubMed] [Google Scholar]
- 93.Dal Moro F, et al. A novel approach for accurate prediction of spontaneous passage of ureteral stones: support vector machines. Kidney Int. 2006;69(1):157–160. doi: 10.1038/sj.ki.5000010. [DOI] [PubMed] [Google Scholar]
- 94.Sun CC, Chang P. Prediction of unexpected emergency room visit after extracorporeal shock wave lithotripsy for urolithiasis - an application of artificial neural network in hospital information system. AMIA Annu Symp Proc. 2006;2006:1113. [PMC free article] [PubMed]
- 95.Bagli DJ, et al. Artificial neural networks in pediatric urology: Prediction of sonographic outcome following pyeloplasty. J Urol. 1998;160(3):980–983. [PubMed] [Google Scholar]
- 96.Seçkiner I, et al. Use of artificial neural networks in the management of antenatally diagnosed ureteropelvic junction obstruction. Can Urol Assoc J. 2011;5(6):E152. doi: 10.5489/cuaj.10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parekattil SJ, et al. Multi-institutional validation study of neural networks to predict duration of stay after laparoscopic radical/simple or partial nephrectomy. J Urol. 2005;174(4):1380–1384. doi: 10.1097/01.ju.0000173921.67597.e8. [DOI] [PubMed] [Google Scholar]
- 98.Vukicevic AM, et al. Evolutionary assembled neural networks for making medical decisions with minimal regret: application for predicting advanced bladder cancer outcome. Expert Syst Appl. 2014;41(18):8092–8100. doi: 10.1016/j.eswa.2014.07.006. [DOI] [Google Scholar]
- 99.Serrano-Durba A, et al. The use of neural networks for predicting the result of endoscopic treatment for vesico-ureteric reflux. BJU Int. 2004;94(1):120–122. doi: 10.1111/j.1464-410X.2004.04912.x. [DOI] [PubMed] [Google Scholar]
- 100.Naguib RNG, Qureshi KN, Hamdy FC, Neal DE. Neural network analysis of prognostic markers in bladder cancer. In: Proceedings of the 19th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Magnificent Milestones and Emerging Opportunities in Medical Engineering (Cat. No.97CH36136), 1997, vol.3, pp. 1007–9.
- 101.Qureshi KN, et al. Neural network analysis of clinicopathological and molecular markers in bladder cancer. J Urol. 2000;163(2):630–633. doi: 10.1016/S0022-5347(05)67948-7. [DOI] [PubMed] [Google Scholar]
- 102.Fujikawa K, et al. Predicting disease outcome of non-invasive transitional cell carcinoma of the urinary bladder using an artificial neural network model: results of patient follow-up for 15 years or longer. Int J Urol. 2003;10(3):149–152. doi: 10.1046/j.1442-2042.2003.00589.x. [DOI] [PubMed] [Google Scholar]
- 103.Catto JWF, et al. Artificial intelligence in predicting bladder cancer outcome: a comparison of neuro-fuzzy modeling and artificial neural networks. Clin Cancer Res. 2003;9(11):4172–4177. [PubMed] [Google Scholar]
- 104.Abbod MF, et al. Artificial intelligence for the prediction of bladder cancer. Biomed Eng Appl Basis Commun. 2004;16(02):49–58. doi: 10.4015/S1016237204000098. [DOI] [Google Scholar]
- 105.Catto JWF, et al. Neuro-fuzzy modeling: an accurate and interpretable method for predicting bladder cancer progression. J Urol. 2006;175(2):474–479. doi: 10.1016/S0022-5347(05)00246-6. [DOI] [PubMed] [Google Scholar]
- 106.Cai T, et al. Artificial intelligences in urological practice: the key to success? Ann Oncol. 2007;18(3):604–U10. doi: 10.1093/annonc/mdl411. [DOI] [PubMed] [Google Scholar]
- 107.Bassi P, et al. Prognostic accuracy of an artificial neural network in patients undergoing radical cystectomy for bladder cancer: a comparison with logistic regression analysis. BJU Int. 2007;99(5):1007–1012. doi: 10.1111/j.1464-410X.2007.06755.x. [DOI] [PubMed] [Google Scholar]
- 108.Catto JWF, et al. Neurofuzzy Modeling to determine recurrence risk following radical cystectomy for nonmetastatic urothelial carcinoma of the bladder. Clin Cancer Res. 2009;15(9):3150–3155. doi: 10.1158/1078-0432.CCR-08-1960. [DOI] [PubMed] [Google Scholar]
- 109.El-Mekresh M, et al. Prediction of survival after radical cystectomy for invasive bladder carcinoma: risk group stratification, nomograms or artificial neural networks? J Urol. 2009;182(2):466–472. doi: 10.1016/j.juro.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 110.Kolasa M, et al. Application of artificial neural network to predict survival time for patients with bladder cancer. Comput Med Act. 2009;65:113–122. doi: 10.1007/978-3-642-04462-5_11. [DOI] [Google Scholar]
- 111.Buchner A, et al. Prediction of outcome in patients with urothelial carcinoma of the bladder following radical cystectomy using artificial neural networks. Ejso. 2013;39(4):372–379. doi: 10.1016/j.ejso.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 112.Wang G, et al. Prediction of mortality after radical cystectomy for bladder cancer by machine learning techniques. Comput Biol Med. 2015;63:124–132. doi: 10.1016/j.compbiomed.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 113.Cai T, et al. Artificial intelligence for predicting recurrence-free probability of non-invasive high-grade urothelial bladder cell carcinoma. Oncol Rep. 2007;18(4):959–64. [PubMed] [Google Scholar]
- 114.Buchner A, et al. Outcome assessment of patients with metastatic renal cell carcinoma under systemic therapy using artificial neural networks. Clin Genitourin Cancer. 2012;10(1):37–42. doi: 10.1016/j.clgc.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 115.Marszall MP, et al. ANN as a prognostic tool after treatment of non-seminoma testicular cancer. Cent Eur J Med. 2012;7(5):672–679. [Google Scholar]
- 116.Kuo R-J, et al. Application of a two-stage fuzzy neural network to a prostate cancer prognosis system. Artif Intell Med. 2015;63(2):119–133. doi: 10.1016/j.artmed.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 117.Tanthanuch M, Tanthanuch S. Prediction of upper urinary tract calculi using an artificial neural network. J Med Assoc Thai. 2004;87(5):515–8. [PubMed] [Google Scholar]
- 118.Cancer Research UK, https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bladder-cancer#heading-Zero. Accessed May 2021.
- 119.Herr HW, et al. Defining optimal therapy for muscle invasive bladder cancer. J Urol. 2007;177(2):437–443. doi: 10.1016/j.juro.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 120.von der Maase H, et al. Long-term-survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer (Retracted article See vol. 16, pg. 1481, 2011) J Clin Oncol. 2005;23(21):4602–4608. doi: 10.1200/JCO.2005.07.757. [DOI] [PubMed] [Google Scholar]
- 121.Krongrad A, et al. Predictors of general quality of life in patients with benign prostate hyperplasia or prostate cancer. J Urol. 1997;157(2):534–538. doi: 10.1016/S0022-5347(01)65195-4. [DOI] [PubMed] [Google Scholar]
- 122.Han M, et al. A neural network predicts progression for men with Gleason score 3+4 versus 4+3 tumors after radical prostatectomy. Urology. 2000;56(6):994–999. doi: 10.1016/S0090-4295(00)00815-3. [DOI] [PubMed] [Google Scholar]
- 123.Parekattil SJ, Fisher HAG, Kogan BA. Neural network using combined urine nuclear matrix protein-22, monocyte chemoattractant protein-1 and urinary intercellular adhesion molecule-1 to detect bladder cancer. J Urol. 2003;169(3):917–920. doi: 10.1097/01.ju.0000051322.60266.06. [DOI] [PubMed] [Google Scholar]
- 124.Djavan B, et al. Longitudinal study of men with mild symptoms of bladder outlet obstruction treated with watchful waiting for four years. Urology. 2004;64(6):1144–1148. doi: 10.1016/j.urology.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 125.Kshirsagar A, et al. Predicting hypogonadism in men based upon age, presence of erectile dysfunction, and depression. Int J Impot Res. 2006;18(1):47–51. doi: 10.1038/sj.ijir.3901369. [DOI] [PubMed] [Google Scholar]
- 126.Stephan C, et al. Clinical utility of human glandular kallikrein 2 within a neural network for prostate cancer detection. BJU Int. 2005;96(4):521–527. doi: 10.1111/j.1464-410X.2005.05677.x. [DOI] [PubMed] [Google Scholar]
- 127.Abbod MF, et al. Artificial intelligence technique for gene expression profiling of urinary bladder cancer. In: 2006 3rd international IEEE conference on intelligent systems. 2006.
- 128.Stephan C, et al. A (-5,-7) ProPSA based artificial neural network to detect prostate cancer. Eur Urol. 2006;50(5):1014–1020. doi: 10.1016/j.eururo.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 129.Stephan C, et al. Improved prostate cancer detection with a human kallikrein 11 and percentage free PSA-based artificial neural network. Biol Chem. 2006;387(6):801–805. doi: 10.1515/BC.2006.101. [DOI] [PubMed] [Google Scholar]
- 130.Stephan C, et al. An artificial neural network for five different assay systems of prostate-specific antigen in prostate cancer diagnostics. BJU Int. 2008;102(7):799–805. doi: 10.1111/j.1464-410X.2008.07765.x. [DOI] [PubMed] [Google Scholar]
- 131.Cinar M, et al. Early prostate cancer diagnosis by using artificial neural networks and support vector machines. Expert Syst Appl. 2009;36(3):6357–6361. doi: 10.1016/j.eswa.2008.08.010. [DOI] [Google Scholar]
- 132.Stephan C, et al. Internal validation of an artificial neural network for prostate biopsy outcome. Int J Urol. 2010;17(1):62–68. doi: 10.1111/j.1442-2042.2009.02417.x. [DOI] [PubMed] [Google Scholar]
- 133.Catto JWF, et al. The application of artificial intelligence to microarray data: identification of a novel gene signature to identify bladder cancer progression. Eur Urol. 2010;57(3):398–406. doi: 10.1016/j.eururo.2009.10.029. [DOI] [PubMed] [Google Scholar]
- 134.Serati M, et al. Urinary symptoms and urodynamic findings in women with pelvic organ prolapse: is there a correlation? results of an artificial neural network analysis. Eur Urol. 2011;60(2):253–260. doi: 10.1016/j.eururo.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 135.Gil D, et al. Predicting seminal quality with artificial intelligence methods. Expert Syst Appl. 2012;39(16):12564–12573. doi: 10.1016/j.eswa.2012.05.028. [DOI] [Google Scholar]
- 136.Girela JL, et al. Semen parameters can be predicted from environmental factors and lifestyle using artificial intelligence methods. Biol Reprod. 2013;88(4):99-1. doi: 10.1095/biolreprod.112.104653. [DOI] [PubMed] [Google Scholar]
- 137.Stephan C, et al. Multicenter evaluation of -2 proprostate-specific antigen and the prostate health index for detecting prostate cancer. Clin Chem. 2013;59(1):306–314. doi: 10.1373/clinchem.2012.195784. [DOI] [PubMed] [Google Scholar]
- 138.Cai T, et al. Clinical importance of lymph node density in predicting outcome of prostate cancer patients. J Surg Res. 2011;167(2):267–72. doi: 10.1016/j.jss.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 139.Kim M, et al. Factors influencing nonabsolute indications for surgery in patients with lower urinary tract symptoms suggestive of benign prostatic hyperplasia: analysis using causal Bayesian networks. Int Neurourol J. 2014;18(4):198–205. doi: 10.5213/inj.2014.18.4.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Green WJF, et al. KI67 and DLX2 predict increased risk of metastasis formation in prostate cancer-a targeted molecular approach. Br J Cancer. 2016;115(2):236–242. doi: 10.1038/bjc.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Logvinenko T, Chow JS, Nelson CP. Predictive value of specific ultrasound findings when used as a screening test for abnormalities on VCUG. J Pediatr Urol. 2015;11(4):176.e1–176.e7. doi: 10.1016/j.jpurol.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wells DM, Niederer J. A medical expert system approach using artificial neural networks for standardized treatment planning. Int J Radiat Oncol Biol Phys. 1998;41(1):173–182. doi: 10.1016/S0360-3016(98)00035-2. [DOI] [PubMed] [Google Scholar]
- 143.Loch T, et al. Artificial neural network analysis (ANNA) of prostatic transrectal ultrasound. Prostate. 1999;39(3):198–204. doi: 10.1002/(SICI)1097-0045(19990515)39:3<198::AID-PROS8>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 144.Mattfeldt T, et al. Prediction of prostatic cancer progression after radical prostatectomy using artificial neural networks: a feasibility study. BJU Int. 1999;84(3):316–323. doi: 10.1046/j.1464-410x.1999.00209.x. [DOI] [PubMed] [Google Scholar]
- 145.Llobet R, et al. Computer-aided detection of prostate cancer. Int J Med Inform. 2007;76(7):547–556. doi: 10.1016/j.ijmedinf.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 146.Hassanien AE, Al-Qaheri H, El-Dahshan ESA. Prostate boundary detection in ultrasound images using biologically-inspired spiking neural network. Appl Soft Comput. 2011;11(2):2035–2041. doi: 10.1016/j.asoc.2010.07.001. [DOI] [Google Scholar]
- 147.Matulewicz L, et al. Anatomic segmentation improves prostate cancer detection with artificial neural networks analysis of H-1 magnetic resonance spectroscopic imaging. J Magn Reson Imaging. 2014;40(6):1414–1421. doi: 10.1002/jmri.24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gatidis S, et al. Combined unsupervised–supervised classification of multiparametric PET/MRI data: application to prostate cancer. NMR Biomed. 2015;28(7):914–922. doi: 10.1002/nbm.3329. [DOI] [PubMed] [Google Scholar]
- 149.Pantazopoulos D, et al. Comparing neural networks in the discrimination of benign from malignant lower urinary tract lesions. Br J Urol. 1998;81(4):574–579. doi: 10.1046/j.1464-410x.1998.00587.x. [DOI] [PubMed] [Google Scholar]
- 150.Xiao D, et al. 3D detection and extraction of bladder tumors via MR virtual cystoscopy. Int J Comput Assist Radiol Surg. 2016;11(1):89–97. doi: 10.1007/s11548-015-1234-x. [DOI] [PubMed] [Google Scholar]
- 151.Hurst RE, et al. Neural net-based identification of cells expressing the p300 tumor-related antigen using fluorescence image analysis. Cytometry. 1997;27(1):36–42. doi: 10.1002/(SICI)1097-0320(19970101)27:1<36::AID-CYTO5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 152.Volmer M, et al. Artificial neural-network predictions of urinary calculus compositions analyzed with infrared-spectroscopy. Clin Chem. 1994;40(9):1692–1697. doi: 10.1093/clinchem/40.9.1692. [DOI] [PubMed] [Google Scholar]
- 153.Pantazopoulos D, et al. Back propagation neural network in the discrimination of benign from malignant lower urinary tract lesions. J Urol. 1998;159(5):1619–1623. doi: 10.1097/00005392-199805000-00057. [DOI] [PubMed] [Google Scholar]
- 154.Lamb DJ, Niederberger CS. Artificial-intelligence in medicine and male-infertility. World J Urol. 1993;11(2):129–136. doi: 10.1007/BF00182040. [DOI] [PubMed] [Google Scholar]
- 155.Holzinger A, et al. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip Rev Data Min Knowl Discov. 2019;9(4):e1312. doi: 10.1002/widm.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 157.Lakkaraju HKE, Caruana R, Leskovec J. Interpretable and explorable approximations of black box models. 2017. Arxiv 1707.01154.
- 158.Bologna G, Hayashi Y. Characterization of symbolic rules embedded in deep DIMLP networks: a challenge to transparency of deep learning. J Artif Intell Soft Comput Res. 2017;7(4):265–286. doi: 10.1515/jaiscr-2017-0019. [DOI] [Google Scholar]
- 159.Cabitza F, Zeitoun JD. The proof of the pudding: in praise of a culture of real-world validation for medical artificial intelligence. Ann Transl Med. 2019;7(8):161. doi: 10.21037/atm.2019.04.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nagendran M, et al. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ. 2020;368:m689. doi: 10.1136/bmj.m689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Moons KGM, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. 2019;170(1):W1–W33. doi: 10.7326/M18-1377. [DOI] [PubMed] [Google Scholar]
- 162.Cabitza F, Campagner A, Balsano C. Bridging the "last mile" gap between AI implementation and operation: "data awareness" that matters. Ann Transl Med. 2020;8(7):501. doi: 10.21037/atm.2020.03.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kattan MW, Cowen ME, Miles BJ. Computer modeling in urology. Urology. 1996;47(1):14–21. doi: 10.1016/S0090-4295(99)80375-6. [DOI] [PubMed] [Google Scholar]
- 164.Eminaga O, Liao JC. Chapter 16—prospect and adversity of artificial intelligence in urology. In: Xing L, Giger ML, Min JK, editors. Artificial intelligence in medicine. London: Academic Press; 2021. pp. 309–337. [Google Scholar]
- 165.Chang TC, et al. Current trends in artificial intelligence application for endourology and robotic surgery. Urol Clin N Am. 2021;48(1):151–160. doi: 10.1016/j.ucl.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 166.NICE. Prostate cancer: diagnosis and treatment CG175. National Institute for Health and Care Excellence. 2014
- 167.NICE. Prostate cancer: diagnosis and treatment CG28. 2008.
- 168.Eminaga O, et al. Diagnostic classification of cystoscopic images using deep convolutional neural networks. JCO Clin Cancer Inform. 2018;2:1–8. doi: 10.1200/CCI.17.00126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
For data and supporting materials access, please contact authors for data requests.