Abstract
Background
Intervertebral disc (IVD) degeneration is a major cause of low back pain (LBP). Following disc injury, nerve growth factor (NGF) concentrations rise in IVDs, and anti-NGF therapy has been shown to attenuate LBP in humans. Increased levels of tumor necrosis factor-α (TNF-α) and transforming growth factor-β (TGF-β) in degenerative IVDs and in in vitro studies suggest that these factors promote NGF production. However, whether these factors regulate NGF in vivo remains unclear. Thus, we studied NGF regulation in a mouse model of IVD injury.
Methods
After inducing IVD injury, we examined mRNA levels of Tnfa, Tgfb, and Ngf in IVDs from control and IVD-injured mice across 7 days. To do this, we used magnetic cell separation to isolate CD11b ( +) (macrophage-rich) and CD11b (-) (IVD cell-rich) cell fractions from injured IVDs. To study the effect of TNF-α on Ngf expression, we examined Ngf expression in injured IVDs from C57BL/6 J and Tnfa-knockout (KO) mice (C57BL/6 J background). To study the effect of TGF-β on Ngf expression, C57/BL6J mice were given an intraperitoneal injection of either the TGF-β inhibitor SB431542 or DMSO solution (vehicle) one and two days before harvesting IVDs.
Results
mRNA expression of Tnfa, Tgfb, and Ngf was significantly increased in injured IVDs. Tnfa was predominantly expressed in the CD11b ( +) fraction, and Tgfb in the CD11b (-) fraction. Ngf expression was comparable between CD11b ( +) and CD11b (-) fractions, and between wild-type and Tnfa-KO mice at post-injury day (PID) 1, 3, and 7. SB431542 suppressed TGF-β-mediated Ngf expression and NGF production in vitro. Further, administration of SB431542 significantly reduced Ngf expression in IVDs such that levels were below those observed in vehicle-treated animals at PID3 and PID7.
Conclusion
A TGF-β inhibitor reduced Ngf expression in a mouse model of IVD injury, suggesting that TGF-β may regulate NGF expression in vivo.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12891-021-04509-w.
Keywords: Transforming growth factor-β, Nerve growth factor, Intervertebral disc
Background
Persistent low back pain (LBP) is a common chronic ailment [1, 2]. A major cause of LBP is the degeneration of intervertebral discs (IVDs) [3, 4]. According to recent reports, the IVDs of animal disc injury models and humans with herniated IVDs show elevated levels of nerve growth factor (NGF) [5, 6]. Moreover, anti‐NGF therapy induces analgesia and shows efficacy in patients with chronic LBP [7–9]. The mechanisms governing NGF regulation, however, remain unclear.
Previous studies have suggested that tumor necrosis factor-α (TNF-α) and transforming growth factor-β (TGF-β) may be regulators of NGF. TNF-α is a major proinflammatory cytokine with powerful proinflammatory activity and the ability to promote secretion of a range of proinflammatory mediators. TNF-α expression is elevated in degenerated compared to non-degenerated IVDs [10, 11]. TNF-α has been shown to stimulate NGF in mouse and human IVD cells, including nucleus pulposus (NP) and anulus fibrosus (AF) cells, in vitro [6, 12]. Meanwhile, TGF-β may protect IVD tissue during restoration by promoting matrix synthesis and suppressing matrix catabolism, cell loss, and inflammatory response. However, over-activation of TGF-β is harmful to IVDs, with high levels of TGF-β observed in degenerated IVDs in mice and humans [6, 13]. We previously reported that TGF-β stimulates NGF production in mouse IVD cells in vitro [6]. Accumulating evidence from in vitro studies thus suggests that TNF-α and TGF-β contribute to the regulation of NGF during IVD degeneration. However, whether TNF-α and TGF-β also regulate NGF expression in vivo remains unclear.
Here, we investigated the role of TNF-α and TGF-β in relation to NGF expression in a mouse IVD puncture model in vivo.
Methods
Animals
Male C57BL/6 J mice and Tnfa-knockout (KO) mice (C57B/6 J background) aged nine weeks were kept in an animal housing system maintained at 23 °C ± 2 °C and 55% ± 10% humidity under a 12‐hour light/dark cycle for the duration of the study. The experimental protocol was approved by the Kitasato University School of Medicine Animal Care Committee (reference number: 2020–089). The study was conducted according to the ARRIVE guidelines for the reporting of animal experiments. All methods complied with the guidelines for the proper conduct of animal experiments of the Science Council of Japan.
Expression of Tnfa, Tgfb, and Ngf in an IVD injury mouse model
Tnfa, Tgfb, and Ngf expression was examined in 40 C57BL/6 J mice following IVD injury. Ten mice were randomly chosen to form the control group, while the remaining 30 mice formed the IVD injury group. After receiving anesthesia comprising isoflurane followed by an intramuscular injection of a 1:3:1 mixture of midazolam (Product no. KB6762, Sando, Yamagata, Japan), Domitor (medetomidine hydrochloride; Product no. 005130, Orion Corporation, Espoo, Finland), and Vetorphale (butorphanol tartrate; Product no. VETLI5, Meiji Seika Pharma Co., Ltd., Tokyo, Japan) into the upper limbs (0.05 ml/100 g body weight), mice in the IVD injury group were subjected to a puncture injury, where a 27-gauge needle was used to puncture IVDs (coccygeal discs 5–6 and 6–7) 10 times. Control mice were subjected to all parts of the surgery except for the puncture injury. Tnfa, Tgfb, and Ngf expression was subsequently examined on post-injury days 0 (PID0), 1 (PID1), 3 (PID3), and 7 (PID7) using quantitative polymerase chain reaction (qPCR; n = 10 per time point). NP and AF tissues were not separated for analysis.
qPCR
IVDs were harvested and immediately homogenized in TRIzol (Product no. 15596026, Invitrogen, Carlsbad, CA) on ice; the digested sample was used as the template for first‐strand complementary DNA (cDNA) synthesis using SuperScript™ III RT (Product no. 18080085, Invitrogen). PCR reactions (25 μl) comprised 2 μl cDNA, a specific primer set (0.2 μM final concentration), and 12.5 μl SYBR Premix Ex Taq (Product no. RR820, Takara, Kyoto, Japan). Table 1 lists the primers used in this study. PCR product size was confirmed by gel electrophoresis using cDNA extracted from intact IVDs (Supplementary Figure 1). qPCR using a RT‐PCR Detection System (CFX‐96; Bio‐Rad, Hercules, CA) was performed at 95 °C for 1 min, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. mRNA expression of the genes of interest was normalized to Gapdh expression. Relative expression was calculated based on the mean value from control samples (non‐injured discs from the control group or vehicle‐treated disc cells in vitro).
Table 1.
Primer sequences
| Primer | Sequence (5'–3') | Product size (bp) |
|---|---|---|
| Tnfa-F | TGGCAATTCAGGAGAGGCAG | 109 |
| Tnfa-R | AGTGGTTGGAGAAACAGGCA | |
| Tgfb-F | CTCCCGTGGCTTCTAGTGC | 133 |
| Tgfb-R | GCCTTAGTTTGGACAGGATCTG | |
| Ngf-F | ATGGTGGAGTTTTGGCCTGT | 192 |
| Ngf-R | GTACGCCGATCAAAAACGCA | |
| F4/80-F | TGGGATGTACAGATGGGGGA | 189 |
| F4/80-R | CCTGGGCCTTGAAAGTTGGT | |
| Cd11b-F | CTGGCTTTAGACCCTGTCCG | 138 |
| Cd11b-R | GTCCACGCAGTCCGGTAAAA | |
| Col2a1-F | GGAGAGACCATGAACGGTGG | 78 |
| Col2a1-R | CATCTGGACGTTAGCGGTGT | |
| Gapdh-F | AACTTTGGCATTGTGGAAGG | 223 |
| Gapdh-R | ACACATTGGGGGTAGGAACA |
Tnfa, Tgfb and Ngf expression following IVD injury
We previously observed increased Tnfa, Tgfb and Ngf expression levels at PID7 in wild-type mice [6]. We also confirmed that > 90% of CD11b ( +) cells were F4/80high cells (macrophages) at PID7 (Supplementary Figure 2). Therefore, we examined Tnfa, Tgfb and Ngf expression in CD11b ( +) cells (macrophages) and IVD cells isolated from IVDs on PID7. IVDs (n = 5) were digested in 0.25 mg/ml collagenase overnight at 37 °C. Thereafter, cells were passed through a cell strainer and then incubated with biotin-conjugated anti-CD11b antibody (clone M1/70, Product no.101204, BioLegend, CA, USA) for 30 min at 4 °C. The cells were washed twice in PBS before incubating with streptavidin-conjugated magnetic beads (Product no. 557812, BD Biosciences, San Diego, CA, USA) for 30 min at 4 °C. The cells were transferred to a magnetic board (Product no. 552311, BD Biosciences) and incubated for 6 min at room temperature. Unbound CD11b (-) cells (disc cells) were collected before removing the remaining cells from the magnetic board and collecting bound CD11b ( +) cells (macrophages). CD11b (-) and ( +) cells were centrifuged at 300 g for 5 min and dissolved in TRIzol. After cDNA synthesis, as described above, a 10 μl reaction mixture containing SuperMix, a reagent from Perfecta Preamp SuperMix (Product no. 95146–040, Quanta Biosciences, MA, USA), 5 μl primer pool (500 nM forward and 500 nM reverse), and 35 μl cDNA was prepared for preamplification PCR for 3 min at 95 °C, followed by 14 cycles of 15 s at 95 °C and 3 min at 60 °C. Pre-amplification PCR products were immediately diluted (1:16) and used for qPCR. Successful isolation of CD11b cells was determined based on F4/80 and Cd11b mRNA expression in each fraction.
Generation of Tnfa KO mice
We designed two guide RNAs (gRNA 5, 6) in exon 1 and exon 4, comprising 20-nucleotide sequences targeting exons 1–4 of the Tnfa gene (5′- GGAGGGAGATGTGGCGCCTT-3′ and 5′- GAGTCCGGGCAGGTCTACTT-3′). Constant regions of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) were synthesized by Thermo Fisher Scientific (Carlsbad, CA, USA). Electroporation into fertilized C57BL/6 J zygotes was performed based on a previously reported procedure [14]. After incubation of the zygotes, surviving two-cell-stage embryos were transferred to the oviducts of pseudo-pregnant female mice.
Genotyping
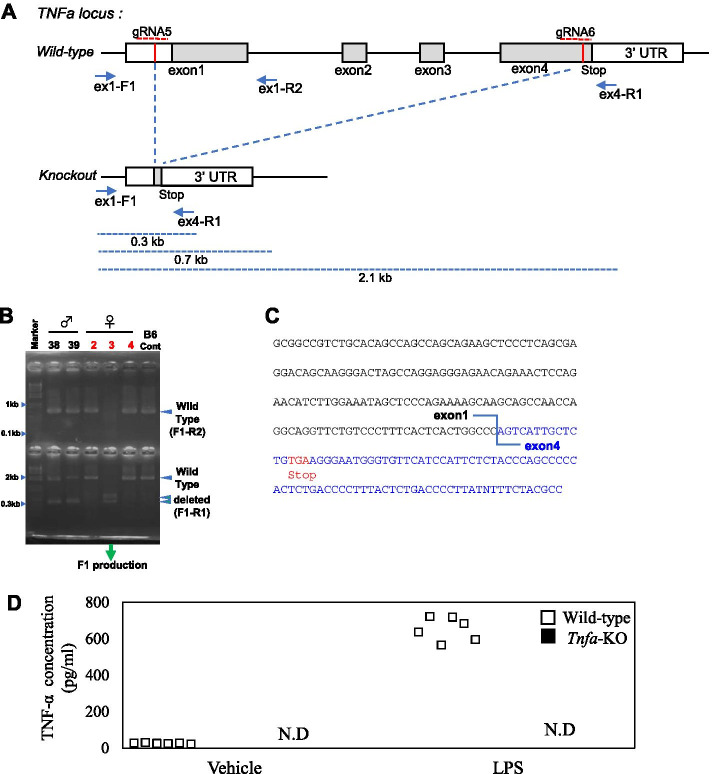
To assess CRISPR/Cas9-mediated deletion of the Tnfa gene, we performed PCR on tissue-extracted genomic DNA using specific primer sets for Tnfa KO mice (ex1-F1: 5′-TTCCTTGATGCCTGGGTGTC-3′, ex1-R2: 5′-TCCGAGGTCCTGACTCTGTCC-3′, and ex4-R1 5′-GTTAGAAGGATACAGACTGGG-3′). PCR was performed using Ex-Taq DNA polymerase (Product no. RR01, Takara Bio, Shiga, Japan) as follows: 35 cycles at 94 °C for 20 s, 61 °C for 10 s, and 72 °C for 20 s. The size of the deleted allele is 0.3 Kbp and that of the wild-type fragments are 0.7 Kbp and 2.1 Kbp (Fig. 1A, B).
Fig. 1.
Generation of Tnfa knockout mice. A Schematic showing the structure and map of the mouse Tnfa locus. The location of the two gRNAs and primer sequences for polymerase chain reaction (PCR) are shown. B PCR screen for Tnfa-KO founder mice. C The gene sequence at the junction of the deleted alleles. D Confirmation of loss of TNF-α protein by ELISA (n = 6)
ELISA
We confirmed the absence of TNF-α protein in Tnfa-KO mice using ELISA. Bone marrow macrophages (BMMs) were harvested from wild-type and Tnfa-KO mice and exposed to 1 μg/ml lipopolysaccharide (LPS; Product no. ALX-581–017-L002, Enzo Life Sciences, Farmingdale, NY) for 24 h (n = 6). Concentrations of TNF-α in cell supernatants were determined using a commercial TNF-α ELISA kit (Product no. 430907, BioLegend).
Effect of Tnfa deficiency on Ngf expression in IVD injury
Forty C57BL/6 J and Tnfa-KO mice were used to study the role of TNF-α in NGF expression. Ten mice were randomly selected to form the control group, while the remaining 30 formed the IVD injury group. IVD injury was induced as described above. Ngf expression were determined using qPCR (n = 10 per time point) on PID0, PID1, PID3, and PID7.
Effect of TGF‐β inhibitor, SB431542, on NGF expression and production in IVD cell culture
To determine whether the TGF‐β inhibitor, SB431542, inhibits Ngf expression and NGF production in IVD cells, we examined the effect of SB431542 on TGF‐β-mediated Ngf expression and NGF production by IVD cells isolated from five mice. IVD cells were isolated using collagenase digestion, as described above. Disc cells were subsequently incubated in α‐minimal essential media (α‐MEM) with 10% fetal bovine serum in six‐well plates. One week later, IVD cells were stimulated with α‐MEM (vehicle), 10 ng/ml recombinant TGF‐β (Product no. 7666-MB, R&D Systems, Minneapolis, MN, USA), or 10 ng/ml recombinant TGF‐β + 1 µM SB431542 (Product no. S4317, Sigma Aldrich, St Louis, MO, USA) for 6 and 24 h. Thereafter, total mRNA was extracted and analyzed using qPCR. NGF concentration in the cell supernatant was determined using a commercial NGF ELISA kit (Product No. DY556, R&D Systems).
Effect of a TGF-β inhibitor in IVD injury model mice
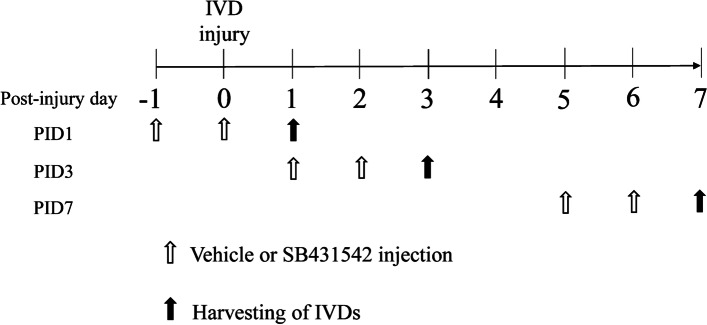
The 60 mice that received the IVD injury described above were randomly assigned to two equal groups: vehicle and treatment groups. The 30 mice in the treatment group received an intraperitoneal (IP) injection of 10 mg/kg SB431542 in 5% DMSO solution (SB43152) 1 and 2 days before harvesting IVDs, and the remaining 30 in the vehicle group received an IP injection of 5% DMSO solution (vehicle) at same time points (Fig. 2). Dosage was chosen according to the optimal inhibitory effect of an IP injection of a 10 mg/kg dose, as reported previously [13, 15]. IVDs were harvested at PID1, PID3, and PID7 and subjected to qPCR analysis (n = 10 for each time point).
Fig. 2.
Administration scheme of transforming growth factor-β (TGF-β) inhibitor. SB431542 or DMSO (vehicle) was intraperitoneally injected into mice with intervertebral disc injury 1 and 2 days before harvesting intervertebral discs at post-injury day 1, 3, and 7
Statistical analysis
Statistical analyses were performed using SPSS software version 11.0 (SPSS Inc., Chicago, IL). Gene and protein expression was analyzed using one‐way analysis of variance followed by Bonferroni’s post hoc multiple comparisons test. After performing a Kolmogorov–Smirnov test, a Wilcoxon signed rank test or paired t-test was used to compare differences between CD11b ( +) and CD11b (-) fractions. Moreover, following a Kolmogorov–Smirnov test, t-test or a Mann–Whitney U test was used to compare differences between wild-type and Tnfa-KO mice or vehicle- and TGF-β inhibitor-treated groups at each time point. P < 0.05 was considered significant.
Results
Tnfa, Tgfb, and Ngf expression after IVD injury
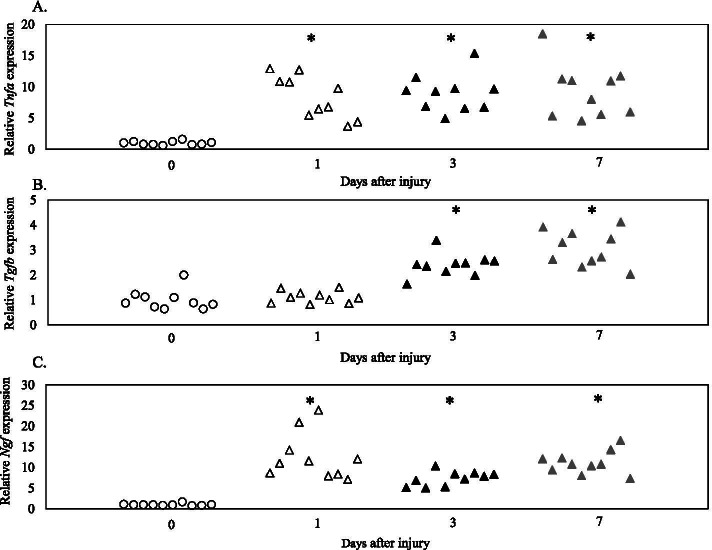
There was a significant increase in Tnfa expression at PID1, 3 and 7 (PID1, P < 0.001; PID3, P < 0.001; PID7, P < 0.001; Fig. 3A) and Tgfb expression at PID3 and 7 (PID1, P = 1.000; PID3, P < 0.001; PID7, P < 0.001; Fig. 3B) in wild-type mice. Similarly, Ngf expression was significantly increased at PID1, 3 and 7 (PID1, P < 0.001; PID3, P = 0.001; PID7, P < 0.001; Fig. 3C) in wild-type mice.
Fig. 3.
Tnfa, Tgfb, Ngf expression following IVD injury. Expression of Tnfa (A), Tgfb (B), and Ngf (C) at post-injury day 0, 1, 3, and 7 (n = 10 for each time point). *P < 0.05 compared to day 0 (control)
Tnfa, Tgfb and Ngf expression in IVD cells
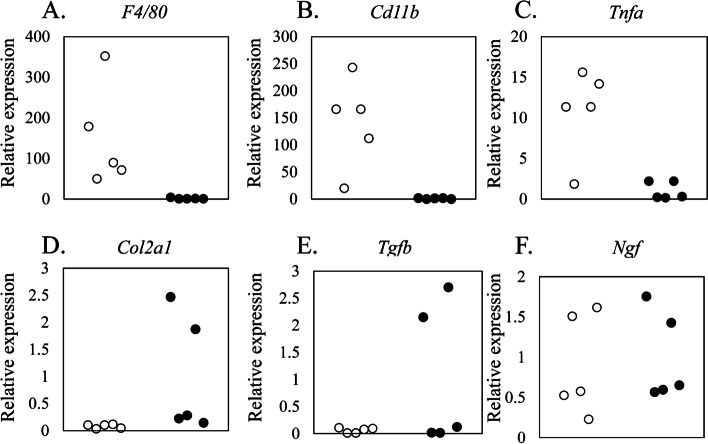
Between CD11b ( +) and (-) cells, Cd11b, F4/80, and Tnfa were predominantly expressed in CD11b ( +) cells (F4/80, P = 0.018; Cd11b, P = 0.018; Tnfa, P = 0.026; Fig. 4A–C). In contrast, Col2a1 and Tgfb were predominantly expressed in CD11b (-) cells (Col2a1, P = 0.026; Tgfb, P = 0.026; Fig. 4D–E). Meanwhile, Ngf expression was comparable between CD11b ( +) and CD11b (-) cells (P = 0.459; Fig. 4F).
Fig. 4.
Gene expression in CD11b-positive and -negative cells following IVD injury. Expression of F4/80 (A), Cd11b (B), Tnfa (C) Col21a (D), Tgfb (E), and Ngf (F) in CD11b ( +) and CD11b (-) cell fractions 7 days after IVD injury (n = 5). *P < 0.05
Effect of Tnfa deficiency on Ngf expression following IVD injury
To knock out the Tnfa gene, we used CRISPR/Cas9 to design two gRNAs to delete exons 1–4 (Fig. 1A). This method led to the successful generation of three independent deletion mutant mouse founders (Fig. 1B). From these, we established an F1 population by crossing with C57BL/6 J mice and checking the genotype of the offspring. We confirmed that all offspring had deletions in exons 1–4 (Fig. 1C). LPS stimulation of BMMs derived from wild-type mice significantly increased supernatant TNF-α levels (P < 0.001; Fig. 1D). However, TNF-α protein levels in vehicle- and LPS-stimulated BMMs derived from Tnfa-KO mice remained below the detection limit (< 7.8 pg/ml). To evaluate the effect of Tnfa on Ngf expression, we compared Ngf expression after IVD injury in wild-type and Tnfa-KO mice. Ngf expression was comparable between wild-type and Tnfa-KO mice at all time points examined (PID1, P = 0.842; PID3, P = 0.578; PID7, P = 0.053; Fig. 5).
Fig. 5.
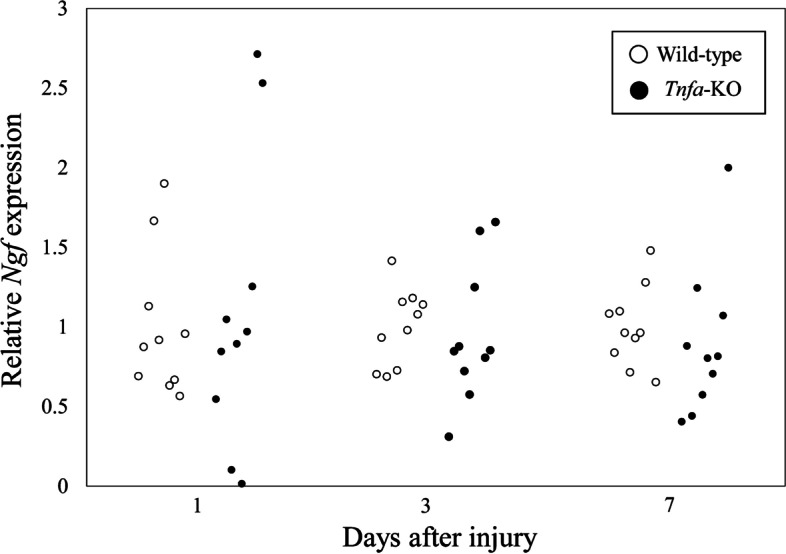
Effect of Tnfa deficiency on Ngf expression in vivo. Expression of Ngf 1, 3, and 7 days after IVD injury in wild-type (C57BL/6 J) and Tnfa-KO mice (n = 10 for each time point). Values represent mean ± standard error. *P < 0.05 compared to wild-type at the same time point. Relative Ngf expression was calculated based on Ngf expression in wild-type mice at each time point
Effect of a TGF-β inhibitor on IVD cell culture
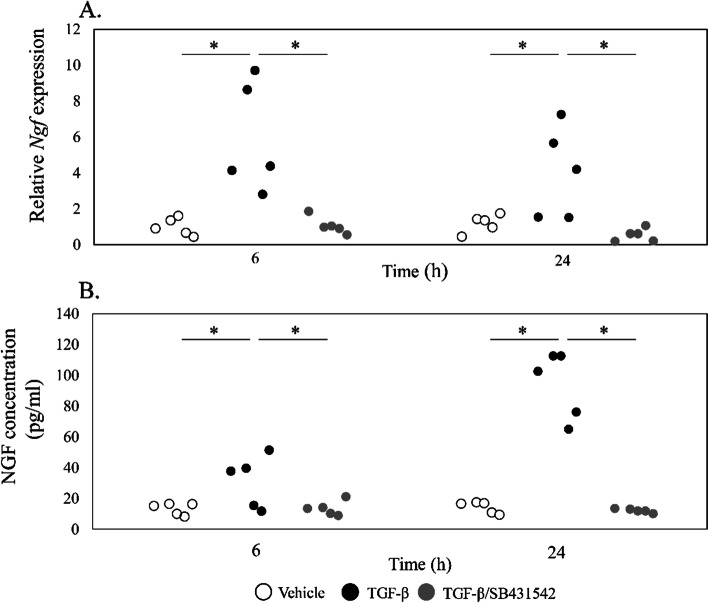
Stimulation of IVD cells with TGF-β for 6 and 24 h significantly increased Ngf mRNA expression (6 h, P = 0.013; 24 h, P = 0.038; Fig. 6A), and addition of the TGF-β inhibitor SB431542 completely reversed this effect (6 h, P = 0.025; 24 h, P = 0.040; Fig. 6A). Likewise, stimulation with TGF-β for 6 and 24 h increased supernatant NGF protein levels compared to vehicle (6 h, P = 0.049; 24 h, P = 0.001; Fig. 6B), and addition of SB431542 completely reversed this effect (6 h, P = 0.036; 24 h, P = 0.002; Fig. 6B).
Fig. 6.
Effect of TGF-β and a TGF-β inhibitor on Ngf expression and NGF production in vitro. A RT-PCR for Ngf. Disc cells were incubated with α-MEM control, TGF-β, or TGF-β/SB431542 for 6 and 24 h. Relative expression was calculated based on expression in control samples. B ELISA for NGF. Disc cells were incubated with α-MEM control, TGF-β, or TGF-β/SB431542 for 6 and 24 h. *P < 0.05 compared to control
Effect of a TGF-β inhibitor in an IVD injury model
Injection of the TGF-β inhibitor SB431542 into mice at PID1 produced no difference in Ngf expression compared to the vehicle-treated group (P = 0.354; Fig. 7). In contrast, injection of SB431542 into mice at PID3 and PID7 led to a significant reduction in Ngf expression compared to the vehicle-treated group (P = 0.038, P = 0.0001, respectively).
Fig. 7.
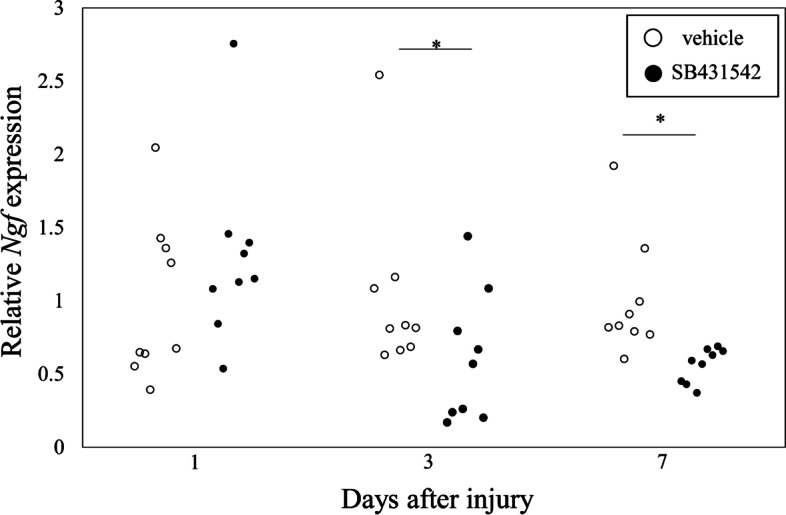
Effect of a TGF-β inhibitor on Ngf expression in vivo. SB431542 or DMSO (vehicle) was intraperitoneally injected into mice with intervertebral disc injury 1 and 2 days before harvesting intervertebral discs at post-injury day 1, 3, and 7 (n = 10 for each time point). *P < 0.05 compared to vehicle. Relative Ngf expression was calculated based on Ngf expression in the vehicle-treated group at each time point
Discussion
Several studies have generated IVD injury models by performing tail puncture under varying conditions [6, 16–22]. A model in which puncture injury was induced using a 31-gauge needle showed a gradual increase in degenerative score and decrease in glycosaminoglycan levels in IVDs at 6 and 12 weeks post-puncture [22]. A model in which puncture injury was induced using a 26-gauge needle displayed lower disc height 8 weeks post-puncture than that in the control group, while a model generated using a 29-gauge needle showed comparable disc height [17]. We generated an IVD injury model by performing 10 punctures with a 27-gauge needle and found that this model was useful for evaluating increased inflammatory cytokine and growth factor levels, and macrophage recruitment across a short time period of 1 to 28 days. [6, 16, 18, 19] Therefore, we used this model to evaluate the effects of TNF-α and TGF-β on NGF expression.
A number of reports have demonstrated that NGF levels rise locally at sites of inflammation [23–26]. TNF-α is a major factor in IVD inflammation [11], and has been shown to promote NGF expression and production in mouse IVD cells [6] and human AF and NP cells in vitro [12]. Therefore, TNF-α is thought to regulate NGF in IVD injury. However, we found that although Tnfa and Ngf expression was immediately elevated in injured IVDs in wild-type mice at PID1, Tnfa deficiency did not alter Ngf expression in injured IVDs. In our IVD puncture model, Tnfa was predominantly expressed in macrophages in injured IVDs. We previously reported that depletion of macrophages using clodronate liposomes in a mouse IVD injury model, generated using 10 punctures with a 27-gauge needle, reduced Tnfa, but not Ngf expression at PID1 [18]. In addition, while we also found that exposure of human synovial fibroblasts and macrophages to recombinant TNF-α led to elevated NGF expression and NGF production in vitro, NGF levels did not correlate with TNFA expression levels in human synovial tissue [27]. Thus, while TNF-α levels may rise in IVDs as a result of an increase in the macrophage population, elevated TNF-α levels may not play a major role in regulating NGF expression after IVD injury.
Previous studies have reported elevated levels of TGF-β in degenerated IVDs and in chondrocyte-like cells compared to fibroblastic cells in IVDs [6, 28]. However, TGF-β can also be produced by macrophages under pathogenic conditions [29, 30]. Macrophages in the kidneys of a kidney injury mouse model express higher levels of TGF‐β in the recovery phase [29]. Similarly, macrophages express TGF‐β in an adriamycin-induced nephrosis mouse model [30]. We compared TGF-β expression in disc cells and macrophages isolated from IVDs and found that TGF-β was predominantly expressed in IVD cells. Further, TGF-β stimulation increased NGF production by IVD cells in vitro and, administration of a TGF-β inhibitor reduced Ngf expression in vivo in an IVD injury mouse model. Our results indicate that TGF-β regulates NGF during IVD degeneration and that changes in Ngf expression may be due to increased autocrine/paracrine activity of IVD cell-derived TGF-β.
There were several limitations in this study. First, neither the TGF-β inhibitor nor Tnfa deficiency suppressed elevated Ngf expression on PID1, suggesting there may be other regulators of NGF in the acute phase. Further investigation is needed to identify these other NGF regulators. Second, previous studies have reported that Tnfa-KO mice do not exhibit an obvious phenotype due to functional redundancy [31] or rewiring of genetic networks [32]. Further investigation is needed to determine the precise mechanisms underlying TNF-α-mediated regulation of Ngf expression in vivo. Third, as we performed 10 needle punctures to induce major injury, other cells besides macrophages (such as endothelial cells) may have migrated into IVDs from the surrounding tissue, and contributed to the observed effects. Finally, we could not separate NP and AF cells due to the major injury induced as a result the 10 needle punctures.
Conclusion
A TGF-β inhibitor reduced Ngf expression in an IVD injury mouse model, suggesting that TGF-β may regulate NGF expression in vivo.
Supplementary Information
Acknowledgements
We are grateful to Mai Matsumoto, Kyoko Muneshige, Yuko Onuki, and Akiko Nakayama for their helpful support during the study.
Abbreviations
- IVD
Intervertebral disc
- TNF-α
Tumor necrosis factor-α
- TGF-β
Transforming growth factor-beta
- LBP
Low back pain
- NGF
Nerve growth factor
- PCR
Polymerase chain reaction
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
Authors’ contributions
YY, KU, AK, and MN contributed to the conception and design of the study. YY and KU wrote the manuscript. YY, KU, AK, MN, TO, MM, GI, MI, HS, and MT participated in data collection and performed statistical analysis. YY and KU edited the manuscript. YY, KU and MT supervised the study. All authors read and approved the final manuscript.
Funding
This investigation was supported in part by Grant-in-Aid for Research Activity Start-up Grant No. 20K22984.
Availability of data and materials
Datasets supporting the conclusions of this article are included within the article. The raw data can be requested from the corresponding author. Data are submitted to DDBJ Sequence Read Archive under accession number LC636331.
Declarations
Ethics approval and consent to participate
The experimental protocol was approved by the Kitasato University School of Medicine Animal Care Committee (reference number: 2020–089). The study was performed according to the ARRIVE guidelines for the reporting of animal experiments. All methods complied with the guidelines for the proper conduct of animal experiments of the Science Council of Japan.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuji Yokozeki, Email: yuji0328yoko@yahoo.co.jp.
Kentaro Uchida, Email: kuchida@med.kitasato-u.ac.jp.
Ayumu Kawakubo, Email: ayumukawakubo0827@gmail.com.
Mitsufumi Nakawaki, Email: mikkun.nakaki.81@gmail.com.
Tadashi Okubo, Email: okubo@med.kitasato-u.ac.jp.
Masayuki Miyagi, Email: masayuki008@aol.com.
Gen Inoue, Email: ginoue@kitasato-u.ac.jp.
Makoto Itakura, Email: mitakura@med.kitasato-u.ac.jp.
Hiroyuki Sekiguchi, Email: guccyon_s@yahoo.co.jp.
Masashi Takaso, Email: mtakaso@kitasato-u.ac.jp.
References
- 1.Simon CB, Hicks GE. Paradigm Shift in Geriatric Low Back Pain Management: Integrating Influences, Experiences, and Consequences. Phys Ther. 2018;98(5):434–446. doi: 10.1093/ptj/pzy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong AY, Karppinen J, Samartzis D. Low back pain in older adults: risk factors, management options and future directions. Scoliosis Spinal Disord. 2017;12:14. doi: 10.1186/s13013-017-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manchikanti L, Hirsch JA. An update on the management of chronic lumbar discogenic pain. Pain Manag. 2015;5(5):373–386. doi: 10.2217/pmt.15.33. [DOI] [PubMed] [Google Scholar]
- 4.Simon J, McAuliffe M, Shamim F, Vuong N, Tahaei A. Discogenic low back pain. Phys Med Rehabil Clin N Am. 2014;25(2):305–317. doi: 10.1016/j.pmr.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Aoki Y, Nakajima A, Ohtori S, Takahashi H, Watanabe F, Sonobe M, Terajima F, Saito M, Takahashi K, Toyone T, et al. Increase of nerve growth factor levels in the human herniated intervertebral disc: can annular rupture trigger discogenic back pain? Arthritis Res Ther. 2014;16(4):R159. doi: 10.1186/ar4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakawaki M, Uchida K, Miyagi M, Inoue G, Kawakubo A, Satoh M, Takaso M. Changes in Nerve Growth Factor Expression and Macrophage Phenotype Following Intervertebral Disc Injury in Mice. J Orthop Res. 2019;37(8):1798–1804. doi: 10.1002/jor.24308. [DOI] [PubMed] [Google Scholar]
- 7.Patel F, Hess DK, Maher DP. Anti-nerve growth factor antibodies for the treatment of low back pain. Expert Rev Clin Pharmacol. 2020;13(6):631–639. doi: 10.1080/17512433.2020.1772052. [DOI] [PubMed] [Google Scholar]
- 8.Schmelz M, Mantyh P, Malfait AM, Farrar J, Yaksh T, Tive L, Viktrup L. Nerve growth factor antibody for the treatment of osteoarthritis pain and chronic low-back pain: mechanism of action in the context of efficacy and safety. Pain. 2019;160(10):2210–2220. doi: 10.1097/j.pain.0000000000001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wise BL, Seidel MF, Lane NE. The evolution of nerve growth factor inhibition in clinical medicine. Nat Rev Rheumatol. 2021;17(1):34–46. doi: 10.1038/s41584-020-00528-4. [DOI] [PubMed] [Google Scholar]
- 10.Hiyama A, Hiraishi S, Sakai D, Mochida J. CCAAT/enhancer binding protein beta regulates the expression of tumor necrosis factor-alpha in the nucleus pulposus cells. J Orthop Res. 2016;34(5):865–875. doi: 10.1002/jor.23085. [DOI] [PubMed] [Google Scholar]
- 11.Le Maitre CL, Hoyland JA, Freemont AJ. Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1beta and TNFalpha expression profile. Arthritis Res Ther. 2007;9(4):R77. doi: 10.1186/ar2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abe Y, Akeda K, An HS, Aoki Y, Pichika R, Muehleman C, Kimura T, Masuda K. Proinflammatory cytokines stimulate the expression of nerve growth factor by human intervertebral disc cells. Spine (Phila Pa 1976) 2007;32(6):635–642. doi: 10.1097/01.brs.0000257556.90850.53. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, Liu S, Ma K, Zhao L, Lin H, Shao Z. TGF-beta signaling in intervertebral disc health and disease. Osteoarthritis Cartilage. 2019;27(8):1109–1117. doi: 10.1016/j.joca.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Okubo T, Hara K, Azuma S, Takada S. Effect of retinoic acid signaling on Ripply3 expression and pharyngeal arch morphogenesis in mouse embryos. Dev Dyn. 2021;250(7):1036–50. [DOI] [PubMed]
- 15.Lemos DR, Babaeijandaghi F, Low M, Chang CK, Lee ST, Fiore D, Zhang RH, Natarajan A, Nedospasov SA, Rossi FM. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat Med. 2015;21(7):786–794. doi: 10.1038/nm.3869. [DOI] [PubMed] [Google Scholar]
- 16.Kawakubo A, Uchida K, Miyagi M, Nakawaki M, Satoh M, Sekiguchi H, Yokozeki Y, Inoue G, Takaso M. Investigation of resident and recruited macrophages following disc injury in mice. J Orthop Res. 2020;38(8):1703–1709. doi: 10.1002/jor.24590. [DOI] [PubMed] [Google Scholar]
- 17.Martin JT, Gorth DJ, Beattie EE, Harfe BD, Smith LJ, Elliott DM. Needle puncture injury causes acute and long-term mechanical deficiency in a mouse model of intervertebral disc degeneration. J Orthop Res. 2013;31(8):1276–1282. doi: 10.1002/jor.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyagi M, Uchida K, Takano S, Fujimaki H, Aikawa J, Sekiguchi H, Nagura N, Ohtori S, Inoue G, Takaso M. Macrophage-derived inflammatory cytokines regulate growth factors and pain-related molecules in mice with intervertebral disc injury. J Orthop Res. 2018;36(8):2274-79. [DOI] [PubMed]
- 19.Nakawaki M, Uchida K, Miyagi M, Inoue G, Kawakubo A, Kuroda A, Satoh M, Takaso M. Sequential CCL2 Expression Profile After Disc Injury in Mice. J Orthop Res. 2020;38(4):895–901. doi: 10.1002/jor.24522. [DOI] [PubMed] [Google Scholar]
- 20.Ohnishi T, Sudo H, Iwasaki K, Tsujimoto T, Ito YM, Iwasaki N. In Vivo Mouse Intervertebral Disc Degeneration Model Based on a New Histological Classification. PLoS One. 2016;11(8):e0160486. doi: 10.1371/journal.pone.0160486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian Z, Ma X, Yasen M, Mauck RL, Qin L, Shofer FS, Smith LJ, Pacifici M, Enomoto-Iwamoto M, Zhang Y. Intervertebral Disc Degeneration in a Percutaneous Mouse Tail Injury Model. Am J Phys Med Rehabil. 2018;97(3):170–177. doi: 10.1097/PHM.0000000000000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Leung VY, Luk KD, Chan D, Cheung KM. Injury-induced sequential transformation of notochordal nucleus pulposus to chondrogenic and fibrocartilaginous phenotype in the mouse. J Pathol. 2009;218(1):113–121. doi: 10.1002/path.2519. [DOI] [PubMed] [Google Scholar]
- 23.Aalto K, Korhonen L, Lahdenne P, Pelkonen P, Lindholm D. Nerve growth factor in serum of children with systemic lupus erythematosus is correlated with disease activity. Cytokine. 2002;20(3):136–139. doi: 10.1006/cyto.2002.1991. [DOI] [PubMed] [Google Scholar]
- 24.Aloe L, Tuveri MA, Carcassi U, Levi-Montalcini R. Nerve growth factor in the synovial fluid of patients with chronic arthritis. Arthritis Rheum. 1992;35(3):351–355. doi: 10.1002/art.1780350315. [DOI] [PubMed] [Google Scholar]
- 25.Manni L, Aloe L. Role of IL-1 beta and TNF-alpha in the regulation of NGF in experimentally induced arthritis in mice. Rheumatol Int. 1998;18(3):97–102. doi: 10.1007/s002960050065. [DOI] [PubMed] [Google Scholar]
- 26.Safieh-Garabedian B, Poole S, Allchorne A, Winter J, Woolf CJ. Contribution of interleukin-1 beta to the inflammation-induced increase in nerve growth factor levels and inflammatory hyperalgesia. Br J Pharmacol. 1995;115(7):1265–1275. doi: 10.1111/j.1476-5381.1995.tb15035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takano S, Uchida K, Itakura M, Iwase D, Aikawa J, Inoue G, Mukai M, Miyagi M, Murata K, Sekiguchi H, et al. Transforming growth factor-beta stimulates nerve growth factor production in osteoarthritic synovium. BMC Musculoskelet Disord. 2019;20(1):204. doi: 10.1186/s12891-019-2595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolonen J, Gronblad M, Vanharanta H, Virri J, Guyer RD, Rytomaa T, Karaharju EO. Growth factor expression in degenerated intervertebral disc tissue. An immunohistochemical analysis of transforming growth factor beta, fibroblast growth factor and platelet-derived growth factor. Eur Spine J. 2006;15(5):588–596. doi: 10.1007/s00586-005-0930-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim MG, Kim SC, Ko YS, Lee HY, Jo SK, Cho W. The Role of M2 Macrophages in the Progression of Chronic Kidney Disease following Acute Kidney Injury. PLoS One. 2015;10(12):e0143961. doi: 10.1371/journal.pone.0143961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J, Cao Q, Zheng D, Sun Y, Wang C, Yu X, Wang Y, Lee VW, Zheng G, Tan TK, et al. Discrete functions of M2a and M2c macrophage subsets determine their relative efficacy in treating chronic kidney disease. Kidney Int. 2013;84(4):745–755. doi: 10.1038/ki.2013.135. [DOI] [PubMed] [Google Scholar]
- 31.Tautz D. Redundancies, development and the flow of information. BioEssays. 1992;14(4):263–266. doi: 10.1002/bies.950140410. [DOI] [PubMed] [Google Scholar]
- 32.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets supporting the conclusions of this article are included within the article. The raw data can be requested from the corresponding author. Data are submitted to DDBJ Sequence Read Archive under accession number LC636331.