Abstract
SARS-CoV, MERS-CoV, and potentially SARS-CoV-2 emerged as novel human coronaviruses following cross-species transmission from animal hosts. Although the receptor binding characteristics of human coronaviruses are well documented, the role of carbohydrate binding in addition to recognition of proteinaceous receptors has not been fully explored. Using natural glycan microarray technology, we identified N-glycans in the human lung that are recognized by various human and animal coronaviruses. All viruses tested, including SARS-CoV-2, bound strongly to a range of phosphorylated, high mannose N-glycans and to a very specific set of sialylated structures. Examination of two linked strains, human CoV OC43 and bovine CoV Mebus, reveals shared binding to the sialic acid form Neu5Gc (not found in humans), supporting the evidence for cross-species transmission of the bovine strain. Our findings, revealing robust recognition of lung glycans, suggest that these receptors could play a role in the initial stages of coronavirus attachment and entry.
Keywords: SARS-CoV-2, Human lung glycan microarray, Phosphorylated glycans, Glycan receptors, Coronavirus receptors
1. Introduction
A variety of pathogens circulate within wild and domesticated animals maintaining a potential reservoir of global health threats. Species barriers are mostly effective at stalling or preventing human infection, but viruses that overcome them have the potential to cause devastating pandemics, as seen with coronaviruses and influenza viruses (Bolles et al., 2011; de Wit and Munster, 2013). Most human coronaviruses have been shown to use a proteinaceous receptor - ACE-2 for SARS-CoV, SARS-CoV-2, and HCoV NL63 or DPP4 for MERS-CoV (Hoffmann et al., 2020; Hofmann et al., 2005; Letko et al., 2020; Li, 2016; Li et al., 2003; Raj et al., 2013; Tortorici et al., 2019), however, there is a growing body of evidence, discussed below, for the recognition of a glycan attachment receptor that aids in targeting the virus to cell surface.
The spike protein (S) is a homotrimeric protein containing two domains, S1 which is responsible for receptor binding and S2 which must be proteolytically activated for entry (Li, 2016). Sialic acid has been proposed as a co-receptor for MERS-CoV and is bound by an alternative receptor binding domain from the one that interacts with DPP4 (Li et al., 2017; Wang et al., 2013; Yuan et al., 2017). Predictive structural analysis of SARS-CoV-2 spike reveals, via sequence comparison, a similarity with MERS-CoV in regard to the flexible loop regions responsible for the sialoside binding of that virus (Awasthi et al., 2020). Additionally, studies revealed that SARS-CoV-2 has the ability to bind specifically to only the type I blood group A antigen of the ABO(H) glycan antigens, confirming glycan recognition and inspiring questions regarding blood group and susceptibility to infection (Wu et al., 2021). Recent studies indicate that for the murine hepatitis virus JHM-CoV, receptor usage is not exclusively limited to a carbohydrate receptor via spike domain ‘S1A’ or a proteinaceous one via ‘S1B’. The model suggests that cooperative binding may be important in initiating infection, first with a more abundant, yet less specific receptor that is a sialylated glycan, and then to the more specialized protein interaction (Qing et al., 2020). Heparin and heparan sulfate have been implicated in HCoV NL-63 and SARS-CoV-2 entry (Clausen et al., 2020; Kim et al., 2020; Milewska et al., 2014). Some strains of coronavirus, such as bovine CoV Mebus, HCoV OC43, and HCoV HKU-1, use carbohydrates as their primary receptors. These viruses bind to the sialic acid derivative 9-O-Ac-Sia as a receptor (Huang et al., 2015; Schultze et al., 1991; Vlasak et al., 1988). BCoV Mebus has been linked to HCoV OC43 as a potential ancestral strain that crossed into the humans (Vijgen et al., 2005). The use of a common carbohydrate receptor lowers the species barrier, making cross-species transmission and adaptation more readily achievable.
In order to identify carbohydrate structures that may serve as potential receptors in the human lung tissues, we examined the glycan recognition of a panel of representative coronaviruses (CoV) from human and animal hosts. We studied the human strains SARS-CoV-2, HCoV-OC43, and HCoV-NL63 and the animal strains, bovine coronavirus BCoV Mebus and porcine respiratory coronavirus PRCV ISU-1. Using defined glycan microarrays and the natural human lung shotgun glycan microarray, we were able to characterize the ability of these viruses to bind specific glycan determinants and discovered that each strain displays robust binding to phosphorylated, high mannose glycans from the human lung. Interestingly, we recently demonstrated that influenza A viruses (IAV) could also bind these structures and often did so with greater affinity than to the widely accepted receptor, sialic acid-terminating glycans (Byrd-Leotis et al., 2019a, 2019b). It is possible that coronaviruses can utilize glycan receptors, which are ubiquitous and often homologous between animal and human hosts, to facilitate initial attachment to human host tissues during cross-species transmission events and ‘gain entry’ to the human reservoir, with further adaptive mutations leading to efficient transmission. Additionally, our findings suggest that a possible common mechanism exists between membrane bound respiratory pathogens for the recognition of phosphorylated glycans, which could have implications for a universal approach to antiviral treatment.
2. Results and discussion
2.1. Characterization of CoVs on NCFG array and HL-SGM
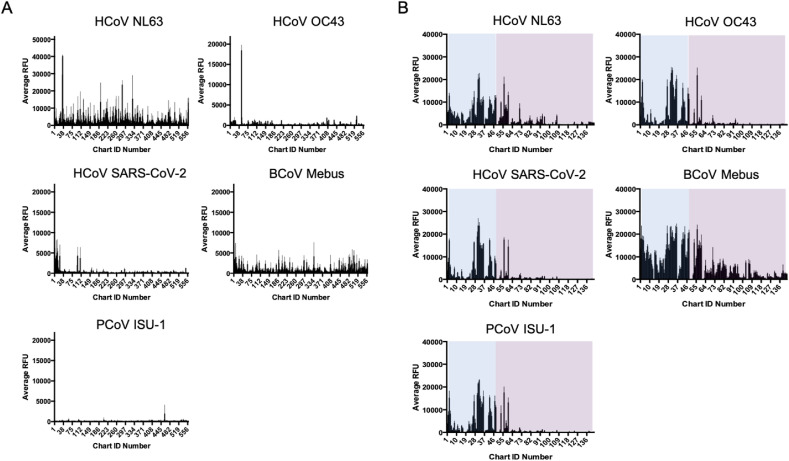
In order to assess the capability of CoV to interact with glycans found at the site of infection in the human lung, we analyzed the binding of each strain to the National Center for Functional Glycomics (NCFG) defined glycan microarray and the human lung shotgun glycan microarray (HL-SGM), a natural glycan microarray prepared from human lung tissue. The NCFG array is a synthetic array, meaning that the glycans and glycan determinants are chemically or enzymatically synthesized. This array provides a range of structures, yet the biological occurrence of many of them is unverified. The NCFG glycan microarray is standard for assaying glycan recognition of proteins and pathogens (http://www.functionalglycomics.org/). The array consists of over 600 glycan structures, representing a variety of combinations. Overall, the CoV strains used in these assays do not exhibit strong binding to the NCFG arrays (Fig. 1 A). The human coronaviruses OC43 and NL63 are the exceptions with very specific binding to isolated glycans- Neu5,9Acα2-Sp8 (Chart ID #48) for OC43 and (3S)Galβ1-4(6S)GlcNAc variants (Chart IDs #34 and 35 with Sp8 and Sp0 linkers, respectively) for NL63. The OC43 binding is not unexpected given prior work suggesting this glycan is the receptor (Vlasak et al., 1988) and our results from the sialic acid derivative array. However, our previous data characterizing the human lung glycome indicate that there is a relatively low abundance of glycans terminating in these structures in the lung tissue (Jia et al., 2020), and results from studies on the HL-SGM reveal binding to typical sialylated complex N-glycan structures. The HCoV NL63 strain binds the sulfated determinants (3S)Galβ1-4(6S)GlcNAc (Chart IDs #34 and 35 with Sp8 and Sp0 linkers, respectively) and Galβ1-4(Fucα1-3) (6S)GlcNAc (Chart ID #283) (Fig. 1A). The HCoV strain SARS-CoV-2 exhibits binding to Chart ID 23 which is 6S(3S)Galb1-4GlcNAcb-Sp0, however this structure is not a strong binder. The recognition of sulfated structures is consistent with recent studies that identified binding to heparin and heparan sulfate (Clausen et al., 2020; Kim et al., 2020; Milewska et al., 2014). Our results indicate that the presence of the sulfate may be more important for recognition than the full glycan structure e.g., N-glycan backbone vs glycosaminoglycans.
Fig. 1.
A. Virus binding to NCFG glycan microarray. Each CoV strain was bound to the NCFG array. The average RFU scale is higher for NL63 and reflects the greater intensity of binding by this strain. B. CoV strains binding on HL-SGM. The human CoV strains NL63, SARS-CoV-2, and OC43, as well as the bovine CoV Mebus and the porcine strain ISU-1 are bound to the HL-SGM. Fractions containing phosphorylated, high mannose glycans are highlighted by the blue box, while sialylated glycans are denoted by the purple box.
To explore the potential of CoVs to bind glycans at the site of infection in the lung, we examined binding to the HL-SGM. These natural glycan microarrays were developed to represent the variety of naturally-occurring N-glycans expressed in the human lung glycome (Byrd-Leotis et al., 2019b). The HL-SGM is comprised of N-glycans isolated from lung tissue via the ORNG method (Song et al., 2016). The shotgun method entails releasing the glycans en masse, derivatizing them with fluorescent linkers, and separating them via two dimensional HPLC. The fractions, grouped largely by charge and mass, are catalogued as the tagged glycan library. Glycans are printed from each fraction and those that are bound by viruses are prioritized for structural characterization. Thus, not all structures on the array are known. Additionally, multiple species can populate the same fraction and therefore be printed in the same spot. General trends about the array have been elucidated by standard lectin binding and enzymatic treatments (Song et al., 2011a) and two distinct populations of glycans have been identified: phosphorylated, high mannose-type N-glycans and sialylated complex N-glycans. We observed that CoV strains bind widely to the phosphorylated, high mannose-type N-glycans with additional high binding to a limited number of sialylated glycans (Fig. 1B). The exception is the BCoV Mebus strain which exhibits a much broader range of binding to sialylated fractions.
In general, all of the CoV strains examined exhibited very similar binding profiles with multiple fractions consistently ranking as top binders for all CoV strains except BCoV Mebus. Fig. 2 panel A shows a plot comparing the top 10 fractions for each virus. Average rank corresponds to the intensity of binding measured by average RFUs and ranked high to low. Frequency relates to the number of viruses that bind to a particular fraction. The structures found in fractions 56, 30, 36, 27, 42 and 33 are known (structural analysis could not be completed on fractions 31 and 32), and the dominant glycan species are represented by the cartoons in Fig. 2 panel B. The structures are predicted based on the mass and compared to known biologically relevant compounds. Interestingly, the predominant structures include two sialylated glycans, each bi-antennary with only one core N-acetylglucosamine. One glycan species is fully sialylated while the other contains one branch that terminates in sialic acid. Of note is the lack of binding to fractions that contain the complete complex N-glycan structure composed of 2 units of N-acetylglucosamine at the core. These structures are among the most abundant in the lung and are present on the array, and yet they are not strongly bound by the CoV strains. Additionally, single chain and tri and tetra-antennary sialylated glycans are not high binding structures. Of the phosphorylated glycans that are recognized, the mannose core sizes range from Man5 to Man7.
Fig. 2.
Top 10 fractions bound by CoV strains and predicted structures. A. For each of the 5 CoV strains tested, the ten fractions with the highest average RFU were compared in frequency (how many of the viruses bound that fraction) and average rank (an average of the ranks of each fraction for each virus). B. Cartoons representing the predicted structures that are dominant in the high rank + high frequency quadrant.
2.2. Coronaviruses versus influenza A viruses on HL-SGM
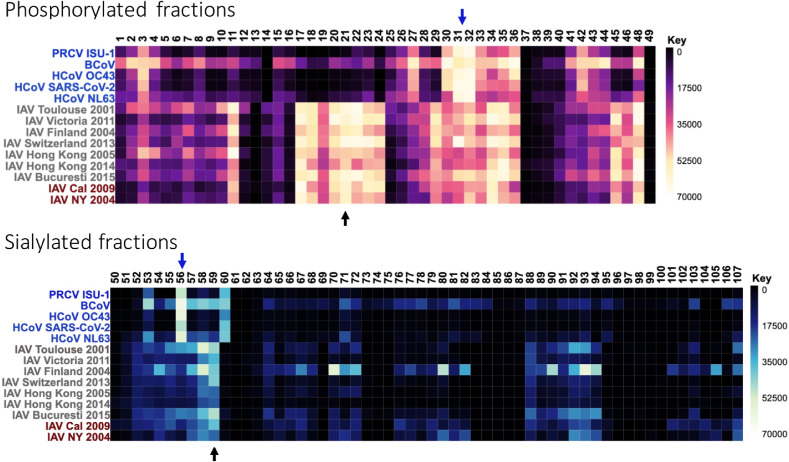
A comparison of these coronavirus results with influenza A virus (IAV) binding to the array reveals that both sets of respiratory pathogens are able to bind to phosphorylated glycans in the lung and, to a more limited extent for CoV, sialylated species. Interestingly, the specific glycan fractions that are highly bound within those broad classifications (phosphorylated vs sialylated) are different (Fig. 3 ). While both pathogens bind to branched N-glycans that are fully or partially sialylated with Neu5Ac, CoV strains prefer a structure without the additional core N-acetylglucosamine. The nearly exclusive binding to a common bi-antennary, disialylated structure minus the core N-acetylglucosamine while the full structure is available is particularly interesting. It could suggest that the structure of the entire glycan is important for recognition, or it could reflect a feature of the array composition in that these fractions may be at different density, or the structures have a specific orientation that makes them more amenable to CoV recognition. The full version of this structure is a consistent high binder for the majority of influenza A virus strains tested and is one of the most abundant structures in the total glycome. The differences in phosphorylated, high mannose glycan recognition are even more striking, with an entire subset of fractions that are bound by IAV but not CoV. While electrostatic interactions with these charged carbohydrates could be important for binding, as is true for many glycan-binding proteins, it is clear that the stereochemistry of the remaining structure play a key role in determining specificity.
Fig. 3.
Comparison of CoV and IAV binding on HL-SGM. The top heatmap represents binding to the phosphorylated fractions, while the bottom one is the sialylated fractions. Not all are shown due to space constraints and lack of binding to upper fraction ID numbers. Strains are listed on the left, with CoV in blue, H3N2 drift IAV strains in gray, and seasonal/pandemic H1N1 IAV strains in red. Arrows point to fractions of interest.
2.3. Comparison of HCoV OC43 and BCoV mebus on modified sialic acid array
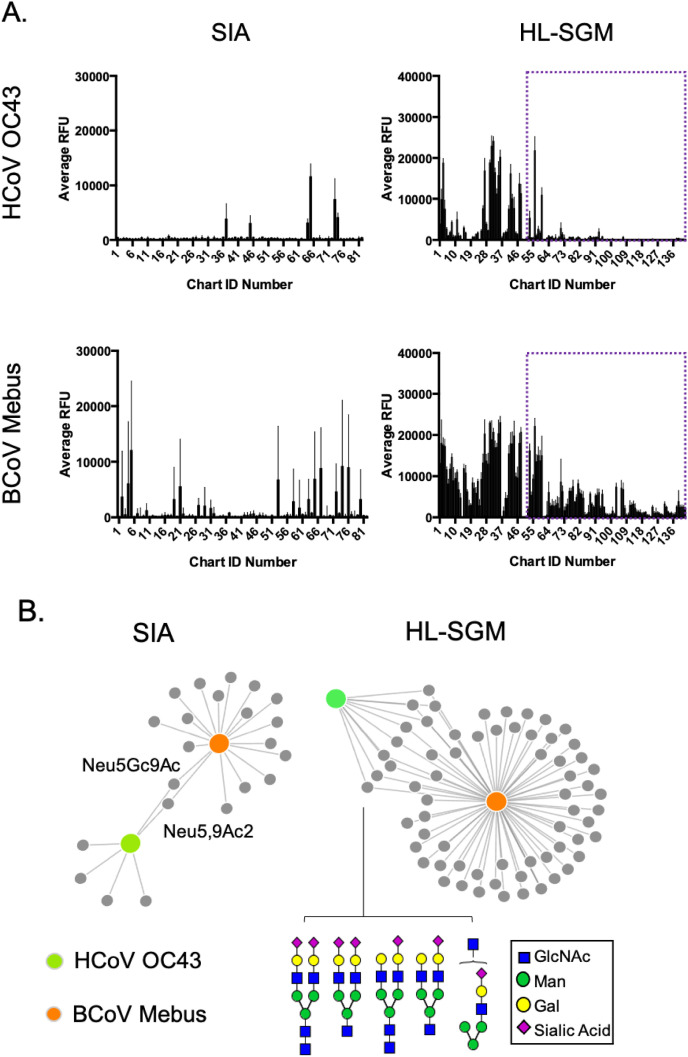
Both the HCoV strain OC43 and BCoV strain Mebus use a sialic acid derivative as their viral receptors (Schultze et al., 1991; Vlasak et al., 1988) and, as these strains are reported to be linked (Vijgen et al., 2005), we sought to specifically identify the glycans from the human lung natural array and a modified sialic acid array (Song et al., 2011b) that may provide clues about cross-species adaptation. We compared the binding of each strain to both arrays and in both cases, the OC43 virus is much more restrictive in the spectrum of glycans bound. BCoV Mebus binds to a wide variety of glycans from the human lung and on the sialic acid derivative array (Fig. 4 panel A). Force plots illustrate the glycan fractions that are bound by one and/or both viruses. The gray dots represent each glycan (from the defined sialic acid derivative array) or fraction potentially containing multiple glycans (on the HL-SGM). The orange circle corresponds to BCoV strain, and the green represents HCoV OC43. A linkage between the circles indicates binding and the length of the link corresponds to binding strength (ie the shorter the link, the higher the RFU). On the sialic acid derivative array, only two species are bound by both viruses, Neu5,9Ac2 and Neu5Gc9Ac. Neu5,9Ac2 is the predicted receptor for OC43 and BCoV Mebus. Interestingly, Neu5Gc structures are not found in humans due to the lack of the appropriate metabolic pathway (Irie et al., 1998) but are prevalent in other mammals. The recognition of this glycan by the HCoV OC43 strain supports proposals that this virus stemmed from a transmission event with the bovine virus. In the human lung repertoire, the predominant species recognized by both are biantennary, fully or partially sialylated N-glycans. These structures make up a significant portion of the human lung glycome based (Jia et al., 2020) and could have eased the transmissibility and adaptation of the bovine strain to humans.
Fig. 4.
A Comparison of HCoV OC43 and its ancestral strain BCoV Mebus. A. Each strain was tested on the sialic acid derivative array (SIA) and the HL-SGM. The sialylated fractions of the HL-SGM are set apart with the purple box. B. The force graphs compare the glycans that are recognized by both strains. On the SIA array, only two structures are common to each virus and are labeled. For the HL-SGM, only the sia-terminating glycans are analyzed and the predicted structures in the fractions that are bound by both viruses are shown.
3. Conclusion
Our results demonstrate that each CoV strain we tested, including SARS-CoV-2, can bind to phosphorylated, high mannose glycans found in the human lung. Additionally, we describe the recognition of specific sialylated glycans by all the viruses tested. The HL-SGM displays the N-glycome of the human lung, and therefore our data reveals interactions that could be biologically relevant for infection. However, the localization of the phosphorylated, high mannose glycans in the cell is currently unknown. While we hypothesize that these interactions may occur at the surface of the cell and function in entry, it is possible that these glycans are intracellular and recognition occurs later in the entry pathway or at other phases of the virus life cycle. While modified sialic acid and sulfated glycans have been implicated in cell culture infection, we find that those interactions do not outweigh the binding to phosphorylated glycans on these arrays. Interestingly, the comparison of CoV binding with IAV reveals similar trends but differences in specific binding partners. Previous work with various strains of CoVs has hinted at the possibility of an attachment receptor, often a carbohydrate, and a more specific entry receptor. Our work indicates that, with the ability to bind a diverse set of glycans from the lungs, the occurrence of a glycan-based interaction is possible and likely. Additionally, comparison of the PRCV ISU-1 with the human strains reveals a very tight binding pattern regardless of animal or human host. The BCoV Mebus strain has a much more diverse and broad binding profile on all the arrays tested, which could be a factor in priming this virus for cross-species transmission.
4. Materials and methods
4.1. Cells and viruses
Each coronavirus strain required a specific cell line for propagation. These cell lines were acquired from ATCC (Manassas, VA): ST (ATCC CRL-1746), LLC-MK2 (ATCC CCL-7.1), Vero C1008 (E6) (ATCC CRL-1586) and MDBK (ATCC® CCL-22). HCT-8 cells were a generous gift from Dr. Mark Tompkins at the University of Georgia. The following coronavirus strains were obtained from BEI Resources, NIAID, NIH: Porcine Respiratory Coronavirus, ISU-1, NR-43286; human coronavirus, NL63, NR-470; and bovine coronavirus, BCoV Mebus, NR-445. Dr. Mehul Suthar provided the UV-inactivated SARS-CoV-2, and Dr. Mark Tompkins shared the human coronavirus OC43 strain. The cell lines VeroE6, LLC-MK2, and ST were maintained with MEM (ThermoFisher) + 10 % FBS (ThermoFisher) growth media, while the MDBK and HCT-8 cell lines were maintained in DMEM (Thermofisher) + 10 % FBS. For virus infection, an MOI of 0.01 to 0.001 was used. Once cell monolayers were 80–90 % confluent, media was removed, and the cells were washed with PBS. The monolayer was overlaid with inoculation media containing the viruses and allowed to incubate for 1 h for adsorption. After the incubation, 20 ml of media was added to the flask. The infected cells were incubated at 34 °C or 37 °C depending on the virus and cells for 4–6 days. The VeroE6 cells were used to amplify SARS-CoV-2, and the infection media was complete DMEM medium consisting of 1x DMEM (VWR, #45000-304), 10 % FBS, 25 mM HEPES Buffer (Corning Cellgro), 2 mM L-glutamine, 1 mM sodium pyruvate, 1x Non-essential Amino Acids, and 1x antibiotics. HCoV NL63 was grown in LLC-MK2 cells, and the infection media was MEM +10 % FBS +10 % tryptose phosphate. ST cells were used to amplify ISU-1, and the infection media was MEM + Earle's salts + NEAA, 1500 mg/L NaHCO3, 1 mm sodium pyruvate, 2 mm L-glutamine. BCoV Mebus and HCoV OC43 were grown in MDBK and HCT-8 cells respectively using serum-free DMEM plus antibiotics for the infection media. SARS-CoV-2 was UV inactivated prior to microarray analysis. The infected cell supernatant was exposed to 1 dose of UV light at 5,000 × 100μJ/cm2. To verify that the virus was inactivated, two sets of plaque assays in duplicate and a limiting dilution assay in triplicate were completed. Influenza A virus (A/Pennsylvania/08/2008) was used as a control for the microarray experiments and was amplified in MDCK cells using serum-free DMEM plus penicillin-streptomycin (Byrd-Leotis et al., 2019b).
4.2. Virus purification and labeling
Supernatants were collected from flasks infected with each CoV and subjected to low-speed centrifugation (3,000 RPM) to pellet the cell debris. The clarified supernatant was laid over a 25 % sucrose in NTE (100 mM NaCl, 10 mM Tris, 1 mM EDTA) cushion, and viruses were pelleted via ultracentrifugation using Ultra-clear centrifuge tubes (Beckman Coulter, Indianapolis IN) and a Beckman Coulter SW32 Ti rotor at 28,000 rpm (133548.5×g) for 2 h at 4 °C. Virus pellets were resuspended in 2 ml of phosphate buffered saline (PBS) and then residual sucrose was cleared by an additional spin using rotor sw41 Ti, 15,000 rpm (55408×g) for 1 h at 4 °C. Viral pellets were resuspended in PBS and aliquoted for downstream applications and storage at −80 °C.
Purified virus was labeled with Alexafluor 488 (Invitrogen) prior to glycan array assays. The virus was mixed with 25 μg of Alexafluor 488 and 20 μl of 1 M NaHCO3 pH 9 and allowed to incubate for 1 h at room temperature. Excess label was dialyzed out of the sample using a 7,000-molecular-weight-cutoff Slide-A-Lyzer mini-dialysis unit (Thermo Scientific) against PBS pH 7.0 overnight. In all cases, labeled viruses were used in experiments the following day. The same procedure was followed for influenza A viruses utilized as controls.
4.3. Microarray assays and analysis
When necessary, viruses were diluted with TSM binding buffer (20 mM Tris-HCl, 150 mM sodium chloride (NaCl), 2 mM calcium chloride (CaCl2) and 2 mM magnesium chloride (MgCl2)+ 0.1%BSA) and then incubated on the glycan microarray for an hour. Influenza A viruses were diluted with PBS as BSA can interfere with the sialic acid binding. For the NCFG arrays with a single array per slide, 70 μl of labeled virus was pipetted onto the slide and covered with a glass coverslip. The HL-SGM and modified sialic acid arrays are printed to have multiple subarrays per slide. For these experiments, ProPlate® Multi-Well chambers (Grace Bio) were used on the slide to separate each array and experiment. While neuraminidase activity is not a concern with these CoV strains, we included IAV as a control for our assays and sought to maintain consistent procedures, so all viruses were incubated at 4 °C. After removal of the virus solutions, the arrays were washed 3 times in TSMT (20 mM Tris-HCl, 150 mM sodium chloride (NaCl), 2 mM calcium chloride (CaCl2) and 2 mM magnesium chloride (MgCl2) + 0.05 % Tween 20), 3 times in TSM (20 mM Tris-HCl, 150 mM sodium chloride (NaCl), 2 mM calcium chloride (CaCl2) and 2 mM magnesium chloride (MgCl2)) and 3 times in water. An InnoScan1100AL (Innopsys, Carbonne, France) fluorescence scanner was used to generate the images using an excitation wavelength of 495 nm and an emission wavelength of 519 nm. The fluorescence of the spots was quantified using GenePix Pro Microarray Analysis software (Molecular Devices, San Jose, CA, USA) or MAPIX analysis software version 8.5.0 (Innopsys, Carbonne, France) and then further processed using Microsoft Excel as previously described (Bradley et al., 2011; Byrd-Leotis et al., 2014; Heimburg-Molinaro et al., 2012). The force graph was generated via the GLAD web application (Mehta and Cummings, 2019) and the following settings were used: Force Range Min 1000 Max 70000, Force Strength −40, Link Distance 50.
CRediT authorship contribution statement
Lauren Byrd-Leotis: Conceptualization, Investigation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. Yi Lasanajak: Supervision, Investigation, Formal analysis. Thomas Bowen: Investigation, Formal analysis. Kelly Baker: Investigation, Formal analysis. Xuezheng Song: Supervision, Writing – review & editing. Mehul S. Suthar: Resources, Investigation. Richard D. Cummings: Supervision, Writing – review & editing. David A. Steinhauer: Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge support by the U.S. Department of Health and Human Services contract HHSN272201400004C (NIAID Centers of Excellence for Influenza Research and Surveillance) and the National Institutes of Health award P41GM103694 and R24GM137763 to RDC and the National Center for Functional Glycomics, which supports the use of the Consortium for Functional Glycomics glycan microarrays. This study was assisted in part by the Emory Glycomics and Molecular Interactions Core (EGMIC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. LBL, YL, and MS performed the experiments, with the assistance of TB and KB. LBL analyzed the data, with the assistance of XS, RC, and DS. LBL, DS, and XS supervised the experiments. LBL wrote the manuscript. The authors declare no competing interests. Correspondence in requests for materials should be addressed to LBL (labyrd@emory.edu).
References
- Awasthi M., Gulati S., Sarkar D.P., Tiwari S., Kateriya S., Ranjan P., Verma S.K. The sialoside-binding pocket of SARS-CoV-2 spike glycoprotein structurally resembles MERS-CoV. Viruses. 2020;12(9):909. doi: 10.3390/v12090909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolles M., Donaldson E., Baric R. SARS-CoV and emergent coronaviruses: viral determinants of interspecies transmission. Curr Opin Virol. 2011;1:624–634. doi: 10.1016/j.coviro.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley K.C., Jones C.A., Tompkins S.M., Tripp R.A., Russell R.J., Gramer M.R., Heimburg-Molinaro J., Smith D.F., Cummings R.D., Steinhauer D.A. Comparison of the receptor binding properties of contemporary swine isolates and early human pandemic H1N1 isolates (Novel 2009 H1N1) Virology. 2011;413:169–182. doi: 10.1016/j.virol.2011.01.027. [DOI] [PubMed] [Google Scholar]
- Byrd-Leotis L., Gao C., Jia N., Mehta A.Y., Trost J., Cummings S.F., Heimburg-Molinaro J., Cummings R.D., Steinhauer D.A. Antigenic pressure on H3N2 influenza virus drift strains imposes constraints on binding to sialylated receptors but not phosphorylated glycans. J. Virol. 2019;93(22) doi: 10.1128/JVI.01178-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd-Leotis L., Jia N., Dutta S., Trost J.F., Gao C., Cummings S.F., Braulke T., Muller-Loennies S., Heimburg-Molinaro J., Steinhauer D.A., Cummings R.D. Influenza binds phosphorylated glycans from human lung. Sci Adv. 2019;5(2) doi: 10.1126/sciadv.aav2554. eaav2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd-Leotis L., Liu R., Bradley K.C., Lasanajak Y., Cummings S.F., Song X., Heimburg-Molinaro J., Galloway S.E., Culhane M.R., Smith D.F., Steinhauer D.A., Cummings R.D. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2241–E2250. doi: 10.1073/pnas.1323162111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Painter C.D., Thacker B.E., Glass C.A., Narayanan A., Majowicz S.A., Zhang Y., Torres J.L., Golden G.J., Porell R., Garretson A.F., Laubach L., Feldman J., Yin X., Pu Y., Hauser B., Caradonna T.M., Kellman B.P., Martino C., Gordts P., Leibel S.L., Chanda S.K., Schmidt A.G., Godula K., Jose J., Corbett K.D., Ward A.B., Carlin A.F., Esko J.D. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. bioRxiv. 2020 doi: 10.1101/2020.07.14.201616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Munster V.J. MERS-CoV: the intermediate host identified? Lancet Infect. Dis. 2013;13:827–828. doi: 10.1016/S1473-3099(13)70193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimburg-Molinaro J., Tappert M., Song X., Lasanajak Y., Air G., Smith D.F., Cummings R.D. Probing virus-glycan interactions using glycan microarrays. Methods Mol. Biol. 2012;808:251–267. doi: 10.1007/978-1-61779-373-8_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Dong W., Milewska A., Golda A., Qi Y., Zhu Q.K., Marasco W.A., Baric R.S., Sims A.C., Pyrc K., Li W., Sui J. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie A., Koyama S., Kozutsumi Y., Kawasaki T., Suzuki A. The molecular basis for the absence of N-glycolylneuraminic acid in humans. J. Biol. Chem. 1998;273:15866–15871. doi: 10.1074/jbc.273.25.15866. [DOI] [PubMed] [Google Scholar]
- Jia N., Byrd-Leotis L., Matsumoto Y., Gao C., Wein A.N., Lobby J.L., Kohlmeier J.E., Steinhauer D.A., Cummings R.D. The human lung glycome reveals novel glycan ligands for influenza A virus. Sci. Rep. 2020;10:5320. doi: 10.1038/s41598-020-62074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Jin W., Sood A., Montgomery D.W., Grant O.C., Fuster M.M., Fu L., Dordick J.S., Woods R.J., Zhang F., Linhardt R.J. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antivir. Res. 2020;181:104873. doi: 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Widjaja I., Raj V.S., McBride R., Peng W., Widagdo W., Tortorici M.A., van Dieren B., Lang Y., van Lent J.W.M., Paulson J.C., de Haan C.A.M., de Groot R.J., van Kuppeveld F.J.M., Haagmans B.L., Bosch B.J. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A.Y., Cummings R.D. GLAD: GLycan array dashboard, a visual analytics tool for glycan microarrays. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska A., Zarebski M., Nowak P., Stozek K., Potempa J., Pyrc K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 2014;88:13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing E., Hantak M., Perlman S., Gallagher T. Distinct roles for sialoside and protein receptors in coronavirus infection. mBio. 2020;11(1) doi: 10.1128/mBio.02764-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B., Gross H.J., Brossmer R., Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J. Virol. 1991;65:6232–6237. doi: 10.1128/JVI.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Ju H., Lasanajak Y., Kudelka M.R., Smith D.F., Cummings R.D. Oxidative release of natural glycans for functional glycomics. Nat. Methods. 2016;13:528–534. doi: 10.1038/nmeth.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Lasanajak Y., Xia B., Heimburg-Molinaro J., Rhea J.M., Ju H., Zhao C., Molinaro R.J., Cummings R.D., Smith D.F. Shotgun glycomics: a microarray strategy for functional glycomics. Nat. Methods. 2011;8(1):85–90. doi: 10.1038/nmeth.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Yu H., Chen X., Lasanajak Y., Tappert M.M., Air G.M., Tiwari V.K., Cao H., Chokhawala H.A., Zheng H., Cummings R.D., Smith D.F. A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J. Biol. Chem. 2011;286(36):31610–31622. doi: 10.1074/jbc.M111.274217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D., Boons G.J., Bosch B.J., Rey F.A., de Groot R.J., Veesler D. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P., Vandamme A.M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasak R., Luytjes W., Spaan W., Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. U. S. A. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., Arledge K.C., Chen Y.H., Zhang L., Wang X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.C., Arthur C.M., Wang J., Verkerke H., Josephson C.D., Kalman D., Roback J.D., Cummings R.D., Stowell S.R. The SARS-CoV-2 receptor-binding domain preferentially recognizes blood group A. Blood Adv. 2021;5:1305–1309. doi: 10.1182/bloodadvances.2020003259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., Zhang X., Gao G.F. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]