Abstract
Deep-learning (DL)-based image processing has potential to revolutionize the use of smartphones in mobile health (mHealth) diagnostics of infectious diseases. However, the high variability in cellphone image data acquisition and the common need for large amounts of specialist-annotated images for traditional DL model training may preclude generalizability of smartphone-based diagnostics. Here, we employed adversarial neural networks with conditioning to develop an easily reconfigurable virus diagnostic platform that leverages a dataset of smartphone-taken microfluidic chip photos to rapidly generate image classifiers for different target pathogens on-demand. Adversarial learning was also used to augment this real image dataset by generating 16,000 realistic synthetic microchip images, through style generative adversarial networks (StyleGAN). We used this platform, termed smartphone-based pathogen detection resource multiplier using adversarial networks (SPyDERMAN), to accurately detect different intact viruses in clinical samples and to detect viral nucleic acids through integration with CRISPR diagnostics. We evaluated the performance of the system in detecting five different virus targets using 179 patient samples. The generalizability of the system was confirmed by rapid reconfiguration to detect SARS-CoV-2 antigens in nasal swab samples (n = 62) with 100% accuracy. Overall, the SPyDERMAN system may contribute to epidemic preparedness strategies by providing a platform for smartphone-based diagnostics that can be adapted to a given emerging viral agent within days of work.
Keywords: deep learning, artificial intelligence, adversarial learning neural networks, smartphones, diagnostics, clustered regularly interspaced short palindromic repeats, severe acute respiratory syndrome coronavirus
Graphical Abstract
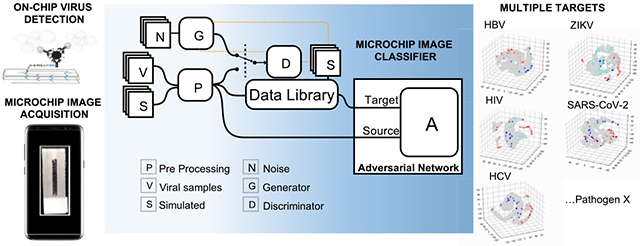
Interfacing efficient image capture and processing on mobile devices with simple visual signal outputs generated upon specific detection of target pathogens is essential to leverage smartphones to real-world mobile health (mHealth) diagnostics for emerging infectious diseases, such as the coronavirus disease 2019 (COVID-19).1,2 This could circumvent common limitations of established centralized diagnostic technologies that often require sophisticated bulky equipment to detect colorimetric or fluorescent outputs generated through enzyme-mediated reactions.2,3 However, limitations that include variabilities in smartphone camera positioning, smartphone model, image distortions, and illumination may hamper the use of mobile phones in mHealth field applications. The recent increment in deep learning (DL) models4 for image processing can aid in tackling these intrinsic constraints of smartphone-based imaging, and the combination with emerging lab-on-chip technologies holds promise to impact mHealth diagnostics in the coming years.5–7
DL image classifiers based on convolutional neural networks (CNNs) have gained great popularity in recent years due to specialist-level performances in medical image diagnosis, mostly in the fields of radiology and pathology.8 CNN architectures are particularly good for medical image classification tasks but suffer from major drawbacks of supervised learning methods, that is, the need for massive amounts of specialist-annotated data and domain dependence.5,8,9 Labeling highly heterogeneous diagnostic images by trained human specialists is a time-consuming and costly procedure, which would not be practical for any smartphone-based diagnostic method aiming at broad applicability to different infectious agents.
Here, we have employed adaptative adversarial learning to reduce the need for human-annotated data and improve the generalizability when reconfiguring a smartphone-based diagnostic system, termed smartphone-based pathogen detection resource multiplier using adversarial networks (SPyDER-MAN), to target different viruses including SARS-CoV-2 (Figure 1). This system leverages a dataset library of unlabeled, heterogeneous, and nonspecific smartphone-taken images to DL model training when developing on-demand image classifiers for each specific target of interest. Deep adversarial learning strategies commonly used for domain adaptation (DA) are good alternatives for situations like this, in which there are high image variability and scarcity of labeled images in the so-called Source domains but sufficient unlabeled images in Target domains, as we can leverage non-annotated data from possibly different, but related, domains and wider distributions to improve the performance of the image classifier.10–12 In our study, the Source domains were composed by limited numbers of smartphone-taken photos of microfluidic chip-based assays to specifically detect intact viruses [hepatitis B virus (HBV), hepatitis C virus (HCV), human immunodeficiency virus-1 (HIV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)] or viral nucleic acids (Zika virus); Target domains, in turn, contained a much larger number of unlabeled microchip images that also included simulated samples and synthetically generated data (Figure S1).
Figure 1.
Overview of the smartphone-based pathogen detection resource multiplier using adversarial networks (SPyDERMAN) system. A resource image library, created with real smartphone-taken microfluidic chip images from virus-specific detection assays (n = 669), nonspecific simulated samples (n = 904), and synthetic non-annotated images generated using style generative adversarial networks (StyleGANs) (n = 16,000), is leveraged to compose Target domains for adversarial neural networks with conditioning-based translation of the SPyDERMAN pathogen detection system to various target viruses on-demand.
RESULTS AND DISCUSSION
To generate the dataset of human-annotated smartphone images for specifically detecting the different viruses, we first standardized an assay to consistently generate a simple, non-enzymatic, visual output in a microfluidic chip upon recognition of specific target viral particles or nucleic acids. This visual output could be any colorimetric or fluorescent signals, for instance. Here, the signal is achieved through conjugation of metal nanocatalysts (i.e., platinum nanoparticles, PtNPs) with target-specific recognition antibodies, hereafter referred to as nanoprobes (Figure 2a and Figure S2). The system is composed of three major steps: (i) capture of the target intact viruses or nucleic acids; (ii) on-chip signal generation using nanoprobes and imaging using a smartphone; and (iii) image analysis using adversarial neural networks with conditioning. Figure S2 shows the basics of the fabrication and functioning of target-specific nanoprobes. In the presence of a fuel solution, the catalase-like activity of the PtNPs disproportionates hydrogen peroxide to water and oxygen and generates a signal output based on oxygen bubbles that can be detected in the microfluidic channel.
Figure 2.
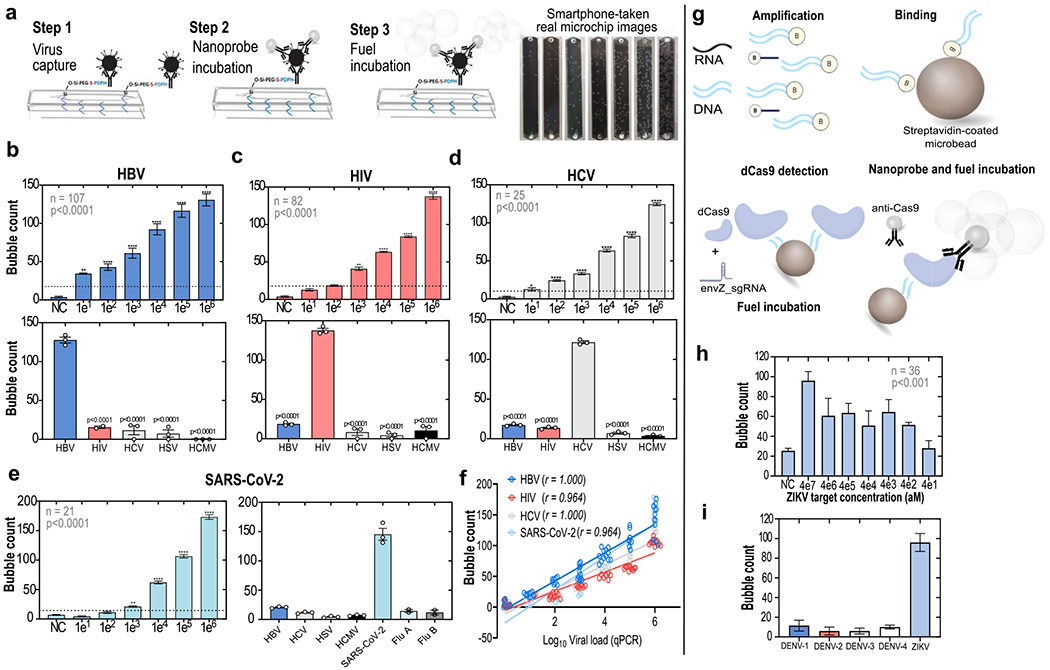
Standardization of the microfluidic chip-based assays to detect intact viruses and viral nucleic acids. (a) General protocol for intact virus detection on-chip through an enzyme-free optical signal output (bubble generation). The bubble outputs observed in HBV microchips incubated with increasing concentrations of spiked viruses are shown. (b–d) Analytical sensitivities (top panels) and specificities (bottom panels) of the intact virus detection assays for HBV, HIV, and HCV, respectively. (b–d) (Top panels) Mean ± SEM of serial 10-fold dilutions of virus spiked samples (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Sample numbers and results of analyses of variance are shown within the charts. (b–d) (Bottom panels) Individually spiked with five different viruses (HBV, HIV, HCV, HSV, HCMV), at concentrations of 1 × 106, and tested in the various assay formats and microchips. (e) Analytical sensitivity and specificity of the assay format to detect the SARS-CoV-2. (f) Bubble counts in the microchip channels were highly correlated with viral loads measured by quantitative real-time PCR (qPCR); r = Spearman’s rho. (g) Translation of the system for viral nucleic acid detection on-chip using CRISPR/dCas9. Shown are mean ± SEM. (h) Sensitivity of detection of a ZIKV synthetic genomic region; aM = attomolar. (i) Specificity of the ZIKV nucleic acid detection assay.
To evaluate the ability of the system to generate a detectable and predictable bubble signal output, we first fabricated nanoprobes using monoclonal antibodies targeting major diagnostic antigens (AgHBs and HCVcAg) of HBV and HCV viruses and also targeting the envelope glycoprotein gp120 of HIV-1. The full characterization of the fabricated nanoprobes is presented in Figure S2. Samples spiked with serial dilutions of laboratory-maintained or commercially available viral strains were then used to standardize on-chip detection assays for these three viruses, using the three-step general protocol described in Figure 2a and Figures S3 and S4. Our microchip functionalization protocol showed significant antibody immobilization and high efficiency of virus capture (Figure S4). The various standardization steps involved in the virus capture assay development are shown in Figure S5.
We found a linear relationship between optical signal output and target virus concentration for the three viral targets that were initially tested (for HBV, coefficient of determination R2 = 0.9931; for HIV, R2 = 0.9112; and for HCV, R2 = 0.9322) (Figure 2b–d, top panels), corroborating recent studies that also used PtNP-based detection of various analytes.13,14 The lowest number of bubbles able to reliably separate positive and negative samples is shown as horizontal dotted lines (Figure 2b–d, top panels). These values were verified by Probit regression analysis, which indicated the 99.0% probability of positive results for the different assays at the following clinically relevant15,16 virus concentrations: 16 IU/mL for HBV; 250 cP/mL for HIV; and 20 IU/mL for HCV (Figure S6). The assays generate specific and strong bubble signals for the respective viruses, while giving significantly lower signals for nonspecific infectious agents (Figure 2b–2d, bottom panels). Additionally, at various virus concentrations, the assays presented repeatability coefficients well above 80.0% (Figure S7) (HBV range: 83.2–90.6%; HIV range: 81.8–96.6%; HCV range: 78.3–97.1%). Figure 2e demonstrates that the same assay format could be easily reconfigured within a few weeks to detect SARS-CoV-2 with similar performance, using a nanoprobe designed with a monoclonal antibody targeting the SARS-CoV-2 spike protein (Figure 2e). A consistent bubble signal was detected for nasal swab samples containing SARS-CoV-2 with viral loads of 1 × 103 copies/mL or higher, which is clinically relevant considering that most COVID-19 patients will present with a viral load of above 105 copies per swab (Figure 2e, left panel).17,18 The signal was also reliably differentiated from results obtained with influenza A and B samples (Figure 2e, right panel). The output signals generated for all four viruses tested were highly correlated with the viral loads measured by real-time qPCR (Figure 2f).
In addition to detecting intact viruses, we explored the possibility of targeting viral nucleic acids, which could also contribute to the rapid translation of the virus detection assay to an emerging target pathogen. For this, we fabricated a nanoprobe targeting the CRISPR/dCas9 protein19 by conjugating an anti-Cas9 monoclonal antibody with PtNPs. Due to the high adaptability of the CRISPR/dCas9 recognition system, which mostly depends on the availability of a specific guide RNA to the nucleic acid target of choice, such an anti-Cas9 nanoprobe could theoretically be employed as a universal nanoprobe to detect different viral nucleic acids using the reported approach (Figure 2g). Full steps involved in the standardization of the dCas9-based detection of a Zika virus (ZIKV) genomic region encoding the envelope protein (Env) are detailed in Figure S8. We were able to reliably detect a specific bubble signal when microbeads were coated with amplified fragments originating from initial target concentrations down to 400 aM of the synthetic ZIKV env fragment (Figure 2h), which approaches the sensitivity levels of state-of-the-art CRISPR-based assays reported for detecting ZIKV.20 The specificity of the detection was confirmed by testing the ZIKV nucleic acid detection assay using microbeads coated with amplified fragments of the Env encoding regions from the four subtypes of Dengue virus (DENV) (Figure 2i).
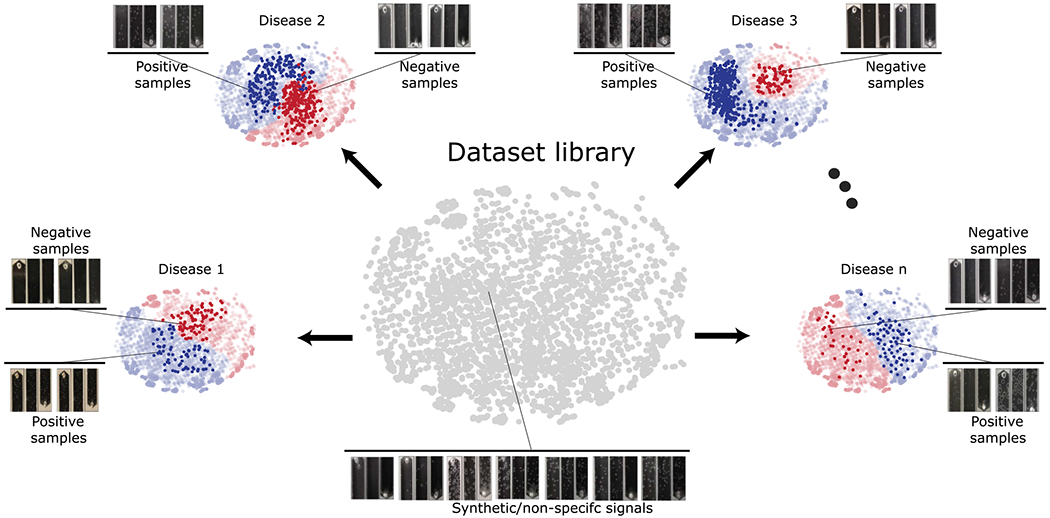
The complete dataset of specialist-labeled microchip images for all five viruses, with viral loads confirmed by real-time qPCR, sums up to 669 reliably annotated photos; additionally, 904 microchip images were taken from simulated samples, and 16,000 images were synthetically generated by GANs to compose an unlabeled image dataset (Figures S1 and S9). Although highly trained specialists (n = 3) showed an overall consistent performance when manually estimating bubble counts (Figure S10), it is clear from the dataset library that the relevant feature representations of the microchip images go beyond bubble counts alone and also include a high variability in bubble sizes, format, and area. This contributed to a suboptimal performance of untrained individuals (n = 4) when blindly classifying microchip images (Figure S10), and most importantly, this reinforces the need for a deep-learning-based image classifier for accurate on-phone image processing.
All specialist-annotated microchip images generated in the intact virus and nucleic acid detection assays were then organized in five different domains—HBV (B), HIV (I), HCV (C), SARS-CoV-2 (C19), and ZIKV (Z)—alongside a sixth domain containing unlabeled simulated, and synthetic samples (S) and five adversarial neural networks with conditioning were developed (Figure 3a,b and Figure S9). The goal was to obtain a classifier network that would perform well in an independent test set of true patient samples for each of the following domains: B, I, C, C19, and Z. The various domains have different feature representations that determine whether a sample is positive or negative, which implies that the joint distributions of feature and class are nonidentical across the domains. Adapting only the feature representations might be insufficient, and therefore, classifier predictions were incorporated as relevant information during the training process as it could potentially provide discriminative information. Using adversarial learning,21 domain discrepancies in the feature representations and the classifier predictions could be modeled simultaneously. For the approach utilized in this work, the domain discriminator is conditioned on the cross-covariance of domain-specific feature representations and classifier predictions, implying that the network focuses on the class-specific features that retain its form when the domain shifts from Source to Target. The data library, comprising both real and synthetic images, was utilized primarily to widen the data distribution by including variabilities of bubble shapes, sizes, concentrations, and positions and thereby improve resultant network generalizability.
Figure 3.
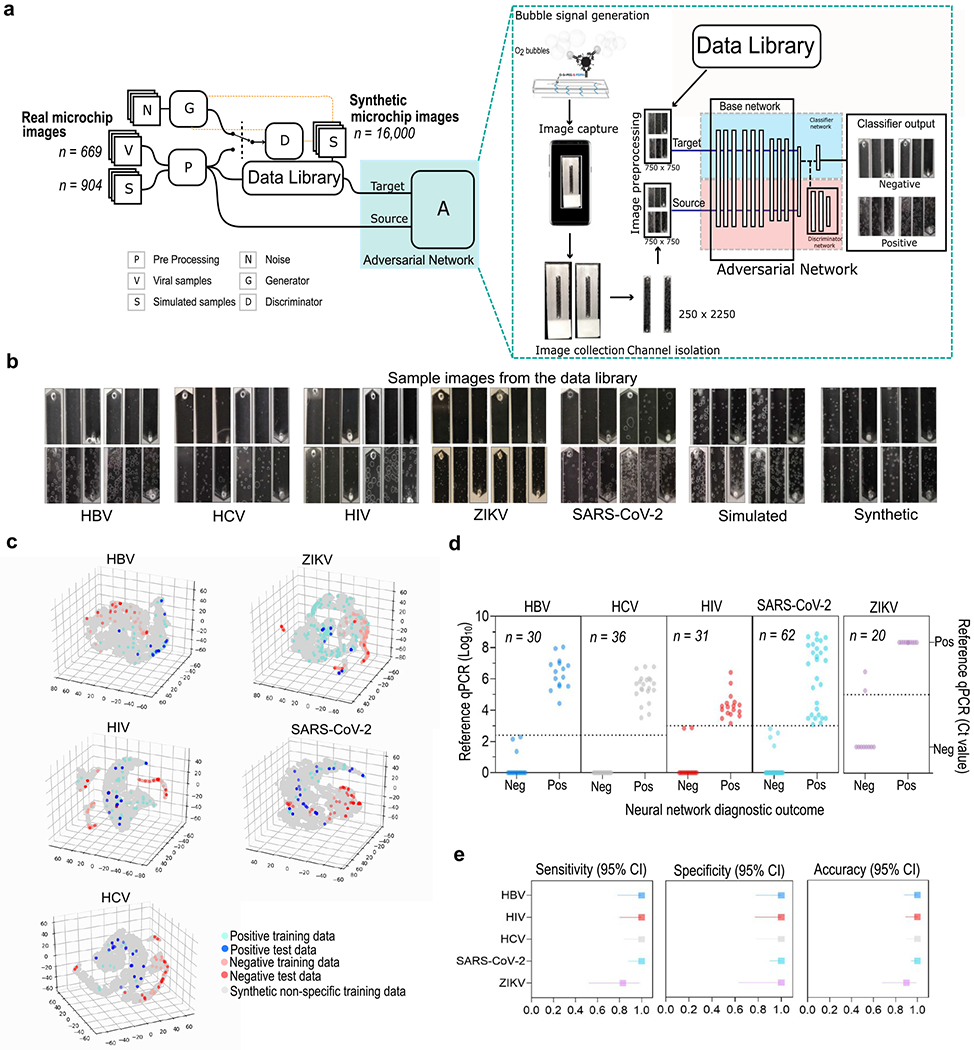
Development of the SPyDERMAN diagnostics platform and performance evaluation with 179 patient samples. (a) Schematic of the development of the adversarial neural networks with conditioning-based image classifier for smartphone-taken microchip images. The general architecture of the adversarial domain adaptive neural network is shown. (b) Examples of real smartphone-taken microfluidic chip images for each assay format alongside synthetic microchip images generated by StyleGANs. (c) Degree of microchip images separability visualized by t-SNE. (d) Scatter plots showing results of the adversarial neural networks with conditioning-based image classifiers on individual patient samples. Cut-off values for defining positive samples are shown as dotted lines. (e) Forest plots demonstrating the overall performance of the network models.
Overall, the SPyDERMAN system rendered a good degree of class separability for all five viruses, as shown by t-distribution stochastic neighbor embedding (t-SNE) (Figure 3c and Movies S1–S5). The image classifiers achieved maximal performances in confidence-based class separation (AUC: ~1.0) (Figure S11) when tested with true patient samples (n = 179) (Figure 3d). Nearly perfect diagnostic accuracies were reached for all targets when clinically relevant thresholds for separating negative and positive samples were considered during model training and testing (Figure 3e and Table S1). Particularly in the SARS-CoV-2 viral antigen detection assay, the COVID adversarial network was able to classify COVID-19 nasopharyngeal swab samples with 100% sensitivity (CI: 87.66–100.00%) and 100% specificity (CI: 89.72–100.00%) (n = 62) in a test panel that also included COVID-19 negative nasal swabs spiked with influenza A and B viruses (Figure 3d,e). Although COVID-19 diagnostics has relied mostly on nucleic acid tests and serologic tests for detecting antibodies,18,22 the availability of an accurate antigen-based test to detect SARS-CoV-2 in swab samples might theoretically provide an alternative for faster and cheaper diagnostics.22 Our COVID diagnostic framework has accuracy and limit of detection comparable to that of RT-PCR and does not require any RNA extraction reagent/kit or bulky equipment for optical signal readout. Our comparison of SPyDERMAN and CDC qRT-PCR assay for detection of SARS-CoV-2 is reported in Table S2. Integration with smartphone-based results readout would, in turn, facilitate case reporting and timely contact tracing to inform isolation programs.7,23 Similarly, the availability of antigen tests that provide proxies of clinically relevant viral loads might be of great value for diagnostics and treatment follow up in HBV, HCV, and HIV infections, which are also highly dependent on nucleic acid testing in the centralized laboratory network.15,16
CONCLUSIONS
In conclusion, using adversarial neural networks with conditioning, we were able to create accurate image classifiers for discriminating positive and negative microchip images taken with smartphones, which worked similarly well for diagnosing infections by five different viruses in true clinical samples: HBV, HCV, HIV, SARS-CoV-2, and ZIKV. To achieve this, we leveraged a single dataset of unlabeled microchip images along with an adversarial learning scheme to account for the scarcity of specialist-annotated images for each target of interest. The use of the built-in smartphone cameras, without reliance on additional hardware, and the ability to process a wide distribution of images, even when acquired in varying environmental conditions, are key features that may expedite the deployment of the SPyDERMAN system to mHealth diagnostics of infectious diseases. Additionally, the simple microchip image acquisition procedure (Figure S12) does not require any specialized training for an inexperienced user. However, mHealth diagnostics technologies still face important challenges for adoption at the technical and regulatory levels, which must be considered.1 A major hurdle that we envision in our system is the need for future data from potential alternative or emerging diseases while maintaining data privacy and security. One potential solution is utilizing the current framework, with federated, secure, and privacy-preserving architectures that allow data connectivity to include additional acquired data in the form of relevant feature representations.24 These may be used as part of the unlabeled data library to further improve and expand its generalizability, especially when it is utilized for other alternative or emerging targets. The generalizability of the system is already extended through combination with CRISPR-mediated detection of nucleic acids in addition to intact viruses. Additionally, our study works as a proof-of-concept for adversarial neural networks with conditioning to be used in other medical problems that are modeled similarly to ours, where reliably labeled image data are scarce.
METHODS
Study Design.
The goal of the study was to interface deep-learning-based processing of smartphone acquired photos with a simple, non-enzymatic signal output (in this specific case, is gas bubbles) generated upon detection of intact viruses or nucleic acids in microfluidic chips, in order to develop a mobile diagnostics system that can be easily and accurately adapted to other alternative or emerging pathogens with minimal DL model training needed for each target. We hypothesized that we could use adaptative adversarial learning with conditioning, using a large unlabeled image data distribution in the Target domains, to account for the data distribution sparsity in the Source domains, prospectively collected for each target virus. To achieve this, we first standardized a protocol that allowed for detecting intact viruses (HBV, HCV, HIV, SARS-CoV-2) or viral nucleic acids (ZIKV) in a microfluidic channel, through bubble counting by trained specialists in photos taken with smartphones. The specialist-annotated photos were composed of a labeled dataset for the different pathogens, and the unlabeled image dataset was composed of photos from simulated samples and synthetically generated samples, containing varying image resolutions, differing lightning, and image distortions. In total, a dataset library with 17,573 microchip images was created, and five different adversarial learning models were generated, using adversarial neural networks with conditioning. Then, the accuracy of the DL models to classify negative and positive samples in the different viral detection assays was evaluated using smartphone photos (n = 179) taken from assays with true patient samples drawn from subjects infected with HIV, HBV, HCV, SARS-CoV-2, and ZIKV.
Target Viruses, Antibodies, and Patient Samples.
Patient plasma/serum samples infected with hepatitis B virus and human immunodeficiency virus-1 were obtained from Discovery Life Sciences (Los Osos, CA, US). Patient serum samples infected with hepatitis C virus were received from the laboratory of Dr. Raymond Chung at Massachusetts General Hospital. Human immunodeficiency virus-1 viral stocks were obtained from coculture supernatants, as previously described25 and received from the laboratory of Drs. Jonathan Li and Daniel Kuritzkes at the Division of Infectious Diseases at Brigham and Women’s Hospital. Spiked Zika virus serum samples were purchased from ZeptoMetrix (Zika virus range verification panel; cat. no. NATZIKV-RV), and ZIKV-infected serum samples were obtained from BocaBiolistics (Pompano Beach, FL, USA). Nasopharyngeal swab samples from COVID-19 patients were obtained from BocaBiolistics (Pompano Beach, FL, USA). Swab samples from healthy individuals and influenza A and B samples were obtained from Discovery Life Sciences (Los Osos, CA, USA). The system evaluation studies using the above human patient samples were reviewed and approved by Brigham and Women’s Hospital institutional review board (IRB#: 2019P002209, 2019P001489, 2019P001996, and 2020P001065).
Whenever required, viral load quantifications by real-time RT-PCR (RT-qPCR) were outsourced at Viracor-Eurofins Clinical Diagnostics, using clinically validated assays. Antibodies targeting HBV surface antigen (HBsAg), HCV core antigen (HCV cAg), HIV envelope glycoprotein (gp120), SARS-CoV-2 spike protein, and CRISPR associated protein 9 (Cas9) were purchased from different sources: anti-HBV B521M mAb (Genetex, cat. no. GTX41736); anti-HCV 6A1 mAb (Abcam, cat. no. ab2582); anti-HIV 2557 mAb (NIH AIDS Reagent Program, cat. no. 13429); goat anti-HIV-1 gp120 Ab (Abcam, cat. no. ab21179); rabbit anti-SARS-CoV-2 mAb (Sino Biological, cat. no. 40150-R007); anti-Cas9 mAb (Applied Biological Materials, abm, cat. no. Y300079). For details on patient samples and antibodies, please see Tables S3 and S4.
Fabrication and Characterization of Platinum Nanoparticles Conjugated with Antibodies (Nanoprobes).
To fabricate specific nanoprobes for different targets, we conjugated citrate-capped PtNPs with periodate-oxidized specific monoclonal antibodies, using the heterobifunctional cross-linking reagent 3-[2-pyridyldithio]-propionyl hydrazide (Thermo Fisher Scientific, cat. no. 22301).25,26 Conjugation of the monoclonal antibodies to the PtNPs and functionality of the nanoprobes were confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), UV—visible spectroscopy, Fourier transform infrared spectroscopy (FT-IR; Cary 630 ATR-FTIR), H2O2 decomposition assay (hydrogen peroxide assay kit, ab102500, Abcam, USA), dynamic light scattering and zeta-potential (Malvern Zetasizer Nano, Malvern Instruments, UK), transmission electron microscopy (JEM-2100Plus, JEOL, Japan), and field-emission scanning electron microscopy (Ultra 55, Zeiss, Germany).
Microchip Fabrication and On-Chip Intact Virus Detection.
To prepare microchips, 3.175 mm thick poly(methyl methacrylate) sheets (8560K239, McMaster-Carr) and double-sided adhesive sheets (76 μm, 8213, 3M; or 125 μm, 8215, 3 M for SARS-CoV-2) were cut using the VLS 2.30 CO2 laser cutter (Universal Laser Systems Inc.), according to a design prepared in CorelDraw X7 graphical editor containing the microfluidic channel and the microchip inlets and outlets (microchannel dimensions: L = 40 mm; W = 5 mm; H = 0.8 mm). Then, all ethanol-cleaned parts were assembled on glass microslides (VWR, 48300-025 corning 75 × 25 mm) previously functionalized for surface immobilization of the virus capture antibodies. In brief, oxygen plasma treatment of the glass surface was done for 3 min at 100 mTorr, and 20 μL of silane-PEG-thiol (20 mg/mL; Nanocs, cat. no. PG2-SLTH-5k) was added for 1 h, followed by ethanol washing. After microchip assembly, specific antibodies (anti-HBV, 45 μg/mL; anti-HCV, 5.2 μg/mL; anti-HIV, 20.4 μg/mL; anti-SARS-CoV-2, 19 μg/mL) previously oxidized and modified with 0.9 mg/mL 3-[2-pyridyldithio]propionyl hydrazide were incubated in the microchannel for antibody immobilization (Figure S3). For intact virus detection, 20 μL (HBV, HCV, HIV) or 30 μL (SARS-CoV-2) of sample was incubated in the microchip for 20 min (HBV, HCV) or 45 min (HIV, SARS-CoV-2), and then the microchannel was washed thoroughly with 0.1 M phosphate buffer solution (pH = 7.45, Sigma-Aldrich). Microchips were incubated with 20 μL of 1:20 specific nanoprobe diluted in phosphate-buffered saline (PBS; pH = 7.4, Caisson Laboratories) for a further 20 min. The nanoprobe solution was then removed, and microchips were washed again with PBS. For bubble development, microchips were filled with 20 μL of a fuel solution (6% hydrogen peroxide + 10% glycerol) and incubated for 10 min at room temperature when photos of bubble development in the microchannels were taken (Figures S3 and S4).
On-Chip Viral Nucleic Acid Detection by CRISPR/dCas9 Assay.
The CRISPR detection assay relied on using dCas9 (null mutant protein, K140, Applied Biological Materials, Canada) associated with a Zika virus specific single guide RNA (envZ sgRNA; see Table S4) to bind a ZIKV amplified genomic region immobilized on a streptavidin-coated microbead surface. Then, an anti-dCas9 nanoprobe (mAb + PtNPs) was used to detect the dCas9 target nucleic acid association in the microfluidic channel through bubble formation (Figure 2g–i and Figure S8). Briefly, isolated ZIKV RNA was reverse transcribed to cDNA and amplified using Platinum SuperScript one-step RT-PCR system (ThermoFisher) and biotinylated oligonucleotide primers (see Table S4) in a Bento Lab portable laboratory setup (Bento Lab, UK). For assay standardization, synthetic genomic fragments of ZIKV or Dengue virus (serotypes DENV 1–4) were also used (Table S4). Following a 2 min cleanup step, 10 μL of the amplified products was bound to 10 μL of Dynabeads MyOne streptavidin C1 microbeads (1 μm; 10 mg/mL; ThermoFisher), previously washed and resuspended in nuclease-free STE buffer (10 mM Tris-HCl, 1 mM EDTA, 2 M NaCl). Microbeads were then incubated with a blocking solution (0.5% biotin; 5% bovine serum albumin) for 20 min before transferring 2.5 μL of the bead solution to a microtube containing a mix of specific sgRNA (100 nM) and dCas9 (100 nM) (in 20 mM HEPES, 5 mM MgCl2, 100 mM NaCl, 0.1 mM EDTA; preincubated for 15 min, 37 °C). Following further incubation for 30 min, at 37 °C, and an additional blocking step, microbeads were finally incubated with an anti-Cas9 nanoprobe solution (1:40), washed twice with 0.05% Triton STE buffer, resuspended in 30 μL of fuel solution (6% hydrogen peroxide + 10% glycerol), and loaded in the microchip. After 15 min, photos of the bubble development in the microchannel were then taken.
Real Microchip Image Acquisition Using Smartphones.
A real microchip image database was created, containing 1573 smartphone photos taken from bubble development in microfluidic channels: HBV assay (n = 129); HCV assay (n = 49); HIV assay (n = 107); SARS-CoV-2 assay (n = 35); ZIKV assay (n = 349); simulated virus-free samples with bubbles generated only by addition of nanoprobes to the microchips (n = 904). All images were obtained (Figure S12) using smartphones with 12-megapixel image sensors (Motorola MotoX and Apple iPhone 8) and were stored as .jpeg or .heic files, with average file sizes of 1.5 megabytes.
Dataset Library.
The full dataset library was composed of microfluidic chip images generated using assays performed with different viral targets and simulated samples along with synthetic images generated using a generative adversarial network (style-GAN). 27 The total number of images in the dataset library was 17,573, which includes 16,000 synthetic images (Figures S1 and S9).
Data Preprocessing.
All images, prior to use with both the GAN and the adversarial network, were preprocessed to maximize the signal-to-noise ratio. The images of the microfluidic chips collected using the smartphone camera were cropped to remove the background and isolate the microfluidic channel. Additionally, the channel images were resized to 250 × 2250 pixels and then split horizontally into three equal parts of size 250 × 750 pixels. The three parts were tiled adjacently into an image of size 750 × 750 pixels.
Synthetic Image Generation Using GANs.
The diversity of the data library was augmented with 16,000 images of synthetic data generated using a generative adversarial network model. StyleGAN was used for image generation of bubbles. Preprocessed images were resized to 256 × 256 before using the StyleGAN model. Next, 1489 images of bubble signals produced using both simulated and viral samples were used in training the model. The dimensionality of the latent space was set to 512. We used an equalized learning rate of 0.0003 and a minibatch size of 16. Images of size 256 × 256 were generated by the network Network training was stopped when the network was stabilized, and images of sufficient quality were consistently generated.
Adversarial Network for Classification.
In this study, adversarial networks with class conditioning were used to develop sufficiently generalizable networks using small sets of labeled training data and large sets of unlabeled data. 21,28,29 The network used the limited labeled datasets as its source data and unlabeled images from the data library as its target. The network uses a discriminator and classifier in unison to effectively utilize both labeled and unlabeled images in its learning process. The discriminator aids in identifying features that tend to overlap between both source and target, and the classifier helps refine these features to maximize the network’s classification performance on the source data. The network was designed as a binary classifier, and image labels were assigned based on the viral concentrations that fall above or below clinically relevant thresholds. All samples that possessed concentrations below those thresholds were classified as negative. Images were resized to 224 × 224 pixels prior to use with the network. The network used the Xception architecture as its base network and was initialized with pretrained ImageNet weights.30 For HBV, HIV, and ZIKV, the source data were loaded in batches by random sampling. For HCV and SARS-CoV-2, batch balancing was performed to minimize the heavy class imbalance in the dataset. Data augmentation was performed to improve the ability of the model to generalize. The following augmentation techniques were applied at random, with a probability of 0.5—horizontal flip, vertical flip, grayscale conversion, rotation between 0 and 360°, and color jitter. Batch sizes of 8 and 16 were used. Adam optimizer was used with initial learning rates of 0.0001, 0.0005, 0.001, 0.005, and 0.001 for HBV, HIV, ZIKV, HCV, and SARS-CoV-2, respectively. A weight decay of 0.0005 with an L2 penalty was used to prevent overfitting. A gamma value of 0.001 and a power of 0.75 were used to adjust the value of the learning rate in the following manner:
where i indicates training progress.
A momentum of 0.9 was used to accelerate the learning process. The optimal batch size and learning rate were determined manually by the selection of the best model based on the lowest validation loss. Early stoppage was used with the patience of 2000 iterations to avoid overfitting.
Adversarial Loss Calculation.
The source classifier is trained by minimizing the classifier loss estimation which is given by
where is the limited labeled source data and is the corresponding labels; Ds is the source data distribution. L() represents cross-entropy loss; C() is the classifier network, and the discriminator loss estimation is maximized, which is given by
where D() is the discriminator network, Dt is the target data distribution, and is the unlabeled target data. Discriminator loss calculation uses weighted entropy conditioning and a multilinear feature map (h), a tensor product of feature representation f and classifier prediction c, given c for k classes vector and feature representation (l dimensional vector) respectively. The result h is expressed as
The combination of f and c is performed as a conditioning step that helps preserving class specific information across domains. Additionally, weighted entropy conditioning w(H()) was used to improve the classification performance on target distribution by encouraging the high confidence predictions in the unlabeled target domain.
Data Analysis and Visualization.
NIH ImageJ software (https://imagej.nih.gov/ij/index.html) was used for manual processing of the microchip photos by trained specialists, using the cell counter plugin. During the standardization of the assays with virus spiked samples, GraphPad Prism v.8 was used for the following analyses: normality test by Shapiro–Wilk and Kolmogorov–Smirnov; nonparametric Kruskal–Wallis test with Dunn’s multiple comparisons test to compare groups or ANOVA with Holm–Sidak’s multiple comparisons for normal data; Spearman correlation for bubble counts and RT-qPCR results; and linear regression for calculating coefficients of determination (R2). MedCalc software v.19.2.0 (MedCalc Software bv, Ostend, Belgium; https://www.medcalc.org; 2019) was used for receiver operating characteristic curve analysis of patient samples tests, Probit regression analysis for limit of detection calculations, Passing–Bablok regression, and Bland–Altman plots for comparing interexperimenter results. Lower limits of detection of the assays were also calculated considering the limit of blank, according to the formula described by Robb et al.31 Repeatability of the assays was calculated with the formula described by Shafiee et al.32 The t-distribution stochastic neighbor embedding was performed to observe the distribution of the test dataset and to verify if our model was able to differentiate negative and positive samples.
Data Availability.
Deidentified patient data used in this study are reported in the Supporting Information, and the actual smartphone-taken microchip image data are available in the online version. The patient samples were purchased from commercial vendors or obtained through our colleagues at Brigham and Women’s Hospital and Massachusetts General Hospital, and the exact same patient sample materials may not be available for share by the authors or by the commercial vendors due to the limited sample volume available from each subject. In addition, the actual patient samples may not be available for share based on the limitations enforced by institutional review board. The codes developed for this study can be accessed via https://github.com/shafieelab/SPyDERMAN. The data analyzed in this study can also be accessed via https://osf.io/et9s6/.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was partially supported by the National Institute of Health under Award Nos. R01AI118502, R01AI138800, and R61AI140489 and Brigham and Women’s Hospital under Brigham Precision Medicine Developmental Grant. L.G.P. was supported through the CAPES/Harvard Junior Visiting Professor/Researcher Program and is currently a recipient of a research fellowship from CNPq-Brazil. F.S. was supported through the CAPES/Print Program and is a recipient of a Ph.D. scholarship from Fapesb.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.0c06807.
Details on the construction of the microchip image dataset using generative adversarial networks (Style-GANs); fabrication and functioning of target-specific nanoprobes; UV—vis, gel electrophoresis, FT-IR, TEM, and SEM of nanoprobes and PtNPs; standardization of the CRISPR/Cas9 nucleic acid detection assay; repeatability of the virus detection assays; overall performance of the image classifiers (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsnano.0c06807
The authors declare the following competing financial interest(s): We have filed a patent on the reported technology through Brigham and Women’s Hospital.
Contributor Information
Ahmed Shokr, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02139, United States.
Luis G. C. Pacheco, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02139, United States; Department of Biotechnology, Institute of Health Sciences, Federal University of Bahia, Salvador, BA 40110-100, Brazil
Prudhvi Thirumalaraju, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02139, United States.
Manoj Kumar Kanakasabapathy, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02139, United States.
Jahnavi Gandhi, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02139, United States.
Deeksha Kartik, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02139, United States.
Filipe S. R. Silva, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02139, United States; Department of Biotechnology, Institute of Health Sciences, Federal University of Bahia, Salvador, BA 40110-100, Brazil
Eda Erdogmus, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02139, United States.
Hemanth Kandula, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02139, United States.
Shenglin Luo, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02139, United States.
Xu G. Yu, The Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology and Harvard University, Boston, Massachusetts 02129, United States; Division of Infectious Diseases, Brigham and Women’s Hospital and , Harvard Medical School, Boston, Massachusetts 02139, United States
Raymond T. Chung, Liver Center, Gastrointestinal Division, Massachusetts General Hospital and , Harvard Medical School, Boston, Massachusetts 02114, United States
Jonathan Z. Li, Division of Infectious Diseases, Brigham and Women’s Hospital and, Harvard Medical School, Boston, Massachusetts 02139, United States
Daniel R. Kuritzkes, Division of Infectious Diseases, Brigham and Women’s Hospital and , Harvard Medical School, Boston, Massachusetts 02139, United States
Hadi Shafiee, Division of Engineering in Medicine, Department of Medicine, Brigham and Women’s Hospital and , Harvard Medical School, Boston, Massachusetts 02139, United States.
REFERENCES
- (1).Wood CS; Thomas MR; Budd J; Mashamba-Thompson TP; Herbst K; Pillay D; Peeling RW; Johnson AM; McKendry RA; Stevens MM Taking Connected Mobile-Health Diagnostics of Infectious Diseases to the Field. Nature 2019, 566 (7745), 467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ding X; Mauk MG; Yin K; Kadimisetty K; Liu C Interfacing Pathogen Detection with Smartphones for Point-of-Care Applications. Anal. Chem 2019, 91 (1), 655–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hernandez-Neuta I; Neumann F; Brightmeyer J; Ba Tis T; Madaboosi N; Wei Q; Ozcan A; Nilsson M Smartphone-Based Clinical Diagnostics: Towards Democratization of Evidence-Based Health Care. J. Intern. Med 2019, 285 (1), 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Jiao L; Zhao J A Survey on the New Generation of Deep Learning in Image Processing. IEEE Access; 2019, 7, 172231. [Google Scholar]
- (5).Riordon J; Sovilj D; Sanner S; Sinton D; Young EWK Deep Learning with Microfluidics for Biotechnology. Trends Biotechnol. 2019, 37 (3), 310–324. [DOI] [PubMed] [Google Scholar]
- (6).Liu J; Geng Z; Fan Z; Liu J; Chen H Point-of-Care Testing Based on Smartphone: The Current State-of-the-Art (2017–2018). Biosens. Bioelectron 2019, 132, 17–37. [DOI] [PubMed] [Google Scholar]
- (7).Farshidfar N; Hamedani S The Potential Role of Smartphone-Based Microfluidic Systems for Rapid Detection of COVID-19 Using Saliva Specimen. Mol. Diagn. Ther 2020, 24, 371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Topol EJ High-Performance Medicine: The Convergence of Human and Artificial Intelligence. Nat. Med 2019, 25 (1), 44–56. [DOI] [PubMed] [Google Scholar]
- (9).Xiang X; Yu Z; Lv N; Kong X; Saddik AE Semi-Supervised Image Classification via Attention Mechanism and Generative Adversarial Network. Eleventh International Conference on Graphics and Image Processing (ICGIP 2019) 2019, 126. [Google Scholar]
- (10).Goodfellow I; Pouget-Abadie J; Mirza M; Xu B; Warde-Farley D; Ozair S; Courville A; Bengio Y Generative Adversarial Networks. Commun. ACM 2020, 63, 139. [Google Scholar]
- (11).Wang M; Deng W Deep Visual Domain Adaptation: A Survey. Neurocomputing 2018, 312, 135. [Google Scholar]
- (12).Lv F; Zhu J; Yang G; Duan L TarGAN: Generating Target Data with Class Labels for Unsupervised Domain Adaptation. Knowl. Based. Syst 2019, 172, 123. [Google Scholar]
- (13).Loynachan CN; Thomas MR; Gray ER; Richards DA; Kim J; Miller BS; Brookes JC; Agarwal S; Chudasama V; McKendry RA; Stevens MM Platinum Nanocatalyst Amplification: Redefining the Gold Standard for Lateral Flow Immunoassays with Ultrabroad Dynamic Range. ACS Nano 2018, 12 (1), 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Chen H; Li Z; Zhang L; Sawaya P; Shi J; Wang P Quantitation of Femtomolar-Level Protein Biomarkers Using a Simple Microbubbling Digital Assay and Bright-Field Smartphone Imaging. Angew. Chem., Int. Ed 2019, 58 (39), 13922–13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Fourati S; Feld JJ; Chevaliez S; Luhmann N Approaches for Simplified HCV Diagnostic Algorithms. J. Int. AIDS Soc 2018, 21 (2), No. e25058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Cornberg M; Wong VW; Locarnini S; Brunetto M; Janssen HLA; Chan HL The Role of Quantitative Hepatitis B Surface Antigen Revisited. J. Hepatol 2017, 66 (2), 398–411. [DOI] [PubMed] [Google Scholar]
- (17).Wolfel R; Corman VM; Guggemos W; Seilmaier M; Zange S; Muller MA; Niemeyer D; Jones TC; Vollmar P; Rothe C; Hoelscher M; Bleicker T; Brunink S; Schneider J; Ehmann R; Zwirglmaier K; Drosten C; Wendtner C Virological Assessment of Hospitalized Patients with COVID-2019. Nature 2020, 581 (7809), 465–469. [DOI] [PubMed] [Google Scholar]
- (18).Weissleder R; Lee H; Ko J; Pittet MJ COVID-19 Diagnostics in Context. Sci. Transl. Med 2020, 12 (546), No. eabc1931. [DOI] [PubMed] [Google Scholar]
- (19).Li Y; Li S; Wang J; Liu G CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019, 37 (7), 730–743. [DOI] [PubMed] [Google Scholar]
- (20).Pardee K; Green AA; Takahashi MK; Braff D; Lambert G; Lee JW; Ferrante T; Ma D; Donghia N; Fan M; Daringer NM; Bosch I; Dudley DM; O’Connor DH; Gehrke L; Collins JJ Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165 (5), 1255–1266. [DOI] [PubMed] [Google Scholar]
- (21).Long CZ; Wang J Conditional Adversarial Domain Adaptation. arXiv:1705.10667v4 2018. [Google Scholar]
- (22).Tang Y-W; Schmitz JE; Persing DH; Stratton CW Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol 2020, 58 (6), No. e00512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Yang T; Wang YC; Shen CF; Cheng CM Point-of-Care RNA-Based Diagnostic Device for COVID-19. Diagnostics 2020, 10 (3), 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kaissis GA; Makowski MR; Ruckert D; Braren RF Secure, Privacy-Preserving and Federated Machine Learning in Medical Imaging. Nat. Mach. Intell 2020, 2, 305–311. [Google Scholar]
- (25).Draz MS; Kochehbyoki KM; Vasan A; Battalapalli D; Sreeram A; Kanakasabapathy MK; Kallakuri S; Tsibris A; Kuritzkes DR; Shafiee H DNA Engineered Micromotors Powered by Metal Nanoparticles for Motion Based Cellphone Diagnostics. Nat. Commun 2018, 9 (1), 4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Draz MS; Venkataramani M; Lakshminarayanan H; Saygili E; Moazeni M; Vasan A; Li Y; Sun X; Hua S; Yu XG; Shafiee G Nanoparticle-Enhanced Electrical Detection of Zika Virus on Paper Microchips. Nanoscale 2018, 10 (25), 11841–11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Karras TLS; Aittala M; Hellsten J; Lehtinen J; Aila T Analyzing and Improving the Image Quality of StyleGAN. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2020, 8107. [Google Scholar]
- (28).Mirza M; Osindero S Conditional Generative Adversarial Nets. arXiv:1411.178v1 2014. [Google Scholar]
- (29).Ganin Y; Ustinova E; Ajakan H; Germain P; Larochelle H; Laviolette F; Marchand M; Lempitsky V Domain-Adversarial Training of Neural Networks. J. Mach. Learn. Res 2017, 17, 35. [Google Scholar]
- (30).Chollet F Xception: Deep Learning with Depthwise Separable Convolutions. ArXiv 2017, 1800. [Google Scholar]
- (31).Robb NC; Taylor JM; Kent A; Pambos OJ; Gilboa B; Evangelidou M; Mentis AA; Kapanidis AN Rapid Functionalisation and Detection of Viruses via a Novel Ca(2+)-Mediated Virus-DNA Interaction. Sci. Rep 2019, 9 (1), 16219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Shafiee H; Jahangir M; Inci F; Wang S; Willenbrecht RB; Giguel FF; Tsibris AM; Kuritzkes DR; Demirci U Acute On-Chip HIV Detection through Label-Free Electrical Sensing of Viral Nano-Lysate. Small 2013, 9 (15), 2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified patient data used in this study are reported in the Supporting Information, and the actual smartphone-taken microchip image data are available in the online version. The patient samples were purchased from commercial vendors or obtained through our colleagues at Brigham and Women’s Hospital and Massachusetts General Hospital, and the exact same patient sample materials may not be available for share by the authors or by the commercial vendors due to the limited sample volume available from each subject. In addition, the actual patient samples may not be available for share based on the limitations enforced by institutional review board. The codes developed for this study can be accessed via https://github.com/shafieelab/SPyDERMAN. The data analyzed in this study can also be accessed via https://osf.io/et9s6/.


