Abstract
Purpose
Differentiated from adult stem cells (ASCs), transit-amplifying cells (TACs) play an important role in tissue homeostasis, development, and regeneration. This study aimed to characterize the gene expression profile of a candidate TAC population in limbal basal epithelial cells using single-cell RNA sequencing (scRNA-seq).
Methods
Single cells isolated from the basal corneal limbus were subjected to scRNA-seq using the 10x Genomics platform. Cell types were clustered by graph-based visualization methods and unbiased computational analysis. BrdU proliferation assays, immunofluorescent staining, and real-time reverse transcription quantitative polymerase chain reaction were performed using multiple culture models of primary human limbal epithelial cells to characterize the TAC pool.
Results
Single-cell transcriptomics of 16,360 limbal basal cells revealed 12 cell clusters. A unique cluster (3.21% of total cells) was identified as a TAC entity, based on its less differentiated progenitor status and enriched exclusive proliferation marker genes, with 98.1% cells in S and G2/M phases. The cell cycle-dependent genes were revealed to be largely enriched by the TAC population. The top genes were characterized morphologically and functionally at protein and mRNA levels. The specific expression patterns of RRM2, TK1, CENPF, NUSAP1, UBE2C, and CDC20 were well correlated in a time- and cycle-dependent manner with proliferation stages in the cell growth and regeneration models.
Conclusions
For the first time, to the best of our knowledge, we have identified a unique TAC entity and uncovered a group of cell cycle-dependent genes that serve as TAC signature markers. The findings provide insight into ASCs and TACs and lay the foundation for understanding corneal homeostasis and diseases.
Keywords: cell cycle-dependent genes, cornea, epithelium, limbal stem cells, single-cell transcriptomics, transit-amplifying cells
Adult stem cells (ASCs), also known as somatic stem cells or tissue-specific stem cells, are present in a variety of human organs and tissues.1 ASCs are small populations of quiescent, slow-cycling, undifferentiated cells with self-renew ability and high proliferative potentiality.2–4
ASCs may generate differentiated progeny through asymmetric or symmetric divisions.5–8 Asymmetric cell division gives rise to one stem cell and one daughter cell committed to progenitor cells or transit-amplifying cells (TACs), which finally differentiate into functionally mature cells, regenerating all of the cell types of the tissue where they are located. Symmetric division yields two identical SCs or two daughter cells committed to differentiation. The essential feature of this “transit” cell population is their capacity to generate many maturing cells from very few cells. The cells entering the transit stage, or TACs, are capable of rapidly producing many differentiated cells, not only during development but also during regeneration. Thus, TACs have been identified as a subpopulation intermediate between ASCs and differentiated cells.6,9,10
This stem cell/TAC model of tissue homeostasis is well recognized in multiple organisms and different organs and tissues, including the skin and hair follicles, hematopoietic system, intestine, nervous system, and corneal epithelium.6,10 The physiological and pathological roles of TACs in health and disease have been increasingly investigated and are broadly recognized in the research areas of tissue homeostasis, wound healing, and regeneration, as well as in malignant diseases such as cancer,9,11–13 although the field of regenerative biology has tended to be stem cell-centric. The importance of TACs and their diverse functions goes beyond merely producing tissues. Defects in regeneration or injury repair are also likely to be caused by problems in TACs, not just in stem cells. The impact of TACs on stem cells and their niche may represent a pathogenesis and potential therapeutic targets for halting or reversing degenerative diseases. Understanding TAC biology is of important significance not only for elucidating the fundamental principles of tissue development and regeneration but also for advancing our current treatment of regenerative and neoplastic disease.
It is worth mentioning, however, that some researchers believe that TACs do not contribute to epidermal homeostasis and tissue formation. The existence of a TAC cell type has been challenged by studies based on a model of mouse tail epidermis,14 raising the question of whether TACs are an entity in their own right or represent a function. This argument has not been solved because most previous research on TAC characterization and identification has been done at tissue and bulk cell population levels.
The cutting-edge, high-throughput, single-cell RNA-sequencing (scRNA-seq) technology holds the promise to revolutionize our understanding of diseases and associated biological processes due to its unprecedented resolution. It has the power to reveal new cell types, identify unique cell states, and dissect underlying heterogeneity in a high-throughput and unbiased manner.15,16 Single-cell transcriptomics opens a new door to uncovering and identifying TAC populations in adult tissues.
The paradigm of limbal stem cells (LSCs), TACs, and differentiated cells in the corneal epithelium has been widely accepted for more than three decades, and many features of TACs have been characterized.17–20 However, this distinct epithelial cell population in the basal cornea limbus has not been well defined at the molecular level. In the present study, we performed scRNA-seq on basal limbal epithelial cells and identified a cell cluster with increased expression of proliferation-associated genes that may represent a TAC population.
Materials and Methods
Donor Corneal Tissues and Single-Cell Isolation From the Corneal Limbal Epithelial Basal Layer
Fresh human corneoscleral tissues were obtained from the Lions Eye Bank of Texas (Houston, TX, USA) for this study. The corneal tissues used for scRNA-seq were obtained from two healthy young donors, one male and one female, who had not undergone any refractive surgery and did not have ocular diseases, chronic diseases, cancer, or chronic infections such as hepatitis B and C or human immunodeficiency virus. After the central cornea was removed and superficial layers were scraped, the remaining limbal tissue with basal cells was incubated in Dispase II (10 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) in supplemented hormonal epithelial medium (SHEM) at 4°C overnight.21 The loose limbal epithelial sheets were gently peeled and digested with 1 mL of 0.05% trypsin/1-mM EDTA at 37°C for 10 minutes. Trypan blue was used to test the viability of the single cells.
Single-Cell RNA-seq
Single-cell RNA-seq was performed at the Single Cell Genomics Core at Baylor College of Medicine. In brief, single-cell cDNA library preparation and sequencing were performed following the manufacturer's protocols (10x Genomics, Pleasanton, CA, USA). Single-cell suspensions at 1000 cells/µL in PBS were loaded onto a 10x Genomics Chromium Controller to generate a single-cell Gel Bead-in-Emulsion (GEM). The scRNA-seq library was prepared with Chromium Single Cell 3′ Reagent Kit, version 2 (10x Genomics). The product was amplified by PCR and sequenced on Illumina NovaSeq 6000 (Illumina, San Diego, CA, USA).
Bioinformatic Analysis of scRNA-seq Data
Cell Ranger 2.1 software (10x Genomics) with default settings was used for alignment, barcode assignment, and unique molecular identifier (UMI) counting of the raw sequencing data with genome reference hg19 (Supplementary Table S1).
After generating the UMI count profile, we applied Seurat 3.0 for quality control and downstream analysis.22 A global-scaling normalization method, LogNormalize, was employed to normalize the gene expression measurements for each cell by the total expression, then the result was multiplied by a scale factor (10,000 by default) and log-transformed in each dataset. For data alignment, we selected 1000 highly variable genes in each data matrix and performed the FindIntegrationAnchors and IntegrateData functions. Next, we performed clustering using FindClusters to identify sub-cell-type clusters. The Uniform Manifold Approximation and Projection (UMAP) results for each dataset were visualized if a cluster was derived from both donors.
To identify differentially expressed genes (DEGs) in each cluster, we used the FindAllMarkers function based on the Wilcoxon rank-sum test in Seurat. Cell-cycle scoring was performed with cell-cycle phase marker genes using the CellCycleScoring function in Seurat.23,24
Primary Human Limbal Epithelial Cultures and In Vitro Cell Growth and Regeneration Models
Primary human limbal epithelial cells (HLECs) were cultured using explants from donor corneal limbal rims for multiple in vitro models by our previous methods.21,25 In brief, for growth stages, cultures were collected at about 70% or 100% confluence for marker staining and reverse transcription and quantitative polymerase chain reaction (RT-qPCR). For the regeneration model, wound incisions were made by scraping cells in 2-mm-wide areas in confluent primary HLEC cultures. Cultures at different time periods of epithelial regeneration after wounding were used for RNA extraction or immunofluorescent staining.
BrdU Proliferation Assays and Immunofluorescent Staining
HLECs were cultured until 70% or 100% confluence, or during different time points after wounding on eight-chamber slides. Each chamber was incubated with fresh SHEM medium with 10-µM 5-bromo-2′-deoxyuridine (BrdU) for 60 minutes at 37°C and 5% CO2 to detect cell proliferation or regeneration. After being labeled with BrdU, slides were washed three times with washing buffer and fixed with 70% acidic ethanol for 20 minutes at –20°C, followed by blocking with 20% normal goat serum in PBS for 60 minutes and incubation with primary anti-BrdU antibody (1:10) for 2 hours. Alexa Fluor 594 (1:200; Thermo Fisher Scientific, Waltham, MA, USA) was used as secondary antibody with 4′,6-diamidino-2-phenylindole (DAPI) for nuclear counterstaining.
Total RNA Extraction, and RT-qPCR
As previously described,25,26 total RNA was isolated from cells using the RNeasy Plus Micro Kit (QIAGEN, Hilden, Germany) and quantified using the NanoDrop ND-1000 (Thermo Fisher Scientific), and stored at –80°C. The first-strand cDNA was synthesized using Ready-To-Go You-Prime First-Strand Beads (Cytiva Life Sciences, Marlborough, MA, USA), and real-time RT-qPCR was performed using the QuantStudio 3 Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) with TaqMan qPCR reagents (Applied Biosystems). The TaqMan gene expression assays included human glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Hs99999905-ml), MKI67 (Hs04260396_g1), RRM2 (Hs00357247_g1), TK1 (Hs01062125_m1), CENPF (Hs00193201_m1), NUSAP1 (Hs01006195_m1), UBE2C (Hs00964100_g1), and CDC20 (Hs00415851_g1) (Applied Biosystems). The results were analyzed by the comparative threshold cycle method and normalized by GAPDH.
Immunofluorescent Staining and Laser Scanning Confocal Microscopy
Immunofluorescent staining was performed using protocols as previously described.25,27 In brief, primary HLECs in different models were fixed with cold acetone at –30°C for 3 minutes or 4% paraformaldehyde at 4°C for 10 minutes. Cultured cells were permeabilized with 0.2% Triton X-100 (Sigma-Aldrich) in PBS at room temperature for 10 minutes. After the samples were blocked with 20% normal goat serum in PBS for 30 minutes, they were incubated with primary antibodies overnight at 4°C. The primary antibodies against markers for TACs were Ki67, RRM2, TK1, CENPF, NUSAP1, UBE2C, and CDC20 (see details in Supplementary Table S2). Alexa Fluor 488 (1:200; Thermo Fisher Scientific) was used as secondary antibody with DAPI for nuclear counterstaining. The digital images were captured with a laser scanning confocal microscope (A1 RMP; Nikon, Melville, NY, USA) at wavelengths from 400 to 750 nm and 1-μm z-steps. The images were processed using NIS Elements 4.20 software (Nikon).
Statistical Analysis
In addition to bioinformatics analysis for scRNA-seq data, Student's t-test or the Mann–Whitney U test was used for all biological experiments to make comparisons between two groups. Analysis of variance or the Kruskal–Wallis test was used to evaluate three or more groups with the appropriate post-test to compare pairs of group means.
Data and Code Availability
All of the raw data are available through the Gene Expression Omnibus (GEO) with accession number GSE153515 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153515). R scripts used to analyze are freely available at github (https://github.com/sangbaekim/10xRNAseq_LSC).
Results
Single-Cell Transcriptomics Revealed a Unique Cell Type Representing TACs in Heterogeneous Limbal Basal Epithelium of Human Cornea
To identify TACs, we isolated single cells from basal limbus where LSCs and their progenitors reside. These cells are relatively small at 10 to 20 µm in diameter, and in our study they accounted for about 15% of total limbal epithelial cells, with 93% viability on average from donor tissues (Supplementary Fig. S1).28
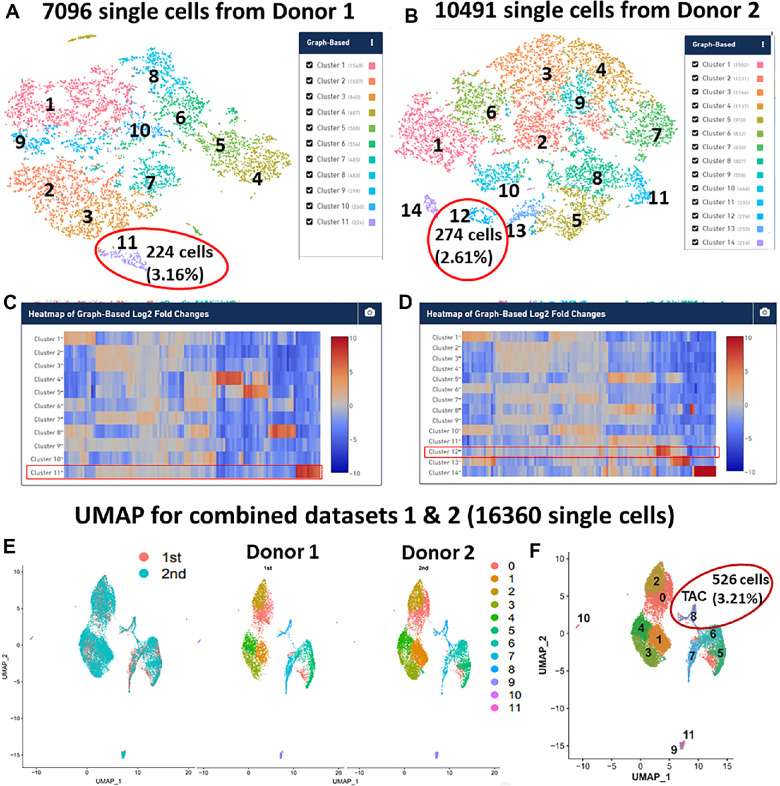
We constructed two scRNA-seq libraries of limbal cells from the two healthy young donors and generated transcriptomic profiles for 7096 and 10,491 cells with 823,020,840 and 861,610,927 total reads, respectively, using the 10x Genomics platform. Mean reads per cell were 115,983 and 82,128, and median genes per cell were 3257 and 2731, respectively (Supplementary Fig. S2). After quality assessment and filtering,22,29 11 and 14 cell clusters were generated from these two donors. Interestingly, the scRNA-seq data revealed a unique cluster with an exclusive gene expression pattern for cell proliferation cluster 11 (C11) from donor 1 (Figs. 1A, 1C) and C12 from donor 2 (Figs. 1B, 1D), which represented a unique cell type with TAC features.
Figure 1.
scRNA-seq revealed the TAC cell type in limbal basal epithelium of human cornea. (A, B) scRNA-seq profiles of basal limbus from both donors showed a unique cluster representing the TAC population. (C, D) Heatmaps from scRNA-seq showed the exclusive gene expression pattern of cluster 11 from donor 1 (C) and cluster 12 from donor 2 (D). (E) UMAP showed similar transcriptomic profiles for cell type clustering for each donor. (F) In a combined dataset of 16,360 single-cell transcriptomic profiles, UMAP revealed that cluster 8 was a TAC population.
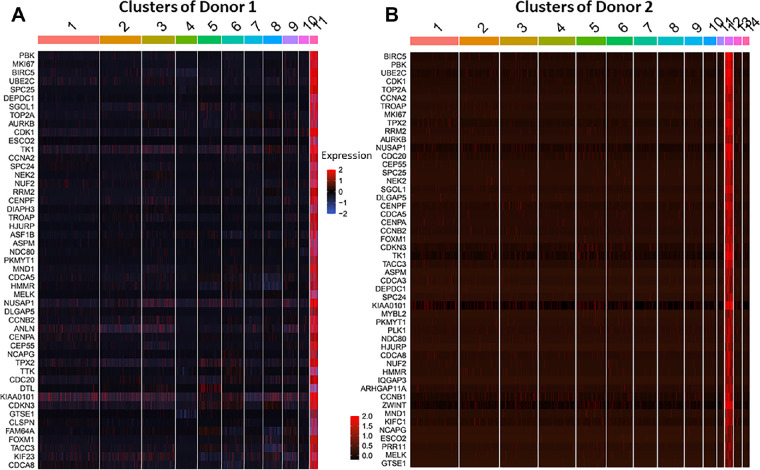
C11 of donor 1 contained 224 cells, which accounted for 3.16% of the total single cells from the limbus, and C12 of donor 2 contained 274 cells (2.61%). Their transcriptomic profiles show a very similar pattern, as 89% of the top 100 DEGs are equal, as shown in their heatmaps (Fig. 2). In contrast, these top DEGs were mostly not or weakly expressed by all other cell clusters in these two datasets.
Figure 2.
Heatmaps revealed a similar gene expression profile of TAC populations from two donors. Heatmaps from scRNA-seq compare the expression patterns of the top 50 DEGs for cluster 11 in donor 1 (A) and cluster 12 in donor 2 (B).
Based on the high similarity, we merged these two scRNA-seq datasets using the graph-based visualization method UMAP in Seurat 3.1.029 (Fig. 1E). A total number of 16,360 cells with good quality data were retained, and 12 cell clusters were identified (Fig. 1E). We identified DEGs in individual clusters using the Wilcoxon rank-sum test in Seurat. As anticipated, among the 12 clusters only cluster 8 displayed an exclusive gene expression pattern (Fig. 1F) like C11 of donor 1 and C12 of donor 2. In the combined dataset, C8 (526 cells) accounted for 3.21% of the total limbal single cells (Fig. 1F), similar to 3.16% and 2.61%, respectively, in the unmerged datasets from donors 1 and 2. The following analysis and TAC identification are based on the combined dataset with the transcriptomic profiles of 16,360 limbal single cells.
A Unique Cell Type Was Identified as a TAC Population Entity Based on Expression Patterns of Marker Genes Known to be Related to Differentiation and Proliferation of Epithelial Cells
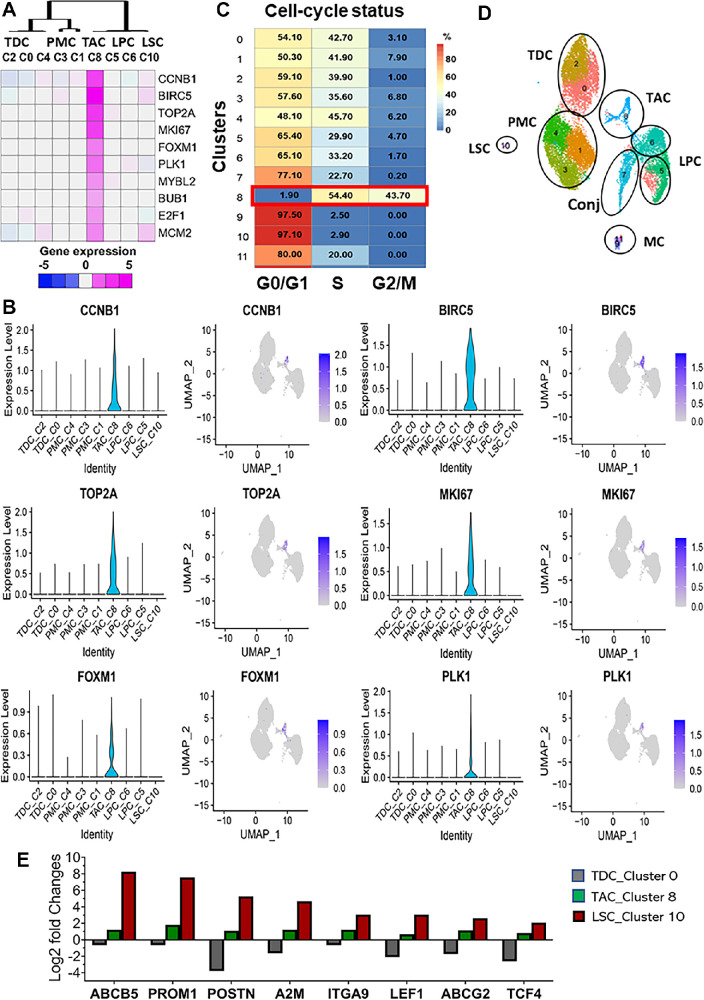
TACs are known to serve as intermediate progenitors between SCs and differentiated cells.6,9,10 To identify the progenitor feature of C8, all nine clusters of limbal epithelial cells were further analyzed; the other three clusters were determined to be conjunctival cells (C7) and melanocytes (C9 and C11).28 Ten markers that have been reported to be related to cell proliferation19,26,30–32 were examined through heatmap visualization (Fig. 3A).
Figure 3.
Cluster 8 was identified as a unique entity representing TAC progenitor cells in the combined scRNA-seq dataset of basal limbus from two donors. (A) Heatmap shows the expression features of C8 TACs using 10 genes known as proliferation markers. (B) Violin and feature plots show the top six proliferation markers enriched by C8. (C) Seurat cell cycle scoring revealed the cell cycle status of each cluster. Most cells (98.1%) in C8 were in cell proliferation cycles. (D) Annotation of 12 clusters showing the different cell types based on the single-cell transcriptomic profiles of 16,360 cells and biological validation. (E) The log2 fold changes of putative stem and progenitor marker genes in three cell types, TDC_C0, TAC_C8, and LSC_C10, as revealed by scRNA-seq. TDC, terminally differentiated cell.
Among the nine clusters, C8 was the only cluster highly expressing major proliferation markers, including CCNB1, BIRC5, TOP2A, MKI67, FOXM1, and PLK1 (Figs. 3A, 3B). The analysis of cell cycle status indicated that most cells (98.1%) in C8 were in cell proliferation cycles, including 54.4% cells at S phase and 43.7% in the G2/M phases (Fig. 3C), suggesting that C8 was the only cell type with proliferative properties in human corneal limbal epithelium. Thus, we identified C8 as a unique cell type of TACs.
In our previous report, we identified C10 as putative LSCs and mapped the trajectory of LSC development stages from LSCs (C10) to limbal progenitor cells (C5 and C6) and TACs (C8), ultimately differentiating to post-mitotic cells (C1, C3, and C4) and terminally differentiated cells (C0 and C2) (Fig. 3D).28
Interestingly, TACs in C8 were found to express moderately high levels of putative stem/progenitor markers such as ABCB5, PROM1, POSTN, A2M, ITGA9, LEF1, ABCG2, and TCF4, whereas these marker genes were strongly upregulated in LSCs (C10) but dramatically downregulated in TDCs (C0), as determined by analyzing the log2 fold changes (all P < 0.05) of these markers in the three cell types from scRNA-seq data (Fig. 3E).
Single-Cell Transcriptomic Profiles Revealed That Many Cell Cycle-Dependent Genes Were Exclusively Enriched by TACs
Interestingly, most of the top TAC population DEGs in C8 not only are proliferation-related genes but are also cell cycle-dependent genes, which directly control and regulate cell cycles during proliferation. The Table lists the top 50 DEGs, among which about 86% are cell cycle-dependent genes; the others are proliferation regulators or related genes. This finding indicates that these cell cycle-dependent genes may serve as the signature markers for TAC populations.
Table.
Top 50 DEGs by TAC Entity in Corneal Limbal Basal Epithelium
| Gene Name | Protein Name | Cell Cycle | Function | |
|---|---|---|---|---|
| 1 | UBE2C | Ubiquitin-conjugating enzyme E2C | M | Controls progression through mitosis |
| 2 | KIAA0101 | Proliferating cell nuclear antigen-associated factor | — | Regulator of DNA repair during DNA replication |
| 3 | TK1 | Thymidine kinase 1 | S | DNA salvage enzyme involved in the synthesis of thymidine triphosphate |
| 4 | BIRC5 | Baculoviral IAP repeat-containing protein 5 | — | Negative regulation of apoptosis or programmed cell death |
| 5 | CENPW | Centromere protein W | G2/M | Assembly of kinetochore proteins, mitotic progression, and chromosome segregation |
| 6 | NUSAP1 | Nucleolar and spindle-associated protein 1 | G2/M | Associated with chromosomes; promotes the organization of mitotic spindle microtubules |
| 7 | ZWINT | ZW10 interacting kinetochore protein | M | Required for kinetochore formation and spindle checkpoint activity |
| 8 | PBK | Lymphokine-activated killer T-cell-originated protein kinase | — | Attenuation of G2/M checkpoint during doxorubicin-induced DNA damage |
| 9 | CENPF | Centromere protein F | G2/M | Required for kinetochore function and chromosome segregation in mitosis |
| 10 | CDC20 | Cell division cycle 20 | M | Required for sister chromatid separation and disassembly of the mitotic spindle |
| 11 | CDK1 | Cyclin-dependent kinase 1 | G1/S, G2/M | Modulating the centrosome cycle and mitotic onset; promotes G2/M transition and G1/S transition |
| 12 | PRC1 | Protein regulator of cytokinesis 1 | S, G2/M | Controlling spatiotemporal formation of the midzone and successful cytokinesis |
| 13 | MAD2L1 | Mitotic spindle assembly checkpoint protein MAD2A | M | Prevents the onset of anaphase until all chromosomes are aligned at the metaphase plate |
| 14 | TYMS | Thymidylate synthetase | — | Catalyzes the conversion of dUMP to dTMP |
| 15 | CENPH | Centromere protein H | M | Plays a role in centromere structure, kinetochore formation, and sister chromatid separation |
| 16 | RRM2 | Ribonucleoside diphosphate reductase subunit M2 | G1/S | Catalyzes the biosynthesis of deoxyribonucleotides from the corresponding ribonucleotides |
| 17 | TOP2A | DNA topoisomerase II alpha | S, G2 | Controls topological states of DNA; essential for proper segregation of daughter chromosomes |
| 18 | TPX2 | Targeting protein for Xklp2 | M | Spindle assembly factor required for normal assembly of mitotic spindles |
| 19 | MKI67 | Marker of proliferation Ki67 | — | Nuclear protein associated with cellular proliferation |
| 20 | CCNB1 | G2/mitotic-specific cyclin-B1 | G2/M | Essential for the control of the cell cycle at the G2/M (mitosis) transition |
| 21 | ANLN | Anillin actin binding protein | G2/M | Plays a role in bleb assembly during metaphase and anaphase of mitosis |
| 22 | STMN1 | Stathmin 1 | G2/M | Regulation of the microtubule filament system; promotes disassembly of microtubules |
| 23 | CENPK | Centromere protein K | M | Assembly of kinetochore proteins, mitotic progression, and chromosome segregation |
| 24 | CDKN3 | Cyclin-dependent kinase inhibitor 3 | G1/S | May play a role in cell cycle regulation |
| 25 | UBE2T | Ubiquitin-conjugating enzyme E2T | G2/M | Promotes proliferation via G2/M checkpoint in hepatocellular carcinoma |
| 26 | GINS2 | DNA replication complex GINS protein PSF2 | G1 | Initiation of DNA replication and progression of DNA replication forks |
| 27 | TROAP | Trophinin-associated protein | — | Involved with bystin and trophinin in a cell adhesion molecule complex |
| 28 | CDT1 | Chromatin licensing and DNA replication factor 1 | G1 | Required for pre-replication complex; promotes stable kinetochore–microtubule attachments |
| 29 | HELLS | Lymphoid-specific helicase | G2/M | Plays a role in DNA methylation, chromatin packaging, control of Hox genes, and stem cell proliferation |
| 30 | CCNB2 | Cyclin B2 | G2/M | Essential component of the cell cycle regulatory machinery |
| 31 | CENPM | Centromere protein M | G2/M | Plays a role in the assembly of kinetochore proteins, mitotic progression, and chromosome segregation |
| 32 | SGOL1 | Shugoshin-like 1 | M | Plays a central role in chromosome cohesion during mitosis |
| 33 | KIF23 | Kinesin family member 23 | M | Transports organelles within cells and moves chromosomes during cell division |
| 34 | DHFR | Dihydrofolate reductase | G1/S | A methyl group shuttle required for the de novo synthesis of purines, thymidylic acid, and certain amino acids |
| 35 | HMMR | Hyaluronan-mediated motility receptor | G2/M | Involved in cell motility |
| 36 | SPC24 | Spindle pole body component 24 | G1/S | Required for chromosome segregation and spindle checkpoint activity |
| 37 | CENPA | Centromere protein A | G2 | Required for recruitment and assembly of kinetochore proteins |
| 38 | KIFC1 | Kinesin family member C1 | M | Minus end-directed microtubule-dependent motor required for bipolar spindle formation |
| 39 | TACC3 | Transforming acidic coiled-coil-containing protein 3 | G2/M | Motor spindle protein that may play a role in stabilization of the mitotic spindle |
| 40 | AURKB | Aurora kinase B | G2/M | Associate with microtubules during chromosome movement and segregation |
| 41 | ASF1B | Anti-silencing function 1B histone chaperone | S | Play a key role in modulating the nucleosome structure of chromatin |
| 42 | NUF2 | Kinetochore protein Nuf2 | G2/M | Associated with centromeres of mitotic HeLa cells |
| 43 | CCNA2 | Cyclin A2 | G2/M | Cyclin that controls both the G1/S and the G2/M transition phases of the cell cycle |
| 44 | CDCA8 | Cell division cycle associated 8 | M | Required for centromeres to ensure correct chromosome alignment, segregation, and chromatin-induced microtubule stabilization and spindle assembly |
| 45 | RMI2 | RecQ-mediated genome instability 2 | — | Plays a role in homologous recombination-dependent DNA repair and is essential for genome stability |
| 46 | CDCA5 | Cell division cycle associated 5 | M | Regulator of sister chromatid cohesion in mitosis, stabilizing cohesin complex association with chromatin |
| 47 | PLK1 | Polo-like kinase 1 | G2/M | Regulation of centrosome maturation, spindle assembly, mitotic exit, and cytokinesis |
| 48 | CEP55 | Centrosomal protein of 55 kDa | M | Mitotic phosphoprotein that plays a key role in cytokinesis |
| 49 | CKAP2L | Cytoskeleton-associated protein 2-like | G2/M | Required for mitotic spindle formation and cell-cycle progression |
| 50 | ARHGAP11A | Rho GTPase activating protein 11A | — | Interacts with p53 and stimulates its tetramerization, which results in cell cycle arrest and apoptosis |
dTMP, deoxythymidine monophosphate; dUMP, deoxyuridine monophosphate.
These genes are known to control and regulate cell cycles in different phases from initiation of DNA synthesis to cell division in proliferation. It has been proposed that many of these genes could serve as disease biomarkers for the diagnosis and prognosis of a variety of cancers. For example, RRM2 may serve as a proliferation marker and pharmaceutical target in adrenocortical cancer33; TK1 overexpression is associated with poor outcomes of patients with lung cancer34; protein expression of BIRC5, TK1, and TOP2A may provide predictive information regarding malignant peripheral nerve sheath tumors after surgical resection35; CCNB2, NUSAP1, and TK1 are associated with the prognosis and progression of hepatocellular carcinoma; UBE2C may serve as a novel prognostic biomarker and a potential therapeutic target for melanoma36; and CDC20 is frequently overexpressed in human cancers.37,38
To identify the signature marker genes of TACs, studies have focused on cell cycle-dependent genes among the top 20 DEGs to explore their expression patterns and functional roles in growth, proliferation, and regeneration of human corneal epithelial cells.
Cell Cycle-Dependent Genes May Serve as TAC Signature Markers Based on Their Expression Patterns in Human Limbal Epithelial Cultures
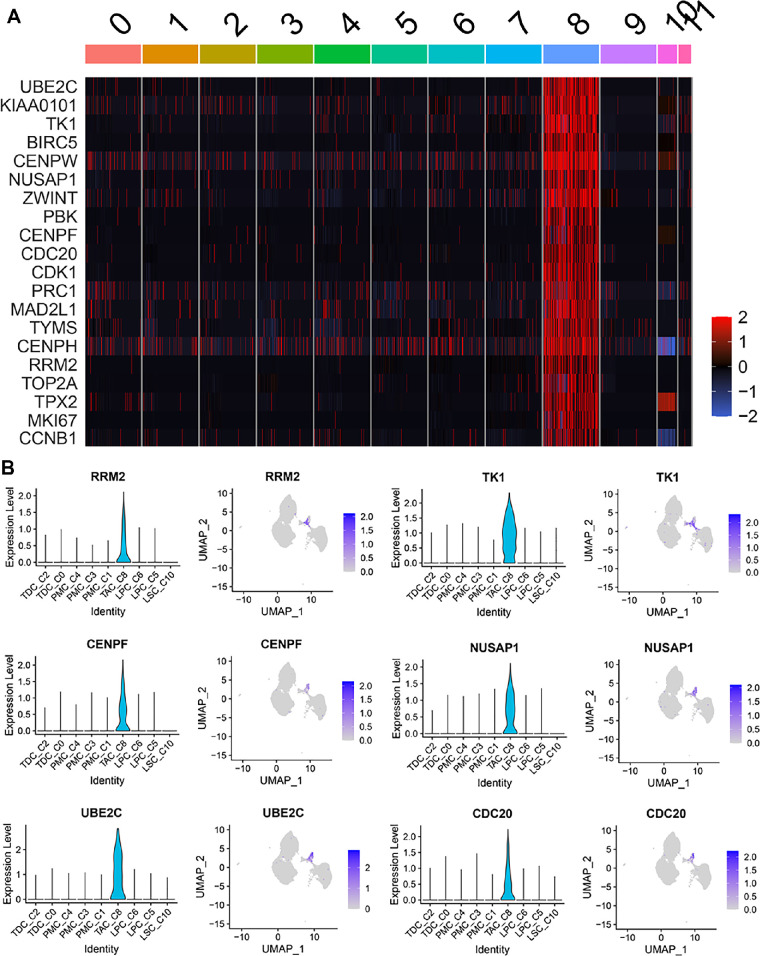
Among the top 20 DEGs (Fig. 4A), the cell cycle-dependent genes RRM2, TK1, CENPF, NUSAP1, UBE2C, and CDC20 (Fig. 4B) were selected for further characterization due to their distinguishable roles in different cell cycle phases. MKI67, a well-known proliferation marker, was used as a positive control.39
Figure 4.
scRNA-Seq revealed the exclusive DEGs in the C8 TAC population. (A) Heatmap shows an exclusive expression pattern for the top 20 DEGs in C8. (B) Violin and feature plots show the expression patterns of six cell cycle-dependent genes in C8 that were selected for TAC characterization.
RRM2 encodes the ribonucleotide reductase regulatory subunit M2, a rate-limiting enzyme for DNA synthesis, by catalyzing the conversion of ribonucleotides into deoxyribonucleotides. RRM2 is expressed only during the late G1/early S phase and is degraded in the late S phase.33,40 TK1 encodes thymidine kinase 1, a pyrimidine salvage enzyme involved in the synthesis and repair of DNA. Its activity is high in proliferating cells and peaks during the S phase of the cell cycle, but it is very low in resting cells.41,42 Both RRM2 and TK1 are genes that initiate proliferation by inducing cells to enter the S phase.
CENPF and NUSAP1 play major roles in the G2/M phase. CENPF encodes centromere protein F, which is involved in chromosome segregation during cell division. In late G2 phase, the protein forms part of the kinetochore, a disc-shaped protein complex that allows the centromere of two sister chromatids to attach to microtubules (forming the spindle apparatus).43,44 NUSAP1 encodes a nucleolar and spindle–associated protein that plays key roles in spindle microtubule organization. The NUSAP1 gene yields the highest protein abundance in M phase. It plays key roles in mitotic progression, spindle formation, and stability.45–47
UBE2C and CDC20 appear to play major roles at M phase, particularly in anaphase, when a cell is dividing and ready for mitotic exit. UBE2C encodes ubiquitin-conjugating enzyme E2C, which is a member of the anaphase-promoting complex/cyclosome (APC/C), promoting the degradation of several target proteins during the metaphase/anaphase transition.36,48 CDC20 encodes the cell division cycle protein 20 homolog and is an essential regulator of cell division; it activates APC/C, which initiates chromatid separation and entrance into the anaphase.49,50 UBE2C ubiquitinates CDC20 to facilitate its binding to APC/C, leading to spindle checkpoint inactivation.51
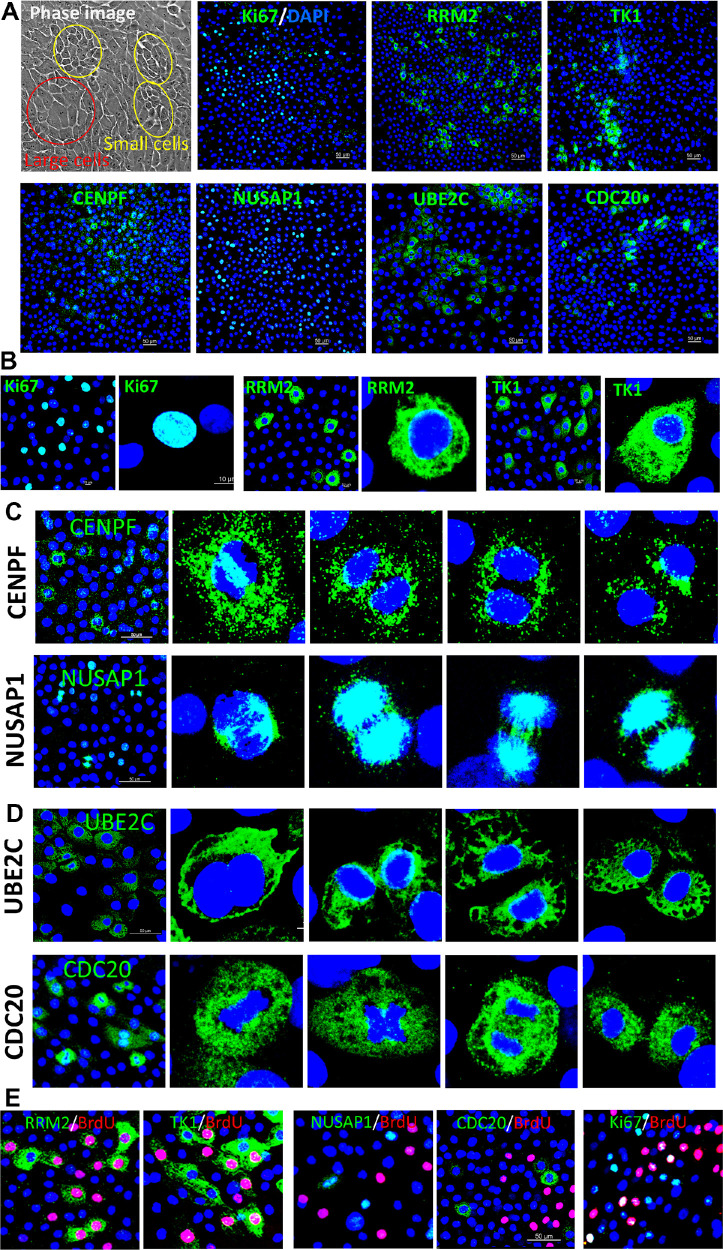
It is well known that primary HLECs cultured from limbal explants display heterogeneous cells ranging from small progenitor cells to bigger differentiated cells,52 as shown in a phase image in Figure 5A. In immunofluorescent staining images, the dense nuclei stained with DAPI indicate small cells, whereas the loose nuclei represent larger cells (Fig. 5A). We observed that most of these cell cycle-dependent proteins were preferentially expressed by small size cells with stronger immunoreactivity in primary HCECs, supporting the notion that TAC cells are progenitor cells.
Figure 5.
Representative immunofluorescent staining images showing the differential expression patterns of six cell cycle-dependent genes at the protein level in HLECs. (A) Six antibodies (green) stained more strongly and with greater frequency in small cells than larger cells, with Ki67 as control. DAPI (blue) was used for nuclear counterstaining. Yellow circle: small cells; red circle: large cells. (B–D) Protein distribution patterns of seven TAC markers. Ki67 showed nuclear staining, and RRM2 and TK1 were entirely cytoplasmic (B). CENPF was partially cytoplasmic surrounding the nucleus and often positive in the dividing cells. NUSAP1 was located at chromosomes in the nucleus; the figure shows the different stages of spindle formation (C). UBE2C and CDC20 were mainly located in the cytoplasm of dividing cells (D). (E) BrdU incorporation with TAC markers. RRM2 and TK1 were partially co-stained with BrdU, but NUSAP1 and CDC20 did not co-stain with BrdU.
Ki67 protein showed nuclear staining, and RRM2 and TK1 immunoreactivities were entirely cytoplasmic (Fig. 5B). Interestingly, special patterns were observed for CENPF, NUSAP1 (Fig. 5C), UBE2C, and CDC20 (Fig. 5D). The CENPF protein was partially cytoplasmic surrounding the nucleus; the positive cells were often in different dividing phases, from beginning to near the end. The NUSAP1 protein appears to be located in chromosomes inside the nucleus, and the different stages of spindle formation can be easily observed in the proliferative cell culture (Fig. 5C). The UBE2C and CDC20 staining was cytoplasmic, mainly in dividing and divided cells, indicating their role in cell cycle anaphase, leading to mitotic exit (Fig. 5D).
To confirm the correlation between cell cycles and these markers, proliferation assays with BrdU labeling for 30 minutes were performed in HLECs, which showed BrdU incorporation in cells in S phase with DNA synthesis. As shown in Figure 5E, double staining with TAC markers clearly showed that cytoplasmic RRM2 and TK1 were often co-stained with BrdU in the nucleus, supporting their role in S phase. Of note, NUSAP1 and CDC20 did not co-stain with BrdU because these two markers are expressed during the mitotic phase. Ki67 nuclear protein was partially co-stained with BrdU, because Ki67 is a general proliferation marker expressed at all cycle phases during cell proliferation, except the resting phase, G0.53
TAC Population Expansion and Activation of Signature Marker Genes Were Correlated With Cell Growing Conditions and Proliferation Stages
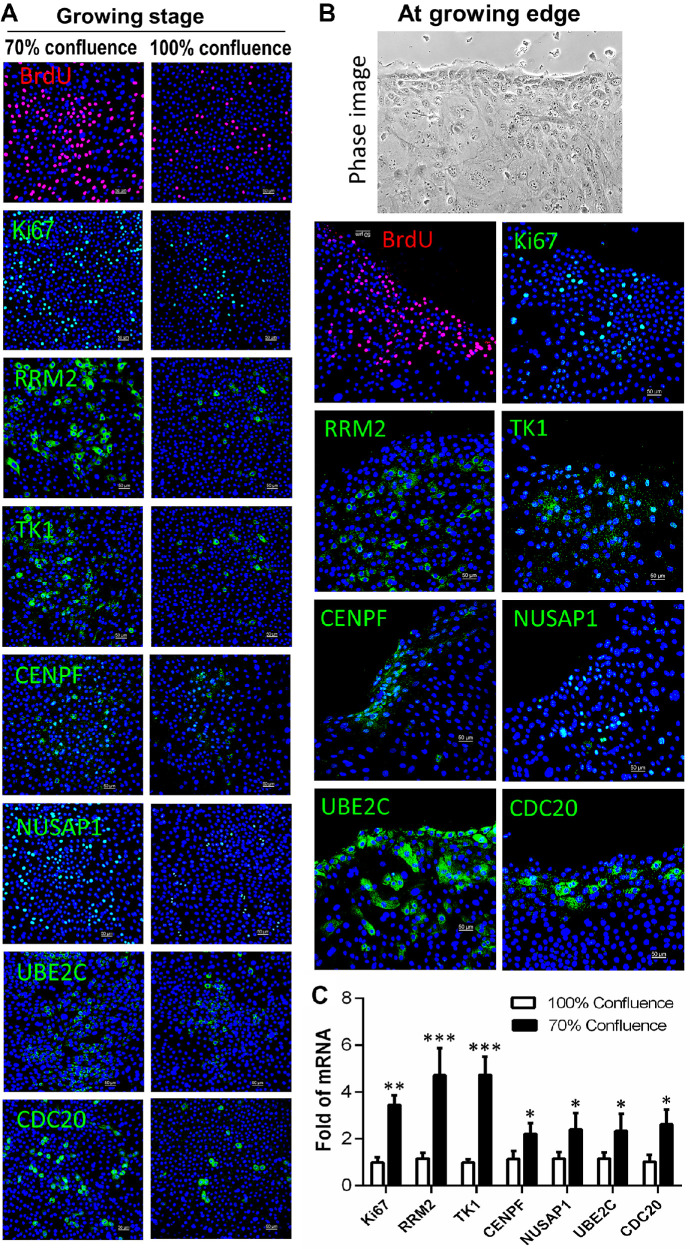
To evaluate the role of these markers in cell growth, the primary HCECs were collected in different growing stages with about 70% or 100% confluence for BrdU proliferation assays, immunofluorescent staining, and RT-qPCR. As shown in Figure 6A, the cultured cells were growing rapidly or exponentially when they reached about 70% confluence, but there was slow or no growth after 100% confluence, as evidenced by the BrdU proliferation assays and general proliferation marker Ki67 with immunofluorescent staining. RRM2, TK1, CENPF, NUSAP1, UBE2C, and CDC20 were all expressed much more strongly in the 70% confluent culture during a rapid growing stage than in the 100% confluent cultures.
Figure 6.
The expression levels of TAC markers were correlated with cell growing stages in HLECs. (A) Representative images show that the immunoreactivity of BrdU, Ki67, RRM2, TK1, CENPF, NUSAP1, UBE2C, and CDC20 was much stronger in the 70% confluent cells than in the 100% confluent cultures. (B) TAC markers showed different patterns at the edges of growing cells. (C) RT-qPCR showed that these seven genes were expressed at a higher rate in the 70% confluent cultures than in the 100% confluent cultures (n = 5). Data are shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
Particularly, we observed that BrdU+ cells and Ki67+ cells were located primarily at or near the growing edges of the 70% confluence rapidly growing cultures. Interestingly, the positive cells of these markers were also preferentially localized near the growing edges (Fig. 6B). TK1 protein was found to translocate from the cytoplasm into the nucleus in some cells at growing edges, indicating activation of TK1 protein during rapidly growing stages.54
The RT-qPCR results shown in Figure 6C confirmed these findings not only at protein but also at mRNA levels. The transcripts of these six marker genes were expressed at significantly higher rates, ranging from 2- to 5-fold, by rapidly growing cells in the 70% confluent cultures than that in the 100% confluent cultures, which contained more differentiated cells. All of these results suggest that these cell cycle-dependent markers are significantly activated during cell cultures, and the activation levels were closely related to cell proliferation conditions and stages. These results also indicate that TAC cells are largely amplified during cell growth.
Functional Roles of TACs With Signature Marker Genes Were Characterized Using the Corneal Epithelial Regeneration Model
Using an in vitro epithelial regeneration model,25 we observed that as the TAC population expanded, the signature marker genes were significantly activated, and their expression levels were well correlated with the epithelial regeneration course after wounding.
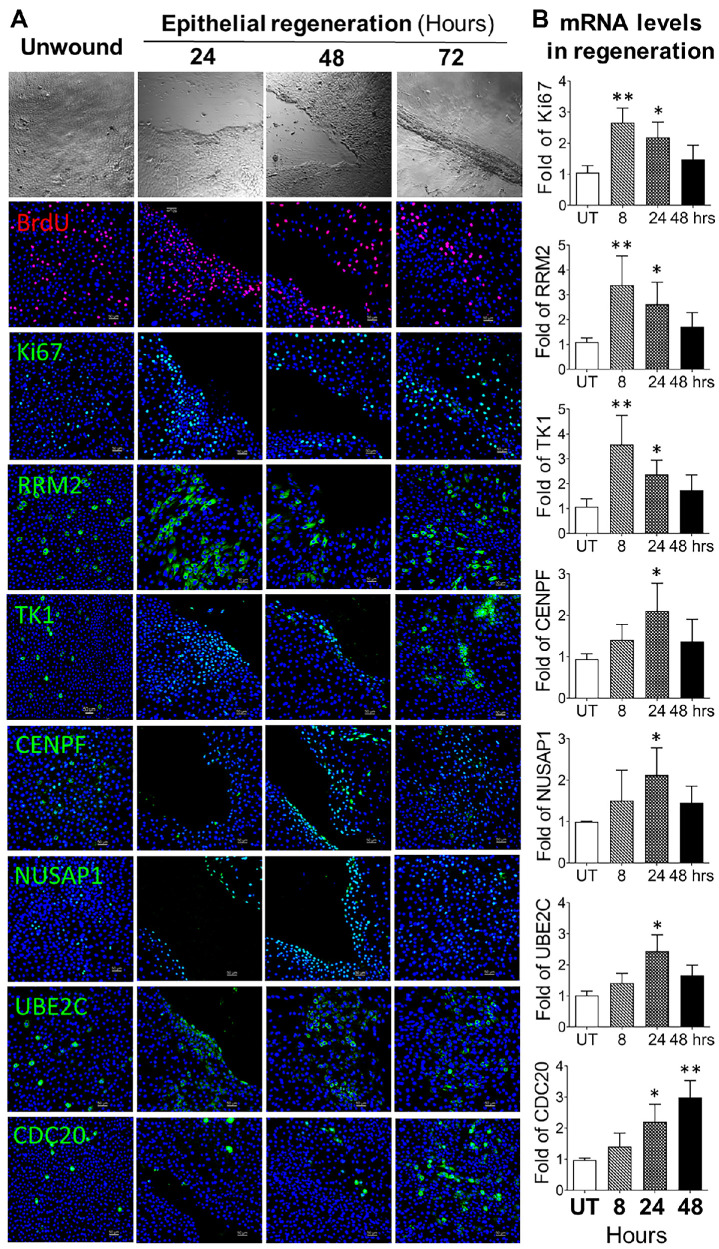
Figure 7A shows the phase images indicating the course of epithelial regeneration after a 2-mm-wide wound was scratched, with untreated cultures serving as controls. Immunofluorescent staining showed that all TAC signature markers were expressed at low levels in the unwounded controls. BrdU-incorporated cells largely increased at the wound edge area during the healing process, with peak levels at 24 hours after wounding, and Ki67 staining showed patterns similar to those for BrdU, which represented cell proliferation levels during the wound closure period.
Figure 7.
TAC markers were highly activated during limbal epithelial regeneration in vitro. (A) Representative images show the BrdU-incorporated cells and protein levels of TAC markers during epithelial regeneration after wounding in HLECs. (B) RT-qPCR determined the mRNA expression of Ki67, RRM2, TK1, CENPF, NUSAP1, UBE2C, and CDC20 during the regeneration process (n = 5). Data are shown as mean ± SD. *P < 0.05; **P < 0.01.
The six cell cycle markers were significantly activated in 24 to 72 hours with different patterns. The increase of positive cells was mostly observed near the wound edges with stronger immunofluorescent intensity. Some cells showed activated TK1 expression with nuclear translocation. Interestingly, the immunoreactivity of RRM2 and TK1 reached peak levels at 24 hours, whereas the cells positive for CENPF, NUSAP1, and UBE2C increased to peak levels at 48 hours, and the highest number of CDC20+ cells occurred after 72 hours, when the wound was closed. These time-differential expression patterns are closely related to the specific roles of the cell cycle markers in cell cycle phases.
Furthermore, RT-qPCR results confirmed the expression pattern of these markers at mRNA levels. The expression of these marker genes increased significantly during the regeneration process. The mRNA expression of RMM2 and TK1 genes reached peak levels at 8 hours, whereas CENPF, NUSAP1, and UBE2C reached peak levels at 24 hours. The highest levels of CDC20 mRNA were observed at 48 hours after wounding (Fig. 7B). These results provide evidence that the TAC population was largely expanded by the strong activation of these cell cycle-dependent genes at protein and mRNA levels during the tissue regeneration process after wounding. The findings suggest that TACs play a vital role in regenerating lost cells and repairing corneal epithelial tissues.
Discussion
Differentiated from ASCs at early stages and serving as the progenitors intermediate between ASCs and differentiated cells,6,9,10 TACs play an important role in tissue homeostasis, development, and regeneration, the most important functional feature of stem cells.5 However, it is still not clear whether a TAC cell type is a real entity or just a functional concept.
The LSCs and TACs paradigm in human corneal epithelium is an excellent model for studying ASCs and TACs because these cell populations are well known to reside at the basal limbus in the human cornea.18,19 Using scRNA-seq, we discovered that single-cell transcriptomic profiles that were generated from two different donors at different times, 4 months apart, revealed the same unique cell type with TAC features in heterogeneous limbal basal epithelium of human cornea (Figs. 1, 2). These cell populations from two different donors showed 89% similarity with regard to the top 100 DEGs in their gene expression profiles. These top DEGs, enriched by TACs, are distinguishable from all other clusters or cell types.
In the combined dataset from 16,360 single cells, C8 (3.21% of total cells) displayed the progenitor feature with a less-differentiated status, as evidenced by the low expression levels of the differentiation markers. C8 was the only cluster exclusively expressing major proliferation markers, with 98.01% of the cells in the proliferative phases of the cell cycle (Fig. 3). Undoubtedly, C8 is a cell type unique to TACs in human corneal limbus.
Interestingly, most of the top DEGs in the C8 TACs are not only proliferation-related genes but also cell cycle-dependent genes, directly controlling and regulating cell cycles during proliferation. In the top 50 DEGs, about 86% of the genes are cell cycle-dependent. The finding that the cell cycle-dependent genes are largely enriched by TACs well supports the role of TACs in maintaining corneal homeostasis. Defects in regeneration or injury repair are also likely to be caused by problems in TACs, not just in stem cells.9
This finding also suggests that the cell cycle-dependent genes may serve as signature markers for TACs. We selected six novel genes to characterize the features of TACs, with general proliferation marker Ki6739 serving as control, from the top 20 DEGs of C8 (Fig 4). Based on their roles in the cell cycle, RRM2 and TK1 are S phase dependent, whereas CENPF, NUSAP1, UBE2C, and CDC20 are specific for the G2/M phase. RRM2 converts ribonucleotides into deoxyribonucleotides,33,55 and TK1 promotes DNA synthesis.34,42 Both are highly expressed at peak levels in S phase for cells coming from G1 phase. The other four genes are G2/M phase specific, but UBE2C and CDC20 appear to play more vital roles at M phase, particularly in anaphase, when the cell is being divided and is ready for mitotic exit.36,43,48,49,56
Multiple culture models were used for characterizing the signature features of these six marker genes for TAC identity. In the confluent HLECs, we observed that most of these cell cycle-dependent proteins were preferentially expressed by small size cells with stronger immunoreactivity, supporting the notion that TAC cells are progenitor cells. The differential patterns of protein distribution of these markers are observed to well represent their specific roles in different cell cycle phases. In particular, the beautiful immunofluorescent images of NUSAP1 showing the spindle formation process and those of UBE2C and CDC20 showing cytoplasmic staining of the dividing and divided cells indicate their roles at mitotic anaphase, before mitotic exit (Fig. 5).
In a cell growth model, we observed that these cell cycle-dependent markers were expressed much more greatly and strongly in the 70% confluent cultures during the rapid growing stages, especially near the growing edges, than in the 100% confluent cultures (Fig. 6).
TAC entities with signature marker genes were further characterized using the corneal epithelial regeneration model. The immunoreactivity of RRM2 and TK1 reached peak levels at 24 hours after wounding, whereas the cells positive for CENPF, NUSAP1, UBE2C, and CDC20 increased to peak levels at 48 to 72 hours, when the wound was closing. These time- and cycle-differential expression patterns at protein levels were also confirmed at mRNA levels with this model, suggesting that their expression levels are closely related to their specific roles in cell cycle phases, as RRM2 and TK1 are mostly activated in S phase34,55 and the others work in the mitotic phase.36,43,49,56
All of these functional assays provide evidence that the TAC population was largely expanded with strong activation of these cell cycle-dependent genes at protein and mRNA levels during the cell growth and regeneration processes. These results also suggest that the cell cycle-dependent genes may serve as signature markers of TACs. Among the six, the four genes in G2/M phase appeared to be more representative of cell cycle status compared with other clusters. As shown in Figure 3C, the huge difference in cell cycle status between TACs and all others is the percentage of cells in G2/M phase, such that TACs represented 43.7% of the cells, and the others accounted for only 0% to 7.9%. In contrast, TACs had 54.4% cells in S phase when other clusters had 2.5% to 45.7%. We propose that CENPF, NUSAP1, UBE2C, and CDC20 may serve as better marker genes to identify TACs in human cornea. Among these four, UBE2C and CDC20 may be the best marker genes because these two may represent the mitotic exit checkpoint genes.
Identification of a TAC entity and signature markers not only advances our understanding of the roles of LSCs and TACs in maintaining corneal homeostasis, but also has an important impact on tumor pathology and treatment. All cell cycle-dependent genes in this study are known to serve as biomarkers that aid in diagnosing disease, determining prognoses and responses to chemotherapy, and identifying metastases, as well as serving as a potential therapeutic target for a variety of malignant diseases.34,36,38,57–59 TACs and cell cycle genes may provide new targets for studying tumor stem cells and TACs in our quest to conquer cancer.
Conclusions
This human corneal scRNA-seq study has identified a unique TAC entity in the heterogeneous limbal basal epithelium. Cell cycle-dependent genes were found to be largely and exclusively enriched by the TAC population. Six novel marker genes were well characterized morphologically and functionally at mRNA and protein levels through the use of multiple cell culture models. We suggest that CENPF, NUSAP1, UBE2C, and CDC20 would serve as better marker genes to identify TACs in human cornea epithelium. The many other cell cycle genes in the list of top DEGs require further investigation for TAC characterization.
Supplementary Material
Acknowledgments
The authors thank Marshall Bowes Hamill, MD, and Alice Matoba, MD, for their kind support and the Lions Eye Bank of Texas for providing human corneoscleral tissues. We also thank Ruthie McNeill for the initial editing of this article.
Supported by grants from the National Eye Institute, National Institutes of Health (EY023598 to D.Q.L.; EY011915 to S.C.P.), by a Core Grant for Vision Research (EY002520), and by the Lions Foundation for Sight, Research to Prevent Blindness, Oshman Foundation, and William Stamps Farish Fund.
Disclosure: J.-M. Li, None; S. Kim, None; Y. Zhang, None; F. Bian, None; J. Hu, None; R. Lu, None; S.C. Pflugfelder, None; R. Chen, None; D.-Q. Li, None
References
- 1. Blau HM, Brazelton TR, Weimann JM.. The evolving concept of a stem cell: entity or function? Cell. 2001; 105: 829–841. [DOI] [PubMed] [Google Scholar]
- 2. Tseng SC. Concept and application of limbal stem cells. Eye. 1989; 3: 141–157. [DOI] [PubMed] [Google Scholar]
- 3. Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM.. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell. 1989; 57: 201–209. [DOI] [PubMed] [Google Scholar]
- 4. Watt FM. Epidermal stem cells as targets for gene transfer. Hum Gene Ther. 2000; 11: 2261–2266. [DOI] [PubMed] [Google Scholar]
- 5. Post Y, Clevers H.. Defining adult stem cell function at its simplest: the ability to replace lost cells through mitosis. Cell Stem Cell. 2019; 25: 174–183. [DOI] [PubMed] [Google Scholar]
- 6. Rangel-Huerta E, Maldonado E.. Transit-amplifying cells in the fast lane from stem cells towards differentiation. Stem Cells Int. 2017; 2017: 7602951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zieske JD. Perpetuation of stem cells in the eye. Eye. 1994; 8: 163–169. [DOI] [PubMed] [Google Scholar]
- 8. Silva-Vargas V, Delgado AC, Doetsch F.. Symmetric Stem Cell Division at the Heart of Adult Neurogenesis. Neuron. 2018; 98: 246–248. [DOI] [PubMed] [Google Scholar]
- 9. Zhang B, Hsu YC.. Emerging roles of transit-amplifying cells in tissue regeneration and cancer. Wiley Interdiscip Rev Dev Biol. 2017; 6: 10.1002/wdev.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diaz-Flores L Jr, Madrid JF, Gutierrez R, et al.. Adult stem and transit-amplifying cell location. Histol Histopathol. 2006; 21: 995–1027. [DOI] [PubMed] [Google Scholar]
- 11. Witte K, Jurchott K, Christou D, et al.. Increased presence and differential molecular imprinting of transit amplifying cells in psoriasis. J Mol Med (Berl). 2020; 98: 111–122. [DOI] [PubMed] [Google Scholar]
- 12. Liu S, Zhou J, Zhang X, et al.. Strategies to optimize adult stem cell therapy for tissue regeneration. Int J Mol Sci. 2016; 17: 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rezza A, Wang Z, Sennett R, et al.. Signaling networks among stem cell precursors, transit-amplifying progenitors, and their niche in developing hair follicles. Cell Rep. 2016; 14: 3001–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones PH, Simons BD, Watt FM.. Sic transit gloria: farewell to the epidermal transit amplifying cell? Cell Stem Cell. 2007; 1: 371–381. [DOI] [PubMed] [Google Scholar]
- 15. Shekhar K, Lapan SW, Whitney IE, et al.. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016; 166: 1308–1323.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Plass M, Solana J, Wolf FA, et al.. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science. 2018; 360: eaaq1723. [DOI] [PubMed] [Google Scholar]
- 17. O'Sullivan F, Clynes M.. Limbal stem cells, a review of their identification and culture for clinical use. Cytotechnology. 2007; 53: 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaplan N, Wang J, Wray B, et al.. Single-cell RNA transcriptome helps define the limbal/corneal epithelial stem/early transit amplifying cells and how autophagy affects this population. Invest Ophthalmol Vis Sci. 2019; 60: 3570–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lehrer MS, Sun TT, Lavker RM.. Strategies of epithelial repair: modulation of stem cell and transit amplifying cell proliferation. J Cell Sci. 1998; 111: 2867–2875. [DOI] [PubMed] [Google Scholar]
- 20. Li DQ, Wang Z, Yoon KC, Bian F.. Characterization, isolation, expansion and clinical therapy of human corneal epithelial stem/progenitor cells. J Stem Cells. 2014; 9: 79–91. [PubMed] [Google Scholar]
- 21. Li D-Q, Chen Z, Song XJ, de Paiva CS, Kim HS, Pflugfelder SC. Partial enrichment of a population of human limbal epithelial cells with putative stem cell properties based on collagen type IV adhesiveness. Exp Eye Res. 2005; 80: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018; 36: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kowalczyk MS, Tirosh I, Heckl D, et al.. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015; 25: 1860–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tirosh I, Izar B, Prakadan SM, et al.. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016; 352: 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu R, Qu Y, Ge J, et al.. Transcription factor TCF4 maintains the properties of human corneal epithelial stem cells. Stem Cells. 2012; 30: 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bian F, Liu W, Yoon KC, et al.. Molecular signatures and biological pathway profiles of human corneal epithelial progenitor cells. Int J Biochem Cell Biol. 2010; 42: 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Z, de Paiva CS, Luo L, Kretzer FL, Pflugfelder SC, Li D-Q.. Characterization of putative stem cell phenotype in human limbal epithelia. Stem Cells. 2004; 22: 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li DQ, Kim S, Li JM, et al.. Single-cell transcriptomics identifies limbal stem cell population and cell types mapping its differentiation trajectory in limbal basal epithelium of human cornea. Ocul Surf. 2021; 20: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stuart T, Butler A, Hoffman P, et al.. Comprehensive integration of single-cell data. Cell. 2019; 177: 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Joyce NC, Meklir B, Joyce SJ, Zieske JD.. Cell cycle protein expression and proliferative status in human corneal cells. Invest Ophthalmol Vis Sci. 1996; 37: 645–655. [PubMed] [Google Scholar]
- 31. Lu R, Bian F, Zhang X, et al.. The β-catenin/Tcf4/survivin signaling maintains a less differentiated phenotype and high proliferative capacity of human corneal epithelial progenitor cells. Int J Biochem Cell Biol. 2011; 43: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guzman A, Ramos-Balderas JL, Carrillo-Rosas S, Maldonado E.. A stem cell proliferation burst forms new layers of P63 expressing suprabasal cells during zebrafish postembryonic epidermal development. Biol Open. 2013; 2: 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heidel JD, Liu JY, Yen Y, et al.. Potent siRNA inhibitors of ribonucleotide reductase subunit RRM2 reduce cell proliferation in vitro and in vivo. Clin Cancer Res. 2007; 13: 2207–2215. [DOI] [PubMed] [Google Scholar]
- 34. Wei YT, Luo YZ, Feng ZQ, Huang QX, Mo AS, Mo SX.. TK1 overexpression is associated with the poor outcomes of lung cancer patients: a systematic review and meta-analysis. Biomark Med. 2018; 12: 403–413. [DOI] [PubMed] [Google Scholar]
- 35. Kolberg M, Holand M, Lind GE, et al.. Protein expression of BIRC5, TK1, and TOP2A in malignant peripheral nerve sheath tumours–a prognostic test after surgical resection. Mol Oncol. 2015; 9: 1129–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu G, Zhao J, Pan B, Ma G, Liu L.. UBE2C overexpression in melanoma and its essential role in G2/M transition. J Cancer. 2019; 10: 2176–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Q, Huang H, Liu A, et al.. Cell division cycle 20 (CDC20) drives prostate cancer progression via stabilization of β-catenin in cancer stem-like cells. EBioMedicine. 2019; 42: 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Z, Wan L, Zhong J, et al.. Cdc20: a potential novel therapeutic target for cancer treatment. Curr Pharm Des. 2013; 19: 3210–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou Y, Hu W, Chen P, et al.. Ki67 is a biological marker of malignant risk of gastrointestinal stromal tumors: a systematic review and meta-analysis. Medicine (Baltimore). 2017; 96: e7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li J, Pang J, Liu Y, et al.. Suppression of RRM2 inhibits cell proliferation, causes cell cycle arrest and promotes the apoptosis of human neuroblastoma cells and in human neuroblastoma RRM2 is suppressed following chemotherapy. Oncol Rep. 2018; 40: 355–360. [DOI] [PubMed] [Google Scholar]
- 41. Zhu X, Shi C, Peng Y, et al.. Thymidine kinase 1 silencing retards proliferative activity of pancreatic cancer cell via E2F1-TK1-P21 axis. Cell Prolif. 2018; 51: e12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou J, He E, Skog S.. The proliferation marker thymidine kinase 1 in clinical use. Mol Clin Oncol. 2013; 1: 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shahid M, Lee MY, Piplani H, et al.. Centromere protein F (CENPF), a microtubule binding protein, modulates cancer metabolism by regulating pyruvate kinase M2 phosphorylation signaling. Cell Cycle. 2018; 17: 2802–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Varis A, Salmela AL, Kallio MJ.. Cenp-F (mitosin) is more than a mitotic marker. Chromosoma. 2006; 115: 288–295. [DOI] [PubMed] [Google Scholar]
- 45. Zhang X, Pan Y, Fu H, Zhang J.. Nucleolar and spindle associated protein 1 (NUSAP1) inhibits cell proliferation and enhances susceptibility to epirubicin in invasive breast cancer cells by regulating cyclin D kinase (CDK1) and DLGAP5 expression. Med Sci Monit. 2018; 24: 8553–8564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gordon CA, Gong X, Ganesh D, Brooks JD.. NUSAP1 promotes invasion and metastasis of prostate cancer. Oncotarget. 2017; 8: 29935–29950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Okamoto A, Higo M, Shiiba M, et al.. Down-regulation of nucleolar and spindle-associated protein 1 (NUSAP1) expression suppresses tumor and cell proliferation and enhances anti-tumor effect of paclitaxel in oral squamous cell carcinoma. PLoS One. 2015; 10: e0142252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hao Z, Zhang H, Cowell J.. Ubiquitin-conjugating enzyme UBE2C: molecular biology, role in tumorigenesis, and potential as a biomarker. Tumour Biol. 2012; 33: 723–730. [DOI] [PubMed] [Google Scholar]
- 49. Kapanidou M, Curtis NL, Bolanos-Garcia VM.. Cdc20: at the crossroads between chromosome segregation and mitotic exit. Trends Biochem Sci. 2017; 42: 193–205. [DOI] [PubMed] [Google Scholar]
- 50. Irniger S. Cyclin destruction in mitosis: a crucial task of Cdc20. FEBS Lett. 2002; 532: 7–11. [DOI] [PubMed] [Google Scholar]
- 51. Reddy SK, Rape M, Margansky WA, Kirschner MW.. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007; 446: 921–925. [DOI] [PubMed] [Google Scholar]
- 52. de Paiva CS, Pflugfelder SC, Li D-Q.. Cell size correlates with phenotype and proliferative capacity in human corneal epithelial cells. Stem Cells. 2006; 24: 368–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li LT, Jiang G, Chen Q, Zheng JN.. Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol Med Rep. 2015; 11: 1566–1572. [DOI] [PubMed] [Google Scholar]
- 54. Chen G, He C, Li L, et al.. Nuclear TK1 expression is an independent prognostic factor for survival in pre-malignant and malignant lesions of the cervix. BMC Cancer. 2013; 13: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grolmusz VK, Karaszi K, Micsik T, et al.. Cell cycle dependent RRM2 may serve as proliferation marker and pharmaceutical target in adrenocortical cancer. Am J Cancer Res. 2016; 6: 2041–2053. [PMC free article] [PubMed] [Google Scholar]
- 56. Mills CA, Suzuki A, Arceci A, et al.. Nucleolar and spindle-associated protein 1 (NUSAP1) interacts with a SUMO E3 ligase complex during chromosome segregation. J Biol Chem. 2017; 292: 17178–17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun J, Huang J, Lan J, et al.. Overexpression of CENPF correlates with poor prognosis and tumor bone metastasis in breast cancer. Cancer Cell Int. 2019; 19: 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sun L, Shi C, Liu S, et al.. Overexpression of NuSAP1 is predictive of an unfavourable prognosis and promotes proliferation and invasion of triple-negative breast cancer cells via the Wnt/β-catenin/EMT signalling axis. Gene. 2020; 747: 144657. [DOI] [PubMed] [Google Scholar]
- 59. Zhan Y, Jiang L, Jin X, et al.. Inhibiting RRM2 to enhance the anticancer activity of chemotherapy. Biomed Pharmacother. 2020; 133: 110996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the raw data are available through the Gene Expression Omnibus (GEO) with accession number GSE153515 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153515). R scripts used to analyze are freely available at github (https://github.com/sangbaekim/10xRNAseq_LSC).