Supplemental Digital Content is available in the text.
Keywords: endothelial cells, microcirculation, pulmonary circulation, transcriptome
Background:
Cellular diversity of the lung endothelium has not been systematically characterized in humans. We provide a reference atlas of human lung endothelial cells (ECs) to facilitate a better understanding of the phenotypic diversity and composition of cells comprising the lung endothelium.
Methods:
We reprocessed human control single-cell RNA sequencing (scRNAseq) data from 6 datasets. EC populations were characterized through iterative clustering with subsequent differential expression analysis. Marker genes were validated by fluorescent microscopy and in situ hybridization. scRNAseq of primary lung ECs cultured in vitro was performed. The signaling network between different lung cell types was studied. For cross-species analysis or disease relevance, we applied the same methods to scRNAseq data obtained from mouse lungs or from human lungs with pulmonary hypertension.
Results:
Six lung scRNAseq datasets were reanalyzed and annotated to identify >15 000 vascular EC cells from 73 individuals. Differential expression analysis of EC revealed signatures corresponding to endothelial lineage, including panendothelial, panvascular, and subpopulation-specific marker gene sets. Beyond the broad cellular categories of lymphatic, capillary, arterial, and venous ECs, we found previously indistinguishable subpopulations; among venous EC, we identified 2 previously indistinguishable populations: pulmonary–venous ECs (COL15A1neg) localized to the lung parenchyma and systemic–venous ECs (COL15A1pos) localized to the airways and the visceral pleura; among capillary ECs, we confirmed their subclassification into recently discovered aerocytes characterized by EDNRB, SOSTDC1, and TBX2 and general capillary EC. We confirmed that all 6 endothelial cell types, including the systemic–venous ECs and aerocytes, are present in mice and identified endothelial marker genes conserved in humans and mice. Ligand-receptor connectome analysis revealed important homeostatic crosstalk of EC with other lung resident cell types. scRNAseq of commercially available primary lung ECs demonstrated a loss of their native lung phenotype in culture. scRNAseq revealed that endothelial diversity is maintained in pulmonary hypertension. Our article is accompanied by an online data mining tool (www.LungEndothelialCellAtlas.com).
Conclusions:
Our integrated analysis provides a comprehensive and well-crafted reference atlas of ECs in the normal lung and confirms and describes in detail previously unrecognized endothelial populations across a large number of humans and mice.
Clinical Perspective.
What Is New?
Single-cell RNA sequencing resolves the identities of previously indistinguishable endothelial populations: pulmonary–venous endothelial cells (ECs) localized to the lung parenchyma, systemic–venous ECs localized to the airways and the visceral pleura, as well as recently described aerocytes and general capillary ECs.
Ligand–receptor connectome analysis highlights the important role of lung ECs in the homeostatic communication with other lung resident cell types.
Novel EC types are conserved in mice, but their marker genes may differ from those of corresponding human EC types.
What Are the Clinical Implications?
Understanding lung endothelial diversity is crucially important to identify new therapeutic approaches in vascular diseases such as pulmonary hypertension.
Single-cell RNA sequencing enables exploration of genes associated with mendelian diseases with involvement of the lung endothelium to gain insights into the contribution of respective EC types.
The most critical function of the circulatory system is to provide tissues throughout the body with a constant supply of oxygenated blood. In mammals, the circulatory system accomplishes this by performing gas exchange with the environment exclusively in the lung. Consistent with its hyperspecialized role, the lung is a highly vascularized organ uniquely composed of 2 circulations.1 The systemic circulation is primarily restricted to the large airways and the pleura, to which it supplies oxygenated blood. In contrast, the pulmonary circulation supplies oxygen-depleted blood to the lung parenchyma to perform gas exchange. The contrasting physiologic functions of these circulations, as well as the distinct cellular niches in which each resides, underscores the diverse and unique roles played by endothelial cells (ECs) in the lung.
The endothelium is more than a simple physical barrier between blood, air, and stromal tissue. The endothelium is metabolically active and has been found to play a key role in processes governing inflammation, leukocyte trafficking, gas and nutrient exchange, hemostasis, angiogenesis, vascular tone, and endocrine signaling.2–6 Lung endothelium experiences constant stretching during breathing and is persistently exposed to substances of the external environment. Dysfunction of the lung endothelium has been described in numerous clinical conditions, including acute respiratory distress syndrome, chronic obstructive pulmonary disease, pulmonary fibrosis, pneumonia, autoimmune conditions, pulmonary hypertension, and others.7
The importance of ECs to lung function in health and disease is clear, but a detailed reference atlas of ECs in the healthy human lung is missing. We created an integrated single-cell atlas of human lungs to identify EC populations, characterize them by prototypic gene expression profiles, and explore their role in tissue homeostasis. To identify cross-species conserved cell populations and markers, as well as key differences between the mouse and the human, we generated an integrated murine endothelial atlas and compared human lung EC populations with those of mice. This article is accompanied by a publicly accessible online tool to interactively explore the data (www.LungEndothelialCellAtlas.com).
Methods
New sequencing data have been made publicly available on Gene Expression Omnibus under the accession number GSE164829 and raw imaging data on Zenodo.8
Complete details can be found in the Methods in the Data Supplement. Briefly, we reprocessed and reanalyzed control lung single-cell RNA sequencing (scRNAseq) data from 5 cohorts9–13 that we used to identify panendothelial and panvascular marker genes. The Yale-Baylor cohort includes 4 previously unreported samples. The corresponding study protocol was approved by the Partners Healthcare institutional review board and the Yale University institutional review board and tissue donors (or next of kin) gave informed consent. We then included a sixth dataset14 of sorted control ECs to interrogate vascular endothelial subpopulations. EC populations were characterized through iterative clustering with subsequent differential expression analysis. EC subpopulations were localized by immunofluorescent, immunohistochemical, and in situ hybridization stains. Using a connectomic analysis, the signaling network between different lung cell types was investigated. For cross-species analysis, we applied the same methods to scRNAseq data obtained from mouse lungs (Figure XII in the Data Supplement).
Statistical Analysis
Cell type–specific marker genes were established using the Wilcoxon rank sum test with P values adjusted for multiple comparisons using the Bonferroni method. Adjusted P values <0.05 were considered significant. For further details, see supplemental methods in the Data Supplement.
Results
Integrated Single-Cell Atlas of the Human Lung
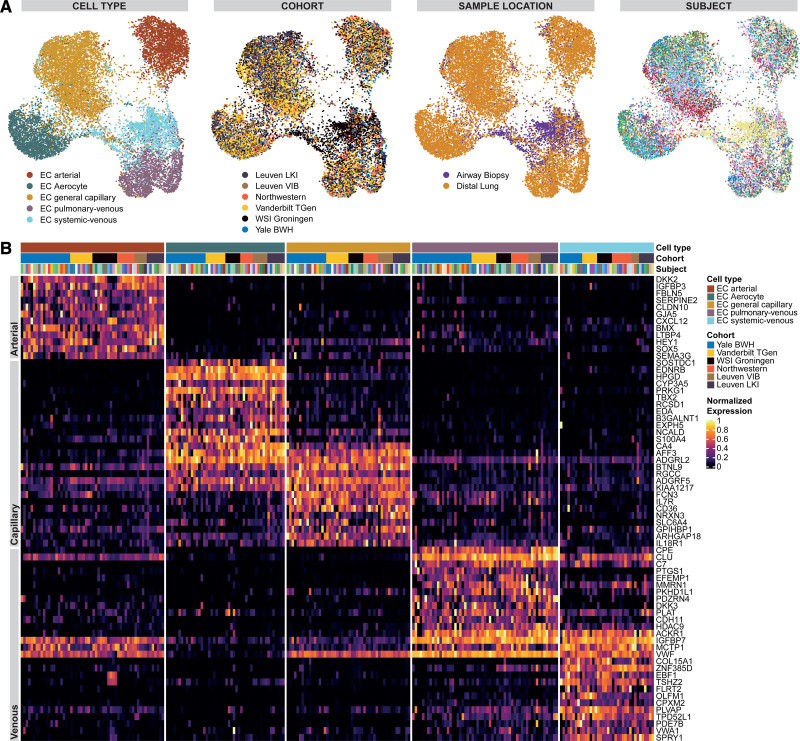
To profile EC populations in the human lung, we collected, reprocessed, and integrated scRNAseq data of 5 publicly available datasets through a common computational pipeline (Methods in the Data Supplement): Vanderbilt–Translational Genomics Research Institute9 (n=10), Wellcome Sanger Institute–Groningen10 (n=10), Northwestern11 (n=8), Leuven Vlaams Instituut voor Biotechnologie12 (n=6), and Yale–Baylor13 (n=34, including samples from 4 previously unpublished additional participants; Figure 1 and Table I in the Data Supplement). The median age of participants was 50 years (interquartile range [IQR], 32–61 years), and 29 out of 68 were female (for basic characteristics by cohort, see Table II in the Data Supplement). In all cohorts, single-cell barcoding had been performed using 10× single-cell RNA 3′ sequencing kits based on v2 Chemistry with the exception of the Vanderbilt–Translational Genomics Research Institute cohort, which had used the 5′ technology. Four samples were derived from airway biopsies and all others from dissociated distal lung samples. Raw scRNAseq data from all 68 human lung samples were aggregated and collectively processed, resulting in the lung dataset consisting of 278 648 cells including 12 563 vascular and lymphatic ECs from 66 of the 68 samples. We identified 37 cell types that aligned with canonical marker gene expression signatures of established cell types (Figures 1B and 2, Figure I in the Data Supplement, and Table III in the Data Supplement). Reported marker genes were universally detected across cell types, all scRNAseq assay schemes, all cohorts, and most participants, and therefore do not represent participant-specific gene expression. Technical summaries of sample preprocessing results can be found in Table IV in the Data Supplement and Figure II in the Data Supplement.
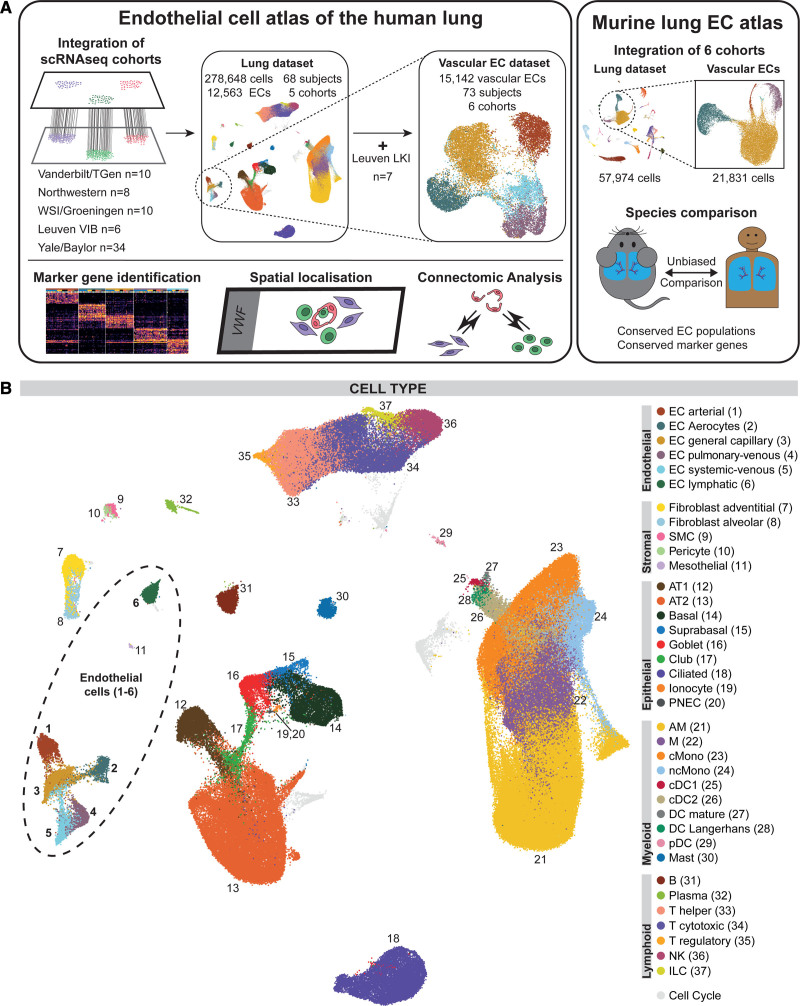
Figure 1.
Overview of the study design and the integrated dataset. A, Overview of study design. Single-cell RNA sequencing (scRNAseq) data from control samples of 5 cohorts (Vanderbilt/Translational Genomics Research Institute [TGen]9: GSE135893, n=10; Northwestern11: phs001750.v1.p1, n=8; Wellcome Sanger Institute [WSI]/Groningen10: EGAD00001005064 and EGAD00001005065, n=10; Leuven Vlaams Instituut voor Biotechnologie [VIB]12; E-MTAB-6149 and E-MTAB-6653, n=6; Yale/Baylor13,39: GSE136831 and GSE133747, n=34) were integrated to form the lung cell dataset. The Yale-Baylor cohort includes 4 previously unreported samples (now published under GSE164829). All vascular endothelial cells (ECs) from samples in which ECs had been profiled (n=66) were integrated with addition of ECs from control samples of a sixth cohort of sorted lung ECs (Leuvens Kankerinstituut [LKI]14: E-MTAB-6308, n=7) to form the vascular EC dataset. Differential expression analysis of ECs revealed signatures corresponding to panendothelial, panvascular, and subpopulation-specific marker gene sets. EC subpopulations were localized by immunofluorescence and immunohistochemical microscopy and in situ hybridization. Potential homeostatic crosstalk of EC with other lung resident cell types was explored by ligand–receptor connectome analysis. scRNAseq data from 6 mouse cohorts were integrated to identify murine marker genes and conserved marker genes in humans and mice. B, Uniform manifold approximation and projection representation of the lung cell dataset of 278 648 cells from 68 control lungs; each dot represents a single cell, and cells are labeled as 1 of 37 discrete cell types. For uniform manifold approximation and projections colored by cohort and participant, refer to Figure IA in the Data Supplement. AM indicates alveolar macrophage; AT1/2, alveolar cell type 1/2; cDC1/2, classical dendritic cell type 1/2; cMono, classical monocyte; DC, dendritic cell; ILC, innate lymphoid cell; M, macrophage; ncMono, nonclassical monocyte; NK, natural killer; pDC, plasmacytoid dendritic cell; PNEC, pulmonary neuroendocrine cell; and SMC, smooth muscle cell.
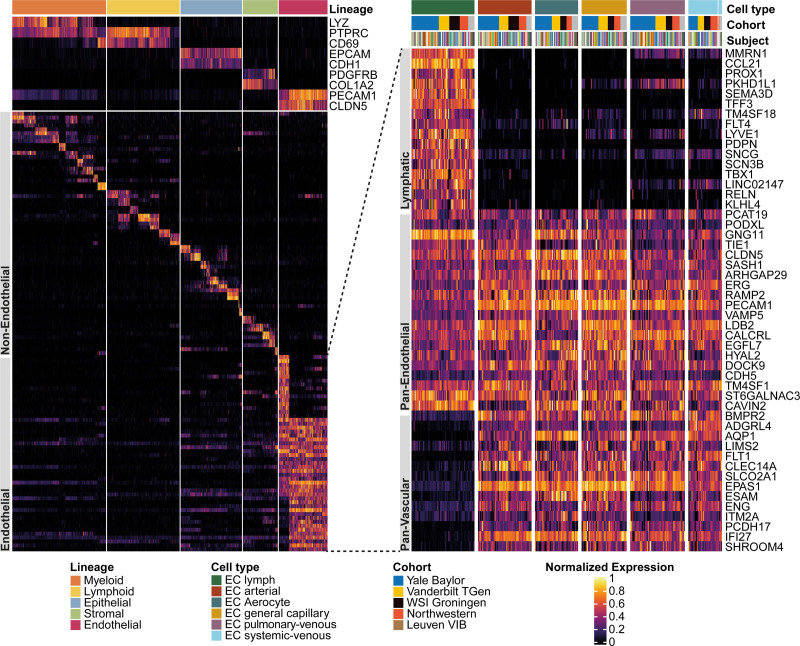
Figure 2.
Assessing coarse-granular endothelial heterogeneity of the human lung by single-cell RNA sequencing. Heat map of marker genes for all 29 non–endothelial cell (EC) cell types and of lymphatic cells as well as panendothelial (specifically expressed in all 6 EC populations) and panvascular (specifically expressed in all EC populations except lymphatic ECs) marker genes. Cell types were grouped into lineages and lineage marker genes are shown at the top of the heat map to the right. Then, 2 marker genes per non-EC cell type are shown, followed by a focus on ECs. The heat map to the left zooms in on the gene expression of ECs and depicts, from top to bottom, sets of lymphatic, panendothelial, and panvascular marker genes in the 6 endothelial populations. Each column represents the average expression value for 1 participant, grouped by cell type and cohort. All gene expression values are unity normalized from 0 to 1 across rows. For an enlarged heat map of non-EC cell types, see Figure I in the Data Supplement. TGen indicates Translational Genomics Research Institute; VIB, Leuven Vlaams Instituut voor Biotechnologie; and WSI, Wellcome Sanger Institute.
We describe all EC populations based on the marker genes with which they were identified followed by a transcriptional/functional characterization of the corresponding populations.
Lung EC Markers, Assessed at Levels of Lineage, Anatomy, and Cell Type
We identified 12 563 ECs within the integrated lung dataset based on the canonical endothelial markers PECAM1 (CD31), CDH5 (VE-cadherin), CLDN5, and ERG (Figures 2, 4, and 5). We identified 147 panendothelial marker genes, defined as genes significantly expressed in all vascular and lymphatic EC populations compared with all other cell types (Table III in the Data Supplement). Panendothelial markers include genes coding for proteins associated with endothelial-intrinsic structures like the glycocalyx (PODXL, ST6GALNAC3, GALNT18), caveolae (CAV1, CAV2, CAVIN1, CAVIN2), focal adhesion structures (ITGA5, ITGB1, LAMB2, AKT3, PXN, MYL12A, MYL12B, PRK2), and adherens junctions (AFDN, TJP1, PTPRM; Figure 2 and Table III in the Data Supplement). All EC subpopulations express common transcription factors (ERG, ETS1, FLI1, SOX18) and regulators of endothelial migration (ANXA3, SASH1, LDB2, PTK2, MEF2C, STARD13). Other panendothelial markers include angiopoietin receptor TIE1, subunits of the receptors for calcitonin gene-related peptide and adrenomedullin CALCRL and RAMP2/RAMP3, as well as general endothelial receptors ROBO4 and S1PR1 (Figure 2 and Table III in the Data Supplement).
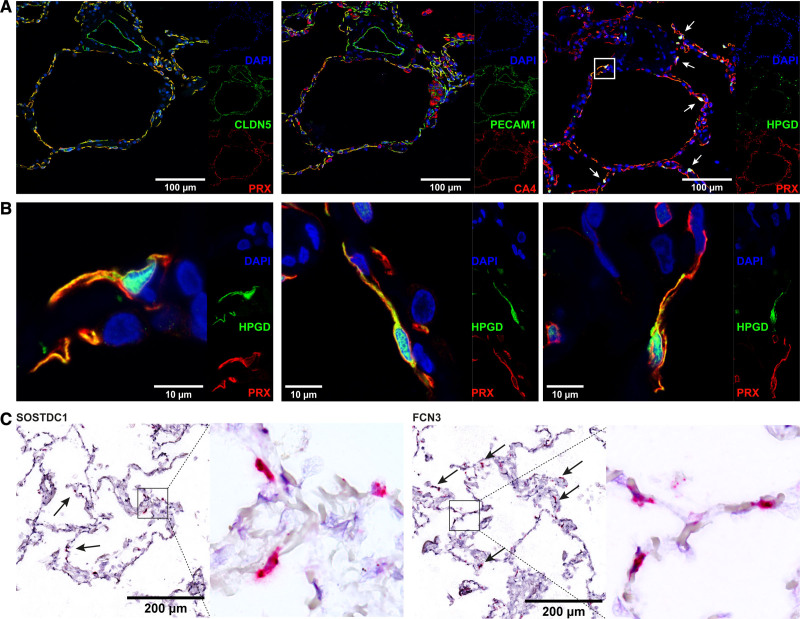
Figure 4.
Localization of aerocytes and general capillary endothelial cells (ECs). A, Representative serial immunofluorescent images of microvascular markers PRX and CA4 with positive red staining in capillaries (CA4 with off-target positive staining in macrophages) and negative staining of a larger central vessel. The general endothelial markers CLDN5 and PECAM1 in green stain the larger central vessels as well, in addition to the microvasculature. The third image shows an immunostain of the aerocyte-specific marker HPGD in green (white arrows), in addition to general microvascular marker PRX in red. Nuclei are counterstained throughout with DAPI (4′,6-diamidino-2-phenylindole). The white box highlights an aerocyte, which is shown at higher magnification in the first image of B. B, Representative immunostains of aerocytes with the specific marker HPGD in green (cytoplasmic and nuclear expression pattern), which colocalizes with the general microvascular marker PRX in red. Nuclei are counterstained throughout with DAPI. C, In situ RNA hybridization stains of markers specific to the capillary subpopulations with SOSTDC1 staining aerocyte ECs (arrows) and FCN3 staining general capillary ECs (arrows) with positive staining in red. The black box highlights an area shown to the right of it in high magnification. Conventional immunohistochemical images of shown markers can be found in Figure IV in the Data Supplement.
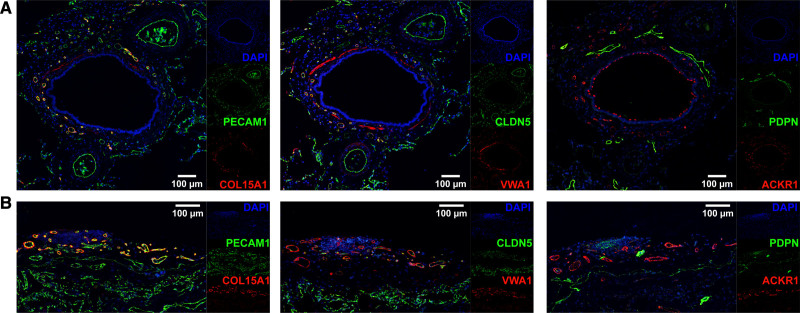
Figure 5.
Systemic–venous endothelial cells (ECs) localize to the bronchial vascular plexus and visceral pleura. Representative, serial immunofluorescent images of systemic–venous EC markers COL15A1 and VWA1, panvenous marker ACKR1 (all in red), panendothelial markers PECAM1 and CLDN5, and lymphatic marker PDPN (all in green) in (A) a bronchus with accompanying arteries and (B) visceral pleura. In (A), COL15A1, VWA1, and ACKR1 stain the small vessels of the bronchial vascular plexus, but not the accompanying arteries; PECAM1 and CLDN5 stain all vessels. In (B), COL15A1, VWA1, and ACKR1 stain the pleural vessels, but not the alveolar microvasculature; PECAM1 and CLDN5 stain all vessels. PDPN identifies bronchial (A) and pleural (B) lymphatic vessels. VWA1 exhibits off-target staining in smooth muscle cells, and ACKR1 in ciliated cells. Nuclei are counterstained throughout with DAPI (4′,6-diamidino-2-phenylindole). Conventional immunohistochemical images of shown markers can be found in Figure IV in the Data Supplement.
Lung ECs separated into 2 broad populations in uniform manifold approximation and projection space (Figures 1B, 2, and 5 and Table III in the Data Supplement): vascular ECs (10 469 cells from 66 participants) and lymphatic ECs (2094 cells from 64 participants). Lymphatic vessels, lined by lymphatic ECs, absorb interstitial fluid and carry it as lymph to a lymph node or collecting vein. Lymphatic ECs were identified on the basis of the expression of canonical lymphatic markers PROX1,15 LYVE1,15 FLT4,15,16 and PDPN16,17 (Figure 2 and Table III in the Data Supplement). Immunofluorescent stains of lymphatic marker PDPN revealed lymphatic vessels in the distal lung (lymphatic capillaries) as well as surrounding large vascular vessels and bronchi (Figure 5 and Figure IV in the Data Supplement). Lymphatic ECs express secreted proteins like CCL2116 mediating homing of T cells and semaphorins SEMA3A and SEMA3D,18 both necessary for lymphatic vessel maturation. Lymphatic marker genes comprise transcription factors TBX1, HOXD3, NR2F1, and NR2F2 and receptors KDR, GPR182, and TEK.
In contrast, blood vessels are lined by vascular ECs and carry blood from and to the heart. Panvascular markers (n=142) expressed across the remaining, nonlymphatic ECs include transmembrane glycoproteins such as ENG, PCDH17, CLEC14A, CLEC1A (C-type lectin domain family 1 member A), ESAM, and ITM2A; the cell surface receptors BMPR2, FLT1, ADGRL4, VIPR1, PLXNA2, FZD4, IL4R, and IL15RA; transporter and channel proteins such as SLCO2A1, SLCO4A1, and AQP1; as well as the transcription factors EPAS1, GATA2, FOXF1, and ETS2 (Figure 2 and Table III in the Data Supplement).
Identification of Arterial, Venous, and Capillary ECs
To focus on vascular ECs, we generated a nonlymphatic EC dataset. To increase the power of our analysis, we included control vascular ECs from a sixth scRNAseq dataset14 (Leuvens Kankerinstituut, n=7 participants) of sorted, CD45neg/CD31pos lung ECs that brought the total number of vascular ECs to 15 142 from 73 participants. The Leuvens Kankerinstituut cohort had not been included in the initial lung dataset to avoid batch effects attributable to missing or selectively sorted non-ECs within this cohort.
We identified 3 broad vascular EC subtypes—arterial (n=2610 cells), capillary (n=7920 cells), and venous (n=4612)—based on expression of canonical marker genes (Figure 3 and Table V in the Data Supplement). Arterial ECs were identified by EFNB2,19–22 SOX17,20,23 BMX,19,24 SEMA3G,19 HEY1,20,21 LTBP4,25 FBLN5,25 GJA5,20 and GJA4.20 Capillary ECs were identified by canonical markers CA416,26 and PRX,27 as well as organism-wide capillary markers RGCC,16 SPARC,16 and SGK1.16 Venous ECs were identified through the canonical transcription factor NR2F220–22 (COUP-TFII [COUP transcription factor 2]), as well as VCAM1,16,22 ACKR128 (Figure 2), and SELP28 expression (Figure 3). Capillary and venous ECs separated further into 2 distinct populations each—venous ECs into pulmonary–venous and systemic–venous ECs and capillary ECs into aerocytes and general capillary ECs—for which a detailed characterization is given in the next 2 paragraphs (for makeup of vascular ECs, see Figure IIIA in the Data Supplement).
Figure 3.
Assessing fine-granular endothelial heterogeneity of the human lung by single-cell RNA sequencing. A, Uniform manifold approximation and projections of 15 142 vascular endothelial cells (ECs) from 73 control lungs of 6 cohorts. Each dot represents a single cell, and cells are labeled, from left to right, by cell type, cohort, sample location, and participant. In the uniform manifold approximation and projection colored by participants, each color represents a distinct participant. B, Heat map of marker genes of all 5 vascular EC populations. Each column represents the average expression value for 1 participant, grouped by cell type and cohort. All gene expression values are unity normalized from 0 to 1 across rows. BWH indicates Brigham and Women’s Hospital; LKI, Leuvens Kankerinstituut; TGen, Translational Genomics Research Institute; VIB, Leuven Vlaams Instituut voor Biotechnologie; and WSI, Wellcome Sanger Institute.
Beyond canonical marker concordance, vascular EC subpopulations were characterized on the basis of their distinct gene signatures associated with their respective cellular roles. Arterial ECs lining pulsating arteries and arterioles are exposed to relevant transmural pressure and shear stress. To withstand these forces, arterial ECs express genes encoding for tight and gap junction proteins such as CLDN10, GJA5, GJA4, and FBLIM1; extracellular matrix proteins that contribute to wall elasticity and strength such as FBLN5, FBLN2, MGP, BGN, LTBP4, LTBP1, and FN1; and the protease inhibitors SERPINE229 and CPAMD8 (Figure 3B and Table V in the Data Supplement). Arterial ECs are active secretory cells and express signaling molecules such as CXCL12, EFBN2, SEMA3G,30 VEGFA, and enzymes of the nitric oxide pathway involved in modulating vascular tonus like NOS1, PDE3A, and PDE4D. Maintenance of their arterial identity is accomplished through the expression of Wnt signaling modulator DKK2, the Notch ligand DLL4, and the transcription factors HEY1, SOX5, SOX17, HES4, and PRDM16 (Figure 3B and Table V in the Data Supplement).
The capillary network surrounds millions of alveoli in the lungs to enable gas exchange. Capillary ECs express genes coding for enzymes related to gas exchange including CA416,26 and CYB5A; multiple receptors including ACVRL1/TMEM100, ADGRF5, ADGRL2, F2RL3, IFNGR1, VIPR1, and ADRB1; intracellular signaling molecules like ARHGAP6, IFI27, PREX1, PRKCE, SGK1,16 SH2D3C, and SORBS1; structural proteins PRX,27 SPARC,16 EMP2, ITGA1, and SLC9A3R2; and transcription regulators AFF3 and MEIS1; and exhibit the highest major histocompatibility complex class I protein expression of all ECs (Figure 3B and Table V in the Data Supplement).
Venules and veins return blood to the heart. In contrast to arteries, veins are thin-walled and subject to low shear forces, which makes in particular the postcapillary venules the primary location for leukocyte extravasation. To facilitate this, venous ECs express genes encoding for proteins associated with diapedesis of leukocytes like VCAM1,16,22 SELP,28 and SELE28 (Figure 3B and Table V in the Data Supplement) as well as ACKR128 for nonspecific transcytosis of chemokines. Venous ECs are further characterized by the major transcription factor NR2F2,20–22 as well as genes coding for secreted (ADAMTS9, IGFBP7), nuclear (HDAC9, RORA), and membrane proteins (ACTN1, LDLRAD3, LDLRAD4, and LRRC1; Table V in the Data Supplement).
scRNAseq Characterization of 2 Previously Indistinguishable Capillary EC Types
scRNAseq reveals that the previously and consistently observed patchy or mosaic histologic staining patterns of vWF (von Willebrand factor), THBD (thrombomodulin), and EMCN (endomucin)14,31–34 as well as EDN1 (endothelin 1) and EDNRB (endothelin receptor B1)35 in lung capillaries can be attributed to 2 discernable capillary populations: a vWFneg/EMCNhigh/EDNRBpos population and a vWFpos/EMCNlow/EDN1pos population (Figure 3A and 3B). Following nomenclature proposed by Gillich et al.,36 we refer to these 2 microvasculature ECs as aerocytes and general capillary ECs.
Aerocytes (n=2317) can be distinguished by their expression of genes encoding for the endothelin receptor EDNRB, the transcription factors TBX2 and FOXP2, the pattern recognition receptors CLEC4E and SPON2, and signaling mediators such as PRKG, CHRM2, S100A4, and EDA (Figure 3B and Table V in the Data Supplement). Aerocytes are the only EC type expressing prostaglandin-degrading HPGD, which is responsible for the first-pass deactivation of most prostaglandins as they pass through the vascular bed of the lung.37 Aerocytes specifically express SOSTDC1, antagonist to BMPR2, which itself is expressed in all ECs and its mutations are associated with pulmonary arterial hypertension. In contrast to all other ECs, aerocytes do not express major components of endothelial-specific Weibel-Palade bodies (vWF, SELP, EDN), storage granules essential for hemostasis and extravasation of leukocytes. Aerocytes are characterized by the absence of expression of genes otherwise expressed in all other ECs including THBD, CD93, PTPRB, ANO2, CRIM1, MECOM, LIFR, PALMD, GNA14, CYYR1, and LEPR. Aerocytes are more rare in the human lung compared with general capillary ECs and were profiled in the median per participant in a ratio of aerocytes to general capillary ECs of 0.48:1 (IQR, 0.26–1.16; Figure IIIB in the Data Supplement).
General capillary ECs (n=5603) are the most generic vascular cell type; they specifically express only 140 genes, compared with arterial (n=373) or venous (n=280) ECs or aerocytes (n=203). Nevertheless, the general capillary EC population can be identified by the expression of genes related to transcytosis of low-density lipoprotein cholesterol and other lipids such as GPIHBP116 and CD3633; innate immune response including FCN3, BTNL9, BTNL8, and CD14; and cytokine receptors such as IL7R and IL18R1 (Figure 3B and Table V in the Data Supplement).
We localized both capillary EC types by immunofluorescent microscopy using CA4 (Carbonic anhydrase 4) and PRX (Periaxin) as common capillary markers and HPGD (15-hydroxyprostaglandin dehydrogenase) as an aerocyte-specific marker, and by in situ hybridization using the subtype-specific markers SOSTDC1 for aerocytes and FCN3 for general capillary ECs (Figure 4). In the control lungs, both consistently localize to and intermingle in the endothelium of the alveolar wall.
Systemically Perfused Vessels of Bronchi and the Visceral Pleura Are Lined by a COL15A1-Positive EC Type
Venous ECs divide into COL15A1pos and COL15A1neg subpopulations. Immunofluorescent microscopy using the markers COL15A1 (Collagen alpha-1(XV) chain) and VWA1 (von Willebrand factor A domain-containing protein 1) revealed that COL15A1pos ECs localized to systemically supplied vessels of the bronchial vascular plexus, as we previously reported,13 and of the visceral pleura (Figure 5), whereas vessels supplied by the pulmonary circulation remained negative. In contrast, the panvenous marker ACKR1 (Atypical chemokine receptor 1) stained both pulmonary and systemic venules (Figure 5). Furthermore, 4 scRNAseq samples from biopsies of the systemically supplied large airways contributed almost exclusively cells to the COL15A1pos population (Figure 3A). On the basis of the immunofluorescent stains and the tissue source of the samples, we were able to assign 2 venous subpopulations to the systemic and pulmonary circulation: systemic–venous ECs (COL15A1pos, n=2291) and pulmonary–venous ECs (COL15A1neg, n= 2321). The integrated dataset allowed us to identify additional genes specific to systemic–venous ECs in addition to COL15A1, a multiplexin collagen linking basement membrane to underlying connective tissue. We also identified the extracellular matrix protein vWA1, as well as PLVAP, which encodes an EC-specific protein that forms the stomatal and fenestral diaphragms of blood vessels and regulates basal permeability, leukocyte migration, and angiogenesis (Figure 3B and Table V in the Data Supplement). Systemic–venous ECs express genes that encode for specific sets of transcription factors, including EBF1, EBF3, MEOX1, MEOX2, and ZNF385D; the cell surface receptors TACR1, ROBO1, and CYSLTR1; the peptidases CPXM2 and MMP16; the phosphodiesterases PDE7B and PDE2A; as well as antagonist of fibroblast growth factor signaling SPRY1.
Whereas aerocytes are characterized by their ability to break down prostaglandins (HPGD) and their lack of hemostatic Weibel-Palade bodies, pulmonary–venous ECs—which are located downstream—express genes encoding for proteins associated with prostaglandin synthesis including PTGS1 and PTGIS, and with complement and coagulation cascades such as C7, PLAT, PROCR, THBD, C1R, and CLU (Figure 3B and Table V in the Data Supplement). Pulmonary–venous ECs express the most specific pulmonary–venous gene CPE as well as Wnt modulator DKK3, which contrasts with the DKK2 expression in arterial ECs. Genes coding for structural proteins specific to pulmonary–venous ECs include EFEMP1 and CDH11. Overlapping with lymphatic ECs, but expressed at lower levels compared with them, pulmonary–venous ECs express factor V/Va binding and extracellular matrix protein MMRN116 and PKHD1L1 (Figure 3B and Table V in the Data Supplement).
EC Subpopulations’ Connectivity With Other Lung Cell Populations
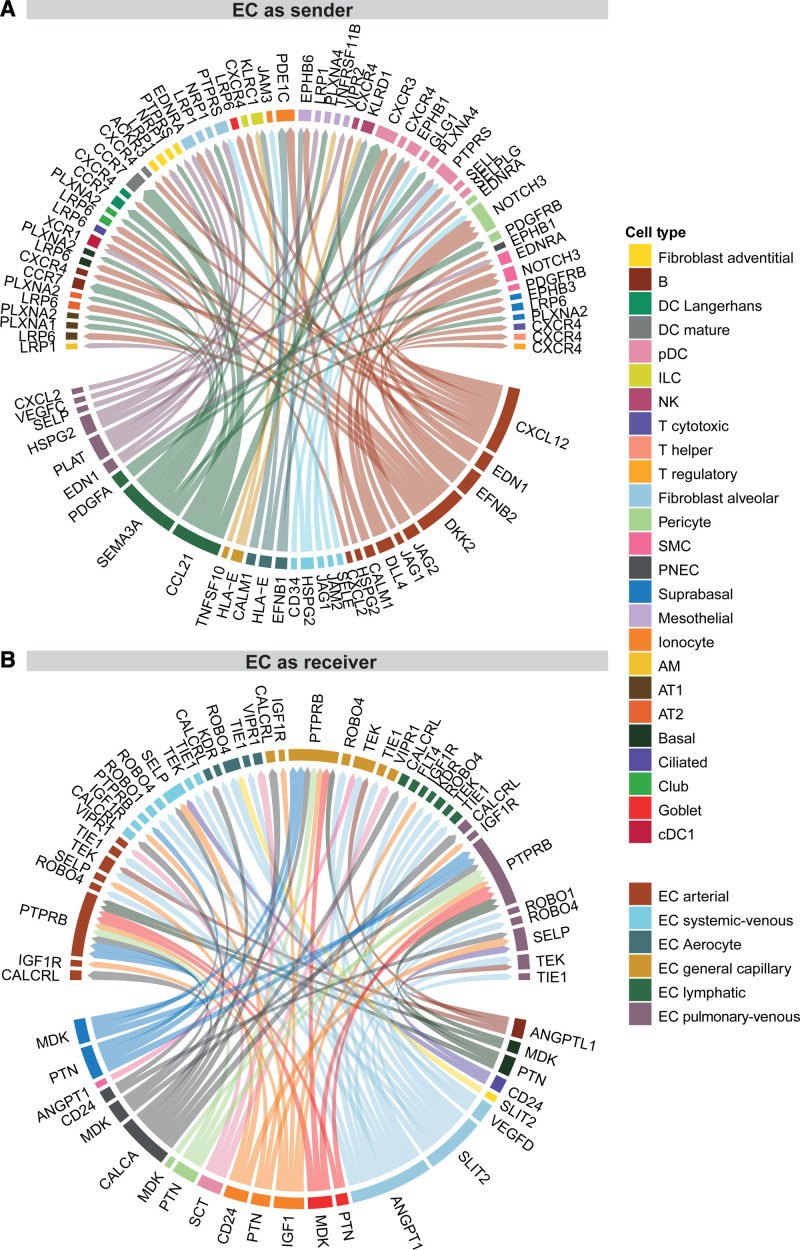
Complex tissues like the lung have distinct cellular niches that are tightly regulated by intercellular communications. To study the role of ECs in tissue homeostasis within the human lung, we performed a connectomic analysis that mapped the averaged gene expression per cell type to known ligand–receptor interactions catalogued in the NicheNet database.38,39 We focused our analysis on potential communication pathways between EC and non-EC cell types. Because of the complexity of the cell–cell signaling connectome, we can only present a limited fraction of our findings here (for the full connectome, see Table VI in the Data Supplement). All ligand–receptor interaction networks can be explored on www.LungEndothelialCellAtlas.com.
We first identified the intercellular signaling patterns wherein ECs are senders of ligands (Figure 6A). Arterial ECs are the most active EC subpopulation in this respect, expressing Notch ligands (DLL4, JAG1, and JAG2), which can be sensed by NOTCH3 in pericytes and smooth muscle cells (SMCs) as a part of a signaling axis that is crucial for arterial EC specification, as well as proliferation and differentiation of supporting pericytes and vascular SMCs.40 Arterial ECs, and to a lesser extent venous and general capillary ECs, express EDN1, which can be sensed by pericytes, SMC, and alveolar fibroblasts through EDNRA; this is a vasoconstrictive axis41 implicated in pulmonary hypertension. Arterial ECs also secrete CXCL12, a chemokine that facilitates homing, migration, and survival of immune cells through binding to CXCR4, which is expressed by most lymphoid cells (T regulatory, T helper, T cytotoxic, B, and innate lymphoid cells) and dendritic cells (DCs; plasmacytoid DCs [pDCs], mature DCs, and Langerhans DCs).42 Lymphatic ECs secrete CCL21, which is sensed by pDCs through CXCR3 and by mature DCs, Langerhans DCs, and B cells through CCR7 (Figure 6A), representing a classic mechanism for guiding matured DCs to secondary lymphoid tissues.43 Lymphatic EC, as well as AT1 (alveolar cell type 1) and pericytes, express PDGFA, with the corresponding receptor PDGFRB expressed in pericytes and SMCs. Lymphatic ECs were found to express the spatial guidance molecule SEMA3A that is critically important for lymphatic vessel maturation.18 Its corresponding receptors are expressed on alveolar and nonparenchymal fibroblasts and vascular ECs (NPR1); AT1, AT2, Club, basal, suprabasal, and mesothelial cells (PLXNA1/PLXNA2); and pDCs (PLXNA4; Figure 6A).
Figure 6.
Endothelial cell (EC)–focused intercellular communication. Visualization of a small subset of the connectomic analysis. Circos plots of top 75 edges by edge weight with EC subpopulations as (A) senders and (B) receivers. Edge thickness is proportional to edge weights. Edge color labels the source cell type. In both Circos plots, ligands occupy the lower semicircle and corresponding receptors the upper semicircle, and ligands and receptors are colored by the expressing cell type. The full results of the connectomic analysis can be found in Table VI. AM indicates alveolar macrophage; AT1/2, alveolar cell type 1/2; cDC1/2, classical dendritic cell type 1/2; cMono, classical monocyte; DC, dendritic cell; ILC, innate lymphoid cell; M, macrophage; ncMono, nonclassical monocyte; NK, natural killer; pDC, plasmacytoid dendritic cell; PNEC, pulmonary neuroendocrine cell; scRNAseq, single cell RNA sequencing; and SMC, smooth muscle cell.
We next identified selected cell–cell signaling patterns with ECs being on the receiving end (Figure 6B). Alveolar fibroblasts strongly express the morphogen SLIT2 and ANGPT1 (to a lesser extent also in pericytes and SMCs). Both SLIT2 and ANGPT1 can be sensed by all 5 EC subpopulations through the receptors ROBO4 and TEK/TIE1, respectively. Furthermore, alveolar fibroblasts are in a position to signal to aerocytes and lymphatic ECs through the VEGFD–KDR and VEGFD-FLT4 axes, respectively (Figure 6B).
Not all ligands sensed by EC receptors are produced by resident lung cells. For example, the receptor VIPR1 is expressed by lung ECs but its main ligand VIP (vasoactive intestinal peptide) was not expressed by any cells profiled in the lungs. This suggests that the VIPR1 ligands targeting lung ECs are mainly derived from extrapulmonary organs. Therefore, alternate sources of ligands beyond those profiled in the present connectomic analysis may contribute to lung EC signaling and function.
Normal Lung Endothelial Subtypes and Pulmonary Hypertension
Whereas we focus on EC subpopulations in the normal lung, we also assessed the relevance of our findings to pulmonary hypertension. Using our integrated scRNAseq dataset, we attributed expression patterns of genes associated with mendelian genetic diseases of the lung to their cells of origin. BMPR2, known to be associated with pulmonary arterial hypertension, was expressed in all ECs, but SOX17 only in arterial ECs (see Results and Figure V in the Data Supplement). Reanalysis of a pulmonary arterial hypertension scRNAseq dataset44 revealed that all vascular endothelial subpopulations could be observed in lungs of patients with pulmonary arterial hypertension (see Results and Figure VI in the Data Supplement). In contrast, scRNAseq revealed that commercially obtained, primary arterial, microvascular, and venous lung ECs lost their respective native lung phenotype (see Results and Figures VII and VIII in the Data Supplement).
For further details, see the Results in the Data Supplement.
Novel EC Populations Found in Humans Are Also Present in Mice
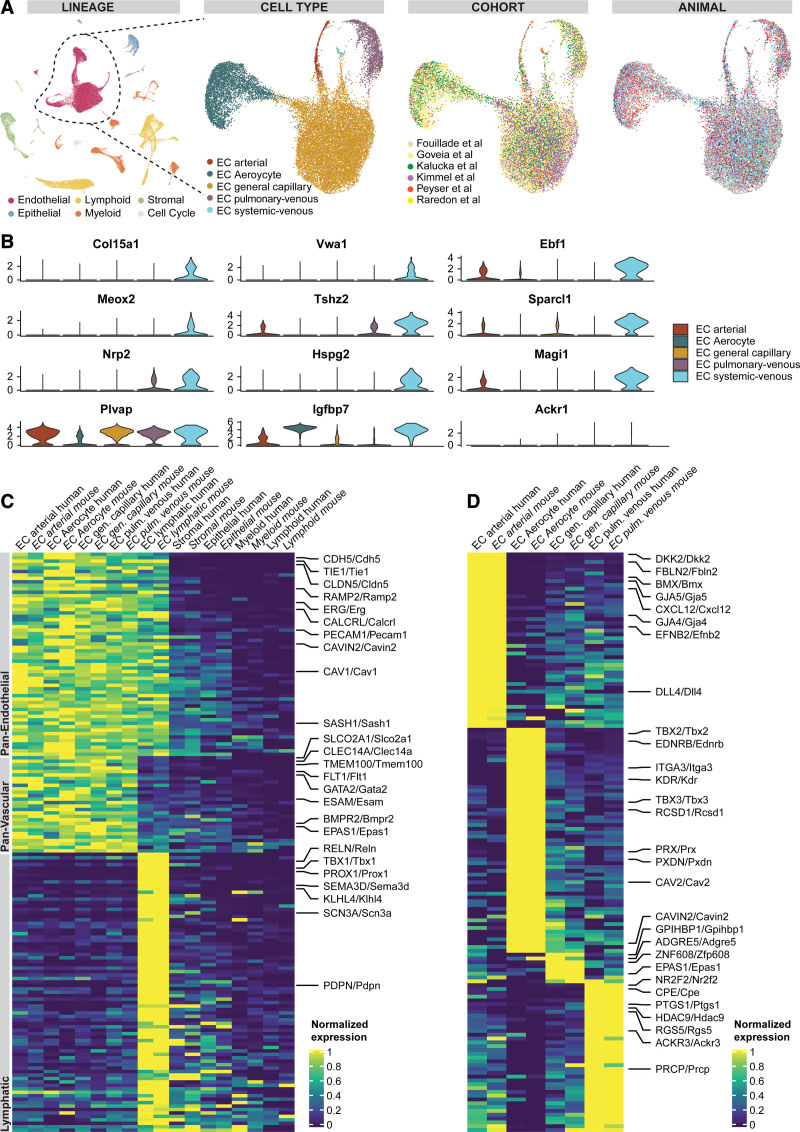
In mammals, the lung is the only organ in which gas exchange between blood and air takes place. Here, we aimed to explore whether cellular and transcriptomic principles of lung vascular biology are preserved and chose the most used mammalian model organism as comparator: the mouse. We applied our human workflow to mouse lung scRNAseq data to generate an integrated dataset of mouse lung cells from 18 control wild-type C57BL/6 mice from 6 cohorts14,16,39,45–47 (Figure IX in the Data Supplement and Table VII in the Data Supplement). All murine scRNAseq data had been generated from whole lung dissociations using 10×3′ scRNAseq kits based on v2 Chemistry. Only 2 out of 18 mice were female and the median age was 12.5 weeks (IQR, 10.75 – 34.0 weeks; for further details by cohort, see Table II in the Data Supplement). We identified 21 831 ECs within the integrated mouse lung dataset containing 57 974 single-cell transcriptomes (Figure 7A and Figure IX in the Data Supplement). Subsetting the ECs from the mouse lung dataset for more specific subtype identification revealed the same EC subpopulations found in humans (Figure 7A). Regarding murine cell type marker genes in the full dataset and the EC subset, see Figures IX and X in the Data Supplement and Tables VIII and IX in the Data Supplement. Technical summaries of sample preprocessing are available in Table X in the Data Supplement.
Figure 7.
Identification of conserved endothelial cell (EC) populations and conserved marker genes in humans and mice. A, Uniform manifold approximation and projection of all 57 974 mouse cells from 18 control mouse lungs colored by lineage membership and of the subset of 21 343 vascular ECs labeled by cell type, cohort, and animal. In the uniform manifold approximation and projection colored by participants, each color represents a distinct mouse. B, Violin plots of marker genes significantly expressed in systemic–venous ECs compared with all other vascular ECs (first to third row) and of homologues to human marker genes of systemic–venous ECs lacking specificity for murine systemic–venous ECs (fourth row). C, Heat map of conserved panendothelial, panvascular, and lymphatic EC marker genes. Each column represents the average expression value per cell type for ECs and per lineage for non-ECs. All gene expression values are unity normalized per species from 0 to 1 across rows. D, Heat map of conserved marker genes of 4 pulmonary EC populations. Each column represents the average expression value per cell type. All gene expression values are unity normalized per species from 0 to 1 across rows. Human genes are indicated by capital letters and mouse genes are indicated by small letters after the first letter, separated by a slash. Labels of mouse, but not human, cell types/lineages are given in italics.
We identified the previously undescribed systemic–venous EC type in mice as well based on the marker genes Col15a1 and Vwa1 (Figure 7B and Figure XA in the Data Supplement and Table IX in the Data Supplement). Systemic–venous ECs are rare in mice and account for only 0.5% of all ECs (n=117 cells; Figure XB in the Data Supplement), probably because—unlike in humans—both the visceral pleura and intraparenchymal airways are supplied by the pulmonary rather than the systemic circulation.48,49 As in humans, murine systemic–venous ECs express transcription factors Ebf1, Ebf3, Meox2, and Tshz2, phosphodiesterase Pde7b, and, overlapping with venous ECs, Selp and Ackr1. However, homologues to some human marker genes of systemic–venous EC show no specificity for murine systemic–venous ECs (Plvap, Spry1, Pkp4), a different specificity profile (Igfbp7), a very low expression (Tacr1, Robo1; Figure 5B), or a homologue does not exist in mice (eg, ZNF385D).
Murine capillary ECs are characterized by the expression of canonical markers Car4,16,26 Prx,27 and Sgk116 and split likewise in 2 subpopulations: aerocytes (n=4853 cells) and general capillary ECs (n=14 119 cells; Figure XA in the Data Supplement and Tables VIII and IX in the Data Supplement). Aerocytes were identified by the expression of Ednrb and Tbx2 and general capillary ECs by Gpihbp1 and Edn1 (Table IX in the Data Supplement). In the median per mouse, capillary ECs were profiled in a ratio of aerocytes to general capillary ECs of 0.24:1 (IQR, 0.15–0.39; Figure IIIB in the Data Supplement), indicating that aerocytes are less common in mice lung capillaries when compared with those of humans (ratio in humans of 0.48:1; IQR, 0.26–1.16). A breakdown of marker genes overlapping with humans for both populations is given in the following section. In contrast to what has observed in humans, murine aerocytes are also characterized by top marker genes like Igfbp7, Emp2, Rgs6, Abcc3, Ackr2, Fibin, and Chst1 and general capillary ECs by genes such as Glp1r, Adgrl3, Plcb1, and Hmcn1 (Table IX in the Data Supplement). Interesting patterns can be observed that distinguish both murine capillary cell types but that are only vaguely recognizable in humans: Apln is specifically expressed in aerocytes, whereas its receptor Aplnr is expressed in general capillary ECs. In contrast, Vegfa is expressed in general capillary ECs and its receptor Kdr on aerocytes (Table IX in the Data Supplement). In contrast to humans, major constituents of Weibel-Palade bodies—Vwf and Selp—are expressed neither in aerocytes nor in general capillary ECs.
Endothelial Marker Genes Are Widely Conserved in Mice
We performed an unsupervised integration of data from both human and mice cells into a single graph embedding and subsequent uniform manifold approximation and projection plot (Figure XI in the Data Supplement). We observed the corresponding overlap of all 5 homologous EC subpopulations and performed an in-depth comparison of corresponding populations between the 2 species. An analysis of the homologous marker genes across human and mouse EC subpopulations revealed a set of conserved marker genes between analogous subpopulations. In the following paragraph, human genes (all capital letters) and mouse genes are separated by a slash.
Conserved marker genes expressed in all EC populations included canonical markers PECAM1/Pecam1, CDH5/Cdh5, and CLDN5/Cldn5 (Figure 7C and Table XI in the Data Supplement). All ECs from both species express the transcription factor ERG/Erg and SOX18/Sox18, angiopoietin receptor TIE1/Tie1, and the adrenomedullin receptor subunits CALCRL/Calcrl and RAMP2/Ramp2. Conserved vascular marker genes—genes not expressed in lymphatic ECs—comprise endothelial receptors (FLT1/Flt1, ADGRL4/Adgrl4, BMPR2/Bmpr2, ACVRL1/Acvrl1), cell adhesion molecules (ESAM/Esam, CLEC14A/Clec14a), transcription factors (GATA2/Gata2, SOX7/Sox7), and a prostaglandin transporter (SLCO2A1/Slco2a1). Lymphatic ECs represent a distinct population in both species and are characterized by canonical PROX1/Prox1, semaphorin SEMD3D/Sema3d, the transcription factor TBX1/Tbx1, podoplanin (PDPN/Pdpn), and matrix protease Reelin (RELN/Reln; Figure 7C and Table XI in the Data Supplement).
We examined the conserved marker genes that are used to distinguish between lung EC subpopulations (Figure 7D and Table XI in the Data Supplement). Arterial ECs share a unique set of genes between both species that includes connexins GJA4/Gja4 and GJA5/Gja5, secreted cytokines CXCL12/Cxcl12 and EFNB2/Efnb2, Wnt modulator DKK2/Dkk2, and Notch ligand DLL4/Dll4. Aerocytes are characterized by the conserved expression of receptors EDNRB/Ednrb and KDR/Kdr, transcription factors TBX2/Tbx2 and TBX3/Tbx3, and peroxidase PXDN/Pxdn (Figure 7D and Table XI in the Data Supplement). Pulmonary–venous EC show conserved expression of transcription factor NR2F2/Nr2f2, receptors IL1R1/Il1r1 and ACKR3/Ackr3, carboxypeptidases CPE/Cpe and PRCP/Prcp, and the marker genes HDAC9/Hdac9, PTGS1/Ptgs1, SELP/Selp, and RGS5/Rgs5. The least overlapping marker genes were observed in general capillary ECs, which included GPIHBP1/Gpihbp1, EPAS1/Epas1, ZNF608/Zfp608, ADGRE5/Adgre5, and NTRK2/Ntrk2 in both species (Figure 7D and Table XI in the Data Supplement).
Discussion
We present a reference atlas of ECs from the human lung on a cellular transcriptomic level derived from 15 142 ECs from 73 participants. We identified prototypic gene expression patterns specific to distinct hierarchies of the endothelial lineage including panendothelial, panvascular, and subpopulation-specific marker gene sets. The expression of all reported marker sets was detected throughout all cohorts and most participants, essentially eliminating the possibility that participant-specific gene expression patterns could be misattributed as cell type–specific patterns. We validated major marker genes on the protein level by immunofluorescent and immunohistochemical microscopy, or at the mRNA level by in situ hybridization, respectively. We describe the identities of 2 previously indistinguishable EC populations: pulmonary–venous ECs localized to the lung parenchyma and systemic–venous ECs localized to the airways and the visceral pleura. We confirmed and characterized the new subclassification of pulmonary capillary ECs into aerocytes and general capillary ECs. We investigated the role of EC subpopulations within the complex cellular signaling networks that occur between different lung cell types, which suggested a supporting role of alveolar fibroblasts with respect to all lung ECs and arterial ECs to mural pericytes/SMCs communication.
The most significant contribution of this study is the unbiased and global-transcriptomic characterization of EC of the human lung on 3 hierarchical levels: we identified 147 genes specifically expressed in all EC populations and 142 genes specifically expressed in vascular ECs and describe in-depth the transcriptional particularities of the 6 EC types. Panendothelial, panvascular, or EC subpopulation–specific gene expression was often associated with a specific function of ECs in general or of endothelial subpopulations in particular. This is exemplified in the expression of genes associated with immune cell homing in lymphatic ECs, gas exchange in capillary ECs, diapedesis of leukocytes and unspecific transcytosis of chemokines in venous ECs or ECM components of the elastic vessel wall, and regulation of vascular tone in arterial ECs. The function of many other genes specifically expressed in ECs is not known; their future study will enhance our understanding of EC biology.
Our integrated analysis confirmed and expanded the description of 2 novel EC cell subpopulations. We had previously noticed a distinct population of COL15A1 pos lung ECs that seemed to appear in the lung parenchyma only in a lung with idiopathic pulmonary fibrosis.13 Here, basing our analysis on many more healthy lungs, we identify this population as a venous EC population that, as revealed through in-depth immunohistologic studies, is usually located in the peribronchial space and the visceral pleura. This discovery is an example of the power of unbiased scRNAseq profiling; it is difficult to imagine how this distinct population could be identified using traditional methods. Whether the specific gene expression profile of systemic–venous ECs reflects adaptation to systemic perfusion of blood vessels of bronchi and the visceral pleura or to the extracellular matrix itself in which they are located will need to be investigated in future studies, but the fact that this EC subtype changes its anatomic distribution in pulmonary fibrosis may suggest involvement in repair and relevance to human lung disease.
Similarly, studies in multiple species on pulmonary scRNAseq datasets identified 2 distinguishable capillary EC populations.13,14,16,36,39,50,51 This consistent but unexpected observation, for which there is no corresponding report from the pre-scRNAseq era, provides an explanation for the mosaic immunohistologic staining patterns of vWF, THBD, EMCN, EDN1, and EDNRB14,31–35 in lung capillaries that had, until recently, remained enigmatic. One of these capillary EC has been recently defined as aerocytes36: the key ECs involved in gas exchange. Intriguingly, aerocytes—unlike all other vascular EC types—lack expression of the major constituents of endothelial-specific Weibel-Palade bodies including vWF, SELP, and EDN. Weibel-Palade bodies are known to be missing in the thinnest lung capillaries, which are thought to be the major location were gas exchange happens.52,53 Vila Ellis et al.50 and Gillich et al.36 showed by lineage-tracing experiments in mice that aerocytes exhibit a much larger surface area and form—together with the AT1 cells and their interposed basement membrane—the blood–air barrier. In addition to confirming their presence in a large cohort of humans, our study adds in-depth transcriptional profiling of aerocytes, thus providing additional clues of their potential functions.
The comparison of human and mouse EC diversity and marker gene expression revealed considerable overlap. All human EC subpopulations could be identified in mice as well, including a very rare systemic–venous Col15a1pos EC population. The rarity of these systemic–venous ECs is probably attributable to the fact that the systemic bronchial circulation in mice is very limited and reaches only the main bronchi.49 Several dozens of panendothelial and panvascular marker genes and of marker genes specific to the EC subpopulations are conserved in mice and humans. Marker genes of general capillary ECs are an exception as they show only a limited overlap, potentially attributable to fewer marker genes per species in general and a lack of homologues to some human marker genes (eg, FCN3) in mice.
Our study has limitations. First, to enable a comprehensive joint analysis of all datasets, a data integration approach had to be chosen (for details, see the Data Supplement). Each cohort entails a batch effect because of slightly different processing of samples and variation in the scRNAseq library preparation and sequencing. A major technical batch effect was alleviated by processing the raw sequencing data through the same computational pipeline and the same reference genome. All differential testing was performed on the nonintegrated data to ensure that it was not distorted by the integration algorithm. However, integration of several datasets has the advantage of leveling out the cell preparation/dissociation bias (ie, the preferential liberation of specific cell types during tissue dissociation). Second, although we validated and localized the expression of major endothelial marker genes on the protein level by immunohistochemistry and on the mRNA level by in situ hybridization, a global spatial localization of endothelial gene expression is lacking in this report. Recent advances in spatial transcriptomics will enable this in the future. Third, the connectomic analysis relies on a limited database of known ligand–receptor interactions and can only infer potential communication between cell types, in addition to other limitations as described elsewhere.39 Therefore, the connectomic analysis should only be considered under hypothesis-generating aspects; a sensible future approach would be to study the molecular crosstalk in functional studies.
The generation of transcriptional profiles of cell types and their intercellular communication within the complex organ lung are tasks that are crucial to understanding the molecular function of the lung and its dysfunction in disease. This integrated lung endothelial atlas will advance lung endothelial research, especially in diseases with vascular involvement. For this purpose, we have created the online tool www.LungEndothelialCellAtlas.com, with which all transcriptomic data of ECs of the lung can be explored in an easily accessible fashion.
Acknowledgments
The authors thank the donors who participated in the studies analyzed in this article and the authors who made these datasets publicly available, M. Zhong at Yale Stem Cell Center for the sequencing service, and A. Brooks and his team at Yale Pathology Tissue Services for tissue processing. This publication is part of the Human Cell Atlas (www.humancellatlas.org/publications). Dr Schupp and Dr Kaminski conceptualized and supervised the study. Dr Schupp, T.S. Adams, and Dr Poli procured and dissociated the lungs. Dr Schupp and T.S. Adams performed single-cell RNA sequencing barcoding and library construction. Data were processed, curated, integrated and visualized by Dr Schupp, T.S. Adams, and Dr Yan and analyzed by Dr Schupp, T.S. Adams, Dr Brickman Raredon, Dr Yan, and Dr Kaminski. The online tool www.LungEndothelialCellAtlas.com was developed by C. Cosme and Dr Neumark. Immunofluorescence stains were performed by Dr Schupp and Dr Chioccioli, immunohistochemistry stains by Dr Schupp and K.-A. Rose, RNA in situ hybridization stains by Dr Omote, and all images were evaluated by Dr Homer, Dr Kaminski, Dr Schupp, Dr Chioccioli, and Dr Omote. Single-cell RNA sequencing of cultured primary ECs was performed by Dr Yuan, Dr Brickman Raredon, and Dr Pierce. The manuscript was drafted by Dr Schupp, T.S. Adams, C. Cosme, Dr Brickman Raredon, and Dr Kaminski and was reviewed and edited by all other authors. Dr Brickman Raredon, Dr Niklason, Dr Yuan, A.C. Habermann, A.J. Gutierrez, Dr Bui, Dr Teichmann, Dr Nawijn, Dr Meyer, Dr Pe’er, Dr Banovich, Dr Kropski, Dr Lafyatis, and Dr Pierce provided datasets and insights. Dr Niklason, Dr Teichmann, Dr Nawijn, Dr Banovich, Dr Pierce, Dr Kropski, Dr Rosas, and Dr Kaminski acquired funding for the integrated datasets.
Sources of Funding
The generation of this integrated single-cell RNA sequencing atlas of endothelial cells of the human lung was supported by Department of Defense Discovery Award W81XWH-19-1-0131 and the German Research Foundation grant SCHU 3147/2-1 to Dr Schupp; National Institutes of Health grant F30HL143906 to Dr Brickman Raredon; National Institutes of Health grant K08HL13689 to Dr Pierce; National Institutes of Health/National Heart, Lung, and Blood Institute grants R01HL127349, R01HL141852, U01HL145567, and UH2HL123886 to Dr Kaminski; a gift from Three Lakes Partners to Dr Kaminski and Dr Rosas; and the Chan Zuckerberg Initiative Seed Networks for the Human Cell Atlas to Dr Kaminski, Dr Pe’er, and Dr Nawijn. The integrated data sets were funded by various sponsors (please refer to the original publications).
Disclosures
Dr Kaminski served as a consultant to Biogen Idec, Boehringer Ingelheim, Third Rock, Pliant, Samumed, NuMedii, Theravance, LifeMax, Three Lake Partners, Optikira, Astra Zeneca, Veracyte, Augmanity, and CSL Behring and over the past 3 years reports equity in Pliant and a grant from Veracyte and nonfinancial support from miRagen and Astra Zeneca. Dr Kaminski has intellectual property on novel biomarkers and therapeutics in idiopathic pulmonary fibrosis licensed to Biotech. The other authors report no conflicts.
Supplemental Materials
Expanded Methods and Materials
Expanded Results
Data Supplement Figures I–XII
Data Supplement Tables I–XI
Supplementary Material
Nonstandard Abbreviations and Acronyms
- DC
- dendritic cell
- EC
- endothelial cell
- IQR
- interquartile range
- pDC
- plasmacytoid dendritic cell
- scRNAseq
- single-cell RNA sequencing
- SMC
- smooth muscle cell
The Data Supplement is available with this article at www.ahajournals.org/doi/suppl/10.1161/circulationaha.120.052318.
For Sources of Funding and Disclosures, see page 300.
Contributor Information
Jonas C. Schupp, Email: jonas.schupp@yale.edu.
Taylor S. Adams, Email: taylor.adams@yale.edu.
Carlos Cosme, Jr, Email: carlos.cosmejr@yale.edu.
Micha Sam Brickman Raredon, Email: michasam.raredon@yale.edu.
Yifan Yuan, Email: yifan.yuan@yale.edu.
Norihito Omote, Email: nori05692000@yahoo.co.jp.
Sergio Poli, Email: sergio.polid@gmail.com.
Maurizio Chioccioli, Email: maurizio.chioccioli@yale.edu.
Kadi-Ann Rose, Email: kadi-ann.rose@yale.edu.
Edward P. Manning, Email: edward.manning@yale.edu.
Maor Sauler, Email: Maor.sauler@yale.edu.
Giuseppe DeIuliis, Email: joseph.deiuliis@yale.edu.
Farida Ahangari, Email: farida.ahangari@yale.edu.
Nir Neumark, Email: nir.neumark@gmail.com.
Arun C. Habermann, Email: ahaberma@uthsc.edu.
Austin J. Gutierrez, Email: agutierrez@tgen.org.
Linh T. Bui, Email: lbui@tgen.org.
Robert Lafyatis, Email: rlafyatis@gmail.com.
Richard W. Pierce, Email: richard.pierce@yale.edu.
Kerstin B. Meyer, Email: km16@sanger.ac.uk.
Martijn C. Nawijn, Email: m.c.nawijn@umcg.nl.
Sarah A. Teichmann, Email: st9@sanger.ac.uk.
Nicholas E. Banovich, Email: nbanovich@tgen.org.
Jonathan A. Kropski, Email: jon.kropski@vumc.org.
Laura E. Niklason, Email: laura.niklason@yale.edu.
Dana Pe’er, Email: peerster@gmail.com.
Xiting Yan, Email: xiting.yan@yale.edu.
Robert J. Homer, Email: robert.homer@yale.edu.
Ivan O. Rosas, Email: ivan.rosas@bcm.edu.
References
- 1.Katz AM. Knowledge of the circulation before William Harvey. Circulation. 1957; 15:726–734 [DOI] [PubMed] [Google Scholar]
- 2.Vita JA. Endothelial function. Circulation. 2011; 124:e906–e912. doi: 10.1161/CIRCULATIONAHA.111.078824 [DOI] [PubMed] [Google Scholar]
- 3.Deanfield JE, Halcox JP, Rabelink TJ. Endothelial function and dysfunction: testing and clinical relevance. Circulation. 2007; 115:1285–1295. doi: 10.1161/CIRCULATIONAHA.106.652859 [DOI] [PubMed] [Google Scholar]
- 4.Flammer AJ, Anderson T, Celermajer DS, Creager MA, Deanfield J, Ganz P, Hamburg NM, Lüscher TF, Shechter M, Taddei S, et al. The assessment of endothelial function: from research into clinical practice. Circulation. 2012; 126:753–767. doi: 10.1161/CIRCULATIONAHA.112.093245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eelen G, de Zeeuw P, Simons M, Carmeliet P. Endothelial cell metabolism in normal and diseased vasculature. Circ Res. 2015; 116:1231–1244. doi: 10.1161/CIRCRESAHA.116.302855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pi X, Xie L, Patterson C. Emerging roles of vascular endothelium in metabolic homeostasis. Circ Res. 2018; 123:477–494. doi: 10.1161/CIRCRESAHA.118.313237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goncharova EA, Chan SY, Ventetuolo CE, Weissmann N, Schermuly RT, Mullin CJ, Gladwin MT. Update in pulmonary vascular diseases and right ventricular dysfunction 2019. Am J Respir Crit Care Med. 2020; 202:22–28. doi: 10.1164/rccm.202003-0576UP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schupp JC, Kaminski N, Homer R, Omote N. Conventional IHC for the integrated single cell atlas of endothelial cells of the human lung. Zenodo. Posted online May 26, 2021. doi: 10.5281/zenodo.4503718 [Google Scholar]
- 9.Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, Peter L, Chung MI, Taylor CJ, Jetter C, et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv. 2020; 6:eaba1972. doi: 10.1126/sciadv.aba1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, Brouwer S, Gomes T, Hesse L, Jiang J, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. 2019; 25:1153–1163. doi: 10.1038/s41591-019-0468-5 [DOI] [PubMed] [Google Scholar]
- 11.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, Fernandez R, Akbarpour M, Chen CI, Ren Z, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019; 199:1517–1536. doi: 10.1164/rccm.201712-2410OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, Bassez A, Decaluwé H, Pircher A, Van den Eynde K, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018; 24:1277–1289. doi: 10.1038/s41591-018-0096-5 [DOI] [PubMed] [Google Scholar]
- 13.Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, Chu SG, Raby BA, DeIuliis G, Januszyk M, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv. 2020; 6:eaba1983. doi: 10.1126/sciadv.aba1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goveia J, Rohlenova K, Taverna F, Treps L, Conradi LC, Pircher A, Geldhof V, de Rooij LPMH, Kalucka J, Sokol L, et al. An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell. 2020; 37:421. doi: 10.1016/j.ccell.2020.03.002 [DOI] [PubMed] [Google Scholar]
- 15.Wigle JT, Harvey N, Detmar M, Lagutina I, Grosveld G, Gunn MD, Jackson DG, Oliver G. An essential role for Prox1 in the induction of the lymphatic endothelial cell phenotype. EMBO J. 2002; 21:1505–1513. doi: 10.1093/emboj/21.7.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalucka J, de Rooij LPMH, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen LA, Veys K, et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020; 180:764–779.e20. doi: 10.1016/j.cell.2020.01.015 [DOI] [PubMed] [Google Scholar]
- 17.Lee JY, Park C, Cho YP, Lee E, Kim H, Kim P, Yun SH, Yoon YS. Podoplanin-expressing cells derived from bone marrow play a crucial role in postnatal lymphatic neovascularization. Circulation. 2010; 122:1413–1425. doi: 10.1161/CIRCULATIONAHA.110.941468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurisic G, Maby-El Hajjami H, Karaman S, Ochsenbein AM, Alitalo A, Siddiqui SS, Ochoa Pereira C, Petrova TV, Detmar M. An unexpected role of semaphorin3a-neuropilin-1 signaling in lymphatic vessel maturation and valve formation. Circ Res. 2012; 111:426–436. doi: 10.1161/CIRCRESAHA.112.269399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018; 554:475–480. doi: 10.1038/nature25739 [DOI] [PubMed] [Google Scholar]
- 20.Su T, Stanley G, Sinha R, D’Amato G, Das S, Rhee S, Chang AH, Poduri A, Raftrey B, Dinh TT, et al. Single-cell analysis of early progenitor cells that build coronary arteries. Nature. 2018; 559:356–362. doi: 10.1038/s41586-018-0288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005; 435:98–104. doi: 10.1038/nature03511 [DOI] [PubMed] [Google Scholar]
- 22.dela Paz NG, D’Amore PA. Arterial versus venous endothelial cells. Cell Tissue Res. 2009; 335:5–16. doi: 10.1007/s00441-008-0706-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corada M, Orsenigo F, Morini MF, Pitulescu ME, Bhat G, Nyqvist D, Breviario F, Conti V, Briot A, Iruela-Arispe ML, et al. Sox17 is indispensable for acquisition and maintenance of arterial identity. Nat Commun. 2013; 4:2609. doi: 10.1038/ncomms3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekman N, Lymboussaki A, Västrik I, Sarvas K, Kaipainen A, Alitalo K. Bmx tyrosine kinase is specifically expressed in the endocardium and the endothelium of large arteries. Circulation. 1997; 96:1729–1732. doi: 10.1161/01.cir.96.6.1729 [DOI] [PubMed] [Google Scholar]
- 25.Noda K, Dabovic B, Takagi K, Inoue T, Horiguchi M, Hirai M, Fujikawa Y, Akama TO, Kusumoto K, Zilberberg L, et al. Latent TGF-β binding protein 4 promotes elastic fiber assembly by interacting with fibulin-5. Proc Natl Acad Sci U S A. 2013; 110:2852–2857. doi: 10.1073/pnas.1215779110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleming RE, Crouch EC, Ruzicka CA, Sly WS. Pulmonary carbonic anhydrase IV: developmental regulation and cell-specific expression in the capillary endothelium. Am J Physiol. 1993; 265:L627–L635. doi: 10.1152/ajplung.1993.265.6.L627 [DOI] [PubMed] [Google Scholar]
- 27.Wang MM, Zhang X, Lee SJ, Maripudi S, Keep RF, Johnson AM, Stamatovic SM, Andjelkovic AV. Expression of periaxin (PRX) specifically in the human cerebrovascular system: PDZ domain-mediated strengthening of endothelial barrier function. Sci Rep. 2018; 8:10042. doi: 10.1038/s41598-018-28190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiriot A, Perdomo C, Cheng G, Novitzky-Basso I, McArdle S, Kishimoto JK, Barreiro O, Mazo I, Triboulet R, Ley K, et al. Differential DARC/ACKR1 expression distinguishes venular from non-venular endothelial cells in murine tissues. BMC Biol. 2017; 15:45. doi: 10.1186/s12915-017-0381-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boulaftali Y, Ho-Tin-Noe B, Pena A, Loyau S, Venisse L, François D, Richard B, Arocas V, Collet JP, Jandrot-Perrus M, et al. Platelet protease nexin-1, a serpin that strongly influences fibrinolysis and thrombolysis. Circulation. 2011; 123:1326–1334. doi: 10.1161/CIRCULATIONAHA.110.000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kutschera S, Weber H, Weick A, De Smet F, Genove G, Takemoto M, Prahst C, Riedel M, Mikelis C, Baulande S, et al. Differential endothelial transcriptomics identifies semaphorin 3G as a vascular class 3 semaphorin. Arterioscler Thromb Vasc Biol. 2011; 31:151–159. doi: 10.1161/ATVBAHA.110.215871 [DOI] [PubMed] [Google Scholar]
- 31.Langenkamp E, Molema G. Microvascular endothelial cell heterogeneity: general concepts and pharmacological consequences for anti-angiogenic therapy of cancer. Cell Tissue Res. 2009; 335:205–222. doi: 10.1007/s00441-008-0642-4 [DOI] [PubMed] [Google Scholar]
- 32.Pusztaszeri MP, Seelentag W, Bosman FT. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli-1 in normal human tissues. J Histochem Cytochem. 2006; 54:385–395. doi: 10.1369/jhc.4A6514.2005 [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto M, Shimokata K, Nagura H. An immunohistochemical study on phenotypic heterogeneity of human pulmonary vascular endothelial cells. Virchows Arch A Pathol Anat Histopathol. 1988; 412:479–486. doi: 10.1007/BF00750582 [DOI] [PubMed] [Google Scholar]
- 34.Kawanami O, Jin E, Ghazizadeh M, Fujiwara M, Jiang L, Nagashima M, Shimizu H, Takemura T, Ohaki Y, Arai S, et al. Heterogeneous distribution of thrombomodulin and von Willebrand factor in endothelial cells in the human pulmonary microvessels. J Nippon Med Sch. 2000; 67:118–125. doi: 10.1272/jnms.67.118 [DOI] [PubMed] [Google Scholar]
- 35.Davenport AP, Hyndman KA, Dhaun N, Southan C, Kohan DE, Pollock JS, Pollock DM, Webb DJ, Maguire JJ. Endothelin. Pharmacol Rev. 2016; 68:357–418. doi: 10.1124/pr.115.011833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillich A, Zhang F, Farmer CG, Travaglini KJ, Tan SY, Gu M, Zhou B, Feinstein JA, Krasnow MA, Metzger RJ. Capillary cell-type specialization in the alveolus. Nature. 2020; 586:785–789. doi: 10.1038/s41586-020-2822-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piper PJ, Vane JR, Wyllie JH. Inactivation of prostaglandins by the lungs. Nature. 1970; 225:600–604. doi: 10.1038/225600a0 [DOI] [PubMed] [Google Scholar]
- 38.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020; 17:159–162. doi: 10.1038/s41592-019-0667-5 [DOI] [PubMed] [Google Scholar]
- 39.Raredon MSB, Adams TS, Suhail Y, Schupp JC, Poli S, Neumark N, Leiby KL, Greaney AM, Yuan Y, Horien C, et al. Single-cell connectomic analysis of adult mammalian lungs. Sci Adv. 2019; 5:eaaw3851. doi: 10.1126/sciadv.aaw3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Travisano SI, Oliveira VL, Prados B, Grego-Bessa J, Piñeiro-Sabarís R, Bou V, Gómez MJ, Sánchez-Cabo F, MacGrogan D, de la Pompa JL. Coronary arterial development is regulated by a Dll4-Jag1-EphrinB2 signaling cascade. Elife. 2019; 8:e49977. doi: 10.7554/eLife.49977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haynes WG, Strachan FE, Webb DJ. Endothelin ETA and ETB receptors cause vasoconstriction of human resistance and capacitance vessels in vivo. Circulation. 1995; 92:357–363. doi: 10.1161/01.cir.92.3.357 [DOI] [PubMed] [Google Scholar]
- 42.García-Cuesta EM, Santiago CA, Vallejo-Díaz J, Juarranz Y, Rodríguez-Frade JM, Mellado M. The role of the CXCL12/CXCR4/ACKR3 axis in autoimmune diseases. Front Endocrinol. 2019; 10:585. doi: 10.3389/fendo.2019.00585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Förster R, Schubel A, Breitfeld D, Kremmer E, Renner-Müller I, Wolf E, Lipp M. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999; 99:23–33. doi: 10.1016/s0092-8674(00)80059-8 [DOI] [PubMed] [Google Scholar]
- 44.Saygin D, Tabib T, Bittar HET, Valenzi E, Sembrat J, Chan SY, Rojas M, Lafyatis R. Transcriptional profiling of lung cell populations in idiopathic pulmonary arterial hypertension. Pulm Circ. 2020; 10. doi: 10.1177/2045894020908782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peyser R, MacDonnell S, Gao Y, Cheng L, Kim Y, Kaplan T, Ruan Q, Wei Y, Ni M, Adler C, et al. Defining the activated fibroblast population in lung fibrosis using single-cell sequencing. Am J Respir Cell Mol Biol. 2019; 61:74–85. doi: 10.1165/rcmb.2018-0313OC [DOI] [PubMed] [Google Scholar]
- 46.Kimmel JC, Penland L, Rubinstein ND, Hendrickson DG, Kelley DR, Rosenthal AZ. Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging. Genome Res. 2019; 29:2088–2103. doi: 10.1101/gr.253880.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fouillade C, Curras-Alonso S, Giuranno L, Quelennec E, Heinrich S, Bonnet-Boissinot S, Beddok A, Leboucher S, Karakurt HU, Bohec M, et al. FLASH irradiation spares lung progenitor cells and limits the incidence of radio-induced senescence. Clin Cancer Res. 2020; 26:1497–1506. doi: 10.1158/1078-0432.CCR-19-1440 [DOI] [PubMed] [Google Scholar]
- 48.Peake JL, Pinkerton KE. Parent RA. Gross and subgross anatomy of lungs, pleura, connective tissue septa, distal airways, and structural units. Comparative Biology of the Normal Lung (Second Edition). 2015, Academic Press; 21–31 [Google Scholar]
- 49.Verloop MC. On the arteriae bronchiales and their anastomosing with the arteria pulmonalis in some rodents: a micro-anatomical study. Acta Anat (Basel). 1949; 7:1–32. doi: 10.1159/000140373 [DOI] [PubMed] [Google Scholar]
- 50.Vila Ellis L, Cain MP, Hutchison V, Flodby P, Crandall ED, Borok Z, Zhou B, Ostrin EJ, Wythe JD, Chen J. Epithelial Vegfa specifies a distinct endothelial population in the mouse lung. Dev Cell. 2020; 52:617–630.e6. doi: 10.1016/j.devcel.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niethamer TK, Stabler CT, Leach JP, Zepp JA, Morley MP, Babu A, Zhou S, Morrisey EE. Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. Elife. 2020; 9:e53072. doi: 10.7554/eLife.53072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuchs A, Weibel ER. Morphometrische Untersuchung der Verteilung einer spezifischen cytoplasmatischen Organelle in Endothelzellen der Ratte. Z Zellforsch Mikrosk Anat. 1966; 73:1–9 [PubMed] [Google Scholar]
- 53.Wu S, Zhou C, King JA, Stevens T. A unique pulmonary microvascular endothelial cell niche revealed by Weibel-Palade bodies and Griffonia simplicifolia. Pulm Circ. 2014; 4:110–115. doi: 10.1086/674879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019; 47:D766–D773. doi: 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Young MD, Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience. 2020; 9:giaa151. doi: 10.1093/gigascience/giaa151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, III, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019; 177:1888, e21–1902. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019; 8:329, e4–337. doi: 10.1016/j.cels.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020; 17:159–162. doi: 10.1038/s41592-019-0667-5 [DOI] [PubMed] [Google Scholar]
- 60.Raredon MSB, Yang J, Garritano J, Wang M, Kushnir D, Schupp JC, Adams TS, Greaney AM, Leiby KL, Kaminski N, et al. Connectome: computation and visualization of cell-cell signaling topologies in single-cell systems data. bioRxiv. Preprint posted online January 21, 2021. doi: 10.1101/2021.01.21.427529
- 61.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize implements and enhances circular visualization in R. Bioinformatics. 2014; 30:2811–2812. doi: 10.1093/bioinformatics/btu393 [DOI] [PubMed] [Google Scholar]
- 62.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009; 4:1184–1191. doi: 10.1038/nprot.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Comhair SA, Xu W, Mavrakis L, Aldred MA, Asosingh K, Erzurum SC. Human primary lung endothelial cells in culture. Am J Respir Cell Mol Biol. 2012; 46:723–730. doi: 10.1165/rcmb.2011-0416TE [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.