Abstract
We have developed a novel bioorthogonal reaction that can selectively displace fluorine substitutions alpha to amide bonds. This fluorine–thiol displacement reaction (FTDR) allows for fluorinated cofactors or precursors to be utilized as chemical reporters, hijacking acetyltransferase-mediated acetylation both in vitro and in live cells, which cannot be achieved with azide- or alkyne-based chemical reporters. The fluoroacetamide labels can be further converted to biotin or fluorophore tags using FTDR, enabling the general detection and imaging of acetyl substrates. This strategy may lead to a steric-free labeling platform for substrate proteins, expanding our chemical toolbox for functional annotation of post-translational modifications in a systematic manner.
Graphical Abstract
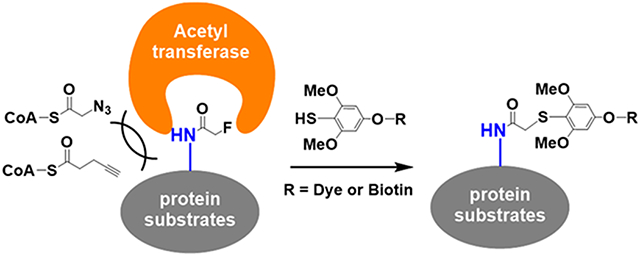
INTRODUCTION
Bioorthogonal reactions have greatly facilitated protein labeling in complex biological systems and led to widespread applications including imaging, enrichment, and identification.1,2 Representative bioorthogonal reactions such as azide–alkyne cycloaddition,3 Staudinger ligation,4 and tetrazine cycloaddition2 have resulted in the successful development of chemical reporters, which in conjunction with detection/affinity tags have advanced our understanding of important biological pathways including post-translational modifications (PTMs).5 For instance, chemical reporters on acetylation have revealed many new protein substrates of p300,6,7 providing a more robust substrate recovery from proteome in comparison to anti-acetyl-lysine antibodies.8,9 Yet, current chemical reporters are bulky in length and size, thereby impeding their general applications. Even alkyne- or azide-based minimalist reporters for copper-catalyzed azide–alkyne cycloaddition (CuAAC) are still significantly larger than the inherent carbon–hydrogen bond (Figure 1A). This intrinsic steric hindrance has largely limited the application of chemical reporters to metabolic incorporation by enzymes possessing spacious active site pockets. For example, alkyne/azide-labeled PTM precursors or cofactors for acetylation10,11 and methylation12,13 were too bulky to be incorporated by many cognate transferases other than p300. While the “bump-hole” protein engineering strategy could work for a given enzyme, careful balances between mutations, structure folding, and function (potency, selectivity, etc.) are required.14 For many acetyltransferases that function as subunits in protein complexes, mutations in the “hole” may alter their substrate specificities, leading to results different from in vivo subacylome,6 which undermines this approach’s broad applications.
Figure 1.
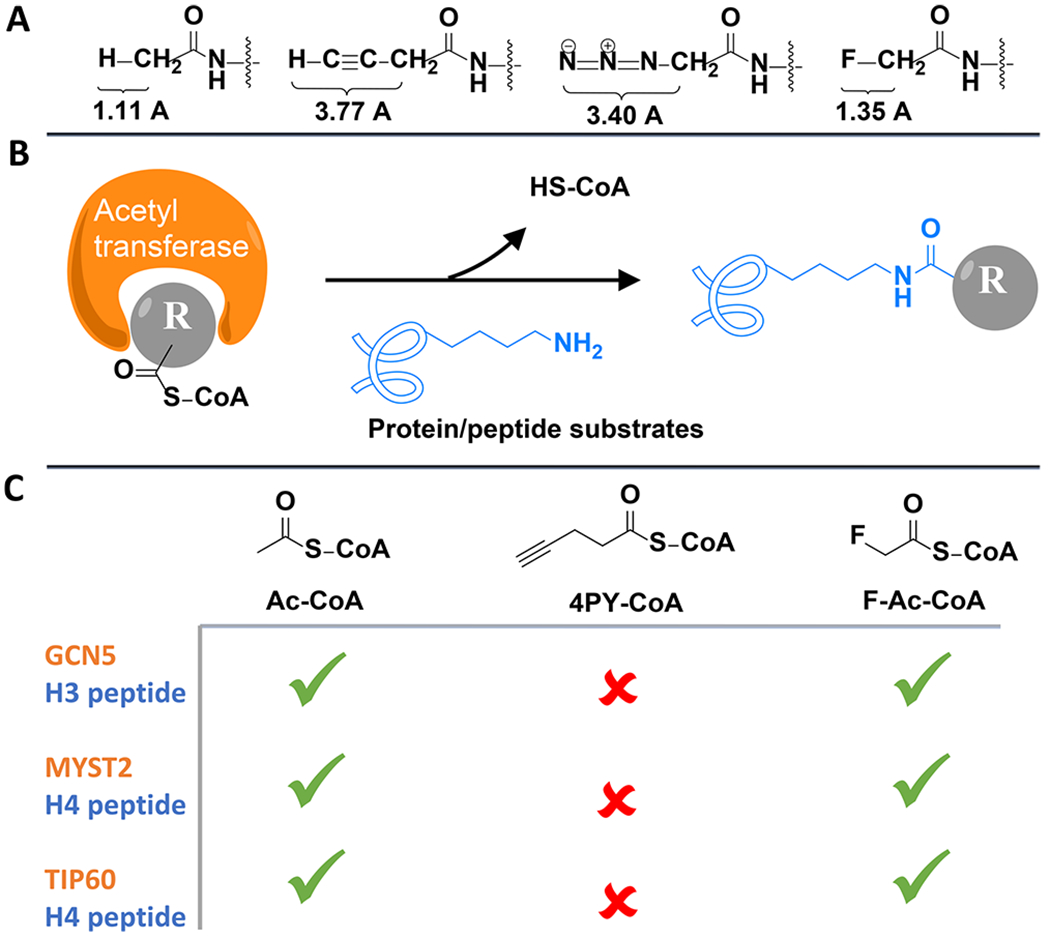
Acetyl–CoA analogs to label acetylation substrates through acetyltransferase assay. (A) Bond lengths of C–H, C–C≡CH, C–N3, and C–F on acetamide. (B) Illustration of the acetyltransferase assay. (C) Summary of the acetyltransferase assay results with Ac–CoA, 4-pentynoyl (4PY)–CoA, and F–Ac–CoA analogs, respectively.
Global profiling of PTM substrates such as those of acetylation can still be achieved by antibody-based detections.15 However, enriched substrates and sites varied significantly per antibody.15 In our hands, a list of commercially available anti-lysine-acetylation antibodies revealed different sensitivity and substrate specificity (Figure S1, Supporting Information). Taken together, elucidating the molecular targets of PTMs such as acetylation have been thereby compromised, despite being a key step toward the systematic dissection of PTMs and their roles in biological and pathology-related cellular signaling regulation.11,13,16 To this end, we asked whether a bioorthogonal reaction can be developed to generate reporters for the steric-free labeling of protein substrates and thereby allow for the global profiling of molecular targets.
RESULTS AND DISCUSSION
The fluorine atom attracted our interest because of its orthogonality from biological molecules,17 similarity in size to hydrogen,17–19 and comparable length of the carbon–fluorine bond to the carbon–hydrogen bond in acetamide, a representative acetylation functional group (Figure 1A).19 Fluorinated amino acids have been exploited to label proteins, bringing in minimal perturbations to protein structure and function.17,18,20 Using acetylation for a proof of concept, we first wanted to investigate whether fluorinated acetyl–CoA can hijack the CoA metabolism and be used by acetyltransferases to label their protein substrates (Figure 1B). Despite the in vivo toxicity of its pro-metabolite fluoroacetate,21 only the late-stage metabolite fluorocitrate was found accountable, mostly damaging organ tissues such as the kidney.21 Fluorinated acetyl–CoA and fluoroacetate have not yet been found to inhibit any enzyme and have been successfully used to study metabolisms.22,23 Ac–CoA analogs with fluorine (F–Ac–CoA)24 or alkyne (4-pentynoyl (4PY)–CoA)11 functional groups were thereby synthesized, and each was mixed with key acetyltransferase GCN5, MYST2, or TIP60 and their corresponding histone peptide substrates.11,25,26 Mass spectrometric analysis (Figure S2) indicated successful acetyl or acetyl analog labeling after either acetyltransferase was incubated with wild-type Ac–CoA or F–Ac–CoA. In contrast, all acetyltransferases failed to incorporate the alkyne-modified 4PY–CoA, which is consistent with previous reports.6,11 Substitution with the electron-withdrawing fluorine also endowed a much higher hydrolysis rate to F–Ac–CoA, which somehow did not display significantly greater nonenzymatic reactivity (Figure S3).27 Taken together (Figure 1C), fluorine modification on the acetyl group afforded a relatively steric-free chemical reporter that can satisfactorily label substrates of acetyltransferases.
Next, we asked whether we can further modify the resulting fluoroacetamides with other tags such as the fluorescent dye “TAMRA”28 or a biotin affinity probe29 for detection, imaging, and future identification of protein substrates. Despite substantial progress in the development of fluorination methodology, there are few efforts aimed toward the replacement of fluorine. One relevant study30 has incorporated an alpha-fluorinated acetophenone moiety (1) into proteins and observed its reaction with a proximal cysteine. This makes us hypothesize that fluorinated acetyl groups may react with thiol derivatives, even at the small-molecule level. To test this, we reacted a representative thiol compound, benzenethiol, with a few fluorinated acetyl substrates in water (Scheme 1). In the presence of a strong base, the primary fluorine alpha to acetophenone (1), ketone (2), and amide (3) were all efficiently displaced (>90% yields). We also observed a similar conversion with the fluoroacetate derivative (4) despite a lower yield, which is likely caused by hydrolysis of the ester given the isolated benzeneethanol side product. The result with 3 particularly encouraged us as it is a model fluoroacetamide substrate for fluorinated peptides/proteins.
Scheme 1.
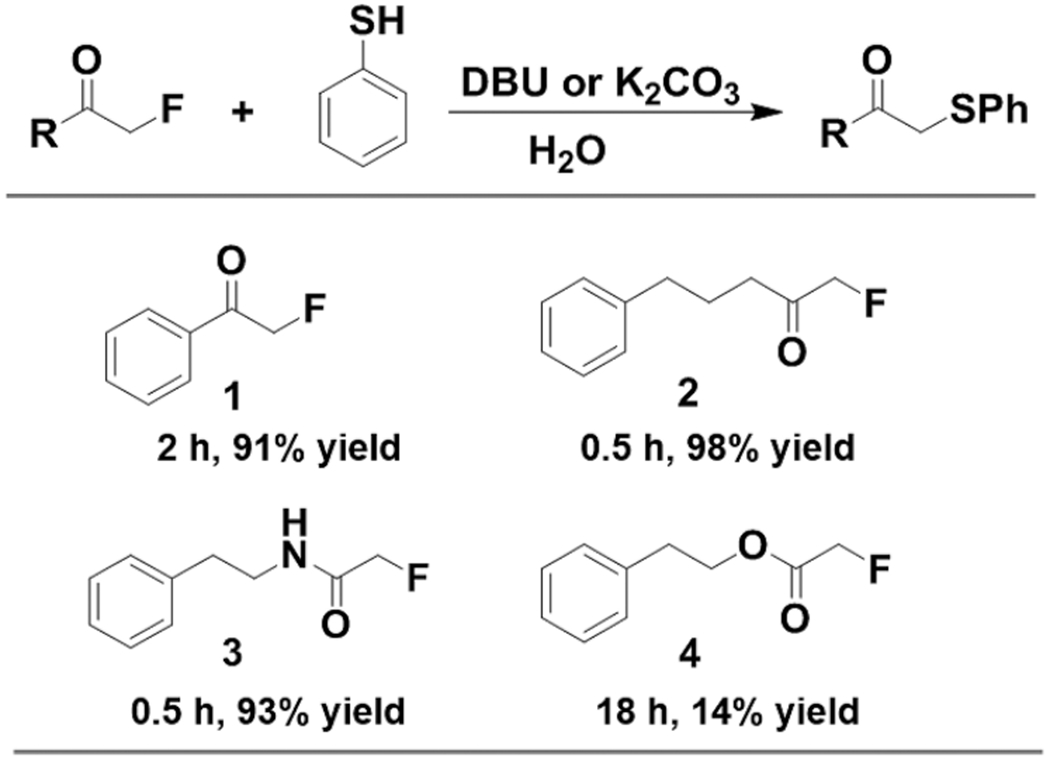
Exploration of Fluorinated Substrate
To identify the optimal pH range for this reaction, we titrated the pH (Figure S4) and found that the reaction rate significantly increased with pH, likely due to the increased deprotonation of benzenethiol. Using the mildly basic condition, pH 8.5, in which the reaction has the best reaction rate, we examined substrate 3’s reactivity with glutathione and cysteine, both as strong intrinsic nucleophiles that exist in cellular environments (Figures S5 and S6). Surprisingly, no reaction was observed for both cases upon incubation for 24 h, suggesting that substrate 3 could be bioorthogonal to other nucleophilic species. We then investigated substitution effects on benzenethiol to efficiently convert fluoroacetamide under the mild physiological reaction condition.31,32 Exploration of a series of benzenethiol derivatives (5–13) found that electron-donating groups at the ortho- and para-positions facilitate the reaction by increasing the nucleophilicity (Figure 2, Figure S7). Given their theoretical pKa values (<7.0) (Figure S8), almost all of these derivatives should be fully deprotonated. The most reactive derivative, 13, is a trimethoxy analog that can reach ~81% reaction conversion within 13 h. Despite potential issues regarding sterics for the ortho-substitutions, the superior reactivity of 13 in comparison to the meta-substituted derivative 12 indicated that the methoxy groups are not bulky enough to perturb reactivity. Likewise, derivative 9, which bears only two substitutions at the electron-donating sites, ended up possessing the same reactivity as 12. Follow-up kinetics studies based on the reported procedures33 suggested that the bimolecular reaction between 3 and 13 followed second-order kinetics and had an observed rate constant of (1.03 ± 0.06) × 10−3 M−1 S−1 (Figure S9), which is similar in rates to the classic Staudinger ligation reaction.33,34
Figure 2.
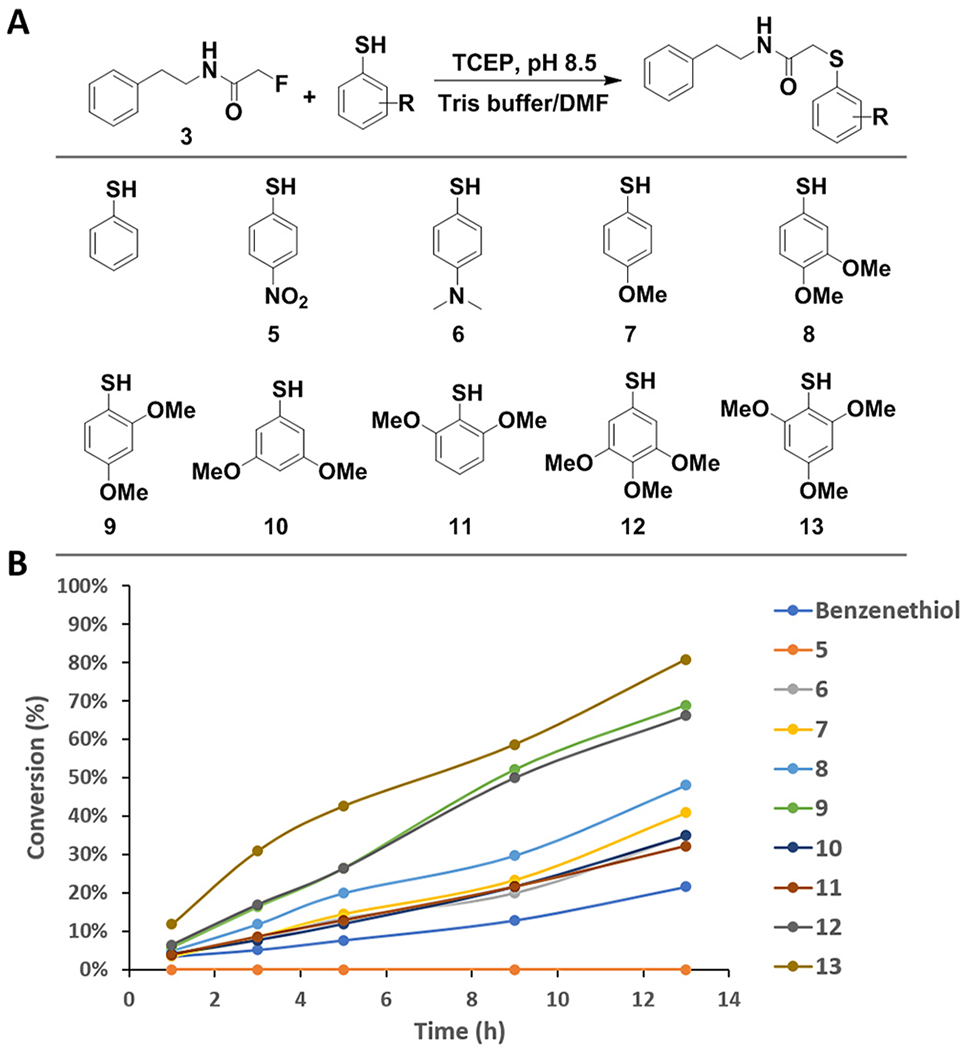
Structure–activity relationship study of benzenethiol derivatives. (A) Scheme of the benzenethiol derivatives. (B) Plot of the conversion against reaction time.
Moreover, the stability of 13 along with substrate 3 were further evaluated in cell lysates (Figure S10), where they remained mostly intact (61.8% and 100%, respectively). In contrast, control 3-Cl, possessing the reported chloroacetamide,35 was not stable, with only 0.15% remaining. This result is consistent with previous observations that chloroacetamide readily reacted with cellular proteins.36 With the aid of LC-MS/MS, the FTDR product of 3 and 13 in cell lysates was confirmed (Figure S11), further corroborating the bioorthogonality of FTDR. Notably, wild-type Ac–CoA has been regarded as a central metabolite that can be also used for ketogenesis, mevalonate pathway, and fatty acid synthesis.37 F–Ac–CoA as a mimic of Ac–CoA was demonstrated to be taken by fatty acid synthetases to make fluoro-fatty acids,38 most of which are also classic PTMs. We thereby tested the FTDR reaction between probe 13 and the alpha-fluorinated model substrates of fatty acids such as butyrate, myristic acid, palmitic acid, malonic acid, and succinic acid (Figure S12). Surprisingly, no FTDR reaction was observed with these substrates, suggesting that because of the increased steric hindrance the secondary fluorides in these fatty acids may not be as readily displaceable as the primary fluoride in fluoroacetamide. On the other hand, these observations also indicated the potential uniqueness of fluoroacetamide and its orthogonality to other fluorine-substituted natural molecules for FTDR-based detection, imaging, and target identification.
Hence, with the most active benzenethiol derivative in hand, we returned to the intriguing question of converting fluoroacetamide to functional detection tags such as fluorescent dyes and biotin probes.29,39 We started by constructing a biotin probe (14, biotin–SH) that contains the benzenethiol structure of 13 as the warhead and the glutamic acid building block (29) as the connecting unit to improve overall solubility (Supplementary Schemes 7–9). Because of the convenience of ESI-MS characterization on peptides, we first demonstrated the successful conversion of the fluorine label to a biotin tag on the aforementioned histone H3-20 peptide substrate (Figure S13). Next, we examined the whole process of labeling and tagging (Figure 3A) on histone protein substrates. Histone H3.1 (Figure 3B) and H4 subunit (Figure S14) were each incubated with F–Ac–CoA, and the corresponding KAT under standard in vitro enzymatic reaction conditions.11 The mixtures were then incubated with the biotin–SH probe for varied time periods, with subsequent in-gel fluorescent imaging to detect the biotin tagging. For both histone proteins, efficient biotinylation occurred within 1 h of incubation and gradually reached saturation after 3 h. Staining with Coomassie Brilliant Blue (CBB) also revealed a complete change of molecular weight, presumably because of the two-step modification. The minimal signal observed from the control group without prior fluorination also indicated the relative specificity of biotin–SH. Concurrently, failure of biotinylation was observed for the group using CuAAC labeling, which was consistent with literature reports and confirmed that many KATs are incapable of uptaking sterically hindered substrates.6,11 To check if the fluoroacetyl label could be recognized by histone deacetylases, we also incubated these fluoroacetylated histone proteins with a few reported deacetylases40 before biotin–SH tagging. Interestingly, most fluoroacetylation can be efficiently removed (Figure S15), suggesting that the label could be a substrate for deacetylases as well.
Figure 3.
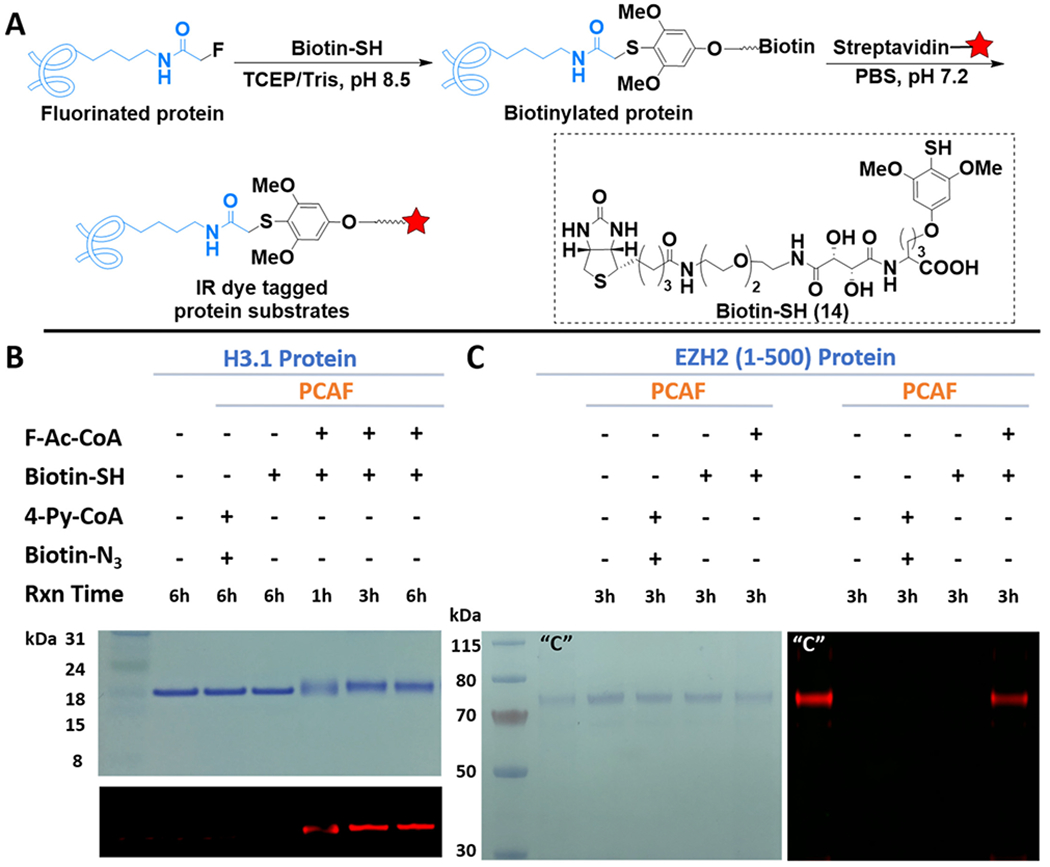
FTDR-based tagging of protein substrates with biotin–SH probe. (A) Reaction scheme. The red star indicates IR dye. (B, C) Labeling of histone protein H3.1 and nonhistone substrate EZH2 (1 – 500), respectively. The top panel for (B) and the left panel for (C) are gel images after CBB staining. The other images are for in-gel fluorescent detection of IR dye. “C” is the positive control for CuAAC, which has been prepared by NHS ester labeling of the lysines on EZH2 with the “click chemistry” tag, followed by CuAAC reaction with the corresponding biotin linker.
In addition to histones, a growing number of nonhistone substrates were recently uncovered, with their roles in protein function regulation being increasingly appreciated.16 For instance, the enhancer of zeste homologue 2 (EZH2) was revealed to be acetylated by PCAF mainly at lysine 348, which improved EZH2’s stability and promoted the migration of lung cancer cells.41 To evaluate whether our labeling strategy is applicable to nonhistone substrates, we specifically constructed and expressed the truncated form of EZH2 (1–500) in Escherichia coli, with a shake-flask yield of approximately 2 mg/L. The protein’s purity and size were confirmed by SDS-PAGE (Figure S16). Reaction of the EZH2 fragment under the same conditions as those for histone proteins resulted in a similar type of tagging, as evident by Figure 3C. Taken together, these observations displayed the steric-free in vitro labeling and tagging of a range of known protein substrates using the FTDR reaction, which could not be achieved by the known CuAAC method.
For labeling proteins in living cells, azide or alkyne analogs of fatty acids have been exploited, which were metabolized intracellularly into CoA derivatives.3–5,10 For an increase of their cellular delivery, pro-metabolites with esters masking the polar carboxylate group constituted an effective strategy in recent years.42 Nevertheless, the intrinsic sterics of these pro-metabolites resulted in varied and suboptimal labeling results,10,42 sometimes requiring extensive structural optimization.42 To explore the utility of our probing system for studying acetylation in the cellular level, we designed the fluorinated version of pro-metabolite, ethyl fluoroacetate (Figure 4A). Given the in vivo toxicity of fluoroacetate,21 we first evaluated the cell cytotoxicity and found that this pro-metabolite exhibited minimal toxicity with doses up to 2 mM after 12 h of incubation (Figure S17). Additional LC-MS/MS studies indeed confirmed its conversion by enzymes to fluoroacetyl–CoA in live cells (Figure S18), which, taken together with the observed minimal toxicity in cell lines, could support the applicability of ethyl fluoroacetate and its CoA metabolite to studies at the cellular level.
Figure 4.
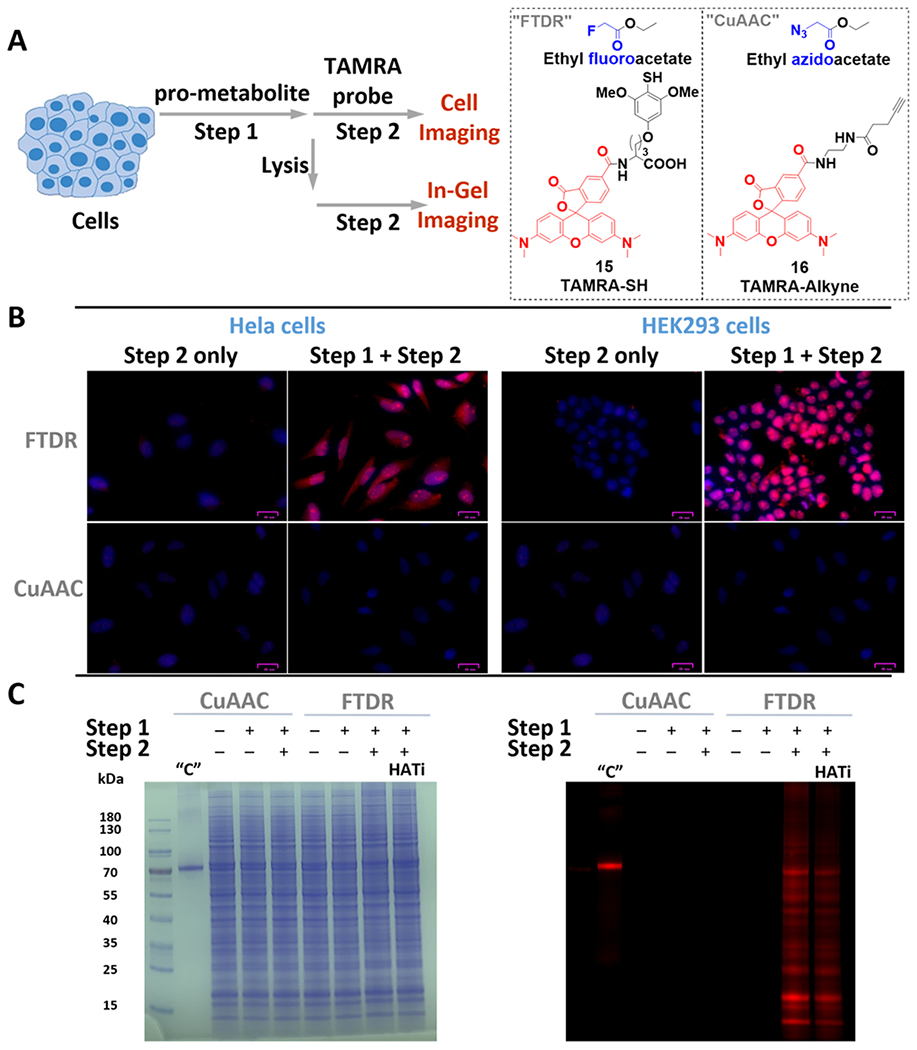
Cellular evaluation of FTDR-based tagging with TAMRA-SH probe. (A) Scheme for cellular pro-metabolite incorporation (1 mM, 6 h, at 37 °C, step 1) and protein substrate detection (step 2). (B) Fluorescent microscopy of fixed and permeabilized cells that were stained by Hoechst 33342 (blue) and TAMRA probes (red); scale bars: 25 μm. (C) Cell lysate protein labeling by pro-metabolites and detection by TAMRA probes (red); left panel, PAGE gel stained by CBB; right panel, in-gel fluorescent detection. “C” is the positive control for CuAAC, which randomly labeled lysines on BSA with azide–NHS ester, followed by CuAAC-mediated conjugation with TAMRA–alkyne. “HATi” indicates the addition of HAT inhibitor (A-485) prior to step 1.
With confidence in the safety profile of our pro-metabolite, we started by treating it with two representative cell lines, HeLa and HEK293, followed by subsequent FTDR with the TAMRA–SH probe (15) as the second step for fluorescent detection (Figure 4A). Treatment with the azido-modified pro-metabolite42 and the TAMRA–alkyne (16) for CuAAC chemistry was performed in parallel as a control, wherein weak signals were previously reported.42 Direct microscope imaging studies (Figure 4B) revealed much stronger intracellular labeling and tagging with TAMRA following the FTDR-based approach, suggesting a drastically improved, more complete profiling of acetylation substrates. We also observed significant fluorescence not only in the nucleus but also in the cytoplasm, which may indicate the successful labeling of both histones and nonhistone proteins. The little background signals emitted from the cells treated with only 15 (step 2) further demonstrated the specificity of the developed–SH probes. Following similar procedures, we also tested the labeling of the cell lysates after metabolic incorporation of fluorine reporters. Lysates following FTDR or CuAAC mediated ligation with TAMRA were separated on SDS-PAGE and visualized by CBB to confirm an equal amount of protein loading (Figure 4C). Yet, multiple labeled protein bands spanning a wide range of molecular weights were observed only for the lysates of cells that underwent a complete two-step process of FTDR (Figure 4C). This discovery was consistent with the microscope imaging results. Furthermore, pretreatment of the cells with A-485 (potent and selective p300/CBP inhibitor)43,44 (Figure 4C) before the two-step process of FTDR also resulted in weaker labeling intensity, which is consistent with the previously reported results using HATi.10,42,45–49 In addition, we exploited the concurrent incubation with HDAC inhibitor cocktails and observed slightly decreased labeling (Figure S19), which was consistent with literature reports,9,10,42 suggesting that incorporation of F-acetylation needs prior deacetylation of intrinsically acetylated lysine residues to make the lysine available. Thus, blocking the removal of wild-type acetylation prevents metabolic incorporation of F-acetylation.9,10,42 Taken together, these observations fully supported that FTDR allowed for profiling the proteome-wide substrates of acetylation from the cellular contexts.
To further validate that the FTDR-based two-step metabolic labeling occurs on acetylation protein substrates, we performed histone extraction (Figure 5A) and confirmed the existence of TAMRA labeling on the primary acetylation substrates, histones (Figure 5B). As controls, both the treatment with HAT inhibitors and the competition with acetate have resulted in decreased TAMRA–SH probe labeling, suggesting that the FTDR-based two-step labeling is acetyltransferase-dependent and relies on F–Ac–CoA metabolites. To test whether the FTDR-based labeling can be used to enrich these protein substrates, we treated cell lysates with the biotin–SH probe in step 2 and pulled down the labeled proteins (Figure 5A). Western blot analysis confirmed the presence of known acetylation substrates including histones, and α-tubulin53,54 only in the protein pool enriched from the cell lysates that have been subjected to the two-step process (Figure 5C). Accordingly, pretreatment with HAT inhibitors decreased the amount of proteins enriched, particularly for H3 and H4. The level of α-tubulin was not significantly perturbed, likely due to the fact that certain acetylation sites such as lysine 40 on α-tubulin are mediated by other acetyltransferases (e.g., αTAT1)53 that were not targeted by the administered HAT inhibitors.
Figure 5.
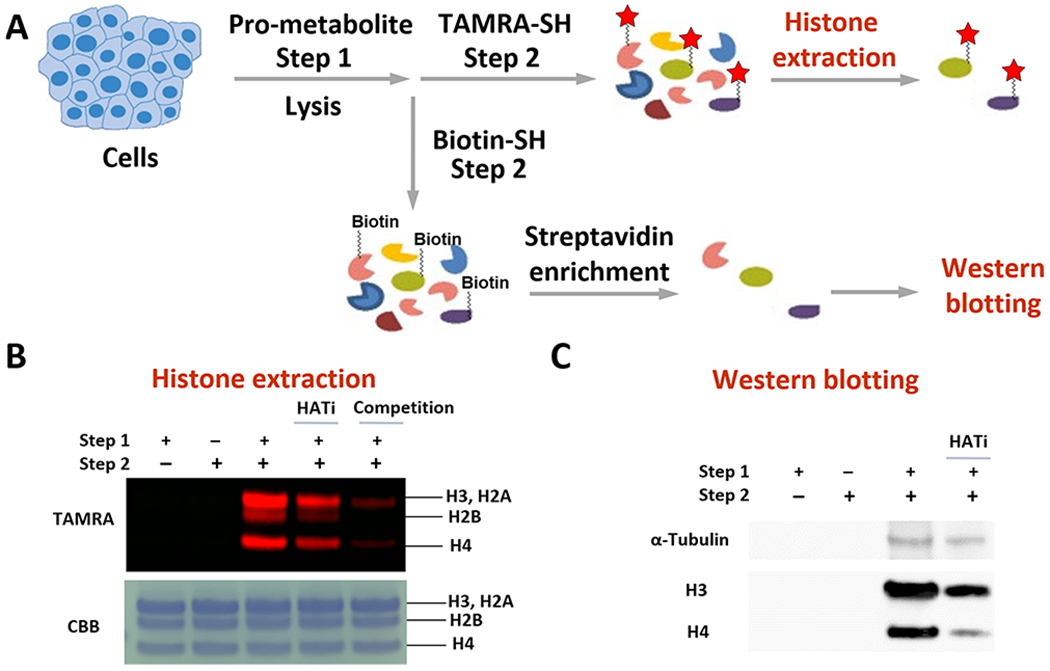
Validation of the FTDR-based labeling of acetylation substrates. (A) Scheme for cellular pro-metabolite incorporation (step 1), protein substrate labeling by TAMRA–SH (step 2), and extraction of the known acetylation substrates histones; or protein substrate labeling by biotin–SH probe (step 2), enrichment with streptavidin beads, followed by Western blot analysis of the proteins pulled down to examine the existence of α-tubulin, histone H3, and histone H4. (B) Histone extraction results. Top panel, in-gel fluorescent detection; bottom panel, CBB. (C) Western blotting results. “HATi” indicates the addition of anacardic acid and MG149.50–52
Lastly, to gain insight into the specific labeling sites by our tagging strategy, we extracted out histones, pulled down α-tubulin from the cells having incorporated the pro-metabolite ethyl fluoroacetate, and carried out proteomics studies (Figure 6A). As shown in Figure 6B and Figures S20–S22, fluoroacetylation has been observed on the following: lysine 30, lysine 34, lysine 43, and lysine 57 of histone H2B; lysine 5, lysine 8, lysine 12, lysine 16, lysine 31, lysine 44, lysine 59, lysine 77, lysine 79, and lysine 91 of histone H4; lysine 40, lysine 60, lysine 163, lysine 164, lysine 166, and lysine 280 of α-tubulin. Almost all of these sites have been consistent with previous literature reports on acetylation sites,54–57 further demonstrating that this labeling strategy can probe lysine acetylation in a general manner. Notably, N-terminal acetylation has recently drawn significant research interest, which is catalyzed by a specific class of N-terminal acetyltransferases using Ac–CoA and has been revealed to play important roles in protein functions and cellular locations, etc.58,59 During the preliminary proteomics studies of histone H2B and H4 (Figures S20–S21), we have not been able to observe any F-acetylation at the N-terminus. Although N-terminal acetylation is widespread and common in human proteins, it may exist only partially within the pool of molecules of a given protein substrate,60 and there are many internal lysine acetylation sites for each N-terminal protein.58 Special enrichment techniques58 will be necessary for our future studies to observe N-terminal acetylation. More importantly, N-terminal acetylation is considered irreversible,58 which could block the incorporation of F-acetylation to the intrinsic N-terminus. Thus, our FTDR-based labeling and imaging may be specific to internal lysine sites.
Figure 6.
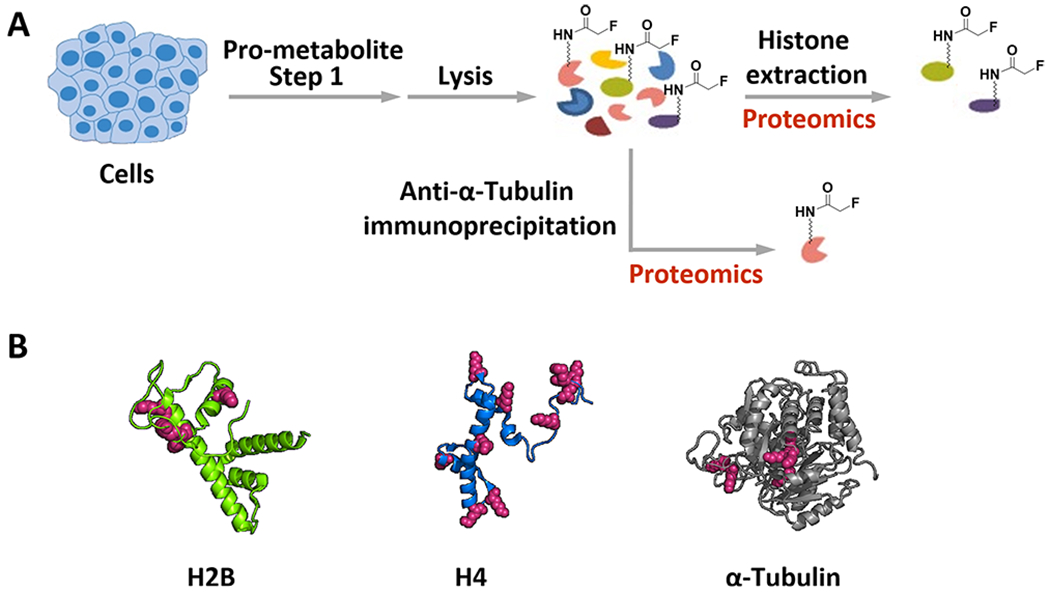
Validation of the fluoroacetyl labeling sites on known protein substrates by proteomics analysis. (A) Scheme for cellular pro-metabolite incorporation, lysis, and the histone proteins extraction; or immunoprecipitation of α-tubulin after cellular pro-metabolite incorporation. (B) Summary of the F–Ac labeling sites (red) on representative histone proteins H2B, H4 (PDB: 1kx5), and α-tubulin (PDB: 1tub). Details of the proteomics results are available in the Supporting Information (Figures S20–S22).
CONCLUSION
In summary, we have developed a fluorine–thiol displacement reaction (FTDR) and demonstrated its use for the steric-free labeling of protein substrates of a representative PTM, acetylation. Along with the benzenethiol-derived functional tags, the FTDR-based imaging and detection of substrates demonstrated great potential for globally profiling acetyl transferases’ substrates, which are of vital significance for understanding the roles of acetylation in physiology and disease. Although the fluorine tag possesses less sterics than the alkyne/azide tags commonly used for CuAAC chemistry, our FTDR appeared to be slower than CuAAC or other tetrazine-based bioorthogonal reactions,61 which could limit this methodology’s certain biological applications such as real-time imaging. Thus, future work would focus on improving and optimizing the FTDR reaction conditions with the exploitation of more reactive probes. Nevertheless, this tool kit, together with future applications to quantitative proteomics studies, is expected to offer versatile probes for identifying targets of acetylation, and possibly many other PTMs that are mediated by transferases with restricted active sites.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the National Cancer Institute of the National Institute of Health (NIH) under Award No. P30 CA006927, R50 CA221838, and National Institute of General Medical Sciences of the NIH under Award 1R35GM133468-01. Support for the Wistar Proteomics and Metabolomics Facility was provided by P30 CA010815 and S10 OD023586. Support for the NMR facility at Temple University by a CURE grant from the Pennsylvania Department of Health is gratefully acknowledged.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.0c05605
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c05605.
Materials and methods, supplementary figures, and NMR spectra (PDF)
The authors declare no competing financial interest.
Contributor Information
Zhigang Lyu, Department of Chemistry, Temple University, Philadelphia, Pennsylvania 19122, United States.
Yue Zhao, Department of Chemistry, Temple University, Philadelphia, Pennsylvania 19122, United States.
Zakey Yusuf Buuh, Department of Chemistry, Temple University, Philadelphia, Pennsylvania 19122, United States.
Nicole Gorman, Proteomics and Metabolomics Facility, The Wistar Institute, Philadelphia, Pennsylvania 19104, United States.
Aaron R. Goldman, Proteomics and Metabolomics Facility, The Wistar Institute, Philadelphia, Pennsylvania 19104, United States.
Md Shafiqul Islam, Department of Chemistry, Temple University, Philadelphia, Pennsylvania 19122, United States.
Hsin-Yao Tang, Proteomics and Metabolomics Facility, The Wistar Institute, Philadelphia, Pennsylvania 19104, United States.
Rongsheng E. Wang, Department of Chemistry, Temple University, Philadelphia, Pennsylvania 19122, United States.
REFERENCES
- (1).Devaraj NK The Future of Bioorthogonal Chemistry. ACS Cent. Sci 2018, 4 (8), 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Elliott TS; Bianco A; Townsley FM; Fried SD; Chin JW Tagging and Enriching Proteins Enables Cell-Specific Proteomics. Cell Chem. Biol 2016, 23 (7), 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Grammel M; Hang HC Chemical reporters for biological discovery. Nat. Chem. Biol 2013, 9 (8), 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Saxon E; Bertozzi CR Cell surface engineering by a modified Staudinger reaction. Science 2000, 287 (5460), 2007–2010. [DOI] [PubMed] [Google Scholar]
- (5).Chuh KN; Pratt MR Chemical methods for the proteome-wide identification of posttranslationally modified proteins. Curr. Opin. Chem. Biol 2015, 24, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Han Z; Chou CW; Yang X; Bartlett MG; Zheng YG Profiling Cellular Substrates of Lysine Acetyltransferases GCN5 and p300 with Orthogonal Labeling and Click Chemistry. ACS Chem. Biol 2017, 12 (6), 1547–1555. [DOI] [PubMed] [Google Scholar]
- (7).Yu-Ying Y; Markus G; Howard HC Identification of lysine acetyltransferase p300 substrates using 4-pentynoyl-coenzyme A and bioorthogonal proteomics. Bioorg. Med. Chem. Lett 2011, 21 (17), 4976–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chuh KN; Batt AR; Pratt MR Chemical Methods for Encoding and Decoding of Posttranslational Modifications. Cell Chem. Biol 2016, 23 (1), 86–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ourailidou ME; Dockerty P; Witte M; Poelarends GJ; Dekker FJ Metabolic alkene labeling and in vitro detection of histone acylation via the aqueous oxidative Heck reaction. Org. Biomol. Chem 2015, 13 (12), 3648–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Yang YY; Ascano JM; Hang HC Bioorthogonal chemical reporters for monitoring protein acetylation. J. Am. Chem. Soc 2010, 132 (11), 3640–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yang C; Mi J; Feng Y; Ngo L; Gao T; Yan L; Zheng YG Labeling lysine acetyltransferase substrates with engineered enzymes and functionalized cofactor surrogates. J. Am. Chem. Soc 2013, 135 (21), 7791–7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Binda O; Boyce M; Rush JS; Palaniappan KK; Bertozzi CR; Gozani O A chemical method for labeling lysine methyltransferase substrates. ChemBioChem 2011, 12 (2), 330–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Grammel M; Yang Y-Y; Hang HC Chemical reporters of protein methylation and acetylation; Cambridge University Press: Cambridge, 2012. [Google Scholar]
- (14).Runcie AC; Chan KH; Zengerle M; Ciulli A Chemical genetics approaches for selective intervention in epigenetics. Curr. Opin. Chem. Biol 2016, 33, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Shaw PG; Chaerkady R; Zhang Z; Davidson NE; Pandey A Monoclonal antibody cocktail as an enrichment tool for acetylome analysis. Anal. Chem 2011, 83 (10), 3623–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Buuh ZY; Lyu Z; Wang RE Interrogating the Roles of Post-Translational Modifications of Non-Histone Proteins. J. Med. Chem 2018, 61 (8), 3239–3252. [DOI] [PubMed] [Google Scholar]
- (17).Tressler CM; Zondlo NJ Perfluoro-tert-butyl Homoserine Is a Helix-Promoting, Highly Fluorinated, NMR-Sensitive Aliphatic Amino Acid: Detection of the Estrogen Receptor.Coactivator Protein-Protein Interaction by (19)F NMR. Biochemistry 2017, 56 (8), 1062–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Gee CT; Koleski EJ; Pomerantz WC Fragment screening and druggability assessment for the CBP/p300 KIX domain through protein-observed 19F NMR spectroscopy. Angew. Chem., Int. Ed 2015, 54 (12), 3735–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).O’Hagan D Understanding organofluorine chemistry. An introduction to the C-F bond. Chem. Soc. Rev 2008, 37 (2), 308–319. [DOI] [PubMed] [Google Scholar]
- (20).Mishra NK; Urick AK; Ember SW; Schonbrunn E; Pomerantz WC Fluorinated aromatic amino acids are sensitive 19F NMR probes for bromodomain-ligand interactions. ACS Chem. Biol 2014, 9 (12), 2755–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Peters RA Croonian Lecture - Lethal synthesis. Proc. R. Soc. B. Biol. Sci 1952, 139 (895), 143–170. [DOI] [PubMed] [Google Scholar]
- (22).Marcus A; Elliott WB Enzymatic reactions of fluoroacetate and fluoroacetyl coenzyme A. J. Biol. Chem 1956, 218 (2), 823–830. [PubMed] [Google Scholar]
- (23).Mori T; Sun LQ; Kobayashi M; Kiyono Y; Okazawa H; Furukawa T; Kawashima H; Welch MJ; Fujibayashi Y Preparation and evaluation of ethyl [(18)F]fluoroacetate as a proradiotracer of [(18)F]fluoroacetate for the measurement of glial metabolism by PET. Nucl. Med. Biol 2009, 36 (2), 155–162. [DOI] [PubMed] [Google Scholar]
- (24).Weeks AM; Chang MC Catalytic control of enzymatic fluorine specificity. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (48), 19667–19672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Marmorstein R Structure and function of histone acetyltransferases. Cell. Mol. Life Sci 2001, 58 (5–6), 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kimura A; Horikoshi M Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells 1998, 3 (12), 789–800. [DOI] [PubMed] [Google Scholar]
- (27).Wagner GR; Bhatt DP; O’Connell TM; Thompson JW; Dubois LG; Backos DS; Yang H; Mitchell GA; Ilkayeva OR; Stevens RD; Grimsrud PA; Hirschey MD A Class of Reactive Acyl-CoA Species Reveals the Non-enzymatic Origins of Protein Acylation. Cell Metab. 2017, 25 (4), 823–837.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Whitaker JE; Haugland RP; Ryan D; Hewitt PC; Haugland RP; Prendergast FG Fluorescent rhodol derivatives: versatile, photostable labels and tracers. Anal. Biochem 1992, 207 (2), 267–279. [DOI] [PubMed] [Google Scholar]
- (29).Yang Y; Hahne H; Kuster B; Verhelst SH A simple and effective cleavable linker for chemical proteomics applications. Mol. Cell. Proteomics 2013, 12 (1), 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Xiang Z; Ren H; Hu YS; Coin I; Wei J; Cang H; Wang L Adding an unnatural covalent bond to proteins through proximity-enhanced bioreactivity. Nat. Methods 2013, 10 (9), 885–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Bos J; Muir TW A Chemical Probe for Protein Crotonylation. J. Am. Chem. Soc 2018, 140 (14), 4757–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zhu X; Lin Q; Chen P; Fu YP; Zhang YM; Wei TB A novel pH sensor which could respond to multi-scale pH changes via different fluorescence emissions. New J. Chem 2016, 40 (5), 4562–4565. [Google Scholar]
- (33).Agard NJ; Baskin JM; Prescher JA; Lo A; Bertozzi C R A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol 2006, 1 (10), 644–648. [DOI] [PubMed] [Google Scholar]
- (34).Sundhoro M; Jeon S; Park J; Ramstrom O; Yan M Perfluoroaryl Azide Staudinger Reaction: A Fast and Bioorthogonal Reaction. Angew. Chem., Int. Ed 2017, 56 (40), 12117–12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Yu M; de Carvalho LP; Sun G; Blanchard JS Activity-based substrate profiling for Gcn5-related N-acetyltransferases: the use of chloroacetyl-coenzyme A to identify protein substrates. J. Am. Chem. Soc 2006, 128 (48), 15356–15357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Barglow KT; Cravatt BF Discovering disease-associated enzymes by proteome reactivity profiling. Chem. Biol 2004, 11 (11), 1523–1531. [DOI] [PubMed] [Google Scholar]
- (37).Pietrocola F; Galluzzi L; Bravo-San Pedro JM; Madeo F; Kroemer G Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015, 21 (6), 805–821. [DOI] [PubMed] [Google Scholar]
- (38).Walker MC; Chang MC Natural and engineered biosynthesis of fluorinated natural products. Chem. Soc. Rev 2014, 43 (18), 6527–6536. [DOI] [PubMed] [Google Scholar]
- (39).Andersen KA; Aronoff MR; McGrath NA; Raines R T. Diazo groups endure metabolism and enable chemoselectivity in cellulo. J. Am. Chem. Soc 2015, 137 (7), 2412–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Seto E; Yoshida M Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor Perspect. Biol 2014, 6 (4), a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wan J; Zhan J; Li S; Ma J; Xu W; Liu C; Xue X; Xie Y; Fang W; Chin YE; Zhang H PCAF-primed EZH2 acetylation regulates its stability and promotes lung adenocarcinoma progression. Nucleic Acids Res. 2015, 43 (7), 3591–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Sinclair WR; Shrimp JH; Zengeya TT; Kulkarni RA; Garlick JM; Luecke H; Worth AJ; Blair IA; Snyder NW; Meier JL Bioorthogonal pro-metabolites for profiling short chain fatty acylation. Chem. Sci 2018, 9 (5), 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Lasko LM; Jakob CG; Edalji RP; Qiu W; Montgomery D; Digiammarino EL; Hansen TM; Risi RM; Frey R; Manaves V; Shaw B; Algire M; Hessler P; Lam LT; Uziel T; Faivre E; Ferguson D; Buchanan FG; Martin RL; Torrent M; Chiang GG; Karukurichi K; Langston JW; Weinert BT; Choudhary C; de Vries P; Kluge AF; Patane MA; Van Drie JH; Wang C; McElligott D; Kesicki E; Marmorstein R; Sun C; Cole PA; Rosenberg SH; Michaelides MR; Lai A; Bromberg KD Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017, 550 (7674), 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhang B; Chen D; Liu B; Dekker FJ; Quax WJ A novel histone acetyltransferase inhibitor A485 improves sensitivity of nonsmall-cell lung carcinoma cells to TRAIL. Biochem. Pharmacol 2020, 175, 113914. [DOI] [PubMed] [Google Scholar]
- (45).Legartova S; Stixova L; Strnad H; Kozubek S; Martinet N; Dekker FJ; Franek M; Bartova E Basic nuclear processes affected by histone acetyltransferases and histone deacetylase inhibitors. Epigenomics 2013, 5 (4), 379–396. [DOI] [PubMed] [Google Scholar]
- (46).Eliseeva ED; Valkov V; Jung M; Jung MO Characterization of novel inhibitors of histone acetyltransferases. Mol. Cancer Ther 2007, 6 (9), 2391–2398. [DOI] [PubMed] [Google Scholar]
- (47).Li S; Peng B; Luo X; Sun H; Peng C Anacardic acid attenuates pressure-overload cardiac hypertrophy through inhibiting histone acetylases. J. Cell. Mol. Med 2019, 23 (4), 2744–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Huang H; Tang S; Ji M; Tang Z; Shimada M; Liu X; Qi S; Locasale JW; Roeder RG; Zhao Y; Li X p300-Mediated Lysine 2-Hydroxyisobutyrylation Regulates Glycolysis. Mol. Cell 2018, 70 (5), 663–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Weinert BT; Narita T; Satpathy S; Srinivasan B; Hansen BK; Scholz C; Hamilton WB; Zucconi BE; Wang WW; Liu WR; Brickman JM; Kesicki EA; Lai A; Bromberg KD; Cole PA; Choudhary C Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/p300 Acetylome. Cell 2018, 174 (1), 231–244.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Deng H; Fujiwara N; Cui H; Whitford GM; Bartlett JD; Suzuki M Histone acetyltransferase promotes fluoride toxicity in LS8 cells. Chemosphere 2020, 247, 125825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Sun Y; Jiang X; Chen S; Price BD Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett. 2006, 580 (18), 4353–4356. [DOI] [PubMed] [Google Scholar]
- (52).Simpson S; Fiches G; Jean MJ; Dieringer M; McGuinness J; John SP; Shamay M; Desai P; Zhu J; Santoso NG Inhibition of Tip60 Reduces Lytic and Latent Gene Expression of Kaposi’s Sarcoma-Associated Herpes Virus (KSHV) and Proliferation of KSHV-Infected Tumor Cells. Front. Microbiol 2018, 9, 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Wei D; Gao N; Li L; Zhu JX; Diao L; Huang J; Han QJ; Wang S; Xue H; Wang Q; Wu QF; Zhang X; Bao L alpha-Tubulin Acetylation Restricts Axon Overbranching by Dampening Microtubule Plus-End Dynamics in Neurons. Cereb. Cortex 2018, 28 (9), 3332–3346. [DOI] [PubMed] [Google Scholar]
- (54).Liu N; Xiong Y; Li S; Ren Y; He Q; Gao S; Zhou J; Shui W New HDAC6-mediated deacetylation sites of tubulin in the mouse brain identified by quantitative mass spectrometry. Sci. Rep 2015, 5, 16869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Hansen BK; Gupta R; Baldus L; Lyon D; Narita T; Lammers M; Choudhary C; Weinert BT Analysis of human acetylation stoichiometry defines mechanistic constraints on protein regulation. Nat. Commun 2019, 10 (1), 1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Barnes CE; English DM; Cowley SM Acetylation & Co: an expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem. 2019, 63 (1), 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Li D; Wan C; Bai B; Cao H; Liu C; Zhang Q Identification of histone acetylation markers in human fetal brains and increased H4K5ac expression in neural tube defects. Mol. Genet. Genomic Med 2019, 7 (12), e1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Ree R; Varland S; Arnesen T Spotlight on protein N-terminal acetylation. Exp. Mol. Med 2018, 50 (7), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Aksnes H; Drazic A; Marie M; Arnesen T First Things First: Vital Protein Marks by N-Terminal Acetyltransferases. Trends Biochem. Sci 2016, 41 (9), 746–760. [DOI] [PubMed] [Google Scholar]
- (60).Aksnes H; Hole K; Arnesen T Molecular, cellular, and physiological significance of N-terminal acetylation. Int. Rev. Cell Mol. Biol 2015, 316, 267–305. [DOI] [PubMed] [Google Scholar]
- (61).Lang K; Chin JW Bioorthogonal reactions for labeling proteins. ACS Chem. Biol 2014, 9 (1), 16–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


