Abstract
Background: Globally, blaCTX-M-15 beta-lactamases are the most popular extended spectrum beta-lactamase alleles that are widely distributed due its mobilisation by mobile genetic elements in several compartments. We aimed to determine the conjugation frequencies and replicon types associated with plasmids carrying blaCTX-M-15 gene from Extended Spectrum Beta-lactamase producing isolates in order to understand the dissemination of resistance genes in different compartments. Material and methods: A total of 51 archived isolates carrying blaCTX-M-15 beta-lactamases were used as donors in this study. Antibiotic susceptibility tests were performed as previously described for both donors and transconjugants. Conjugation experiment was performed by a modified protocol of the plate mating experiment, and plasmid replicon types were screened among donor and transconjugant isolates by multiplex Polymerase Chain Reaction in a set of three primer panels. Results: The conjugation efficiency of plasmids carrying blaCTX-M-15 was 88.2% (45/51) with conjugation frequencies in the order of 10−1 to 10−9 and a 100% transfer efficiency observed among E. coli of animal origin. Majority of donors (n = 21) and transconjugants (n = 14) plasmids were typed as either Inc FIA or Inc FIB. Resistance to non-beta-lactam antibiotics was transferrable in 34/45 (75.6%) of events. Ciprofloxacin, tetracycline and sulphamethoxazole-trimethoprim resistance was co-transferred in 29/34 (85.3%) such events. Gentamicin resistance was transferred in 17/34 (50%) of events. Conclusions: Majority of plasmids carrying blaCTX-M-15 were conjugatively transferred by IncF plasmids along with non-beta lactam resistance. There is a need for more research on plasmids to understand how plasmids especially multi replicon plasmids interact and the effect of such interaction on conjugation. One Health approach is to be intensified to address antimicrobial resistance which is a public health threat.
Keywords: conjugation, CTX-M-15, replicon, plasmid, non-beta lactam antibiotics, One Health
1. Introduction
The increasing trend of antimicrobial resistance is intensified by mobile genetic elements that harbour resistance genes [1]. The effect of these elements is extensively reported among bacteria of the Enterobacteriaceae family where multi drug resistance (MDR) is high. The CTX-M extended-spectrum beta-lactamases (ESBL) are the most successful MDR determinants [2], with over 100 alleles in five distinct phylogenetic groups [3]. The ecological success of CTX-M-ESBL attributes to the enzymes’ spread both clonally and horizontally [3,4,5] in multiple hosts that include Acinetobacter spp., Enterobacter spp., E. coli, P. aeruginosa, K. pneumoniae and P. mirabilis.
In natural environments, ESBL enzymes are chromosomally mediated by the selection pressure induced by beta-lactamase-producing soil organisms [6,7] or the irrational use of third-generation cephalosporins [6,8], however as previously reviewed [9], plasmid-mediated ESBL resulted from transposon-mediated insertion of different blaCTX-M genes in bacteria chromosome. Specifically, the precursors of plasmid-mediated blaCTX-M-15 are environmental Kluyvera spp. whose chromosomal CTX-M clusters are incorporated into the chromosome of host bacteria by mobilising elements such as ISEcp1 or ISCR1. Moreover, the location of ISEcp1 upstream blaCTX-M genes together with multiple inverted repeats downstream the gene facilitates the expression and ongoing transposition of blaKLU genes that result to various CTX-M enzymes, including plasmid-mediated blaCTX-M-15 [9,10]. The mobilisation potential of ISEcp1 for chromosome-linked multi-resistant determinants in other members of enterobacteriaceae increases with the additional possession of ISCR1, another mobile genetic element (MGE) embedded in a Class 1 integron that mobilises unrelated CTX-M groups in similar or different species.
As vectors and carriers of AMR genes, plasmids are responsible for the intracellular accumulation and intercellular transfer of these genes by the process of conjugation [11]. In such cases, high conjugation rates ensure the stable long-term persistence of plasmids and associated AMR genes in minimal fitness costs even in the absence of selection pressure [12,13,14]. A multidrug resistance phenomenon is observed when these plasmids are associated with other MGE possessing different resistance determinants and code for adaptive traits such as virulence or metal resistance genes among host bacteria strains [15,16].
The globally disseminated O25: H4-ST131 E. coli clone producing CTX-M-15 is by conjugative IncF plasmids that are frequently recovered from hospital and community settings [17]. In Tanzania, the prevalence of bacteria producing ESBL ranges between 25 and 50 percent [18], with blaCTX-M-15 as the predominant allele in both community [19] and hospital settings [20]. The gene is also observed among companion and domestic animals and the environment combined with quinolone and aminoglycoside resistance genes [21,22]. Therefore, as demonstrated by its discovery in a novel Enterobacter spp. [23] and location in multiple plasmid types such as IncF, IncY and IncHI1, there is a possibility of an extensive variation in the epidemiology of blaCTX-M-15 carrying plasmids in Tanzania.
The presence of blaCTX-M-15 gene in multiple E.coli clones of human, animal and the environment of Tanzania [24], and limited information on the persistence of the gene’s alleles in any compartments can lead to the acquisition, transmission and evolution of new resistant strains even among non-conjugative bacteria. Since plasmids facilitate the spread of AMR genes in different compartments efforts to understand their spread and establishment in these settings is unquestionable. This study has improved our understanding of the importance of IncF plasmids in disseminating multidrug-resistant determinants among human, animal and environmental settings. It has further highlighted the importance of collaborative One Health based efforts that focus on animal and human health as critical when addressing the global threat.
2. Results
2.1. Isolates Characteristics
Escherichia coli was the only species isolated in both human and animals. The environment included isolates from soil and fresh water fish and comprised of E. coli, K. pneumoniae, C. braakii and E. cloacae species (Table 1).
Table 1.
Bacteria species distributed among donor isolates of human, animal and environment.
| Sample Origin | Sample Type | Frequency n% |
Species | Species n (%) | Total n (%) |
|---|---|---|---|---|---|
| Human | Human | 22 (43.14) | E. coli | 22 (43.1) | 22 (43.14) |
| Goat | 1 (1.96) | E. coli | 1 (1.96) | ||
| Animal | Pig | 3 (5.88) | E. coli | 3 (5.88) | |
| Dog | 6 (11.76) | E. coli | 6 (11.76) | 12 (23.52) | |
| Chicken | 2 (3.92) | E. coli | 2 (3.92) | ||
| Environment | Soil | 6 (11.76) | E. coli | 6 (11.76) | |
| E. coli | 2 (3.92) | ||||
| Fish | 11 (21.57) | K. pneumoniae | 3 (5.88) | 17 (33.32) | |
| C. braakii | 2 (3.92) | ||||
| E. cloacae | 4 (7.84) | ||||
| Total (n) | 51 (100) | 51 (100) |
2.2. Conjugation Efficiency of blaCTX-M-15 Gene among Isolates of Human, Animals and the Environment
Among 51 blaCTX-M-15 positive donor isolates, 45 (88.2%) transferred plasmids by conjugation with a transfer rate (transconjugants per donor cells) ranging from 4.8 × 10−1 to 1.5 × 10−9 as observed from a human and environment isolate, respectively (Table 2).
Table 2.
Conjugation efficiency of human, animal and environment donor isolates.
| Sample ID | Source | Species | Conjugation Frequency |
|---|---|---|---|
| CN1 | Fish | E. cloacae | 8.2 × 10−5 |
| CN2 | Fish | E. cloacae | 2.3 × 10−4 |
| CN3 | Fish | E. cloacae | 5.2 × 10−5 |
| CN4 | Fish | E. cloacae | NIL |
| CN5 | Fish | C. braakii | 7.5 × 10−6 |
| CN6 | Fish | E. coli | 7.6 × 10−3 |
| CN7 | Fish | E. coli | NIL |
| CN8 | Fish | K. pneumoniae | 2.0 × 10−5 |
| CN9 | Fish | K. pneumoniae | 4.2 × 10−4 |
| CN10 | Fish | K. pneumoniae | 3.3 × 10−5 |
| CN11 | Fish | C. braakii | 9.4 × 10−4 |
| CN12 | Pig | E. coli | 4.7 × 10−5 |
| CN13 | Pig | E. coli | 2.6 × 10−6 |
| CN14 | Pig | E. coli | 9.8 × 10−5 |
| CN15 | Local chicken | E. coli | 4.7 × 10−5 |
| CN16 | Local chicken | E. coli | 8.4 × 10−7 |
| CN17 | Goat | E. coli | 4.1 × 10−6 |
| CN18 | Dog | E. coli | 2.1 × 10−5 |
| CN19 | Dog | E. coli | 1.2 × 10−7 |
| CN20 | Dog | E. coli | 5.0 × 10−5 |
| CN21 | Dog | E. coli | 1.1 × 10−6 |
| CN22 | Dog | E. coli | 6.0 × 10−4 |
| CN23 | Dog | E. coli | 9.6 × 10−6 |
| CN24 | Environment | E. coli | 1.5 × 10−9 |
| CN25 | Environment | E. coli | 2.6 × 10−7 |
| CN26 | Environment | E. coli | 3.5 × 10−6 |
| CN27 | Environment | E. coli | 2.9 × 10−7 |
| CN28 | Environment | E. coli | 6.1 × 10−6 |
| CN29 | Environment | E. coli | 7.2 × 10−3 |
| CN30 | Human | E. coli | 1.0 × 10−3 |
| CN31 | Human | E. coli | 4.7 × 10−4 |
| CN32 | Human | E. coli | 2.1 × 10−4 |
| CN33 | Human | E. coli | 4.0 × 10−5 |
| CN34 | Human | E. coli | 5.4 × 10−5 |
| CN35 | Human | E. coli | 4.8 × 10−1 |
| CN36 | Human | E. coli | 1.7 × 10−4 |
| CN37 | Human | E. coli | 3.5 × 10−7 |
| CN38 | Human | E. coli | 8.1 × 10−5 |
| CN39 | Human | E. coli | 1.2 × 10−5 |
| CN40 | Human | E. coli | 2.7 × 10−5 |
| CN41 | Human | E. coli | 2.4 × 10−7 |
| CN42 | Human | E. coli | NIL |
| CN43 | Human | E. coli | 5.5 × 10−6 |
| CN44 | Human | E. coli | 4.4 × 10−6 |
| CN45 | Human | E. coli | 2.9 × 10−6 |
| CN46 | Human | E. coli | NIL |
| CN47 | Human | E. coli | 2.1 × 10−5 |
| CN48 | Human | E. coli | 1.2 × 10−4 |
| CN49 | Human | E. coli | 1.1 × 10−7 |
| CN50 | Human | E. coli | NIL |
| CN51 | Human | E. coli | NIL |
NIL: no conjugation.
2.3. Transferrable Resistance of Non-Beta-Lactam Phenotype among Isolates of Human, Animal and the Environment
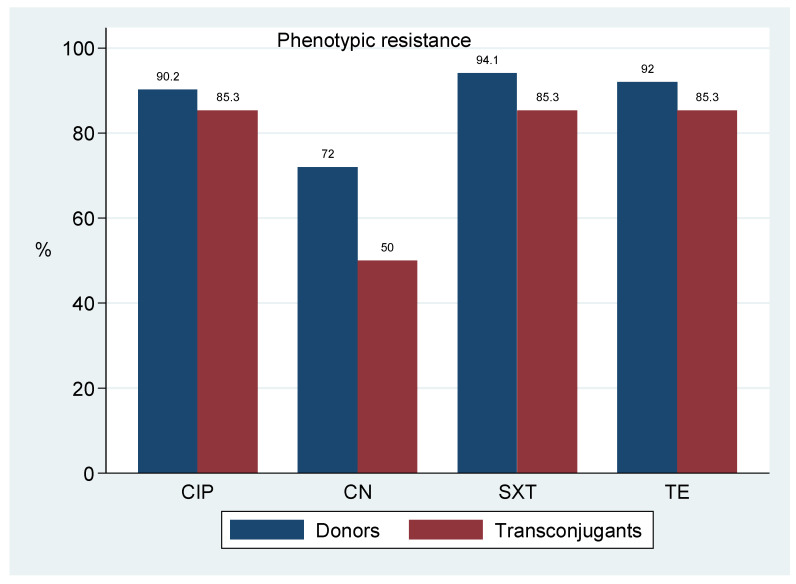
Table 3 and Figure 1 presents a summary of non-beta lactam resistant phenotypes transferred along the blaCTX-M-15 gene. A total of 45 plasmids successful transferred the gene to transconjugants. Non-beta-lactam resistance phenotypes were observed in 34/45(75.6%) transconjugants. Donor resistance to ciprofloxacin (CIP), tetracycline (TE) and trimethoprim-sulphamethoxazole (SXT) was observed in 46/51 (90.2%), 47/51 (92.2%) and 48/51 (94.1%) of events, respectively, and was co-transferred in 29/34 (85.3%) of such events. Gentamicin was the least transferred with a frequency of 17/34 (50.0%).
Table 3.
Antibiotic resistance phenotypes of donors and transconjugants of human animals and the environment.
| Sample No. | Source | Species | Donor’s Non-B-lactam Resistance Phenotype |
|---|---|---|---|
| CN1 | Fish | E. cloacae | SXT *, CIP *,CN *,TE * |
| CN2 | Fish | E. cloacae | CIP, SXT, CN, TE |
| CN3 | Fish | E. cloacae | CIP *, SXT *, TE *, CN* |
| CN4 | Fish | E. cloacae | CIP, CN, TE, SXT |
| CN5 | Fish | C. braakii | CIP *, SXT *, CN *, TE * |
| CN6 | Fish | E. coli | CIP, SXT, CN, TE |
| CN7 | Fish | E. coli | CIP, TE |
| CN8 | Fish | K. pneumoniae | CIP *, SXT *, CN *, TE * |
| CN9 | Fish | K. pneumoniae | CIP *, SXT *, CN *, TE * |
| CN10 | Fish | K. pneumoniae | CIP, SXT, CN, TE |
| CN11 | Fish | C. braakii | CIP, SXT, CN, TE * |
| CN12 | Pig | E. coli | CIP *, SXT *, TE * |
| CN13 | Pig | E. coli | TE, CIP, CN |
| CN14 | Pig | E. coli | CIP *, SXT *, TE *, CN * |
| CN15 | Local chicken | E. coli | CIP, SXT, CN, TE |
| CN16 | Local chicken | E. coli | CIP, SXT, CN, TE |
| CN17 | Goat | E. coli | SXT, TE *, CN, CIP * |
| CN18 | Dog | E. coli | SXT |
| CN19 | Dog | E. coli | SXT *, CIP *, TE, CN |
| CN20 | Dog | E. coli | CIP *, SXT *, TE * |
| CN21 | Dog | E. coli | CIP *, SXT *, TE *, CN * |
| CN22 | Dog | E. coli | CIP *, CN *, TE *, SXT * |
| CN23 | Dog | E. coli | SXT, TE, CN, CIP |
| CN24 | Environment | E. coli | SXT *, CIP *, TE * |
| CN25 | Environment | E. coli | SXT, TE, CIP |
| CN26 | Environment | E. coli | CIP *, SXT *, TE* |
| CN27 | Environment | E. coli | CIP * |
| CN28 | Environment | E. coli | CIP *, SXT *, CN *, TE * |
| CN29 | Environment | E. coli | CN, CIP *, SXT *, TE * |
| CN30 | Human | E. coli | TE *, CIP *, CN, SXT * |
| CN31 | Human | E. coli | CIP *, SXT * |
| CN32 | Human | E. coli | SXT *, CIP * |
| CN33 | Human | E. coli | TE *, CN *, CIP *, SXT * |
| CN34 | Human | E. coli | SXT *, TE *, CN *, CIP |
| CN35 | Human | E. coli | CIP *, CN *, SXT *, TE * |
| CN36 | Human | E. coli | CIP *, CN *, SXT *, TE * |
| CN37 | Human | E. coli | CIP *, CN *, SXT *, TE * |
| CN38 | Human | E. coli | SXT *, TE *, CIP*, CN * |
| CN39 | Human | E. coli | SXT, TE, CIP *, CN * |
| CN40 | Human | E. coli | SXT *, TE * |
| CN41 | Human | E. coli | SXT, TE *, CIP *, CN |
| CN42 | Human | E. coli | SXT, CIP, CN, TE |
| CN43 | Human | E. coli | CN *, CIP *, SXT *, TE * |
| CN44 | Human | E. coli | SXT, TE, CIP |
| CN45 | Human | E. coli | SXT, TE, CIP, CN |
| CN46 | Human | E. coli | TE, SXT |
| CN47 | Human | E. coli | SXT *, TE *, CIP, CN |
| CN48 | Human | E. coli | SXT *, TE *, CIP, CN |
| CN49 | Human | E. coli | CIP *, CN *, SXT *, TE * |
| CN50 | Human | E. coli | SXT, TE |
| CN51 | Human | E. coli | CN, CIP, SXT, TE |
* Transferable resistance; SXT: Trimethoprim-sulphamethoxazole, CIP: Ciprofloxacin, TE: tetracycline, CN: Gentamicin.
Figure 1.
Transfer success of blaCTX-M-15 among E. coli isolates of human, animals and the environment.
2.4. Replicon Types of Plasmids Carrying blaCTX-M-15
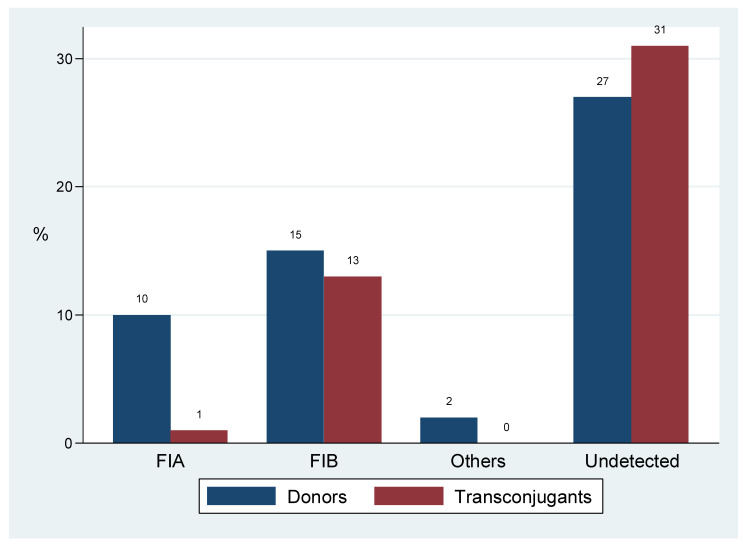
Common replicon types were FIA (n = 11) and FIB (n = 27) that occurred as single transferrable replicons in 14 events. Inc A/C and Y replicons were minor, and each was typed once. Of all the 14 typed transconjugant plasmids, 7 replicons were observed in both donors and transconjugants, while 20/27 donor replicons were not observed in respective transconjugants (Table 4), (Figure 2).
Table 4.
Replicon types of plasmids carrying blaCTX-M-15 among donors and transconjugants.
| Sample Source | Conjugation Efficiency | Conjugation Range | Donor’s Plasmid Replicon | Transconjugant Replicon Type |
|---|---|---|---|---|
| Human | 1.2 × 10−4 | FIB | FIA | |
| Human | 8.1 × 10−5 | FIA, FIB | FIB | |
| Dog | 5.0 × 10−5 | FIB | FIB | |
| Human | 5.4 × 10−5 | 10−6–10−3 | FIB | FIB |
| Human | 2.1 × 10−4 | FIB | FIB | |
| Environment | 7.2 × 10−3 | FIB | FIB | |
| Dog | 1.1 × 10−6 | FIB | FIB | |
| Human | 1.7 × 10−4 | FIB | FIB | |
| Dog | 9.6 × 10−6 | no rep | FIB | |
| Dog | 2.1 × 10−5 | no rep | FIB | |
| Human | 1.2 × 10−5 | 10−7–10−4 | no rep | FIB |
| Human | 4.7 × 10−4 | no rep | FIB | |
| Environment | 2.9 × 10−7 | no rep | FIB | |
| Fish | 2.3 × 10−4 | no rep | FIB | |
| Fish | NIL | FIA, Y | NA | |
| Human | NIL | no rep | NA | |
| Human | NIL | 0 | no rep | NA |
| Human | NIL | no rep | NA | |
| Human | NIL | no rep | NA | |
| Fish | NIL | no rep | NA | |
| Fish | 4.2 × 10−4 | A/C, FIA | no rep | |
| Pig | 2.6 × 10−6 | FIA | no rep | |
| Human | 5.5 × 10−6 | FIA | no rep | |
| Dog | 6.0 × 10−4 | FIA | no rep | |
| Pig | 9.8 × 10−5 | FIA | no rep | |
| Human | 2.9 × 10−6 | FIA | no rep | |
| Human | 4.0 × 10−5 | FIA | no rep | |
| Human | 4.8 × 10−1 | 10−9–10−1 | FIA | no rep |
| Dog | 1.2 × 10−7 | FIB | no rep | |
| Human | 3.5 × 10−7 | FIB | no rep | |
| Environment | 1.5 × 10−9 | FIB | no rep | |
| Environment | 2.6 × 10−7 | FIB | no rep | |
| Human | 4.4 × 10−6 | FIB | no rep | |
| Environment | 3.5 × 10−6 | FIB | no rep | |
| Human | 2.1 × 10−5 | FIB | no rep | |
| Fish | 7.5 × 10−6 | no rep | no rep | |
| Fish | 9.4 × 10−4 | no rep | no rep | |
| Human | 2.7 × 10−5 | no rep | no rep | |
| Local chicken | 4.7 × 10−5 | no rep | no rep | |
| Pig | 4.7 × 10−5 | no rep | no rep | |
| Human | 2.4 × 10−7 | no rep | no rep | |
| Fish | 3.3 × 10−5 | no rep | no rep | |
| Fish | 2.0 × 10−5 | 10−7–10−3 | no rep | no rep |
| Fish | 7.6 × 10−3 | no rep | no rep | |
| Human | 1.1 × 10−7 | no rep | no rep | |
| Fish | 5.2 × 10−5 | no rep | no rep | |
| Goat | 4.1 × 10−6 | no rep | no rep | |
| Environment | 6.1 × 10−6 | no rep | no rep | |
| Local chicken | 8.4 × 10−7 | no rep | no rep | |
| Human | 1.0 × 10−3 | no rep | no rep | |
| Fish | 8.2 × 10−5 | no rep | no rep |
NIL: no conjugation, NA: no transconjugants, no rep: no typable replicon.
Figure 2.
Replicon types of plasmids carrying blaCTX-M-15 among donors and transconjugants.
2.5. Transfer Success of blaCTX-M-15 among Escherichia coli Isolates
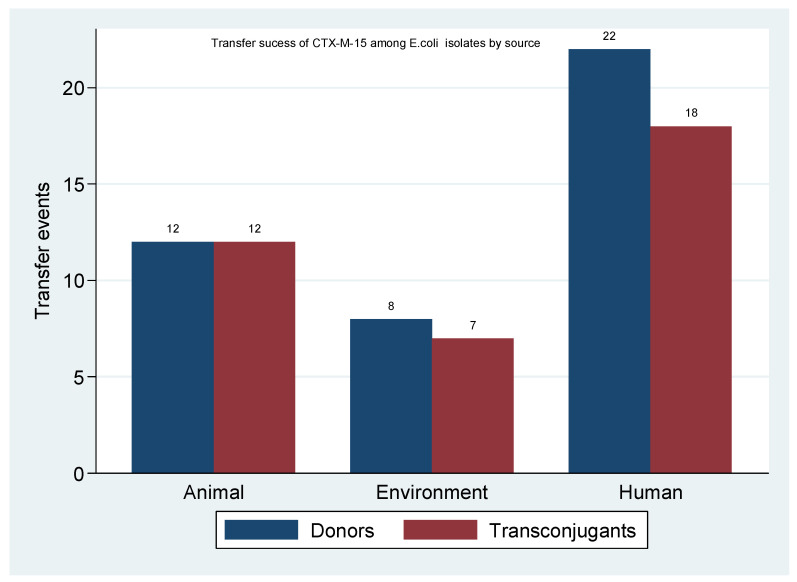
Table 5 shows the percentage transfer of blaCTX-M-15 among E. coli donor isolates. Out of 42 E. coli donors, 37 (88.1%) successfully transferred the gene, accounting for an 82.1% of all transconjugants. All E. coli originating from animals transferred the gene successfully.
Table 5.
Transfer success of blaCTX-M-15 among E. coli isolates of human, animals and the environment.
| Source |
E. coli Donors n |
E. coli Transconjugants n (%) |
|---|---|---|
| Human | 22 | 18 (81.8) |
| Animal | 12 | 12 (100.0) |
| Environment | 8 | 7 (87.5) |
| Total | 42 | 37(88.1) |
3. Discussion
In this study, we aimed to determine conjugation frequencies and type plasmids carrying the blaCTX-M-15 gene from human, animal and environment ESBL producing isolates. The study is epidemiologically important in understanding the pattern and possibly predict the flow of AMR from different sources. As presented in (Table 1), Escherichia coli was the dominant bacteria species from all sourced samples. The successful colonisation of E. coli in human and animal gastrointestinal tract (GIT) have been reported previously [25], the GIT can serve as exchange hotspots and reservoirs of antimicrobial resistance genes. Likewise, Escherichia coli and Klebsiella pneumoniae are frequently isolated in infections associated with CTX-M-15 in hospitals [26] and the community, including households [27], aquatic environment [28] and the soil [29].
The transfer efficiency of blaCTX-M-15 among isolates in this study was higher (88%) [Table 2], than that reported by Zurfluh and colleagues [30], where a 38.3% efficiency was observed, however, despite the varying frequencies of transfer, the reported efficiency is slightly lower than that previously reported (100%) for randomly selected hospital originating isolates [20]. It is reported that high conjugation rates above thresholds compensate fitness costs and establish a long-term persistence of plasmids in multiple hosts [13,31,32] through maintaining successive generations of bacteria with adaptive traits. Therefore, the high transfer efficiency is a fitness advantage that improves the persistence and dissemination potential of blaCTX-M-15 ESBL in human, animal and the environment interface. Moreover, transfer failure for some isolates‒ CN4, CN7, CN42, CN46, CN50 and CN51, could possibly be due to the gene’s integration in the chromosome [33] or transposition events that prevent plasmid mobility.
As summarised in Table 3 and Figure 1, transferable multidrug resistance phenotypes were also observed. The conjugative spread of blaCTX-M-15 gene by IncF plasmids along with tetracycline, aminoglycoside and quinolones have been reported [34]. These plasmids harbor several combinations of resistance determinants and transfer them to human, animals and environment isolates through the ecological interaction of bacteria in these settings. Moreover, the genetic environment of blaCTX-M-15 is dominated by multiple antibiotic resistance genes such as aac (6′)-lb-cr, tet (A, B), qnrS, qnr and sul genes [35,36,37], whose phenotypic expression denotes the existing selection pressure for these antibiotics. Such selection can increase their transfer rate and possibly account for the high co-transfer of non-beta lactam antibiotics observed in this study.
We also observed single replicon IncF plasmids as common vectors of blaCTXM-15, (Table 4), (Figure 2). Replicon typing of plasmids carrying antimicrobial resistance genes is important for detecting, tracing or monitoring the spread of antimicrobial resistance. These observations are in line with findings in the same setting [20] and elsewhere [30,38] where multireplicon FIA and FIB plasmids were reported to carry multiple resistance genes including blaCTX-M-15. As in previous studies, IncY plasmids and Inc A/C carrying blaCTX-M-15 in association with quinolone and aminoglycoside genes were also reported [22], [39,40]. Together these findings support the diverse nature of plasmids adapted to spread and maintain blaCTXM-15 gene.
The absence of donor replicons in respective transconjugants might have resulted from conjugation failure, multi-replicons (undetected by the method used) that destabilise and prevent the transfer of some resident replicons [41], and prior plasmid dependent mutations (which do not occur in transconjugants) that may have altered backbone genes of donor plasmids and obscure the detection of existing replicons [32,39,41]. In addition, and as a shortcoming, the PBRT technique used in detecting plasmid replicons can give false-negative results when replicon sequences go undetected by the primer sets used, target replicon sequences undergo mutation through transpositional alterations and the unknown existence of new replicons in such plasmid [42].
Lastly, we observed that all animal originating E. coli isolates transferred blaCTX-M-15 to respective transconjugants (Table 5), (Figure 3). These findings were also supported by a recent review [34], that human and animal originating E.coli are adapted to disseminate ESBL genes by IncF plasmids. The colonisation and infection of animals by E. coli maximises microbial interactions between non-pathogenic and pathogenic commensal E.coli in either companion or food-producing animals and facilitate the exchange of materials between them through conjugation. In addition, the increasing use of antibiotics in animals could select and transfer resistant pathogenic bacteria from animals to human and the environment with huge cost implications. Since AMR is a public health threat, the highest transfer rate observed in animal originating E. coli calls for integrated efforts to address AMR with experts from veterinary, human and ecological fields. It further implies that animals may serve as dual targets for studies focusing on the horizontal transfer and evolution of antimicrobial resistance.
Figure 3.
Transfer success of blaCTX-M-15 among E. coli isolates of human, animals and the environment.
4. Materials and Methods
4.1. Study Isolates
All isolates used as donors in this study were obtained from the Catholic University of Health and Allied Sciences (CUHAS) in Mwanza Tanzania. A total of 51 blaCTX-M-15 positive isolates were purposively selected and activated overnight in Luria Bertani (LB) broth at 37 °C ready for use in conjugation and PBRT techniques.
Among the 51 isolates, twenty-two blaCTX-M-15 positive isolates were obtained from a study that reported the magnitude of fecal carriage and diversity of ESBL genotypes among human residing in rural communities of Mwanza Tanzania [19], 12 other blaCTX-M-15 positive isolates were from a study that reported the fecal carriage of ESBL among companion and domestic farm animals that included pigs, chicken, dogs and goats [21]. The remaining 17 environmental isolates were obtained from a study that investigated the presence of blaCTX-M-15 from muddy soils and gut contents of freshwater fish from Lake Victoria in Mwanza Tanzania [22].
4.2. Antibiotic Susceptibility Testing
Susceptibility testing of all donor isolates and the resulting transconjugants was performed by the disk diffusion method on Mueller Hinton agar as recommended by the Clinical and Laboratory Standard Institute [43]. Antibiotics tested were tetracycline (30 µg), gentamicin (30 µg), ciprofloxacin (5 µg) and trimethoprim/sulphamethoxazole (1.25/23.75 μg) (Hi-media, India).
4.3. Conjugation Experiment
A total of 51 known blaCTX-M-15 positive isolates and Escherichia coli J53 ((F−, met, pro, Azr)‒a mutant strain of E. coli [44] obtained from the Institute of Medical Microbiology, Giessen, Germany, were used as donors and recipient strain, respectively. As previously described [20], we performed conjugation experiments with some modifications. Shortly, the recipient strain was prepared by streaking Escherichia coli J53 in Luria Bertani (LB) plates supplemented with 100 µg/mL NaN3 (LB++), while donor strains were selected in LB plates supplemented with 2 µg/mL cefotaxime only (LB+). From these, fresh overnight donor and recipient strains were prepared by picking single colonies emulsified in 10 mL LB broth and incubated overnight at 37 °C in a 150 rpm shaking incubator. After exactly 12 h, equal volumes (500 µL) of donor and recipient strains were immediately mixed in 1.5 mL eppendorf tubes previously labeled transconjugant (Tc) while 1000 µL of donor strain were added in fresh tubes of similar volume‒to be separately selected on LB+ and LB++ plates as respective controls. All tubes were incubated at 37 °C for 15 min, vortexed briefly, centrifuged at 12,000 g for 2 min and the pellet re-suspended in fresh 1000 µL LB broth. Finally, 0.1 mL of 10−1 to 10−4 transconjugant cultures were double selected on LB plates supplemented with 100 µg/mL NaN3 and 2 µg/mL cefotaxime. Conjugation efficiency was reported as transconjugants per donor cells, with the denominator obtained from an initial volume of 100 µL.
4.4. Genomic Extraction of Donor and Transconjugants DNA
Donor and transconjugant genomic DNA was extracted using a previously described chelex protocol with slight modifications [45]. First, 5 µL of proteinase K (10 mg/mL) were added into tubes containing 100 µL fresh LB emulsified colonies. In the same tubes, 300 µL of chelex buffer (Qiagen GmbH, Hilden, Germany) was added consecutively. The mixture was incubated for 3 hr at 55 °C before adding 85 µL of 5 M NaCl and vortexed for 15 s to precipitate proteins. The supernatant was centrifuged at 13,000× g for 10 min followed by the addition of 300 µL of 100% cold ethanol and a 5 min centrifugation at 13,000× g that precipitated and pelleted the DNA. Lastly, the pellet was rinsed by pouring off the remaining fluid, adding 500 µL of 70% ethanol, centrifuging at 13,000× g for 5 min and leaving the pellet to air dry at 55 °C for 10 min. The DNA was then re-suspended in 50 µL nuclease-free water. Nanodrop (Thermo Scientific, Wilmington, DE) was used to check the quantity of the DNA, while the quality was confirmed by electrophoresis in 1.5% (w/v) agarose gel using TAE buffer. The obtained DNA samples were used in typing plasmid replicons or stored at −20 °C.
4.5. PCR Based Replicon Typing
Targeted genes were amplified by a simplified version of the previously described PBRT technique [42]. Shortly, the eight Polymerase Chain Reaction (PCR) panels illustrated by Caratolli and colleagues [46], were reduced to three [42], (Table 6). PCR was performed using a readily reconstituted master mix according to manufacturer’s instructions (New England BioLabs, Inc. Beverly, MA) under the following conditions; 5 min at 94 °C; 30 cycles of 30 s at 94 °C, 30 s at 60 °C and 90 s at 72 °C; then a final extension of 5 min at 72 °C. Amplicons were visualised on 1.5% tris-acetate EDTA agarose gels alongside a 100 bp DNA ladder (New England BioLabs, Inc. Beverly, MA). The sample was considered positive for replicon gene (s) if an amplicon of the expected band size was observed.
Table 6.
Primers used in PCR based replicon typing of donor and transconjugant plasmids.
| Primer Panel/Target | Direction | Primer Sequence | Annealing Temp (°C) | Amplicon Size (bp) |
|---|---|---|---|---|
| Panel 1 | ||||
| B/O | F | 5′-gcggtccggaaagccagaaaac-3′ | 60 | 159 |
| R | 5′-tctgcgttccgccaagttcga-3′ | |||
| FIC | F | 5′-gtgaactggcagatgaggaagg-3′ | 60 | 262 |
| R | 5′-ttctcctcgtcgccaaactagat-3′ | |||
| A/C | F | 5′-gagaaccaaagacaaagacctgga3′ | 60 | 465 |
| R | 5′-acgacaaacctgaattgcctcctt-3′ | |||
| P | F | 5′ctatggccctgcaaacgcgccagaaa3′ | 60 | 534 |
| R | 5′-tcacgcgccagggcgcagcc-3′ | |||
| T | F | 5′-ttggcctgtttgtgcctaaaccat-3′ | 60 | 750 |
| R | 5′-cgttgattacacttagctttggac-3′ | |||
| Panel 2 | ||||
| K/B | F | 5′-gcggtccggaaagccagaaaac-3′ | 60 | 160 |
| R | 5′-tctttcacgagcccgccaaa-3 | |||
| W | F | 5′-cctaagaacaacaaagcccccg-3′ | 60 | 242 |
| R | 5′-ggtgcgcggcatagaaccgt-3′ | |||
| FIIS | F | 5′-ctgtcgtaagctgatggc-3′ | 60 | 270 |
| R | 5′-ctctgccacaaacttcagc-3′ | |||
| FIA | F | 5′-ccatgctggttctagagaaggtg-3′ | 60 | 462 |
| R | 5′-gtatatccttactggcttccgcag-3′ | |||
| FIB | F | 5′-ggagttctgacacacgattttctg-3′ | 60 | 702 |
| 5′-ctcccgtcgcttcagggcatt-3′ | ||||
| Y | F | 5′-aattcaaacaacactgtgcagcctg-3′ | 60 | 765 |
| R | 5′-gcgagaatggacgattacaaaacttt-3′ | |||
| Panel 3 | ||||
| I1 | F | 5′-cgaaagccggacggcagaa-3′ | 60 | 139 |
| R | 5′-tcgtcgttccgccaagttcgt-3′ | |||
| FrepB | F | 5′-tgatcgtttaaggaattttg-3′ | 60 | 270 |
| R | 5′-gaagatcagtcacaccatcc-3′ | |||
| X | F | 5′-aaccttagaggctatttaagttgctgat-3′ | 60 | 376 |
| R | 5′-tgagagtcaatttttatctcatgttttagc3′ | |||
| HI1 | F | 5′-ggagcgatggattacttcagtac-3′ | 60 | 471 |
| R | 5′-tgccgtttcacctcgtgagta-3′ | |||
| N | F | 5′-gtctaacgagcttaccgaag-3′ | 60 | 559 |
| R | 5′-gtttcaactctgccaagttc-3′ | |||
| HI2 | F | 5′-tttctcctgagtcacctgttaacac-3′ | 60 | 644 |
| R | 5′-ggctcactaccgttgtcatcct-3′ | |||
| L/M | F | 5′-ggatgaaaactatcagcatctgaag-3′ | 60 | 785 |
| R | 5′-ctgcaggggcgattctttagg-3′ |
5. Conclusions
Majority of plasmids carrying blaCTX-M-15 were conjugatively transferred by IncF plasmids along with non-beta lactam resistance. The heterogeneous nature of these plasmids continuously maintains and reserves the blaCTX-M-15 gene in these settings. The 100% transfer efficiency among E. coli of animal origin is of concern since the networked interaction of animals with human and their environment continuously exchange and reserve resistance determinants in this interface. Therefore, there is a need for more research to understand the interaction and spread of mobile elements circulating in animals, One Health approach is to be intensified to further address AMR as a public health threat.
Acknowledgments
The authors acknowledge the technical support provided by the department of Molecular biology at Nelson Mandela African Institution of Science and Technology. Specifically, we thank Emmanuel and Beatus Lyimo for providing necessary technical assistance.
Abbreviations
| AMR | Antimicrobial resistance |
| CREATES | Centre for Research, Agricultural advancement, Teaching Excellence and Sustainability in Food and Nutritional Security |
| CUHAS | Catholic University of Health and Allied Sciences |
| DNA | Deoxyribonucleic acid |
| EDTA | Ethylene Diaminetetracetic acid |
| ESBL | Extended spectrum beta-lactamase |
| GIT | Gastrointestinal tract |
| LB | Luria Bertani |
| MGE | Mobile genetic element |
| NaCl | Sodium Chloride |
| NaN3 | Sodium azide |
| PCR | Polymerase chain reaction |
| TAE | Tris acetate EDTA |
Author Contributions
Conceptualisation, C.A.M. and S.E.M.; Formal analysis, S.E.M.; Funding acquisition, C.A.M.; Investigation, C.A.M. and G.S.; Supervision, G.S. and S.E.M.; Writing–original draft, C.A.M.; Writing–review and editing, G.S. and S.E.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Centre for Research, Agricultural advancement, Teaching Excellence and Sustainability in Food and Nutritional Security (CREATES) of the Nelson Mandela African Institution of Science and Technology (NM-AIST). The APC was funded by Catholic University of Health and Allied Sciences (CUHAS).
Data Availability Statement
All data supporting presented results is available in this article.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barlow M. What antimicrobial resistance has taught us about horizontal gene transfer. Horiz. Gene Transf. 2009:397–411. doi: 10.1007/978-1-60327-853-9_23. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet R. Growing group of extended-spectrum β-lactamases: The CTX-M enzymes. Antimicrob. Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lahlaoui H., Khalifa A.B.H., Moussa M.B. Epidemiology of Enterobacteriaceae producing CTX-M type extended spectrum β-lactamase (ESBL) Med. Et Mal. Infect. 2014;44:400–404. doi: 10.1016/j.medmal.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Hirai I., Fukui N., Taguchi M., Yamauchi K., Nakamura T., Okano S., Yamamoto Y. Detection of chromosomal blaCTX-M-15 in Escherichia coli O25b-B2-ST131 isolates from the Kinki region of Japan. Int. J. Antimicrob. Agents. 2013;42:500–506. doi: 10.1016/j.ijantimicag.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Coque T.M., Novais Â., Carattoli A., Poirel L., Peixe L., Baquero F., Cantón R., Nordmann P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg. Infect. Dis. 2008;14:195. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin J.F., Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- 7.Allen H.K., Moe L.A., Rodbumrer J., Gaarder A., Handelsman J. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. ISME J. 2009;3:243–251. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 8.Paterson D.L., Bonomo R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantón R., González-Alba J.M., Galán J.C. CTX-M enzymes: Origin and diffusion. Front. Microbiol. 2012;3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naseer U., Sundsfjord A. The CTX-M conundrum: Dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 2011;17:83–97. doi: 10.1089/mdr.2010.0132. [DOI] [PubMed] [Google Scholar]
- 11.Vrancianu C.O., Popa L.I., Bleotu C., Chifiriuc M.C. Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front. Microbiol. 2020;11:761. doi: 10.3389/fmicb.2020.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopatkin A.J., Meredith H.R., Srimani J.K., Pfeiffer C., Durrett R., You L. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat. Commun. 2017;8:1689. doi: 10.1038/s41467-017-01532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin B.R., Rozen D.E. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 2006;4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 14.Benz F., Huisman J.S., Bakkeren E., Herter J.A., Stadler T., Ackermann M., Diard M., Egli A., Hall A.R., Hardt W.-D. Plasmid-and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo. ISME J. 2021;15:862–878. doi: 10.1038/s41396-020-00819-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Q.E., Agouri S.R., Tyrrell J.M., Walsh T.R. Heavy metal resistance genes are associated with blaNDM-1-and blaCTX-M-15-carrying Enterobacteriaceae. Antimicrob. Agents Chemother. 2018;62: e02642-17. doi: 10.1128/AAC.02642-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernández-Ramírez K.C., Reyes-Gallegos R.I., Chávez-Jacobo V.M., Díaz-Magaña A., Meza-Carmen V., Ramírez-Díaz M. A plasmid-encoded mobile genetic element from Pseudomonas aeruginosa that confers heavy metal resistance and virulence. Plasmid. 2018;98:15–21. doi: 10.1016/j.plasmid.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Nicolas-Chanoine M.-H., Blanco J., Leflon-Guibout V., Demarty R., Alonso M.P., Caniça M.M., Park Y.-J., Lavigne J.-P., Pitout J., Johnson J.R. Intercontinental emergence of Escherichia coli clone O25: H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008;61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 18.Mshana S.E., Hain T., Domann E., Lyamuya E.F., Chakraborty T., Imirzalioglu C. Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect. Dis. 2013;13:466. doi: 10.1186/1471-2334-13-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mshana S.E., Falgenhauer L., Mirambo M.M., Mushi M.F., Moremi N., Julius R., Seni J., Imirzalioglu C., Matee M., Chakraborty T. Predictors of bl a CTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infect. Dis. 2016;16:187. doi: 10.1186/s12879-016-1527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mshana S., Imirzalioglu C., Hain T., Domann E., Lyamuya E.F., Chakraborty T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin. Microbiol. Infect. 2011;17:1279–1282. doi: 10.1111/j.1469-0691.2011.03518.x. [DOI] [PubMed] [Google Scholar]
- 21.Seni J., Falgenhauer L., Simeo N., Mirambo M.M., Imirzalioglu C., Matee M., Rweyemamu M., Chakraborty T., Mshana S.E. Multiple ESBL-producing Escherichia coli sequence types carrying quinolone and aminoglycoside resistance genes circulating in companion and domestic farm animals in Mwanza, Tanzania, harbor commonly occurring plasmids. Front. Microbiol. 2016;7:142. doi: 10.3389/fmicb.2016.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moremi N., Manda E.V., Falgenhauer L., Ghosh H., Imirzalioglu C., Matee M., Chakraborty T., Mshana S.E. Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza, Tanzania. Front. Microbiol. 2016;7:1862. doi: 10.3389/fmicb.2016.01862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mshana S.E., Gerwing L., Minde M., Hain T., Domann E., Lyamuya E., Chakraborty T., Imirzalioglu C. Outbreak of a novel Enterobacter sp. carrying blaCTX-M-15 in a neonatal unit of a tertiary care hospital in Tanzania. Int. J. Antimicrob. Agents. 2011;38:265–269. doi: 10.1016/j.ijantimicag.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 24.Seni J., Falgenhauer L., Simeo N., Mirambo M., Imirzalioglu C., Matee M., Rweyemamu M., Chakraborty T., Mshana S.E. Preliminary insights into the occurrence of similar clones of extended-spectrum beta-lactamase-producing bacteria in humans, animals and the environment in Tanzania: A systematic review and meta-analysis between 2005 and 2016. Zoonoses Public Health. 2018;65:1–10. doi: 10.1111/zph.12387. [DOI] [PubMed] [Google Scholar]
- 25.Hosuru Subramanya S., Bairy I., Nayak N., Amberpet R., Padukone S., Metok Y., Bhatta D.R., Sathian B. Detection and characterization of ESBL-producing Enterobacteriaceae from the gut of healthy chickens, Gallus gallus domesticus in rural Nepal: Dominance of CTX-M-15-non-ST131 Escherichia coli clones. PLoS ONE. 2020;15:e0227725. doi: 10.1371/journal.pone.0227725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mshana S.E., Kamugisha E., Mirambo M., Chakraborty T., Lyamuya E. Prevalence of multiresistant gram-negative organisms in a tertiary hospital in Mwanza, Tanzania. BMC Res. Notes. 2009;2:49. doi: 10.1186/1756-0500-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obeng-Nkrumah N., Labi A.-K., Blankson H., Awuah-Mensah G., Oduro-Mensah D., Anum J., Teye J., Kwashie S.D., Bako E., Ayeh-Kumi P.F. Household cockroaches carry CTX-M-15-, OXA-48-and NDM-1-producing enterobacteria, and share beta-lactam resistance determinants with humans. BMC Microbiol. 2019;19:272. doi: 10.1186/s12866-019-1629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyimo B., Buza J., Subbiah M., Temba S., Kipasika H., Smith W., Call D.R. IncF plasmids are commonly carried by antibiotic resistant Escherichia coli isolated from drinking water sources in northern Tanzania. Int. J. Microbiol. 2016;2016:3103672. doi: 10.1155/2016/3103672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gekenidis M.-T., Rigotti S., Hummerjohann J., Walsh F., Drissner D. Long-Term Persistence of blaCTX-M-15 in Soil and Lettuce after Introducing Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli via Manure or Water. Microorganisms. 2020;8:1646. doi: 10.3390/microorganisms8111646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zurfluh K., Glier M., Hächler H., Stephan R. Replicon typing of plasmids carrying blaCTX-M-15 among Enterobacteriaceae isolated at the environment, livestock and human interface. Sci. Total Environ. 2015;521:75–78. doi: 10.1016/j.scitotenv.2015.03.079. [DOI] [PubMed] [Google Scholar]
- 31.San Millan A., Peña-Miller R., Toll-Riera M., Halbert Z.V., McLean A.R., Cooper B.S., MacLean R.C. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 2014;5:5208. doi: 10.1038/ncomms6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dionisio F., Conceicao I.C., Marques A.C.R., Fernandes L., Gordo I. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 2005;1:250–252. doi: 10.1098/rsbl.2004.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragupathi N.K.D., Sethuvel D.P.M., Gajendran R., Anandan S., Walia K., Veeraraghavan B. Horizontal transfer of antimicrobial resistance determinants among enteric pathogens through bacterial conjugation. Curr. Microbiol. 2019;76:666–672. doi: 10.1007/s00284-019-01676-x. [DOI] [PubMed] [Google Scholar]
- 34.Rozwandowicz M., Brouwer M.S.M., Fischer J., Wagenaar J.A., Gonzalez-Zorn B., Guerra B., Mevius D.J., Hordijk J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 35.Yousfi M., Mairi A., Touati A., Hassissene L., Brasme L., Guillard T., De C.C. Extended spectrum β-lactamase and plasmid mediated quinolone resistance in Escherichia coli fecal isolates from healthy companion animals in Algeria. J. Infect. Chemother. 2016;22:431–435. doi: 10.1016/j.jiac.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Rafaï C., Frank T., Manirakiza A., Gaudeuille A., Mbecko J.-R., Nghario L., Serdouma E., Tekpa B., Garin B., Breurec S. Dissemination of IncF-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from surgical-site infections in Bangui, Central African Republic. BMC Microbiol. 2015;15:15. doi: 10.1186/s12866-015-0348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiiru J., Butaye P., Goddeeris B.M., Kariuki S. Analysis for prevalence and physical linkages amongst integrons, ISE cp 1, IS CR 1, Tn 21 and Tn 7 encountered in Escherichia coli strains from hospitalized and non-hospitalized patients in Kenya during a 19-year period (1992–2011) BMC Microbiol. 2013;13:109. doi: 10.1186/1471-2180-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mshana S.E., Imirzalioglu C., Hossain H., Hain T., Domann E. Chakraborty, T. Conjugative IncFI plasmids carrying CTX-M-15 among Escherichia coli ESBL producing isolates at a University hospital in Germany. BMC Infect. Dis. 2009;9:97. doi: 10.1186/1471-2334-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maherault A.-C., Kemble H., Magnan M., Gachet B., Roche D., Le Nagard H., Tenaillon O., Denamur E., Branger C., Landraud L. Advantage of the F2: A1: B-IncF pandemic plasmid over IncC plasmids in in vitro acquisition and evolution of blaCTX-M gene-bearing plasmids in Escherichia coli. Antimicrob. Agents Chemother. 2019;63:e01130-19. doi: 10.1128/AAC.01130-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee M.Y., Ko K.S., Kang C.–I., Chung D.R., Peck K.R., Song J.-H. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: Diverse clones and clonal dissemination. Int. J. Antimicrob. Agents. 2011;38:160–163. doi: 10.1016/j.ijantimicag.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 41.Dionisio F., Zilhão R., Gama J.A. Interactions between plasmids and other mobile genetic elements affect their transmission and persistence. Plasmid. 2019;102:29–36. doi: 10.1016/j.plasmid.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Johnson T.J., Wannemuehler Y.M., Johnson S.J., Logue C.M., White D.G., Doetkott C., Nolan L.K. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl. Environ. Microbiol. 2007;73:1976–1983. doi: 10.1128/AEM.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clinical Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI Standard M100; Wayne, PA, USA: 2018. [Google Scholar]
- 44.Jacoby G.A., Han P. Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J. Clin. Microbiol. 1996;34:908–911. doi: 10.1128/jcm.34.4.908-911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casquet J., Thebaud C., Gillespie R.G. Chelex without boiling, a rapid and easy technique to obtain stable amplifiable DNA from small amounts of ethanol-stored spiders. Mol. Ecol. Resour. 2012;12:136–141. doi: 10.1111/j.1755-0998.2011.03073.x. [DOI] [PubMed] [Google Scholar]
- 46.Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K., Threlfall E. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting presented results is available in this article.