Abstract
The emergence of multi-drug resistant (MDR) strains and even pan drug resistant (PDR) strains is alarming. In this study, we studied the resistance pattern of E. coli pathogens recovered from patients with different infections in different hospitals in Minia, Egypt and the co-existence of different resistance determinants. E. coli was the most prevalent among patients suffering from urinary tract infections (62%), while they were the least isolated from eye infections (10%). High prevalence of MDR isolates was found (73%) associated with high ESBLs and MBLs production (89.4% and 64.8%, respectively). blaTEM (80%) and blaNDM (43%) were the most frequent ESBL and MBL, respectively. None of the isolates harbored blaKPC and blaOXA-48 carbapenemase like genes. Also, the fluoroquinolone modifying enzyme gene aac-(6′)-Ib-cr was detected in 25.2% of the isolates. More than one gene was found in 81% of the isolates. Azithromycin was one of the most effective antibiotics against MDR E. coli pathogens. The high MAR index of the isolates and the high prevalence of resistance genes, indicates an important public health concern and high-risk communities where antibiotics are abused.
Keywords: MDR E. coli, ESBLs, MBLs, MAR index
1. Introduction
Escherichia coli, belongs to the family Enterobacteriaceae, is the most common human gastrointestinal commensal as well as important etiological agent of many hospital and community-acquired infections. Pathogenic strains are capable of causing a wide variety of diseases including diarrhea, dysentery, overwhelming sepsis, and the hemolytic-uremic syndrome and neonatal meningitis. E. coli can be sorted into intestinal or extraintestinal according to the site of infection [1].
Antibiotics have been the most successful form of chemotherapy developed in the 20th century, saving human lives every day [2]. The evolution of pathogens resistant to antibiotics limits their clinical use, making such infections difficult to control. The antimicrobial resistance (AMR) can be of chromosomal or mobile genetic elements origin [3]. The most common resistance mechanism is the production of the β-lactamase hydrolytic enzymes, which specifically have an inactivated β-lactam ring so that they cannot inhibit the bacterial transpeptidases [4].
β-lactamases are classified into four classes. Serine classes (A, C and D) have serine residue at the hydrolysis active sites. Metallo- β-lactamases (MBLs) (class B) in which the hydrolytic action is promoted by one or two zinc ions at the active site [5]. Class A enzymes include blaTEM which is the first identified plasmid-encoded β-lactamase; blaSHV which has similar activity to blaTEM; blaCTX-M (cefotaximase) and blaKPC which confers carbapenem resistance [6]. Class A Extended-spectrum β-lactamase (ESBL) producing strains (blaTEM, blaSHV and blaCTX-M types) are of the most clinically significant pathogens which can resist all β-lactam drugs including monobactams [5,6]. The most clinically significant class B enzymes are blaVIM, blaIMP and blaNDM. MBLs are a group of carbapenemases that resist most β-lactam drugs except the monobactams. Monobactams (e.g., aztreonam) are intrinsically stable to MBLs, but their susceptibility to other serine β-lactamases which are often co-expressed with the MBL limit their usage against MBL expressing strains [7]. Another group that able to hydrolyze carbapenems in addition to other β-lactams are class D β-lactamases (e.g., blaOXA-48 like enzymes) [8].
Another example for enzymatic inactivation of antibiotics is the enzymatic modification at different -OH or -NH2 groups of aminoglycosides. As a result of the induced steric and/or electrostatic interactions, the modified antibiotic is unable to bind to the target RNA. They can be nucleotidyltransferases (ANTs), phosphotransferases (APHs), or acetyltransferases (AACs) [4]. In addition, the enzyme variant aac(6′)-Ib-cr has two amino acid changes that allow the enzyme to inactivate quinolones as well [9,10].
Being plasmid-encoded, hydrolyzing enzymes are likely to be transmissible and widespread. As a single plasmid may encode more than one enzyme, a strain may express many different enzymes, as each one deactivates a different antibiotic [5]. As a result, MDR, or even PDR strains, arising and returning to the pre-antibiotic era has become a nightmare for medical professionals.
The present study aimed to report the resistance pattern of E. coli pathogens, detect the co-existence of different resistance determinants and their correlations to the resistance of E. coli pathogens of different infection origins, which would help in identifying local effective therapeutic options and infection control.
2. Results
2.1. Prevalence of E. coli Among Samples
In the present study, 200 (47%) E. coli pathogens were isolated from 425 patients suffering from different infections attending three hospitals in EL-Minia, Egypt. The highest prevalence was among urinary tract infections (62%) while it was lowest among eye infections (10%) (Table 1). Among the three hospitals, E. coli isolates were most prevalent in Minia University Hospital samples (51.37%), followed by Minia General Hospital (40.8%) (Table 2).
Table 1.
Prevalence of E. coli among different clinical samples.
| Infection | No. of Samples | No. of E. coli Isolates | E. coli (%) * |
|---|---|---|---|
| Wound infections (burns, diabetic foot, surgery wound, cuts) |
150 | 66 | 44% |
| Urinary tract infection | 100 | 62 | 62% |
| Gastro-enteritis | 50 | 24 | 48% |
| Blood | 75 | 39 | 52% |
| Chest infection | 20 | 4 | 20% |
| Ear infection | 20 | 4 | 20% |
| Eye | 10 | 1 | 10% |
| Total | 425 | 200 | 47% |
* Percent of E. coli were correlated to the number of samples of each infection.
Table 2.
Distribution of E. coli among samples collected from different hospitals.
| Hospitals | No. of Samples | E. coli | |
|---|---|---|---|
| No. | % * | ||
| Minia University Hospitals | 290 | 149 | 51.37 |
| Minia Chest Hospital | 20 | 4 | 20 |
| Minia General Hospital | 115 | 47 | 40.8 |
| Total | 425 | 200 | |
* Percent of E. coli were correlated to the number of collected samples from each hospital.
2.2. Antibiotic Resistance of the E. coli Pathogens
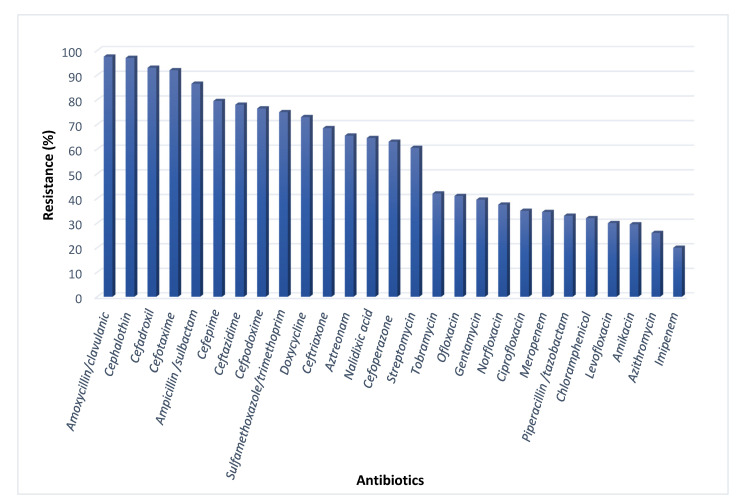
The antibiotic susceptibility was tested in 27 antibiotics that cover most of the available antibiotics in the Egyptian market. Table S1 (Supplementary Data) indicates the different used antibiotics and their different targets. The test revealed that 73% of E. coli were MDR. The pathogens were approximately totally resistant to Amoxycillin/clavulanic (97.5%), cephalothin (97%) and cefadroxil (93%). Also, high resistance levels were observed for Ceftazidime (78%) and Aztreonam (65.5%). Imipenem was the most effective antibiotic (20%), followed by Azithromycin (26%) (Figure 1). Our supplementary spread sheet indicates the resistance patterns of the isolates. One hundred MDR isolates were subjected for further investigation.
Figure 1.
Antibiotic resistance of the total E. coli isolates.
2.3. Serotyping of the Intestinal E. coli
Out of the selected 100 isolates, 20 were isolated from stool. Since E. coli normally inhabit the intestine, stool isolates were serotyped to confirm its pathogenicity. Out of 20 intestinal E. coli isolates 15 (75%) isolates were diarrheagenic E. coli (DEC). Three isolates (20%) were identified as Enterohaemorrhagic E. coli (EHEC) O157:H7. Different O serotypes were observed as O115, O158, O55, O126, O125 and O86a. The identified pathotypes are listed in Table S3 (Supplementary Data). Untyped five isolates were excluded from the further testing so that 95 isolates were further tested phenotypically and genotypically.
2.4. Multiple Antibiotic Resistance Index MDR E. coli Pathogens
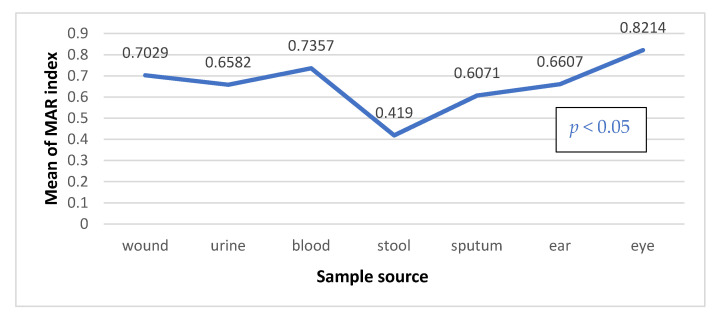
The multiple antibiotic resistance index (MARI) ratio between the number of antibiotics that an isolate is resistant to and the total number of antibiotics the organism is exposed to, have been calculated for 95 MDR E. coli. It was found that 98.9% of isolates have showed MAR index higher than 0.2, indicating high risk communities where antibiotics are abused. However, there was a statistically significant difference in MARI mean among E. coli isolates of different sources (p value < 0.05). Eye and blood isolates showed highest MARI mean of 0.82 and 0.74, respectively. On the other hand, stool samples had the lowest MARIs (Figure 2).
Figure 2.
MAR index mean values (), p value calculated by One-way ANOVA test.
2.5. Phenotypic Characteristics
ESBL production phenotypically tested by combined disk test (CDT). It was found that 89.4% (85/95) of the tested strains were ESBL producers. Carbapenemase production was tested by Modified Hodge test (MHT), then carbapenemase producers were tested for MBL using combined-disk synergy test. MBL producers accounted for 50.50% of the isolates (64.8% of carbapenemase producers). Positive significant association was found between ESBL and MBL phenotypes (p = 0.001) as all MBL producers were ESBL producers. Statistically significant difference in distribution of ESBL and MBL producers between different infection groups was observed (p < 0.001). Regardless of the eye infection, MBL producers were mostly frequent in UTIs (71%) while no intestinal E. coli was reported as MBL producers (Table 3).
Table 3.
Distribution of ESBLs and MBLs producing isolates among clinical specimens.
| Type of Infection | β-Lactam Resistant Isolates | ESBLs | MBL | ||
|---|---|---|---|---|---|
| No. | % * | No. | % * | ||
| Wound infections | 25 | 25 | 100% | 12 | 48% |
| Urinary tract infection | 28 | 28 | 100% | 20 | 71.4% |
| Gastro-enteritis | 15 | 5 | 33.3% | 0 | 0% |
| Blood | 20 | 20 | 100% | 12 | 46.67% |
| Respiratory infection | 4 | 4 | 100% | 2 | 50% |
| Ear infection | 2 | 2 | 100% | 1 | 50% |
| Eye | 1 | 1 | 100% | 1 | 100% |
| Total | 95 | 85 | 89.4% ** | 48 | 50.5% ** |
* percent was correlated to the total number of samples of each infection type; ** percent was correlated to total number of samples. Significant if p ≤ 0.05.
2.6. Antimicrobial Resistance of ESBLs and MBLs Producers
Resistance patterns of ESBL-producers revealed that ESBL producers were highly resistant to β-lactam antibiotics such as amoxycillin/clavulanic (99%), cefadroxil (95%), ceftazidime (85%) and meropenem (69%). MBL producers showed higher resistance rates to same antibiotics (100%, 97.9%, 93.75% and 93.75%, respectively). Azithromycin and chloramphenicol were the most effective drugs against ESBLs and MBLs producers. Table S4 (Supplementary Data) indicates the antibiogram of ESBLs and MBLs producers.
2.7. Prevalence of Resistance Genotypes Among the Tested Isolates
There was a statistically significant difference in the distribution of the different genotypes between different infections. The most prevalent gene was the blaTEM (80%) followed by blaSHV, blaCTX-M and blaNDM (54.7%, 42% and 44.2%, respectively). All isolates were negative for blaKPC or blaoxa-48 genes. Also, the aac-(6′)-Ib-cr gene was observed in 26.3% of the tested pathogens (Table 4).
Table 4.
Distribution of detected MBLs and ESBLs genotypes.
| Type of Infection | β-Lactam Resistant Isolates N |
blaNDM | blaTEM | blaCTX-M | blaSHV | blaIMP | aac-(6′)-Ib-cr |
|---|---|---|---|---|---|---|---|
| N (%) * | N (%) * | N (%) * | N (%) * | N (%) * | N (%) * | ||
| Wound infections | 25 | 14 (56%) |
23 (92%) |
14 (56%) |
15 (60%) |
12 (48%) |
9 (36%) |
| UTI | 28 | 17 (60.7%) |
27 (96.4%) |
11 (39.2%) |
12 (42.8%) |
16 (57%) |
6 (21%) |
| Gastro-enteritis | 15 | 1 (7.6%) |
7 (46.6%) |
0 (0%) |
0 (0%) |
0 (0%) |
0 (0%) |
| Blood | 20 | 7 (35%) |
20 (100%) |
13 (65%) |
20 (100%) |
7 (35%) |
10 (50%) |
| Chest infection | 4 | 1 (25%) |
3 (75%) |
0 (0%) |
4 (100%) |
0 (0%) |
0 (0%) |
| Ear infection | 2 | 1 (50%) |
2 (100%) |
1 (50%) |
0 (0%) |
0 (0%) |
0 (0%) |
| Eye | 1 | 1 (100%) |
1 (100%) |
1 (100%) |
1 (100%) |
0 (0%) |
0 (0%) |
| Total | 95 | 42 (44.2%) |
76 (80%) |
40 (42%) |
52 (54.7%) |
35 (36.8%) |
25 (26.3%) |
| p value ** | 0.11 | <0.001 | 0.011 | <0.001 | 0.011 | 0.042 |
* percent was correlated to the total number of β-lactam resistant isolates in each type of infection. ** Significant p value at p ≤ 0.05.
2.8. Genotypic-Phenotypic Agreement of the Tested Genes
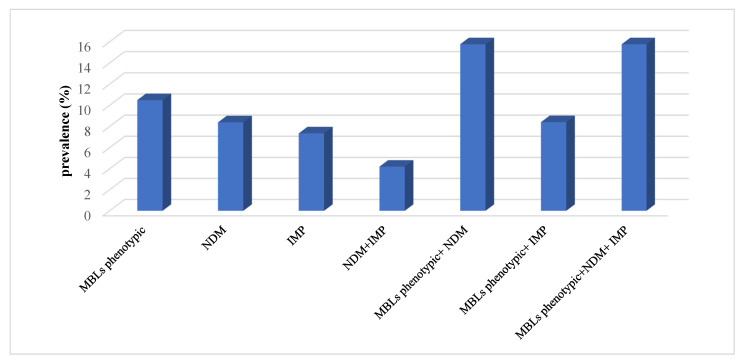
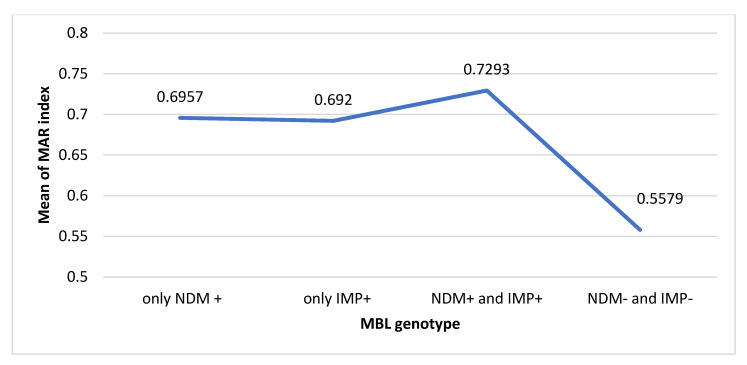
Out of 48 MBL phenotypic positive samples, 38 (79%) isolates were confirmed genotypically as MBL producers. It was found that 15.78% of isolates were phenotypically positive and harbored both blaNDM and blaIMP (Figure 3). Furthermore, there was a significant decrease in MAR index when isolates were both blaNDM and blaIMP negative (p value < 0.001). The blaNDM producers had higher MAR index than those which were only blaIMP producers. However, isolates harbored both blaNDM and blaIMP was observed resisting higher number of antibiotics (Figure 4). Moreover, positive correlations between MBLs phenotype, genotypes and carbapenem resistance were observed. Statistically significant correlations between detected MBL genotypes and meropenem resistance were observed. The MBLs phenotypes were more significantly associated with blaNDM than blaIMP (Table 5).
Figure 3.
Phenotypic and genotypic agreement of MβL tested genes.
Figure 4.
Distribution of MAR index mean values () among the detected MBLs genotypes.
Table 5.
Relation between MBLs phenotype, detected genotype and carbapenem resistance across the E. coli isolates.
| MBLs Phenotype | blaNDM | blaIMP | Imipenem Resistance | Meropenem Resistance | |
|---|---|---|---|---|---|
| MBLs phenotype | 1 | 0.372 ** | 0.232 * | 0.237 * | 0.465 ** |
| blaNDM | 1 | 0.155 | 0.054 | 0.212 * | |
| blaIMP | 1 | 0.122 | 0.275 ** | ||
| Imipenem resistance | 1 | 0.292 ** | |||
| Meropenem resistance | 1 |
* Correlation is significant at the 0.05 level (2-tailed). ** Correlation is significant at the 0.01 level (2-tailed). p values calculated by Fisher’s exact test.
ESBLs phenotypes showed strong positive correlation with the presence of blaTEM. However, all correlations were significant at the 0.01 level. Among the detected ESBL genes, presence of blaTEM and blaCTX-M types had the upper hand on the isolates’ resistance followed by aac(6′)Ib-cr gene (Table 6).
Table 6.
Correlation matrix (r2) between phenotypes, genotypes and antibiotic resistance across the E. coli isolates.
| ESBLs Production | MBLs Production | blaNDM | blaIMP | blaTEM | blaCTX-M | blaSHV | aac(6′)1b-cr | |
|---|---|---|---|---|---|---|---|---|
| MAR index | 0.611 ** | 0.342 ** | 0.330 ** | 0.289 ** | 0.366 ** | 0.365 ** | 0.251 * | 0.360 ** |
| ESBLs phenotype | 1 | 0.347 ** | 0.305 ** | 0.262 * | 0.696 ** | 0.293 ** | 0.377 ** | 0.205 * |
* Correlation is significant at the 0.05 level (2-tailed). ** Correlation is significant at the 0.01 level (2-tailed). p values calculated by Fisher’s exact test.
The current study identified that aac(6′)Ib-cr gene was mainly related to aminoglycoside antibiotics than fluoroquinolones. It was found that 47.2% of amikacin resistant isolates and 32.4% of ciprofloxacin resistant isolates harbored aac(6′)-Ib-cr gene. The presence of aac(6′)-Ib-cr gene was least correlated to ofloxacin resistance. Significant moderate positive correlation was observed between aac(6′)-Ib-cr and the resistance to amikacin and tobramycin, p values < 0.001 and 0.003, respectively (Table 7).
Table 7.
Correlation between aac(6′)Ib-cr, aminoglycoside and fluoroquinolone resistance.
| Antibiotics | Number of Resistant Isolates | Number of aac(6′)Ib-cr Positive Isolates (%) * | Person Correlation (r2) | p Value |
|---|---|---|---|---|
| Streptomycin | 61 | 19 (31) | 0.135 | 0.193 |
| Tobramycin | 43 | 17 (39.5) | 0.301 | 0.003 ** |
| Gentamycin | 43 | 13 (30.2) | 0.044 | 0.670 |
| Amikacin | 36 | 17 (47.2) | 0.374 | <0.01 *** |
| Ofloxacin | 39 | 9 (23) | 0.018 | 0.866 |
| Norfloxacin | 42 | 14 (33.3) | 0.1450 | 0.162 |
| Ciprofloxacin | 37 | 12 (32.4) | 0.152 | 0.142 |
* percent correlated to no. of resistant isolates of each antibiotic. p values were calculated by Fisher’s exact test. ** p value is significant at 0.05 level (2-tailed), *** p value is significant at 0.01 level (2-tailed).
2.9. Association of Different Resistance Genotypes
Most isolates harbored more than one resistance gene (81%). The resistance frequency has significantly increased with the increased number of the co-existed genes (p < 0.01). The most frequent association was of the five genes blaNDM, blaIMP, blaTEM, blaCTX-M and blaSHV (8.4%) (Table 8).
Table 8.
Co-existence of different genotypes.
| Number of Isolates (%) | |
|---|---|
| One gene | 9 (9.5) |
|
blaTEM
blaSHV |
8 (8.5) 1 (1) |
| Two genes | 20 (21.1) |
|
blaTEM, aac(6′)Ib-cr
blaIMP, blaTEM blaTEM, blaSHV blaNDM, blaTEM blaTEM, blaCTX-M |
1 (1) 5 (5.2) 5 (5.2) 7 (7.3) 2 (2.1) |
| Three genes | 21 (22.1) |
|
blaIMP, blaTEM, blaSHV
blaIMP, blaTEM, blaCTX-M blaNDM, blaTEM, aac(6′)Ib-cr blaTEM, blaCTX-M, blaSHV blaTEM, blaSHV, aac(6′)Ib-cr blaNDM, blaCTX-M, blaSHV blaNDM, blaIMP, blaTEM blaNDM, blaTEM, blaSHV |
4 (4.2) 1 (1) 2 (2.1) 5 (5.2) 1 (1) 1 (1) 2 (2.1) 5 (5.2) |
| Four genes | 21(22.1) |
|
blaTEM, blaCTX-M, blaSHV, aac(6′)Ib-cr
blaNDM, blaIMP, blaTEM, aac(6′)Ib-cr blaNDM, blaTEM, blaCTX-M, blaSHV blaIMP, blaTEM, blaCTX-M, blaSHV blaIMP, blaTEM, blaSHV, aac(6′)Ib-cr blaNDM, blaIMP, blaTEM, blaCTX-M blaNDM, blaTEM, blaSHV, aac(6′)Ib-cr blaNDM, blaCTX-M, blaSHV, aac(6′)Ib-cr blaNDM, blaTEM, blaCTX-M, aac(6′)Ib-cr |
5 (5.2) 3 (3.1) 3 (3.1) 3 (3.1) 1 (1) 1 (1) 1 (1) 1 (1) 3 (3.1) |
| Five genes | 12 (12.6) |
|
blaNDM, blaIMP,, blaTEM, blaCTX-M, blaSHV
blaNDM, blaIMP, blaTEM, blaCTX-M, aac(6′)Ib-cr blaIMP, blaTEM, blaCTX-M, blaSHV, aac(6′)Ib-cr |
8 (8.5) 2 (2.1) 2 (2.1) |
| Six genes | 3 (3.2) |
| Total | 95 |
Percentages were correlated to the total number of isolates.
The correlation matrix of the detected genes indicated overall positive correlations. The strongest and most significant correlation was observed between blaCTX-M and blaSHV (r = 0.519). Moreover, aac(6′)Ib-cr gene was significantly associated with blaCTX-M (Table 9).
Table 9.
Correlation matrix (r2) between the different genotypes.
| blaNDM | blaIMP | blaTEM | blaCTX-M | blaSHV | aac(6′)1b-cr | |
|---|---|---|---|---|---|---|
| blaNDM | 1 | 0.155 | 0.211 * | 0.185 | 0.086 | 0.190 |
| blaIMP | 1 | 0.290 ** | 0.233 * | 0.081 | 0.089 | |
| blaTEM | 1 | 0.196 | 0.227 * | 0.155 | ||
| blaCTX-M | 1 | 0.519 ** | 0.265 ** | |||
| blaSHV | 1 | 0.159 | ||||
| aac(6′)Ib-cr | 1 |
* Correlation is significant at the 0.05 level (2-tailed). ** Correlation is significant at the 0.01 level (2-tailed). p values were calculated by Fisher’s exact test.
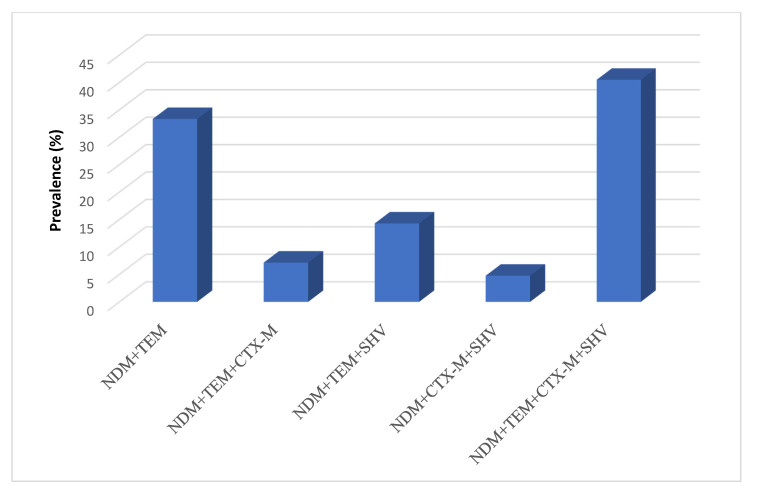
Studying the association of blaNDM gene with class A ESBL genes (blaTEM, blaCTX-M and blaSHV) among the blaNDM positive isolates indicated that the association of the blaNDM with the three ESBLs (blaTEM + blaSHV + blaCTX-M) genes was the highest, accounting for 40.47% of the isolates harboring blaNDM (Figure 5).
Figure 5.
blaNDM association with class A ESBLs genes.
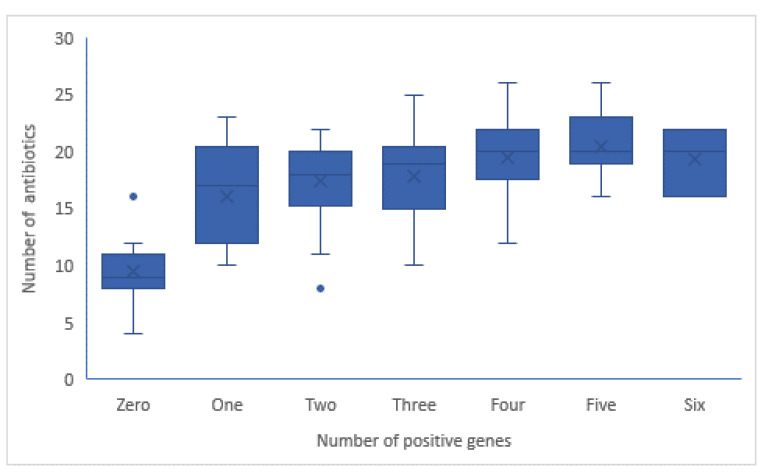
Furthermore, the spectrum of antibiotics to which the isolates were resistance is significantly increased (p value < 0.01) with the number of positive genes. The isolate harbored five genes showed the highest mean of antibiotic resistance (21 antibiotics), as indicted in Figure 6. As indicated in Table S5 (Supplementary Data) the resistance of β-lactam drugs is significantly associated with presence of higher number of genes. Also, significant moderate correlation of number of positive genes with ciprofloxacin, norfloxacin and aminoglycoside were observed.
Figure 6.
Compatibility of number of detected genes and number of antibiotics to which isolates were resistant among 95 isolates.
3. Discussion
The last two decades have witnessed a conspicuous increase in the number of infections caused by multi-drug resistant strains of E. coli, and this has impacted the outcomes of different infections [11].
The present study demonstrated that the prevalence of E. coli pathogens isolated from patients suffering from different infections in El-Minya hospitals accounted for 47%. This result was in accordance with results reported by Amer et al. (45%) [12] and Fam et al. (56%) [13].
Among extraintestinal infections, E. coli was the most common among urine isolates (62%) followed by blood infections (52%). Likewise, in Saudi Arabia Alanazi et al. [14] reported that E. coli was isolated from 60.24% of the urine samples and in Greece Koupetori et al. [15] reported high incidence of E. coli accounting for 48% of blood isolates. In contrary, many studies showed lower E. coli incidence [16,17]. On the other hand, E. coli was least isolated from eye infections (10%). This was higher than results reported in USA by Miller et al. (5.9%) [18].
Moreover, intestinal E. coli was serotyped to ensure its pathogenicity. Diarrheagenic E. coli (DEC) serotypes accounted for 54.1% which was considered very high in comparison to results obtained by Zhou et al. (7.9%) [19]. E. coli O157:H7 accounted for 12.5% of total DEC serotypes. Comparably, in the same region of study Abd El Gany et al. [20] reported incidence rate of 15.72%. The variations between the different studies may be ascribed to many socioeconomical, demographical and geographical factors. It was obviously noted that E. coli were relatively high in the present study compared to other studies indicates poor hygienic attitudes that correlated to the mentioned factors.
Antimicrobial resistance (AMR) emphasizes an overwhelming health and economic burden in both developed and developing countries. As Resistance narrows the therapeutic options leading to increased morbidity and mortality [21]. Our results showed high prevalence of MDR E. coli (73%) which were higher than results obtained by Siwakoti et al. [22] and Abdelaziz et al. [23] (28% and 60%, respectively).
Although carbapenem resistance is considered low, it is higher than previous studies done at the same government [9,20,24]. This may be attributed to the availability and the usage of the drugs when the studies were held. Concerning our results, the drugs were more available and highly used but in previous studies there was a shortage in many antibiotics inside the hospitals. Moreover, the resistance to meropenem was higher than imipenem which may be attributed to that meropenem is cheaper than imipenem, so it is commonly used while some other studies reported complete meropenem sensitivity [9,23].
The continuous spread of ESBLs and carbapenemase mediated resistance has dramatically increased in both hospital and community infections. It was found that 89.4% of the tested isolates were ESBL producers. Abd El-Baky et al. [24] in earlier study in our area reported that 46.8% of isolates were phenotypically ESBL producers. It seems very alerting as the prevalence is almost doubled in short period.
The incidence of the carbapenemase and MBLs producers accounted for 77.8% and 64.8%, respectively. These rates were very high when compared to results obtained by Ibrahim, et al. [25] who reported that carbapenemase and MBLs incidences were 37.6% and 46.3% respectively. A previous study in our area showed that 52.3% of P. aeruginosa were MBL producers [26]. The differences in prevalence may be due to strains and time variations but overall indicate high incidence of MBLs among bacteria in our area.
There was a significant difference in the distribution of detected genes among the different sample sources p values < 0.05 which was in agreement with many studies [27,28].
The most prevalent genotype was blaTEM (80%) followed by blaSHV (54.7%), blaNDM (44.2%) and blaCTX-M (42%). Similarly, blaTEM was predominant in results reported by Mohamed et al. [28] and Maamoun et al. [29]. On the other hand, a study on Escherichia coli causing sepsis among Egyptian children reported that blaSHV was the most common ESBL (61.22%), followed by blaTEM (38.78%) and blaCTX-M (20.41%) [30]. Furthermore, the higher incidence of blaTEM gene reported by the current study or other studies in our region suggests that blaTEM gene may be endemic. In contrast to our study, several studies in Asia reported that blaCTX-M was the most frequent indicating that blaCTX-M is a predominant genotype in Asia [31,32,33]. Also, reports from Qatar stated that blaCTX-M type genes evolved through mutations in blaTEM and blaSHV genes and it is a recent endemic [34].
Similar to our study, studies in UK have reported 44% of isolates as blaNDM producers, most of them were from urine samples [35]. The number of blaNDM producers is increasing in Egypt which is reflected by many studies conducted in this area [30,36,37]. Lower blaIMP incidence reported in previous studies in Egypt compared to this study (36.8%) suggesting an increasing rate of MBLs producers [36,37]. However, the current reported high prevalence of MBLs may be attributed to the ability of E. coli to acquire novel resistance genes through horizontal transfer or the increased use of carbapenems in the clinical treatment.
blaOxa-48 like and blaKPC were not detected in any isolate. Quite higher prevalence of blaOxa-48 like and blaKPC (38.46% and 23%, respectively) was reported in Bangladesh [38]. In accordance with the current study, blaKPC wasn’t detected in several previous studies in Egypt or detected in very low rate [37,38]. In addition, blaKPC wasn’t detected in countries such as Saudi Arabia [39] or those of the Arabian Peninsula [40]. These data confirmed that blaKPC genes does not predominate in this geographical region, where it is frequently detected in the United States [41] and endemic in Israel [42]. The aac(6′)-Ib-cr gene prevailed in 26.3% of the isolates which were mostly isolated from wounds. This rate is lower than rates previously reported by Al-Agamy et al. [43] and Mohamed et al. [28]. The differences across studies may be attributed to differences in geographical locations, age groups, or clinical criteria.
Most of the isolates (81%) harbored more than one resistance determinant. Co-harboring of multiple ESBL genes was detected previously in Egypt [28,44] and some other countries; Burkina Faso [45], Qatar [33] and Iran [46].The co-existence of blaNDM, blaIMP, blaTEM, blaCTX-M and bla-SHV was the most frequent, accounting for 8.5% of the isolates. There was a significant association between blaCTX-M and blaSHV, which agree with other studies [28,30,46]. There was a significant positive correlation between blaNDM and blaIMP. This was comparable to Zaki et al. [30] and Kamel et al. [47] where single E. coli isolate had more than one type of metallo β-lactamase. The association between aac(6′)-Ib-cr and blaCTX-M genes was statistically significant, agreeing with previous studies [48,49,50]. None of the isolates harbored aac(6′)Ib-cr alone. Moreover, there was significant association of aac(6′)-Ib-cr gene with ESBL phenotypes. This may be due to the common presence of ESBL genes and PMQR genes on the same plasmid in Enterobacteriaceae [51]. Moreover, the aac(6′)-Ib-cr gene showed significant positive correlation with amikacin and tobramycin resistance in ESBL producers. Similarly, Mohamed et al. [28] reported significant association of ESBL genes with aac(6′)-Ib-cr gene that resulted in increased ciprofloxacin, gentamicin and amikacin resistance in ESBL producers.
The resistance rates were significantly increased in ESBL producers than non-producers that reported by several studies [28,52,53,54]. In agreement with our results, many studies reported higher resistance rate of MBL producers in comparison to MBLs non-producers. In accordance with the current work, previous studies have reported significant high resistance rates in MBL producers [55,56]. It was reported that blaTEM and blaSHV are important factors in increased resistance of ESBL E. coli producers to third-generation cephalosporin [57].
Finally, variations in rate and predominance of resistance genes between different countries and even among the same country institutions may be due to difference in locally prescribed antibiotics and if the infection control guidelines are followed or not in different health institutes. In Egypt, the high rate of ESΒLs and MBLs is a reflection of the inappropriate use of antimicrobials due to the over counter availability of antibiotics without prescription and patients incompliance or the wide use of antibiotics in veterinary care and farms [58,59].
4. Material and Methods
4.1. Bacterial Isolates
Two hundred E. coli isolates were isolated from 425 patients attending different hospitals in El-Minia with different infections. All clinical samples were obtained as part of the routine hospital laboratory procedures. Samples were processed and cultured on trypticase soy agar (Lab M, Hewwood, UK) at 37 °C for overnight. E. coli colonies gave pink color on MacConkey agar and green metallic sheen on Eosin methylene blue (EMB) (lab M, Hewwood, UK). Colonies were further identified by regular microbiological biochemical tests [60].
4.2. Antimicrobial-Susceptibility Testing
The antimicrobial susceptibility of the isolates was tested by the Kirby-Bauer Disk Diffusion method [61]. The used antibiotics discs were ready cartilages purchased from Oxoid; Basingstoke, UK. The following antibiotic discs were used Cefpodoxime (10 μg), Streptomycin (10 μg), Aztreonam (30 μg), Ceftriaxone (30 μg), Gentamycin (10 μg), Amoxycillin/clavulanic (20/10 μg), Piperacillin/Tazobactam (100/10 μg), Ceftazidime (30 μg), Imipenem (10 μg), Meropenem (10 μg), Cefoperazone (75 μg), Doxycycline (30 μg), Ciprofloxacin (5 μg), Amikacin (30 μg), Nalidixic acid (30 μg), Cefotaxime (30 μg), Cefepime (30 μg), Ampicillin/sulbactam (10/10 μg), Norfloxacin (10 μg), Tobramycin (10 μg), Sulfamethoxazole/trimethoprim (23.75/1.25 μg), Chloramphenicol (30 μg). Isolates classified as sensitive, intermediate and resistant according to inhibition zones interpretation standards of Clinical Laboratory standards Institute (CLSI) 2018 (Table S2) [62].
4.3. Serotyping of Intestinal E. coli
Escherichia coli recovered from gastroenteritis infections were sent to the Animal Health Research Institute, Giza, Egypt to be serotyped. The isolates serotyped through detection of isolates agglutination with O and H antisera using the slide agglutination method according to the manufacturer instructions (Pro-Lab Diagnostics, Round Rock, TX, USA).
4.4. Phenotypic Detection of ESBLs and MBLs Production
Detection of ESBL in E. coli isolates was carried out by combined disc test (CDT). Isolates defined positive when the difference between the inhibition zones of cefotaxime and cefotaxime/clavulanic or ceftazidime and ceftazidime/clavulanic disks is ≥5 mm [62]. Carbapenemases were detected in carbapenem resistant isolates by Modified Hodge test (MHT). MHT positive isolates w ere further tested for MBL production using EDTA-combined disk synergy test. An increase in zone diameter of at least 7 mm around the imipenem–EDTA or meropenem–EDTA disks were recorded positive result [63].
4.5. Amplification of Resistance Genes
The DNA templet was extracted by available commercial kit QIAprep® Spin Miniprep Kit (QIAGEN, Germany) by following the manufacturer instruction. Resistance genes were detected using conventional PCR technique. Amplification was done using 25 μL PCR reaction mixture consisting of 12.5 μL master mix (BIOMATIk, Kitchener, Canada) 1 μL of each forward and reverse primers (BIOMATIk, Canada), 2 μL DNA template and 8.5 μL nuclease-free water. PCR cycling conditions are indicated in Table S5 (Supplementary Data).
4.6. Statistical Analysis
Data were analyzed using IBM SPSS version 20.0. First, normal distribution of data was tested by normality tests as Kolmogorov–Smirnov and Shapiro–Wilk p-values in addition to histograms. Descriptive analysis was done to analyze prevalence of E. coli isolates among different infections and hospitals, percentage of resistance and prevalence of ESBL and MBL producers and prevalence of the different genes. To compare differences in distribution between different groups chi-square (X2) test was done but when more than 20% of cells were less than 5, Fisher’s exact test was done to be more accurate. One-way ANOVA tests was done to compare mean values between different groups as MAR index mean values in different sample sources. Non parametric tests were used for non-parametric data as Kruskal–Wallis. To study associations between phenotypes, genotypes and resistance, correlations were established using Pearson’s correlation coefficient (r2) in bivariate. p-values are significant if they are ≤0.05.
5. Conclusions
High resistance reported in our study indicates poor awareness of the microbiological laboratory test importance, high empirical antimicrobial prescription and high patient incompliance. Moreover, the massive co-existence of the detected genes strongly supports the presence of one or more circulating plasmids that harbors different resistance genes. Finally, the study highlighted the importance of continuous surveillance of the resistance trends and the direct need to strictly apply the infection control policies, implementing a national antimicrobial stewardship plan.
Acknowledgments
The authors received no financial support for the research or publication of this article.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10070835/s1, Table S1: The different tested antibiotics and their different biological processes targets, Table S2: The different thresholds of inhibition zones according to CLSI (2018), Table S3: Different serotypes of the intestinal E. coli, Table S4: Resistance pattern of ESBLs and MBLs producers, Table S5: Correlation coefficient of the number of detected genes in isolates with the resistance of the antibiotics, Table S6: Primers used in the current study. References [63,64,65,66,67,68,69,70] are cited in Supplementary File.
Author Contributions
Formal analysis, S.M.M., S.A.A. and R.A.I.; Methodology, R.M.A.E.-B., S.M.M., S.A.A. and R.A.I.; Supervision, R.M.A.E.-B., S.A.A. and R.A.I.; Validation, S.A.A.; Visualization, R.A.I.; Writing—original draft, S.M.M.; Writing—review & editing, R.M.A.E.-B. and S.M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Commission on the Ethics of Scientific Research of Faculty of Pharmacy, Minia University (protocol code HV06/2020 and date of approval 17-6-2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abdelwahab R., Yasir M., Godfrey R.E., Christie G.S., Element S.J., Saville F., Hassan E.A., Ahmed E.H., Abu-Faddan N.H., Daef E.A.J.V. Antimicrobial Resistance and Gene Regulation in Enteroaggregative Escherichia Coli from Egyptian Children with Diarrhoea: Similarities and Differences. Virulence. 2021;12:57–74. doi: 10.1080/21505594.2020.1859852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banin E., Hughes D., Kuipers O.P. Bacterial Pathogens, Antibiotics and Antibiotic Resistance. FEMS Microbiol. Rev. 2017;41:450–452. doi: 10.1093/femsre/fux016. [DOI] [PubMed] [Google Scholar]
- 3.Sultan I., Rahman S., Jan A.T., Siddiqui M.T., Mondal A.H., Haq Q.M.R. Antibiotics, Resistome and Resistance Mechanisms: A Bacterial Perspective. Front. Microbiol. 2018;9:2066. doi: 10.3389/fmicb.2018.02066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christaki E., Marcou M., Tofarides A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020;88:26–40. doi: 10.1007/s00239-019-09914-3. [DOI] [PubMed] [Google Scholar]
- 5.Matlashewski G., Berghuis A., Sheppard D., Wainberg M.A., Gotte M. Handbook of Antimicrobial Resistance. Springer; Berlin/Heidelberg, Germany: 2017. [Google Scholar]
- 6.Tooke C.L., Hinchliffe P., Bragginton E.C., Colenso C.K., Hirvonen V.H., Takebayashi Y., Spencer J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019;431:3472–3500. doi: 10.1016/j.jmb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reck F., Bermingham A., Blais J., Capka V., Cariaga T., Casarez A., Colvin R., Dean C.R., Fekete A., Gong W., et al. Optimization of Novel Monobactams with Activity against Carbapenem-Resistant Enterobacteriaceae—Identification of LYS228. Bioorg. Med. Chem. Lett. 2018;28:748–755. doi: 10.1016/j.bmcl.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Bush K., Bradford P.A. Interplay between β-Lactamases and New β-lactamase Inhibitors. Nat. Rev. Genet. 2019;17:295–306. doi: 10.1038/s41579-019-0159-8. [DOI] [PubMed] [Google Scholar]
- 9.Ramírez-Castillo F.Y., Moreno-Flores A.C., Avelar-González F.J., Márquez-Díaz F., Harel J., Guerrero-Barrera A.L. An Evaluation of Multidrug-Resistant Escherichia Coli Isolates in Urinary Tract Infections from Aguascalientes, Mexico: Cross-Sectional Study. Ann. Clin. Microbiol. Antimicrob. 2018;17:34. doi: 10.1186/s12941-018-0286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park C.H., Robicsek A., Jacoby G.A., Sahm D., Hooper D.C. Prevalence in the United States of aac(6′)-Ib-cr Encoding a Ciprofloxacin-Modifying Enzyme. Antimicrob. Agents Chemother. 2006;50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peralta G., Sanchez M.B., Garrido J.C., De Benito I., Cano M.E., Martínez-Martínez L., Roiz M.P. Impact of Antibiotic Resistance and of Adequate Empirical Antibiotic Treatment in the Prognosis of Patients with Escherichia coli bacteraemia. J. Antimicrob. Chemother. 2007;60:855–863. doi: 10.1093/jac/dkm279. [DOI] [PubMed] [Google Scholar]
- 12.Amer R., El-Baghdady K., Kamel I., El-Shishtawy H. Prevalence of Extended Spectrum Beta- Lactamase Genes among Escherichia Coli and Klebsiella Pneumoniae Clinical Isolates. Egypt. J. Microbiol. 2019 doi: 10.21608/ejm.2019.16460.1113. [DOI] [Google Scholar]
- 13.Fam N., Leflon-Guibout V., Fouad S., Aboul-Fadl L., Marcon E., Desouky D., El-Defrawy I., Abou-Aitta A., Klena J., Nicolas-Chanoine M.-H. CTX-M-15-Producing Escherichia Coli Clinical Isolates in Cairo (Egypt), Including Isolates of Clonal Complex ST10 and Clones ST131, ST73, and ST405 in Both Community and Hospital Settings. Microb. Drug Resist. 2011;17:67–73. doi: 10.1089/mdr.2010.0063. [DOI] [PubMed] [Google Scholar]
- 14.Alanazi M.Q., Alqahtani F.Y., Aleanizy F.S. An Evaluation of E. Coli in Urinary Tract Infection in Emergency Department at KAMC in Riyadh, Saudi Arabia: Retrospective Study. Ann. Clin. Microbiol. Antimicrob. 2018;17:3. doi: 10.1186/s12941-018-0255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koupetori M., Retsas T., Antonakos N., Vlachogiannis G., Perdios I., Nathanail C., Makaritsis K., Papadopoulos A., Sinapidis D., Giamarellos-Bourboulis E.J., et al. Bloodstream Infections and Sepsis in Greece: Over-Time Change of Epidemiology and Impact of de-escalation on final outcome. BMC Infect. Dis. 2014;14:272. doi: 10.1186/1471-2334-14-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohammed M.A., Alnour T.M., Shakurfo O.M., Aburass M.M. Prevalence and antimicrobial resistance pat-tern of bacterial strains isolated from patients with urinary tract infection in Messalata Central Hospital, Libya. Asian Pac. J. Trop. Med. 2016;9:771–776. doi: 10.1016/j.apjtm.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Musicha P., Cornick J.E., Bar-Zeev N., French N., Masesa C., Denis B., Kennedy N., Mallewa J., Gordon M.A., Msefula C.L., et al. Trends in Antimicrobial Resistance in Bloodstream Infection Isolates at a Large Urban Hospital in Malawi (1998–2016): A Surveillance Study. Lancet Infect. Dis. 2017;17:1042–1052. doi: 10.1016/S1473-3099(17)30394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller D. Update on the Epidemiology and Antibiotic Resistance of Ocular Infections. Middle E. Afr. J. Ophthalmol. 2017;24:30. doi: 10.4103/meajo.MEAJO_276_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y., Zhu X., Hou H., Lü Y., Yu J., Mao L., Mao L., Sun Z. Characteristics of Diarrheagenic Escherichia Coli among Children under 5 Years of Age with Acute Diarrhea: A Hospital Based Study. BMC Infect. Dis. 2018;18:63. doi: 10.1186/s12879-017-2936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Gany S.A., Gad G.M., Mousa S., Ibrahem R. Characterization of Verotoxigenic E.Coli and Enteropathogenic E.Coli Isolated from Infants with Diarrhea in Combination with Antimicrobial Resistance Pattern in Minia, Egypt. J. Adv. Biomed. Pharm. Sci. 2020;3:101–109. doi: 10.21608/jabps.2020.24153.1070. [DOI] [Google Scholar]
- 21.Jit M., Ng D.H.L., Luangasanatip N., Sandmann F., Atkins K.E., Robotham J.V., Pouwels K.B. Quantifying the Economic cost of Antibiotic Resistance and the Impact of Related Interventions: Rapid Methodological Review, Conceptual Framework and Recommendations for Future Studies. BMC Med. 2020;18:38. doi: 10.1186/s12916-020-1507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siwakoti S., Subedi A., Sharma A., Baral R., Bhattarai N.R., Khanal B. Incidence and Outcomes of Multidrug-Resistant Gram-negative Bacteria Infections in Intensive Care Unit from Nepal- a Prospective Cohort Study. Antimicrob. Resist. Infect. Control. 2018;7:114. doi: 10.1186/s13756-018-0404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdelaziz S.M., Aboshanab K.M., Yahia I.S., Yassien M.A., Hassouna N.A.J.A. Correlation between the Antibiotic Resistance Genes and Susceptibility to Antibiotics among the Carbapenem-Resistant Gram-Negative Pathogens. Antibiotics. 2021;10:255. doi: 10.3390/antibiotics10030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abd El-Baky R.M., Ibrahim R.A., Mohamed D.S., Ahmed E.F., Hashem Z.S.J.I., Resistance D. Prevalence of Virulence Genes and Their Association with Antimicrobial Resistance Among Pathogenic E. Coli Isolated from Egyptian Patients with Different Clinical Infections. Infect. Drug Resist. 2020;13:1221. doi: 10.2147/IDR.S241073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ibrahim Y., Sani Y., Saleh Q., Saleh A., Hakeem G. Phenotypic Detection of Extended Spectrum Beta Lactamase and Carbapenemase Co-Producing Clinical Isolates from Two Tertiary Hospitals in Kano, North West Nigeria. Ethiop. J. Health Sci. 2017;27:3–10. doi: 10.4314/ejhs.v27i1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farhan S.M., A Ibrahim R., Mahran K.M., Hetta H.F., El-Baky R.M.A. Antimicrobial Resistance Pattern and Molecular Genetic Distribution of Metallo-β-Lactamases Producing Pseudomonas aeruginosa Isolated from Hospitals in Minia, Egypt. Infect. Drug Resist. 2019;12:2125–2133. doi: 10.2147/IDR.S198373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fazlul M., Deepthi S., Farzana Y., Najnin A., Rashid M., Munira B., Srikumar S. Detection of Metal-lo-Β-Lactamases-Encoding Genes among Clinical Isolates of Escherichia Coli in a Tertiary Care Hospital, Malaysia. BMC Res. Notes. 2019;11:291–298. [Google Scholar]
- 28.Mohamed E.S., Khairy R.M.M., Abdelrahim S.S. Prevalence and Molecular Characteristics of ESBL and AmpC β -Lactamase Producing Enterobacteriaceae Strains Isolated from UTIs in Egypt. Antimicrob. Resist. Infect. Control. 2020;9:198. doi: 10.1186/s13756-020-00856-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Sherif R.H., Maamoun H.A.H. Molecular Characteristics of Extended-Spectrum Beta-Lactamases Among Gram-Negative Isolates Collected in Cairo University Hospital. Comp. Haematol. Int. 2012;22:733–739. doi: 10.1007/s00580-012-1475-9. [DOI] [Google Scholar]
- 30.Zaki M., El-Halaby H., Elmansoury E., Zeid M., Khaled K., Nomir M. Genetic Study of Extended Spectrum Beta-Lactamase and Carbapenemase Producing Escherichia Coli Causing Sepsis among Egyptian Children. Open Microbiol. J. 2019;13:128–137. doi: 10.2174/1874285801913010128. [DOI] [Google Scholar]
- 31.Myat T.O., Hannaway R.F., Zin K.N., Htike W.W., Win K.K., Crump J.A., Murdoch D.R., Ussher J.E. ESBL-and Carbapenemase-Producing Enterobacteriaceae in Patients with Bacteremia, Yangon, Myanmar. Emerg. Infect. Dis. 2017;23:857. doi: 10.3201/eid2305.161100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abrar S., Ain N.U., Liaqat H., Hussain S., Rasheed F., Riaz S. Distribution of blaCTX—M, blaTEM, blaSHV and blaOXA Genes in Extended-Spectrum-β-Lactamase-Producing Clinical Isolates: A Three-Year Multi-Center Study from Lahore, Pakistan. Antimicrob. Resist. Infect. Control. 2019;8:80. doi: 10.1186/s13756-019-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eltai N.O., Al Thani A.A., Al-Ansari K., Deshmukh A.S., Wehedy E., Al-Hadidi S.H., Yassine H.M. Molecular Characterization of Extended Spectrum β -Lactamases Enterobacteriaceae Causing Lower Urinary Tract Infection among Pediatric Population. Antimicrob. Resist. Infect. Control. 2018;7:90. doi: 10.1186/s13756-018-0381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed M.A.S., Bansal D., Acharya A., Elmi A.A., Hamid J.M., Ahmed A.M.S., Chandra P., Ibrahim E., Sultan A.A., Doiphode S., et al. Antimicrobial Susceptibility and Molecular Epidemiology of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae from Intensive Care Units at Hamad Medical Corporation, Qatar. Clin. Microbiol. Rev. 2016;5 doi: 10.1128/CMR.00096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumarasamy K.K., Toleman M.A., Walsh T.R., Bagaria J., Butt F., Balakrishnan R., Chaudhary U., Doumith M., Giske C.G., Irfan S., et al. Emergence of A New Antibiotic Resistance Mechanism in India, Pakistan, and the UK: A Molecular, Biological, and Epidemiological Study. Lancet Infect. Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amer W.H., Khalil H.S., Abd EL Wahab M.A. Risk Factors, Phenotypic and Genotypic Characterization of Carbapenem Resistant Enterobacteriaceae in Tanta University Hospitals, Egypt. Int. J. Infect. Control. 2016;12 doi: 10.3396/IJIC.v12i2.012.16. [DOI] [Google Scholar]
- 37.Khalifa H., Soliman A.M., Ahmed A.M., Shimamoto T., Hara T., Ikeda M., Kuroo Y., Kayama S., Sugai M., Shimamoto T. High Carbapenem Resistance in Clinical Gram-Negative Pathogens Isolated in Egypt. Microb. Drug Resist. 2017;23:838–844. doi: 10.1089/mdr.2015.0339. [DOI] [PubMed] [Google Scholar]
- 38.Begum N., Shamsuzzaman S. Emergence of Carbapenemase-Producing Urinary Isolates at A Tertiary Care Hospital in Dhaka, Bangladesh. Tzu Chi Med. J. 2016;28:94–98. doi: 10.1016/j.tcmj.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Memish Z.A., Assiri A., Almasri M., Roshdy H., Hathout H., Kaase M., Gatermann S.G., Yezli S. Molecular Characterization of Carbapenemase Production Among Gram-Negative Bacteria in Saudi Arabia. Microb. Drug Resist. 2015;21:307–314. doi: 10.1089/mdr.2014.0121. [DOI] [PubMed] [Google Scholar]
- 40.Sonnevend A., Ghazawi A.A., Hashmey R., Jamal W., Rotimi V.O., Shibl A.M., Al-Jardani A., Al-Abri S.S., Tariq W.U.Z., Weber S., et al. Characterization of Carbapenem-Resistant Enterobacteriaceae with High Rate of Autochthonous Transmission in the Arabian Peninsula. PLoS ONE. 2015;10:e0131372. doi: 10.1371/journal.pone.0131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekyere J.O., Govinden U., Essack S. The Molecular Epidemiology and Genetic Environment of Carbapenemases Detected in Africa. Microb. Drug Resist. 2016;22:59–68. doi: 10.1089/mdr.2015.0053. [DOI] [PubMed] [Google Scholar]
- 42.Chen L., Anderson D., Paterson D. Overview of the Epidemiology and the Threat of Klebsiella Pneumoniae Carbapenemases (KPC) Resistance. Infect. Drug Resist. 2012;5:133–141. doi: 10.2147/IDR.S26613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Agamy M.H., Aljallal A., Radwan H.H., Shibl A.M. Characterization of Carbapenemases, ESBLs, and Plasmid-Mediated Quinolone Determinants in Carbapenem-Insensitive Escherichia coli and Klebsiella pneumoniae in Riyadh hospitals. J. Infect. Public Health. 2018;11:64–68. doi: 10.1016/j.jiph.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Hassuna N.A., Khairalla A.S., Farahat E.M., Hammad A.M., Abdel-Fattah M. Molecular Characterization of Extended-Spectrum β Lactamase- Producing E. coli Recovered from Community-Acquired Urinary Tract Infections in Upper Egypt. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-59772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouedraogo A.-S., Sanou M., Kissou A., Sanou S., Solaré H., Kaboré N.F., Poda A., Aberkane S., Bouzinbi N., Sano I., et al. High Prevalence of Extended-Spectrum ß-Lactamase producing Enterobacteriaceae among Clinical Isolates in Burkina Faso. BMC Infect. Dis. 2016;16 doi: 10.1186/s12879-016-1655-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faghri J., Maleki N., Tahanasab Z., Mobasherizadeh S., Rezaei A. Prevalence of CTX-M and TEM β-lactamases in Klebsiella pneumoniae Isolates from Patients with Urinary Tract Infection, Al-Zahra Hospital, Isfahan, Iran. Adv. Biomed. Res. 2018;7:10. doi: 10.4103/abr.abr_17_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamel N.A., El-Tayeb W.N., El-Ansary M.R., Mansour M.T., Aboshanab K.M. Phenotypic Screening and Molecular Characterization of Carbapenemase-Producing Gram-Negative Bacilli Recovered from Febrile Neutropenic Pediatric Cancer Patients in Egypt. PLoS ONE. 2018;13:e0202119. doi: 10.1371/journal.pone.0202119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bado I., Gutiérrez C., Garcia-Fulgueiras V., Cordeiro N.F., Pirez L.A., Seija V., Bazet C., Rieppi G., Vignoli R. CTX-M-15 in Combination with aac(6′)-Ib-cr is the Most Prevalent Mechanism of Resistance both in Escherichia coli and Klebsiella pneumoniae, including K. pneumoniae ST258, in an ICU in Uruguay. J. Glob. Antimicrob. Resist. 2016;6:5–9. doi: 10.1016/j.jgar.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Vignoli R., García-Fulgueiras V., Cordeiro N.F., Bado I., Seija V., Aguerrebere P., Laguna G., Araújo L., Bazet C., Gutkind G., et al. Extended-Spectrum β-lactamases, Transferable Quinolone Resistance, and Virulotyping in Extra-Intestinal E. coli in Uruguay. J. Infect. Dev. Ctries. 2016;10:43–52. doi: 10.3855/jidc.6918. [DOI] [PubMed] [Google Scholar]
- 50.Azargun R., Sadeghi M.R., Barhaghi M.H.S., Kafil H.S., Yeganeh F., Oskouee M.A., Ghotaslou R. The Prevalence of Plasmid-mediated Quinolone Resistance and ESBL-Production in Enterobacteriaceae Isolated from Urinary tract Infections. Infect. Drug Resist. 2018;ume 11:1007–1014. doi: 10.2147/IDR.S160720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue G., Li J., Feng Y., Xu W., Li S., Yan C., Zhao H., Sun H. High Prevalence of Plasmid-Mediated Quinolone Resistance Determinants in Escherichia coli and Klebsiella pneumoniae Isolates from Pediatric Patients in China. Microb. Drug Resist. 2017;23:107–114. doi: 10.1089/mdr.2016.0004. [DOI] [PubMed] [Google Scholar]
- 52.Mahato S., Mahato A., Yadav J. Prevalence and identification of uropathogens in eastern Nepal and under-standing their antibiogram due to multidrug resistance and Esbl. Asian Pac. J. Microbiol. Res. 2018;2:09–17. [Google Scholar]
- 53.Alfola M.M.H.R., Kamel Z., Nada M.G.E.D., Rashed L.A., El-Awady B.A. Phenotypic and Genotypic Characterization of ESBL-Producing Escherichia coli and Klebsiella pneumonia Isolates from Patient’s Urine Specimens. Int. Arab. J. Antimicrob. Agents. 2016;6:6. doi: 10.3823/798. [DOI] [Google Scholar]
- 54.Abdel-Moaty M.M., Mohamed W.S., Abdel-All S.M., El-Hendawy H.H. Prevalence and Molecular Epidemiology of Extended Spectrum β-Lactamase Producing Escherichia coli from Hospital and Community Settings in Egypt. J. Appl. Pharm. Sci. 2016;6:042–047. doi: 10.7324/JAPS.2016.600107. [DOI] [Google Scholar]
- 55.Nepal K., Pant N.D., Neupane B., Belbase A., Baidhya R., Shrestha R.K., Lekhak B., Bhatta D.R., Jha B. Extended Spectrum Beta-Lactamase and Metallo Beta-Lactamase Production among Escherichia coli and Klebsiella pneumoniae Isolated from Different Clinical Samples in A Tertiary Care Hospital in Kathmandu, Nepal. Ann. Clin. Microbiol. Antimicrob. 2017;16:62. doi: 10.1186/s12941-017-0236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Javed H., Ejaz H., Zafar A., Rathore A.W., Haq I.U. Metallo-Beta-Lactamase Producing Escherichia coli and Klebsiella pneumoniae: A Rising Threat for Hospitalized Children. J. Pak. Med. Assoc. 2016;66:1068–1072. [PubMed] [Google Scholar]
- 57.Wu T.-L., Siu L., Su L.-H., Lauderdale T., Lin F., Leu H.-S., Lin T.-Y., Ho M. Outer membrane protein change combined with co-existing TEM-1 and SHV-1 β-lactamases lead to false identification of ESBL-producing Klebsiella pneu-moniae. J. Antimicrob. Chemother. 2001;47:755–761. doi: 10.1093/jac/47.6.755. [DOI] [PubMed] [Google Scholar]
- 58.WHO . Egypt: Pharmaceutical Country Profile. WHO; Geneva, Switzerland: 2011. [Google Scholar]
- 59.El-Shazly D.A., Nasef S., Mahmoud F.F., Jonas D. Expanded Spectrum β–Lactamase Producing Escherichia coli Isolated from Chickens with Colibacillosis in Egypt. Poult. Sci. 2017;96:2375–2384. doi: 10.3382/ps/pew493. [DOI] [PubMed] [Google Scholar]
- 60.Tille P. Bailey & Scott’s Diagnostic Microbiology-E-Book. Elsevier Health Sciences; Amsterdam, The Netherlands: 2015. [Google Scholar]
- 61.Hudzicki J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. Am. Soc. Microbiol. 2009;3189:1–23. [Google Scholar]
- 62.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI supplement M100; Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. [Google Scholar]
- 63.Lee K., Chong Y., Shin H., Kim Y., Yong D., Yum J. Modified Hodge and EDTA-Disk Synergy Tests to Screen Metallo-β-Lactamase-Producing Strains of Pseudomonas and Acinetobactet Species. Clin. Microbiol. Infect. 2001;7:88–91. doi: 10.1046/j.1469-0691.2001.00204.x. [DOI] [PubMed] [Google Scholar]
- 64.Nordmann P., Poirel L., Carrër A., Toleman M.A., Walsh T.R. How to detect NDM-1 producers. J. Clin. Microbiol. 2011;49:718–721. doi: 10.1128/JCM.01773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stürenburg E., Kühn A., Mack D., Laufs R. A novel extended-spectrum β-lactamase CTX-M-23 with a P167T substitution in the active-site omega loop associated with ceftazidime resistance. J. Antimicrob. Chemother. 2004;54:406–409. doi: 10.1093/jac/dkh334. [DOI] [PubMed] [Google Scholar]
- 66.Ghorbani-Dalini S., Kargar M., Doosti A., Abbasi P., Sarshar M. Molecular epidemiology of ESBL genes and multi-drug resistance in diarrheagenic Escherichia coli strains isolated from adults in Iran. Iran. J. Pharm. Res. 2015;14:1257. [PMC free article] [PubMed] [Google Scholar]
- 67.Ellington M.J., Kistler J., Livermore D.M., Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J. Antimicrob. Chemother. 2007;59:321–322. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 68.Gröbner S., Linke D., Schütz W., Fladerer C., Madlung J., Autenrieth I.B., Witte W., Pfeifer Y.J. Emergence of carbapenem-non-susceptible extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tübingen, Germany. J. Med. Microbiol. 2009;58:912–922. doi: 10.1099/jmm.0.005850-0. [DOI] [PubMed] [Google Scholar]
- 69.Poirel L., Héritier C., Tolün V., Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. J. Antimicrob. Agents. Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang Y., Zhou Z., Qian Y., Wei Z., Yu Y., Hu S., Li L. Plasmid-mediated quinolone resistance determinants qnr and aac (6′)-Ib-cr in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 2008;61:1003–1006. doi: 10.1093/jac/dkn063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.