Abstract
Tedizolid and linezolid have antibacterial activity against the most important acute bacterial skin and skin-structure infection (ABSSSIs) pathogens. The objective of this work was to apply PK/PD analysis to evaluate the probability of attaining the pharmacodynamic target of these antimicrobials based on the susceptibility patterns of different clinical isolates causing ABSSSI. Pharmacokinetic and microbiological data were obtained from the literature. PK/PD breakpoints, the probability of target attainment (PTA) and the cumulative fraction of response (CFR) were calculated by Monte Carlo simulation. PTA and CFR are indicative of treatment success. PK/PD breakpoints of tedizolid and linezolid were 0.5 and 1 mg/L, respectively. Probability of treatment success of tedizolid was very high (>90%) for most staphylococci strains, including MRSA and coagulase-negative staphylococci (CoNS). Only for methicillin- and linezolid-resistant S. aureus (MLRSA) and linezolid resistant (LR) CoNS strains was the CFR of tedizolid very low. Except for LR, daptomycin-non-susceptible (DNS), and vancomycin-resistant (VRE) E. faecium isolates, tedizolid also provided a high probability of treatment success for enterococci. The probability of treatment success of both antimicrobials for streptococci was always higher than 90%. In conclusion, for empiric treatment, PK/PD analysis has shown that tedizolid would be adequate for most staphylococci, enterococci, and streptococci, even those LR whose linezolid resistance is mediated by the cfr gene.
Keywords: tedizolid, linezolid, PK/PD, acute bacterial skin and skin-structure infections
1. Introduction
Linezolid, the first commercialized oxazolidinone antibiotic, has activity against a wide variety of Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus [1]. Regardless of the patient’s hepatic or renal function, the authorized dose of linezolid is 600 mg every 12 h. Tedizolid phosphate is the second commercially available oxazolidinone antibiotic, with antibacterial activity against the most important acute bacterial skin and skin-structure infection (ABSSSIs) pathogens, including some linezolid-resistant Staphylococcus aureus and enterococci [2]. Tedizolid, like linezolid, inhibits bacterial protein synthesis by binding to 23S rRNA of the 50S subunit of the ribosome. It has been approved in several countries, including the United States, the European Union, and Canada [3].
Although it is similar to linezolid, tedizolid has a modified side chain at the C-5 position of the oxazolidinone nucleus that confers activity against some linezolid-resistant strains and possesses an optimized C- and D-ring system that increases potency through additional binding site interactions. Tedizolid offers theoretical advantages: on the one hand, it is dosed once daily [4]; on the other hand, it is a more potent protein synthesis inhibitor, enabling it to overcome chloramphenicol-florfenicol resistance mechanisms (cfr gene) [5]. Like linezolid, the dose of tedizolid does not need to be modified in patients with renal impairment, hepatic impairment, on hemodialysis, or whenever switching from intravenous to oral administration [6].
Tedizolid is four- to eightfold more potent in vivo than linezolid against all species of staphylococci, enterococci, and streptococci, including drug-resistant phenotypes such as methicillin-resistant S. aureus (MRSA) and vancomycin-resistant enterococci (VRE) and linezolid-resistant (LR) phenotypes whose linezolid resistance is mediated by the cfr gene [7]. The cfr gene, due to its ubiquity, plays an important role in the spread of drug resistance. In terms of drug resistance mechanism, the cfr gene confers resistance through a non-mutated mechanism, while linezolid resistance is mainly due to gene mutations. Specifically, the cfr gene belongs to the methylated transferases, which can act on the binding site of linezolid and methylate at position 2503 of the 23S rRNA gene, thus making bacteria resistant to linezolid. The cfr gene has been identified in clinical linezolid-resistant Gram-positive bacteria, indicating that the presence of the cfr gene is an important mechanism of bacterial resistance to linezolid [8,9].
In multicenter, randomized, double-blind, and non-inferiority trials, the efficacy and safety of tedizolid and linezolid were compared for the treatment of Gram-positive ABSSSI infections (intravenous tedizolid 200 mg once daily for 6 days, or intravenous linezolid 600 mg twice daily for 10 days) [10,11]. In these studies (Establish-1 and Establish-2), clinical response was high (>70%) and similar with both antibiotics.
Although tedizolid and linezolid have a similar safety profile, tedizolid seems to present a lower incidence of gastrointestinal adverse effects and bone marrow suppression than linezolid [12]. Tolerability in clinically important subpopulations (obese, elderly, renal impairment, hepatic disease/impairment) appears to be comparable to the overall population [13].
In order to preserve both new and older antimicrobials, it is critical to evaluate if the dose administered to the patient is sufficient to provide the necessary exposure for optimal clinical response. The success of the therapy depends on the characteristics of the patient, the pharmacokinetic profile of the antibiotic, and the susceptibility of the microorganism responsible for the infection [14]. In fact, inadequate dose selection is one of the most common reasons why drug development programs fail to achieve New Drug Application (NDA) approval [15].
To maximize the likelihood of a favorable clinical/microbiological response, and to minimize the probability of exposure-related toxicity, pharmacokinetic/pharmacodynamic (PK/PD) modelling represents a very useful tool for dose regimen decision-making. The use of Monte Carlo simulation provides an estimate of the antibiotic dosing regimen’s probability of achieving the targeted pharmacodynamic exposure, given uncertainty in patient pharmacokinetics and the minimum inhibitory concentration (MIC) distribution of the bacterial population [16]. The study of PK/PD is an iterative process whereby pre-clinical (in vitro and in vivo) experiments, population PK models, and in silico simulations are used to investigate potential dosing regimens and PK/PD targets. These studies can be used to inform regimen selection for clinical studies [17]. In the field of antibacterial agents, the guidelines of the European Medicine Agency (EMA, Amsterdam, The Netherlands) and its Committee for Medicinal Products for Human Use (CHMP, London, UK) on the evaluation of medicinal products include PK/PD studies [18].
PK/PD analysis integrates information about the concentration of the drug that reaches the infection site and induces the therapeutic response, and the susceptibility of the pathogen against the antibiotic, expressed as the MIC. The quantitative relationship between a pharmacokinetic parameter and a microbiological parameter is known as a PK/PD index. For tedizolid and linezolid, the PK/PD index that best correlates with efficacy is the ratio of the area under the free drug concentration–time curve at steady state over 24 h to the MIC (fAUC24/MIC). The magnitude required for antimicrobial efficacy has been established as 3 for tedizolid and 80 for linezolid [19,20,21].
In this work, we evaluated by PK/PD analysis and Monte Carlo simulation the activity of tedizolid and compared it to linezolid, with the objective of predicting the probability of treatment success considering the susceptibility profile of staphylococci, enterococci, and streptococci reported in Europe and the United States.
2. Results
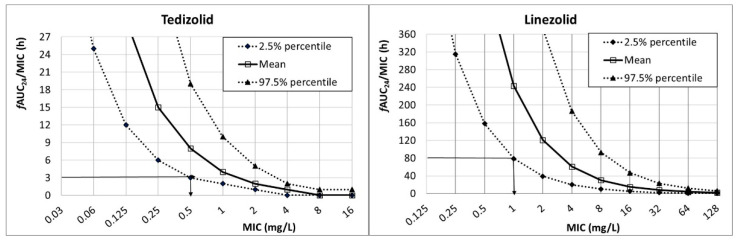
Figure 1 shows the fAUC24/MIC for tedizolid (200 mg q24h) and linezolid (600 mg q12). The PK/PD breakpoint, which is the highest MIC value at which there is a high probability of target attainment, can be read directly from the figure at the intersection of the horizontal line at the PK/PD target and the lower limit of the 95% confidence interval (2.5% percentile). For tedizolid (PK/PD target: fAUC24/MIC ≥ 3), the PK/PD breakpoint was 0.5 mg/L, and for linezolid (PK/PD target: fAUC24/MIC ≥ 80) it was 1 mg/L.
Figure 1.
Relationship between fAUC24/MIC and MIC for tedizolid (200 mg q24h) and linezolid (600 mg q12h).
Table 1 features the PTA values of both antimicrobials for an MIC range from 0.12 to 16 mg/L. PTA is defined as the probability that the specific value of the PK/PD index associated with the efficacy is achieved at a certain MIC value. In other words, it corresponds to the percentage of simulated patients with the estimated PK/PD index equal to or higher than the value related to efficacy against a pathogen with a certain MIC [14]. Simulation results confirm that for the selected targets, standard doses of tedizolid and linezolid cover infections with MIC values up to 0.5 and 1 mg/L, respectively.
Table 1.
Probability of target attainment (PTA) for tedizolid (200 mg q24h) and linezolid (600 mg q12h).
| Probability of Target Attainment, PTA (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC (mg/L) | ||||||||||
| PK/PD Target | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | |
| Tedizolid | fAUC24/MIC ≥ 3 | 100 | 100 | 100 | 97 | 55 | 11 | 1 | 0 | 0 |
| Linezolid | fAUC24/MIC ≥ 80 | 100 | 100 | 100 | 100 | 97 | 62 | 19 | 4 | 0 |
In grey PTA values ≥ 90% (high probability of success).
Table 2, Table 3 and Table 4 show the susceptibility pattern of tedizolid and linezolid for staphylococci, enterococci, and streptococci compiled from different studies who reported MIC data of these microorganisms for the two antibiotics. The tables include resistance phenotype, number of isolates, collection period, and antimicrobial activity. The rates of isolates inhibited at every MIC value were used for Monte Carlo simulations.
Table 2.
Resistance phenotype, number of isolates, collection period, and antimicrobial activity of staphylococci for tedizolid and linezolid.
| Microorganism/Study | Antimicrobial | Resistance Phenotype | Nº Isolates | Collection Period | Percent of Isolates (%) Inhibited at MIC (mg/L) of: | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | >8 | |||||
| S. aureus | ||||||||||||
| Sahm et al. [22] | Tedizolid | 4499 | 2011–2012 | 10.8 | 55.2 | 33.9 | ||||||
| Linezolid | 0.2 | 0.3 | 27.2 | 67.8 | 4.2 | 0.0 | 0.3 | |||||
| Zurenko et al. [23] | Tedizolid | 7187 | 2008–2013 | 7.7 | 62.3 | 29.8 | 0.2 | 0.1 | 0.1 | |||
| Linezolid | 0.1 | 0.3 | 21.6 | 72.8 | 4.9 | 0.0 | 0.2 | |||||
| Rodríguez-Avial et al. [24] | Tedizolid | LR | 5 | 2010–2011 | 100 | |||||||
| Linezolid | 60.0 | 40.0 | ||||||||||
| Pfaller [25] | Tedizolid | 4364 | 2014–2015 | 95.2 | 4.8 | |||||||
| MRSA | ||||||||||||
| Sahm et al. [22] | Tedizolid | 1770 | 2011–2012 | 12.7 | 53.2 | 33.8 | 0.1 | 0.1 | 0.1 | |||
| Linezolid | 0.2 | 0.3 | 30.8 | 65.7 | 2.7 | 0.0 | 0.3 | |||||
| Barber et al. [26] | Tedizolid | hVISA | 120 | Before 2016 | 20.8 | 45.9 | 31.6 | 1.7 | ||||
| VISA | 100 | 59.0 | 25.0 | 16.0 | ||||||||
| DNS | 75 | 30.7 | 50.6 | 18.7 | ||||||||
| LR | 7 | 28.6 | 28.5 | 0 | 42.9 | |||||||
| Peñuelas et al. [27] | Tedizolid | MRSA | 18 | Before 2016 | 11.1 | 61.1 | 27.8 | |||||
| MLRSA | 11.1 | 11.1 | 72.2 | 5.6 | ||||||||
| Linezolid | MRSA | 18 | 38.9 | 61.1 | ||||||||
| MLRSA | 16.7 | 83.3 | ||||||||||
| Pfaller et al. [25] | Tedizolid | 1006 | 2014–2015 | 96.6 | 3.4 | |||||||
| CoNS | ||||||||||||
| Sahm et al. [22] | Tedizolid | 537 | 2011–2012 | 51.4 | 39.1 | 8.6 | 0 | 0 | 0.7 | |||
| Zurenko et al. [23] | Tedizolid | 674 | 2008–2013 | 47.9 | 40.6 | 10.0 | 0.7 | 0.0 | 0.6 | 0.2 | ||
| Linezolid | 0.9 | 23.2 | 61.2 | 12.6 | 1.2 | 0.1 | 0.8 | |||||
| Rodríguez-Avial et al. [24] | Tedizolid | LR | 164 | 2010–2011 | 1.2 | 2.5 | 12.2 | 6.1 | 7.3 | 49.4 | 20.1 | 1.2 |
| Linezolid | LR | 4.3 | 95.7 | |||||||||
| Pfaller et al. [25] | Tedizolid | 729 | 2014–2015 | 98.6 | 1.2 | 0 | 0 | 0.1 | ||||
Light grey: values under the susceptibility range. DNS: daptomycin-non-susceptible; LR: linezolid resistant; hVISA: heterogeneous vancomycin intermediate; VISA: vancomycin intermediate; MRSA: methicillin-resistant S. aureus, MLRSA: methicillin- and linezolid-resistant S. aureus.
Table 3.
Resistance phenotype, number of isolates, collection period, and antimicrobial activity of enterococci for tedizolid and linezolid.
| Microorganism/Study | Antimicrobial | Resistance Phenotype | Nº Isolates | Collection Period | Percent of Isolates (%) Inhibited at MIC (mg/L) of: | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | >8 | |||||
| E. faecalis | ||||||||||||
| Sahm et al. [22] | Tedizolid | 634 | 2011–2012 | 5.8 | 54.8 | 39.1 | 0.1 | 0.2 | ||||
| Barber et al. [26] | Tedizolid | 100 | Before 2016 | 30.0 | 69.0 | 1.0 | ||||||
| Pfaller et al. [25] | Tedizolid | 559 | 2014–2015 | 55.6 | 44.0 | 0.4 | ||||||
| E. faecium | ||||||||||||
| Sahm et al. [22] | Tedizolid | 221 | 2011–2012 | 10.4 | 52.5 | 35.7 | 0.9 | 0.5 | ||||
| Barber et al. [26] | Tedizolid | 120 | Before 2016 | 5.0 | 42.5 | 26.7 | 20.8 | 2.5 | 2.5 | |||
| LR | 10 | 40.0 | 30.0 | 30.0 | ||||||||
| DNS | 25 | 44.0 | 12.0 | 32.0 | 8.0 | 4.0 | ||||||
| Pfaller et al. [25] | Tedizolid | 311 | 2014–2015 | 80.4 | 18.6 | 0.6 | 0.0 | 0.3 | ||||
| VRE | 245 | 79.2 | 20.0 | 0.8 | ||||||||
| VSE | 66 | 84.8 | 13.6 | 0.0 | 1.5 | |||||||
| MIC (mg/L) | ||||||||||||
| Klupp et al. [28] | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | ||||
| Tedizolid | VRE | 30 | 2012–2015 | 20.0 | 60.0 | 10.0 | 3.3 | 6.7 | ||||
| Linezolid | VRE | 26.7 | 20.0 | 20.0 | 33.3 | |||||||
| Enterococcus spp. (predominantly E. faecalis and E. faecium without differentiating species) | MIC (mg/L) | |||||||||||
| ≤0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | >8 | |||||
| Zurenko et al. [23] | Tedizolid | 1241 | 2008–2013 | 5.7 | 50.7 | 42.2 | 1.0 | 0.3 | 0.1 | |||
| Linezolid | 0.4 | 2.7 | 40.7 | 54.4 | 1.4 | 0.1 | 0.3 | |||||
| Other enterococci | ||||||||||||
| Sahm et al. [22] | Tedizolid | 18 | 2011–2012 | 33.3 | 38.9 | 27.8 | ||||||
| Tedizolid | VRE | 163 | 10.4 | 52.2 | 36.2 | 0.6 | 0.6 | |||||
| Linezolid | VRE | 1.2 | 40.5 | 57.2 | 0.5 | 0 | 0.6 | |||||
| Tedizolid | VSE | 705 | 14.2 | 73.2 | 12.6 | |||||||
| Linezolid | VSE | 0.1 | 0.5 | 5.9 | 51.9 | 41.3 | 0.3 | |||||
Light grey: values under the susceptibility range. DNS: daptomycin-non-susceptible; LR: linezolid resistant; VRE: vancomycin resistant; VSE: vancomycin susceptible.
Table 4.
Resistance phenotype, number of isolates, collection period, and antimicrobial activity of streptococci for tedizolid and linezolid.
| Microorganism/Study | Antimicrobial | Resistance Phenotype | Nº Isolates | Collection Period | Percent of Isolates (%) Inhibited at MIC (mg/L) of: | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. pneumoniae | 0.094 | 0.125 | 0.19 | 0.25 | 0.38 | ||||||
| Hipp et al. [29] | Tedizolid | Penicillin-susceptible | 56 | 2009–2016 | 1.8 | 3.6 | 57.1 | 35.7 | 1.8 | ||
| MIC (mg/L) | |||||||||||
| 0.064 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | |||||
| EUCAST [30] | Linezolid | 60,180 | 0.1 | 1.0 | 7.8 | 65.3 | 25.9 | ||||
| MIC (mg/L) | |||||||||||
| Pfaller et al. [25] | 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | ||||
| Tedizolid | 1273 | 2014–2015 | 0.3 | 0.5 | 6.4 | 72.5 | 20.2 | ||||
| Streptococcus spp. | MIC (mg/L) | ||||||||||
| Zurenko et al. a [23] | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | ||||
| Tedizolid | 1600 | 2008–2013 | 5.00 | 44.38 | 50.13 | 0.50 | |||||
| Linezolid | 3.50 | 26.13 | 65.13 | 5.19 | 0.06 | ||||||
| S. anginosus group | |||||||||||
| Zurenko et al. b [23] | Tedizolid | 91 | 2008–2013 | 38.46 | 43.96 | 17.58 | |||||
| Linezolid | 38.46 | 35.16 | 26.37 | ||||||||
| S. viridans group | |||||||||||
| Pfaller et al. c [25] | Tedizolid | 218 | 2014–2015 | 38.53 | 59.17 | 2.29 | |||||
| β-hemolytic streptococci | |||||||||||
| Pfaller et al. d [25] | Tedizolid | 2014–2015 | 93.5 | 4.5 | |||||||
a Includes S. pyogenes, S. agalactiae, S. groups C, F, and G, and S. viridans group; b Includes isolates of S. anginosus, S. intermedius, and S. constellatus; c Includes S. anginosus, S. anginosus group, S. australis, S. constellatus, S. cristatus, S. gordonii, S. infantis, S. lutetiensis, S. massiliensis, S. mitis, S. mitis group, S. mitis/oralis, S. mutans, S. oralis, S. parasanguinis, S. salivarius, S. salivarius group, and S. sanguinis. d Includes S. agalactiae, S. canis, S. dysgalactiae, and S. pyogenes. Light grey: values under the susceptibility range.
Monte Carlo simulation allowed us to estimate the cumulative fraction of response (CFR), defined as the expected population PTA for a specific drug dose and a specific population of microorganisms. This index must be understood as the expected probability of success of a dosing regimen against bacteria in the absence of the specific value of MIC, and thus the population distribution of MICs is used. Table 5 shows the probability of success (expressed as CFR) of tedizolid and linezolid for staphylococci considering the susceptibility rates reported in the different studies. CFR values close to 100% were obtained with tedizolid for S. aureus, and for all strains of MRSA except linezolid-resistant (LR) isolates reported by Barber et al. [26] and methicillin- and linezolid-resistant S. aureus (MLRSA) isolates reported by Peñuelas et al. [27]. CFR with tedizolid was close to 100% for LR CoNS isolates reported by Rodríguez-Avial et al. [24]. Linezolid only provided CFR values higher than 90% for CoNS strains from the study of Zurenko et al. [23].
Table 5.
Probability of success (expressed as CFR or cumulative fraction of response) of tedizolid and linezolid for staphylococci.
| Microorganism/Study | Resistance Phenotype | Probability of Success (CFR, %) | |
|---|---|---|---|
| Tedizolid | Linezolid | ||
| S. aureus | |||
| Sahm et al. [22] | 99 | 69 | |
| Zurenko et al. [23] | 99 | 67 | |
| Rodríguez-Avial et al. [24] | LR | 97 | 3 |
| Pfaller et al. [25] | 100 | n.c. | |
| MRSA | |||
| Sahm et al. [22] | 71 | ||
| Barber et al. [26] | hVISA | 99 | n.c. |
| VISA | 100 | n.c. | |
| DNS | 100 | n.c. | |
| LR | 80 | n.c. | |
| Peñuelas et al. [27] | MRSA | 99 | 75 |
| MLRSA | 62 | n.c. | |
| Pfaller et al. [25] | 100 | n.c. | |
| CoNS | |||
| Sahm et al. [22] | 94 | n.c. | |
| Zurenko [23] | 99 | 93 | |
| Rodríguez-Avial et al. [24] | LR | 20 | 0 |
| Pfaller et al. [25] | 100 | n.c. | |
DNS: daptomycin-non-susceptible; LR: linezolid resistant; hVISA: heterogenous vancomycin intermediate; VISA: vancomycin intermediate; MRSA: methicillin-resistant S. aureus, MLRSA: methicillin- and linezolid-resistant S. aureus. n.c.: not calculated due to lack of MIC data.
Table 6 features the CFR values of tedizolid and linezolid for enterococci. For E. faecalis and other enterococci, tedizolid provided CFR ranging from 98% to 100%. Regarding E. faecium, CFR was higher than 85% except for the daptomycin-non-susceptible (DNS) and vancomycin-resistant (VR) isolates from the study of Barber et al. [26] and VR isolates from the study of Pfaller et al. [25]. CFR values obtained for linezolid was 80% when using the MIC distribution provided by Zurenko et al. [23], who reported the susceptibility rates of enterococci without differentiating species, although E. faecium and E. faecalis were the most represented. When considering the MIC distribution reported by Klupp et al. [28], CFR was 0 for both linezolid and tedizolid. For other enterococci, the CFR obtained for linezolid ranged from 79% to 85%.
Table 6.
Probability of success (expressed as CFR) of tedizolid and linezolid for enterococci.
| Microorganism/Study | Resistance Phenotype | Probability of Success (CFR, %) | |
|---|---|---|---|
| Tedizolid | Linezolid | ||
| E. faecalis | |||
| Sahm et al. [22] | 99 | n.c. | |
| Barber et al. [26] | 100 | n.c. | |
| Pfaller et al. [25] | 100 | n.c. | |
| E. faecium | |||
| Sahm et al. [22] | 99 | n.c. | |
| Barber et al. [26] | 86 | n.c. | |
| LR | 25 | n.c. | |
| DNS | 75 | n.c. | |
| Klupp et al. [28] | VRE | 0 | 0 |
| Pfaller et al. [25] | 100 | n.c. | |
| VRE | 100 | n.c. | |
| VSE | 99 | n.c. | |
| Enterococcus spp. (predominantly E. faecalis and E. faecium) | |||
| Zurenko et al. [23] | 98 | 80 | |
| Other enterococci | |||
| Sahm et al. [22] | 99 | n.c. | |
| VRE | 98 | 79 | |
| VSE | 100 | 85 | |
DNS: daptomycin-non-susceptible; LR: linezolid resistant; VRE: vancomycin resistant; VSE: vancomycin susceptible. n.c.: not calculated due to lack of MIC data.
The CFR values of both antimicrobials for streptococci are presented in Table 7. Irrespective of the microorganism, the CFR of tedizolid was always 100%, and the CFR of linezolid varied from 91% to 98%.
Table 7.
Probability of success (expressed as CFR or cumulative fraction of response) of tedizolid and linezolid for streptococci.
| Microorganism/Study | Resistance Phenotype | Probability of Success (CFR, %) | |
|---|---|---|---|
| Tedizolid | Linezolid | ||
| S. pneumoniae | |||
| Hipp et al. [29] | Penicillin-susceptible | 100 | n.c. |
| EUCAST et al. [30] | 91 | ||
| Pfaller [25] | 100 | n.c. | |
| Streptococcus spp. | |||
| Zurenko et al. [23] | 100 | 98 | |
| S. anginosus group | |||
| Zurenko et al. [23] | 100 | 98 | |
| S. viridans group | |||
| Pfaller et al. [25] | 100 | n.c. | |
| β-hemolytic streptococci | |||
| Pfaller et al. [25] | 100 | n.c. | |
n.c.: not calculated due to lack of MIC data.
3. Discussion
Linezolid, the first representative member of the oxazolidinone family introduced into the pharmaceutical market, shows excellent activity; however, in recent years, resistance to microorganisms (mainly MRSA) has emerged [31]. Due to the limited number of therapeutic options available for ABSSSIs, new therapeutic alternatives have been developed, especially for resistant Gram-positive microorganisms. In fact, the development of new generations of antimicrobials is one of the most accepted strategies to mitigate the current and future impact of antimicrobial resistance. One of the new antibiotics developed to treat ABSSSIs is tedizolid, which has greater potency, a better spectrum of activity, and a lower resistance profile [2].
PK/PD analysis and Monte Carlo simulation have proved to be very useful tools to select adequate antibiotic treatments with the goal of increasing efficacy and reducing the risk of selecting multidrug-resistant isolates [32]. The use of PK/PD analyses can ameliorate this risk and improve the likelihood of selecting an effective dose regimen, thereby increasing the likelihood of success. These tools have also been applied to identify changes in the antimicrobial activity of antibiotics, providing complementary information to the simple assessment of MIC values [33,34,35], and to establish PK/PD breakpoints [14] as well.
In this work, we calculated the PK/PD breakpoint of tedizolid and linezolid based on the likelihood of obtaining a targeted exposure. PK/PD breakpoints can be estimated as the highest MIC value at which a high probability of target attainment is obtained (PTA ≥ 90%). Another option to estimate the breakpoints is by graphical representation of the PK/PD index as a function of the MIC [36], which provides a much more restrictive PK/PD breakpoint. In our study, the PK/PD breakpoint for tedizolid was 0.5 mg/L, and it was 1 mg/L for linezolid. For S. aureus, the tedizolid PK/PD breakpoint matched the epidemiologic cutoff value ECOFF—that is, the highest MIC for organisms devoid of phenotypically detectable acquired resistance mechanisms [21]. At present, there is no clinical nor ECOFF breakpoint defined for enterococci, so the PK/PD breakpoint may be especially useful when considering this antibiotic as a treatment option. An important advantage of PK/PD breakpoints, as opposed to the clinical ones, is that they are species-independent. In addition, they consider the antibiotic exposure—that is, they are calculated for a specific dosage regimen, and therefore they help to optimize the selection of doses. Regarding streptococci, tedizolid clinical breakpoints have only been defined for β-hemolytic groups A, B, C, and G streptococci and the S. anginosus group. For these species, the PK/PD breakpoint agrees with the current clinical breakpoint defined at 0.5 mg/L. Regarding linezolid, the PK/PD breakpoint we calculated (1 mg/L) is lower than the clinical breakpoints (4 mg/L for staphylococci and enterococci, and 2 mg/L for streptococci). However, according to the linezolid-EUCAST rationale document for clinical breakpoints [20], Monte Carlo simulations and target attainment rates for 600 mg twice a day support a susceptible breakpoint of less than 1 or 2 mg/L, which is in agreement with our PK/PD breakpoint. According to Mouton [37], the use of pharmacokinetic parameters from different populations in Monte Carlo simulations for established dosing regimens results in different breakpoints. When discrepancies in breakpoints are observed, the PK/PD breakpoints are generally lower than those defined by the CLSI or EUCAST [32], as seen in the present study. Consequently, and compared to tedizolid, the qualification of an isolate as susceptible for linezolid according to the MIC value and the clinical breakpoint may be less useful in guiding therapy.
The highest MIC values that provided a PTA value ≥90% (0.5 mg/L for tedizolid and 1 mg/L for linezolid, see Table 1) agree with the PK/PD breakpoints considering the EUCAST approach [36]. According to our results, a high probability of treatment success is expected with tedizolid if infection is due to microorganisms with MIC ≤ 0.5 mg/L. Looking through the MIC values reported in Europe and the USA, irrespective of the microorganism (Table 2, Table 3 and Table 4), most isolates would be covered by the standard dose of tedizolid. In the case of linezolid, a high probability of treatment success would be achieved for MIC ≤ 1 mg/L.
CFR is an index that estimates the probability of target attainment for an MIC distribution, and it is very useful to predict the probability of treatment success when applied empirically. For staphylococci (Table 5), tedizolid was adequate for most isolates, including LR, heterogenous vancomycin intermediate (hVISA), vancomycin intermediate (VISA), CoNS, and MRSA. Delpech et al. [38] demonstrated that tedizolid remains active against staphylococci strains harboring the cfr gene, probably due the sterically compact nature of the hydroxymethyl group of this drug. The potential spread of cfr-mediated linezolid resistance in S. aureus makes tedizolid very useful for treating infections due to multidrug-resistant Gram-positive pathogens. Only for MLRSA reported by Peñuelas et al. [27] (18 isolates) and LR CoNS isolates reported by Rodríguez-Avial et al. [24] (164 isolates), the probability of treatment success is low (62% and 20%, respectively). The genetic basis for linezolid resistance in isolates reported by Rodríguez-Avial et al. [24] was not studied, but the susceptibility rates and CFR value indicate that resistance to tedizolid was not related to the cfr gene.
Regarding enterococci (Table 6), tedizolid was adequate for E. faecalis, regardless of the MIC provided by the different studies (CFR or probability of treatment success of 99–100%). The same is applicable to other enterococci. For E. faecium, tedizolid also provides a high probability of treatment success, except for the LR and DNS isolates reported by Barber et al. [26] and the VRE isolates reported by Klupp et al. [28], in which the cfr gene was not present. In fact, resistance associated with the cfr gene is not predominant in enterococci; alterations in 23S rRNA remain the main oxazolidinone-resistance mechanism in E. faecium, while the optrA gene prevails in E. faecalis [39,40]. Tedizolid represents a therapeutic option only for a limited subset of LR-VRE strains [28]. Regarding linezolid, only Sahm et al. [22] and Zurenko et al. [23] provide susceptibility rates for enterococci, and according to them, linezolid provides a moderate probability of treatment success when used for infections due to enterococci (even VRE and VSE).
Finally, both tedizolid and linezolid are expected to provide a high probability of treatment success when used for infections due to streptococci (Table 7). These results were expected due to the high susceptibility profile of streptococci to oxazolidinones.
Our results are based on simulations, and a number of considerations are assumed. A previous work [41] reports the main limitations of PK/PD analysis and Monte Carlo simulation. First, pharmacokinetic parameters in the specific population must be used since it is known that pathophysiological conditions affect the distribution and elimination of antimicrobials. In our study, the CL of tedizolid was obtained from pooled data from seven densely and sparsely sampled clinical trials, most subjects were healthy, and the CL of linezolid used was from healthy subjects. Second, pharmacokinetic equations used to estimate drug exposure are usually simple models that facilitate calculations. Third, the simulations in our work were based on serum pharmacokinetics, and therefore the results are mainly applicable to bloodstream infections. Moreover, PK/PD modelling is based on prior studies that have identified correlations between PK/PD indices and health outcomes. These relationships have typically been derived from animal models; however, a good correlation and validation between animal models and the clinic has been established.
4. Materials and Methods
This study was performed in different steps: (1) acquisition of pharmacokinetic parameters, PK/PD targets, and susceptibility data; and (2) PK/PD analysis and Monte Carlo simulation.
4.1. Pharmacokinetic Parameters, PK/PD Targets, and Susceptibility Data
The PK parameters of linezolid and tedizolid, as well as the PK/PD targets, were obtained from published studies [19,20,21,42,43]. These data are presented in Table 8.
Table 8.
Pharmacokinetic parameters and PK/PD targets of tedizolid and linezolid used for Monte Carlo simulations. CL data are expressed as mean ± standard deviation.
| Antibiotic | Dosing Regimen |
CL (L/h) | Fu | PK/PD Target | References |
|---|---|---|---|---|---|
| Tedizolid | 200 mg q24h | 6.69 ± 2.07 | 0.10 | fAUC24/MIC ≥ 3 | [19,21,42] |
| Linezolid | 600 mg q12h | 4.80 ± 1.74 | 0.69 | fAUC24/MIC ≥ 80 | [19,20,43] |
CL: total body clearance; Fu: unbound fraction; fAUC24: area under the unbound concentration–time curve over a period of 24 h.
The CL value of tedizolid was obtained with a population PK model developed with data from four phase 1 studies, one phase 2 study, and three phase 3 studies, in which tedizolid was administered by either intravenous or oral route. The CL of linezolid is for healthy volunteers.
The antimicrobial activity of linezolid and tedizolid for staphylococci, enterococci, and streptococci were obtained from the literature [22,23,24,25,26,27,28,29,30]. We selected those studies that provided MIC values for both antibiotics. Clinical isolates from Europe and the United States were collected between 2011 and 2016. Table 2, Table 3 and Table 4 show the MIC values of staphylococci, enterococci, and streptococci, respectively, obtained from the abovementioned studies. The MIC distribution of the isolates from the different studies was used for the PK/PD analysis.
4.2. Pharmacokinetic/Pharmacodynamic Analysis and Monte Carlo Simulation
Monte Carlo simulation is an advanced statistical modeling tool that makes it possible to expand the sample size considering the variability of the PK and PD parameters on the estimation of the PK/PD indices in order to provide predictions of the likely result of different therapeutic approaches, or the achievement of therapeutic targets [14]. A 10,000-subject Monte Carlo simulation was conducted for each antibiotic using Oracle® Crystal Ball Fusion Edition v.11.1.2.3.500 (Oracle USA Inc., Redwood City, CA). The values of fAUC24/MIC (the ratio of the area under the free drug concentration–time curve at steady state over 24 h to MIC) were calculated for tedizolid (200 mg q24h) and linezolid (600 mg q12h) over an MIC range of serial two-fold dilutions from 0.06 to 16 mg/L and from 0.06 to 128 mg/L, respectively. The following equation was used [32]:
| fAUC24/MIC = D × Fu/CL | (1) |
where D is the daily dose, Fu is the unbound fraction, and CL is the total clearance.
For simulations, a log-normal distribution was assumed for CL, according to statistical criteria. Unbound fraction was included as a fixed value [44].
For every MIC value and considering the variability of the CL, the output of the simulation consisted of a probability distribution, and the mean value and the 95% CI (expressed as percentiles) of the fAUC24/MIC were extracted. The PK/PD breakpoint, considered as the highest MIC value at which fAUC24/MIC is ≥3 or ≥80 for tedizolid and linezolid, respectively, were estimated. According to EUCAST, the PK/PD breakpoint was obtained from the lower limit of the 95% CI (2.5% percentile) [36].
The PTA [45], defined as the probability that a specific value of a PK/PD index associated with the efficacy of the antimicrobial treatment is achieved at a certain MIC, was estimated. PTA ≥80% but <90% was associated with moderate probabilities of success, whereas PTA ≥ 90% was considered as optimal against that bacterial population [32].
CFR values, which allowed us to calculate the probability of success of an empiric treatment, were calculated considering the PTA for each MIC value and the bacterial population MIC distribution, according to the following equation [45]:
| (2) |
where i indicates the MIC category, PTAi is the PTA of each MIC category, and Fi is the fraction of the microorganism population in each MIC category. As with PTA, CFR ≥80% but <90% was associated with moderate probabilities of success, whereas CFR ≥ 90% was considered as optimal against that bacterial population [32].
5. Conclusions
In conclusion, PK/PD breakpoints calculated for tedizolid and linezolid were 0.5 and 1 mg/L, respectively. For empiric treatment, tedizolid seems to be adequate for the treatment of infections due to most staphylococci, enterococci, and streptococci. For LR isolates not related to the cfr gene, tedizolid does not guarantee efficacy. This study confirms the importance of considering the susceptibility profile of the geographical area or hospital setting and the PK/PD analysis to guide empiric therapy.
Author Contributions
Conceptualization, A.R.-G. and A.C.-B.; data curation, A.R.-G., and A.C.-B.; investigation, A.R.-G., A.A.-Q., M.A.S.A., and A.C.-B.; methodology, A.R.-G., A.A.-Q., M.A.S.A., and A.C.-B.; resources, A.A.-Q. and A.C.-B.; software, A.R.-G. and A.A.-Q.; supervision, M.A.S.A.; writing—original draft, A.R.-G., A.A.-Q., M.A.S.A., and A.C.-B.; writing—review and editing, A.R.-G., A.A.-Q., M.A.S.A. and A.C.-B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of the Basque Country UPV/EHU (GIU17/032).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are applicable in the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.MacGowan A.P. Pharmacokinetic and pharmacodynamic profile of linezolid in healthy volunteers and patients with Gram-positive infections. J. Antimicrob. Chemother. 2003;51:ii17–ii25. doi: 10.1093/jac/dkg248. [DOI] [PubMed] [Google Scholar]
- 2.Carena A.A., Stryjewski M.E. Tedizolid (torezolid) for the treatment of complicated skin and skin structure infections. Expert Rev. Clin. Pharmacol. 2020;13:577–592. doi: 10.1080/17512433.2020.1774362. [DOI] [PubMed] [Google Scholar]
- 3.Singh K.V., Arias C.A., Murray B.E. Efficacy of Tedizolid against Enterococci and Staphylococci, Including cfr + Strains, in a Mouse Peritonitis Model. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.02627-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burdette S.D., Trotman R. Tedizolid: The First Once-Daily Oxazolidinone Class Antibiotic. Clin. Infect. Dis. 2015;61:1315–1321. doi: 10.1093/cid/civ501. [DOI] [PubMed] [Google Scholar]
- 5.Bouza E., Muñoz P., Burillo A. The role of tedizolid in skin and soft tissue infections. Curr. Opin. Infect. Dis. 2018;31:131–140. doi: 10.1097/QCO.0000000000000439. [DOI] [PubMed] [Google Scholar]
- 6.European Medicines Agency Sivextro, EPAR—Product Information. [(accessed on 14 June 2021)]; Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/002846/WC500184802.pdf.
- 7.Zhanel G.G., Love R., Adam H., Golden A., Zelenitsky S., Schweizer F., Gorityala B., Lagacé-Wiens P.R.S., Rubinstein E., Walkty A., et al. Tedizolid: A Novel Oxazolidinone with Potent Activity Against Multidrug-Resistant Gram-Positive Pathogens. Drugs. 2015;75:253–270. doi: 10.1007/s40265-015-0352-7. [DOI] [PubMed] [Google Scholar]
- 8.Liu B.G., Yuan X.L., He D.D., Hu G.Z., Miao M.S., Xu E.P. Research progress on the oxazolidinone drug linezolid re-sistance. Eur. Rev. Med. Pharmacol. Sci. 2020;24:9274–9281. doi: 10.26355/eurrev_202009_23009. [DOI] [PubMed] [Google Scholar]
- 9.Rodríguez-Lucas C., Rodicio M., Càmara J., Domínguez M., Alaguero M., Fernández J. Long-term endemic situation caused by a linezolid- and meticillin-resistant clone of Staphylococcus epidermidis in a tertiary hospital. J. Hosp. Infect. 2020;105:64–69. doi: 10.1016/j.jhin.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Prokocimer P., De Anda C., Fang E., Mehra P., Das A. Tedizolid phosphate vs. linezolid for treatment of acute bacterial skin and skin structure infections: The ESTABLISH-1 randomized trial. JAMA. 2013;309:559–569. doi: 10.1001/jama.2013.241. [DOI] [PubMed] [Google Scholar]
- 11.Moran G.J., Fang E., Corey G.R., Das A.F., De Anda C., Prokocimer P. Tedizolid for 6 days versus linezolid for 10 days for acute bacterial skin and skin-structure infections (ESTABLISH-2): A randomised, double-blind, phase 3, non-inferiority trial. Lancet Infect. Dis. 2014;14:696–705. doi: 10.1016/S1473-3099(14)70737-6. [DOI] [PubMed] [Google Scholar]
- 12.Lan S.-H., Lin W.-T., Chang S.-P., Lu L.-C., Chao C.-M., Lai C.-C., Wang J.-H. Tedizolid Versus Linezolid for the Treatment of Acute Bacterial Skin and Skin Structure Infection: A Systematic Review and Meta-Analysis. Antibiotics. 2019;8:137. doi: 10.3390/antibiotics8030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardalo C., Lodise T.P., Bidell M., Flanagan S., De Anda C., Anuskiewicz S., Prokocimer P. Clinical safety and tolerability of tedizolid phosphate in the treatment of acute bacterial skin and skin structure infections. Expert Opin. Drug Saf. 2018;17:359–367. doi: 10.1080/14740338.2018.1446939. [DOI] [PubMed] [Google Scholar]
- 14.Asín-Prieto E., Rodríguez-Gascón A., Isla A. Applications of the pharmacokinetic/pharmacodynamic (PK/PD) analysis of antimicrobial agents. J. Infect. Chemother. 2015;21:319–329. doi: 10.1016/j.jiac.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Lodise T.P., Drusano G.L. Use of Pharmacokinetic/Pharmacodynamic Systems Analyses to Inform Dose Selection of Tedizolid Phosphate. Clin. Infect. Dis. 2014;58:S28–S34. doi: 10.1093/cid/cit615. [DOI] [PubMed] [Google Scholar]
- 16.Bradley J.S., Dudley M.N., Drusano G.L. Predicting efficacy of antiinfectives with pharmacodynamics and Monte Carlo simulation. Pediatr. Infect. Dis. J. 2003;22:982–992. doi: 10.1097/01.inf.0000094940.81959.14. [DOI] [PubMed] [Google Scholar]
- 17.McAleenan A., Ambrose P.G., Bhavnani S.M., Drusano G.L., Hope W.W., Mouton J.W., Higgins J.P.T., MacGowan A.P. Methodological features of clinical pharmacokinetic–pharmacodynamic studies of antibacterials and antifungals: A systematic review. J. Antimicrob. Chemother. 2020;75:1374–1389. doi: 10.1093/jac/dkaa005. [DOI] [PubMed] [Google Scholar]
- 18.EMA-CHMP . Guideline on the Use of Pharmacokinetics and Pharmacodynamics in the Development of Antimicrobial Medicinal Products. European Medicines Agency; London, UK: 2016. EMA/CHMP/594085/2015. [Google Scholar]
- 19.Flanagan S., Passarell J., Lu Q., Fiedler-Kelly J., Ludwig E., Prokocimer P. Tedizolid Population Pharmacokinetics, Exposure Response, and Target Attainment. Antimicrob. Agents Chemother. 2014;58:6462–6470. doi: 10.1128/AAC.03423-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linezolid: Rationale for the EUCAST Clinical Breakpoints, Version 1.0. [(accessed on 14 June 2021)];2005 Nov 18; Available online: http://www.eucast.org.
- 21.Tedizolid: Rationale for EUCAST Clinical Breakpoints, Version 1.0. [(accessed on 14 June 2021)];2018 Apr 19; Available online: http://www.eucast.org.
- 22.Sahm D.F., Deane J., Bien P.A., Locke J.B., Zuill D.E., Shaw K.J., Bartizal K.F. Results of the Surveillance of Tedizolid Activity and Resistance Program: In vitro susceptibility of Gram-positive pathogens collected in 2011 and 2012 from the United States and Europe. Diagn. Microbiol. Infect. Dis. 2015;81:112–118. doi: 10.1016/j.diagmicrobio.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Zurenko G., Bien P., Bensaci M., Patel H.N., Thorne G. Use of linezolid susceptibility test results as a surrogate for the susceptibility of Gram-positive pathogens to tedizolid, a novel oxazolidinone. Ann. Clin. Microbiol. Antimicrob. 2014;13:46. doi: 10.1186/s12941-014-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez-Avial I., Culebras E., Betriu C., Morales G., Pena I., Picazo J.J. In vitro activity of tedizolid (TR-700) against linezolid-resistant staphylococci. J. Antimicrob. Chemother. 2011;67:167–169. doi: 10.1093/jac/dkr403. [DOI] [PubMed] [Google Scholar]
- 25.Pfaller M., Sader H.S., Shortridge D., Castanheira M., Flamm R.K., Mendes R.E. Activity of tedizolid against gram-positive clinical isolates causing infections in Europe and surrounding areas (2014–2015) J. Chemother. 2019;31:188–194. doi: 10.1080/1120009X.2019.1609740. [DOI] [PubMed] [Google Scholar]
- 26.Barber K.E., Smith J.R., Raut A., Rybak M.J. Evaluation of tedizolid against Staphylococcus aureus and enterococci with reduced susceptibility to vancomycin, daptomycin or linezolid. J. Antimicrob. Chemother. 2016;71:152–155. doi: 10.1093/jac/dkv302. [DOI] [PubMed] [Google Scholar]
- 27.Peñuelas M., Candel F.J., Lejarraga C., Gonzalez L.L., Viñuela-Prieto J.M., De Mendoza D.L. Activity of linezolid and tedizolid against clinical isolates of methicillin-resistant and methicillin and linezolid resistant Staphylococcus aureus: An in vitro comparison. Rev. Espanola Quimioter. Publicacion Soc. Espanola Quimioter. 2016;29:255–258. [PubMed] [Google Scholar]
- 28.Klupp E.-M., Both A., Campos C.B., Büttner H., König C., Christopeit M., Christner M., Aepfelbacher M., Rohde H. Tedizolid susceptibility in linezolid- and vancomycin-resistant Enterococcus faecium isolates. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:1957–1961. doi: 10.1007/s10096-016-2747-0. [DOI] [PubMed] [Google Scholar]
- 29.Hipp M., Burckhardt I. In vitro activity of newer antimicrobials against penicillin non-susceptible strains of Streptococcus pneumoniae. Infect. Drug Resist. 2019;12:1889–1893. doi: 10.2147/IDR.S202789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The European Committee on Antimicrobial Susceptibility Testing—EUCAST Clinical Breakpoints-Bacteria (v 11.0) [(accessed on 14 June 2021)];2021 Jan 1; Available online: https://www.eucast.org/clinical_breakpoints/
- 31.Grau S., Ferrández O., Urbina O., Espona M., Salas E., Ferrández I. Potential role of tedizolid phosphate in the treatment of acute bacterial skin infections. Drug Des. Dev. Ther. 2013;7:243–265. doi: 10.2147/DDDT.S30728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asín E., Isla A., Canut A., Gascón A.R. Comparison of antimicrobial pharmacokinetic/pharmacodynamic breakpoints with EUCAST and CLSI clinical breakpoints for Gram-positive bacteria. Int. J. Antimicrob. Agents. 2012;40:313–322. doi: 10.1016/j.ijantimicag.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Ibar-Bariain M., Rodríguez-Gascón A., Isla A., Solinís M.Á., Canut-Blasco A. Application of pharmacokinetic/pharmacodynamic analysis to evaluate the adequacy of antimicrobial therapy for pediatric acute otitis media in Spain before and after the introduction of the PCV7 vaccine. Rev. Esp. Quimioter. Publ. Soc. Esp. Quimioter. 2019;32:121–129. [PMC free article] [PubMed] [Google Scholar]
- 34.Valero A., Isla A., Rodríguez-Gascón A., Calvo B., Canut A., Solinís M.Á. Pharmacokinetic/pharmacodynamic analysis as a tool for surveillance of the activity of antimicrobials against Pseudomonas aeruginosa strains isolated in critically ill patients. Enferme. Infecc. Microbiol. Clín. 2019;37:380–386. doi: 10.1016/j.eimc.2018.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Zelenitsky S.A., Rubinstein E., Ariano R.E., Zhanel G.G., Hoban D.J., Adam H.J., Karlowsky J.A., Baxter M.R., Nichol K.A., Lagacé-Wiens P.R.S., et al. Integrating pharmacokinetics, pharmacodynamics and MIC distributions to assess changing antimicrobial activity against clinical isolates of Pseudomonas aeruginosa causing infections in Canadian hospitals (CANWARD) J. Antimicrob. Chemother. 2013;68:i67–i72. doi: 10.1093/jac/dkt028. [DOI] [PubMed] [Google Scholar]
- 36.Mouton J., Brown D., Apfalter P., Cantón R., Giske C., Ivanova M., MacGowan A., Rodloff A., Soussy C.-J., Steinbakk M., et al. The role of pharmacokinetics/pharmacodynamics in setting clinical MIC breakpoints: The EUCAST approach. Clin. Microbiol. Infect. 2012;18:E37–E45. doi: 10.1111/j.1469-0691.2011.03752.x. [DOI] [PubMed] [Google Scholar]
- 37.Mouton J.W. Setting clinical MIC breakpoints from a PK/PD point of view: It is the dose that matters. In: Vinks A.A., Derendorf H., Mouton J.W., editors. Fundamentals of Antimicrobial Pharmacokinetics and Pharmacodynamics. Springer; New York, NY, USA: 2014. pp. 45–61. [Google Scholar]
- 38.Delpech P., Aleryan M., Jones B., Gemmell C., Lang S. An in vitro evaluation of the efficacy of tedizolid: Implications for the treatment of skin and soft tissue infections. Diagn. Microbiol. Infect. Dis. 2018;91:93–97. doi: 10.1016/j.diagmicrobio.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Deshpande L.M., Castanheira M., Flamm R.K., Mendes R. Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: Results from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 2018;73:2314–2322. doi: 10.1093/jac/dky188. [DOI] [PubMed] [Google Scholar]
- 40.Moure Z., Lara N., Marín M., Sola-Campoy P.J., Bautista V., Gómez-Bertomeu F., Gómez-Dominguez C., Pérez-Vázquez M., Aracil B., Campos J., et al. Interregional spread in Spain of linezolid-resistant Enterococcus spp. isolates carrying the optrA and poxtA genes. Int. J. Antimicrob. Agents. 2020;55:105977. doi: 10.1016/j.ijantimicag.2020.105977. [DOI] [PubMed] [Google Scholar]
- 41.Frei C.R., Wiederhold N., Burgess D.S. Antimicrobial breakpoints for Gram-negative aerobic bacteria based on pharmacokinetic–pharmacodynamic models with Monte Carlo simulation. J. Antimicrob. Chemother. 2008;61:621–628. doi: 10.1093/jac/dkm536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rybak J.M., Marx K., Martin C.A. Early Experience with Tedizolid: Clinical Efficacy, Pharmacodynamics, and Resistance. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2014;34:1198–1208. doi: 10.1002/phar.1491. [DOI] [PubMed] [Google Scholar]
- 43.Dryden M.S. Linezolid pharmacokinetics and pharmacodynamics in clinical treatment. J. Antimicrob. Chemother. 2011;66:iv7–iv15. doi: 10.1093/jac/dkr072. [DOI] [PubMed] [Google Scholar]
- 44.Owens R.C., Jr., Bhavnani S.M., Ambrose P.G. Assessment of pharmacokinetic-pharmacodynamic target attainment of gemifloxacin against Streptococcus pneumoniae. Diagn Microbiol. Infect Dis. 2005;51:45–49. doi: 10.1016/j.diagmicrobio.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 45.Mouton J.W., Dudley M.N., Cars O., Derendorf H., Drusano G.L. Standardization of pharmacokinetic/pharmacodynamic (PK/PD) terminology for anti-infective drugs: An update. J. Antimicrob. Chemother. 2005;55:601–607. doi: 10.1093/jac/dki079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are applicable in the paper.