Abstract
Acinetobacter baumannii has become a serious threat to human health due to its extreme antibiotic resistance, environmental persistence, and capacity to survive within the host. Two A. baumannii strains, A118 and AB5075, commonly used as model systems, and three carbapenem-resistant strains, which are becoming ever more dangerous due to the multiple drugs they can resist, were exposed to 3.5% human serum albumin (HSA) and human serum (HS) to evaluate their response with respect to antimicrobial resistance, biofilm formation, and quorum sensing, all features responsible for increasing survival and persistence in the environment and human body. Expression levels of antibiotic resistance genes were modified differently when examined in different strains. The cmlA gene was upregulated or downregulated in conditions of exposure to 3.5% HSA or HS depending on the strain. Expression levels of pbp1 and pbp3 tended to be increased by the presence of HSA and HS, but the effect was not seen in all strains. A. baumannii A118 growing in the presence of HS did not experience increased expression of these genes. Aminoglycoside-modifying enzymes were also expressed at higher or lower levels in the presence of HSA or HS. Still, the response was not uniform; in some cases, expression was enhanced, and in other cases, it was tapered. While A. baumannii AB5075 became more susceptible to rifampicin in the presence of 3.5% HSA or HS, strain A118 did not show any changes. Expression of arr2, a gene involved in resistance to rifampicin present in A. baumannii AMA16, was expressed at higher levels when HS was present in the culture medium. HSA and HS reduced biofilm formation and production of N-Acyl Homoserine Lactone, a compound intimately associated with quorum sensing. In conclusion, HSA, the main component of HS, stimulates a variety of adaptative responses in infecting A. baumannii strains.
Keywords: Acinetobacter baumannii, antibiotic resistance, quorum sensing, biofilm, human serum, albumin
1. Introduction
Infections caused by antibiotic-resistant bacteria have increased in frequency, resulting in significant patient morbidity and mortality [1,2]. Acinetobacter baumannii is a multidrug resistant (MDR) nosocomial pathogen that causes a variety of infections, many of them life-threatening, such as pneumonia, bacteremia, meningitis, and wound infections. A. baumannii strains resistant to carbapenems (CRAB) have frequently been reported [3], and this characteristic warranted the acknowledgemnt of CRAB as an “urgent threat” by the Centers for Disease Control and Prevention (CDC) in the 2019 Antibiotic Resistance Threats Report [4,5].
Few active antimicrobials are left to treat CRAB infections [6,7,8]. A laboratory- and population-based surveillance showed that most of the CRAB isolates also presented non-susceptibility to antibiotics generally considered “second-line” agents for treatment [9,10]. For example, ~94% demonstrated resistance against fluoroquinolones, both for levofloxacin and ciprofloxacin. Around 80% of the isolates possessed extendeded-spectrum β-lactamases (ESBLs), with 97% resistant to piperacillin/tazobactam and between 84 and 88% resistant to cefepime and ceftazidime. In contrast, tigecycline and colistin demonstrated resistance rates of approximately 40% and 7%, respectively [9,10]. However, these agents have significant limitations, such as a narrow therapeutic range (colistin, tigecycline), insufficient levels of free antibiotic in blood (colistin, tigecycline), significant adverse effects (colistin, tigecycline), and limited or no bactericidal activity (tigecycline) [10,11,12]. It is worth noting that there is only one orally administered antibiotic, minocycline, currently in use to treat CRAB infections. This not only limits treatment options but also complicates therapy by the need for hospitalization [10,12].
In addition to resistance, A. baumannii possesses intrinsic versatility in response to changing environments, within and outside the human host, and to acquire or change the expression of resistance and virulence genes [13,14,15,16,17,18,19]. When exposed to different human fluids or proteins, A. baumannii can modify the expression of a wide variety of genes that are needed to persist and survive [15,16,19,20,21,22]. Also, antibiotics can trigger DNA acquisition and affect the expression of resistance genes (Le et al., 2021 submitted manuscript [23,24,25]).
We recently demonstrated that human serum albumin (HSA) increases natural transformation as well as expression of competence associated and carbapenem resistance genes in both susceptible and resistant strains (Le et al., 2021 submitted manuscript). Cells exposed to HSA in combination with meropenem increased their capability to uptake DNA. This mix was also associated to enhanced expression of competence-associated genes. Similar changes were induced in cells cultured in the presence of human serum (HS) (Le et al., 2021 submitted manuscript). All these features can contribute to the increase of antibiotic resistance. To further understand how A. baumannii responds to HSA and human serum (HS), we focused on non-carbapenem antibiotic resistance genes, quorum sensing, and biofilm formation.
2. Results and Discussion
2.1. HSA and HS Modulate the Expression of Antibiotic Resistance Genes from Different Antibiotic Families
To further evaluate the role of HSA and HS in the expression level of antibiotic resistance genes, we performed quantitative RT-PCR (qRT-PCR) assays using total RNA extracted from two A. baumannii model strains (A118 and AB5075), and three carbapenem-resistant strains (AB0057, AMA16, and ABUH702). Cells were cultured in LB, LB supplemented with 3.5% HSA, or cultured in 100% pooled normal HS.
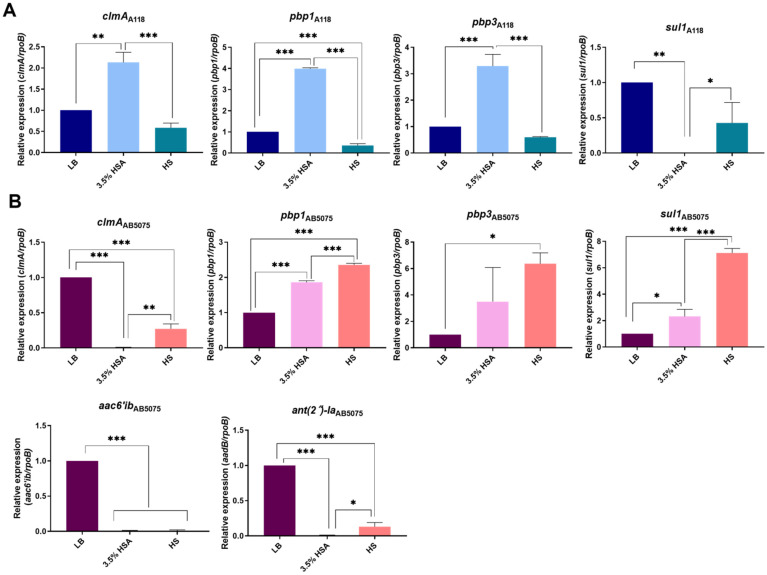
A. baumannii A118 showed an increase in expression of the chloramphenicol resistance clmA gene [26] when LB was supplemented with 3.5% HSA. Conversely, HS did not produce changes in the expression level of this gene (Figure 1A). In the case of A. baumannii AB5075 (Figure 1B) and AB0057 (Supplementary Figure S1), levels of expression of clmA were significantly down-regulated in LB plus 3.5% HSA and HS conditions. In contrast, in the A. baumannii AMA16 and ABUH702 strains, clmA was up-regulated when growing under these conditions (Figure S1).
Figure 1.
Effect of human serum albumin (HSA) and pooled normal human serum (HS) on the expression of antibiotic resistance genes of A. baumannii A118 and AB5075 strains. qRT-PCR of A118 (A) and AB5075 (B) strains genes associated with antibiotic resistance expressed in LB, LB supplemented with 3.5% HSA, or in HS. Fold changes were calculated using double ΔΔCt analysis. At least three independent samples were used. LB was used as the reference condition. Statistical significance (p < 0.05) was determined by ANOVA followed by Tukey’s multiple-comparison test, one asterisks: * p < 0.05; two asterisks: ** p < 0.01, and three asterisks: *** p < 0.001.
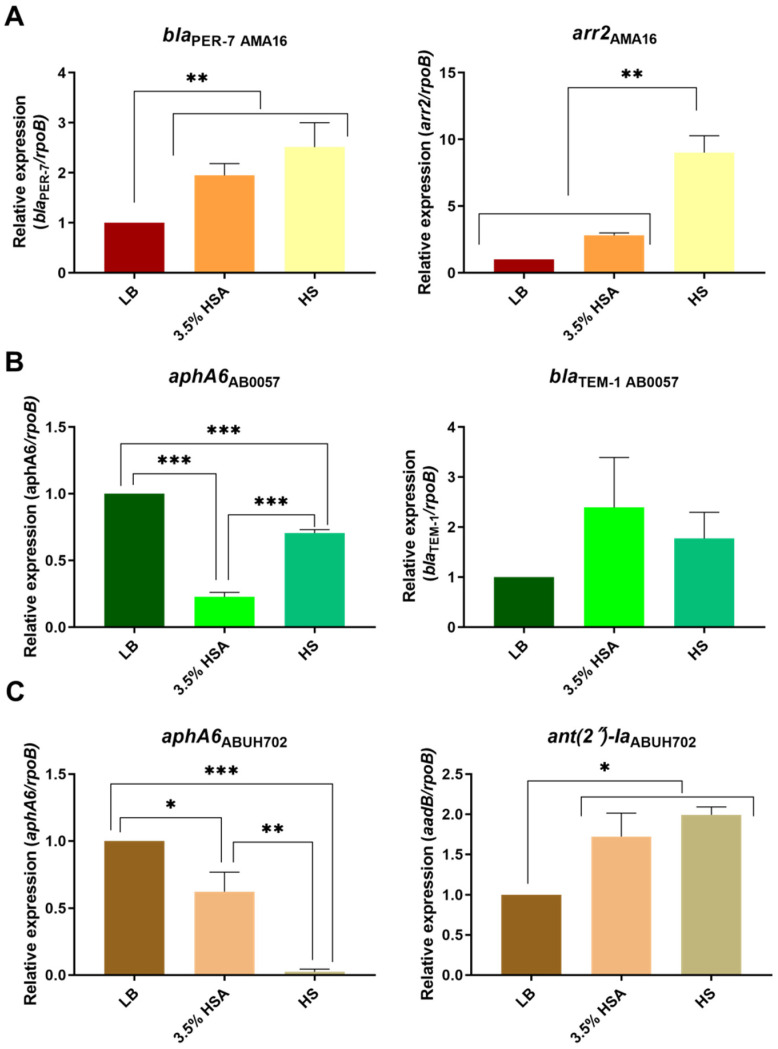
We previously observed that the expression levels of carbapenem-resistance genes were affected when LB was supplemented with 3.5% HSA or HS (Le et al., 2021 submitted). Further examination of the effect of these supplements on other genes related to β-lactam resistance, showed that the expression levels of pbp1 and pbp3, coding for penicillin-binding proteins [27,28], were up-regulated in LB supplemented with 3.5% HSA in A. baumannii A118, AB5075 (Figure 1A,B), and ABUH702 (Figure S1). All strains but A118 underwent an increase in levels of expression of pbp1 and pbp3 when cultured in HS (Figure 1, Figure 2 and Figure S1). The blaPER-7 gene, present only in strain AMA16, was also up-regulated in the presence of 3.5% HSA or HS (Figure 2A).
Figure 2.
Effect of human serum albumin (HSA) and pooled normal human serum (HS) on the expression of antibiotic resistance genes of A. baumannii strains. qRT-PCR of AMA16: (A) AB0057 (B) and ABUH702 (C) strains genes associated with antibiotic resistance expressed in LB, LB supplemented with 3.5% HSA, or in HS. Fold changes were calculated using double ΔΔCt analysis. At least three independent samples were used. LB was used as the reference condition. Statistical significance (p < 0.05) was determined by ANOVA followed by Tukey’s multiple-comparison test, one asterisks: * p < 0.05; two asterisks: ** p < 0.01; and three asterisks: *** p < 0.001.
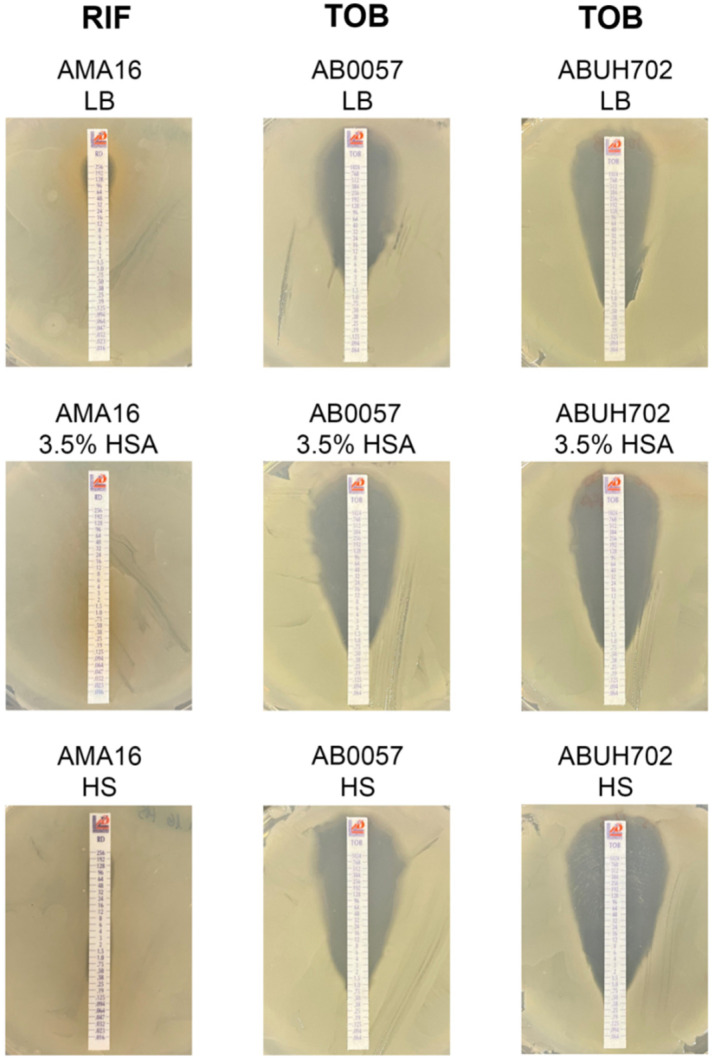
A. baumannii AB5075 strain is resistant to most aminoglycoside antibiotics (gentamicin, amikacin and tobramycin (Table 1) and posseses the aac(6a)-Ib and ant(2n)-Ia (aadB), which encode an aminoglycoside 6′-N-acetyltransferase and an aminoglycoside O-nucleotydyltransferase, respectively [29]. qRT-PCR revealed a significant reduction in the expression of both genes in the presence of 3.5% HSA or HS (Figure 1B). In contrast, ant(2n)-Ia, also present in A. baumannii ABUH702, was expressed at significantly higher levels in cells cultured in LB supplemented with 3.5% HSA or HS (Figure 2C). The armA gene present in A. baumannii AMA16, which encodes a 16S rRNA methylase that confers resistance to amikacin, plazomicin, gentamicin and tobramycin [30], did not exhibit any significant changes in the tested conditions (Figure S1). The expression of aphA6, a gene present in A. baumannii AB0057 and ABUH702 that codes for an aminoglycoside O-phosphotransferase [29], was significantly down-regulated in the presence of 3.5% HSA or HS (Figure 2B,C). This effect could be related to an increase observed in the susceptibility of A. baumannii AB0057 to tobramycin (Figure 3 and Table S1). However, we did not observe any decrease in the MIC values of amikacin, tobramycin, and gentamicin in the AB5075, AB0057 and ABUH702 strains (Figure 3 and Figures S2 and S3).
Table 1.
MICs of A118 or AB5075 grew in LB, LB plus 3.5 % HSA, or HS.
| Strain | A118 | AB5075 | ||||
|---|---|---|---|---|---|---|
| Antibiotic (μg/mL) | LB | 3.5% HSA | HS | LB | 3.5% HSA | HS |
| AK | 2 (S) | 2 (S) | 2 (S) | 32 (I) | 32 (I) | 32 (I) |
| CN | 1 (S) | 1 (S) | 0.75 (S) | 64 (R) | 512 (R) | 96 (R) |
| TOB | 0.75 (S) | 0.75 (S) | 0.75 (S) | 32 (R) | 512 (R) | 32 (R) |
| RIF | 2 (S) | 1.5 (S) | 2 (S) | 64 (R) | 0.38 (S) | 3 (S) |
| CLO | >256 (R) | >256 (R) | >256 (R) | >256 (R) | >256 (R) | >256 (R) |
AK: amikacin; CN: gentamicin; TOB: tobramycin, RIF: rifampicin, CLO: chloramphenicol. R: resistant; I: intermediate; S: susceptible.
Figure 3.
Effect of human serum albumin (HSA) and pooled normal human serum (HS) on the antimicrobial susceptibility of A. baumannii strains. AMA16, AB0057, and ABUH702 strains grew in LB broth, LB broth plus 3.5 % HSA, or in HS were used to performed rifampicin (RIF) and tobramycin (TOB) susceptibility. Minimum inhibitory concentration (MIC) was performed by E-test (Liofilchem, Italy) following CLSI recommendations.
Modifications in levels of resistance to rifampicin, a drug that alone or in combination with colistin has been used for severe infections due to MDR A. baumannii [31], were also evaluated. An increase in the susceptibility to rifampicin was observed in the case of A. baumannii AB5075 in the presence of 3.5% HSA or HS, while changes were not observed in strain A118 (Table 1). In the case of A. baumannii AMA16, which posseses the arr2 gene, related to rifampicin resistance, a significant increase in the expression level of arr2 and a concomitant increase in MIC were observed when growing in HS (Figure 2A and Figure 3).
The sulfonamine resistance gene sul1 [32], present in A. baumannii A118, AB5075, AMA16, and AB0057, responded differently to the additions. It was down-regulated when growing in medium supplemented with 3.5% as determined using qRT-PCR analysis in A. baumannii A118, AMA16, and AB0057 (Figure 1A and Figure S1). Conversely, this gene was expressed at higher levels in A. baumannii AB5075 exposed to 3.5% HSA or HS, and in A. baumannii AB0057 cultured in HS (Figure S1).
The impact of human fluids and/or human proteins on the expression of antibiotic resistance genes and the concomitant levels of resistance are largely unknown [18,22,33]. Previous work showed that 0.2% HSA and 4% Human Pleural Fluid altered the expression of genes related to antibiotic resistance [17,21]. Moreover, Huang et al. demostrated the ability of mucin to bind antibiotics, such as colistin, which leads to a reduction in the efficacy of antibiotics used to treat A. baumannii infections [34]. The results described in this analysis indicate that in five different strains, the expression levels of resistance genes are affected by HSA and HS. Resistant and susceptible strains behaved identically with respect to the β-lactam and sulfonamide resistance-associated genes, which were upregulated in the presence of 3.5% HSA. However, while the addition of 3.5% HSA to A. baumannii A118 cultures produced an increase in expression levels of a chloramphenicol resistance gene, the opposite effect was observed in carbapenem-resistant strains. For some genes or groups of genes encoding resistance to an antibiotic class, the effect in the expression levels varied depending on the strain and/or the condition. In the case of β-lactam resistance-associated genes, an increase in the expression was observed for most of the genes in all the strains in both conditions (Figure 1 and Figure 2). The aminoglycoside resistance genes were mostly down-regulated in all strains and conditions, with the only exception of ant(2n)-Ia in the ABUH702 strains exposed to 3.5% HSA or HS (Figure 1B and Figure 2C). Future studies will increase the understanding of how the processes used by bacteria to respond to the presence of human fluids and/or human proteins contribute to antibiotic treatment failure.
2.2. HSA and HS Negatively Affects Biofilm Formation
Since biofilm formation has a direct impact in antibiotic resistance [33,35,36,37], leading in some cases to treatment failure, we investigated the effect of 3.5% HSA and HS in the expression of genes involved in biofilm formation and the corresponding impact on phenotype.
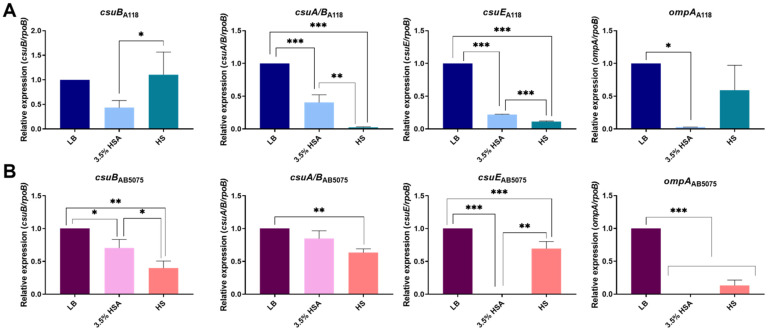
qRT-PCR analysis of csuA/B, csuB, and csuE genes belonging to CsuA/BABCDE chaperone-usher secretion system [38] was carried out in A118 and AB5075 cells grown in LB, LB supplemented with 3.5% HSA, and HS (Figure 4). The transcriptional level of A118 and AB5075 csuA/B, csuB, and csuE showed a significant down-regulation under 3.5% HSA and HS conditions, with the exception of csuB for the A118 strain (Figure 4A,B). Moreover, analysis of mRNA extracted from A. baumannii AMA16, AB0057, and ABUH702 strains cultured in medium containing 3.5% HSA or in HS showed that the expression of csuA/B was significantly reduced in the presence of both conditions, with the exception of AB0057 strain grown in LB plus 3.5% HSA (Figure S4). In addition, the outer membrane protein A (OmpA) of A. baumannii is reported as playing a role in the development of robust biofilms on plastic [39]. The transcriptional analysis of ompA was also investigated and we observed a down-regulation in the presence of 3.5% HSA or HS with respect to LB condition for all strains with the exception of ABUH702 strain (Figure 4 and Figure S4).
Figure 4.
Effect of human serum albumin (HSA) and pooled normal human serum (HS) on the expression of biofilm formation genes in A. baumannii strains. qRT-PCR of A118 (A) and AB5075 (B) strains of the csuB, csuA/B, csuE, and ompA genes biofilm formation related in cells cultured in LB, LB supplemented with 3.5 % HSA, or in HS. Fold changes were calculated using double ΔΔCt analysis. At least three independent samples were used. LB was used as the reference condition. Statistical significance (p < 0.05) was determined by ANOVA followed by Tukey’s multiple-comparison test, one asterisks: * p < 0.05; two asterisks: ** p < 0.01 and three asterisks: *** p < 0.001.
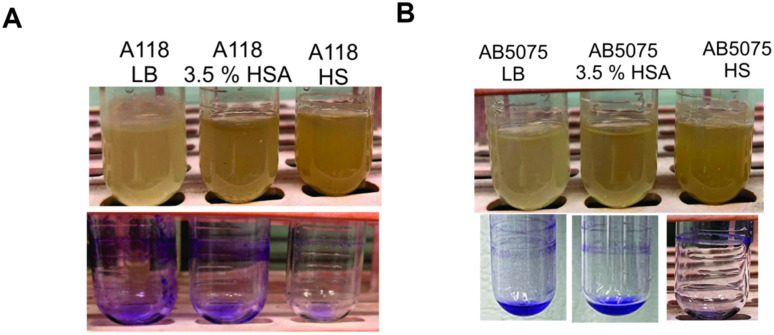
These results were in agreement with biofilm formation in tubes, since a reduction or absence of biofilm was observed (Figure 5). To further examine the inhibitory effect of 3.5% HSA and HS on other A. baumannii strains, AMA16, AB0057, and ABUH702 strains were grown in LB, LB plus 3.5% HSA, or HS. Less biofilm production was observed supporting the previous observation (Figure S4). Our results showed a strong negative effect of HSA and HS in biofilm formation in all the included strains, leading us to conclude that this inhibitory effect occurs in the different A. baumannii strains.
Figure 5.
Effect of human serum albumin (HSA) and pooled normal human serum (HS) on biofilm formation in A. buamannii. Biofilm assays performed in A118 (A) and AB5075 (B) strains grown in LB, LB supplemented with 3.5% HSA, or in HS. Tubes were stained with 1% crystal violet.
A previous work analysing the biofilm formation of different A. baumannii isolates that recovered from blood or sputum, showed that blood isolates formed less biofilm compared to respiratory isolates [40]. Our results are in agreement with this observation [40]. Another human molecule that also exhibited biofilm formation inhibition in A. baumannii strains was L-Adrenaline [41]. In addition, a recent study reported that lactoferrin has antimicrobial and antibiofilm activities against A. baumannii clinical strains that were isolated from wounds, blood, urine, and sputum/bronchial wash [42]. Lactoferrin is an innate immune glycoprotein produced in high concentrations in both human and bovine milk, and is considered one of the most effective iron chelators. Lactoferrin’s binding affinity for iron is 300 times higher than that of transferrin. These observations provide evidence that indicates the complex interplay between this pathogen’s and both human and bovine isoforms of host products [42]. Together, these observations demonstrate how human proteins can have an important and significant impact on biofilm formation.
2.3. Quorum Network Is Altered by HSA and HS
Many bacteria communicate with each other and respond collectively to a changing environment through quorum sensing (QS) [43]. QS permits bacteria to monitor one another’s presence and to modulate gene expression in response to changes in population density, controlling survival and virulence [43,44].
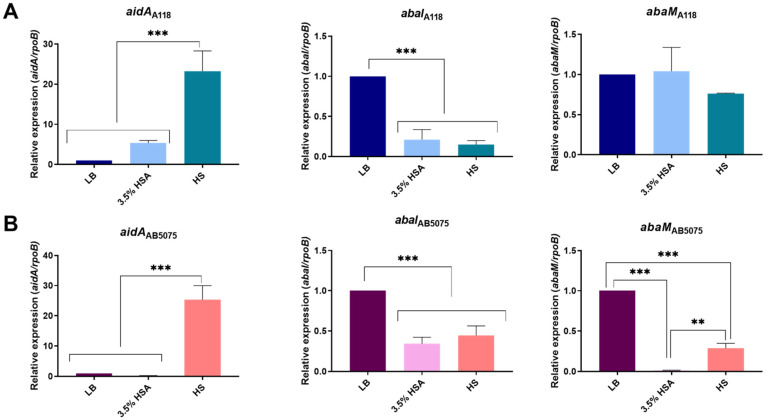
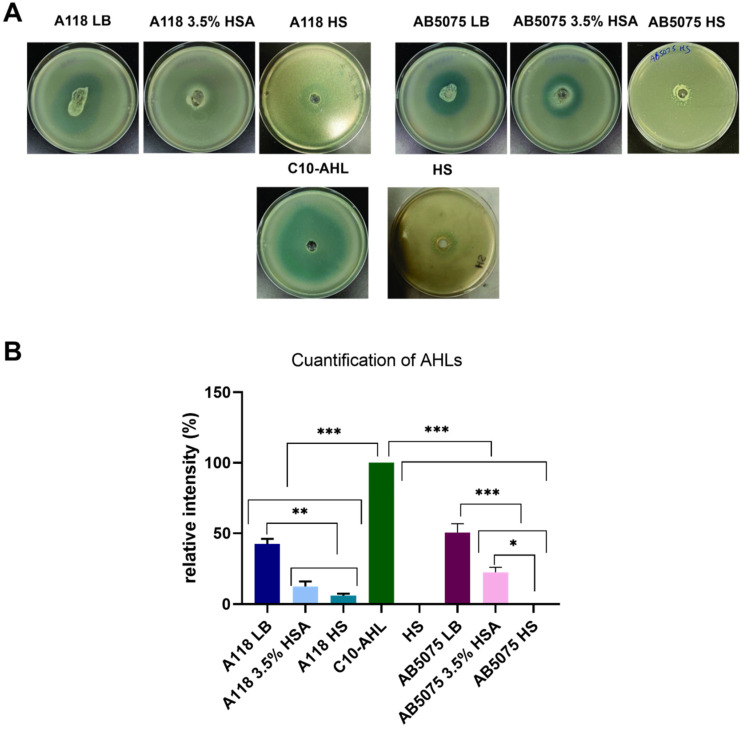
To explore the QS network in A. baumannii A118 and AB5075 strains growing in LB plus 3.5% HSA or in HS, qRT-PCR analysis of important genes of QS/quorum quenching (abaI, aidA, and abaM) were carried out [45,46,47]. The results showed that aidA expression was up-regulated and abaI expression was down-regulated under both treatments in both strains (Figure 6). Differences were not observed in the expression levels of abaM between LB, LB plus 3.5% HSA, and HS in A. baumannii A118 (Figure 6A). In contrast, in A. baumannii AB5075, the levels of expression of abaM were significantly reduced under both treatments (Figure 6B). These results suggest that when A. baumannii is exposed to HSA, the cells modulate the expression of QS network genes to reduce AHL synthesis or increase its degradation. We confirmed these findings by using A. tumefaciens as a bacterial biosensor. Supernatants from A. baumannii A118 and AB5075 cultures in LB showed the highest AHL production, suggesting down-regulation of QS and/or up-regulation of quorum quenching in the presence of HSA molecules or HS components (Figure 7). These results are consistent with a previous observation that A. baumannii AB5075 cultured in human pleural fluid produced reduced levels of AHLs [16].
Figure 6.
Effect of human serum albumin (HSA) and pooled normal human serum (HS) on the expression of quorum sensing genes in A. baumannii. qRT-PCR of A118 (A) and AB5075 (B) strains of the aidA, abaI, and abaM genes associated with quorum sensing and quorum quenching expressed in LB, LB supplemented with 3.5% HSA, or in HS. Fold changes were calculated using double ΔΔCt analysis. At least three independent samples were used. LB was employed as the reference condition. Statistical significance (p < 0.05) was determined by ANOVA followed by Tukey’s multiple-comparison test, two asterisks: ** p < 0.01 and three asterisks: *** p < 0.001.
Figure 7.
Effect of human serum albumin (HSA) and pooled normal human serum (HS) on quorum sensing in A. baumannii. (A) N-Acyl Homoserine Lactone (AHL) detection in A. baumannii A118 and AB5075 supernatants of bacterial cultured in LB, LB supplemented with 3.5% HSA, or HS, using Agrobacterium tumefaciens as reported strain. The presence of AHL was determined by the development of the blue color. (B) Quantification of 5,5′-dibromo-4,4′-dichloro-indigo was estimated as the percentage relative to C10-AHL standard, measured with ImageJ (NIH). As negative control, HS was used. The mean ± SD is informed. Statistical significance (p < 0.05) was determined by ANOVA followed by Tukey’s multiple-comparison test. one asterisks: * p < 0.05; two asterisks: ** p < 0.01 and three asterisks: *** p < 0.001.
In addition, considering the role of the AHL molecules in the upregulation of bfmS and bfmR genes involved in robust biofilm formation on abiotic surfaces in A. baumannii [48], and the evidence that abaI mutants are impaired in biofilm development [49], our results agreed with reduced biofilm formation observed when the cells were exposed to 3.5% HSA and HS.
Taken together, the results described herein indicate that both strains coordinate and modulate the activity of many genes that regulate the response to a variety of environments and conditions. These responses likely play a major role in persistence, virulence, and colonization.
3. Materials and Methods
3.1. Bacterial Strains
The model A. baumannii strains A118 and AB5075, which show a different degree of susceptibility and virulence, were used [16,21,50,51]. Additional strains containing metallo β-lactamases, such as New Delhi metallo- β-lactamase (NDM) or oxacilinases (OXA) were used. The used strains were AMA 16 strain (NDM-1 positive strains) [52], ABUH702 (carbapenem resistance due to increase expression of blaOXA-66 by ISAba1) [53], and AB0057 (blaOXA-23) [52].
3.2. RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Overnight cultures of A. baumannii strains were diluted 1:10 in fresh LB medium; 3.5% HSA containing LB medium or pooled normal human serum (HS) and incubated with agitation for 5 h at 37 °C. Pure HSA (Sigma-Aldrich, St. Louis, MO, USA) and pooled normal human serum from a certified vendor (Innovative Research Inc, Novi, MI, USA) were used in the cultures.
RNA was extracted from each strain using the Direct-zol RNA Kit (Zymo Research, Irvine, CA, USA) following manufacturer’s instructions. Total RNA extractions were performed in three biological replicates for each condition. The extracted and DNase-treated RNA was used to synthesize cDNA using the manufacturer’s protocol provided within the iScriptTM Reverse Transcription Supermix for qPCR (Bio-Rad, Hercules, CA, USA). The cDNA concentrations were adjusted to 50 ng/μL and qPCR was conducted using the qPCRBIO SyGreen Blue Mix Lo-ROX following manufacturer’s protocol (PCR Biosystems, Wayne, PA, USA). At least three biological replicates of cDNA were used in triplets and were run using the CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA).
Transcriptional levels of each sample were normalized to the transcriptional level of rpoB. The relative quantification of gene expression was performed using the comparative threshold method 2−ΔΔCt. The ratios obtained after normalization were expressed as folds of change compared with cDNA samples isolated from bacteria cultures on LB. Asterisks indicate significant differences as determined by ANOVA followed by Tukey’s multiple comparison test (p < 0.05), using GraphPad Prism (GraphPad software, San Diego, CA, USA).
3.3. Antimicrobial Susceptibility Testing
Antibiotic susceptibility assays were performed following the procedures recommended by the Clinical and Laboratory Standards Institute (CLSI) [51]. After OD adjustment, 100 μL of cells grown in fresh LB medium, 3.5% HSA containing LB medium or HS, were inoculated on Mueller-Hinton agar plates as previously described [17,21]. Antimicrobial commercial E-strips (Liofilchem S.r.l., Roseto degli Abruzzi, Italy) for rifampicin (RIF), tobramycin (TOB), amikacin (AK), and gentamicin (CN) were used. Mueller-Hinton agar plates were incubated at 37 °C for 18 h. CLSI breakpoints were used for interpretation.
3.4. Biofilm Assays
Biofilms assays were performed as previously described [21]. A. baumannii A118 and AB5075 cells were cultured in fresh LB medium, 3.5% HSA containing LB medium or HS, with agitation for 18 h at 37 °C. Tubes were emptied, washed three times with 1× phosphate-buffered saline (PBS) and stained with 1% crystal violet (CV) for 15 m. Excess CV was removed by washing three more with 1× PBS. Experiments were performed in triplicate, with at least three technical replicates per biological replicate.
3.5. N-Acyl Homoserine Lactone (AHL) Detection
Agrobacterium tumefaciens-based solid plate assays were carried out to detect AHL production [54] as previously described [55]. Briefly, 500 µL of the homogenate were loaded in a central well of 0.7% LB agar plates supplemented with 40 µg of 5-bromo-3-indolyl-β-D-galactopyranoside (X-Gal) per mL and 250 µL (OD = 2.5) of overnight cultures of Agrobacterium tumefaciens biosensor. The presence of AHL was determined by the development of the blue color [56]. As a positive control, 100 µL of N-Decanoyl-DL-homoserine lactone (C10-AHL) 12.5 mg/mL, and as negative control, 100 µL of pooled normal HS was utilized. Quantification of 5,5′-dibromo-4,4′-dichloro-indigo production in different conditions was determined by measuring the intensity of each complete plate and substracting the intensity measured in the negative control, using ImageJ software (NIH). The values were normalized to the positive control, which received the arbitrary value of 100.
3.6. Statistical Analysis
All experiments were performed at least in three technical and biological replicates. Data were expressed as means ± standard deviation. Statistical analysis using one way-ANOVA followed by Tukey’s multiple comparison test were performed using GraphPad Prism (GraphPad software, San Diego, CA, USA), and a p value < 0.05 was considered statistically significant.
4. Conclusions
Our results reveal that in A. baumannii a variety of proteins present in human fluids that are encountered during infection significantly modulate a transcriptional response resulting in opportunities for increasing survival. Changes at the transcriptional level affecting genes involved in antimicrobial resistance were observed in the five different A. baumannii strains used, despite a differential behavior depending on the studied gene and strain. Results of biofilm formation were in accordance with previous observations obtained with other human fluids [16], where a reduced expression of genes involved in biofilm formation, supported by a substantial decrease in biofilm formation, was observed in all the strains. The transcriptional level of quorum sensing network genes, known to control different virulence factors, was also affected. A reduction in quorum sensing molecules was observed, suggesting a switch towards a commensal state facilitating its persistence. The inhibition of the quorum sensing network is in agreement with the reduced biofilm formation, allowing A. baumannii to survive in the bloodstream, where HSA, the main component of HS, is the predominant protein.
Together, the results presented provide important clues that a clear adaptative response of A. baumannii is induced by HSA, opening a new venue to explore alternative therapeutic approaches [15,16,17,21,57].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10070833/s1, Figure S1: Effect of human serum albumin (HSA) and pooled normal human serum (HS) on the expression of antibiotic resistance genes of A. baumannii AMA16, AB0057 and ABUH702 strains, Figure S2: Effect of human serum albumin (HSA) and pooled normal human serum (HS) on the antimicrobial susceptibility of A. baumannii AB5075 strain, Figure S3. Effect of human serum albumin (HSA) and pooled normal human serum (HS) on the antimicrobial susceptibility of A. baumannii AB0057 and ABUH702 strains, and Figure S4: Effect of human serum albumin (HSA) and pooled normal human serum (HS) on biofilm formation in A. baumannii AMA16, AB0057 and ABUH702 strains.
Author Contributions
C.P. and C.L. have equal contribution to the work. C.P., C.L., M.R.T., T.S., R.A.B. and M.S.R. conceived the study and designed the experiments. C.P., C.L., M.R.T., T.S. and M.S.R. performed the experiments and genomics and bioinformatics analyses. C.P., C.L., M.R.T., T.S., K.M.P.-W., R.A.B., M.E.T. and M.S.R. analyzed the data and interpreted the results. K.M.P.-W., R.A.B., M.E.T. and M.S.R. contributed reagents/materials/analysis tools. M.R.T., T.S., K.M.P.-W., R.A.B., M.E.T. and M.S.R. wrote and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The authors’ work was supported by NIH SC3GM125556 to MSR, R01AI100560 to RAB, R01AI063517, R01AI072219 to RAB, and 2R15 AI047115 to MET. CP was supported by grant MHRT 2T37MD001368 from the National Institute on Minority Health and Health Disparities, National Institute of Health. This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Number 1I01BX001974 to RAB and 1I01 BX002872 to KMPW from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 to RAB. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs. MRT and TS are recipient of a postdoctoral fellowship from CONICET.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Gilbert D., Rice L.B., Scheld M., Spellberg B., Bartlett J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 2.Rice L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008;197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez C.H., Nastro M., Famiglietti A. Carbapenemases in Acinetobacter baumannii. Review of their dissemination in Latin America. Rev. Argent. Microbiol. 2018;50:327–333. doi: 10.1016/j.ram.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 4.CDC . Antibiotic Resistance Threats in the United States. Department of Health and Human Services, CDC; Atlanta, GA, USA: 2019. [Google Scholar]
- 5.Mulani M.S., Kamble E.E., Kumkar S.N., Tawre M.S., Pardesi K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019;10:539. doi: 10.3389/fmicb.2019.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karaiskos I., Lagou S., Pontikis K., Rapti V., Poulakou G. The “Old” and the “New” Antibiotics for MDR Gram-Negative Pathogens: For Whom, When, and How. Front. Public Health. 2019;7:151. doi: 10.3389/fpubh.2019.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassetti M., Echols R., Matsunaga Y., Ariyasu M., Doi Y., Ferrer R., Lodise T.P., Naas T., Niki Y., Paterson D.L., et al. Efficacy and safety of cefiderocol or best available therapy for the treatment of serious infections caused by carbapenem-resistant Gram-negative bacteria (CREDIBLE-CR): A randomised, open-label, multicentre, pathogen-focused, descriptive, phase 3 trial. Lancet Infect. Dis. 2021;21:226–240. doi: 10.1016/S1473-3099(20)30796-9. [DOI] [PubMed] [Google Scholar]
- 8.Bassetti M., Peghin M., Vena A., Giacobbe D.R. Treatment of Infections Due to MDR Gram-Negative Bacteria. Front. Med. 2019;6:74. doi: 10.3389/fmed.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulens S.N., Yi S.H., Walters M.S., Jacob J.T., Bower C., Reno J., Wilson L., Vaeth E., Bamberg W., Janelle S.J., et al. Carbapenem-Nonsusceptible Acinetobacter baumannii, 8 US Metropolitan Areas, 2012–2015. Emerg. Infect. Dis. 2018;24:727–734. doi: 10.3201/eid2404.171461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isler B., Doi Y., Bonomo R.A., Paterson D.L. New treatment options against carbapenem-resistant Acinetobacter baumannii infections. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.01110-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paterson D.L., Isler B., Stewart A. New treatment options for multiresistant gram negatives. Curr. Opin. Infect. Dis. 2020;33:214–223. doi: 10.1097/QCO.0000000000000627. [DOI] [PubMed] [Google Scholar]
- 12.Wong D., Nielsen T.B., Bonomo R.A., Pantapalangkoor P., Luna B., Spellberg B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017;30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller G.L., Tuttobene M., Altilio M., Martinez Amezaga M., Nguyen M., Cribb P., Cybulski L.E., Ramirez M.S., Altabe S., Mussi M.A. Light Modulates Metabolic Pathways and Other Novel Physiological Traits in the Human Pathogen Acinetobacter baumannii. J. Bacteriol. 2017;199 doi: 10.1128/JB.00011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuttobene M.R., Fernandez-Garcia L., Blasco L., Cribb P., Ambroa A., Muller G.L., Fernandez-Cuenca F., Bleriot I., Rodriguez R.E., Barbosa B.G.V., et al. Quorum and Light Signals Modulate Acetoin/Butanediol Catabolism in Acinetobacter spp. Front. Microbiol. 2019;10:1376. doi: 10.3389/fmicb.2019.01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez J., Razo-Gutierrez C., Le C., Courville R., Pimentel C., Liu C., Fung S.E., Tuttobene M.R., Phan K., Vila A.J., et al. Cerebrospinal fluid (CSF) augments metabolism and virulence expression factors in Acinetobacter baumannii. Sci. Rep. 2021;11:4737. doi: 10.1038/s41598-021-81714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pimentel C., Le C., Tuttobene M.R., Subils T., Martinez J., Sieira R., Papp-Wallace K.M., Keppetipola N., Bonomo R.A., Actis L.A., et al. Human Pleural Fluid and Human Serum Albumin Modulate the Behavior of a Hypervirulent and Multidrug-Resistant (MDR) Acinetobacter baumannii Representative Strain. Pathogens. 2021;10:471. doi: 10.3390/pathogens10040471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez J., Fernandez J.S., Liu C., Hoard A., Mendoza A., Nakanouchi J., Rodman N., Courville R., Tuttobene M.R., Lopez C., et al. Human pleural fluid triggers global changes in the transcriptional landscape of Acinetobacter baumannii as an adaptive response to stress. Sci. Rep. 2019;9:17251. doi: 10.1038/s41598-019-53847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez-Fraga L., Vazquez-Ucha J.C., Martinez-Guitian M., Vallejo J.A., Bou G., Beceiro A., Poza M. Pneumonia infection in mice reveals the involvement of the feoA gene in the pathogenesis of Acinetobacter baumannii. Virulence. 2018;9:496–509. doi: 10.1080/21505594.2017.1420451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez M., Blasco L., Gato E., Perez A., Fernandez-Garcia L., Martinez-Martinez L., Fernandez-Cuenca F., Rodriguez-Bano J., Pascual A., Bou G., et al. Response to Bile Salts in Clinical Strains of Acinetobacter baumannii Lacking the AdeABC Efflux Pump: Virulence Associated with Quorum Sensing. Front. Cell Infect. Microbiol. 2017;7:143. doi: 10.3389/fcimb.2017.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodman N., Martinez J., Fung S., Nakanouchi J., Myers A.L., Harris C.M., Dang E., Fernandez J.S., Liu C., Mendoza A.M., et al. Human Pleural Fluid Elicits Pyruvate and Phenylalanine Metabolism in Acinetobacter baumannii to Enhance Cytotoxicity and Immune Evasion. Front. Microbiol. 2019;10:1581. doi: 10.3389/fmicb.2019.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn B., Rodman N., Jara E., Fernandez J.S., Martinez J., Traglia G.M., Montana S., Cantera V., Place K., Bonomo R.A., et al. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci. Rep. 2018;8:14741. doi: 10.1038/s41598-018-33072-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohneck E.J., Arivett B.A., Fiester S.E., Wood C.R., Metz M.L., Simeone G.M., Actis L.A. Mucin acts as a nutrient source and a signal for the differential expression of genes coding for cellular processes and virulence factors in Acinetobacter baumannii. PLoS ONE. 2018;13:e0190599. doi: 10.1371/journal.pone.0190599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin H., Lo N.W., Loo J.F., Lin X., Yim A.K., Tsui S.K., Lau T.C., Ip M., Chan T.F. Comparative transcriptomics of multidrug-resistant Acinetobacter baumannii in response to antibiotic treatments. Sci. Rep. 2018;8:3515. doi: 10.1038/s41598-018-21841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinn B., Martinez J., Liu C., Nguyen M., Ramirez M.S. The effect of sub-inhibitory concentrations of antibiotics on natural transformation in Acinetobacter baumannii. Int. J. Antimicrob. Agents. 2018;51:809–810. doi: 10.1016/j.ijantimicag.2018.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarshar M., Behzadi P., Scribano D., Palamara A.T., Ambrosi C. Acinetobacter baumannii: An Ancient Commensal with Weapons of a Pathogen. Pathogens. 2021;10:387. doi: 10.3390/pathogens10040387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karalewitz A.P., Miller S.I. Multidrug-Resistant Acinetobacter baumannii Chloramphenicol Resistance Requires an Inner Membrane Permease. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.00513-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vashist J., Tiwari V., Das R., Kapil A., Rajeswari M.R. Analysis of penicillin-binding proteins (PBPs) in carbapenem resistant Acinetobacter baumannii. Indian J. Med. Res. 2011;133:332–338. [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Cuenca F., Martinez-Martinez L., Conejo M.C., Ayala J.A., Perea E.J., Pascual A. Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 2003;51:565–574. doi: 10.1093/jac/dkg097. [DOI] [PubMed] [Google Scholar]
- 29.Ramirez M.S., Tolmasky M.E. Aminoglycoside modifying enzymes. Drug Resist. Updat. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong S.B., Shin K.S., Ha J., Han K. Co-existence of blaOXA-23 and armA in multidrug-resistant Acinetobacter baumannii isolated from a hospital in South Korea. J. Med. Microbiol. 2013;62:836–844. doi: 10.1099/jmm.0.055384-0. [DOI] [PubMed] [Google Scholar]
- 31.Park H.J., Cho J.H., Kim H.J., Han S.H., Jeong S.H., Byun M.K. Colistin monotherapy versus colistin/rifampicin combination therapy in pneumonia caused by colistin-resistant Acinetobacter baumannii: A randomised controlled trial. J. Glob. Antimicrob. Resist. 2019;17:66–71. doi: 10.1016/j.jgar.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Dominguez M., Miranda C.D., Fuentes O., de la Fuente M., Godoy F.A., Bello-Toledo H., Gonzalez-Rocha G. Occurrence of Transferable Integrons and sul and dfr Genes Among Sulfonamide-and/or Trimethoprim-Resistant Bacteria Isolated From Chilean Salmonid Farms. Front. Microbiol. 2019;10:748. doi: 10.3389/fmicb.2019.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozkul C., Hazirolan G. Oxacillinase Gene Distribution, Antibiotic Resistance, and Their Correlation with Biofilm Formation in Acinetobacter baumannii Bloodstream Isolates. Microb. Drug Resist. 2020 doi: 10.1089/mdr.2020.0130. [DOI] [PubMed] [Google Scholar]
- 34.Huang J.X., Blaskovich M.A., Pelingon R., Ramu S., Kavanagh A., Elliott A.G., Butler M.S., Montgomery A.B., Cooper M.A. Mucin Binding Reduces Colistin Antimicrobial Activity. Antimicrob. Agents Chemother. 2015;59:5925–5931. doi: 10.1128/AAC.00808-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassan P.A., Khider A.K. Correlation of biofilm formation and antibiotic resistance among clinical and soil isolates of Acinetobacter baumannii in Iraq. Acta Microbiol. Immunol. Hung. 2020;67:161–170. doi: 10.1556/030.66.2019.026. [DOI] [PubMed] [Google Scholar]
- 36.Qi L., Li H., Zhang C., Liang B., Li J., Wang L., Du X., Liu X., Qiu S., Song H. Relationship between Antibiotic Resistance, Biofilm Formation, and Biofilm-Specific Resistance in Acinetobacter baumannii. Front. Microbiol. 2016;7:483. doi: 10.3389/fmicb.2016.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senobar Tahaei S.A., Stajer A., Barrak I., Ostorhazi E., Szabo D., Gajdacs M. Correlation Between Biofilm-Formation and the Antibiotic Resistant Phenotype in Staphylococcus aureus Isolates: A Laboratory-Based Study in Hungary and a Review of the Literature. Infect. Drug Resist. 2021;14:1155–1168. doi: 10.2147/IDR.S303992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomaras A.P., Flagler M.J., Dorsey C.W., Gaddy J.A., Actis L.A. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology. 2008;154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 39.Gaddy J.A., Tomaras A.P., Actis L.A. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 2009;77:3150–3160. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vijayakumar S., Rajenderan S., Laishram S., Anandan S., Balaji V., Biswas I. Biofilm Formation and Motility Depend on the Nature of the Acinetobacter baumannii Clinical Isolates. Front. Public Health. 2016;4:105. doi: 10.3389/fpubh.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiwari V., Patel V., Tiwari M. In-silico screening and experimental validation reveal L-Adrenaline as anti-biofilm molecule against biofilm-associated protein (Bap) producing Acinetobacter baumannii. Int. J. Biol. Macromol. 2018;107:1242–1252. doi: 10.1016/j.ijbiomac.2017.09.105. [DOI] [PubMed] [Google Scholar]
- 42.Avery T.M., Boone R.L., Lu J., Spicer S.K., Guevara M.A., Moore R.E., Chambers S.A., Manning S.D., Dent L., Marshall D., et al. Analysis of Antimicrobial and Antibiofilm Activity of Human Milk Lactoferrin Compared to Bovine Lactoferrin against Multidrug Resistant and Susceptible Acinetobacter baumannii Clinical Isolates. ACS Infect. Dis. 2021 doi: 10.1021/acsinfecdis.1c00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camilli A., Bassler B.L. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seleem N.M., Abd El Latif H.K., Shaldam M.A., El-Ganiny A. Drugs with new lease of life as quorum sensing inhibitors: For combating MDR Acinetobacter baumannii infections. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1687–1702. doi: 10.1007/s10096-020-03882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saipriya K., Swathi C.H., Ratnakar K.S., Sritharan V. Quorum-sensing system in Acinetobacter baumannii: A potential target for new drug development. J. Appl. Microbiol. 2020;128:15–27. doi: 10.1111/jam.14330. [DOI] [PubMed] [Google Scholar]
- 46.Lopez M., Mayer C., Fernandez-Garcia L., Blasco L., Muras A., Ruiz F.M., Bou G., Otero A., Tomas M., Geih G. Quorum sensing network in clinical strains of A. baumannii: AidA is a new quorum quenching enzyme. PLoS ONE. 2017;12:e0174454. doi: 10.1371/journal.pone.0174454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez-Martin M., Dubern J.F., Alexander M.R., Williams P. AbaM Regulates Quorum Sensing, Biofilm Formation and Virulence in Acinetobacter baumannii. J. Bacteriol. 2021 doi: 10.1128/JB.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eze E.C., Chenia H.Y., El Zowalaty M.E. Acinetobacter baumannii biofilms: Effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments. Infect. Drug Resist. 2018;11:2277–2299. doi: 10.2147/IDR.S169894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niu C., Clemmer K.M., Bonomo R.A., Rather P.N. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 2008;190:3386–3392. doi: 10.1128/JB.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams M.D., Goglin K., Molyneaux N., Hujer K.M., Lavender H., Jamison J.J., MacDonald I.J., Martin K.M., Russo T., Campagnari A.A., et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 2008;190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.C.L.S.I. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Nine Informational Supplement. Clinical Lab Standards Institute; Wayne, PA, USA: 2019. CLSI Document M100-S29: 2019. [Google Scholar]
- 52.Adams M.D., Pasteran F., Traglia G.M., Martinez J., Huang F., Liu C., Fernandez J.S., Lopez C., Gonzalez L.J., Albornoz E., et al. Distinct mechanisms of dissemination of NDM-1 metallo- beta-lactamase in Acinetobacter spp. in Argentina. Antimicrob. Agents Chemother. 2020 doi: 10.1128/AAC.00324-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams M.D., Wright M.S., Karichu J.K., Venepally P., Fouts D.E., Chan A.P., Richter S.S., Jacobs M.R., Bonomo R.A. Rapid Replacement of Acinetobacter baumannii Strains Accompanied by Changes in Lipooligosaccharide Loci and Resistance Gene Repertoire. mBio. 2019;10 doi: 10.1128/mBio.00356-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paulk Tierney A.R., Rather P.N. Methods for Detecting N-Acyl Homoserine Lactone Production in Acinetobacter baumannii. Methods Mol. Biol. 2019;1946:253–258. doi: 10.1007/978-1-4939-9118-1_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pimentel C.B., Snow A.L., Carnes S.L., Shah N.R., Loup J.R., Vallejo-Luces T.M., Madrigal C., Hartmann C.W. Huddles and their effectiveness at the frontlines of clinical care: A scoping review. J. Gen. Intern. Med. 2021 doi: 10.1007/s11606-021-06632-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cha C., Gao P., Chen Y.C., Shaw P.D., Farrand S.K. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant. Microbe. Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- 57.Martinez J., Liu C., Rodman N., Fernandez J.S., Barberis C., Sieira R., Perez F., Bonomo R.A., Ramirez M.S. Human fluids alter DNA-acquisition in Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 2018 doi: 10.1016/j.diagmicrobio.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article.