Abstract
Significant progress has been made in addressing the scientific challenges of biomedical text mining. However, the transition from a demonstration of scientific progress to the production of tools on which a broader community can rely requires that fundamental software engineering requirements be addressed. In this paper we characterize the state of biomedical text mining software with respect to software testing and quality assurance. Biomedical natural language processing software was chosen because it frequently specifically claims to offer production-quality services, rather than just research prototypes.
We examined twenty web sites offering a variety of text mining services. On each web site, we performed the most basic software test known to us and classified the results. Seven out of twenty web sites returned either bad results or the worst class of results in response to this simple test. We conclude that biomedical natural language processing tools require greater attention to software quality.
We suggest a linguistically motivated approach to granular evaluation of natural language processing applications, and show how it can be used to detect performance errors of several systems and to predict overall performance on specific equivalence classes of inputs.
We also assess the ability of linguistically-motivated test suites to provide good software testing, as compared to large corpora of naturally-occurring data. We measure code coverage and find that it is considerably higher when even small structured test suites are utilized than when large corpora are used.
1. Introduction
Biomedical natural language processing tools and data generated by their application are beginning to gain widespread use in biomedical research. Significant progress has been made recently in addressing the scientific challenges of creating computer programs that can properly handle the complexities of human language. However, the transition from a demonstration of scientific progress to the production of tools on which a broader community can depend requires that fundamental software engineering requirements be addressed. Software for medical devices has the benefit of explicit quality assurance requirements per Section 201(h) of the Federal Food, Drug, and Cosmetic Act; Title 21 of the Code of Federal Regulations Part 820; and 61 Federal Register 52602 [8] (p. 7). However, unless it is embedded in a medical device, biomedical natural language processing software is not currently subject to federal quality assurance requirements.
This paper represents the first attempt to characterize the state of one portion of the diverse world of computational bioscience software, specifically biomedical natural language processing applications, with respect to software testing and quality assurance. We assay a broad range of biomedical natural language processing services that are made available via web sites for evidence of quality assurance processes. Our findings suggest that at the current time, software testing and quality assurance are lacking in the community that produces biomedical natural language processing tools. For the tool consumer, this finding should come as a note of caution.
2. Approach to Assessing the State of Natural Language Processing Applications with Respect to Software Testing and Quality Assurance
We looked at twenty web sites offering a variety of text-mining-related services. In the body of this work, we never identify them by name: following the tradition in natural language processing, we do not want to punish people for making their work freely available. Our purpose is not to point fingers—indeed, one of our own services is every bit as lacking in most or all of the measures that we describe below as any. Rather, our goal is to allow the community to make a realistic assessment of the state of the art with respect to software testing and quality assurance for biomedical natural language processing systems, with the hope of stimulating a healthy change.
The claim to have produced a useful tool is a commonplace in the biomedical natural language processing literature. The explicitly stated motivation for much work in the field is to assist in the understanding of disease or of life, not to advance the state of computer science or of understanding of natural (i.e., human) language. (In this, the biomedical natural language processing community differs from the mainstream NLP community, which at least in theory is motivated by a desire to investigate hypotheses about NLP or about natural language, not to produce tools.) Software is widely known to be characterized by “bugs,” or undesired behaviors—[15] reviews a wide range of studies that suggest an industry average of error rates of 1 to 25 bugs per thousand lines of code in a wide variety of types of software, and a Food and Drug Administration analysis of 3,140 medical device recalls in the 1990s concluded that 7.7% of them (242/3,140) were due to software errors [8] (p. 7). Given the stated intent to provide “mission-critical” tools to doctors and researchers, one might expect due diligence with regard to the quality of software artifacts to be a commonplace in the biomedical natural language processing community and an established subfield of its research milieu. Surprisingly, that is not the case: on the widest imaginable definition of quality assurance, there are fewer than a dozen published studies on quality assurance for biomedical natural language processing software, despite the high (and rapidly growing) level of activity in the biomedical natural language processing area reported in [24] and reviewed in work such as [25]. Given the apparently urgent need for biomedical natural language processing tools that many papers claim in an introductory paragraph citing the latest count of papers in PubMed/MEDLINE, it seems plausible that although researchers in the area are exercising due diligence with respect to the artifacts that they produce, they simply are not taking the time to do research on quality assurance per se. We assayed the extent to which this might be the case, and report the results here.
3. Methods and results for assessing natural language processing applications with respect to software testing and quality assurance
Our methodology was simple. We examined 20 web sites that either provide some form of text mining service (e.g. gene name identification or protein-protein interaction extraction) or provide access to the output of text mining (e.g. a textmining-produced database). On each web site, we tried the most basic software test imaginable. This test, which our experience suggests is probably the first action performed by a typical professional software tester presented with any new application to test, consists of passing the application an empty input. For many web sites, the test consisted of simply hitting the “Submit” button or its equivalent. For some web sites, this was first preceded by clearing sample input from a text box. This is indeed the simplest and most basic software test of which we are aware. We make the following (undoubtedly simplified) assumption: if the system builders paid any attention to software testing and quality assurance at all, they will have run this test; evidence that they tried the test will be that the system responds to a blank input by prompting the user to populate the empty field.
What constitutes an appropriate response to an empty input? We propose that the best response to an empty input where a non-empty input was expected is to give the user helpful feedack—to prompt the user to provide an input. For a GUI-based application, the next-best response is probably Google’s strategy—to do nothing, and present the user with the exact same input screen. (For an API, the second-best response may well be to throw an uncaught exception—this has the advantage of making it clear to the programmer that something is amiss.) This second-best response is not necessarily bad.
A bad response would be to return something. For example, we found that in response to an empty input, many systems will return something along the lines of “0 results found”. This is bad in that it does not allow the user (or the calling routine) to differentiate between the situation where no results were found because the input is empty and the situation where no results were found because there truly should not have been any results—the user is given no indication whatsoever that anything was amiss with the input in the former case. (A less common phenomenon that we observed was that a system might return results that are clearly invalid if examined by a human. For example, one system returned a table full of SQL error messages when passed an empty input. This may not be a problem when called from a GUI, but if called by an API, the results might not be noticed until much later in the processing pipeline, if at all, and are likely to be difficult to track down to their origin.) Finally, the worst possible response is to return something that looks like a legitimate response. For example, one application that we examined returns a perfectly valid-looking list of disease-associated quantitative trait loci (multiple gene locations that contribute to a single physical manifestation) when passed an empty input. This program may seriously mislead an application that calls it.
In total, we examined 23 web sites, selecting them in alphabetical order from a web page that lists biomedical natural language processing applications1. Two of them were down completely, and one of them crashed every time that we attempted to use it, whether or not the input field was empty. Table 1 summarizes the results: of the 20 that were operative and did not crash, a full 7/20 returned either the “bad” or the “worst” type of results, and one of those seven returned the worst.
Table 1.
Summary of behaviors. 7 of 20 sites returned the “bad” or “worst” type of results.
| Response type | Sites |
|---|---|
| Good (prompt or input screen displayed) | 13 |
| Bad (invalid-appearing or false 0 returned) | 6 |
| Worst (valid-appearing data returned) | 1 |
This test assesses the user interface, not the underlying functionality. However, we suspect that the testing applied to the interface that authors use to showcase their work to the world may be better than the testing applied to their underlying application. And, it is certainly the case that this test can reveal real problems with the underlying application, as in the system described above which returned a table of SQL error messages in response to the test.
As a reviewer pointed out, the test is not repeatable—web sites go down, their functionality is changed, and their authors sometimes respond to bug reports. However, the survey captures a snapshot of the state of the biomedical natural language processing world at one point in time, and the general approach is applicable to any application.
4. A Linguistically Motivated Approach to Testing Natural Language Processing Applications
Although the natural language processing community has a long tradition of global evaluation of applications in terms of global metrics like precision, recall, and F-measure, there has been much less work on granular evaluation of the performance of such applications. (In the remainder of the paper, there is a deliberate blurring or mixing of metaphors between what Palmer and Finin have called glass-box evaluation (which I refer to as granular evaluation), or fine-grained evaluation of specific linguistic features [18]; and finding errors in performance, or bugs. As will be seen, it is fruitful to blur this distinction.) There has been correspondingly little research on methods for doing so. We describe here a methodology for granular evaluation of the performance of natural language processing applications using techniques from traditional software testing and from linguistics. Software testing conventionally makes use of test suites. A test suite is a set of test inputs with known desired outputs that is structured so as to explore the feature space of a specified type of input. Test cases are built by determining the set of features that a type of input might have and the contexts in which those features might be found. For a simple example, a function that takes numbers as inputs might be tested with a test suite that includes integers, real numbers, positive numbers, negative numbers, and zero. Good testing also includes a suite of “dirty” or unexpected inputs—for example, the function that takes numbers as inputs might be passed a null variable, a non-null but empty variable, and letters.
There is a theoretical background for test suite construction. It turns out to be overwhelmingly similar to the formal foundations of linguistics. Indeed, if one examines the table of contents of a book on the theory of software testing (see several listed below) and Partee et al.’s textbook on the formal foundations of linguistics [19], one finds very similar chapters. The table of contents of [19] includes chapters on basic concepts of set theory, relations and functions, properties of relations, basic concepts of logic and formal systems, statement logic, predicate logic, finite automata, formal languages, and Type 3 grammars. Similarly, if we look at the contents of a good book on software testing, we see coverage of set theory [2], graphs and relations [3], logic [2], and automata [2,3, 14].
The theoretical similarities between software testing and linguistics turn out to translate into practical methodologies, as well. In particular, the techniques of software testing have much in common with the techniques of descriptive or field linguistics—the specialty of determining the structures and functioning of an unknown language. In the case of software testing, an application is approached by determining the features of inputs and combinations of inputs (both “clean” and “dirty”) that an application might be presented with, and constructing test suites to explore this feature space. In field linguistics, an unknown language is approached by constructing questions to be answered about the language that allow us to determine the elements of the language on all levels—phonemic and phonetic (sounds), morphological (word formation), lexicon (words), syntactic (phrasal structure)—and the ways in which they can combine. These questions are formulated in sets called schedules that are assembled to elucidate specific aspects of the language, in a procedure known as scheduled elicitation. The software tester’s test suites have a clear analogue in the “schedules” of the field linguist. Like test suites, schedules include “dirty” data, as well—for example, in studying the syntax of a language, the linguist will test the acceptability of sentences that his or her theory of the language predicts to be ungrammatical. Thus, even though there has not been extensive research into the field of software testing of natural language processing applications, we already have a well-developed methodology available to us for doing so, provided by the techniques of descriptive linguistics.
An example of how the techniques of software testing and descriptive linguistics can be merged in this way is provided in [6]. This paper looked at the problem of testing named entity recognition systems. Named entity recognition is the task of finding mentions of items of a specific semantic type in text. Commonly addressed semantic types have been human names, company names, and locations (hence the term “named entity” recognition). [6] looked at the application of named entity recognition to gene names. They constructed a test suite based on analyzing the linguistic characteristics of gene names and the contexts in which they can appear in a sentence. Linguistic characteristics of gene names included orthographic and typographic features on the level of individual characters, such as letter case, the presence or absence of punctuation marks (gene names may contain hyphens, parentheses, and apostrophes), and the presence or absence of numerals. (Gene names and symbols often contain numbers or letters that indicate individual members of a family of genes. For example, the HSP family of genes contains the genes HSP1, HSP2, HSP3, and HSP4.) Morphosyntactic features addressed characteristics of the morpheme or word, such as the presence or absence of participles, the presence or absence of genitives, and the presence or absence of function words. The contextual features included whether or not a gene name was an element of a list, its position in the sentence, and whether or not it was part of an appositive construction. (Gene names can have a dizzying variety of forms, as they may reflect the physical or behavioral characteristics of an organism in which they are mutated, the normal function of the gene when it is not mutated, or conditions with which they are associated. Thus, we see gene names like pizza (reflecting the appearance of a fly’s brain when the gene is mutated), heat shock protein 60 (reflecting the function of the gene), and muscular dystrophy (reflecting a disease with which the gene is associated). This high range of variability adds greatly to the difficulty of gene name recognition.)
Five different gene name recognition systems were then examined. These features of gene names and features of contexts were sufficient to find errors in every system. One system missed every one-word gene name. One system missed lower-case-initial gene names when they occurred in sentence-initial position. (Sentences in genomics articles can begin with a lower-case letter if they begin with the name of a gene and the mutant form of the gene, commonly named with a lower-case-initial name, is being discussed.) One system only found multi-word gene names if every word of the name was upper-case-initial. One system only found multi-word names if they ended with an alphanumeric modifier (e.g. the gene names alcohol dehydrogenase 6 or spindle A). One system missed all numerals at the right edge of gene names (see preceding example). One system missed names, but not symbols, containing hyphens (catching symbols like Nat-1 but missing names like the corresponding N-acetyltransferase 1). One system missed names containing apostrophes just in the case where they were genitives (catching names like 5’ nucleotidase precursor but missing names like corneal dystrophy of Bowman’s layer type II (Thiel-Behnke)). Two systems had failures related to the format of Greek letters. One system performed well on symbols but did not recognize any names at all. (Genes typically have both a name, such as white or N-acetyltransferase 1, and a “symbol,” similar to an abbreviation, such as w for white and Nat-1 for N-acetyltransferase.)
Test suites are effective for granular evaluation of performance, but should not be able to predict global measures such as precision, recall, and F-measure, since the proportions of named entity types in the test suite do not reflect the distribution of those types in naturally occurring data. (In fact, this is one of their advantages—as pointed out by [17], an advantage of test suites is that they limit the redundancy of common entity types and address the scarcity of rare entity types that are observed in naturally occurring data.) However, it was hypothesized that test suites might be able to predict performance on specific equivalence classes of inputs (where an equivalence class is a set of inputs that are all expected to test the same functionality and reveal the same bugs; they are similar to what linguists call natural classes). To test this hypothesis, the authors built a number of simple test suites, varying only the length of the gene name, letter case, hyphenation, and sentence position. They then ran a single gene name recognition system on all of these test suites. Based on the results obtained from the test suites, they made the following predictions:
Recall should be poor for gene names with initial numerals, such as 12-LOX and 18-wheeler.
Recall should be poor for gene names that contain function words, such as Pray for elves and ken and barbie.
Recall should be poor for upper-case-initial gene names in sentence-medial position
Recall should be poor for 3-character-long symbols.
Recall should be good for numeral-final gene names such as yeast heat shock protein 60.
The system was then used to process two corpora containing gene names—the BioCreative I corpus [23] and the PMC corpus [22]. Overall performance for the BioCreative I corpus was a precision of 0.65 and recall of 0.68. Overall performance for the PMC corpus was a precision of 0.71 and recall of 0.62.
The performance of the system for the various equivalence classes was as shown in Table 2.
Table 2.
Performance on two corpora for the predictable categories [6].
| Prediction | BioCreative | ||||
|---|---|---|---|---|---|
| TP | FP | FN | P | R | |
| 1 | 12 | 57 | 17 | 0.17 | 0.41 |
| 2 | 0 | 1 | 38 | 0.0 | 0.0 |
| 4 | 556 | 278 | 512 | 0.67 | 0.52 |
| 5 | 284 | 251 | 72 | 0.53 | 0.80 |
| PubMed Central | |||||
| TP | FP | FN | P | R | |
| 1 | 8 | 10 | 0 | 0.44 | 1.0 |
| 2 | 1 | 0 | 2 | 1.0 | 0.33 |
| 4 | 163 | 64 | 188 | 0.72 | 0.46 |
| 5 | 108 | 54 | 46 | 0.67 | 0.70 |
The predictions based on the test suites were almost entirely supported. The single anomaly was the high recall observed on the PMC corpus for prediction 1, where low recall was predicted. In all other cases, the predictions were correct—recall for the equivalence class was predicted to be low for 1, 2, and 4 and it was lower than the recall for the corpus as a whole for these equivalence classes; recall was predicted to be high for 5, and it was higher than the recall for the corpus as a whole for this equivalence class.
It will be noted that there are no results given for prediction 3. This is because it concerns letter case, and letter case had been normalized to lower case in the corpora. This points out again an advantage of test suites—we know that such gene names exist in the literature, but they were not represented in these corpora at all, making the corpora unsuitable for assessing the performance of a system on this type of name.
It should be noted that these findings are significant (in the non-statistical sense of that term) because of the small numbers of items in some of the cells, not in spite of it. These details of performance would likely be lost in an evaluation that only assessed precision, recall, and F-measure, and are the difference between finding or missing elusive statements that are of crucial interest to the biologist, perhaps precisely because of their rarity.
5. An Engineering Perspective on the Use of Test Suites Versus Corpora
To the extent that testing is considered in the natural language processing community, there is an implicit assumption that the way to test an application is by running it on a large corpus. We tested this assumption by measuring code coverage when a natural language processing application was run with a large corpus as its input and with a small structured test suite as its input. The natural language processing application was a semantic parser known as OpenDMAP [11]. It allows free mixing of terminals and non-terminals, and semantically typed phrasal constituents, such as “gene phrases.” It has been applied to a variety of information extraction tasks in the biomedical domain and has achieved winning results in two shared tasks [1,9].
Code coverage is a measure of the percentage of code in an application that is executed during the running of a test suite. The goal is to maximize coverage—bugs in code will not be found if the code is not executed. Various kinds of coverage can be measured. Line coverage is the percentage of lines of code that have been executed. It is the weakest indicator of code coverage. Branch coverage is the percentage of branches within conditionals that have been traversed. It is more informative than line coverage.
The corpus that we employed was the largest biomedical corpus available at the time. It consisted of 3,947,200 words. The test suite that we used was much smaller. It contained altogether 278 test cases constructed by the application developer. He did not monitor code coverage while designing the test suite.
Table 3 (next page) shows the results of running the application on the corpus and on the test suite. As can be seen, the small test suite yielded higher code coverage for every component of the system and every measure of code coverage—sometimes much higher coverage, as in the case of branch coverage for the rules components, where the corpus achieved 24% code coverage and the test suite achieved 71% code coverage. The last three columns show the results of an experiment in which we varied the size of the rule set. As can be seen from the fact that the coverage for the entire rule set, a partition of the rule set that only covered nominals, and a partition of the rule set that covered only verbs, are all equal, the number of rules processed was not a determiner of code coverage.
Table 3.
Application- and package-level coverage statistics using the test suite, the full corpus with the full set of rules, and the full corpus with two reduced sets of rules. The highest value in a row is bolded. The last three columns are intentionally identical [7].
| Metric | Functional tests Corpus, all rules nominal rules verbal rules | |||
|---|---|---|---|---|
| Overall line coverage | 56% | 41% | 41% | 41% |
| Overall branch coverage | 41% | 28% | 28% | 28% |
| Parser line coverage | 55% | 41% | 41% | 41% |
| Parser branch coverage | 57% | 29% | 29% | 29% |
| Rules line coverage | 63% | 42% | 42% | 42% |
| Rules branch coverage | 71% | 24% | 24% | 24% |
| Parser class coverage | 88% (22/25) | 80% (20/25) | ||
| Rules class coverage | 100% (20/20) | 90% (18/20) | ||
In a further experiment, we examined how code coverage is affected by variations in the size of the corpus. We monitored coverage as increasingly larger portions of the the corpus were processed. The results for line coverage are shown in Figure 1. (The results for branch coverage are very similar and are not shown.) The x axis shows the number of sentences processed. The thick solid line indicates line coverage for the entire application. The thin solid line indicates line coverage for the rules package. The broken line and the right y axis indicate the number of pattern matches.
Fig. 1.
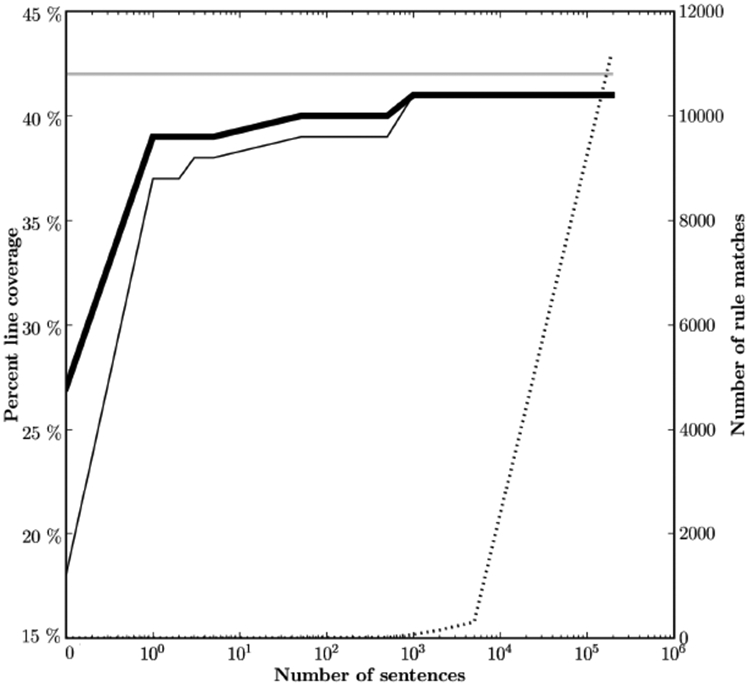
Increase in percentage of line coverage as increasing amounts of the corpus are processed. The left y axis is the percent coverage. The right y axis is the number of rule matches [7].
As the figure shows quite clearly, increasing the size of the corpus does not lead to increasing code coverage. It is 39% when a single sentence has been processed, 40% when 51 sentences have been processed, and 41%—the highest value that it will reach—when 1,000 sentences have been processed. The coverage after processing 191,478 sentences—the entire corpus of almost 4,000,000 words—is no higher than it was at 1,000 sentences, and is barely higher than after processing a single sentence.
Thus, we see that the “naturally occurring data assumption” does not hold—from an engineering perspective, there is a clear advantage to using structured test suites.
This should not be taken as a claim that running an application against a large corpus is bad. In fact, we routinely do this, and have found bugs that were not uncovered in other ways. However, testing with a structured test suite should remain a primary element of natural language processing software testing.
It will be noted that even with the structured test suite, our code coverage was less than 60% overall, as predicted by Wieger’s work, which shows that when software is developed without monitoring code coverage, typically only 50-60% of the code is executed by test suites [15] (p. 526). However, as soon as we tried to increase our code coverage, we almost immediately uncovered two “showstopper” bugs.
6. Discussion
Although our assay of the software testing status of biomedical natural language processing applications was crude, the findings are consistent with the claim that 7/20 biomedical natural language processing web sites have not been subjected to even the lowest, most superficial level of software testing. For the rest, we cannot conclude that they have been adequately tested—only that they appear to have benefited from at least the lowest, most superficial level of testing.
This absence of software testing and quality assurance comes despite the fact that like the mainstream NLP community, the biomedical natural language processing community has paid considerable attention to software evaluation. Some clarification of terminology is useful here. [10] distinguish between gold-standard-based evaluation and feature-based evaluation. This is completely analogous to the distinction between what we are referring to as evaluating software with respect to some metric (gold-standard-based evaluation) and what we are referring to as testing it, or attempting to find bugs (feature-based evaluation). The biomedical natural language processing community has participated enthusiastically in software evaluation via shared tasks—agreed-upon task definitions used to evaluate systems against a shared data set using centralized, third-party evaluation with a corpus (or a document collection) as input and with an agreed-upon implementation of a scoring metric. However, the community’s investment in testing its products has apparently been much smaller. It has been suggested [20] that biomedical natural language processing applications are ready for use by working bioscientists. If this is the case, we argue that there is a moral obligation on the part of biomedical natural language processing practitioners to exercise due diligence and ensure that their applications do not just perform well against arbitrary metrics, but also behave as intended.
We showed in our experiments with building linguistically motivated test suites that such test suites, informed by the techniques of descriptive linguistics, are effective at granular characterization of performance across a wide variety of named entity recognition systems. We also demonstrated the surprising finding that such test suites could be used to predict global performance scores such as precision, recall, and F-measure (although only recall was predicted in our experiment) for specific equivalence classes (or, as linguists call them, natural classes) of inputs.
Drawing directly on a software engineering technique, we used a test suite to test the commonly held, if tacit, assumption that large corpora are the best testing material for natural language processing applications. We demonstrated that in fact even a small test suite can achieve much better code coverage than a very large corpus.
As a reviewer pointed out, most linguistic phenomena are Zipfian in nature. How far must we go in evaluating and handling the phenomena in the Zipfian tail? Steedman has an insightful observation on this question:
We have come to believe that the linguists have forgotten Zipf’s law, which says that most of the variance in linguistic behavior can be captured by a small part of the system.
The linguists, on the other hand, think that it is we who have forgotten Zipf’s law, which also says that most of the information about the language system as a whole is in the Long Tail.
It is we who are at fault here, because the machine learning techniques that we rely on are actually very bad at inducing systems for which the crucial information is in rare events…
One day… the Long Tail will come back to haunt us.
[21]
Even for work whose goal is not application-building but basic research, the costs of failing to attend to basic software testing and quality assurance issues can be quite severe. As Rob Knight has put it, “For scientific work, bugs don’t just mean unhappy users who you’ll never actually meet: they mean retracted publications and ended careers. It is critical that your code be fully tested before you draw conclusions from the results it produces.” The recent case of Geoffrey Chang (see [16] for a succinct discussion) is illustrative. In 2006, he was a star of the protein crystallography world. That year he discovered a simple software error in his code which led to a reversal of the sign (positive versus negative) of two columns of numbers in his output. This led to a reversed prediction of handedness in the ABC transporter gene MsbA. This error had implications for the work of many other scientists in addition to his own. The story is an object lesson in the potential consequences of failure to attend to basic software testing and quality assurance issues, although his principled response to the situation suggests that in his case, those consequences will be limited to retracted publications and will not be career-ending (see [5] for the retractions). For the sorts of standard software testing techniques that we looked for in the work reported here, a considerable amount of good material is available, ranging from cookbook-like how-to manuals (e.g. [13]) to theoretical work [3,14,4]. Language processing presents a number of specific testing issues related to unique characteristics of the input data, and the literature on it is quite limited (but see [6,12, 7] for some attempts to address this topic in the biomedical natural language processing domain, specifically). No non-trivial application is ever likely to be completely free of bugs, but that does not free us of the obligation to test for them. As we have shown here, approaches to doing so that are inspired by linguistic techniques are effective at granular characterization of performance, finding bugs, and achieving high code coverage.
Acknowledgements
We would like to thank our co-authors on some of the work cited in this paper. Lynette Hirschman (The MITRE Corporation) and Bob Carpenter (Alias-I) have discussed the general issues of software testing and quality assurance with us extensively. Martin Krallinger’s web page at biocreative.sourceforge.net listing web-based biomedical NLP services was invaluable in performing the work reported here. Jonathan M. Cohen (Monterey County Public Defender’s Office) and John Pestian (Cincinnati Children’s Hospital Medical Center) helped us understand regulatory issues regarding medical software. This work was funded in part by grants NIH 5 R01 LM009254-06, NIH 5 R01 LM008111-07, NIH 5 R01 GM083649-04, and NIH 5 R01 LM009254-03 to Lawrence E. Hunter.
Footnotes
References
- 1.Baumgartner William A. Jr.; Zhiyong Lu; Johnson Helen L.; Caporaso J. Gregory; Paquette Jesse; Lindemann Anna; White Elizabeth K.; Medvedeva Olga; Cohen K. Bretonnel; and Hunter Lawrence E. (2008) Concept recognition for extraction protein interaction relations from biomedical text. Genome Biology 9(Suppl. 2):S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beizer Boris (1990) Software testing techniques, 2nd edition. International Thomson Computer Press. [Google Scholar]
- 3.Beizer Boris (1995) Black-box testing: Techniques for functional testing of software and systems. Wiley. [Google Scholar]
- 4.Binder Robert V. (1999) Testing object-oriented systems: models, patterns, and, tools. Addison-Wesley Professional. [Google Scholar]
- 5.Geoffrey Chang; Roth Christopher R.; Reyes Christopher L.; Pornillos Owen; Chen Yen-Ju; and Chen Andy P. (2006) Letters: Retraction. Science 314:1875. [DOI] [PubMed] [Google Scholar]
- 6.Cohen K. Bretonnel; Tanabe Lorraine; Kinoshita Shuhei; and Hunter Lawrence (2004) A resource for constructing customized test suites for molecular biology entity identification systems. BioLINK 2004: Linking biological literature, ontologies, and, databases: tools for users, pp. 1–8. Association for Computational Linguistics. [Google Scholar]
- 7.Cohen K. Bretonnel; Baumgartner William A. Jr.; and Hunter Lawrence (2008) Software testing and the naturally occurring data assumption in natural language processing. Software engineering, testing, and quality assurance for natural language processing, pp. 23–30. Association for Computational Linguistics. [Google Scholar]
- 8.Food and Drug Administration, US Department of Health and Human Services (2002) General principles of software validation: Final guidance for industry and FDA staff.
- 9.Joerg Hakenberg; Leaman Robert; Vo Nguyen Ha; Jonnalagadda Siddhartha; Sullivan Ryan; Miller Christopher; Tari Luis; Baral Chitta; and Gonzalez Graciela (2010) Efficient extraction of protein-protein interactions from full-text articles. IEEE/ACM Transactions on Computational Biology and Bioinformatics, July 2010. [DOI] [PubMed] [Google Scholar]
- 10.Lynette Hirschman; and Mani Inderjeet (2003) Evaluation. Chapter 23 of Ruslan Mitkov (editor), The Oxford Handbook of Computational Linguistics, Oxford University Press. [Google Scholar]
- 11.Hunter Lawrence E.; Zhiyong Lu; James Firby; Baumgartner William A. Jr.; Johnson Helen L.; Ogren Philip V.; and Bretonnel Cohen K (2008) OpenDMAP: An open-source, ontology driven concept analysis engine, with applications to capturing knowledge regarding protein transport, protein interactions and cell-specific gene expression. BMC Bioinformatics 9(78). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson Helen L.; Bretonnel Cohen K; and Hunter Lawrence (2007) A fault model for ontology mapping, alignment, and linking systems. Pacific Symposium on Biocomputing 2007, pp. 233–244. World Scientific Publishing. [PMC free article] [PubMed] [Google Scholar]
- 13.Cem Kaner; Nguyen Hung Quoc; and Falk Jack (1999) Testing computer software, 2nd edition. John Wiley and Sons. [Google Scholar]
- 14.Marick Brian (1997) The craft of software testing: subsystem testing including object-based and object-oriented testing. Prentice Hall. [Google Scholar]
- 15.McConnell Steve (2004) Code complete, 2nd edition. Microsoft Press. [Google Scholar]
- 16.Miller Greg (2006) A scientist’s nightmare: software problem leads to five retractions. Science 314:1856–1857. [DOI] [PubMed] [Google Scholar]
- 17.Oepen S; Netter K; and Klein J (1998) TSNLP - test suites for natural language processing. Nerbonne John, editor, Linguistic Databases, Chapter 2, pp. 13–36. CSLI Publications. [Google Scholar]
- 18.Palmer Martha; and Tim Finin (1990) Workshop on the evaluation of natural language processing systems. Computational Linguistics 16(3):175–181. [Google Scholar]
- 19.Partee Barbara H.; Alice ter Meulen; and Wall Robert E. (1990) Mathematical methods in linguistics. Springer. [Google Scholar]
- 20.Dietrich Rebholz-Schuhmann; Kirsch Harald; and Couto Francisco (2005) Facts from text—is text mining ready to deliver? PLoS Biology 3(2):188–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steedman Mark (2008) On becoming a discipline. Computational Linguistics 34(1):137–144. [Google Scholar]
- 22.Lorraine Tanabe; and Wilbur W. John (2002) Tagging gene and protein names in biomedical text. Bioinformatics 18(8):1124–1132. [DOI] [PubMed] [Google Scholar]
- 23.Lorraine Tanabe; Xie Natalie; Thom Lynne H.; Matten Wayne; and Wilbur W. John (2005) GENETAG: a tagged corpus for gene/protein name recognition. BMC Bioinformatics 6(Suppl. 1):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karin Verspoor; Cohen K. Bretonnel; Mani Inderjeet; and Goertzel Benjamin (2006) Introduction to BioNLP’06. Linking natural language processing and biology: towards deeper biological literature analysis, pp. iii–iv. Association for Computational Linguistics. [Google Scholar]
- 25.Pierre Zweigenbaum; Demner-Fushman Dina; Yu Hong; and Cohen K. Bretonnel (2007) Frontiers for biomedical text mining: current progress. Briefings in Bioinformatics 8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]


