Abstract
Oxidative stress has been postulated to play a role in several diseases, including cardiovascular diseases, diabetes, and stress-related disorders (anxiety/depression). Presently, natural plant-derived phytochemicals are an important tool in reducing metabolomic disorders or for avoiding the side effects of current medicinal therapies. Brown Rice (Oryza sativa L.) is an important part of Asian diets reported as a rich source of bioactive phytonutrients. In our present study, we have analyzed the effect of different lactic acid bacteria (LABs) fermentation on antioxidant properties and in the enhancement of bioactive constituents in Korean brown rice. Therefore, the antioxidant activities and phytochemical analysis were investigated for raw brown rice (BR) and different fermented brown rice (FBR). BR fermented with Limosilactobacillus reuteri, showed the highest antioxidant activities among all samples: DPPH (121.19 ± 1.0), ABTS (145.80 ± 0.99), and FRAP (171.89 ± 0.71) mg Trolox equiv./100 g, dry weight (DW). Total phenolic content (108.86 ± 0.63) mg GAE equiv./100 g, DW and total flavonoids content (86.79 ± 0.83) mg catechin equiv./100 g, DW was also observed highest in Limosilactobacillus reuteri FBR. Furthermore, phytochemical profiling using ultra-high-performance liquid tandem chromatography quadrupole time-of-flight mass spectrometry (UHPLC-QTOF/MS) and cell antioxidant assay (CAA) revealed L. reuteri FBR as a strong antioxidant with an abundance of bioactive compounds such as gamma-aminobutyric acid, coumarin, cinnamic acid, butanoic acid, ascorbic acid, nicotinic acid, and stearic acid. This study expanded current knowledge on the impact of fermentation leading to the enhancement of antioxidant capacity with an abundance of health-related bioactive compounds in BR. The results obtained may provide useful information on functional food production using fermented brown rice.
Keywords: brown rice, fermentation, antioxidants, oxidative stress, untargeted metabolomics, UHPLC-QTOF/MS, health benefits
1. Introduction
Oxidative stress is a condition that is caused by an imbalance between antioxidants and free radicals of living organisms. This imbalance occurs due to the excessive production of reactive oxygen species (ROS) or antioxidant deficiency that leads to the damage of aerobic organisms as well as chronic inflammation; referred to as oxidative stress [1]. Lower ROS concentration is important for normal cellular signaling, while excess ROS can cause oxidative damage to DNA, lipids, proteins, and is associated with several chronic diseases [2,3]. The current definition of oxidative stress includes metabolic stress-related pathways that participate in both cellular and extracellular metabolic events. The biology of oxidative stress is extremely complex, with multiple mechanisms at work [4]. Regardless of the mechanism, oxidative stress causes the onset of many diseases including cardiovascular diseases, diabetes, and anxiety or depression which are considered a major public health issue worldwide. As a result, consuming antioxidants to prevent oxidative stress is becoming important for health. Moreover, an increase in the health-consciousness of consumers has increased the demand for nutritional and disease-preventing functional foods, probiotics, prebiotics, and postbiotics. Numerous studies have focused on probiotics, specifically lactobacilli strains, that have the potential to act as antioxidants to protect the host from oxidative stress [5]. Some lactobacilli strains have been found to quench oxygen free radicals using a chemical antioxidant method.
Many studies have been reported that phytochemicals (e.g., polyphenols and phenolic acids) derived from natural plants have the potential to target oxidative stress and inflammatory pathways [6,7]. Rice is a staple food (in many countries) that belongs to the grass family (Oryza sativa). The total worldwide production of rice was about 769,657,791 tonnes in an area of 167,249,103 ha. Epidemiological studies have shown that the low incidence of chronic diseases in rice-consuming regions can be correlated with rice antioxidants [8,9]. The antioxidant activity and phytochemical content of brown rice (BR) have been recorded in several studies. Components such as as γ-oryzanol, phenolic acids, gamma-aminobutyric acid (GABA), flavonoids, and γ-tocotrienol contribute to the health-promoting properties of brown rice [10].
Evidence supports the effect of solid-state fermentation (SSF) techniques using lactic acid bacteria (LABs) and fungal strains on antioxidant levels and bioactive properties in a variety of substrates, including barley [11], pearl millet [12], and rice [2]. Many researchers, food scientists, and industrialists use the SSF process to enhance the nutritional quality of food and food products. Biological methods are environmentally friendly, relatively safe, and rely on the use of appropriate and specific microorganisms [13]. Our study aimed to provide knowledge to quantify the quality of these phytochemical antioxidants in whole brown rice to meet the needs of food producers and consumers of rice: (1) to analyze the antioxidant properties of differently fermented brown (FBR) rice over raw brown rice (BR); (2) detection of bioactive compounds in raw BR and different LABs fermented brown rice using ultra-high performance liquid chromatography quadrupole time-of-flight mass spectrometry (UHPLC-QTOF/MS), and (3) detection of cellular antioxidant activity of the best LAB fermenting bacterial strain (L. reuteri FBR).
2. Materials and Methods
2.1. Rice Samples
BR (Oryza sativa L. Var. Japonica) used in this experiment were obtained from the local market of Chuncheon, Gangwon-do, South Korea. Using an electric mill, raw BR and brown rice after processing (fermentation) were pulverized into a fine powder and sifted through a 40 mesh sieve. Before further extraction, samples were kept at −20 °C.
2.2. Chemicals
Acetonitrile, ethanol, methanol, sodium carbonate, sodium hydroxide, anhydrous sodium acetate, hydrochloric acid, potassium persulfate, acetic acid and sulfuric acid, Dulbecco’s Modified Eagle Medium (DMEM), L-glutamine, 4-(2-hydroxyethyl)piperazine-1-ethane sulfonic acid (HEPES), phosphate-buffered saline (PBS), fetal bovine serum (FBS), penicillin-streptomycin, Hanks’ Balanced Salt Solution (HBSS), and trypsin EDTA were purchased from Daejung chemicals and metals Co., Ltd., South Korea. The phenolic standards and other chemicals such as Folin–Ciocalteu reagent, ABTS (2,2′-Azino-bis (3-ethyl benzothiazoline-6-sulfonic acid), DPPH (2,2-diphenyl-1-picrylhydrazyl); Trolox (6-hydroxy-2,5,7,8-tetramethyl chroman-2-carboxylic acid), p-Coumaric acids, dimethyl sulfoxide (DMSO), TPTZ (2,4,6-Tris (2-pyridyl)-s-triazine), gallic, ferulic, (+)-catechin hydrate (≥98% purity by HPLC), and quercetin dehydrate (≥98% purity by HPLC), 2,2′-azobis (2-amidino propane) dihydrochloride (ABAP); and 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA, ≥97% purity), were obtained from Sigma, Seoul, South Korea.
2.3. Microorganisms
Limosilactobacillus reuteri AKT1 and all other lactic acid bacterial strains used for fermentation in our study were obtained from the Department of Food Science and Biotechnology, Kangwon National University, Korea. LABs were chosen for fermentation in the current study because they demonstrated high GABA (inhibitory neurotransmitter) content and fermentation efficiency in our last study (data not shown) [2]. The bacteria stock culture was kept at −80 °C in MRS broth (Difco), which contained 20% glycerol (v/v).
2.4. Sample Preparation
2.4.1. Brown Rice Fermentation
The LAB’s growth medium consists of sterilized rice powder in distilled water. Before inoculation with lactic acid bacteria, the growth media were autoclaved for 15 min at 121 °C. Different bacterial strains used for fermentation (L. reuteri (AKT1), L. fermentum (AKT2), and L. plantarum (FMP2)) (2 × 107 cfu/mL) were transferred from 12 h (overnight) incubated culture to 100 mL of autoclaved growth media. The media was then incubated for 48 h at 37 °C with 150 rpm agitation, then centrifuged for 10 min at 10,000× g and the supernatant was freeze-dried and stored at −20 °C for further study.
2.4.2. Preparation of Extracts
Extraction was done by the method used by Pradeep et al. [14] with some modification. To remove lipids, samples were defatted with hexane using soxhlet equipment. Grounded samples were defatted three times in an orbital shaker at room temperature with hexane (1:5, w/v, 2 h). Defatted flours were kept at −20 °C till further use. Soluble phenolics from defatted samples (5 g) were extracted in an orbital shaker (RK-2D, DAIHAN scientific, Wonju, Korea) for 1 h at 50 °C with 50% ethanol (1:20 w/v). The extracts were centrifuged (Union 32R plus, Hanil Science Industrial, Incheon, Korea) at 4000 rpm for 10 min and supernatants were collected and this process was repeated until the third extraction. The supernatants were evaporated at 50 °C and freeze-dried. Before being reconstituted in ethanol, the lyophilized solids were stored at −20 °C. The stock solution of samples was prepared at a concentration of 1 mg/mL that was used throughout the analysis.
2.5. Determination of Total Phenolic Content (TPC)
Folin–Ciocalteu colorimetric method was used to measure the TPC reported by Pradeep et al. [15] with few modifications. In brief, 100 μL of the sample extract or standard (gallic acid solution) or 95% (v/v) methanol as blank was treated with 200 μL of Folin–Ciocalteu reagent for a short duration of 6 min. The mixture was then alkalized with 1 mL of Na2CO3 700 mM. After being kept in dark conditions for 90 min, the SpectraMax i3 plate reader (Molecular Devices Korea, LLC, Seoul, Korea) was used to measure the absorbance at 760 nm wavelength. Gallic acid was used as a standard to calculate the TPC and results were expressed as milligrams of gallic acid equivalent per 100 g of sample (mg GAE equiv./100 g, DW).
2.6. Determination of Total Flavonoid Content (TFC)
TFC was analyzed using the method of Pradeep et al. [15] with few modifications. Briefly, a 200 μL sample extract was combined with 75 μL of NaNO2 (50 gL−1) followed by the addition of 1 mL of distilled water. Then, the reaction mixture was allowed to settle for 5 min and then 75 μL of AlCl3 (100 gL−1) was added. After 6 min, 600 μL of distilled water followed by 500 μL of 1 M NaOH were added. The SpectraMax i3 plate reader (Molecular Devices Korea, LLC) was used to measure the absorbance at 510 nm wavelength. Catechin was used as a standard and results were expressed as milligram catechin equivalents per 100 g of sample (mg catechin equiv./100 g, DW).
2.7. Determination of Antioxidant Activities of BR
2.7.1. DPPH Radical Scavenging Activity
DPPH activity was determined by the methods of Chang et al. [16] after slight modifications. In short, 100 μL of the sample extract or standard (Trolox) or blank (methanol) was mixed with freshly prepared 100 μL of 500 μM DPPH solution (dissolved in methanol) in a 24-well microplate and incubated at room temperature for 30 min. The absorbance was measured at 515 nm wavelength. The Trolox concentration plot with DPPH radical scavenging activity was used as a baseline curve. DPPH values were expressed as mg Trolox equivalent per 100 g of sample (mg Trolox equiv./100 g, DW) using the following formula:
| DPPH Radical Scavenging Activity (%) = (Ac − Ae)/Ab × 100 |
where Ae represents the absorbance of the extract or standard and Ac represents the absorbance value of the blank sample.
2.7.2. ABTS Radical Scavenging Activity
ABTS assay was carried out as described by Chang et al. [16] with little modifications. ABTS stock solution was prepared by mixing 2.45 mmol/L of potassium persulfate with 7 mmol/L of ABTS solution (1:1, v/v) and kept in the dark for 12–16 h at room temperature. The ABTS + reagent was constantly diluted with methanol until 0.700 ± 0.020 absorbance at 734 nm wavelength. Afterwards, 100 μL of extracts or standards were mixed with ABTS + solution (1 mL) and absorbance was measured at 734 nm. The per cent inhibition of ABTS was measured using the same formula as for the DPPH assay (mentioned above). ABTS values were expressed as mg Trolox equiv./100 g, DW using the Trolox standard curve.
2.7.3. Ferric Reducing Antioxidant Power (FRAP)
FRAP assay was analyzed using the method as documented by Xiang et al. [17] with little modifications. In short, 0.1 mL of extracts were combined with a FRAP reagent of 3.9 mL that was prepared using acetate buffer (50 mL, 0.3 M, pH 3.6), Tripyridyl Triazine (5 mL, TPTZ) solution (10 mM of TPTZ in 40 mM of HCl) and FeCl3 · 6H2O (5 mL, 20 mM) and kept for 10 min at 37 °C, then absorbance was taken at 593 nm wavelength. These findings were expressed as mg Trolox equiv./100 g, DW.
2.8. Identification of BR Bioactive Compounds Using UHPLC-Q-TOF-MS/MS
The bioactive compounds of BR samples were analyzed using UHPLC Q-TOF-MS/MS (SCIEX Exion LC AD system, Framingham, MA, USA) according to the protocol previously conducted in our laboratory by Tyagi et al., and Daliri et al. [2,18]. The mass spectrometric analysis was conducted in both positive (ESI+) and negative (ESI−) ion modes. In summary, UHPLC Q-TOF-MS/MS system was fitted with different components including an autosampler, photodiode array detector, and controller. The analytical column used in the analysis was 100 mm × 3 mm Accucore C18 column (Thermo Fisher Scientific, Waltham, MA, USA). Later, the sample (10 µL) was injected by autosampler and eluted into the column with a binary mobile phase consisting of solvent A (water with 0.1% of formic acid) and solvent B (methanol). A flow rate (0.4 mL/min) with a linear gradient programmed for 25 min was used in this analysis. Under these conditions, the scanning time was approximately 1 s. The bioactive compounds of BR were identified by using a metabolomics workbench.
2.9. Cell Viability Assay
After performing antioxidant assays and untargeted metabolomics of raw BR and different LABs treated BR samples, the best (L. reuteri FBR) sample was selected for further cell line analysis. To test the viability in Caco-2 cell lines, the L. reuteri FBR sample was analyzed using a laboratory EZ-cytox assay. WST of EZ-cytox exists in the respiratory chain of mitochondria and is active only in living cells. Briefly, Caco-2 cells in the growth medium were seeded on a 96-well plate at a density of 4 × 104 cells/well. The growth medium was removed and the cells were washed using PBS after 24 h of incubation at 37 °C with 5 per cent CO2. Then, 100 μL of growth medium with various sample extract concentrations was applied. In the control group, a medium without sample extract was added. After 12 h of incubation at 37 °C with 5 per cent CO2, 10 μL WST-1 solution was added to each sample. The 96-well plate was left for 10 min at 37 °C and the absorbance was measured at 455 nm by SpectraMax i3 plate reader. If a concentration of sample extract reduced the cell viability by >10%, then the extract at this concentration was cytotoxic.
2.10. Cellular Antioxidant Activity (CAA)
The formation of ROS within cells was investigated using oxidation sensitive DCFH-DA probes by the method of Ti et al. [19] with slight modifications. In brief, Caco-2 cells were grown overnight at a density of 6 × 104 cells per well in black 96-well microplates. Later, PBS (50 μL) was used to wash the cells after 2 h of pretreatment with various concentrations of sample extracts (0.5–5 mg/mL) and 100 μL of DCFH-DA (25 μmol/L). Then, 100 μL of DMEM medium (composed of 600 µmol L−1 ABAP) was inoculated in each well excluding the blank well, which received 100 μL of DMEM medium without ABAP. The fluorescence was measured using a plate reader SpectraMax i3 plate reader (Seoul, South Korea) and wavelengths of 485 nm excitation and 538 nm emission for 13 cycles (5 min each). The formula used for calculating the CAA unit was as follows:
| CAA unit = % reduction = (1 − (∫ SA/∫CA)) × 100 |
where ∫SA and ∫CA denote the integral areas under the sample and control time-fluorescence value curves, respectively.
2.11. Statistical Analysis
GraphPad Prisma 8.0 was used to analyze the data. Using the SPSS program and GraphPad Prism 8.0, differences in mean values between brown rice samples of phenolic extracts were calculated using one-way variance analysis (ANOVA) followed by a Tukey test at p < 0.05 significance stage. The findings were referred to as mean standard deviation (SD).
The empirical formula was used to identify compounds such as PubChem (https://pubchem.ncbi.nlm.nih.gov/ accessed on 10 June 2021) or ChemSpider (http://www.chemspider.com/ accessed on 10 June 2021). ClustVis program (http://biit.cs.ut.ee/clustvis/ accessed on 10 June 2021), XCMS online (Metlin) (https://xcmsonline.scripps.edu accessed on 10 June 2021), Paleontological Statistics (PAST), and clustVis (https://biit.cs.ut.ee/clustvis/ accessed on 10 June 2021) were used in multivariate statistical analyses, including heat maps [20].
3. Results and Discussion
3.1. TPC and TFC
The TPC and TFC of all four samples are shown in (Table 1) as mean ± SD of triplicate analyses with statistically significant differences (Tukey and Duncan test p ≤ 0.05). TPC ranged between 16.08 ± 0.49 to 108.86 ± 0.63 mg GAE equiv./100 g, DW. TPC content was found lowest in raw BR samples 16.08 ± 0.49 and highest in L. reuteri FBR 108.86 ± 0.63. Hydrolysis by enzymes during fermentation usually increases total phenolic content, as observed in this study. TPC increases with fermentation, L. fermentum FBR (75.00 ± 0.017 mg GAE/100 g, DW), L. plantarum FBR (96.87 ± 0.94 mg GAE equiv./100 g, DW), and L. reuteri FBR (108.86 ± 0.63 mg GAE equiv./100 g, DW), compared with raw BR. This study shows higher TPC content of BR than reported by [21,22].
Table 1.
Total antioxidants DPPH, ABTS, FRAP, total phenolic content (TPC), and total flavonoid content (TFC) of raw and different LABs fermented BR samples.
| S.NO | Sample | DPPH (mg Trolox Equiv./100 g, DW) | ABTS (mg Trolox Equiv./100 g, DW) | FRAP (mg Trolox Equiv./100 g, DW) | TPC (mg Gallic Acid Equiv./100 g, DW) | TFC (mg Catechin Equiv./100 g, DW) |
|---|---|---|---|---|---|---|
| 1 | Raw BR | 17.342 ± 0.44 d | 19.86 ± 0.86 d | 18.01 ± 0.74 d | 16.08 ± 0.49 d | 13.42 ± 0.80 d |
| 2 | L. fermentum FBR | 90.93 ± 0.74 c | 92.10 ± 0.81 c | 99.59 ± 1.60 b | 75.00 ± 0.01 c | 54.77 ± 1.02 c |
| 3 | L. reuteri FBR | 121.19 ± 1.0 a | 145.80 ± 0.99 a | 171.89 ± 0.71 a | 108.86 ± 0.63 a | 86.79 ± 0.83 a |
| 4 | L. plantarum FBR | 95.03 ± 0.81 b | 97.73 ± 0.47 b | 104.31 ± 0.48 b | 96.87 ± 0.94 b | 66.28 ± 0.71 b |
BR—Brown rice, FBR—fermented brown rice. Results were expressed as mean ± SD of triplicate analyses. Different alphabetical letters in each column represent statistically significant differences (Tukey and Duncan test p ≤ 0.05) DW, dry weight sample.
TFC was found higher in L. reuteri FBR 86.79 ± 0.83 mg catechin equiv./100 g, DW, followed by L. plantarum FBR and L. fermentum FBR (66.28 ± 0.71 and 54.77 ± 1.02 mg catechin equiv./100 g, DW) (Table 1). TFC content was lower than TPC in BR samples. TFC in our study was higher than previously reported by Huang et al. [23], similar to TPC, but both TPC and TFC were found lower than reported by Gong et al. [24]. These differences in BR values of different researchers could be because of genotype, cultivation landscape, and climate conditions. In addition, it is worth noting that the phenolic content can be significantly influenced by different extraction solvents and procedures.
3.2. Antioxidant Assay (DPPH, ABTS, FRAP)
Various methodologies including reducing capacity, free radical scavenging, lipid peroxidation inhibition, and metal ion chelation have been studied to explain how rice extracts have shown effective antioxidant potential [25]. In recent years, the fermentation process is thought to be an effective method for increasing antioxidant activity in cereals. The antioxidant activity of raw and different LABs fermented brown rice samples were assessed using the DPPH, ABTS, and FRAP assays in the current study. The antioxidant values of DPPH, ABTS, and FRAP of phenolic extracts of BR and differently treated BR samples were presented in Table 1, respectively.
The detection of scavenging activity of the 2,2′-diphenyl-1-picrylhydrazyl (DPPH) radical by spectrophotometry is one of the most commonly used assays in the determination of antioxidant activity in natural products. The DPPH radical scavenging activity was found highest in the L. reuteri FBR (121.19 ± 1.0 mg Trolox equiv./100 g, DW), followed by L. plantarum and L. fermentum FBR (95.03 ± 0.81, 90.93 ± 0.74 mg Trolox equiv./100 g, DW). The lowest values were observed in raw BR (17.342 ± 0.44 mg Trolox equiv./100 g, DW).
Similarly, ABTS is considered an important method for determining radical scavenging activity in grains and plant materials. Furthermore, FRAP assay was originally designed to assess plasma antioxidant ability but has also been commonly used in a wide variety of pure compounds and biological samples to determine antioxidant capacity. It measures absorption changes caused by the formation of blue iron (II) from colorless iron oxide (III). In the present research, the same trend as DPPH was observed in ABTS and FRAP assays. ABTS activity was measured highest in L. reuteri FBR (145.80 ± 0.99 mg Trolox equiv./100 g, DW) (Table 1). In ABTS, the lowest activity was also by raw BR. Similarly, FRAP was found highest in FBR (171.89 ± 0.71 mg Trolox equiv./100 g, DW) followed by L. plantarum and L. fermentum FBR. As a result of different antioxidants assays, L. reuteri FBR showed the highest activity among all samples. These findings were higher than earlier reports by Lin et al. [26] and IIowefah et al. [21] in fermented BR. Furthermore, other studies have reported the high antioxidant activity of BR [27].
3.3. Untargeted Metabolomics Using UHPLC Q-TOF-MS/MS in Brown Rice Samples
UHPLC Q-TOF-MS/MS detection is considered a gold standard technique for the precise detection and quantification of a wide variety of components. Therefore, in this study, we have used this detection technique for the identification of phenolic compounds in brown rice.
3.3.1. Phenolic Compounds
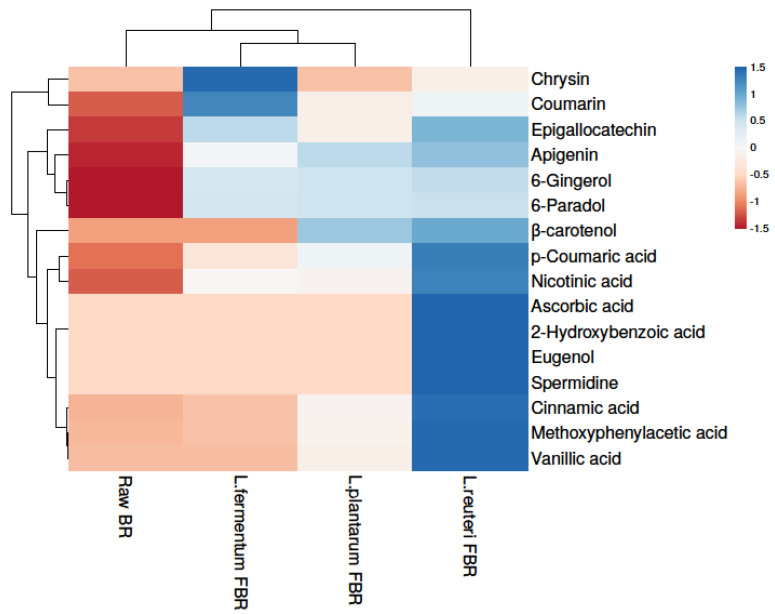
In the present research, the phenolic compositions of BR treated with different fermentation bacteria were selected and positively or tentatively identified by UHPLCQ-TOF-MS/MS. Phenolic identification and characterization were achieved by comparing our results with mass spectral literature evidence and cross-referencing it with other available spectral databases, such as Metlin and Metabolomics Workbench. A total of 15 phenolic compounds were tentatively found from our soluble extracts of raw BR, L. reuteri FBR, L. fermentum FBR, and L. plantarum FBR respectively, as shown in Table 2. In the ethanol extract, we identified compounds 1 to 14 at different adduct charges [M − H]− and [M − H]+ which are identified by comparing with mass spectral libraries, XCMS online (Metlin), and Metabolomics Workbench. Heat map analysis was used for clustering phenolic compounds based on their concentrations (Figure 1) where the color scheme from blue to red shows concentration in decreasing order.
Table 2.
Phenolic compounds detected in raw and LABs fermented BR.
| S. No | Sample Name | Retention Time (min) | Peak Area | Adduct/Charge | Precursor Mass | Found at Mass | Molecular Formula |
Tentative Phenolic Compound |
|---|---|---|---|---|---|---|---|---|
| 1 | Raw BR | Nd | Nd | [M − H]− | Nd | Nd | C25H36O | Beta-carotenol |
| L. plantarum FBR | 45.56 | 4.53 × 105 | [M − H]− | 353.268 | 353.187 | |||
| L. fermentum FBR | Nd | Nd | [M − H]− | Nd | Nd | |||
| L. reuteri FBR | 45.45 | 5.19 × 105 | [M − H]− | 353.268 | 353.284 | |||
| 2 | Raw BR | Nd | Nd | [M + H]+ | Nd | Nd | C10H12O2 | Eugenol |
| L. plantarum FBR | Nd | Nd | [M + H]+ | Nd | Nd | |||
| L. fermentum FBR | Nd | Nd | [M + H]+ | Nd | Nd | |||
| L. reuteri FBR | 20.81 | 2.24 × 105 | [M + H]+ | 179.107 | 179.1067 | |||
| 3 | Raw BR | 33.80 | 2.20 × 105 | [M − H]− | 293.177 | 293.1761 | C17H26O4 | 6-Gingerol |
| L. plantarum FBR | 33.80 | 2.08 × 106 | [M − H]− | 293.177 | 293.176 | |||
| L. fermentum FBR | 33.81 | 2.03 × 106 | [M − H]− | 293.177 | 293.1761 | |||
| L. reuteri FBR | 33.80 | 2.13 × 106 | [M − H]− | 293.177 | 293.1762 | |||
| 4 | Raw BR | Nd | Nd | [M + H]+ | Nd | Nd | C15H10O4 | Chrysin |
| L. plantarum FBR | Nd | Nd | [M + H]+ | Nd | Nd | |||
| L. fermentum FBR | 14.78 | 4.09 × 105 | [M + H]+ | 253.052 | 253.0524 | |||
| L. reuteri FBR | 14.81 | 1.01 × 105 | [M + H]+ | 253.052 | 253.0527 | |||
| 5 | Raw BR | Nd | Nd | [M + H]+ | Nd | Nd | C16H8N2O5 | Apigenin |
| L. plantarum FBR | 14.79 | 4.68 × 105 | [M + H]+ | 269.047 | 269.0457 | |||
| L. fermentum FBR | 14.78 | 3.38 × 105 | [M + H]+ | 269.047 | 269.0457 | |||
| L. reuteri FBR | 14.78 | 5.15 × 105 | [M + H]+ | 269.047 | 269.0458 | |||
| 6 | Raw BR | Nd | Nd | [M + H]+ | Nd | Nd | C9H6O2 | Coumarin |
| L. plantarum FBR | 1.92 | 1.24 × 105 | [M + H]+ | 147.044 | 147.0444 | |||
| L. fermentum FBR | 1.87 | 2.94 × 105 | [M + H]+ | 147.044 | 147.0447 | |||
| L. reuteri FBR | 1.90 | 1.55 × 105 | [M + H]+ | 147.044 | 147.0445 | |||
| 7 | Raw BR | Nd | Nd | [M + H]+ | Nd | Nd | C15H14O7 | Epigallocatechin |
| L. plantarum FBR | 12.26 | 7.44 × 105 | [M + H]+ | 305.071 | 305.067 | |||
| L. fermentum FBR | 12.28 | 1.20 × 106 | [M + H]+ | 305.071 | 305.067 | |||
| L. reuteri FBR | 12.28 | 1.38 × 106 | [M + H]+ | 305.071 | 305.067 | |||
| 8 | Raw BR | Nd | Nd | [M + H]+ | Nd | Nd | C7H19N3 | Spermidine |
| L. plantarum FBR | Nd | Nd | [M + H]+ | Nd | Nd | |||
| L. fermentum FBR | Nd | Nd | [M + H]+ | Nd | Nd | |||
| L. reuteri FBR | 0.96 | 8.75 × 105 | [M + H]+ | 188.176 | 188.1761 | |||
| 9 | Raw BR | 38.06 | 3.21 × 104 | [M − H]− | 277.182 | 277.1812 | C17H26O3 | 6-Paradol |
| L. plantarum FBR | 38.06 | 4.38 × 105 | [M − H]− | 277.182 | 277.1812 | |||
| L. fermentum FBR | 38.08 | 4.33 × 105 | [M − H]− | 277.182 | 277.1813 | |||
| L. reuteri FBR | 38.06 | 4.42 × 105 | [M − H]− | 277.182 | 277.1812 | |||
| 10 | Raw BR | Nd | Nd | [M − H]− | Nd | Nd | C9H8O2 | Cinnamic acid |
| L. plantarum FBR | 4.01 | 3.26 × 105 | [M − H]− | 147.046 | 147.0455 | |||
| L. fermentum FBR | 4.03 | 4.46 × 104 | [M − H]− | 147.046 | 147.0456 | |||
| L. reuteri FBR | 3.98 | 1.09 × 106 | [M − H]− | 147.046 | 147.0454 | |||
| 11 | Raw BR | Nd | Nd | [M + NH4]+ | Nd | Nd | C9H8O3 | p-Coumaric acid |
| L. plantarum FBR | 1.86 | 1.06 × 106 | [M + NH4]+ | 182.081 | 182.0813 | |||
| L. fermentum FBR | 1.92 | 6.90 × 105 | [M + NH4]+ | 182.081 | 182.0813 | |||
| L. reuteri FBR | 1.87 | 2.13 × 106 | [M + NH4]+ | 182.081 | 182.0812 | |||
| 12 | Raw BR | Nd | Nd | [M − H]− | Nd | Nd | C9H10O3 | Methoxyphenylacetic acid |
| L. plantarum FBR | 15.28 | 5.20 × 106 | [M − H]− | 165.057 | 165.0558 | |||
| L. fermentum FBR | 15.29 | 4.09 × 105 | [M − H]− | 165.057 | 165.056 | |||
| L. reuteri FBR | 15.27 | 1.92 × 107 | [M − H]− | 165.057 | 165.0557 | |||
| 13 | Raw BR | Nd | Nd | [M − H]− | Nd | Nd | C7H6O3 | Sesamol/2-Hydroxybenzoic acid |
| L. plantarum FBR | Nd | Nd | [M − H]− | Nd | Nd | |||
| L. fermentum FBR | Nd | Nd | [M − H]− | Nd | Nd | |||
| L. reuteri FBR | 19.63 | 3.24 × 105 | [M − H]− | 137.025 | 137.0249 | |||
| 14 | Raw BR | Nd | Nd | [M − H]− | Nd | Nd | C8H8O | Vanillic acid |
| L. plantarum FBR | 15.28 | 3.18 × 105 | [M − H]− | 119.051 | 119.0504 | |||
| L. fermentum FBR | Nd | Nd | [M − H]− | Nd | Nd | |||
| L. reuteri FBR | 15.27 | 1.27 × 106 | [M − H]− | 119.051 | 119.0504 | |||
| 15 | Raw BR | Nd | Nd | Nd | Nd | Nd | C6H8O6 | Ascorbic acid (Vitamin C) |
| L. plantarum FBR | Nd | Nd | Nd | Nd | Nd | |||
| L. fermentum FBR | Nd | Nd | Nd | Nd | Nd | |||
| L. reuteri FBR | 1.02 | 3.05 × 103 | [M + H]+ | 209.009 | 209.0107 |
Nd—not detected, BR—brown rice, and FBR—fermented brown rice.
Figure 1.
Heat map showing levels of phenolic compounds in raw and LABs fermented BR samples.
Results showed that the highest phenolic compounds were detected in the L. reuteri FBR sample. Because phenolic compounds are not readily available, they typically occur in cereals in esterified linkages to the cereal wall matrix [28]. Fermentation is considered to be a possible strategy to release insoluble or bound phenolic compounds and thus leading to improve the poor bioavailability of grain phenolics. Comparing different fermenting bacteria in the present study we found that L. reuteri fermentation releases most of the phenolic compounds compared with other bacterial strains and thus improves the bioavailability and bioaccessibility of cereal grains such as brown rice phenolics [29]. Many phenolic compounds detected in the current study such as p-coumaric acid [30], ascorbic acid [31], cinnamic acid [32], and vanillic acid [33] are already reported in the literature for their strong antioxidant capacities.
3.3.2. Levels of Amino Acid in Brown Rice
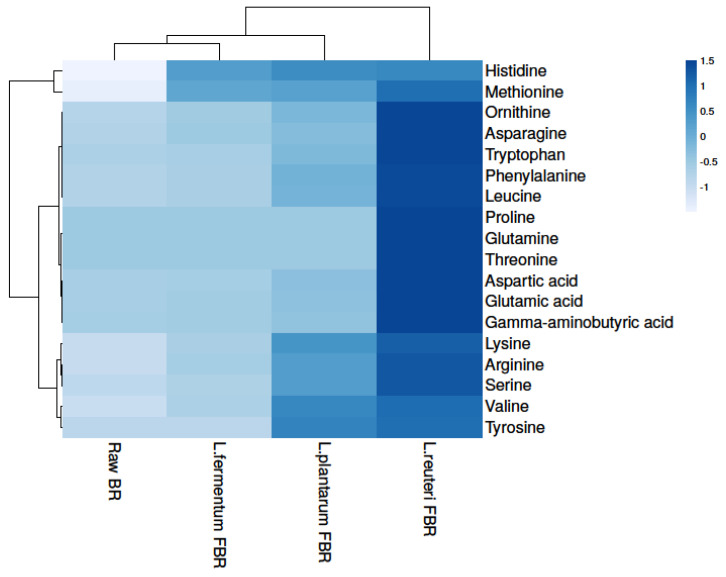
In the growth and development of organisms, amino acids play an important role and can also improve the taste of food. In our present study, a total of 18 amino acids were detected in raw and differently fermented BR samples (Figure 2 and Table 3) which shows statistically significant differences from each other after comparing their levels. Raw BR contained the least number of amino acids, which may be due to more bound molecules with the parent, whereas fermentation leads to an increase in amino acid content. The levels of amino acids were detected highest in the L. reuteri FBR sample which might strain-specific as fermentation microorganisms produce enzymes that lead to the formation of several metabolites and bioactive compounds from the food matrix [34]. In the ethanol extract, we found levels of some essential amino acids (tryptophan, lysine, methionine, and histidine), as well as certain conditionally essential amino acids (arginine, ornithine, serine, and glutamine), increased drastically after fermentation (Figure 2 and Table 3). The identification was done by comparing with mass spectral libraries, XCMS online (Metlin) and Metabolomics Workbench. In amino acids, L. reuteri FBR also shows the highest number of amino acids content as observed in phenolic compounds.
Figure 2.
Heat map showing levels of amino acids in raw and different fermented BR samples.
Table 3.
Amino acids detected in raw and LABs fermented brown rice.
| S. No | Sample Name | Retention Time (Min) | Peak Area | Adduct/ Charge |
Precursor Mass | Found at Mass | Formula Finder Result | Amino Acid | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Raw BR | 1.00 | 1.52 × 103 | [M + H]+ | 156.077 | 156.077 | C6H9N3O2 | Histidine | |
| L. plantarum FBR | 1.00 | 7.31 × 105 | [M + H]+ | 156.077 | 156.077 | ||||
| L. fermentum FBR | 1.02 | 6.33 × 105 | [M + H]+ | 156.077 | 156.0771 | ||||
| L. reuteri FBR | 1.02 | 7.45 × 105 | [M + H]+ | 156.077 | 156.0771 | ||||
| 2 | Raw BR | ND | ND | [M − H]− | ND | ND | C6H14N2O2 | Lysine | |
| L. plantarum FBR | 1.02 | 9.16 × 105 | [M − H]− | 145.099 | 145.0982 | ||||
| L. fermentum FBR | 1.02 | 2.17 × 105 | [M − H]− | 145.099 | 145.0984 | ||||
| L. reuteri FBR | 1.02 | 1.40 × 106 | [M − H]− | 145.099 | 145.0983 | ||||
| 3 | Raw BR | ND | ND | [M + H]+ | ND | ND | C5H11NO3S |
Methionine | |
| L. plantarum FBR | 1.17 | 5.66 × 105 | [M + H]+ | 166.053 | 166.0536 | ||||
| L. fermentum FBR | 1.17 | 5.33 × 105 | [M + H]+ | 166.053 | 166.0537 | ||||
| L. reuteri FBR | 1.18 | 8.41 × 105 | [M + H]+ | 166.053 | 166.0536 | ||||
| 4 | Raw BR | 1.17 | 2.56 × 102 | [M − H]− | 146.047 | 146.0457 | C5H9NO4 | Glutamic acid | |
| L. plantarum FBR | 1.47 | 3.18 × 105 | [M − H]− | 146.047 | 146.046 | ||||
| L. fermentum FBR | 1.47 | 4.58 × 104 | [M − H]− | 146.047 | 146.046 | ||||
| L. reuteri FBR | 1.47 | 2.65 × 106 | [M − H]− | 146.047 | 146.0458 | ||||
| 5 | Raw BR | ND | ND | [M + H]− | ND | ND | C4H9NO2 | Gamma-aminobutyric acid | |
| L. plantarum FBR | 1.16 | 1.38 × 105 | [M − H]− | 102.057 | 102.056 | ||||
| L. fermentum FBR | 1.16 | 2.76 × 104 | [M − H]− | 102.057 | 102.0563 | ||||
| L. reuteri FBR | 1.16 | 1.28 × 106 | [M − H]− | 102.057 | 102.0561 | ||||
| 6 | Raw BR | ND | ND | [M + H]+ | ND | ND | C6H14N4O2 | Arginine | |
| L. plantarum FBR | 1.11 | 2.81 × 106 | [M + H]+ | 175.118 | 175.1183 | ||||
| L. fermentum FBR | 1.14 | 9.09 × 105 | [M + H]+ | 175.118 | 175.1194 | ||||
| L. reuteri FBR | 1.11 | 4.99 × 106 | [M + H]+ | 175.118 | 175.1184 | ||||
| 7 | Raw BR | ND | ND | [M − H]− | ND | ND | C5H11NO2 | Valine | |
| L. plantarum FBR | 1.49 | 6.15 × 105 | [M − H]− | 116.073 | 116.0717 | ||||
| L. fermentum FBR | 1.50 | 1.37 × 105 | [M − H]− | 116.073 | 116.0718 | ||||
| L. reuteri FBR | 1.45 | 7.61 × 105 | [M − H]− | 116.073 | 116.0719 | ||||
| 8 | Raw BR | 1.14 | 1.19 × 103 | [M − H]− | 132.031 | 132.0307 | C4H7NO4 | Aspartic acid | |
| L. plantarum FBR | 1.15 | 7.26 × 104 | [M − H]− | 132.031 | 132.0303 | ||||
| L. fermentum FBR | 1.15 | 8.76 × 103 | [M − H]− | 132.031 | 132.0305 | ||||
| L. reuteri FBR | 1.12 | 4.89 × 105 | [M − H]− | 132.031 | 132.0302 | ||||
| 9 | Raw BR | ND | ND | [M − H]− | ND | ND | C9H11NO2 | Phenylalanine | |
| L. plantarum FBR | 4.01 | 2.26 × 106 | [M − H]− | 164.072 | 164.0718 | ||||
| L. fermentum FBR | 4.03 | 3.50 × 105 | [M − H]− | 164.072 | 164.072 | ||||
| L. reuteri FBR | 3.98 | 7.21 × 106 | [M − H]− | 164.072 | 164.0718 | ||||
| 10 | Raw BR | ND | ND | [M − H]− | ND | ND | C5H12N2O2 | Ornithine | |
| L. plantarum FBR | 1.01 | 1.53 × 105 | [M − H]− | 131.084 | 131.0827 | ||||
| L. fermentum FBR | 1.01 | 6.67 × 104 | [M − H]− | 131.084 | 131.0828 | ||||
| L. reuteri FBR | 1.01 | 5.35 × 105 | [M − H]− | 131.084 | 131.0827 | ||||
| 11 | Raw BR | 1.12 | 3.31 × 102 | [M − H]− | 104.036 | 104.0353 | C3H7NO3 | Serine | |
| L. plantarum FBR | 1.12 | 3.06 × 105 | [M − H]− | 104.036 | 104.0353 | ||||
| L. fermentum FBR | 1.13 | 4.70 × 104 | [M − H]− | 104.036 | 104.0356 | ||||
| L. reuteri FBR | 1.12 | 5.73 × 105 | [M − H]− | 104.036 | 104.0353 | ||||
| 12 | Raw BR | ND | ND | [M − H]− | ND | ND | C6H13NO2 | Leucine | |
| L. plantarum FBR | 2.46 | 1.95 × 106 | [M − H]− | 130.088 | 130.0874 | ||||
| L. fermentum FBR | 2.48 | 3.24 × 105 | [M − H]− | 130.088 | 130.0875 | ||||
| L. reuteri FBR | 2.41 | 6.63 × 106 | [M − H]− | 130.088 | 130.0875 | ||||
| 13 | Raw BR | ND | ND | [M − H]− | ND | ND | C5H10N2O3 | Glutamine | |
| L. plantarum FBR | ND | ND | [M − H]− | ND | ND | ||||
| L. fermentum FBR | ND | ND | [M − H]− | ND | ND | ||||
| L. reuteri FBR | 1.10 | 1.62 × 104 | [M − H]− | 145.063 | 145.0619 | ||||
| 14 | Raw BR | ND | ND | [M − H]− | ND | ND | C9H11NO3 | Tyrosine | |
| L. plantarum FBR | 1.98 | 3.38 × 105 | [M − H]− | 180.068 | 180.0667 | ||||
| L. fermentum FBR | ND | ND | [M − H]− | ND | ND | ||||
| L. reuteri FBR | 1.89 | 4.08 × 105 | [M − H]− | 180.068 | 180.0667 | ||||
| 15 | Raw BR | ND | ND | [M − H]− | ND | ND | C4H9NO3 | Threonine | |
| L. plantarum FBR | ND | ND | [M − H]− | ND | ND | ||||
| L. fermentum FBR | ND | ND | [M − H]− | ND | ND | ||||
| L. reuteri FBR | 1.14 | 1.14 × 105 | [M − H]− | 118.052 | 118.051 | ||||
| 16 | Raw BR | ND | ND | [M − H]− | ND | ND | C4H8N2O3 | Asparagine | |
| L. plantarum FBR | 1.10 | 6.8 × 104 | [M − H]− | 131.047 | 131.0462 | ||||
| L. fermentum FBR | 1.13 | 3.41 × 104 | [M − H]− | 131.047 | 131.0464 | ||||
| L. reuteri FBR | 1.13 | 3.01 × 105 | [M − H]− | 131.047 | 131.0461 | ||||
| 17 | Raw BR | ND | ND | [M − H]− | ND | ND | C11H12N2O2 | Tryptophan | |
| L. plantarum FBR | 7.64 | 6.19 × 105 | [M − H]− | 203.084 | 203.0829 | ||||
| L. fermentum FBR | 7.66 | 7.77 × 104 | [M − H]− | 203.084 | 203.0832 | ||||
| L. reuteri FBR | 7.61 | 2.78 × 106 | [M − H]− | 203.084 | 203.0829 | ||||
| 18 | Raw BR | ND | ND | [M + H]+ | ND | ND | C5H9NO2 | Proline | |
| L. plantarum FBR | ND | ND | [M + H]+ | ND | ND | ||||
| L. fermentum FBR | ND | ND | [M + H]+ | ND | ND | ||||
| L. reuteri FBR | 0.86 | 7.90 × 105 | [M +H]+ | 116.07 | 116.0704 | ||||
ND—not detected, BR—brown rice, and FBR—fermented brown rice.
3.3.3. Level of Fatty Acid in Brown Rice Samples
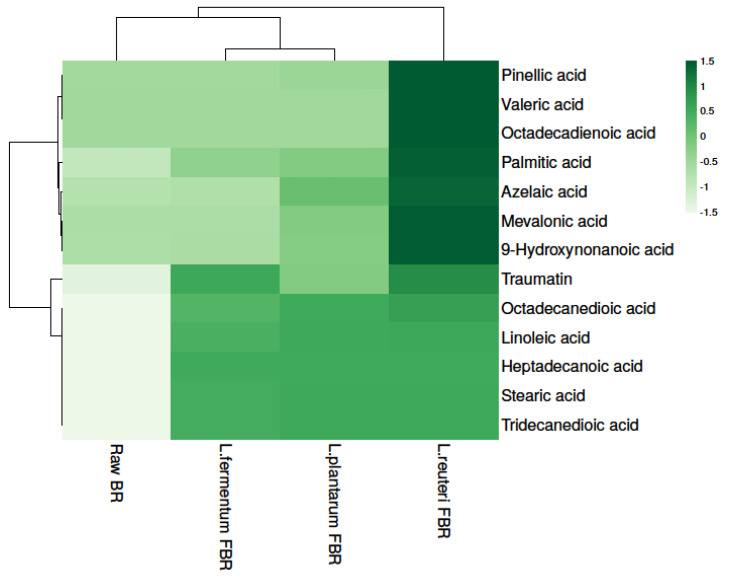
In particular, fermentation has been proposed as a tool for enhancing foods’ nutritional values, both in terms of enhanced bioavailability of bioactive components as well as the production of health-promoting end-products. Due to their proven benefit, in the last decade, short-chain fatty acids (SCFAs) have emerged as some of the most researched compounds. In the present study, 13 fatty acids were detected in raw and different LABs fermented BR samples (Table 4) and fatty acid levels were found to be significantly different in all samples. The results show that the highest levels of fatty acids were found in L. reuteri FBR. Heat map analysis was used for separating fatty acids based on the different concentrations, represented in different shades of green (dark to light) in decreasing order (Figure 3).
Table 4.
Fatty acids detected in raw and LABs fermented brown rice.
| S. No | Sample Name | Retention Time | Peak Area | Adduct/Charge | Precursor Mass | Found at Mass | Formula Finder Result | Fatty Acid |
|---|---|---|---|---|---|---|---|---|
| 1 | Raw BR | 39.97 | 1.91 × 102 | [M − H]− | 255.234 | 255.2331 | C16H32O2 | Palmitic Acid |
| L. plantarum FBR | 1.94 | 5.15 × 103 | [M − H]− | 255.234 | 255.2331 | |||
| L. fermentum FBR | 28.03 | 4.28 × 103 | [M − H]− | 255.234 | 255.2332 | |||
| L. reuteri FBR | 23.73 | 1.53 × 104 | [M − H]− | 255.234 | 255.2332 | |||
| 2 | Raw BR | ND | ND | [M + H]+ | ND | ND | C5H10O2 | Valeric acid |
| L. plantarum FBR | ND | ND | [M + H]+ | ND | ND | |||
| L. fermentum FBR | ND | ND | [M + H]+ | ND | ND | |||
| L. reuteri FBR | 22.88 | 1.02 × 103 | [M + H]+ | 185.066 | 185.0663 | |||
| 3 | Raw BR | 46.24 | 5.29 × 103 | [M − H]− | 279.234 | 279.2332 | C18H32O2 | Linoleic Acid |
| L. plantarum FBR | 46.26 | 6.56 × 105 | [M − H]− | 279.234 | 279.2332 | |||
| L. fermentum FBR | 46.24 | 6.16 × 105 | [M − H]− | 279.234 | 279.2334 | |||
| L. reuteri FBR | 46.25 | 6.64 × 105 | [M − H]− | 279.234 | 279.2334 | |||
| 4 | Raw BR | 47.28 | 1.45 × 105 | [M + H]+ | 271.264 | 271.2637 | C17H34O2 | Heptadecanoic acid |
| L. plantarum FBR | 47.28 | 1.44 × 106 | [M + H]+ | 271.264 | 271.2636 | |||
| L. fermentum FBR | 47.26 | 1.44 × 106 | [M + H]+ | 271.264 | 271.2637 | |||
| L. reuteri FBR | 47.27 | 1.45 × 106 | [M + H]+ | 271.264 | 271.2638 | |||
| 5 | Raw BR | 27.69 | 5.28 × 103 | [M − H]− | 283.265 | 283.2644 | C18H36O2 | Stearic acid |
| L. plantarum FBR | 49.10 | 1.23 × 106 | [M − H]− | 283.265 | 283.2645 | |||
| L. fermentum FBR | 49.09 | 1.20 × 106 | [M − H]− | 283.265 | 283.2644 | |||
| L. reuteri FBR | 49.10 | 1.24 × 106 | [M − H]− | 283.265 | 283.2645 | |||
| 6 | Raw BR | 34.57 | 3.18 × 104 | [M − H]− | 243.161 | 243.1605 | C13H24O4 | Tridecanedioic acid |
| L. plantarum FBR | 34.58 | 2.98 × 105 | [M − H]− | 243.161 | 243.1606 | |||
| L. fermentum FBR | 34.58 | 2.90 × 105 | [M − H]− | 243.161 | 243.1604 | |||
| L. reuteri FBR | 34.57 | 3.01 × 105 | [M − H]− | 243.161 | 243.1605 | |||
| 7 | Raw BR | ND | ND | [M + H]+ | ND | ND | C12H20O3 | Traumatin |
| L. plantarum FBR | 32.80 | 2.64 × 105 | [M + H]+ | 213.149 | 213.1491 | |||
| L. fermentum FBR | 32.82 | 4.26 × 105 | [M + H]+ | 213.149 | 213.1492 | |||
| L. reuteri FBR | 32.82 | 5.20 × 105 | [M + H]+ | 213.149 | 213.1492 | |||
| 8 | Raw BR | ND | ND | [M − H]− | ND | ND | C18H32O5 | Octadecadienoic acid/Corchorifatty acid F |
| L. plantarum FBR | ND | ND | [M − H]− | ND | ND | |||
| L. fermentum FBR | ND | ND | [M − H]− | ND | ND | |||
| L. reuteri FBR | 30.87 | 4.08 × 105 | [M − H]− | 327.219 | 327.2181 | |||
| 9 | Raw BR | ND | ND | [M − H]− | ND | ND | C6H12O4 | Mevalonic Acid |
| L. plantarum FBR | 3.49 | 7.39 × 104 | [M − H]− | 147.067 | 147.0667 | |||
| L. fermentum FBR | ND | ND | [M − H]− | ND | ND | |||
| L. reuteri FBR | 3.47 | 3.13 × 105 | [M − H]− | 147.067 | 147.0667 | |||
| 10 | Raw BR | 22.59 | 3.99 × 104 | [M − H]− | 187.099 | 187.0979 | C9H16O4 | Azelaic Acid |
| L. plantarum FBR | 22.50 | 2.77 × 105 | [M − H]− | 187.099 | 187.0979 | |||
| L. fermentum FBR | 22.61 | 6.12 × 104 | [M − H]− | 187.099 | 187.0978 | |||
| L. reuteri FBR | 22.50 | 6.25 × 105 | [M − H]− | 187.099 | 187.0979 | |||
| 11 | Raw BR | ND | ND | [M − H]− | ND | ND | C9H18O3 | 9-Hydroxynonanoic acid |
| L. plantarum FBR | 23.49 | 3.10 × 104 | [M − H]− | 173.119 | 173.1187 | |||
| L. fermentum FBR | 23.50 | 3.49 × 103 | [M − H]− | 173.119 | 173.1188 | |||
| L. reuteri FBR | 23.48 | 1.39 × 105 | [M − H]− | 173.119 | 173.1186 | |||
| 12 | Raw BR | 39.06 | 2.45 × 103 | [M − H]− | 313.24 | 313.2389 | C18H34O4 | Octadecanedioic acid |
| L. plantarum FBR | 39.07 | 3.52 × 105 | [M − H]− | 313.24 | 313.2388 | |||
| L. fermentum FBR | 39.06 | 3.22 × 105 | [M − H]− | 313.24 | 313.2389 | |||
| L. reuteri FBR | 39.06 | 3.83 × 105 | [M − H]− | 313.24 | 313.2386 | |||
| 13 | Raw BR | ND | ND | [M − H]− | ND | ND | C18H34O5 | Pinellic acid |
| L. plantarum FBR | 33.48 | 3.12 × 105 | [M − H]− | 329.234 | 329.2337 | |||
| L. fermentum FBR | 33.48 | 3.54 × 104 | [M − H]− | 329.234 | 329.2339 | |||
| L. reuteri FBR | 32.82 | 7.37 × 106 | [M − H]− | 329.234 | 329.2331 |
ND—not detected, BR—brown rice, and FBR—fermented brown rice.
Figure 3.
Heat map showing levels of fatty acids in raw and different LABs fermented BR samples.
3.4. Cell Viability Assay and Cellular Antioxidant Activity (CAA)
3.4.1. Cell Viability Assay
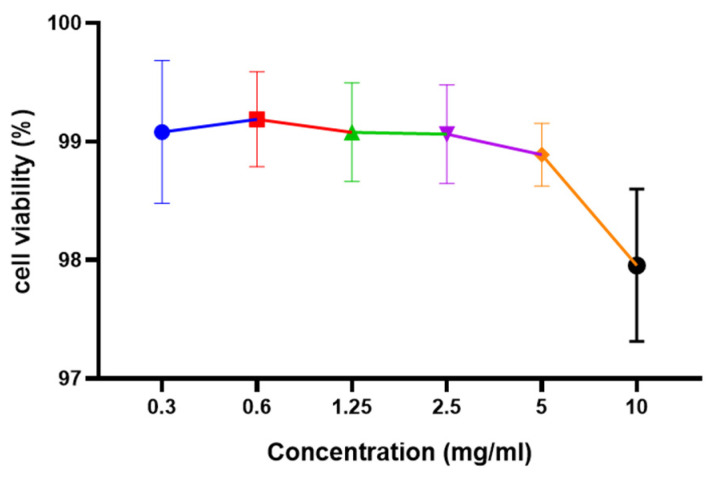
Cytotoxicity is regarded as an important step in determining the suitability and further applications of any food extract. Using the Ez cytox assay kit, the cytotoxic effect of L. reuteri FBR extracts at 0.3–10 mg/mL concentrations was investigated using Caco-2 cell lines. Figure 4 depicts the cell viability results of the extract after 12 h of incubation. It was observed that cell viability was not much decreased after increasing the concentration up to 10 mg/mL. No significant differences were observed in cytotoxicity assay by using 0.3–10 mg/mL concentrations (Figure 4). The extract was observed to be non-toxic after 12 h assay as extract still shows about 97 per cent of cell viability. Our results were found similar to the results presented by Yue et al. [35].
Figure 4.
Effect of L. reuteri FBR extracts on viability of Caco-2 cells analyzed by Ez cytox assay kit. Cells were treated with an increased concentration of L. reuteri FBR extracts for 12 h. Data are represented as means ± standard deviations (n = 3).
3.4.2. Cellular Antioxidant Activity (CAA)
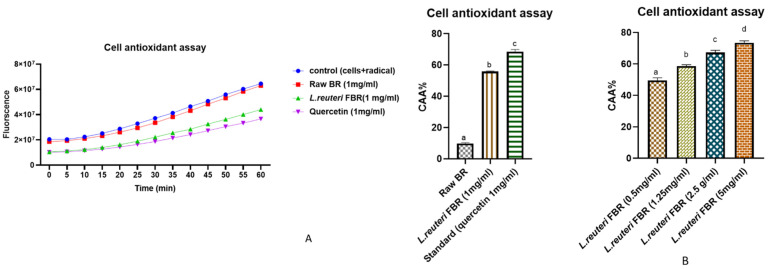
The effect of pretreatment of Caco-2 cells with L. reuteri fermented extract of brown rice on intracellular reactive oxygen species (ROS) was determined using a cell-based assay. The fluorescent probe DCFH-DA is used as an indicator of ROS and oxidative stress in our study. The nonionic and nonpolar DCFH-DA probe diffuses passively into cells before being hydrolyzed by intracellular esterases to form nonfluorescent 2′,7′-dichlorofluorescein (DCFH). Later in the presence of ROS, DCFH that is trapped inside cells is oxidized into fluorescent 2′,7′-dichlorofluorescein (DCF) [36]. When the cellular antioxidant defense system fails to compensate for ROS production, oxidative stress occurs. This reaction can be slowed down using bioactive compounds, preventing the generation of DCF. Following the uptake of antioxidant compounds can be accomplished on the cell membrane surface or within the cell [37]. We evaluated the effect of our L. reuteri FBR extract against oxidative stress in Caco-2 cells. In our study, ABAP was chosen as an intracellular oxidizing agent to simulate oxidative stress in cells. 600 µmol L−1 ABAP was chosen as the optimal concentration to induce oxidation. As represented in Figure 5A, CAA values in L. reuteri FBR extract were observed to be 5.7 times higher than the raw BR sample at a concentration of 1mg/mL. Our results indicate that extracts reduced ROS levels at rest in a dose-dependent manner (Figure 5B); CAA values were increased with concentration (0.5 mg/mL to 5 mg/mL) from 49.50 ± 1.67% to 72.49 ± 1.23%. The strength of inhibition strongly followed a curvilinear pattern as L. reuteri FBR extract concentrations increased. A similar effect was previously observed in the study of Grauzdytė et al., where they observed Phyllanthus phillyreifolius extracts in HEK-293 cells [38], and the study of Kellett et al. [39] in Caco-2 cells.
Figure 5.
In Caco-2 cells, peroxyl radical-induced oxidation of DCFH to DCF and ROS inhibition by raw BR and L. reuteri FBR extract (A,B) showing the effect of dose-dependent inhibition of L. reuteri FBR extracts (0.5–5 mg/mL). Data were represented as means ± standard deviations (n = 3) with one way ANOVA. The columns with different letters (a–d) show significant differences using Tukey’s test at p < 0.05.
4. Conclusions
In our study, we discovered that L. reuteri FBR had higher antioxidant activity as well as a higher concentration of phenolics and flavonoids among all LABs used for the study. This shows the ability of L. reuteri as a promising fermentation strain to increase the bioavailability of cereals or grains in producing health-promoting functional materials. L. reuteri fermentation improves phenolic constituents and antioxidant activity of BR, improves food quality, and confers organoleptic characteristics. Furthermore, we discovered that L. reuteri FBR enhanced the production of essential amino acids and fatty acids using untargeted metabolomics. The present study has provided information on bioactive compounds and antioxidant activities as well as the cellular antioxidant capacities of L. reuteri FBR. These data are required for the processing of the whole BR and its products for the pharmaceutical and food markets. As a result, new strategies and collaborations among industry, researchers, and relevant agencies are required to publicize whole grain consumption. Additionally, the current research is part of ongoing efforts to increase the added value of brown rice production and use in the prevention of human chronic diseases caused by oxidative stress. Moreover, these findings also make this sample a promising material for the development of health-promoting functional food. Whereas, it is necessary to perform more research into the mechanisms of different types of fermentation (solid and liquid-state) on single/pure phenolic compounds and antioxidant properties. Furthermore, in vivo models should be used to study the bioavailability and absorption of phenolic compounds in the gut.
Abbreviations
| BR | Raw brown rice |
| FBR | Fermented brown rice |
| ROS | Reactive oxygen species |
| GABA | Gamma-aminobutyric acid |
Author Contributions
Conceptualization, A.T. and D.-H.O.; data curation, A.T. and U.S.; formal analysis; A.T. and E.B.-M.D.; funding acquisition, D.-H.O.; investigation, U.S. and R.C.; methodology, X.C., software, R.C.; supervision, D.-H.O.; roles/writing—original draft, A.T.; writing—review and editing: A.T., U.S., X.C., and E.B.-M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research work was supported by a grant from the 4th Brain Korea (B.K.) 21 Plus Project (Grant No. 4299990913942) funded by the Korean Government, Republic of Korea.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Poljsak B., Šuput D., Milisav I. Achieving the Balance between ROS and Antioxidants: When to Use the Synthetic Antioxidants. Oxidative Med. Cell. Longev. 2013;2013:1–11. doi: 10.1155/2013/956792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyagi A., Yeon S.-J., Daliri E., Chen X., Chelliah R., Oh D.-H. Untargeted Metabolomics of Korean Fermented Brown Rice Using UHPLC Q-TOF MS/MS Reveal an Abundance of Potential Dietary Antioxidative and Stress-Reducing Compounds. Antioxidants. 2021;10:626. doi: 10.3390/antiox10040626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyagi A., Daliri E.B.-M., Ofosu F.K., Yeon S.-J., Oh D.-H. Food-Derived Opioid Peptides in Human Health: A Review. Int. J. Mol. Sci. 2020;21:8825. doi: 10.3390/ijms21228825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sack M.N., Fyhrquist F.Y., Saijonmaa O.J., Fuster V., Kovacic J.C. Basic biology of oxidative stress and the cardiovascular system: Part 1 of a 3-part series. J. Am. Coll. Cardiol. 2017;70:196–211. doi: 10.1016/j.jacc.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Wu Y., Wang Y., Xu H., Mei X., Yu D., Wang Y., Li W. Antioxidant Properties of Probiotic Bacteria. Nutrients. 2017;9:521. doi: 10.3390/nu9050521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callcott E., Blanchard C., Oli P., Santhakumar A.B. Pigmented Rice-Derived Phenolic Compounds Reduce Biomarkers of Oxidative Stress and Inflammation in Human Umbilical Vein Endothelial Cells. Mol. Nutr. Food Res. 2018;62:e1800840. doi: 10.1002/mnfr.201800840. [DOI] [PubMed] [Google Scholar]
- 7.Shabbir U., Arshad M., Sameen A., Oh D.-H. Crosstalk between Gut and Brain in Alzheimer’s Disease: The Role of Gut Microbiota Modulation Strategies. Nutrients. 2021;13:690. doi: 10.3390/nu13020690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okarter N., Liu R.H. Health Benefits of Whole Grain Phytochemicals. Crit. Rev. Food Sci. Nutr. 2010;50:193–208. doi: 10.1080/10408390802248734. [DOI] [PubMed] [Google Scholar]
- 9.Imam M.U., Musa S.N.A., Azmi N.H., Ismail M. Effects of White Rice, Brown Rice and Germinated Brown Rice on Antioxidant Status of Type 2 Diabetic Rats. Int. J. Mol. Sci. 2012;13:12952–12969. doi: 10.3390/ijms131012952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pang Y., Ahmed S., Xu Y., Beta T., Zhu Z., Shao Y., Bao J. Bound phenolic compounds and antioxidant properties of whole grain and bran of white, red and black rice. Food Chem. 2018;240:212–221. doi: 10.1016/j.foodchem.2017.07.095. [DOI] [PubMed] [Google Scholar]
- 11.Sandhu K.S., Punia S. Enhancement of bioactive compounds in barley cultivars by solid substrate fermentation. J. Food Meas. Charact. 2017;11:1355–1361. doi: 10.1007/s11694-017-9513-6. [DOI] [Google Scholar]
- 12.Salar R.K., Purewal S.S., Sandhu K.S. Fermented pearl millet (Pennisetum glaucum) with in vitro DNA damage protection activity, bioactive compounds and antioxidant potential. Food Res. Int. 2017;100:204–210. doi: 10.1016/j.foodres.2017.08.045. [DOI] [PubMed] [Google Scholar]
- 13.Salar R.K., Purewal S.S. Improvement of DNA damage protection and antioxidant activity of biotransformed pearl millet (Pennisetum glaucum) cultivar PUSA-415 using Aspergillus oryzae MTCC 3107. Biocatal. Agric. Biotechnol. 2016;8:221–227. doi: 10.1016/j.bcab.2016.10.005. [DOI] [Google Scholar]
- 14.Pradeep P., Sreerama Y.N. Soluble and bound phenolics of two different millet genera and their milled fractions: Comparative evaluation of antioxidant properties and inhibitory effects on starch hydrolysing enzyme activities. J. Funct. Foods. 2017;35:682–693. doi: 10.1016/j.jff.2017.06.033. [DOI] [Google Scholar]
- 15.Chang X., Ye Y., Pan J., Lin Z., Qiu J., Guo X., Lu Y. Comparative assessment of phytochemical profiles and antioxidant activities in selected five varieties of wampee (Clausena lansium) fruits. Int. J. Food Sci. Technol. 2018;53:2680–2686. doi: 10.1111/ijfs.13877. [DOI] [Google Scholar]
- 16.Xiang J., Zhang M., Apea-Bah F.B., Beta T. Hydroxycinnamic acid amide (HCAA) derivatives, flavonoid C-glycosides, phenolic acids and antioxidant properties of foxtail millet. Food Chem. 2019;295:214–223. doi: 10.1016/j.foodchem.2019.05.058. [DOI] [PubMed] [Google Scholar]
- 17.Zeng Z., Hu X., McClements D.J., Luo S., Liu C., Gong E.S., Huang K. Hydrothermal stability of phenolic extracts of brown rice. Food Chem. 2019;271:114–121. doi: 10.1016/j.foodchem.2018.07.180. [DOI] [PubMed] [Google Scholar]
- 18.Daliri E.B.-M., Ofosu F.K., Chelliah R., Kim J.-H., Kim J.-R., Yoo D., Oh D.-H. Untargeted Metabolomics of Fermented Rice Using UHPLC Q-TOF MS/MS Reveals an Abundance of Potential Antihypertensive Compounds. Foods. 2020;9:1007. doi: 10.3390/foods9081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ti H., Guo J., Zhang R., Wei Z., Liu L., Bai Y., Zhang M. Phenolic profiles and antioxidant activity in four tissue fractions of whole brown rice. RSC Adv. 2015;5:101507–101518. doi: 10.1039/C5RA15963F. [DOI] [Google Scholar]
- 20.Metsalu T., Vilo J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilowefah M., Bakar J., Ghazali H.M., Muhammad K. Enhancement of Nutritional and Antioxidant Properties of Brown Rice Flour Through Solid-State Yeast Fermentation. Cereal Chem. J. 2017;94:519–523. doi: 10.1094/CCHEM-08-16-0204-R. [DOI] [Google Scholar]
- 22.Gong E.S., Liu C., Li B., Zhou W., Chen H., Li T., Wu J., Zeng Z., Wang Y., Si X., et al. Phytochemical profiles of rice and their cellular antioxidant activity against ABAP induced oxidative stress in human hepatocellular carcinoma HepG2 cells. Food Chem. 2020;318:126484. doi: 10.1016/j.foodchem.2020.126484. [DOI] [PubMed] [Google Scholar]
- 23.Huang S.-H., Ng L.-T. Quantification of polyphenolic content and bioactive constituents of some commercial rice varieties in Taiwan. J. Food Compos. Anal. 2012;26:122–127. doi: 10.1016/j.jfca.2012.03.009. [DOI] [Google Scholar]
- 24.Gong E.S., Luo S.J., Li T., Liu C.M., Zhang G.W., Chen J., Zeng Z.C., Liu R.H. Phytochemical profiles and antioxidant activity of brown rice varieties. Food Chem. 2017;227:432–443. doi: 10.1016/j.foodchem.2017.01.093. [DOI] [PubMed] [Google Scholar]
- 25.Ghasemzadeh A., Jaafar H.Z.E., Juraimi A.S., Tayebi-Meigooni A. Comparative Evaluation of Different Extraction Techniques and Solvents for the Assay of Phytochemicals and Antioxidant Activity of Hashemi Rice Bran. Molecules. 2015;20:10822–10838. doi: 10.3390/molecules200610822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin C.-C. Antioxidant properties and antibacterial activity of fermented Monascus purpureus extracts. Food Biosci. 2019;35:100605. [Google Scholar]
- 27.Rao S., Santhakumar A.B., Chinkwo K., Snell P., Oli P., Blanchard C. Rice phenolic compounds and their response to variability in growing conditions. Cereal Chem. J. 2020;97:1045–1055. doi: 10.1002/cche.10327. [DOI] [Google Scholar]
- 28.Dai T., Chen J., McClements D.J., Hu P., Ye X., Liu C., Li T. Protein–polyphenol interactions enhance the antioxidant capacity of phenolics: Analysis of rice glutelin–procyanidin dimer interactions. Food Funct. 2019;10:765–774. doi: 10.1039/C8FO02246A. [DOI] [PubMed] [Google Scholar]
- 29.Janarny G., Gunathilake K. Changes in rice bran bioactives, their bioactivity, bioaccessibility and bioavailability with solid-state fermentation by Rhizopus oryzae. Biocatal. Agric. Biotechnol. 2020;23:101510. doi: 10.1016/j.bcab.2020.101510. [DOI] [Google Scholar]
- 30.Zeki E., KÖSE S.B.E., GARLI S., ŞAHİNDOKUYUCU F.K. The Antioxidant Effect of p-Coumaric Acid Against Toluene-Induced Oxidative Stress in Rats. Kahramanmaraş Sütçü İmam Üniversitesi Tarım ve Doğa Dergisi. 2021;24:473–478. [Google Scholar]
- 31.El-Beltagi H.S., Mohamed H.I., Sofy M.R. Role of Ascorbic acid, Glutathione and Proline Applied as Singly or in Sequence Combination in Improving Chickpea Plant through Physiological Change and Antioxidant Defense under Different Levels of Irrigation Intervals. Molecules. 2020;25:1702. doi: 10.3390/molecules25071702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anlar H.G. In: Chapter 23-Cinnamic Acid as a Dietary Antioxidant in Diabetes Treatment. 2nd ed. Preedy V.R., editor. Academic Press; Cambridge, MA, USA: 2020. pp. 235–243. [Google Scholar]
- 33.Ahmadi N., Safari S., Mirazi N., Karimi S.A., Komaki A. Effects of vanillic acid on Aβ1-40-induced oxidative stress and learning and memory deficit in male rats. Brain Res. Bull. 2021;170:264–273. doi: 10.1016/j.brainresbull.2021.02.024. [DOI] [PubMed] [Google Scholar]
- 34.Adebo O.A., Njobeh P.B., Adebiyi J.A., Gbashi S., Phoku J.Z., Kayitesi E. Functional Food-Improve Health through Adequate Food. InTechOpen; London, UK: 2017. Fermented Pulse-Based Food Products in Developing Nations as Functional Foods and Ingredients; pp. 77–109. [Google Scholar]
- 35.Gao Y., Guo X., Liu Y., Zhang M., Zhang R., Abbasi A.M., You L., Li T., Liu R.H. Comparative assessment of phytochemical profile, antioxidant capacity and anti-proliferative activity in different varieties of brown rice (Oryza sativa L.) LWT. 2018;96:19–25. doi: 10.1016/j.lwt.2018.05.002. [DOI] [Google Scholar]
- 36.Girard-Lalancette K., Pichette A., Legault J. Sensitive cell-based assay using DCFH oxidation for the determination of pro- and antioxidant properties of compounds and mixtures: Analysis of fruit and vegetable juices. Food Chem. 2009;115:720–726. doi: 10.1016/j.foodchem.2008.12.002. [DOI] [Google Scholar]
- 37.Chen P.X., Zhang H., Marcone M.F., Pauls K.P., Liu R., Tang Y., Zhang B., Renaud J.B., Tsao R. Anti-inflammatory effects of phenolic-rich cranberry bean ( Phaseolus vulgaris L.) extracts and enhanced cellular antioxidant enzyme activities in Caco-2 cells. J. Funct. Foods. 2017;38:675–685. doi: 10.1016/j.jff.2016.12.027. [DOI] [Google Scholar]
- 38.Grauzdytė D., Pukalskas A., Viranaicken W., El Kalamouni C., Venskutonis P.R. Protective effects of Phyllanthus phillyreifolius extracts against hydrogen peroxide induced oxidative stress in HEK293 cells. PLoS ONE. 2018;13:e0207672. doi: 10.1371/journal.pone.0207672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kellett M.E., Greenspan P., Pegg R.B. Modification of the cellular antioxidant activity (CAA) assay to study phenolic antioxidants in a Caco-2 cell line. Food Chem. 2018;244:359–363. doi: 10.1016/j.foodchem.2017.10.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is contained within the article.