Abstract
Type‐2 (T2) inflammation is a characteristic feature of asthma. Biological therapies have been developed to target T2‐inflammation in asthma. IL‐13 is a key component of T2‐inflammation in asthma, driving mucus hypersecretion, IgE‐induction, and smooth muscle contraction. Early phase clinical trials for treatments that target T2‐inflammation require biomarkers to assess pharmacological effects. The aim of this study was to examine levels of IL‐13 inducible biomarkers in the airway epithelium of patients with mild asthma compared to healthy controls. Ten patients with mild asthma with high blood eosinophil and high fractional exhaled nitric oxide (FeNO) were recruited, and six healthy subjects. Blood eosinophil and FeNO reproducibility was assessed prior to bronchoscopy. Epithelial brushings were collected and assessed for IL‐13 inducible gene expression. Blood eosinophil and FeNO levels remained consistent in both patients with asthma and healthy subjects. Of the 11 genes assessed, expression levels of 15LOX1, POSTN, CLCA1, SERPINB2, CCL26, and NOS2 were significantly higher in patients with asthma compared to healthy controls. These six genes, present in patients with mild asthma with T2 inflammation, have the potential to be used in translational early phase asthma clinical trials of novel therapies as bronchial epithelial biomarkers.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Type 2 (T2) inflammation is found in many patients with asthma and is not always controlled by inhaled corticosteroids. T2‐specific biomarkers may be useful for measuring the pharmacological effects of novel anti‐T2 treatments.
WHAT QUESTION DID THIS STUDY ADDRESS?
We sought to identify IL‐13 associated biomarkers in the airways of patients with asthma with T2 inflammatory phenotype.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
Six genes were identified in airway epithelium whose expressions were elevated in patients with T2‐high asthma compared to healthy subjects.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
The six genes identified have the potential to be used as target engagement biomarkers in early phase clinical development for novel asthma treatments targeting T2‐inflammation.
INTRODUCTION
Asthma is a heterogeneous disease associated with increased Type 2 (T2) inflammation, which is characterized by eosinophilic infiltration and/or increased levels of the cytokines interleukin (IL)‐4, IL‐5, and IL‐13. 1 Whereas T2‐inflammation is a prominent feature in many patients with asthma, there are also patients who do not express T2 features. 2 T2‐inflammation can be suppressed by the use of inhaled corticosteroids (ICS), but T2‐inflammation may remain persistently elevated in some patients despite the use of high‐dose ICS. 3 Biological therapies targeting T2 inflammation are currently used in the treatment of severe asthma, and novel anti‐T2 biologics are in clinical development (reviewed in ref. 4).
The early phase clinical development of novel asthma treatments targeting T2‐inflammation requires biomarkers to measure pharmacological effects. 5 , 6 , 7 , 8 These biomarkers can help with dose selection decisions and provide information regarding mechanism of action. These early phase studies often involve patients with mild asthma in order to provide initial safety assessment before proceeding to larger studies in patients with more severe asthma.
IL‐13 levels are increased in sputum and mucosal tissue from patients with mild, moderate, and severe asthma. 9 IL‐13 has different roles in asthma pathophysiology, causing small airway remodeling by increasing extracellular matrix formation and myofibroblast differentiation, inducing epithelial mucus hypersecretion and B‐cell derived IgE, and enhancing airway smooth muscle contraction (reviewed in ref. 10). Woodruff identified three IL‐13‐induced genes expressed in bronchial epithelial cells, including POSTN, SERPINB2, and CLCA1, which are elevated in asthma and downregulated by corticosteroid treatment. 11 , 12 Bhakta et al. (2013) showed that an average of POSTN, SERPINB2, and CLCA1 expression correlated well with blood eosinophil numbers and forced exhaled nitric oxide (FeNO) levels in patients with mild to moderate asthma. POSTN, SERPINB2, and CLCA1 are therefore IL‐13 pathway biomarkers expressed in the bronchial epithelium that may be useful for measuring the effects of anti‐T2 biologics in early phase asthma studies.
This paper reports a further investigation of bronchial epithelial T2 biomarkers in asthma, focusing on a subgroup with high FeNO levels and blood eosinophil counts in order to enrich the population for individuals with “T2‐high” asthma. The main aim was to investigate the gene expression of IL‐13 pathway biomarkers (11 genes, including POSTN, CLCA1, and SERPINB2) in bronchial epithelial samples from patients with T2 high mild asthma compared to healthy controls. Patients with mild asthma were studied because this subgroup is often enrolled in early phase asthma studies for reasons of safety. 6 , 8 The validation of POSTN, CLCA1, and SERPINB2 and the identification of other T2 bronchial epithelial biomarkers in this asthma subgroup would enable future early asthma clinical trials to use these biomarkers to evaluate novel anti‐T2 treatments.
METHODS
Subject recruitment and sample collection
Ten steroid naive patients with mild asthma using short acting beta agonists only, with blood eosinophil counts (>0.3 × 109/L) and high FeNO levels (>50 ppb), were recruited for bronchoscopy at the Medicines Evaluation Unit, Manchester, UK, along with six healthy subjects. All subjects were never smokers. Subjects with any other lung disease, inflammatory disorders, or an acute respiratory illness within 4 weeks of recruitment were excluded. At the initial visit, subjects answered the asthma control questionnaire and underwent spirometry, skin prick test, FeNO assessment (Aerocrine, Solna, Sweden), and full blood count (Manchester University NHS Foundation Trust, Manchester, UK). Two repeat full blood counts and FeNO measurements were taken 1–2 weeks apart, prior to bronchoscopy, to assess variability. The study was approved by the local research ethics committee (NRES Committee North West – Greater Manchester South; REC Ref: 06/Q1403/156) and all subjects provided written informed consent.
Gene expression analysis
Bronchial brush samples were collected from the lower lobes, stored in RNA‐Later (Invitrogen, Paisley, UK) and analyzed by Almac, Craigavon, UK. RNA was isolated using RNASTAT60 (Amsbio, Abingdon, UK). Following DNase‐1 treatments and purification, concentration and purity was evaluated using spectrophotometry, and integrity by an Agilent Bioanalyzer 2100. Reverse transcription was performed using the ABI High capacity kit and quantitative polymerase chain reaction (qPCR) performed using a Roche Lightcycler 480 system using ABI Taqman Universal Mastermix for the following genes: DPP4, MUC5AC, 15LOX1, POSTN, CLCA1, SERPINB2, TARC, MUC5B, CCL11, CCL26, and NOS2 were analyzed by qPCR; these epithelial genes are all induced by IL‐13. 11 , 13 , 14 , 15 Gene expression levels were quantified using the 2−Δct method and normalized to the mean expression of the following housekeeping genes: GAPDH1, TOP1, PPIA, CYC1, HMBS, SDHA, GUSB, HPRT1, ACTB, and TUBB2A. The three‐gene‐mean for POSTN, CLCA1, and SERPINB2 was calculated as per Bhakta et al. (2013).
Statistical analysis
All results were assessed using Graphpad Prism version 8.4.3 (San Diego, CA, USA). Comparisons between patients with asthma and healthy subjects were Mann‐Whitney test, with distribution of data assessed by D'Agostino and Pearson normality test. Correction for multiple testing, using methods such as Bonferoni or Šidák, are more appropriate for independent variables. 16 The genes investigated are all IL‐13 inducible; corrections for multiple testing were not applied due to the dependency of the genes, which was assessed by Spearman’s rank analysis. Any p values less than 0.05 was taken as statistically significant.
RESULTS
Patients with asthma had lower lung function compared to the healthy subjects (FEV1% predicted 77.2 ± 11.3 vs. 106 ± 12.8; p < 0.001); both groups had a similar age (mean (range) age: asthma 36 (21–55) years; healthy 42 (32–46) years, and were predominantly men (asthma: 10 men, 0 women; healthy 5 men, 1 woman). All patients with asthma were atopic, assessed by the skin prick test to house dust mites, cat hair, and grass pollen; all healthy subjects were non‐atopic. Blood neutrophil and basophil levels were significantly higher in the patients with asthma. The patients with asthma had partly controlled (Asthma Control Questionnaire [ACQ] score 0.75–1.49 17 ) or uncontrolled asthma (ACQ score >1.5); mean ACQ score of 1.3 (range: 0.7–2.0). All subjects were White.
Healthy subjects had lower blood eosinophil counts and FeNO levels than the patients with asthma (mean eosinophils: 0.13 vs. 0.46 × 109/L, respectively; mean FeNO: 13.1 vs. 78.7 ppb, respectively; both p < 0.0001). Three repeat measurements of blood eosinophils and FeNO, over a 2–4‐week period, showed that levels remain relatively stable (Figure 1); in healthy subjects, levels were consistently below the cutoffs of 0.3 × 106 eosinophils/ml and 50 ppb FeNO, whereas 93% of measurements in the patients with asthma were above these two cutoffs.
FIGURE 1.
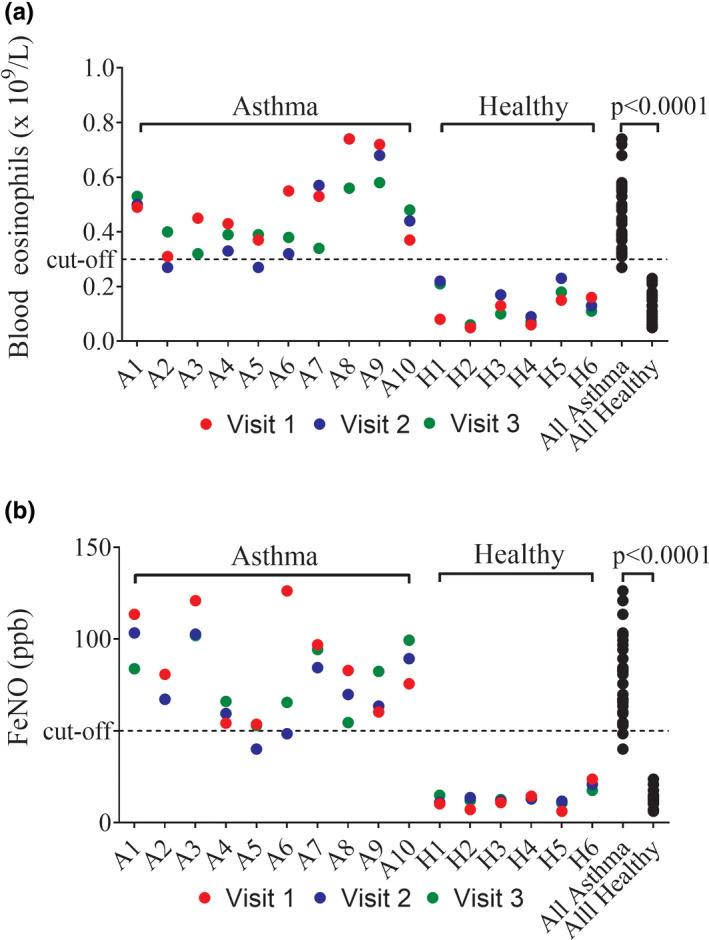
Blood eosinophil and FeNO reproducibility in patients with asthma (n = 10) and healthy subjects (n = 6). Three repeat blood eosinophil (a) and FeNO (b) measurements were taken 1–2 weeks apart. Asthma visit 1 inclusion cutoffs for blood eosinophils (>0.3 × 109/L) and FeNO (>50 ppb) are illustrated by dotted lines. Comparisons between asthma and healthy levels were by Mann‐Whitney test. FeNO, fractional exhaled nitric oxide
Bronchial epithelial gene expression results from three of the 10 patients with asthma were excluded from the analysis due to insufficient RNA quantity or integrity. Of the 11 genes assessed, expression levels of 15LOX1, POSTN, CLCA1, SERPINB2, CCL26, and NOS2 were significantly higher in the patients with asthma compared to the healthy controls (Figure 2a). Analyzing the data using the three‐gene‐mean method for POSTN, CLCA1, and SERPINB2 showed significantly higher expression levels in the patients with asthma versus controls, with results for all patients with asthma being above the 99% confidence interval for healthy subjects (Figure 2b). A potential three‐gene mean cutoff value of 0.0037 was calculated to identify T2‐high patients, based on the mean healthy score plus three SDs; all asthma scores were above this cutoff.
FIGURE 2.
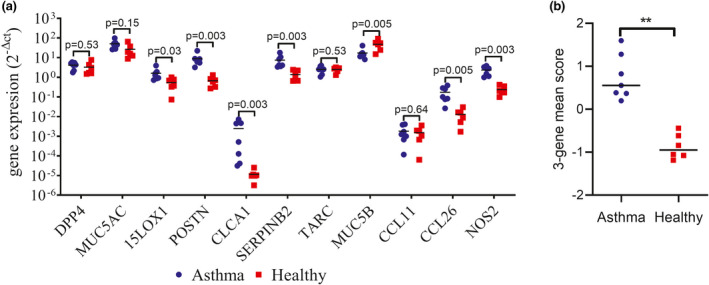
Bronchial epithelial expression of IL‐13 inducible genes in patients with asthma (n = 7) and healthy subjects (n = 6). (a) Epithelial brushings were collected during bronchoscopy and gene expression assessed by qPCR. Expression was quantified using the 2−Δct method, with results normalized to housekeeping gene expression. (b) The three‐gene‐mean composite score for CLCA1, POSTN, and SERPINB2 epithelial gene expression to compensate for variability in expression. Bars illustrate median expression levels. Comparisons between patients with asthma and healthy were by Mann‐Whitney test: **p < 0.01. qPCR, quantitative polymerase chain reaction
To evaluate the interdependency of the genes, associations with POSTN levels were assessed; there were strong positive correlations with 15LOX1 (rho = 0.790, p = 0.002), CLCA1 (rho = 0.853, p = 0.0004), SERPINB2 (rho = 0.937, p < 0.0001), CCL26 (rho = 0.944, p < 0.0001), and NOS2 (rho = 0.825, p = 0.001).
DISCUSSION
These results demonstrate that, in patients with mild asthma with clinical evidence of T2 inflammation (blood eosinophil counts and FeNO), the gene expression levels of POSTN, CLCA1, and SERPINB2 are increased in the bronchial epithelium compared to healthy subjects, confirming the findings of Bhakta et al. We now show that levels of 15LOX1, CCL26, and NOS2 are also elevated. The 11 genes measured in this study are all induced by IL‐13 11 , 13 , 14 , 15 ; we demonstrate that six of these can serve as epithelial biomarkers of T2 inflammation, with potential to be used in early phase clinical trials to measure the effects of drugs targeting T2 inflammation, and in particular IL‐13 signaling. CLCA1 and SERPINB2 have recently been used as biomarkers to demonstrate the effects of the anti‐IL13 monoclonal antibody lebrikizumab in uncontrolled asthma. 5
The six genes upregulated in patients with T2 high mild asthma are associated with asthma severity, increased corticosteroid response, or other T2 biomarkers. The 15LOX1 plays a key regulatory role in the production of lipid mediators that regulate inflammation in asthma and is associated with asthma severity. 18 CCL26 induces eosinophil migration, 19 and CCL26 epithelial expression is associated with sputum eosinophilia and asthma severity. 20 In vitro induction of CCL26 by IL‐13 in airway epithelial cells has been shown to be corticosteroid sensitive. 21 Bronchial gene expression of NOS2 is elevated in patients with atopic mild asthma compared to controls and is further increased following allergen challenge with levels correlating with FeNO. 22 Inhaled corticosteroid treatment reduces FeNO levels, and budesonide treatment reduces iNOS expression in the airways of patients with asthma. 23 Higher epithelial expression of CLCA1, POSTN, and SERPINB2 are associated with increased corticosteroid sensitivity. 11 Bhakta et al. (2013) developed a composite score for CLCA1, POSTN, and SERPINB2 gene expression to compensate for variability in expression; levels of this three‐gene‐mean correlated with FeNO and blood eosinophil levels. 12 Our analysis of CLCA1, POSTN, and SERPINB2 gene expression provides further evidence of an association of this three‐gene panel with FeNO and blood eosinophils.
Our results showing that POSTN, SERPINB2, CLCA1 have higher expression in the airways of patients with asthma compared to healthy subjects agree with previous studies. However, these earlier studies showed considerable overlap in expression levels between patients with asthma and controls. 12 , 24 This heterogeneity in previous studies could be attributed to the inclusion of patients with asthma with low or absent T2 inflammation. Here, we studied a homogeneous asthma population who were not using inhaled corticosteroids and who all had evidence of T2‐high asthma based on FeNO levels and blood eosinophil counts. It is striking from our results that expression of POSTN, SERPINB2, CLCA1, and NOS2, as well as the three‐gene mean, all showed perfect separation in expression levels between healthy subjects and patients with T2‐high mild asthma. Ideally, validation in a larger population would be valuable. Nevertheless, these results indicate that the FeNO (>50 ppb) and blood eosinophil (>0.3 × 109/L) cutoffs can identify patients with mild asthma with high levels of T2 bronchial epithelial biomarkers. This high bronchial epithelial T2 signal enhances the opportunity to observe biological effects of pharmacological interventions directed against T2 inflammation.
The three‐gene mean has been used in different studies to assess T2 inflammation. 24 The method used to calculate the three‐gene mean using a cohort‐specific average and differences in PCR methodology means that direct comparisons of three‐gene mean scores between studies is not possible. The accuracy of cutoff values generated is also limited by the small number of subjects involved in many studies.
Recent changes to GINA now recognize that initial asthma treatment may be better treated with intermittent ICS use by combining ICS with formoterol as a reliever treatment. 25 Our results, in patients who were being treated with short acting beta agonist treatment only, show excessive T2 inflammation (based on blood eosinophils and FeNO), which was associated with airflow obstruction (FEV1 = 77% predicted) and suboptimal asthma control (ACQ = 1.3). These patients would likely benefit from ICS treatment targeted at the T2 inflammation present.
Although numerically clear and statistically significant differences for epithelial gene expression between groups was observed, this study was limited by small sample size. The usefulness of using airway gene expression to measure T2 inflammation in a clinical setting is limited by the need to perform bronchoscopy. However, early phase clinical trials regularly involve bronchoscopies and such biomarkers are highly applicable. Our study is potentially limited by recruitment of a predominantly male, White population; any follow‐up study should study a wider population.
In conclusion, we have demonstrated a group of bronchial epithelial biomarkers present in patients with mild asthma with T2 inflammation, identified clinically by high blood eosinophil counts and FeNO levels. Expression of these genes may be useful for translational early phase asthma clinical trials.
CONFLICTS OF INTEREST
D.S. reports personal fees from AstraZeneca, personal fees from Boehringer Ingelheim, personal fees from Chiesi, personal fees from Cipla, personal fees from Genentech, personal fees from GlaxoSmithKline, personal fees from Glenmark, personal fees from Gossamerbio, personal fees from Menarini, personal fees from Mundipharma, personal fees from Novartis, personal fees from Peptinnovate, personal fees from Pfizer, personal fees from Pulmatrix, personal fees from Theravance, personal fees from Verona. M.V.G. was an employee of AstraZeneca when this work was carried out and also holds stock in AstraZeneca. M.V.G.is currently an employee of Trinet Pharma. T.S. declares no competing interests for this work.
AUTHOR CONTRIBUTIONS
T.S. and D.S. wrote the manuscript. T.S., M.V.G., and D.S. designed the research. T.S. performed the research. T.S., M.V.G., and D.S. analyzed the data.
Southworth T, Van Geest M, Singh D. Type‐2 airway inflammation in mild asthma patients with high blood eosinophils and high fractional exhaled nitric oxide. Clin Transl Sci. 2021;14:1259–1264. 10.1111/cts.13078
Funding information
Funding for the study was provided by the North West Lung Centre Charity, Manchester, UK and by AstraZeneca, Gothenburg, Sweden. This report is independent research, and the views expressed in this publication are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
REFERENCES
- 1. Dunican EM, Fahy JV. The role of type 2 inflammation in the pathogenesis of asthma exacerbations. Ann Am Thorac Soc. 2015;12(Suppl 2):S144‐S149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fahy JV. Type 2 inflammation in asthma–present in most, absent in many. Nat Rev Immunol. 2015;15(1):57‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peters MC, Kerr S, Dunican EM, et al. Refractory airway type 2 inflammation in a large subgroup of asthmatic patients treated with inhaled corticosteroids. J Allergy Clin Immunol. 2019;143(1):104‐13 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McGregor MC, Krings JG, Nair P, Castro M. Role of biologics in asthma. Am J Respir Crit Care Med. 2019;199(4):433‐445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Austin CD, Gonzalez Edick M, Ferrando RE, et al. A randomized, placebo‐controlled trial evaluating effects of lebrikizumab on airway eosinophilic inflammation and remodelling in uncontrolled asthma (CLAVIER). Clin Exp Allergy. 2020;50(12):1342‐1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Braithwaite IE, Cai F, Tom JA, et al. Inhaled JAK inhibitor GDC‐0214 reduces exhaled nitric oxide in patients with mild asthma: a randomized, controlled, proof‐of‐activity trial [published online ahead of print March 17, 2021]. J Allergy Clin Immunol. https://doi.org/ 10.1016/j.jaci.2021.02.042 [DOI] [PubMed] [Google Scholar]
- 7. Burgess G, Boyce M, Jones M, et al. Randomized study of the safety and pharmacodynamics of inhaled interleukin‐13 monoclonal antibody fragment VR942. EBioMedicine. 2018;35:67‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hodsman P, Ashman C, Cahn A, et al. A phase 1, randomized, placebo‐controlled, dose‐escalation study of an anti‐IL‐13 monoclonal antibody in healthy subjects and mild asthmatics. Br J Clin Pharmacol. 2013;75(1):118‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saha SK, Berry MA, Parker D, et al. Increased sputum and bronchial biopsy IL‐13 expression in severe asthma. J Allergy Clin Immunol. 2008;121(3):685‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marone G, Granata F, Pucino V, et al. The intriguing role of interleukin 13 in the pathophysiology of asthma. Front Pharmacol. 2019;10:1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Woodruff PG, Boushey HA, Dolganov GM, et al. Genome‐wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA. 2007;104(40):15858‐15863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhakta NR, Solberg OD, Nguyen CP, et al. A qPCR‐based metric of Th2 airway inflammation in asthma. Clin Transl Allergy. 2013;3(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsukura S, Stellato C, Georas SN, et al. Interleukin‐13 upregulates eotaxin expression in airway epithelial cells by a STAT6‐dependent mechanism. Am J Respir Cell Mol Biol. 2001;24(6):755‐761. [DOI] [PubMed] [Google Scholar]
- 14. Faffe DS, Whitehead T, Moore PE, et al. IL‐13 and IL‐4 promote TARC release in human airway smooth muscle cells: role of IL‐4 receptor genotype. Am J Physiol Lung Cell Mol Physiol. 2003;285(4):L907‐L914. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Y, Derycke L, Holtappels G, et al. Th2 cytokines orchestrate the secretion of MUC5AC and MUC5B in IL‐5‐positive chronic rhinosinusitis with nasal polyps. Allergy. 2019;74(1):131‐140. [DOI] [PubMed] [Google Scholar]
- 16. Sidak Z. Rectangular confidence regions for the means of multivariate normal distributions. J Am Stat Assoc. 1967;62(318):626‐633. [Google Scholar]
- 17. Juniper EF, Bousquet J, Abetz L, Bateman ED, Committee G. Identifying ‘well‐controlled’ and ‘not well‐controlled’ asthma using the Asthma Control Questionnaire. Respir Med. 2006;100(4):616‐621. [DOI] [PubMed] [Google Scholar]
- 18. Lindley AR, Crapster‐Pregont M, Liu Y, Kuperman DA. 12/15‐lipoxygenase is an interleukin‐13 and interferon‐gamma counterregulated‐mediator of allergic airway inflammation. Mediators Inflamm. 2010;2010:727305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Provost V, Larose MC, Langlois A, Rola‐Pleszczynski M, Flamand N, Laviolette M. CCL26/eotaxin‐3 is more effective to induce the migration of eosinophils of asthmatics than CCL11/eotaxin‐1 and CCL24/eotaxin‐2. J Leukoc Biol. 2013;94(2):213‐222. [DOI] [PubMed] [Google Scholar]
- 20. Larose M‐C, Chakir J, Archambault A‐S, et al. Correlation between CCL26 production by human bronchial epithelial cells and airway eosinophils: Involvement in patients with severe eosinophilic asthma. J Allergy Clin Immunol. 2015;136(4):904‐913. [DOI] [PubMed] [Google Scholar]
- 21. Banwell ME, Tolley NS, Williams TJ, Mitchell TJ. Regulation of human eotaxin‐3/CCL26 expression: modulation by cytokines and glucocorticoids. Cytokine. 2002;17(6):317‐323. [DOI] [PubMed] [Google Scholar]
- 22. Roos AB, Mori M, Grönneberg R, et al. Elevated exhaled nitric oxide in allergen‐provoked asthma is associated with airway epithelial iNOS. PLoS One. 2014;9(2):e90018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saleh D, Ernst P, Lim S, Barnes PJ, Giaid A. Increased formation of the potent oxidant peroxynitrite in the airways of asthmatic patients is associated with induction of nitric oxide synthase: effect of inhaled glucocorticoid. FASEB J. 1998;12(11):929‐937. [PubMed] [Google Scholar]
- 24. Mo Y, Zhang K, Feng Y, et al. Epithelial SERPINB10, a novel marker of airway eosinophilia in asthma, contributes to allergic airway inflammation. Am J Physiol Lung Cell Mol Physiol. 2019;316(1):L245‐L254. [DOI] [PubMed] [Google Scholar]
- 25. Global Initiative for Asthma . Global Strategy for Asthma Management and Prevention; 2020. Available from: www.ginasthma.org [Google Scholar]


