Abstract
The frequencies of genetic variants in the CYP3A4 and CYP3A5 genes differ greatly across global populations, leading to profound differences in the metabolic activity of these enzymes and resulting drug metabolism rates, with important consequences for therapeutic safety and efficacy. Yet, the impact of genetic variants on enzyme activity are incompletely described, particularly in American Indian and Alaska Native (AIAN) populations. To characterize genetic variation in CYP3A4 and CYP3A5 and its effect on enzyme activity, we partnered with AIAN people living in two regions of Alaska: Yup’ik Alaska Native people living in the Yukon‐Kuskokwim Delta region of rural southwest Alaska and AIAN people receiving care at the Southcentral Foundation in Anchorage, Alaska. We identified low frequencies of novel and known variation in CYP3A4 and CYP3A5, including low frequencies of the CYP3A4*1G and CYP3A5*1 variants, and linkage disequilibrium patterns that differed from those we previously identified in an American Indian population in western Montana. We also identified increased activity of the CYP3A4*1G allele in vitro and in vivo. We demonstrated that the CYP3A4*1G allele confers increased protein content in human lymphoblastoid cells and both increased protein content and increased activity in human liver microsomes. We confirmed enhanced CYP3A4‐mediated 4β‐vitamin D hydroxylation activity in Yup’ik people with the CYP3A4*1G allele. AIAN people in Alaska and Montana who carry the CYP3A4*1G allele—coupled with low frequency of the functional CYP3A5*1 variant—may metabolize CYP3A substrates more rapidly than people with the reference CYP3A4 allele.
Study highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Alleles in CYP3A4 and CYP3A5 are highly variable in populations globally, but have not been well‐characterized in American Indian and Alaska Native (AIAN) populations. The functional effects of some variants are not well known, including the CYP3A4*1G allele, which we identified in unique linkage disequilibrium patterns in prior work with an American Indian population in Montana.
WHAT QUESTION DID THIS STUDY ADDRESS?
This study characterizes the CYP3A4*1G allele using in vitro and in vivo models. It also improves our understanding of genetic variation across AIAN populations in North America.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
We show increased activity of the CYP3A4*1G allele, and identify low frequencies of CYP3A4 and CYP3A5 variants, including CYP3A4*1G and functional CYP3A5*1, in Alaska Native people.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Inclusion of diverse populations in estimating diplotype frequencies and their function is important for ensuring equitable access to the benefits of personalized medicine for all. We showed that people with the CYP3A4*1G allele may require dose escalation of CYP3A4‐metabolized medications due to increased activity of the CYP3A4 enzyme.
INTRODUCTION
The human cytochrome P450 3A enzymes, CYP3A4 and CYP3A5, metabolize many xenobiotics and dominate the clearance of about one‐third of all medications. The frequencies of genetic variants in the CYP3A4 and CYP3A5 genes differ exceptionally across global populations, leading to profound differences in the metabolic activity of these enzymes and resulting drug metabolism rates, with important consequences for therapeutic safety and efficacy. 1 , 2 , 3 Understanding CYP3A4 and CYP3A5 genetic variation, and the resulting effects on enzyme activity across diverse populations and allele combinations, is essential for improving the equity and utility of preemptive pharmacogenetic testing.
Compared with the reference CYP3A4*1A allele (herein referred to as CYP3A4*1), a number of single nucleotide variants (SNVs) have been associated with a change in the metabolic clearance of CYP3A drug substrates. 4 SNVs that have received the most research attention are CYP3A4*1B, CYP3A4*1G, and CYP3A4*22, which arise from noncoding variants thought to affect gene transcription or the stability of mRNA. 5 , 6 , 7 Whereas the CYP3A4*22 allele has been associated consistently with reduced enzyme activity and reduced clearance of substrates, such as tacrolimus, simvastatin, cyclosporine, and atorvastatin, CYP3A4*1B and CYP3A4*1G remain poorly characterized, each having been associated with both decreased and increased enzyme activity. 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 Most variation in CYP3A5 is associated with loss‐of‐function alleles, conferred primarily by three alleles, including a common variant (CYP3A5*3) that results in aberrant mRNA splicing. The other two, CYP3A5*6 and CYP3A5*7, are splicing defect and frameshift variants, respectively, and are seen more often in populations of African descent. 31 , 32 The frequency of the functional CYP3A5*1 reference allele varies widely between populations: ~ 7% in Europeans and 47% in Sub‐Saharan African populations. 33
In prior work, we identified a unique linkage disequilibrium pattern with the CYP3A4*1G allele in participants from the Confederated Salish and Kootenai Tribes (CSKTs) in western Montana 34 ; we observed a relatively high frequency of CYP3A4*1G, but a low level of linkage disequilibrium between CYP3A4*1G and CYP3A5*1, the functional allele of CYP3A5. These findings are unique because CYP3A4 protein expressed as a consequence of the CYP3A4*1G variant exists in the absence of functional CYP3A5 protein for a majority of CSKT people compared with the minority of people in many world populations. The goal of this study was to assess the functional effects of the CYP3A4*1G allele in both in vitro and in vivo models. Additionally, we also sought to characterize genetic variation in CYP3A4 and CYP3A5 genes in other American Indian and Alaska Native (AIAN) populations. Through collaborative partnerships with the Southcentral Foundation (SCF), a tribal health organization in Anchorage, AK, serving AIAN people, and with Yup’ik Alaska Native people from the Yukon‐Kuskokwim (YK) Delta, we identified novel variation and population allele frequencies within the CYP3A4 and CYP3A5 genes in AIAN populations living in different regions of Alaska.
METHODS
Institutional review board approvals and research settings
The Alaska Area Institutional Review Board (AAIRB) and SCF tribal review boards, Yukon‐Kuskokwim Health Corporation (YKHC) Executive Board of Directors, and institutional review boards from the University of Alaska Fairbanks and University of Washington approved this research. Research questions were developed in partnership with tribal representatives through community‐based participatory research; the partnerships with SCF, YKHC, and CSKT have been described previously in detail. 35 , 36
CYP3A4 and CYP3A5 genetic variant discovery and frequency assessment
A convenience sample of 363 SCF and 350 YK participants, aged greater than or equal to 18 years contributed DNA. To discover any common novel SNVs, we resequenced CYP3A4 and CYP3A5 in a subset of these participants: 188 and 94 from the SCF and YK cohorts, respectively (referred to as the resequencing study populations). Participants from the YK cohort were confirmed to be unrelated through pedigree analysis. The methods for blood collection, buffy coat isolation, and DNA extraction of samples are described previously. 37 Exons and adjacent intronic regions of CYP3A4 and CYP3A5 were resequenced, as described previously. 34
Next, we assessed population frequencies of the SNVs. Among all participants within the SCF (n = 363) and YK (n = 350) cohorts (referred to as the genotyping study populations), we genotyped named star alleles, previously identified nonsynonymous variants, and previously unreported nonsynonymous variants that we identified during the resequencing phase. As a result, we genotyped 8 SNVs in CYP3A4 and 8 SNVs in CYP3A5 using the Fluidigm Specific Target Amplification protocol and Genotyping assays (South San Francisco, CA) as described previously. 37 , 38 DNA samples with call rates below 95% across all SNVs were removed from further analysis. Of samples initially selected for genotyping, 17 from the SCF (4.5%) were excluded; all samples from the YK met quality thresholds. Concordance was 100% between samples assessed by both resequencing and genotyping.
CYP3A4 protein content in human lymphoblastoid cell lines
Human lymphoblastoid cell lines (LCLs) from the Peruvian in Lima, Peru (PEL) population in the 1000 Genomes Collection (Coriell Institute, Camden, NJ) varied by CYP3A4 diplotypes CYP3A4*1/*1 (n = 4), CYP3A4*1/*1G (n = 4), and CYP3A4*1G/*1G (n = 4). LCLs from the PEL population were chosen because—similar to our observations in the CSKT population—there is low linkage disequilibrium between CYP3A4*1G and CYP3A5*1. Therefore, we could select LCL samples with desired CYP3A4*1G diplotypes that were also all homozygous CYP3A5*3/*3, which eliminated contribution from CYP3A5. The CYP3A4*1B and CYP3A4*22 variants were also not present in these samples. LCLs were grown according to Coriell’s recommended protocols. Briefly, cells were cultured at 37°C in RPMI 1640 media (with 2 mM L‐gln, 15% FBS, and 1% penicillin/streptomycin) and grown at 37°C in the presence of 5% CO2. Approximately 1 × 106 cells were pelleted, lysed (40 mM Tris, 120 mM NaCl, 1% Triton X‐100, 0.1% SDS, pH 7.3, and protease inhibitors), and lysates collected and stored at −80°C. Protein concentration of cell lysates was determined using Bio‐Rad Protein Assay Dye according to manufacturer’s protocol using BSA as a standard.
CYP3A4 protein content in LCLs was measured using a standard Bio‐Rad (Hercules, CA) immunoblot protocol according to manufacturer’s protocol with SDS‐PAGE Mini‐PROTEAN TGX gels (Bio‐Rad). Recombinant CYP3A4 enzyme from baculovirus‐infected insect cells was used as a positive control (BD Gentest; Corning, Corning, NY). CYP3A4 was detected with primary antibody, CYP3A4 Mouse MaxPab (Abnova, Taipei City, Taiwan), diluted (1:1,000) in blocking buffer (Tris‐buffered saline +0.2% Tween +7% nonfat milk), and secondary antibody, HRP‐conjugated anti‐mouse (Thermo Fisher) diluted (1:100,000) in blocking buffer +1% goat. Endogenous control, glyceraldehyde 3‐phosphate dehydrogenase (GAPDH), was measured with HRP‐conjugated anti‐human GAPDH antibody (Thermo Fisher) diluted (1:200,000) in blocking buffer +1% goat. The membrane was developed with SuperSignal West Femto kit, according to its manufacturer’s protocol (Thermo Fisher). The membrane was visualized using a FluorChem M system (Protein Simple, San Jose, CA). CYP3A4 protein was quantitated relative to GADPH in each lane.
DNA isolation, CYP3A4 and CYP3A5 sequencing, and mRNA and protein quantitation in human liver microsomes
Human liver tissue samples (n = 319) were obtained from the University of Washington Human Liver Bank (n = 59) and the St. Jude Liver Resource at St. Jude Children’s Research Hospital (n = 260). Preparation of human liver microsome (HLM) and quantitation of total protein were described previously. 39 Methods for DNA isolation, quantitation of aldo‐keto reductase 1D1 (AKR1D1) mRNA content, and quantitation of CYP3A4 and cytochrome P450 oxidoreductase (POR) protein content were as described previously. 39 , 40 , 41 , 42 , 43 CYP3A4 and CYP3A5 sequencing was performed using the PGRNseq platform (version 1.1), which captures all exons as well as 2 kb of upstream and 1 kb of downstream sequence. 44 CYP3A4 and CYP3A5 alleles are defined according to the Pharmacogene Variation Consortium. 4
CYP3A4 activity in human liver microsomes
CYP3A4 activity in HLMs was determined with the P450‐Glo CYP3A4 Assay System (Promega, Madison, WI) according to manufacturer’s specifications with the CYP3A4‐specific substrate, Luciferin‐IPA, which does not measure any CYP3A5 activity. Microsomal incubation conditions were optimized using pooled HLMs (Xenotech, Lenexa, KS) to ensure experiments were in the linear range of substrate concentration, total HLM protein content, and incubation time (data not shown). Five µg of total HLM protein were preincubated with 3 µM P450‐Glo CYP3A4 substrate (Luciferin‐IPA) and 100 mM potassium phosphate buffer for 10 min in a water bath at 37°C. An equal volume of 2 mM NADPH was added to initiate the reaction and HLMs were incubated for 10 min in a water bath at 37°C. An equal volume of Luciferin Detection Reagent was added and incubated for 15–20 min at room temperature. All incubations were completed in triplicate. Pooled HLMs were used as a positive control in every incubation set. Luminescence was detected on a SynergyMX microplate reader (Biotek, Winooski, VT).
Endogenous CYP3A4‐mediated vitamin D hydroxylation
We recruited a convenience sample of 743 self‐identified Yup’ik participants, greater than or equal to 14 years of age from the YK Delta, and selected 526 who were unrelated to the third degree to reduce nonindependence, as described previously. 45 From this subset, 514 participants were successfully genotyped for CYP3A4*1G using the Fluidigm genotyping platform described above. These participants included the 350 that also participated in the gene variant discovery and population allele frequency studies, described above. The unrelated subset was representative of the overall dataset. 45 We collected serum samples and quantified serum 4β,25‐dihydroxyvitamin D3 (4ß,25[OH]2D3) and 25‐hydroxyvitamin D3 (25[OH]D3), as described previously. 46 Adjusting for gender, we compared serum 4ß,25(OH)2D3/25(OH)D3 between participants with at least one CYP3A4*1G allele and those with no copies of the allele because we only identified 3 participants homozygous for the CYP3A4*1G allele.
Statistical analysis
We performed all analyses in R programming language. We used pedigrees and the best linear unbiased estimator, 47 as described in ref. 45, to calculate allele frequencies and confidence intervals for the SCF and YK cohorts. We tested all SNVs for deviations from Hardy–Weinberg equilibrium using a χ2 test, with a significance level of 0.05. A p value less than 0.05 (p < 0.05) was considered to be statistically significant for all analyses.
The association of the CYP3A4*1G diplotype and CYP3A4 protein content in LCLs was estimated using one‐way analysis of variance with GraphPad Prism 7.0 software (GraphPad, San Diego, CA). CYP3A4 protein content and activity in HLMs were tested for skewness and kurtosis, and log transformed to improve normal distribution. We used multivariable linear regression to assess the association of CYP3A4*1G diplotype and CYP3A4 protein content and activity in HLMs. All analyses of HLMs were adjusted for site of sample collection, which has been consistently associated with differences in outcomes. 39 , 40 , 43 Primary analysis used linear regression, adjusting for site of sample collection, to assess the association of the CYP3A4*1G/*1G diplotype on log transformed CYP3A4 protein content and activity, compared with livers that did not have this diplotype. Secondary analysis using multiple linear regression was used to assess the influence of other potential covariates on log transformed CYP3A4 protein and activity, including age (continuous), sex (binary: male vs. female), POR protein content (continuous), presence of the CYP3A4*22 allele (binary: presence or absence), and AKR1D1 mRNA content (continuous). Covariates with significant unadjusted associations with CYP3A4 protein content and activity in the primary analysis were kept in the final multivariable model to determine the association between CYP3A4*1G diplotype and both log CYP3A4 protein content and activity. We used an unpaired t‐test to compare serum 4ß,25(OH)2D3/25(OH)D3 between participants with at least one CYP3A4*1G allele and those with no copies of the allele.
RESULTS
CYP3A4 and CYP3A5 genetic variant discovery and frequency assessment
Population demographics
Table 1 summarizes the demographics for the SCF and YK participants included in the cohorts for the DNA genotyping study and the YK participants in the vitamin D hydroxylation study. All participants self‐identified as Yup’ik in the YK cohort or as AIAN in the SCF cohort. Donor demographics for the HLM study are also summarized in Table 1.
TABLE 1.
Demographics of study populations
| Study populations | |
|---|---|
| SCF genotyping (n = 363) | |
| Sex, n | |
| Male | 127 (35.0%) |
| Female | 236 (65.0%) |
| YK genotyping (n = 350) | |
| Sex, n | |
| Male | 163 (46.6%) |
| Female | 187 (53.4%) |
| HLM set (n = 319) | |
| Age, years a | |
| Mean ± SD | 39.6 ± 22.2 |
| Range | 0–87 |
| Sex, n | |
| Male | 185 (58.0%) |
| Female | 129 (40.4%) |
| Unknown | 5 (1.6%) |
| Race/ethnicity, n | |
| European | 288 (90.3%) |
| African | 5 (1.6%) |
| Asian | 1 (0.3%) |
| Hispanic | 1 (0.3%) |
| Unknown | 24 (7.5%) |
| YK vitamin D hydroxylation (n = 526) | |
| Sex, n | |
| Male | 282 (53.6%) |
| Female | 244 (46.4%) |
Abbreviations: HLM: human liver microsome; SCF, Southcentral Foundation; YK, Yukon Kuskokwim.
Age was recorded in years for all subjects and an age of zero was given to donors under 1 year old; more granular data (e.g., weeks, months) were not available.
Resequencing for SNV discovery
Tables S1 and S2 present the SNVs—known and previously unreported—identified in resequencing CYP3A4 and CYP3A5, respectively, from a convenience sample cohort of SCF (n = 188) and YK (n = 94) participants from the genotyping study. In addition to previously identified SNVs, we identified two variants in CYP3A5 that had not been described previously: a nonsynonymous variant at position chr7:99245986 resulting in a proline to leucine change at amino acid 484 that was identified in the SCF cohort and a deletion at chr7:99247737 resulting in a frameshift at amino acid 458 that was identified in the YK cohort, and has since been assigned rs547253411.
Genotyping for assessing population frequencies
All SNVs were in Hardy–Weinberg equilibrium in a larger cohort of SCF (n = 363) and YK (n = 350) participants. Table 2 presents the frequencies of the novel and known important alleles that were genotyped in the SCF and YK cohorts in relation to other populations around the globe.
TABLE 2.
Allele frequencies in SCF (n = 363) and YK (n = 350) populations compared with other populations globally
| rs number | Allele name | Reference allele | Variant allele | Protein effect | SCF | YK | CSKT | AMR | AFR | EAS | EUR | SAS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 726 | 700 | 188 | 694 | 1322 | 1008 | 1006 | 978 | |||||
| Number of alleles: | ||||||||||||
| CYP3A4 Allele frequency (%) a | ||||||||||||
| rs2242480 | CYP3A4*1G | C | T | Intron 10 | 12.7 (10.3–15.1) | 4.0 (2.5–5.5) | 26.8 | 39.3 | 85.0 | 26.8 | 8.2 | 37.1 |
| rs2740574 | CYP3A4*1B | T | C | 5’ flanking | 3.0 (1.8–4.2) | 0 | 2.2 | 10.5 | 76.6 | 0.4 | 2.8 | 4.0 |
| rs55785340 | CYP3A4*2 | T | C | S222P | 0 | 0 | 0 | 0 | 0 | 0 | 0.2 | 0 |
| rs4646438 | CYP3A4*6 | Del | A | 277 frameshift | 0.6 (0.0–1.2) | 0 | 0 | ND | ND | ND | ND | ND |
| rs56324128 | CYP3A4*7 | G | A | G56D | 0 | 0 | 0 | 0 | 0 | 0 | 0.2 | 0 |
| rs12721627 | CYP3A4*16 | C | G | T185S | 0 | 0 | 0 | 0 | 0 | 0.4 | 0 | 0 |
| rs28371759 | CYP3A4*18 | T | C | L293P | 0 | 0 | 0 | 0 | 0 | 1.1 | 0 | 0.6 |
| rs35599367 | CYP3A4*22 | C | T | Intron 6 | 2.4 (1.3–3.5) | 0.3 (0.0–0.7) | 2.4 | 2.6 | 0.1 | 0 | 5.0 | 0 |
| CYP3A5 Allele frequency (%) a | ||||||||||||
| rs28365083 | CYP3A5*2 | C | A | T398 N | 0.3 (0.0–0.7) | 0.0 | 0 | 0 | 0 | 0 | 0.4 | 0 |
| rs776746 | CYP3A5*3 | T | C | Splice | 89.8 (87.6–92.0) | 95.9 (94.4–97.4) | 92.5 | 79.9 | 18.0 | 71.3 | 94.3 | 37.1 |
| rs10264272 | CYP3A5*6 | C | T | Splice | 0.6 (0.0–1.2) | 0.0 | 0 | 2.3 | 15.4 | 0 | 0.3 | 4.0 |
| rs41303343 | CYP3A5*7 | Del | A | 346 frameshift | 0.1 (0.0–0.3) | 0.0 | 0 | 0.3 | 11.8 | 0 | 0 | 0 |
| rs28383468 | C | T | H30Y | 0.4 (0.0–0.9) | 0.0 | 0 | 0.1 | 0 | 0 | 1.2 | 0 | |
| rs28365085 | T | C | I488 T | 0.4 (0.0–0.9) | 0.0 | 0 | 0 | 0 | 0.8 | 0 | 0 | |
| rs547253411 | G | Del | 458 frameshift | 0.3 (0.0–0.7) | 0.0 | 0 | ND | ND | ND | ND | ND | |
| rsNA | C | T | P484L | 0.1 (0.0–0.3) | 0.0 | 0 | ND | ND | ND | ND | ND | |
Abbreviations: AFR, African, including Yoruba in Ibadan, Nigeria; Luhya in Webuye, Kenya; Americans of African Ancestry in SW USA; Gambian in Western Divisions in the Gambia; Mende in Sierra Leone; Esan in Nigeria; AMR, Ad Mixed American, including Puerto Ricans from Puerto Rico; Colombians from Medellin, Colombia; Peruvians from Lima, Peru; Mexican Ancestry from Los Angeles, USA; CSKT, Confederated Salish Kootenai Tribes; EAS, East Asian, including Han Chinese in Beijing, China; Southern Han Chinese; Japanese in Tokyo, Japan; Chinese Dai in Xishuangbanna, China; Kinh in Ho Chi Minh City, Vietnam; EUR, European, including Utah Residents with Northern and Western European ancestry; Finnish in Finland; British in England and Scotland; Iberian population in Spain; Toscani in Italia; ND, not determined; rsNA, indicates a SNV without a reference identification number; SAS, South Asian, including Gujarati Indians from Houston Texas; Punjabi from Lahore, Pakistan; Bengali from Bangladesh; Sri Lankan Tamil from the UK; Indian Telugu from the UK; SCF, Southcentral Foundation; YK, Yukon Kuskokwim.
Global frequencies are from the 1,000 genomes Phase 3 database, as accessed through Ensembl (http://uswest.ensembl.org/index.html) on July 15, 2020, and compared with the Confederated and Salish Kootenai Tribes (CSKT). 34
The CYP3A4*1G allele was infrequent in the SCF cohort (12.7%) relative to other populations and the frequency in the YK cohort (4.0%) was the lowest reported in any population to date. The nonfunctional CYP3A5*3 allele was found at high frequencies in both the SCF (89.8%) and YK (95.9%) cohorts. The linkage disequilibrium between CYP3A4*1G and CYP3A5*1 was r 2 = 0.55 in the SCF cohort and r 2 = 0.90 in the YK cohort (Figure S1). Importantly, the CYP3A5*3/*3 diplotype is associated with very low expression of CYP3A5 enzyme compared with either the CYP3A5*1/*1 and CYP3A5*1/*3 diplotypes. Based on these phenotypes, only 19.6% of the SCF cohort and 8.9% of the YK cohort are expected to have functional CYP3A5 activity (i.e., people with at least one copy of the CYP3A5*1 allele).
CYP3A4 protein content in human lymphoblastoid cell lines
CYP3A4*1G was significantly associated with increased CYP3A4 protein content, normalized to endogenous control GAPDH (p < 0001; Figure 1). Without normalization to GAPDH, CYP3A4 protein content was highest in LCLs with the CYP3A4*1G/*1G diplotype (2.37 ± 0.23 relative fluorescent units), intermediate for heterozygote CYP3A4*1/*1G (1.48 ± 0.27), and lowest for CYP3A4*1/*1 (0.92 ± 0.05, p < 0.01). GAPDH was not statistically different between CYP3A4 diplotypes (p = 0.16).
FIGURE 1.
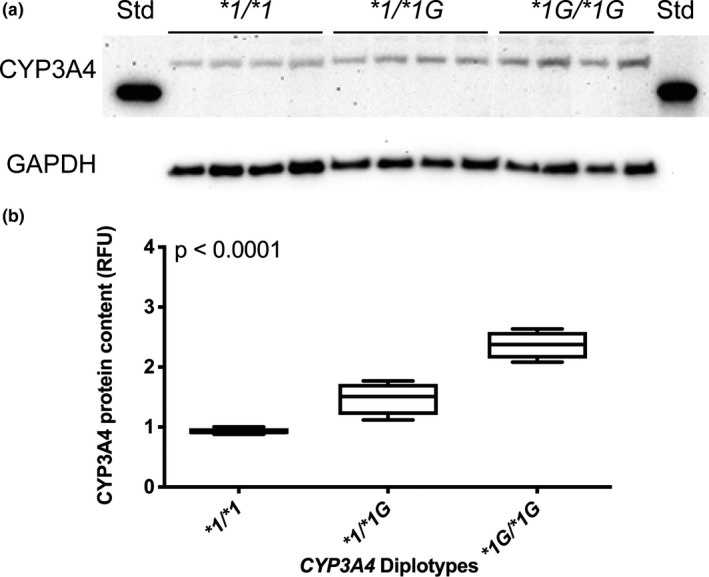
CYP3A4 protein content in lymphoblastoid cell lines (LCLs). Immunoblot of CYP3A4 and glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) in LCLs with CYP3A4*1/*1, *1/*3, and *3/*3 diplotypes (panel a). Quantitation of CYP3A4 normalized to GAPDH endogenous control (panel b). One‐way analysis of variance was used to compare CYP3A4 diplotypes
CYP3A4 protein content and activity in human liver microsomes
Adjusting for site of sample collection, HLM samples with a CYP3A4*1G/*1G diplotype exhibited a statistically significant higher level of log transformed CYP3A4 protein content (p < 0.01; Figure 2A) and log transformed CYP3A4 activity measured with a CYP3A4‐specific substrate (p = 0.04; Figure 2B) than did samples homozygous or heterozygous for the reference allele at that position. CYP3A4 protein content (mean ± SD) was 63.1 ± 62.9, 54.6 ± 56.0, and 221 ± 325 pmol/mg protein for CYP3A4*1/*1, *1/*1G, and *1G/*1G diplotypes, respectively. CYP3A4 activity (mean ± SD) was 0.046 ± 0.076, 0.040 ± 0.065, and 0.116 ± 0.148 pmol/mg protein/min for CYP3A4*1/*1, *1/*1G, and *1G/*1G diplotypes, respectively. Log POR content (p < 0.01), AKRD1 content (p < 0.01), site of sample collection (p = 0.02), age (p < 0.01), sex (p < 0.01), and CYP3A4*22 genotype (p = 0.01) each were individually significantly associated with log CYP3A4 protein content. Log POR content (p < 0.01), AKRD1 content (p < 0.01), site of sample collection (p < 0.01), and age (p = 0.04) each were individually significantly associated with log CYP3A4 activity, but sex (p = 0.14) and CYP3A4*22 (p = 0.11) genotype were not.
FIGURE 2.
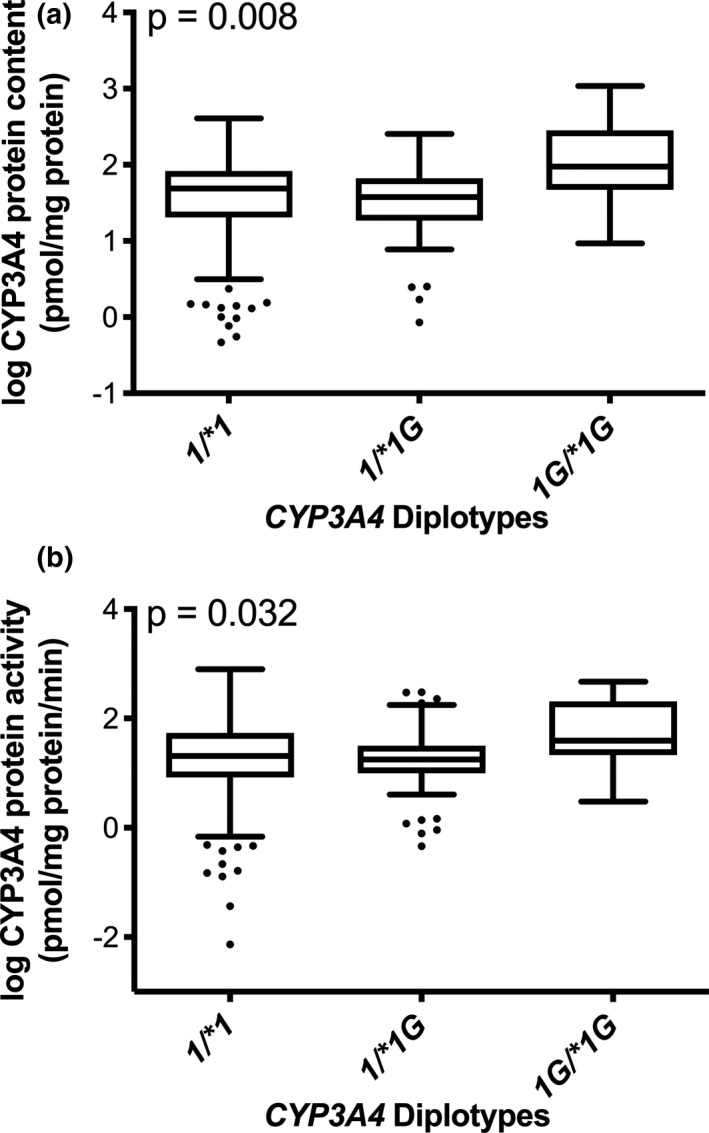
CYP3A4 protein content and activity in human liver microsomes (HLMs). CYP3A4 protein content (panel a) and CYP3A4 metabolic activity (panel b) by CYP3A4 diplotypes: CYP3A4*1/*1 (n = 245), *1/*1G (n = 64), and *1G/*1G (n = 10). All incubations were completed in triplicate. Multivariable linear regression was used to compare protein content and activity between HLMs based on genotype, adjusting for site of sample collection
Using a recessive inheritance model and adjusting for the factors found to be significant in individual models, the CYP3A4*1G/1G was no longer significantly associated with increased CYP3A4 protein content and metabolic activity in HLMs (Table 3). As expected, CYP3A4 protein content correlated strongly with CYP3A4 protein activity (p < 2 × 10−16). Liver collection site was a significant predictor of CYP3A4 activity, which is consistent with previous studies with CYP2A6, CYP2C19, and CYP2D6 function in these liver samples. 39 , 40 , 43 POR protein content and AKR1D1 mRNA content were also significant predictors of CYP3A4 protein content and activity, consistent with previous studies from these liver samples. 39 , 40 Interestingly, the CYP3A4*22 allele that has been associated with altered CYP3A4 activity elsewhere, was not a significant predictor of CYP3A4 activity, but was a significant predictor of CYP3A4 protein.
TABLE 3.
Multiple linear regression analysis of covariates with log CYP3A4 protein content and log CYP3A4 activity in human liver microsomes, adjusting for variables independently associated with log CYP3A4 protein content and activity
| Covariate | β value | Standard error | p value |
|---|---|---|---|
| Log CYP3A4 protein content (pmol/mg protein) | |||
| CYP3A4*1G/*1G diplotype | 0.12 | 0.09 | 0.18 |
| log POR protein content | 1.06 | 0.13 | 5.0 × 10−14 |
| 100*AKR1D1 mRNA content | 0.30 | 0.97 | 2.5 × 10−3 |
| UW sample collection | 0.04 | 0.06 | 0.52 |
| 100*Age by year | 0.34 | 0.13 | 0.01 |
| Male | −0.06 | 0.06 | 0.10 |
| CYP3A4*22 present | −0.21 | 0.09 | 0.03 |
| Log CYP3A4 activity (pmol/mg protein/min) | |||
| CYP3A4*1G/*1G diplotype | 0.18 | 0.11 | 0.12 |
| Log POR protein content | 1.33 | 0.16 | 1.1 × 10−14 |
| 100*AKR1D1 mRNA content | 0.27 | 0.12 | 2.3 × 10−2 |
| UW sample collection | 0.30 | 0.08 | 3.7 × 10−4 |
| 100*Age by year | 0.51 | 0.16 | 1.9 × 10−3 |
Abbreviations: POR, cytochrome P450 oxidoreductase; AKR1D1, aldo‐keto reductase 1D1; UW, University of Washington.
Endogenous CYP3A4‐mediated vitamin D hydroxylation
To confirm our in vitro data showing increased function of the CYP3A4*1G allele, we measured the ratio of serum 4β,25(OH)2D3 to 25(OH)D3 in vivo. This ratio is proportional to an individual’s CYP3A4‐dependent intrinsic metabolite formation clearance. Participants with at least one CYP3A4*1G exhibited an increased ratio of serum 4β,25(OH)2D3 to 25(OH)D3 (Figure 3), indicating that CYP3A4 activity was 17.2% higher in individuals with the CYP3A4*1G allele (n = 57) than in individuals without the CYP3A4*1G allele (n = 457, p < 0.01). We did not have enough participants with homozygous CYP3A4*1G diplotypes to analyze separately.
FIGURE 3.
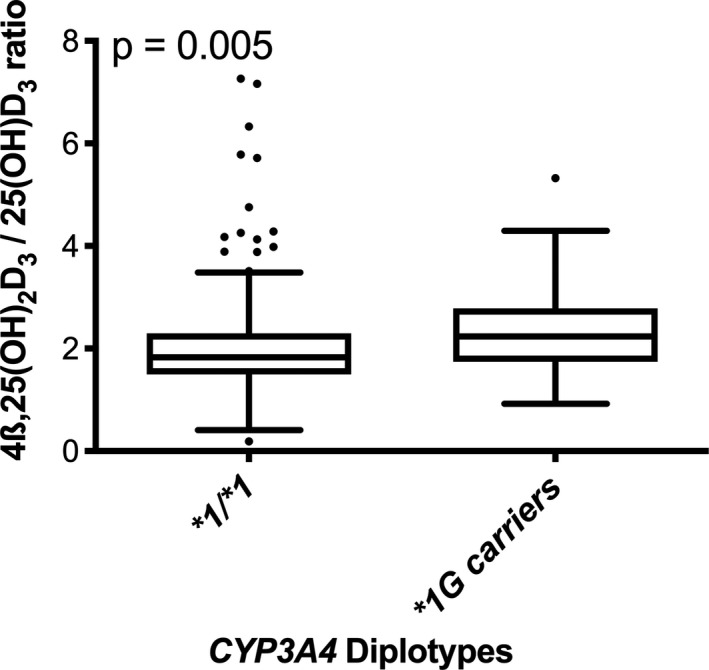
CYP3A4‐mediated vitamin D hydroxylation. Ratio of serum 4ß,25(OH)2D3 and 25(OH)D3 in unrelated participants from the Yukon‐Kuskokwim (YK) Delta (n = 514) by CYP3A4 diplotypes: individuals with no CYP3A4*1G (n = 457) and individuals who at least one copy of CYP3A4*1G (n = 57). A t‐test was used to compare the two groups
DISCUSSION
Using both in vitro and in vivo data, our research demonstrates that the CYP3A4*1G allele is associated with increased CYP3A4 protein content and metabolic activity compared with the reference allele. We also identified relatively low frequencies of novel and known CYP3A4 and CYP3A5 alleles, including low frequencies of the functional CYP3A5*1 variant, in AIAN participants from 2 regions of Alaska. Our work illustrates broad heterogeneity across American indigenous populations and suggests that AIAN people with the CYP3A4*1G allele may exhibit, on average, higher metabolic activity toward CYP3A4 substrates than people from other populations worldwide.
To identify novel variants and determine the frequency of these and known variants, we resequenced CYP3A4 and CYP3A5 genes in AIAN participants from 2 regions of Alaska, one representing southcentral Alaska and the Aleutian Islands, and the other a region of western Alaska in the YK Delta. We identified the CYP3A4*1G at lower frequencies in the SCF cohort (12.7%) and YK cohort (4.0%) than what we previously reported in the CSKT cohort (26.8%). 34 We also found other CYP3A4 variants at low frequencies in both Alaskan cohorts, including the CYP3A4*22 allele, which has been associated with reduced CYP3A4 mRNA and protein expression, and with poor metabolic activity toward tacrolimus and statins. 5 , 17 , 18 , 19 We identified very low frequencies of the functional CYP3A5 allele, CYP3A5*1, in both the SCF and YK cohorts, similar to our prior work with the CSKT. 34 These findings contrast with reported frequencies of CYP3A5*1 in other North and South American indigenous populations, including ~ 60% among Pima Indians (n = 6) and 23.7% in 2 admixed Central American indigenous populations (n = 232). 32 , 48 Of note, the linkage disequilibrium between CYP3A4*1G and CYP3A5*1 was r 2 = 0.55 in the SCF cohort and r 2 = 0.90 in YK cohort, compared with r 2 = 0.158 in the CSKT cohort. 34 Collectively, these differences in allele frequencies and linkage disequilibrium patterns across CYP3A4 and CYP3A5, and the heterogeneity they represent, highlight the importance of introducing comprehensive genetic testing of individuals, without relying on preconceived racial and ethnic assumptions, if genetic information is to be used optimally to guide drug therapy decisions.
The CYP3A4*1G allele has been associated with increased clearance of amfepramone, 20 tacrolimus, 21 , 22 imatinib mesylate, 23 and finasteride. 24 Additionally, the CYP3A4*1G allele has been associated with lower rates of clopidogrel resistance 25 , 26 and higher rates of sodium valproate‐response in pediatric patients. 27 Data are conflicting for vitamin D, however, with CYP3A4*1G associated with higher levels of 25‐hydroxyvitamin D in pregnant women in one study, 28 but lower levels in another. 29 Furthermore, clinical data emerging primarily from eastern Asia, where the CYP3A4*1B allele is not present or occurs at a very low frequency, suggest lower metabolic activity toward fentanyl 11 , 13 , 14 , 15 and atorvastatin 7 , 30 attributed to the CYP3A4*1G allele. Our in vitro and in vivo data support the increased expression and activity of CYP3A4 with the CYP3A4*1G allele. LCLs heterozygous or homozygous for CYP3A4*1G had higher CYP3A4 protein content relative to LCLs homozygous for CYP3A4*1. We were not able to quantitate activity in LCLs with CYP3A4 activity below the limit of detection of the assay, presumably due to relatively low levels of CYP3A4 expression. Although the CYP3A4 bands in the immunoblot from LCL lysates migrated more slowly than those from the recombinant CYP3A4 standard from baculovirus‐infected insect cells, it is likely that the differences in co‐migration are due to different matrices between the cell lysates and recombinant enzyme, which is consistent with the manufacturer’s findings that CYP3A4 migration patterns differ by matrices.
Importantly, livers containing the CYP3A4*1G/*1G diplotype had increased CYP3A4 protein content and metabolic activity measured with a CYP3A4‐specific substrate. After adjusting for covariates, including AKR1D1 mRNA content, POR protein content, liver collection site, and age for both protein content and activity, and also sex and CYP3A4*22 in the case of CYP3A4 protein content, however, the association with CYP3A4*1G was not significant, although still trending toward an association with increased CYP3A4 protein and activity. The association between POR protein content and CYP3A4 protein content and activity dominates any signal from the CYP3A4*1G allele, which may result in a loss of power to detect the effects of CYP3A4*1G. Interestingly, the CYP3A4*1G allele does not appear to be an expression quantitative trait loci in the liver according to the Genotype‐Tissue Expression (GTEx) project. 49 Thus, whereas gene expression may not be affected, changes in protein synthesis or stability may affect protein and activity levels.
We further corroborated our in vitro results using vitamin D hydroxylation as an in vivo biomarker for CYP3A4 activity. Participants from the YK Delta with the CYP3A4*1G variant exhibited increased metabolic activity as measured by the ratio of serum 4β,25(OH)2D3 to 25(OH)D3. Our group has previously shown data to validate the use of this ratio as an in vivo biomarker of CYP3A4 activity. We have shown that 4ß,25(OH)2D3 is produced from 25(OH)D3 predominantly by CYP3A4, with only minor (<10%) contribution from CYP3A5. 50 Additionally, we showed that the 4β,25(OH)2D3/25(OH)D3 ratio is strongly correlated with midazolam hydroxylation in HLMs, and 4β,25(OH)2D3 formation was induced by rifampin and inhibited by CYP3A4‐specific inhibitors in human hepatocytes. 50 We also showed that 4β,25(OH)2D3 plasma concentration increased 60% after short‐term rifampin administration in healthy volunteers. 51 The enhanced activity we observed in individuals with the CYP3A4*1G allele may extend toward other CYP3A4 substrates, such as some of the statins and tacrolimus that are of interest for genetically guided drug therapy. 33 The presence of the CYP3A4*1G allele may necessitate increased dose requirements in individuals with this allele relative to people with the reference allele.
In summary, allele frequencies of CYP3A4 and CYP3A5 vary widely across subgroups of AIAN people and across the globe. Characterizing unique genetic factors that contribute to alterations in CYP3A4 and CYP3A5 function is critical for the equitable implementation of pharmacogenetic‐guided therapies. 52 By better understanding the functional effects of these alleles, such as the CYP3A4*1G allele that we demonstrate conferring increased activity, we make pharmacogenetic testing more valuable for people with understudied variants.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
AUTHOR CONTRIBUTIONS
A.E.F., K.E.T., and E.L.W. wrote the manuscript. K.E.T., and E.L.W. designed the research. A.E.F., R.D., K.S., K.J., S.E.H., R.R., B.A.K., D.A.D., and B.B.B. performed the research. A.E.F., R.D., K.S., T.A.T., and E.L.W. analyzed the data. K.G.C., B.P., E.S., and D.A.N. contributed new reagents/analytical tools.
DISCLAIMER
As an Associate Editor of Clinical and Translational Science, Erica Woodahl was not involved in the review or decision process for this paper.
Supporting information
Fig S1
Table S1
Table S2
ACKNOWLEDGMENTS
The authors thank all of the recruitment personnel in the YK Delta, research staff at SCF, and Barbara Kavanaugh, Program Manager for the Northwest Alaska – Pharmacogenomics Research Network (NWA‐PGRN), for directing programmatic collaborations. We would also like to thank Patricia L. Stapleton and Jesse M. Tsai in the Functional Genomics Laboratory at the University of Washington for performing the genotyping assay. This work was supported by National Institutes of Health (NIH) grant funding to the NWA‐PGRN (U01GM092676 and P01GM116691), R01GM63666, and the EDGE center grant (P30ES007033). Finally, the authors would like to express their deep appreciation to all of the American Indian and Alaska Native people who participated in this research study and the communities in which they live.
Funding Information
This work was supported by NIH grant funding to the Northwest Alaska ‐ Pharmacogenomics Research Network (NWA‐PGRN) (U01GM092676 and P01GM116691), R01GM63666, and the EDGE center grant (P30ES007033).
REFERENCES
- 1. Rodríguez‐Antona C, Sayi JG, Gustafsson LL, et al. Phenotype‐genotype variability in the human CYP3A locus as assessed by the probe drug quinine and analyses of variant CYP3A4 alleles. Biochem Biophys Res Commun. 2005;338(1):299‐305. [DOI] [PubMed] [Google Scholar]
- 2. Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A‐mediated metabolism. Adv Drug Deliv Rev. 2002;54(10):1271‐1294. [DOI] [PubMed] [Google Scholar]
- 3. Werk AN, Cascorbi I. Functional gene variants of CYP3A4. Clin Pharmacol Ther. 2014;96(3):340‐348. [DOI] [PubMed] [Google Scholar]
- 4. Gaedigk A, Ingelman‐Sundberg M, Miller NA, et al. The pharmacogene variation (PharmVar) consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin Pharmacol Ther. 2018;103(3):399‐401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2011;11(4):274‐286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Amirimani B, Ning B, Deitz AC, Weber BL, Kadlubar FF, Rebbeck TR. Increased transcriptional activity of the CYP3A4*1B promoter variant. Environ Mol Mutagen. 2003;42(4):299‐305. [DOI] [PubMed] [Google Scholar]
- 7. Gao Y, Zhang LR, Fu Q. CYP3A4*1G polymorphism is associated with lipid‐lowering efficacy of atorvastatin but not of simvastatin. Eur J Clin Pharmacol. 2008;64(9):877‐882. [DOI] [PubMed] [Google Scholar]
- 8. Hesselink DA, van Schaik RHN, van der Heiden IP, et al. Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR‐1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin Pharmacol Ther. 2003;74(3):245‐254. [DOI] [PubMed] [Google Scholar]
- 9. Garcia‐Martin E, Martínez C, Pizarro RM, et al. CYP3A4 variant alleles in white individuals with low CYP3A4 enzyme activity. Clin Pharmacol Ther. 2002;71(3):196‐204. [DOI] [PubMed] [Google Scholar]
- 10. Wandel C, Witte JS, Hall JM, Stein CM, Wood AJ, Wilkinson GR. CYP3A activity in African American and European American men: population differences and functional effect of the CYP3A4*1B5'‐promoter region polymorphism. Clin Pharmacol Ther. 2000;68(1):82‐91. [DOI] [PubMed] [Google Scholar]
- 11. Dong ZL, Li H, Chen Q‐X, et al. Effect of CYP3A4*1G on the fentanyl consumption for intravenous patient‐controlled analgesia after total abdominal hysterectomy in Chinese Han population. J Clin Pharm Ther. 2012;37(2):153‐156. [DOI] [PubMed] [Google Scholar]
- 12. Miura M, Satoh S, Kagaya H, et al. Impact of the CYP3A4*1G polymorphism and its combination with CYP3A5 genotypes on tacrolimus pharmacokinetics in renal transplant patients. Pharmacogenomics. 2011;12(7):977‐984. [DOI] [PubMed] [Google Scholar]
- 13. Yuan R, Zhang X, Deng Q, Wu Y, Xiang G. Impact of CYP3A4*1G polymorphism on metabolism of fentanyl in Chinese patients undergoing lower abdominal surgery. Clin Chim Acta. 2011;412(9–10):755‐760. [DOI] [PubMed] [Google Scholar]
- 14. Zhang W, Chang Y‐Z, Kan Q‐C, et al. CYP3A4*1G genetic polymorphism influences CYP3A activity and response to fentanyl in Chinese gynecologic patients. Eur J Clin Pharmacol. 2010;66(1):61‐66. [DOI] [PubMed] [Google Scholar]
- 15. Zhang W, Yuan J‐J, Kan Q‐C, et al. Influence of CYP3A5*3 polymorphism and interaction between CYP3A5*3 and CYP3A4*1G polymorphisms on post‐operative fentanyl analgesia in Chinese patients undergoing gynaecological surgery. Eur J Anaesthesiol. 2011;28(4):245‐250. [PubMed] [Google Scholar]
- 16. Elens L, Becker ML, Haufroid V, et al. Novel CYP3A4 intron 6 single nucleotide polymorphism is associated with simvastatin‐mediated cholesterol reduction in the Rotterdam Study. Pharmacogenet Genomics. 2011;21(12):861‐866. [DOI] [PubMed] [Google Scholar]
- 17. Elens L, Bouamar R, Hesselink DA, et al. A new functional CYP3A4 intron 6 polymorphism significantly affects tacrolimus pharmacokinetics in kidney transplant recipients. Clin Chem. 2011;57(11):1574‐1583. [DOI] [PubMed] [Google Scholar]
- 18. Elens L, Bouamar R, Hesselink DA, Haufroid V, van Schaik RHN, van Gelder T. The new CYP3A4 intron 6 C>T polymorphism (CYP3A4*22) is associated with an increased risk of delayed graft function and worse renal function in cyclosporine‐treated kidney transplant patients. Pharmacogenet Genomics. 2012;22(5):373‐380. [DOI] [PubMed] [Google Scholar]
- 19. Elens L, Hesselink DA, van Schaik RHN, van Gelder T. The CYP3A4*22 allele affects the predictive value of a pharmacogenetic algorithm predicting tacrolimus predose concentrations. Br J Clin Pharmacol. 2013;75(6):1545‐1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gomez‐Silva M, Piñeyro‐Garza E, Vargas‐Zapata R, et al. Pharmacogenetics of amfepramone in healthy Mexican subjects reveals potential markers for tailoring pharmacotherapy of obesity: results of a randomised trial. Sci Rep. 2019;9(1):17833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tamashiro EY, Felipe CR, Genvigir FDV, et al. Influence of CYP3A4 and CYP3A5 polymorphisms on tacrolimus and sirolimus exposure in stable kidney transplant recipients. Drug Metab Pers Ther. 2017;32(2):89‐95. [DOI] [PubMed] [Google Scholar]
- 22. Liu MZ, He H, Zhang Y, et al. IL‐3 and CTLA4 gene polymorphisms may influence the tacrolimus dose requirement in Chinese kidney transplant recipients. Acta Pharmacol Sin. 2017;38(3):415‐423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu J, Chen Z, Chen H, et al. Genetic polymorphisms contribute to the individual variations of imatinib mesylate plasma levels and adverse reactions in Chinese GIST patients. Int J Mol Sci. 2017;18(3):603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chau CH, Price DK, Till C, et al. Finasteride concentrations and prostate cancer risk: results from the Prostate Cancer Prevention Trial. PLoS One. 2015;10(5):e0126672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu R, Zhou Z, Chen Y, et al. Associations of CYP3A4, NR1I2, CYP2C19 and P2RY12 polymorphisms with clopidogrel resistance in Chinese patients with ischemic stroke. Acta Pharmacol Sin. 2016;37(7):882‐888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yi X, Lin J, Wang Y, et al. Association of cytochrome P450 genetic variants with clopidogrel resistance and outcomes in acute ischemic stroke. J Atheroscler Thromb. 2016;23(10):1188‐1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feng W, Mei S, Zhu L, et al. Effects of UGT2B7, SCN1A and CYP3A4 on the therapeutic response of sodium valproate treatment in children with generalized seizures. Seizure. 2018;58:96‐100. [DOI] [PubMed] [Google Scholar]
- 28. Shao B, Jiang S, Muyiduli X, et al. Vitamin D pathway gene polymorphisms influenced vitamin D level among pregnant women. Clin Nutr. 2018;37(6):2230–2237. [DOI] [PubMed] [Google Scholar]
- 29. Robien K, Butler LM, Wang R, et al. Genetic and environmental predictors of serum 25‐hydroxyvitamin D concentrations among middle‐aged and elderly Chinese in Singapore. Br J Nutr. 2013;109(3):493‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng C, Ding Y, Yi X, et al. Polymorphisms in CYP450 genes and the therapeutic effect of atorvastatin on ischemic stroke: a retrospective cohort study in Chinese population. Clin Ther. 2018;40(3):469‐477 e2. [DOI] [PubMed] [Google Scholar]
- 31. Hustert E, Haberl M, Burk O, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11(9):773‐779. [DOI] [PubMed] [Google Scholar]
- 32. Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27(4):383‐391. [DOI] [PubMed] [Google Scholar]
- 33. Birdwell KA, Decker B, Barbarino JM, et al. Clinical pharmacogenetics implementation consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin Pharmacol Ther. 2015;98(1):19‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fohner A, Muzquiz LI, Austin MA, et al. Pharmacogenetics in American Indian populations: analysis of CYP2D6, CYP3A4, CYP3A5, and CYP2C9 in the Confederated Salish and Kootenai Tribes. Pharmacogenet Genomics. 2013;23(8):403‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Woodahl EL, Lesko LJ, Hopkins S, Robinson RF, Thummel KE, Burke W. Pharmacogenetic research in partnership with American Indian and Alaska Native communities. Pharmacogenomics. 2014;15(9):1235‐1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boyer BB, Dillard D, Woodahl EL, Whitener R, Thummel KE, Burke W. Ethical issues in developing pharmacogenetic research partnerships with American Indigenous communities. Clin Pharmacol Ther. 2011;89(3):343‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fohner AE, Robinson R, Yracheta J, et al. Variation in genes controlling warfarin disposition and response in American Indian and Alaska Native people: CYP2C9, VKORC1, CYP4F2, CYP4F11. GGCX Pharmacogenet Genomics. 2015;25(7):343‐353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Khan BA, Robinson R, Fohner AE, et al. Cytochrome P450 genetic variation associated with tamoxifen biotransformation in American Indian and Alaska Native People. Clin Transl Sci. 2018;11(3):312‐321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shirasaka Y, Chaudhry AS, McDonald M, et al. Interindividual variability of CYP2C19‐catalyzed drug metabolism due to differences in gene diplotypes and cytochrome P450 oxidoreductase content. Pharmacogenomics J. 2016;16(4):375‐387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tanner JA, Prasad B, Claw KG, et al. Predictors of variation in CYP2A6 mRNA, protein, and enzyme activity in a human liver bank: influence of genetic and nongenetic factors. J Pharmacol Exp Ther. 2017;360(1):129‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prasad B, Unadkat JD. Optimized approaches for quantification of drug transporters in tissues and cells by MRM proteomics. AAPS J. 2014;16(4):634‐648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vrana M, Whittington D, Nautiyal V, Prasad B. Database of optimized proteomic quantitative methods for human drug disposition‐related proteins for applications in physiologically based pharmacokinetic modeling. CPT Pharmacometrics Syst Pharmacol. 2017;6(4):267‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dalton R, Lee S‐B, Claw KG, et al. Interrogation of CYP2D6 structural variant alleles improves the correlation between CYP2D6 genotype and CYP2D6‐mediated metabolic activity. Clin Transl Sci. 2020;13(1):147‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gordon AS, Fulton RS, Qin X, Mardis ER, Nickerson DA, Scherer S. PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet Genomics. 2016;26(4):161‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fohner AE, Wang Z, Yracheta J, et al. Genetics, diet, and season are associated with serum 25‐Hydroxycholecalciferol concentration in a Yup'ik study population from Southwestern Alaska. J Nutr. 2016;146(2):318‐325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Z, Senn T, Kalhorn T, et al. Simultaneous measurement of plasma vitamin D(3) metabolites, including 4beta,25‐dihydroxyvitamin D(3), using liquid chromatography‐tandem mass spectrometry. Anal Biochem. 2011;418(1):126‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McPeek MS, Wu X, Ober C. Best linear unbiased allele‐frequency estimation in complex pedigrees. Biometrics. 2004;60(2):359‐367. [DOI] [PubMed] [Google Scholar]
- 48. Sinues B, Vicente J, Fanlo A, et al. CYP3A5*3 and CYP3A4*1B allele distribution and genotype combinations: differences between Spaniards and Central Americans. Ther Drug Monit. 2007;29(4):412‐416. [DOI] [PubMed] [Google Scholar]
- 49. Harvard Biomedical Interdisciplinary Occupations and Medical Exposure (BIOME) . Genotype‐Tissue Expression (GTEx) portal. 2019.
- 50. Wang Z, Lin YS, Dickmann LJ et al. Enhancement of hepatic 4‐hydroxylation of 25‐hydroxyvitamin D3 through CYP3A4 induction in vitro and in vivo: implications for drug‐induced osteomalacia. J Bone Miner Res. 2013;28(5):1101‐1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Z, Lin YS, Zheng XE, et al. An inducible cytochrome P450 3A4‐dependent vitamin D catabolic pathway. Mol Pharmacol. 2012;81(4):498‐509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fohner AE, Volk KG, Woodahl EL. Democratizing precision medicine through community engagement. Clin Pharmacol Ther. 2019;106(3):488‐490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1
Table S2


