Abstract
Fluorescent proteins based on light, oxygen, and voltage (LOV) sensing photoreceptors are among the few reporter gene technologies available for studying living systems in oxygen-free environments that render reporters based on the green fluorescent protein nonfluorescent. LOV reporters develop fluorescence by binding flavin mononucleotide (FMN), which they endogenously obtain from cells. As FMN is essential to cell physiology as well as for determining fluorescence in LOV proteins, it is important to be able to study and characterize flavin binding in LOV reporters. To this end, we report a method for reversibly separating FMN from two commonly used LOV reporters to prepare stable and soluble apoproteins. Using fluorescence titration, we measured the equilibrium dissociation constant for binding with all three cellular flavins: FMN, flavin adenine dinucleotide, and riboflavin. Finally, we exploit the riboflavin affinity of apo LOV reporters, identified in this work, to develop a fluorescence turn-on biosensor for vitamin B2.
1 |. INTRODUCTION
Fluorescence imaging using genetic reporters is one of the best established techniques for monitoring dynamic biological processes in live cells.1–3 However, flagship reporters based on the green fluorescent protein (GFP) depend on environmental oxygen to emit light,4 which renders them unusable in gut microbes, archaea, anaerobic communities, marine bacteria, and other biological systems that thrive in oxygen-free environments.5–9 Unlike GFP, chemogenetic reporters function by pairing proteins with synthetic dyes or biogenic fluorophores such as bilins or flavin mononucleotide (FMN), thereby retaining the ability to fluoresce even in oxygen-free conditions.10–15 Compared to bilins and synthetic dyes, FMN is an essential metabolite found in all living systems,16 which makes it possible to use FMN based fluorescent proteins to label cells without requiring external agents to be delivered across the largely impenetrable bacterial and fungal cell walls. FMN-based reporters are derived from light, oxygen, and voltage sensing (LOV) domains that are found in certain photoactive proteins, which use FMN to initiate cell signaling by absorbing light and forming a covalent bond with a cysteine residue located in the FMN binding cavity. Mutating the cysteine to alanine blocks this photochemistry and results in a noncovalent complex that exhibits fluorescence with excitation and emission peaks centered at 450 and 495 nm respectively.17,18 Previously, we have successfully used saturation mutagenesis of the FMN binding pocket to improve quantum yield of a bacterially sourced LOV reporter.19 We have also discovered brighter LOV reporters from algae by implementing a genome mining-based approach,20 which was recently extended to identify a new and highly thermostable LOV reporter from thermophilic bacteria.21 Taken together with additional benefits such as small size (~12 kDa), acid tolerance,22 and metal-responsive fluorescence,23–25 LOV reporters are making it possible to extend fluorescence imaging to several anaerobes, including gut bacteria, oral flora, parasitic protists, and pathogenic fungi.26–32
As the fluorescence properties of LOV reporters are determined by protein-bound FMN, it would be useful to develop a method for separating FMN from the protein and reconstituting the apo protein with FMN (or other flavins) in order to characterize the equilibrium dissociation constant that is, Kd, specificity, reaction kinetics, binding energetics, and related thermodynamic properties. Such a platform is important for engineering and evolving new fluorescent LOV reporters as well as deciphering mechanisms that lead to improvements in fluorescence properties. However, flavoproteins are generally known to tightly bind flavins, with dissociation constants in the sub-micromolar to nanomolar range, which makes them challenging to deflavinate without affecting stability or solubility of the flavin-free apo protein.33,34 As a result, deflavination techniques need to be carefully tailored for each unique flavoprotein in order to achieve maximum flavin removal, while producing reconstitutable forms of the apo-protein.34 Although several methods have been developed for resolving flavins33 from flavodoxins, photolyases, flavin-based redox sensing proteins, oxidases, reductases, and photoreceptors, to our knowledge, these methods have not been tested in existing fluorescent LOV reporters. In this work, we develop a method to reversibly dissociate FMN from two widely used LOV reporters, iLOV17 (from A. thaliana) and EcFbFP18 (from B. subtilis) and use the distinctive fluorescence properties of flavin bound LOV reporters to measure the Kd for FMN binding. We also demonstrate that LOV reporters are capable of binding riboflavin and flavin adenine dinucleotide (FAD), which represent two other forms of flavins commonly found in biological contexts. Finally, given the demand for easy-to-use riboflavin testing methods in pharmaceutical and nutritional sectors, we make use of deflavinated LOV reporters to develop a simple turn-on fluorescent biosensor for riboflavin and apply it to quantify riboflavin content in complex mixtures, including a commercial multivitamin dietary supplement.
2 |. MATERIALS AND METHODS
2.1 |. Cloning of LOV reporter genes
LOV reporter genes were synthesized by GenScript (Piscataway, NJ) or Integrated DNA Technologies (Coralville, IA), based on the originally published sequences of EcFbFP18 and iLOV.33 The genes were cloned in the pQE80L expression vector using BamHI and HindIII restriction enzymes, which appended a His6 tag at the N terminus of each LOV protein. PCR amplification, restriction digestion, and ligation were accomplished using standard protocols. Briefly, PCR was carried out in 50 μl reaction volume using 1–10 ng of template DNA and 0.5 μM primers, 0.2 mM dNTPs, and 2.5 units Taq DNA polymerase. The PCR cycle consisted of an initial denaturation at 94°C for 2 min followed by 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s. A final extension step at 72°C for 5–10 min was employed to complete synthesis of full-length templates. In some cases, the PCR products were digested with 10 units of DpnI at 37°C for 1 hour in order to remove the methylated template DNA. Amplicons were digested with 10 units each of BamHI and HindIII restriction endonucleases at 37°C for 1 hour and subsequently ligated into pQE80L expression vector digested with BamHI and HindIII using similar reaction conditions. Ligation reactions were performed using 400 units T4 DNA ligase in a 20 μl reaction volume at room temperature for 1 hour. All plasmid constructs were propagated by transformation in E. coli DH5α cells using heat shock at 42°C or electroporation at 1.8 kV using a GenePulser electroporator (Biorad). Cells were plated on LB-agar supplemented with 100 μg/ml ampicillin for selection. Plasmids were isolated from E. coli DH5α transformants and used to transform E. coli MG1655 cells for protein expression. All plasmid constructs were verified by sequencing (Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign).
2.2 |. Expression and purification of apo and holo LOV reporters
Single colonies of E. coli MG1655 transformants expressing the LOV reporter constructs were inoculated in 5 ml Lennox broth supplemented with ampicillin at 100 μg/ml and grown for 16 hours at 37°C with vigorous (300 r.p.m.) orbital shaking. Cells from the overnight cultures were diluted in 0.5 L medium in a 2 L shake flask and grown under similar conditions as before. Protein expression was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM when the culture reached an optical density of 0.4–0.6 at 600 nm (typically, 2 hours after inoculation). Protein expression was continued for another 4–6 hours at 37°C before harvesting cells by centrifugation at 5000 × g for 15 min at 4°C. Pellets were stored at − 80°C until use. For protein purification, frozen pellets were thawed at room temperature and resuspended in 10–15 ml lysis buffer (20 mM Tris hydrochloride, 200 mM sodium chloride, 10 mM imidazole, pH 8.0, 1 mg/ml lysozyme) and incubated for 30 minutes at room temperature, followed by ultrasonication (five cycles of 10 1-second pulses of 17–20 W each). Cell debris was removed by centrifuging the lysate at 10,000 × g for 20 min at 4°C and the supernatant was incubated with 2–4 ml of nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) on a rocker for 1 hour at 4°C. The Ni-NTA resin and supernatant were loaded onto a gravity flow chromatographic column (Fischer Scientific) and washed with 40–50 ml of wash buffer (20 mM Tris hydrochloride, 200 mM sodium chloride, 40 mM imidazole, pH 8.0) to remove nonspecifically bound proteins. Finally, the nickel-bound protein was eluted with 20 ml elution buffer (20 mM Tris hydrochloride, 200 mM sodium chloride, 500 mM imidazole, pH 8.0) and further purified using anion exchange chromatography. For anion exchange, proteins were loaded onto an anion exchange column (HiTrap, GE Life Sciences) by flowing with the Ni-NTA elution buffer followed by step elution using a high salt buffer (20 mM Tris hydrochloride, 1 M sodium chloride, 500 mM imidazole, pH 8.0). The resulting protein was exchanged into phosphate buffered saline (PBS) by dialyzing (10 kDa MWCO Slide-A-Lyzer cassette) against ~150 volumes of PBS for 2–3 hours following which a fresh volume of PBS was added and dialysis continued overnight. In some cases, we were able to bypass the anion exchange step, proceeding straight from Ni-NTA chromatography to dialysis without noticeable loss of protein quality or yield.
For preparing apo protein, the Ni-NTA loaded column was first washed with 15 ml of denaturing buffer (20 mM Tris hydrochloride, 20 mM sodium chloride, pH 8.0, 6 M guanidine hydrochloride) at 4°C for 2 hours. Following the first wash, the nickel-bound protein was incubated overnight with fresh denaturing buffer (15 ml) at 4°C under mild agitation. At the end of overnight incubation, the bound protein was washed again (1–2 times) using fresh denaturing buffer with 1 hour incubation at each wash step. At this stage, the protein turned visibly nonfluorescent, indicating removal of bound flavin. The resulting apo protein was rinsed with 20 ml imidazole-free wash buffer, to wash out the denaturant, and further incubated in the same buffer for an hour at 4°C before eluting with 20 ml elution buffer. Deflavinated apo protein was dialyzed as before to remove imidazole and remaining traces of denaturant. Protein fractions were run on denaturing PAGE gels and quantified using the Bradford assay. Purified holo and apo proteins were stored at 4°C and typically used within 1–2 days.
2.3 |. Analytical gel filtration
We determined oligomeric states of holo and apo LOV reporters using gel filtration chromatography. To this end, a Superdex 200 (GE Healthcare) size exclusion chromatography column was calibrated with globular proteins standards — bovine thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa), and vitamin B12 (1.35 kDa). Purified LOV proteins were loaded in the column, washed with 50 ml PBS, and elution volumes (Velution) corresponding to peaks in the 280 nm absorption chromatogram were recorded. The void volume (Vvoid) and interstitial pore volume (Vpore) were calculated based on the elution volumes of thyroglobulin and vitamin B12 respectively. The distribution coefficient (K) of each protein between the mobile phase and stationary phase was calculated as: . Molar mass was estimated by calibrating the logarithm of molecular weight of protein standards against K. Oligomeric state was then determined by dividing the estimated molar mass by the known molecular weight of LOV monomers.
2.4 |. Fluorescence and circular dichroism spectroscopy
Fluorescence intensity measurements were performed using 150 μl quartz cuvettes (Starna Cells) in a Cary Varian Eclipse fluorometer. Excitation and emission slit widths were set at 5 nm and the photomultiplier tube gain was set to medium (600 V). For emission scans, excitation was typically performed at 450 nm, and emission spectra were scanned between 470 nm and 600 nm. Excitation spectra were obtained by monitoring emission at 540 nm and scanning excitation wavelengths between 300 nm and 520 nm. For circular dichroism spectroscopy, 150 μl of iLOV (40 μM) or EcFbFP (20 μM) were loaded in a 1 cm path length quartz cuvette (Starna Cells). IFar-UV CD spectra were recorded using a Jasco 720 spectrometer between with a resolution of 1 nm and a scan rate of 50 nm/min, averaged over 10 individual spectral scans, and expressed in molar ellipticity. Typically, the HT voltage was maintained at <600 V to minimize noise. Prediction of secondary structure from far-UV CD spectra was made using BeStSel.35
2.5 |. Analysis of flavin binding by LOV reporters
For fluorescence titration experiments, apo proteins were diluted to a concentration of 10–20 μM in PBS and stock solutions of FMN, FAD, and riboflavin were prepared at a final concentration of 200 μM (also in PBS). Titration was carried out by adding flavin in 1 μl steps to 200 μl apo protein in quartz microcuvettes. After each addition, the solution was gently mixed using a pipette and incubated for 3 min in the dark to ensure complete equilibration. Fluorescence measurements were performed as described before. Flavin binding was assumed to proceed by simple bimolecular interaction as follows:
A dynamic mass balance model for the above reaction can be constructed as follows:
| (1) |
| (2) |
| (3) |
Here, k1 and k2 represent on and off rates for complex formation and n is a Hill-like parameter that we initially included to account for possible cooperative effects. As fits derived with n as a free parameter essentially resulted in Hill coefficients ~1, we assumed noncooperative binding for our final fits. The equilibrium dissociation constant can be calculated as Kd = k2/k1. Flavin concentrations were varied between 1–20 μM and Equations (1)–(3) were solved using the ode23s in Matlab (version R2020a) to calculate equilibrium concentrations and determine the fraction of total, that is, holo + apo LOV protein, that converts into the fluorescent holo form in the presence of flavin. We fitted these results to experimental data from the fluorescence titration experiments and estimated Kd using nonlinear least squares regression, implemented using lsqcurvefit (Matlab R2020a). We treated the starting concentration of the protein as an unknown, similar to prior studies on apo flavodoxins.36,37 Concentrations of the apo protein derived in this way were found to be within 50–70% of the actual starting concentration, measured independently using the Bradford assay, indicating that some fraction of the apo protein could not be reconstituted into a fluorescent form.
2.6 |. Fluorescent riboflavin biosensor
To estimate riboflavin concentration in complex mixtures, calibration curves were first constructed by titrating known concentrations of riboflavin in apo LOV and measuring fluorescence using a Cary Varian Eclipse fluorometer. Excitation and emission wavelengths were set to 450 and 495 nm, slit widths were set at 5 nm and the photomultiplier tube gain was kept at medium (600 V). We fixed the concentration of apo proteins at 10 μM, which resulted in linear calibration curves for riboflavin titration from ~0.1–1 μM (concentration range commonly found in vitamin pills, flavin-fortified food and beverages) while avoiding aggregation that we observed at higher concentrations (Figure S3). 100X minimal essential medium (MEM) vitamin solution, a mixture of eight water soluble vitamins, was purchased from Thermo Fisher and used directly in fluorometric assays after dilution. Multivitamin supplements were purchased from a commercial source (Nature Made®), ground to a fine powder (~1 g per pill), mixed in PBS, mildly sonicated for 30–40 min, and clarified using centrifugation at 5000 × g for 15 min, before using the supernatant in the riboflavin assay. In each case, fluorescence recordings were acquired as described above and compared with the standard calibration curve to determine riboflavin content, which was finally validated by comparing with the manufacturer specified concentrations.
2.7 |. Data analysis
Fluorescence titration measurements were conducted using N ≥ 4 independent replicates from 1 (iLOV + FMN, Ec + FAD titrations) or 2 distinct batches of purified proteins (iLOV + FAD, riboflavin; Ec + FMN, riboflavin titrations). Quality of model fits was judged by inspecting residuals (Figure S3). All Kd values are reported as mean ± standard error. Pairwise comparisons of Kd values were performed using the 2-tailed Student’s t-test with significance level set at 0.05. A 95% confidence interval for the measured riboflavin content in multivitamin tablets was computed using the Student’s t-distribution with 2 (for iLOV) or 3° of freedom (for EcFbFP).
3 |. RESULTS AND DISCUSSION
3.1 |. Preparation of flavin-free LOV reporters
In our earlier work,22 we observed that most LOV reporters lost fluorescence following thermal treatment at 70°C for 1 hour, but fluorescence recovered rapidly upon cooling. Based on this observation, we speculated that it should be possible to develop a method for reversibly separating the flavin fluorophore from LOV reporters, while maintaining the ability of the apo protein to recover fluorescence upon supplying flavin. We first attempted to separate bound flavin from the plant-derived LOV reporter (iLOV) using several deflavination techniques that are well-established in flavin enzymology,34 including protein precipitation with trichloroacetic acid, ammonium sulfate, treatment with halide salts (up to 2 M KBr), and chaotropic agents (up to 8 M urea). However, these methods either failed to remove flavin or produced apo proteins that aggregated and could not be converted into the fluorescent holo protein by supplying FMN. Attachment of flavoproteins to a chromatographic support has previously been shown to improve chances of generating stable preparations of apo proteins, presumably by preventing the apo proteins from aggregating in solution.34,38 To implement this technique, we introduced a hexa-histidine motif at the N terminus of iLOV, which enabled binding to a chromatographic nickel column. Using columnimmobilized iLOV, we were able to successfully remove flavin after overnight treatment with a strong chaotropic denaturant, guanidine hydrochloride. Following extensive on-column washing, imidazole based elution, and dialysis (see Table S1 for yields and percent recovery), we obtained purified apo preparations (Figure S1) that were nonfluorescent (Figure 1(A)) and did not form visible precipitates even after prolonged storage (> 7 days) at 4°C. Using analytical size exclusion chromatography (Figures 1(B) and S2), we verified that the purified apo preparation retains the parent holo-protein’s monomeric state (although higher concentrations tended to aggregate, Figure S3) but lacked the characteristic excitation and emission spectra of LOV reporters (Figure 1(A)), indicating removal of protein-bound flavin. Far-UV circular dichroism spectroscopy revealed a shallower trough at ~220 nm for the apo protein compared to holo iLOV, suggesting a partial loss of α helical content in the apo protein (Figure 1(C) &Table S2). Importantly, addition of FMN to apo iLOV immediately (within seconds) restored the characteristic fluorescence spectrum of LOV reporters (Figure 1(D)), indicating successful reconstitution. We were also able to extend this approach to separate flavin from the bacterial LOV reporter, EcFbFP (Figure 1(E)), which retained its dimeric state (Figure 1(F)) but, as before, displayed a partial loss of α helical secondary structure in the far-UV CD spectrum (Figure 1(G) & Table S2). As in the case of iLOV, apo EcFbFP readily reverts into the fluorescent holo form upon supplying FMN (Figure 1(H)).
FIGURE 1.
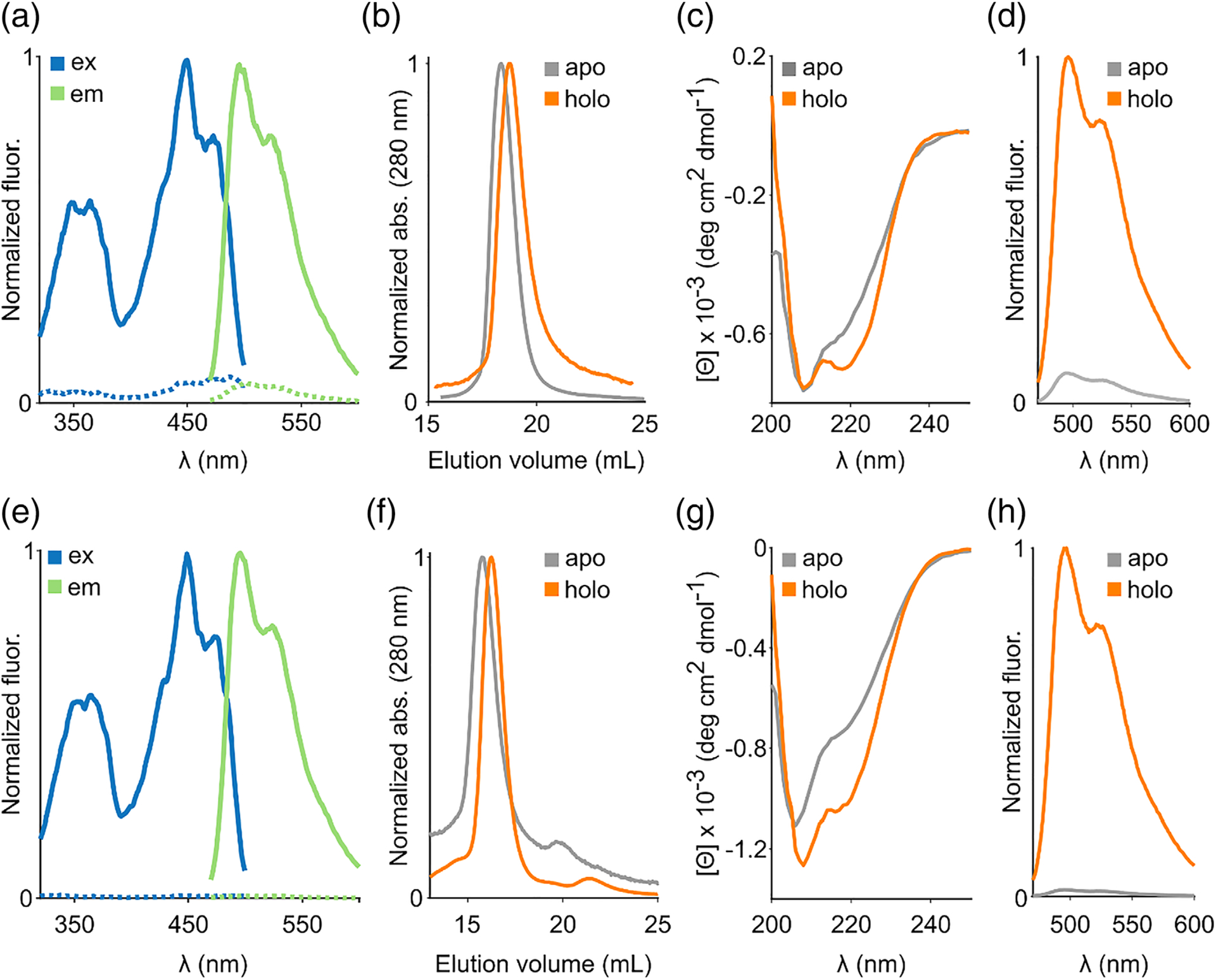
Flavin removal in fluorescent LOV reporters. A, Excitation and emission spectra of holo (solid lines) and deflavinated (dashed lines) iLOV. B, Elution profiles of apo and holo iLOV from gel filtration chromatography. C, Far UV CD spectra of apo and holo iLOV. D, Recovery of fluorescence emission upon adding FMN to apo iLOV. E, Excitation/emission spectra, F, elution profiles, G, far UV CD spectra and (H) fluorescence recovery in the bacterial fluorescent protein EcFbFP
3.2 |. Flavin binding in LOV reporters
Free FMN demonstrates negligible fluorescence at 495 nm, which corresponds to peak emission in flavin-bound fluorescent LOV proteins. Thus, the distinct fluorescence properties of LOV-bound FMN compared to free FMN, make it possible to use fluorescence spectroscopy to monitor FMN binding in LOV proteins. Similar methods, based on changes in fluorescence intensity between free and protein-bound FMN,39 have been widely used to study flavin binding in enzymes, including flavodoxin,37,40 L-amino acid oxidase,41 and cytochromes,42,43 albeit in these cases the protein environment quenches FMN fluorescence, unlike for LOV reporters. To measure the equilibrium dissociation constant (Kd), we titrated aliquots of FMN into solutions containing apo iLOV. After each titration, we allowed the solution to equilibrate in the dark before recording emission at 495 nm. We fitted the fluorescence measurements to a simple bimolecular binding model, assuming noncooperative binding behavior (see Figure S4(A)–(C) for fit residuals) and derived a Kd of 0.23 ± 0.02 μM between iLOV and FMN (Figure 2(A)). Using a similar approach, we determined a Kd of 0.18 ± 0.03 μM for EcFbFP (Figure 2(A)), suggesting that both monomeric (iLOV) and dimeric (EcFbFP) LOV reporters bind FMN with similar affinities (p-value = 0.2, N = 4 for iLOV, 5 for Ec) (Figure 2(D)). Notably, the calculated Kd values reveal that these LOV reporters bind their fluorophore with a stronger affinity than most biliverdin based fluorescent reporters (Kd ~ 490 nM–4.84 μM44), but similar to recently evolved fluorogenic reporters such as YFAST, which binds its synthetic fluorophore with a Kd ~ 0.13 μM.45
FIGURE 2.
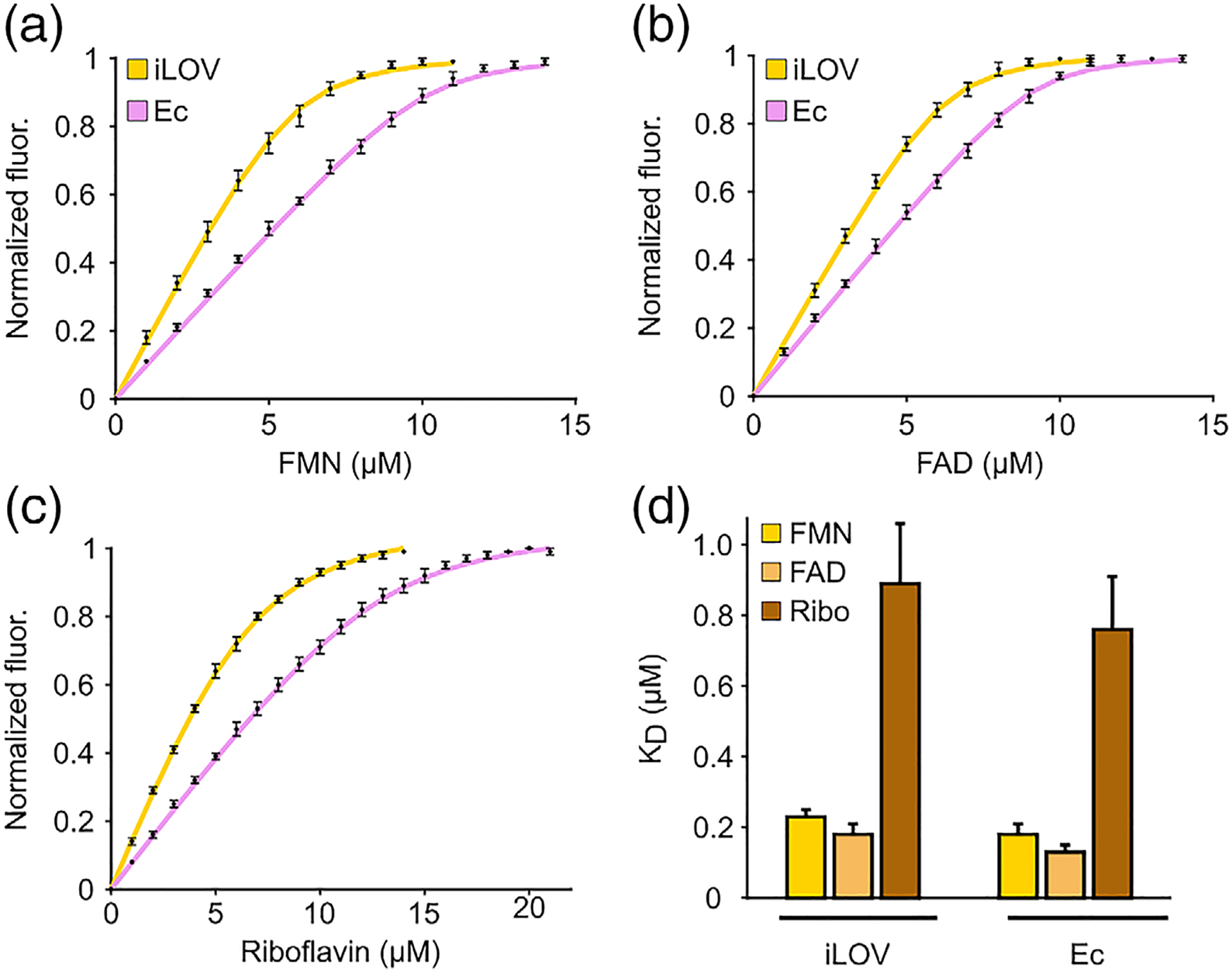
Flavin-binding in LOV reporters. Fluorescence titration of, A, FMN, B, FAD and, C, riboflavin in purified preparations of apo iLOV and apo EcFbFP. D, Equilibrium dissociation constant (KD) for binding of LOV reporters to various flavins. All measurements represent means of at least four independent replicates from 1–2 purification batches and error bars correspond to the standard error of mean
Some flavoproteins are known to exhibit nonspecific binding to physiological flavins other than their specific prosthetic group.46–49 To this end, we tested iLOV for its ability to bind riboflavin, the precursor molecule to FMN, as well as FAD, which is a bulkier, adenylated form of FMN. Interestingly, we found that iLOV can be reconstituted into a fluorescent form using either FAD or riboflavin and the resulting spectrum is indistinguishable from the holo iLOV protein. As free FAD is nearly nonfluorescent due to intramolecular stacking between the adenine and isoalloxazine rings,50 the recovery of FAD fluorescence upon LOV binding suggests that the adenine and isoalloxazine rings are unstacked as FAD is inserted in the binding pocket of iLOV. Using fluorescence titration we determined a Kd of 0.18 ± 0.03 μM for FAD (Figure 2(B)) and 0.89 ± 0.17 μM for riboflavin (Figure 2(C)), suggesting that iLOV can bind FAD with a similar affinity as FMN, but its interaction with riboflavin is almost four-fold weaker (p = .01, N = 4 for FMN, 5 for riboflavin) (Figure 2(D)). We observed a similar trend in the binding properties of bacterial EcFbFP, which was found to bind FAD with Kd = 0.13 ± 0.02 μM and riboflavin with Kd = 0.76 ± 0.15 μM (Figure 2(B)–(D)). The lower affinity for riboflavin has been previously observed in other flavoproteins (e.g., dodecins)46 and may be caused by the lack of a phosphate group that anchors FMN (and presumably, FAD) in the flavin binding pocket. Consistent with this notion, increasing ionic strength was found to weaken the affinity for FMN (Figure S5), which may point to electrostatic screening51 of interactions involving the phosphate group of FMN and positively charged amino acids (e.g., arginine) in the flavin binding pocket.
3.3 |. Fluorescence “turn-on” biosensors for riboflavin
Riboflavin, (vitamin B2) is an essential nutrient as it serves as the precursor for synthesizing FMN and FAD within cells, which then serve as prosthetic groups for several flavoproteins involved in metabolism. Consequently, riboflavin deficiency in humans is associated with conditions such as fatigue, cheilosis, growth retardation, and night blindness.52 Animals lack riboflavin biosynthesis pathways and thus typically obtain riboflavin through external means such as dairy products, fortified cereals, bread, energy drinks, and multivitamin pills. For these reasons, the development of easy-to-use assays to estimate riboflavin content in food, infant formulations, dairy products, and bodily fluids (e.g., urine) is of considerable importance to nutritional and pharmaceutical sectors. Existing approaches for detecting riboflavin make use of liquid chromatography,37,53 electrochemical techniques such as voltammetry,54,55 immunoassays,56 surface plasmon resonance,57 and culture based tests involving riboflavin-dependent microbes grown using the test sample as the only source of riboflavin.58 These methods, while widely used, suffer from limitations such as slow response, interference from other components in complex mixtures, and limited scalability to a medium or high throughput format. Fluorescent sensors can overcome many of these challenges, which has prompted the development of riboflavin sensors based on Förster Resonance Energy Transfer (FRET) between riboflavin and doped carbon dots59 or graphene60 as well as quenching of fluorescence using PEG-dispersed graphene.61 However, to our knowledge, a fluorescence “turn-on” biosensor for riboflavin has not been reported. As apo LOV reporters are capable of binding riboflavin with low micromolar affinity (Figure 2(D)) and exhibit both visually detectable (Figure 3(A)) and quantifiable gain of fluorescence, we reasoned that purified apo iLOV or EcFbFP should enable simple, rapid, and quantitative determination of riboflavin content in complex mixtures without a need for extensive sample processing steps such as liquid chromatography. To test this idea, we first verified that LOV fluorescence is specific to flavins and cannot be reconstituted using other biological cofactors such as NAD, deoxynucleotides, and amino acids (Figure S6). Next, we used apo iLOV to quantify riboflavin content in a mixture consisting of eight water-soluble vitamins (riboflavin, choline chloride, calcium pantothenate, folic acid, nicotinamide, inositol, thiamine hydrochloride, and pyridoxal hydrochloride) that is commonly used as a nutrient supplement in animal cell culture. We treated various dilutions of the 8-vitamin mixture with a fixed concentration of apo iLOV and estimated the riboflavin content by measuring fluorescence and comparing against a calibration of known concentration standards. We found that apo iLOV could be used to estimate riboflavin content in the vitamin mixture over a concentration span of 0.04–0.4 μg/ml (Figure 3(B)) with reasonable accuracy and precision. Building on these results, we proceeded to use apo iLOV to assay riboflavin content in a more complex milieu involving a commercial multivitamin supplement comprising all water soluble B-vitamins, vitamins C, E, trace metals, several organic, and inorganic salts. We extracted the pill contents using mild ultrasonication and dissolved it in PBS to achieve a final riboflavin concentration of 41.8 μg/ml (calculated based on manufacture specification). Following treatment with apo iLOV and comparison with calibration standards, we obtained a riboflavin concentration of 47.9 μg/ml (95% CI: 38.7–57.1 ug/ml). Similar results were obtained using apo EcFbFP as a sensor, which determined the riboflavin concentration as 35.5 μg/ml (95% CI: 32.1–38.9 ug/ml) for a multivitamin supplement preparation comprising 32.2 μg/ml riboflavin. Taken together, these results establish the general utility of apo LOV reporters for simple, rapid, and quantitative riboflavin testing in real-world samples.
FIGURE 3.
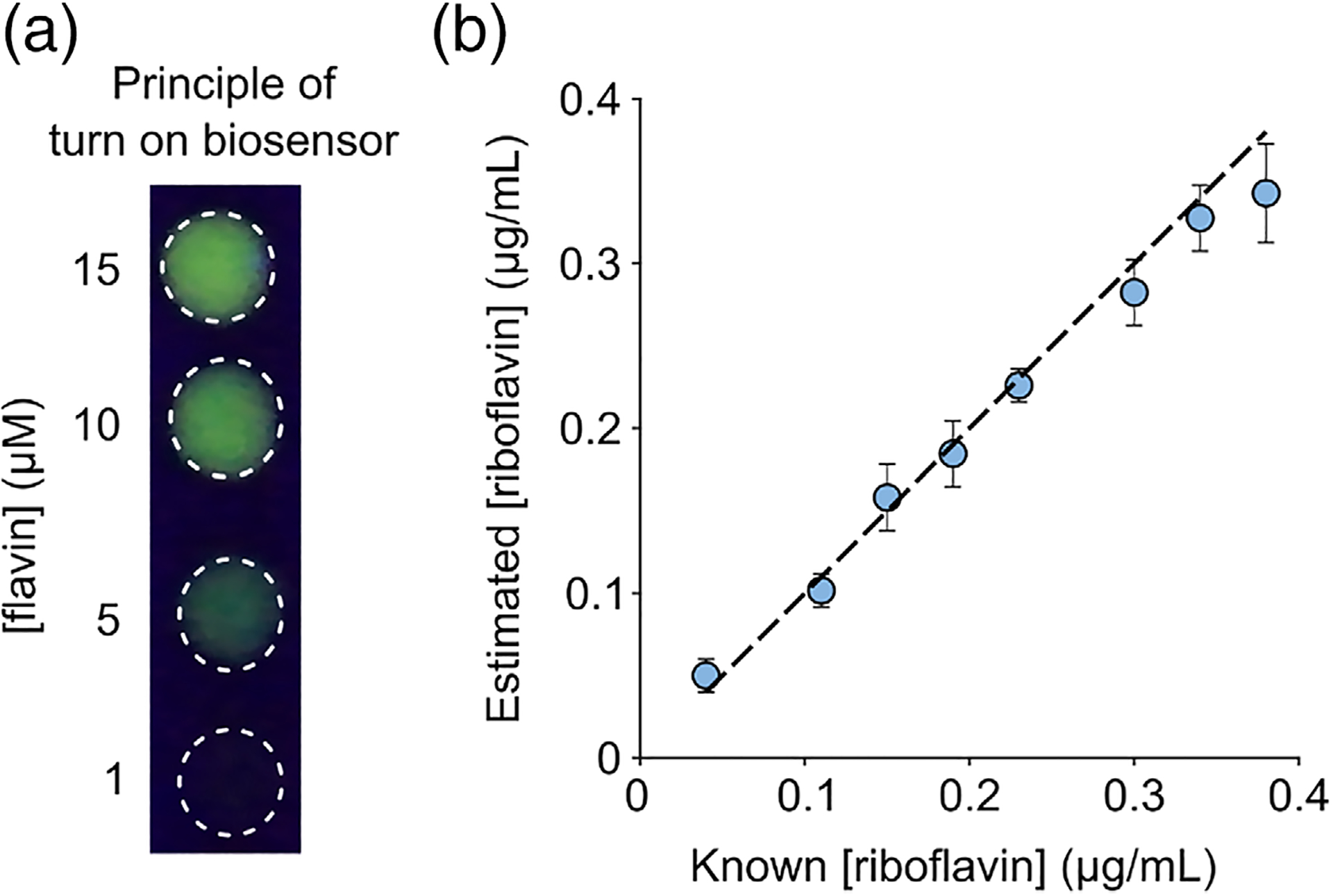
Turn-on fluorescent biosensor for riboflavin detection. A, Apo iLOV exhibits a visually detectable gain of fluorescence upon titrating flavin. Images were acquired by spotting ~ 10 μM apo iLOV on a clear UV-transparent plastic surface, exciting with a 320 nm light source in trans-illumination mode and recording with a digital camera. B, Agreement between manufacture-specified and estimated riboflavin concentrations (determined using apo iLOV assay) present in various dilutions of an 8-vitamin cell culture supplement
4 |. CONCLUSIONS
In this work, we develop an approach for the reversible deflavination of fluorescent LOV proteins and apply it to establish the first (to our knowledge) measurements of equilibrium binding between engineered LOV reporters and the three physiological forms of flavins: riboflavin, FMN, and FAD. Although we have developed and validated this method in the context of two commonly used LOV reporters, it should be possible, in principle, to adapt this technique for binding studies in other fluorescent LOV proteins, including brighter proteins such as CreiLOV,20 photostable variants such as phiLOV,62 and recently discovered thermostable LOV proteins such as CagFbFP.21 Our work has important implications for engineering and applying LOV proteins as self-contained (i.e., no external agent needed) reporter genes in anaerobic microbes. For example, the relatively strong flavin-binding affinities that we measured in this work suggest that over-expression of LOV reporters may cause intracellular flavin (typically present in cells at low micromolar levels63,64) to drop, thereby metabolically burdening cells. To this end, we have observed varying levels of growth retardation22 associated with the expression of different LOV reporters in E. coli. Future studies may reveal whether and to what extent growth retardation correlates with flavin affinity of various LOV reporters and if certain organisms can offset this resource constraint by activating feedback regulated flavin biosynthesis65 in response to LOV expression. From the standpoint of reporter protein engineering, our binding studies suggest that a potential avenue for increasing cellular fluorescence obtained with LOV reporters could be to engineer variants with even tighter Kd (ideally, low nM), which could increase their flavin occupancy by allowing LOV reporters to effectively compete with the native cellular flavoproteome that comprises several tight binding flavoproteins. Notably, equilibrium dissociation constants in the sub-nM range have proved beneficial for developing reporters based on efficient occupancy with fluorescent cofactors, such as the bilirubin binding fluorescent protein, UnaG.13 As before, potential effects on cell physiology resulting from increased flavin affinity will need to be carefully characterized and mitigated.
In addition to measuring the equilibrium dissociation constants, we also demonstrated that apo iLOV (and apo EcFbFP) could be used to optically assay for riboflavin (i.e., vitamin B2) based on gain of fluorescence. Apo preparations of some flavoproteins such as the riboflavin-binding protein (RfBP) from egg white and apo flavodoxin have been previously used to quantify vitamin B2 content in cell lysates, dairy products, and beverages – however, these methods either relied on liquid chromatography47 or quenching of fluorescence for riboflavin detection.66–68 To our knowledge, the methodology introduced here represents the only known fluorescence turn on biosensor for riboflavin. One potential limitation of this technique is that it cannot distinguish riboflavin from FMN or FAD, which are enzymatically synthesized from riboflavin within cells. While this can be problematic for monitoring specific flavins intracellularly or in lysed cells, we do not believe this will become a major limitation for using the sensors in extracellular or noncellular contexts such as food materials and vitamin supplements where FMN and FAD are typically present in negligible amounts. To this end, we envision that purified preparations of apo LOV proteins could be used to evaluate riboflavin content in common dietary and pharmaceutical sources of vitamin B2 with minimal sample processing, while avoiding time and equipment-intensive methods such as chromatography, mass spectrometry, and microbial culture based assays. Another potential application of our sensor would be for monitoring bioproduction of vitamin B2 in commercial flavin-overproducing microbes such as Lactobacillus plantarum, which secrete riboflavin at concentrations (> 1 μg/ml) that should be readily detectable with our sensor. As fluorimetry is readily scalable to medium or high throughput screening applications, one important benefit of these sensors could be in optimizing natural or bioengineered strains for riboflavin overproduction. Although further studies will be required to rigorously characterize apo LOV-based sensors to determine limit of detection, limit of quantification, reusability, response kinetics, and long-term stability as well as validate in a broader set of real-world samples, our results provide a proof-of-concept for a simple, rapid, and scalable fluorescence turn on assay for riboflavin.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Charles M. Schroeder, Dr. Joshua Walker, current members of the Mukherjee lab, and all our collaborators for helpful discussions. Related work in the Mukherjee lab has been supported by the Institute for Collaborative Biotechnologies, National Institutes of Health Maximizing Investigators’ Research Award (1R35GM133530-01), and a Department of Defense Peer Reviewed Medical Research Program Discovery Award (PR191154). N.T.A. is supported by the UCSB Chancellor’s Fellowship. We extend our deepest gratitude to the anonymous reviewers whose comments and suggestions greatly improved and strengthened the quality of this work and the manuscript.
Funding information
Institute of Collaborative Biotechnologies; National Institute of General Medical Sciences, Grant/Award Number: 1R35GM133530-01; U.S. Department of Defense, Grant/Award Number: PR191154
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Terai T, Nagano T. Fluorescent probes for bioimaging applications. Curr Opin Chem Biol. 2008;12(5):515–521. [DOI] [PubMed] [Google Scholar]
- 2.Dean KM, Palmer AE. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat Chem Biol. 2014;10(7):512–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorn K. Genetically encoded fluorescent tags. Mol Biol Cell. 2017;28 (7):848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci. 1994;91(26):12501–12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozbakir HF, Anderson NT, Fan K-C, Mukherjee A. Beyond the green fluorescent protein: biomolecular reporters for anaerobic and deep-tissue imaging. Bioconjug Chem. 2020;31:293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guglielmetti S, Santala V, Mangayil R, Ciranna A, Karp MT. O2-requiring molecular reporters of gene expression for anaerobic microorganisms. Biosens Bioelectron. 2019;123:1–6. [DOI] [PubMed] [Google Scholar]
- 7.Scott KP, Mercer DK, Glover LA, Flint HJ. The green fluorescent protein as a visible marker for lactic acid bacteria in complex ecosystems. FEMS Microbiol Ecol. 1998;26(3):219–230. [Google Scholar]
- 8.Coralli C, Cemazar M, Kanthou C, Tozer GM, Dachs GU. Limitations of the reporter green fluorescent protein under simulated tumor conditions. Cancer Res. 2001;61(12):4784–4790. [PubMed] [Google Scholar]
- 9.Zhang C, Xing X-H, Lou K. Rapid detection of a gfp-marked Enterobacter aerogenes under anaerobic conditions by aerobic fluorescence recovery. FEMS Microbiol Lett. 2005;249(2):211–218. [DOI] [PubMed] [Google Scholar]
- 10.Buckley AM, Petersen J, Roe AJ, Douce GR, Christie JM. LOV-based reporters for fluorescence imaging. Curr Opin Chem Biol. 2015;27: 39–45. [DOI] [PubMed] [Google Scholar]
- 11.Chia HE, Zuo T, Koropatkin NM, Marsh ENG, Biteen JS. Imaging living obligate anaerobic bacteria with Bilin-binding fluorescent proteins. Curr Res Microbial Sci. 2020;1:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jullien L, Gautier A. Fluorogen-based reporters for fluorescence imaging: a review. Meth Appl Fluores. 2015;3(4):042007. [DOI] [PubMed] [Google Scholar]
- 13.Kumagai A, Ando R, Miyatake H, et al. A bilirubin-inducible fluorescent protein from eel muscle. Cell. 2013;153(7):1602–1611. [DOI] [PubMed] [Google Scholar]
- 14.Mukherjee A, Schroeder CM. Flavin-based fluorescent proteins: emerging paradigms in biological imaging. Curr Opin Biotechnol. 2015; 31:16–23. [DOI] [PubMed] [Google Scholar]
- 15.Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu Rev Biochem. 2015;84:519–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abbas CA, Sibirny AA. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust bio-technological producers. Microbiol Mol Biol Rev. 2011;75(2):321–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman S, Faulkner C, Kaiserli E, et al. The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc Natl Acad Sci. 2008;105(50):20038–20043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drepper T, Eggert T, Circolone F, et al. Reporter proteins for in vivo fluorescence without oxygen. Nat Biotechnol. 2007;25(4):443–445. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee A, Weyant KB, Walker J, Schroeder CM. Directed evolution of bright mutants of an oxygen-independent flavin-binding fluorescent protein from pseudomonas putida. J Biol Eng. 2012;6(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee A, Weyant KB, Agrawal U, Walker J, Cann IK, Schroeder CM. Engineering and characterization of new LOV-based fluorescent proteins from Chlamydomonas reinhardtii and Vaucheria frigida. ACS Syn Biol. 2015;4(4):371–377. [DOI] [PubMed] [Google Scholar]
- 21.Nazarenko VV, Remeeva A, Yudenko A, et al. A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: a framework for ultra-high resolution structural studies. Photochem Photobiol Sci. 2019;18(7):1793–1805. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee A, Walker J, Weyant KB, Schroeder CM. Characterization of flavin-based fluorescent proteins: an emerging class of fluorescent reporters. PLoS One. 2013;8(5):e64753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou W, Le K, Zastrow ML. Live cell copper-induced fluorescence quenching of the flavin-binding fluorescent protein CreiLOV. Chembiochem. 2020;21:1356–1363. [DOI] [PubMed] [Google Scholar]
- 24.Ravikumar Y, Nadarajan SP, Lee C-S, Jung S, Bae D-H, Yun H. FMN-based fluorescent proteins as heavy metal sensors against mercury ions. J Microbiol Biotechnol. 2016;26(3):530–539. [DOI] [PubMed] [Google Scholar]
- 25.Ravikumar Y, Nadarajan SP, Lee C-S, Rhee J-K, Yun H. A new generation fluorescent based metal sensor-iLOV protein. J Microbiol Biotechnol. 2015;25(4):503–510. [DOI] [PubMed] [Google Scholar]
- 26.Buckley AM, Jukes C, Candlish D, et al. Lighting up Clostridium difficile: reporting gene expression using fluorescent Lov domains. Sci Rep. 2016;6:23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobo LA, Smith CJ, Rocha ER. Flavin mononucleotide (FMN)-based fluorescent protein (FbFP) as reporter for gene expression in the anaerobe Bacteroides fragilis. FEMS Microbiol Lett. 2011;317(1):67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang SE, Brooks AE, Cann B, Simoes-Barbosa A. The fluorescent protein iLOV outperforms eGFP as a reporter gene in the microaerophilic protozoan Trichomonas vaginalis. Mol Biochem Parasitol. 2017;216:1–4. [DOI] [PubMed] [Google Scholar]
- 29.Choi CH, DeGuzman JV, Lamont RJ, Yilmaz Ö. Genetic transformation of an obligate anaerobe, P. gingivalis for FMN-green fluorescent protein expression in studying host-microbe interaction. PLoS One. 2011;6(4):e18499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo S-O, Lu T, Jin Y-S, Blaschek HP. Development of an oxygen-independent flavin mononucleotide-based fluorescent reporter system in Clostridium beijerinckii and its potential applications. J Biotechnol. 2018;265:119–126. [DOI] [PubMed] [Google Scholar]
- 31.Teng L, Wang K, Xu J, Xu C. Flavin mononucleotide (FMN)-based fluorescent protein (FbFP) as reporter for promoter screening in clostridium cellulolyticum. J Microbiol Methods. 2015;119:37–43. [DOI] [PubMed] [Google Scholar]
- 32.Tielker D, Eichhof I, Jaeger K-E, Ernst J. Flavin mononucleotide-based fluorescent protein as an oxygen-independent reporter in Candida albicans and Saccharomyces cerevisiae. Eukaryot Cell. 2009;8(6): 913–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapman SK, Reid GA. Flavoprotein protocols. Vol 131. Springer Science & Business Media; 1999. [Google Scholar]
- 34.Hefti MH, Vervoort J, van Berkel WJH. Deflavination and reconstitution of flavoproteins. Eur J Biochem. 2003;270(21):4227–4242. [DOI] [PubMed] [Google Scholar]
- 35.Micsonai A, Wien F, Kernya L, et al. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc Natl Acad Sci. 2015;112(24):E3095–E3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bollen YJM, Westphal AH, Lindhoud S, van Berkel WJH, van Mierlo CPM. Distant residues mediate picomolar binding affinity of a protein cofactor. Nat Commun. 2012;3(1):1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lostao A, El Harrous M, Daoudi F, Romero A, Parody-Morreale A, Sancho J. Dissecting the energetics of the Apoflavodoxin-FMN complex. J Biol Chem. 2000;275(13):9518–9526. [DOI] [PubMed] [Google Scholar]
- 38.Hefti MH, Milder FJ, Boeren S, Vervoort J, van Berkel WJH. A his-tag based immobilization method for the preparation and reconstitution of apoflavoproteins. Biochim Biophys Acta. January 20, 2003/ 2003; 1619(2):139–143. [DOI] [PubMed] [Google Scholar]
- 39.VISSER AJWG, GHISLA S, MASSEY V, MÜLLER F, VEEGER C. Fluorescence properties of reduced Flavins and Flavoproteins. Eur J Biochem. 1979;101(1):13–21. [DOI] [PubMed] [Google Scholar]
- 40.Mayhew SG. Studies on flavin binding in flavodoxins. Biochim Biophys Acta. 1971;235(2):289–302. [DOI] [PubMed] [Google Scholar]
- 41.CASALIN P, POLLEGIONI L, CURTI B, SIMONETTA MP. A study on apoenzyme from Rhodotorula gracilis D-amino acid oxidase. Eur J Biochem. 1991;197(2):513–517. [DOI] [PubMed] [Google Scholar]
- 42.Shumyantseva VV, Bulko TV, Petushkova NA, Samenkova NF, Kuznetsova GP, Archakov AI. Fluorescent assay for riboflavin binding to cytochrome P450 2B4. J Inorg Biochem. 2004;98(2):365–370. [DOI] [PubMed] [Google Scholar]
- 43.Strittmatter P. The nature of the Flavin binding in microsomal cytochrome b5 Reductase. J Biol Chem. 1961;236(8):2329–2335. [Google Scholar]
- 44.Yu D, Baird MA, Allen JR, et al. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat Meth. 2015;12(8): 763–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plamont M-A, Billon-Denis E, Maurin S, et al. Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo. Proc Natl Acad Sci U S A. 2016;113(3):497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bourdeaux F, Hammer CA, Vogt S, et al. Flavin storage and sequestration by Mycobacterium tuberculosis Dodecin. ACS Infect Dis. 2018;4(7):1082–1092. [DOI] [PubMed] [Google Scholar]
- 47.Dorn M, Jurk M, Wartenberg A, Hahn A, Schmieder P. LOV takes a pick: thermodynamic and structural aspects of the flavin-LOV-interaction of the blue-light sensitive photoreceptor YtvA from Bacillus subtilis. PloS One. 2013;8(11):e81268–e81268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fitzpatrick PF, Ghisla S, Massey V. 8-Azidoflavins as photoaffinity labels for flavoproteins. J Biol Chem. 1985;260(14):8483–8491. [PubMed] [Google Scholar]
- 49.Mathes T, Vogl C, Stolz J, Hegemann P. Vivo generation of Flavoproteins with modified cofactors. J Mol Biol. 2009;385(5):1511–1518. [DOI] [PubMed] [Google Scholar]
- 50.Stanley RJ, MacFarlane. Ultrafast excited state dynamics of oxidized Flavins: direct observations of quenching by purines. J Phys Chem A. 2000;104(30):6899–6906. [Google Scholar]
- 51.Papaneophytou CP, Grigoroudis AI, McInnes C, Kontopidis G. Quantification of the effects of ionic strength, viscosity, and hydrophobicity on protein–ligand binding affinity. ACS Med Chem Lett. 2014;5(8): 931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thakur K, Tomar SK, Singh AK, Mandal S, Arora S. Riboflavin and health: a review of recent human research. Crit Rev Food Sci Nutr. 2017;57(17):3650–3660. [DOI] [PubMed] [Google Scholar]
- 53.Zempleni J. Determination of riboflavin and flavocoenzymes in human blood plasma by high-performance liquid chromatography. Annals Nutri Metab. 1995;39(4):224–226. [DOI] [PubMed] [Google Scholar]
- 54.Hou W, Wang E. Liquid chromatography with series dual-electrode electrochemical detection for riboflavin. Analyst. 1990;115(2): 139–141. [Google Scholar]
- 55.Ly SY, Yoo HS, Ahn JY. Pico molar assay of riboflavin in human urine using voltammetry. Food Chem. 2011;127(1):270–274. [Google Scholar]
- 56.Ravi G, Venkatesh YP. Immunoassays for riboflavin and flavin mononucleotide using antibodies specific to d-ribitol and d-ribitol-5-phosphate. J Immunol Methods. 2017;445:59–66. [DOI] [PubMed] [Google Scholar]
- 57.Caelen I, Kalman A, Wahlström L. Biosensor-based determination of riboflavin in Milk samples. Analytic Chem. 2004;76(1):137–143. [DOI] [PubMed] [Google Scholar]
- 58.Salvetti S, Celandroni F, Ghelardi E, Baggiani A, Senesi S. Rapid determination of vitamin B2 secretion by bacteria growing on solid media. J Appl Microbiol. 2003;95(6):1255–1260. [DOI] [PubMed] [Google Scholar]
- 59.Lin L, Wang Y, Xiao Y, Chen X. Ratiometric fluorescence detection of riboflavin based on fluorescence resonance energy transfer from nitrogen and phosphorus co-doped carbon dots to riboflavin. Analytic Bioanalytic Chem. May 1, 2019. 2019;411(13):2803–2808. [DOI] [PubMed] [Google Scholar]
- 60.Kundu A, Nandi S, Layek RK, Nandi AK. Fluorescence resonance energy transfer from Sulfonated Graphene to riboflavin: a simple way to detect vitamin B2. ACS Appl MaterInterf. 2013;5(15):7392–7399. [DOI] [PubMed] [Google Scholar]
- 61.Díez-Pascual AM, García-García D, San Andrés MP, Vera S. Determination of riboflavin based on fluorescence quenching by graphene dispersions in polyethylene glycol. RSC Adv. 2016;6(24):19686–19699. [Google Scholar]
- 62.Christie JM, Hitomi K, Arvai AS, et al. Structural tuning of the fluorescent protein iLOV for improved photostability. J Biol Chem. 2012;287(26):22295–22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pedrolli D, Langer S, Hobl B, Schwarz J, Hashimoto M, Mack M. The ribB FMN riboswitch from Escherichia coli operates at the transcriptional and translational level and regulates riboflavin biosynthesis. FEBS J. 2015;282(16):3230–3242. [DOI] [PubMed] [Google Scholar]
- 64.WILSON AC, PARDEE AB. Regulation of Flavin synthesis by Escherichia coli. Microbiology. 1962;28(2):283–303. [DOI] [PubMed] [Google Scholar]
- 65.Krauss U, Svensson V, Wirtz A, Knieps-Grünhagen E, Jaeger K-E. Cofactor trapping, a new method to produce flavin mononucleotide. Appl Environ Microbiol. 2011;77(3):1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duyvis MG, Hilhorst R, Laane C, Evans DJ, Schmedding DJ. Role of riboflavin in beer flavor instability: determination of levels of riboflavin and its origin in beer by fluorometric apoprotein titration. J Agric Food Chem. 2002;50(6):1548–1552. [DOI] [PubMed] [Google Scholar]
- 67.Wassink JH, Mayhew SG. Fluorescence titration with apoflavodoxin: a sensitive assay for riboflavin 5′-phosphate and flavin adenine dinucleotide in mixtures. Anal Biochem. 1975;68(2):609–616. [DOI] [PubMed] [Google Scholar]
- 68.Zandomeneghi M, Carbonaro L, Zandomeneghi G. Biochemical fluorometric method for the determination of riboflavin in milk. J Agric Food Chem. 2007;55(15):5990–5994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


