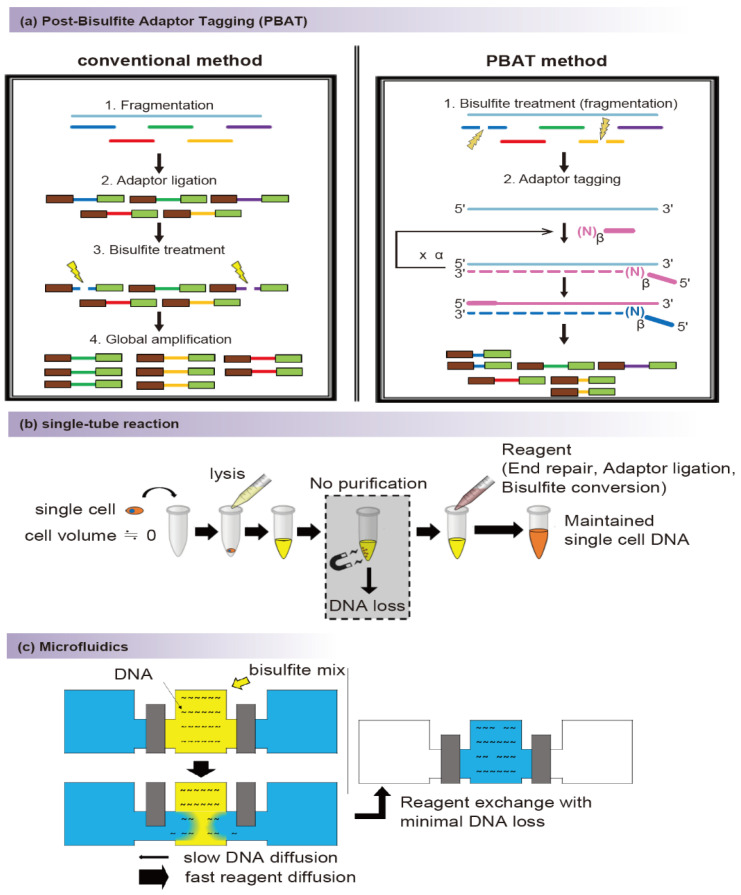
Figure 1.
Key strategies to minimize DNA loss in single-cell DNA methylation profiling methods. Because a single cell contains a small amount of DNA, several methods are used to minimize loss. (a) Overview of the post-bisulfite adaptor tagging (PBAT) method to prevent loss due to degradation during the bisulfite conversion process. Unlike the conventional method (left panel), loss of shortened DNA fragments is prevented in PBAT (right panel). Each single-cell methylation profiling method using the PBAT strategy differs in the number of amplifications of the bisulfite conversion product (α) and the number of random sequences in the primer (β). (b) Overview of single-tube reaction. Common to several methods, reagents are continuously added, without purification, to the tube or well containing cell lysate or nuclei. In this way, DNA loss during the purification process can be prevented. (c) Use of microfluidics demonstrated in the microfluidic diffusion (MID)-based reduced representation bisulfite sequencing (RRBS) process. DNA loss can be minimized during purification using microfluidics. This figure is based on the figure of a previous article (Ma, S., de la Fuente Revenga, M., Sun, Z. et al. 2018) [41] and adapted with permission from 2018 Springer Nature.