Genetic analysis in zebrafish reveals conserved and novel aspects of nuclear receptor Fxr function and bile salt signaling.
Abstract
Bile salt synthesis, secretion into the intestinal lumen, and resorption in the ileum occur in all vertebrate classes. In mammals, bile salt composition is determined by host and microbial enzymes, affecting signaling through the bile salt–binding transcription factor farnesoid X receptor (Fxr). However, these processes in other vertebrate classes remain poorly understood. We show that key components of hepatic bile salt synthesis and ileal transport pathways are conserved and under control of Fxr in zebrafish. Zebrafish bile salts consist primarily of a C27 bile alcohol and a C24 bile acid that undergo multiple microbial modifications including bile acid deconjugation that augments Fxr activity. Using single-cell RNA sequencing, we provide a cellular atlas of the zebrafish intestinal epithelium and uncover roles for Fxr in transcriptional and differentiation programs in ileal and other cell types. These results establish zebrafish as a nonmammalian vertebrate model for studying bile salt metabolism and Fxr signaling.
INTRODUCTION
Bile salts are the end product of cholesterol catabolism in the liver of all vertebrates (1). Upon lipid ingestion, bile salts are released into the duodenum as emulsifiers to solubilize lipids and are then reabsorbed by the ileum into the portal vein to return to the liver, a process known as enterohepatic circulation. Bile salts also act as signaling molecules that exert diverse effects by activating nuclear or membrane-bound receptors (2). This includes the nuclear receptor farnesoid X receptor (FXR/NR1H4), an evolutionarily conserved transcription factor (TF) that uses bile salts as endogenous ligands (3). Upon binding with bile salts, FXR regulates a large number of target genes involved in bile salt, lipid, and glucose metabolism (4). FXR activity can be modulated by the chemical structure of bile salts, which differ considerably across vertebrate species (5). For example, fish and amphibians contain predominantly 27-carbon (C27) bile alcohols, whereas mammals mainly have 24-carbon (C24) bile acids (1). Even within the same species, there can be substantial diversity in bile salt structures. One key contributor to this diversity is the gut microbiota, which can modify the side chain(s) or stereostructure of the conjugated primary bile salts synthesized by the liver (6). This leads to the production of various unconjugated or secondary bile salts in the intestine with different activities toward FXR, therefore altering FXR-mediated signaling pathways. Although bile salts and FXR are present in diverse vertebrate species (7), our knowledge about bile salt–FXR signaling has been almost entirely limited to humans and rodents. It remains unclear when this signaling axis arose and whether its functions changed over the course of vertebrate evolution. Furthermore, while mice have been an effective model at revealing FXR functions relevant to humans, there are substantial differences unique to mice, including the presence of endogenous bile salt Fxr antagonist, and Fxr-mediated metabolic activities distinct from humans (8, 9). Therefore, additional vertebrate models are needed to provide complementary perspectives into the mechanistic relationships between microbiota, bile salts, and FXR signaling across vertebrates and to potentially reveal previously unidentified functions of FXR.
The zebrafish (Danio rerio) has emerged as a powerful model for studying bile salt–related liver diseases due to their conserved mechanisms of liver and intestinal development and bile secretion, facile genetic and transgenic manipulations, and ease of monitoring host-microbiota interactions and other physiological processes in vivo (10–13). The genome of zebrafish has orthologs of many mammalian genes known to be involved in bile salt homeostasis, including bile salt transporters, bile salt synthesis enzymes, and FXR (7, 11, 14, 15). Furthermore, genes involved in bile salt absorption are expressed in a conserved ileal region of the zebrafish intestine (16). However, the requirement for those zebrafish genes in enterohepatic circulation and bile salt signaling remains largely untested. In addition, although primary bile salt composition in zebrafish has been assessed (17, 18), microbial metabolism of zebrafish bile salts has not been explored.
Here, we establish zebrafish as a nonmammalian vertebrate model to study the bile salt–Fxr signaling axis. We establish the evolutionary conservation of key components of this axis between zebrafish and mammals and assess the contribution of zebrafish gut microbes to the modulation of the bile salt–Fxr signaling. Furthermore, we uncover the requirements of zebrafish Fxr in gene expression and differentiation in multiple intestinal epithelial cell (IEC) types using single-cell transcriptomics.
RESULTS
Key components of the Fxr signaling pathway are conserved in zebrafish
We used CRISPR-Cas9 to generate fxr mutant zebrafish harboring a 10-bp (base pair) deletion in the Fxr DNA binding domain (designated rdu81, homozygous mutant hereafter referred to as fxr−/−) (Fig. 1A and fig. S1, A and B). These fxr−/− zebrafish were viable, exhibited normal development, and were externally indistinguishable from their fxr+/+ siblings (fig. S1C). To define the impacts of fxr mutation on gene expression, we evaluated predicted Fxr targets. Fatty acid binding protein 6 (fabp6), the gene encoding the ileal bile acid binding protein, is a known Fxr target in mammals and is highly expressed in the zebrafish ileum (16, 19). Using a new reporter line Tg(-1.7fabp6:GFP) that expresses green fluorescent protein (GFP) in the ileal epithelium under control of the 1.7-kb fabp6 promoter, we observed notable attenuation of GFP fluorescence in fxr−/− zebrafish larvae compared to fxr+/+ wild-type (wt) controls (Fig. 1B). This suggested that expression of fabp6 in the ileum is dependent on Fxr in larval zebrafish as it is in mammals (19), and that this reporter line can be used to monitor Fxr activity in vivo. Examination of a larger panel of predicted Fxr target genes involved in bile salt homeostasis revealed similar transcriptional changes in fxr−/− zebrafish as seen in Fxr knockout mice (Fig. 1C) (11, 19–21). This includes reduced expression of fabp6, the fibroblast growth factor fgf19, and the bile salt export pump abcb11b, along with induction of cyp7a1 that encodes the rate-limiting enzyme cholesterol 7α-hydroxylase in hepatic bile salt synthesis. Notably, the apical sodium–dependent bile acid transporter slc10a2, which is indirectly repressed by FXR in mice and humans, appeared to be positively regulated by Fxr in zebrafish, as slc10a2 expression was reduced in fxr−/− zebrafish (Fig. 1B). This could lead to increased fecal excretion of bile salt in the fxr−/− zebrafish and may explain why the total bile salt levels remained similar in larvae of both genotypes (fig. S1D). Nonetheless, these data reveal that Fxr is critical for the coordinated expression of bile salt metabolism genes in zebrafish larvae as in mammals.
Fig. 1. Genetic analyses reveal conserved key components of the bile salt–Fxr signaling axis in zebrafish.
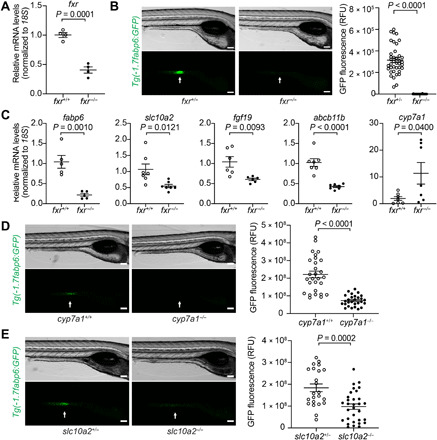
(A) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) comparing fxr expression in whole 7-day post-fertilization (dpf) fxr wild-type (wt) (fxr+/+) or fxr homozygous mutant (fxr−/−) zebrafish larvae. (B) Imaging and quantification of GFP fluorescence of the ileal region of 7-dpf fxr+/+ and fxr−/− Tg(-1.7fabp6:GFP) larvae. The ileal region is indicated by arrows. RFU, relative fluorescence unit. (C) qRT-PCR comparing expression of genes related to bile salt metabolism in dissected digestive tracts (including intestine, liver, pancreas, and gall bladder) of 7-dpf fxr+/+ or fxr−/− larvae (for fabp6, slc10a2, and fgf19) or dissected liver of gender- and size-matched adult male fxr+/+ or fxr−/− zebrafish (for abcb11b and cyp7a1). The results are represented as relative expression levels normalized to 18S (mean ± SEM). (D and E) Imaging and quantification of GFP fluorescence of the ileal region of 7-dpf Tg(-1.7fabp6:GFP) cyp7a1+/+ and cyp7a1−/− larvae (D) or slc10a2+/− and slc10a2−/− larvae (E). The zebrafish ileal region is indicated by arrows. Scale bars, 100 μm in (B), (D), and (E). Statistical significance was calculated by unpaired t test. Representative data from at least three independent experiments are shown.
In mammals, Fxr-mediated regulation of host physiology can occur in a sex- and developmental stage–specific manner (22–24). When examining the expression of host bile metabolism genes in adult zebrafish, we found that several genes also exhibited sex-specific responsiveness toward Fxr regulation that is distinct from larval zebrafish (fig. S1E). For example, in adult male fxr−/− zebrafish, only slc10a2 and abcb11b, but not fabp6, showed reduced expression upon fxr mutation, whereas in adult female fxr−/− zebrafish, none of these bile salt transporters displayed altered expression. The causes of these developmental stage– and sex-specific phenotypes in fxr mutant zebrafish remain unclear. Transcript levels of fxr are strongly reduced in fxr−/− zebrafish as expected due to nonsense-mediated decay (fig. S1F), and its closest homologs lxr/nr1h3 and fxrb/nr1h5 show no compensatory increases in expression in fxr−/− zebrafish (fig. S1F) (25). Because many Fxr targets can be co-regulated by Fxr and other TFs (26), we speculate that the altered responsiveness of these bile salt transporters to Fxr in adult zebrafish may be due to increased roles for other TFs at those stages.
Bile salt–mediated Fxr activation is conserved in zebrafish as in mammals
Having identified conservation of the signaling pathways downstream of Fxr between zebrafish and mammals, we next sought to test whether the activity of Fxr is regulated by bile salts in zebrafish. To do so, we first defined the level and diversity of zebrafish bile salts by analyzing the biliary bile extracted from pooled adult zebrafish gallbladders using electrospray ionization liquid chromatography–mass spectrometry (ESI-LC-MS). On the basis of the mass ion, the major component (83.4%) of the purified zebrafish bile was determined to be 5α-cyprinol sulfate (5αCS), a C27 bile alcohol species commonly present in fishes (Fig. 2A) (27). This was further validated by examining the 1H, 13C, COSY (correlation spectroscopy), and HSQC NMR (heteronuclear single-quantum coherence nuclear magnetic resonance) spectra of this compound (fig. S2A). We also identified several minor bile salt species, including 8.8% taurocholic acid (TCA), a C24 bile acid commonly found in mammals, 7.8% 5α-cholestane-3α,7α,12α,26-tetrol sulfate, a precursor of 5αCS (1, 27), and a trace amount of the dehydrogenated form of 5αCS (Fig. 2A).
Fig. 2. Zebrafish and carp bile salts consist of both C24 bile alcohols and C27 bile acids.
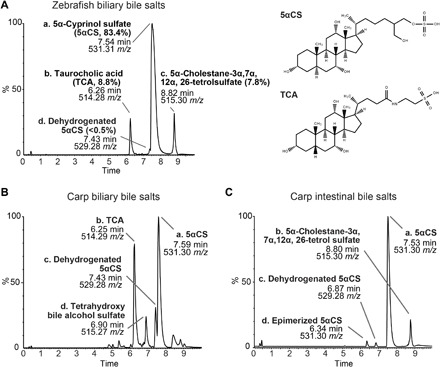
(A) Mass spectrometry chromatograms of the bile salts extracted from adult zebrafish gallbladders. The identity and proportion of major peaks are listed. The height of each peak is shown as the relative percentage that was normalized to the highest peak of the metabolites detected. Structures of 5αCS and TCA are shown. (B and C) MS chromatograms of the bile salts extracted from adult carp gallbladders (B) or adult carp intestinal contents (C). The identities of major peaks are listed accordingly.
The predominant zebrafish bile salt, the C27 bile alcohol 5αCS, differs markedly from the common mammalian bile salts, the C24 bile acids, in both the stereostructure and the number of carbon atoms. Therefore, we asked whether such distinct bile salt composition results in differential regulation of Fxr signaling between fishes and mammals. We thus modulated the zebrafish bile salt levels by disrupting the hepatic synthesis or ileal uptake of bile salts and monitored the impacts on Fxr activity using the Tg(-1.7fabp6:GFP) reporter. To reduce hepatic bile salt synthesis, we generated a new cyp7a1 mutant zebrafish (16 bp, designated rdu84, homozygous mutant hereafter referred to as cyp7a1−/−), which exhibited a significant reduction in the total bile salt levels as compared to its wt counterparts (fig. S2, C to E, and Supplementary Results). Using the reporter assay, we observed an over 50% decrease in GFP fluorescence in cyp7a1 mutant zebrafish as compared to wt, suggesting that Fxr activity was reduced as a result of bile salt deficiency in zebrafish (Fig. 1D). To reduce bile salt uptake in the ileum, we used slc10a2 mutant (slc10a2sa2486/sa2486, designated as slc10a2−/−) zebrafish (fig. S2F) (28), which also showed significantly decreased ileal GFP fluorescence, consistent with compromised Fxr activity due to insufficient bile salt uptake (Fig. 1E). Together, these results suggest that despite the compositional differences in bile salts between zebrafish and mammals, bile salts still activate Fxr and the downstream signaling in the ileal epithelium of zebrafish.
Next, we asked if zebrafish Fxr can respond to chemical agents other than its natural ligand bile salts. We therefore treated the Tg(-1.7fabp6:GFP) reporter zebrafish larvae with chemicals known to regulate FXR activities in mammals and monitored changes in GFP fluorescence. We observed a strong reduction of GFP fluorescence when larval zebrafish were treated with the Fxr antagonist guggulsterone (29) and an increase of fluorescence with the Fxr agonist GW4064 (fig. S2G) (30). This suggests that zebrafish Fxr signaling can be tuned through chemical manipulations. Therefore, while genetic mutation of fxr or other bile metabolism genes in zebrafish is efficient to study Fxr signaling, the zebrafish model can also be used in a more physiologically relevant setting by controlling Fxr activities.
Fish microbiota modulate bile salt diversity in vivo and in vitro
Primary bile salts can be modified by intestinal microbiota into various unconjugated primary and secondary bile salts and then cycled back to the liver through enterohepatic circulation (5). Our findings on zebrafish biliary bile salt diversity demonstrated the presence of a dehydrogenated 5αCS (Fig. 2A). However, it is not clear if this modified 5αCS is an intermediate derived from de novo 5αCS biosynthesis or a recycled bile salt that has been modified by gut microbiota. Furthermore, biliary bile salts do not accurately reflect the full spectrum of microbial modification that occurs in the intestine. We therefore examined bile salt diversity in the intestinal contents of adult zebrafish, aiming to determine whether microbial modifications of bile salts occur. 5αCS and TCA were present in zebrafish intestinal contents (fig. S3); however, the low biomass of the zebrafish luminal contents limited our ability to accurately detect and/or quantify these bile salts and their derivatives. Thus, we turned to a larger cyprinid fish species closely related to zebrafish, the Asian grass carp (Ctenopharyngodon idella), and compared the bile salt diversity between the carp biliary bile and gut contents to determine whether bile salts are modified by carp gut microbiota (Fig. 2, B and C). Carp have a similar biliary bile salt profile to zebrafish, as all major peaks found in zebrafish were also present in carp (Fig. 2, A and B), except that it produces a different tetrahydroxy bile alcohol sulfate (Fig. 2B, peak d), likely a 5β-isomer of the cholestane-3α,7α,12α,26-tetrolsulfate (1, 31). Notably, in the bile salts isolated from carp intestinal contents, we observed a new peak sharing the same mass ratio but a different retention time with 5αCS, indicative of an epimerized 5αCS. This suggests that carp microbiota can oxidize and epimerize an α- to a β-hydroxyl group of the primary bile alcohol 5αCS (Fig. 2C). To our knowledge, this is the first evidence demonstrating the ability of microbes to metabolize bile alcohols in vertebrates.
To test whether similar and/or additional microbial modifications might be present in the zebrafish gut, we developed an in vitro bile salt modification assay using LC-MS (Fig. 3A). Complex microbiota or individual microbes isolated from the zebrafish intestine were first enriched under aerobic or anaerobic conditions and then incubated with the bile salts of interest. This assay system was validated through successful detection of common modifications of primary bile salts upon treating with microbes known to perform these modifications (fig. S4, A and B). We then used this system to test whether zebrafish microbiota modify 5αCS and TCA, the primary bile alcohol and acid in zebrafish. For 5αCS cultured with zebrafish microbiota, two newly emerged peaks, representing the microbial metabolites of 5αCS, were detected in both aerobic and anaerobic conditions. One peak showed a mass ion of 529.3 mass/charge ratio (m/z) with an elution time of 7.4 min, suggesting a keto-5αCS variant (Fig. 3B). The loss of two mass units observed in this product is consistent with bacterial hydroxysteroid dehydrogenase (HSDH) activity found in human gut microbiota (32). The other peak, with a mass ion of 531.3 m/z and an elution time of 6.4 min, corresponded to an epimerized 5αCS, a downstream product of the keto-5αCS variant, therefore further confirming the presence of HSDHs in zebrafish microbiota (Fig. 3B). Both peaks were also present in carp intestinal content (Fig. 2C), suggesting that microbiota-mediated 5αCS dehydrogenation and epimerization are conserved in these cyprinid fishes.
Fig. 3. Microbiota modulate bile salt diversity in zebrafish.
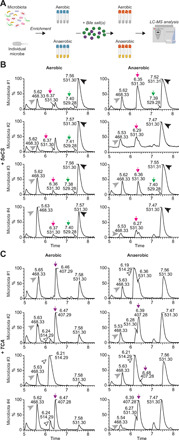
(A) Schematic diagram of the in vitro bile salt metabolism assay. Complex microbiota or individual bacterial strains were enriched in modified tryptic soy broth (TSB) media under both aerobic and anaerobic conditions, and then incubated with 50 μM bile salts of interest, after which the total bile salts were extracted from individual bacterial cultures and subjected to LC-MS to identify modification of the supplemented bile salts. The diagram is created with BioRender.com. (B and C) LC-MS chromatographs of bile salt metabolites extracted from enriched zebrafish microbiota cultures supplemented with 50 μM 5αCS (B) or TCA (C) under aerobic (left) or anaerobic (right) conditions. The arrowheads indicate the supplemented bile salts and internal control: black arrowhead, 5αCS; white arrowhead, TCA; gray arrowhead, internal standard D4-glycocholic acid (D4-GCA). The arrows indicate that bile salt metabolites resulted from microbial modification of the supplemented bile salts: green arrow, dehydrogenated 5αCS; magenta arrow, epimerized 5αCS; purple arrow, CA. The height of each peak is shown as the relative percentage that was normalized to the highest peak of the metabolites detected. Representative chromatographs of zebrafish microbiota from four independent biological replicate cultures are shown.
Cultures incubated with TCA resulted in a new peak corresponding to cholic acid (CA), the deconjugated product of TCA, in several microbiota cultures prepared from pooled adult zebrafish guts, although the extent of deconjugation varied across biological replicate cultures (Fig. 3C). For example, aerobic microbiota #1 exhibited a complete deconjugation of TCA to CA, whereas aerobic microbiota #3 showed no sign of deconjugation. This likely indicates the variable distribution of microbes containing bile salt hydrolase (BSH), the enzyme catalyzing the deconjugation of TCA, among zebrafish. The presence or absence of BSH activity in a given zebrafish microbial community does not always match between aerobic and anaerobic conditions. For instance, zebrafish microbiota #3 deconjugated TCA only under anaerobic conditions, whereas microbiota #1 catalyzed deconjugation only under aerobic conditions. This suggests that the bacteria responsible for deconjugation are likely different among distinct zebrafish microbiota and that more than one deconjugating bacterium is present in zebrafish. No other transformations of 5αCS or CA were detected in cultures. Collectively, our results indicate that microbial modification of bile salts is a conserved feature between zebrafish and mammals.
Having shown that zebrafish microbiota modify both 5αCS and TCA, we sought to determine the bacterial specificity of bile salt modification in zebrafish. We screened a panel of zebrafish gut isolates representing several major bacterial taxa in the zebrafish gut toward 5αCS and TCA (Fig. 4A and fig. S4C). None of the tested strains modified 5αCS. Yet, we identified one Gammaproteobacteria strain, Acinetobacter sp. ZOR0008, capable of deconjugating TCA (Fig. 4A). After overnight incubation with Acinetobacter sp., we observed complete conversion of 25 μM TCA to CA, suggesting robust BSH activity (Fig. 4A). To our knowledge, this the first zebrafish gut bacterium confirmed to have bile salt metabolizing activity.
Fig. 4. A zebrafish gut bacterium modifies primary bile acid producing a metabolite that augments Fxr signaling in zebrafish.
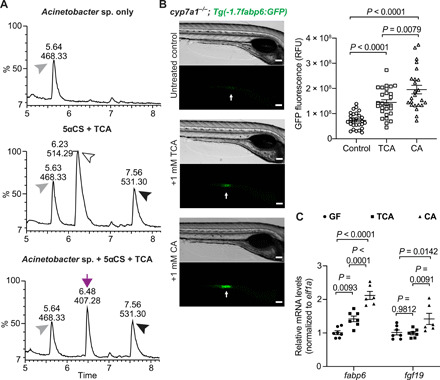
(A) LC-MS chromatographs of medium only control (top), bile salts only control (middle), and Acinetobacter sp. ZOR0008 culture supplemented with bile salts (bottom) under aerobic conditions. The arrowheads indicate the supplemented bile salts and internal control: black arrowhead, 5αCS; white arrowhead, TCA; gray arrowhead, internal standard D4-GCA. The purple arrow indicates that the liberated CA resulted from bacterial deconjugation of TCA. Results from other bacterial strains are shown in fig. S4C. (B) Imaging and quantification of GFP fluorescence of the ileal region of 7-dpf Tg(-1.7fabp6:GFP) cyp7a1−/− larvae that were untreated or treated with either 1 mM TCA or CA for 4 days. The zebrafish ileal region is indicated by arrows. Scale bars, 100 μm. (C) qRT-PCR comparing the expression of Fxr target genes in 7-dpf wt germ-free larvae that were untreated or treated with either 1 mM TCA or CA for 4 days. The results are represented as relative expression levels normalized to the housekeeping gene ef1a (mean ± SEM). Statistical significance was calculated by one-way (B) and two-way (C) analysis of variance (ANOVA) with Tukey’s multiple comparisons test. Representative data from at least three independent experiments are shown.
Microbial modifications of bile salts in zebrafish modulate Fxr activity
In mammals, microbial modification of bile salts can alter the signaling property of bile salts and modulate host physiology. Given that the primary bile acid TCA can be metabolized into CA by gut microbes in zebrafish, we investigated the potential impact of that modification on Fxr signaling in vivo by monitoring Tg(-1.7fabp6:GFP) zebrafish treated with exogenous TCA or CA. To reduce the influences of Fxr activity caused by endogenous bile salts, we performed the reporter assay in the cyp7a1−/− background. Physiological concentrations of TCA or CA were supplemented to larval zebrafish, and GFP fluorescence was monitored after 4 days (27). Zebrafish larvae treated with CA exhibited increased GFP fluorescence as compared to those with TCA, and both showed higher fluorescence than the nontreated controls (Fig. 4B). This suggests that both TCA and CA activate Fxr and that CA is more potent than TCA, consistent with observations in mammals (33). We further validated these findings under a more stringent setting using wt germ-free (GF) zebrafish, which permit competition between endogenous versus exogenous bile salts and eliminate potential influences of microbiota on metabolizing the supplemented bile salts. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) results suggested that CA treatment increased expression of Fxr targets, such as fabp6 and fgf19, as compared to TCA, confirming that CA displays higher potency than TCA in activating Fxr (Fig. 4C). Together, our observations suggest that zebrafish gut microbiota have the potential to regulate Fxr-mediated signaling through modification of primary bile salts.
Fxr regulates diverse cell types identified in zebrafish intestine by single-cell RNA sequencing
Having established that bile salts and gut microbes interactively regulate Fxr activity, we next sought to discern how Fxr, in turn, contributes to intestinal functions. Gross intestinal morphology appeared normal in zebrafish and mice lacking Fxr function (Fig. 1B) (34), but strong attenuation of the fabp6 reporter in fxr mutant zebrafish (Fig. 1B) suggested potential effects of fxr mutation on functional specification of IECs. To test this possibility, we performed single-cell RNA sequencing (scRNA-seq) on 12,543 IECs sorted from 6-day post-fertilization (dpf) fxr+/+ or fxr−/− zebrafish larvae on a TgBAC(cldn15la-GFP) transgenic background that expresses GFP in all IECs (35). After quality control, 4710 cells from fxr+/+ and 5208 cells from fxr−/− samples were used for downstream analyses (fig. S5, A to E). Twenty-seven distinct clusters were generated by unsupervised clustering of these cells using the Seurat R package as described previously (Fig. 5A and fig. S5, A to E) (36). The cell types represented by these clusters were inferred through integrative analysis of published expression data of previously identified gene markers, novel markers of each cluster identified in this study by differential gene expression, and functional predictions from the gene expression data generated in this study (datasets S1 to S3 and Supplementary Results). The resulting annotation revealed a range of IEC types including absorptive enterocytes, goblet cells (including those that resemble mammalian tuft cells and microfold cells), enteroendocrine cells, secretory precursors, ionocytes (including those that resemble mammalian BEST4/OTOP2 cells) (37), and foregut epithelial cells, as well as low levels of several other apparent contaminating cell types (e.g., exocrine pancreas cells, epidermis cells, mesenchymal cells, leukocytes, and red blood cells) (Fig. 5A, table S1, and Supplementary Results). These results, combined with our extended annotation of this scRNA-seq dataset provided in the Supplementary Materials, provide a useful new resource for zebrafish intestinal biology.
Fig. 5. ScRNA-seq reveals extensive cellular diversity in larval zebrafish intestinal epithelium.
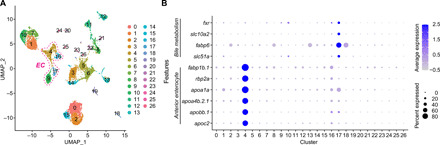
(A) Uniform manifold approximation and projection (UMAP) visualization of cell type clusters using combined fxr+/+ and fxr−/− single-cell transcriptomes (4710 cells from fxr+/+ and 5208 cells from fxr−/−). Clusters were hierarchically ordered in principal components analysis spaces and were numbered accordingly. Detailed annotations of each cluster are shown in table S1 and Supplementary Results. Clusters 4, 16, 17, and 9 are absorptive enterocytes (EC). (B) Dot plots showing relative expression of bile salt metabolism genes and anterior enterocyte markers in fxr+/+ cells throughout all clusters.
We next leveraged our scRNA-seq data to test the requirement for fxr in different IEC types. Supporting the notion that Fxr regulates diverse aspects of intestinal physiology, we found that nearly one-third of all clusters exhibited over 50% change in cell abundance with an average of ~500 genes displaying over 1.5-fold changes in expression in response to Fxr mutation (fig. S6, A and B, and dataset S4). To further evaluate conservation of Fxr-mediated gene expression between zebrafish and mammals, we compared these results to an existing dataset of 489 mouse genes differentially regulated in the ileum or colon in response to intestinal Fxr agonism (38). We identified 583 zebrafish genes that were determined by BioMart to be homologous to those 489 mouse genes and also detected in our zebrafish dataset. Of the 583 zebrafish genes, 213 of them were differentially expressed in response to fxr mutation in at least one cluster (dataset S5). In those instances where 1 of those 213 genes was differentially expressed in a cluster, the directionality of change due to Fxr function was consistent with mouse ileum in 59.5% (72 of 121) of cases and with mouse colon in 49.7% (251 of 499) of cases. Although it remains unknown if these gene expression changes are due to direct or indirect effects of Fxr activity, these results do suggest substantial differences between the gene regulons influenced by Fxr activity in the zebrafish and mouse intestines. This further underscores the importance of using multiple animal models to gain complementary insights into bile salt–Fxr signaling pathways. Although we already showed that loss of fxr function in zebrafish results in reduction of several conserved Fxr target genes (Fig. 1C), this comparative functional genomic analysis identified potential additional targets of Fxr regulation that are conserved between zebrafish and mice such as Pck1 (39), Akr1b7 (40), and Apoa1 (dataset S5) (41). Collectively, our scRNA-seq results unveil extensive cellular diversity in the larval zebrafish intestine and highlight the broad impacts of Fxr on gene expression in diverse cell types.
Fxr regulates functional specialization of ileal epithelial cells
Given the substantial attenuation of the ileal fabp6 reporter in fxr mutant zebrafish (Fig. 1B), we next examined how Fxr affects zebrafish ileal epithelial cells in our scRNA-seq analysis. We discerned cluster 17 as enriched for zebrafish ileal epithelial cells based on the abundant expression of bile transporters fabp6, slc10a2, and slc51a (Fig. 5B). This cluster exhibited a higher level of fxr expression as compared to all other clusters, consistent with the notion that fxr displays spatially patterned expression along the intestine with highest levels in the ileal epithelium (42, 43). We also observed heterogeneity in the expression of these bile transporter genes in cluster 17, raising the possibility that multiple subcell types are present in this cluster (fig. S7A). For clarity, we operationally defined cells located in cluster 17 as “ileal epithelial cells” and the subset that expresses one or more bile transporters as “ileocytes.”
Mutation of fxr markedly affected gene expression in cluster 17 cells (fig. S6B and dataset S4). As expected, many down-regulated genes in cluster 17 fxr mutant cells were related to bile salt metabolism, such as fabp6, slc10a2, and slc51a, consistent with the strong reduction in the fabp6 reporter activity upon Fxr mutation (Figs. 1B and 6A). On the other hand, among the up-regulated cluster 17–enriched markers, lysosome process (dre: 04142) was the most enriched pathway in fxr mutant cells (Fig. 6A and fig. S7B). Lysosome-mediated degradation is a hallmark function of a specific type of vacuolated enterocytes named lysosome-rich enterocytes (LREs) (44). LREs are found in the ileum of fishes and suckling mammals and are known to internalize dietary macronutrients for intracellular digestion (44, 45). Therefore, our data confirm that LREs are a key type of ileal epithelial cell, and further suggest that Fxr normally represses LRE gene expression. We observed increased expression of known LRE markers in fxr mutant cells in cluster 17. This includes multiple classes of digestive enzymes involved in macromolecule degradation and transporters responsible for dietary protein uptake in LREs (Fig. 6A) (44). To confirm these results, we used qRT-PCR to examine expression of several LRE markers including amn, which encodes Amnionless, the major component of the multiligand endocytic machinery in LREs, and ctsbb, which encodes cathepsin B commonly found in lysosomes. Both genes exhibited higher expression in the zebrafish intestine upon fxr mutation, validating our scRNA-seq observations (Fig. 6B). To identify potential transcriptional regulatory pathways involved in the induction of LRE genes upon fxr mutation, we searched for TF binding sites (TFBSs) that are overrepresented within accessible chromatin regions (16) near genes up-regulated in fxr mutant cells in cluster 17. The top 3 enriched TFBS were ZBTB33, Atf2, and TATA-box, raising the possibility that Fxr may interact with TFs that bind at these TFBSs to regulate LRE functions such as lysosomal-mediated degradation (fig. S7C). Collectively, these results establish that Fxr promotes expression of bile absorption genes and represses expression of lysosomal degradation genes in ileal epithelial cells in zebrafish.
Fig. 6. Fxr determines specific cell functions of ileal epithelial cells but is not required for establishment of regional boundaries.
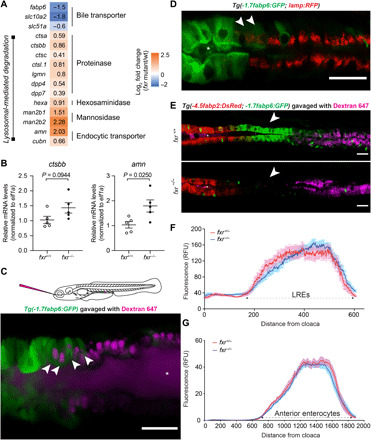
(A) Differential expression of genes related to bile metabolism and lysosomal functions in fxr−/− cells as compared to fxr+/+ cells in cluster 17. (B) qRT-PCR comparing expression of LRE markers in dissected intestines of gender- and size-matched adult fxr+/+ or fxr−/− zebrafish. The results are represented as relative expression levels normalized to the housekeeping gene ef1a (mean ± SEM). Statistical significance was calculated by unpaired t test. Representative data from two independent experiments are shown. (C) Confocal single-plane image of the intestinal epithelium in 7-dpf live larvae expressing the Tg(-1.7fabp6:GFP) transgene (green) following gavage with Alexa Fluor 647 dextran (magenta). White arrowheads mark GFP-expressing cells that uptake dextran. The schematic diagram depicting gavage approach for labeling lysosome-rich enterocytes (LREs) in the larval zebrafish intestine using fluorescent luminal cargo is shown in the top panel. Image is representative of six larvae examined. (D) Confocal three-dimensional projections of the intestinal epithelium of 7-dpf larvae expressing the Tg(-1.7fabp6:GFP) (green) and TgBAC(lamp2:RFP) (red) transgenes. White arrowheads mark cells showing both GFP and RFP (red fluorescent protein) expression. Image is representative of five larvae examined. (E) Confocal single-plane image of the intestinal epithelium in 7-dpf fxr+/+ (top) and fxr−/− (bottom) larvae expressing the Tg(-4.5fabp2:DsRed) (red) and Tg(-1.7fabp6:GFP) (green) transgenes following gavage with Alexa Fluor 647 dextran (magenta). White arrowheads mark the ileocyte region, which persists in fxr−/− larvae despite loss of GFP expression. Images are representative of five fxr+/+ and five fxr−/− larvae examined. Asteroids mark zebrafish lumen in (C) to (E). Scale bars, 25 μm. (F) Uptake profiles along LRE region following gavage with Alexa Fluor 647 dextran (1.25 mg/ml) in 7-dpf fxr+/− (n = 14) and fxr−/− (n = 22) larvae. (G) DsRed fluorescence along the intestine of 7-dpf fxr+/− (n = 18) and fxr−/− (n = 24) Tg(-4.5fabp2:DsRed) larvae.
Mice lacking Fxr function display increased inflammation linked with gut microbial overgrowth and compromised intestinal epithelial barrier function (46); therefore, we speculated that the observed increase in LRE lysosomal gene expression could be due to similar phenotypes in zebrafish fxr mutants. However, fxr mutant zebrafish larvae displayed no evidence of gut microbial overgrowth (fig. S7D), loss of intestinal barrier function (fig. S7E), or significant increases in inflammatory gene expression (dataset S4) or transgene reporters (fig. S7, F and G). This suggests that the increased expression of lysosomal genes in fxr mutant larvae is not associated with intestinal inflammation.
The altered gene expression seen in fxr mutant cluster 17 cells could be explained by Fxr regulating the relative abundance of different ileal cell types such as ileocytes and LREs, or regulating expression of genes characteristic of those cell types in cluster 17. We therefore carried out subclustering of cluster 17, aiming to distinguish ileocytes from LREs and to delineate the heterogeneity within these ileal epithelial cells (fig. S8A). To our surprise, we could not cleanly separate these two subtypes, as many cluster 17 cells expressed both bile transporter genes and LRE markers (fig. S8, A to C). This indicates an overlap between the bile salt absorption and lysosomal degradation programs in some cluster 17 ileal epithelial cells, and is in agreement with previous bulk RNA-seq study showing that LREs can also express ileocyte markers such as fabp6 and slc10a2 (44). To test this overlap in vivo, we took advantage of the high endocytotic property of LREs and labeled them in Tg(-1.7fabp6:GFP) zebrafish by gavaging with fluorescent dextran, which is internalized by LREs (Fig. 6C) (44). Some ileal epithelial cells were labeled by both GFP and dextran, whereas cells anterior to this region were only GFP+ and cells posterior to this region were only dextran+. This was confirmed with a second LRE reporter TgBAC(lamp2:RFP) (Fig. 6D), further establishing the partial overlap of these two functionally distinct transcriptional programs. Collectively, these findings demonstrate that cluster 17 represents cells located in the zebrafish ileum that include at least three subtypes that we operationally define as (i) ileocytes, which express bile metabolism genes and are responsible for bile salt absorption; (ii) LREs, which express lysosomal enzymes and are responsible for macromolecule degradation; and (iii) bifunctional cells, which exhibit both of those programs.
We next sought to determine whether Fxr regulates the abundance or location of these ileal cell types. In contrast to the notable changes in the gene expression of cluster 17 cells upon fxr mutation (Fig. 6A and fig. S6B), the relative abundance of this cluster remained similar between fxr mutant and wt (fig. S6A), suggesting that Fxr deficiency does not prevent the establishment of a zebrafish ileum. To validate this observation in vivo, we measured the length and the spatial location of the ileal epithelium, including ileocytes and LREs, in fxr wt or mutant zebrafish larvae. The LRE region was evaluated by gavaging dextran into fxr wt and mutant zebrafish followed with in vivo imaging. No significant difference was observed in the length of the dextran-positive region or the intensity of the absorbed dextran after gavaging, suggesting that the abundance of LREs remains unchanged in the absence of Fxr (Fig. 6, E and F). This is further validated in adult zebrafish intestine, which exhibited similar length of the LRE region in both genotypes (fig. S8D). To assess ileocyte abundance and positioning, we gavaged dextran into the double transgenic reporter Tg(-4.5fabp2:DsRed, -1.7fabp6:GFP) in either fxr wt or mutant background (47). The proximal intestinal region, labeled by DsRed, demarcates the anterior boundary of the ileocytes, while the LRE region, labeled by dextran, demarcates the posterior boundary. The anterior boundary remained intact in the fxr mutant zebrafish, as the DsRed region did not expand or contract (Fig. 6, E and G). We did observe a nearly complete loss of GFP fluorescence in the fxr mutant animals, consistent with the findings that fabp6 is under strong regulation by Fxr (Figs. 1A and 6E). However, this nonfluorescent region, flanked by the anterior enterocytes and LREs in the fxr mutant, shares similar length and spatial position as the GFP-positive region in the fxr wt larvae (Fig. 6, E to G). These findings are in agreement with our scRNA-seq data and confirm that Fxr affects the gene expression program of the ileal epithelial cells without overtly affecting the number of those cells or the segmental boundaries that organize that region of the intestine. Together, our data suggest that Fxr is not required for developmental organization of the ileal region; instead, it is involved in distinct physiological aspects of the cell types in this region.
Fxr promotes differentiation of anterior absorptive enterocytes
Because fxr is expressed along the length of the intestine in zebrafish and mammals (Fig. 5B) (42), we next examined how Fxr contributes to the functions of absorptive enterocytes other than the ileal epithelial cells. We focused on cluster 4, which represents mature enterocytes in the anterior intestine based on their expression of known jejunal markers such as fabp1b.1 and rbp2a, as well as genes involved in lipid metabolism, a hallmark function of mammalian jejunum (Fig. 5B, table S1, and datasets S2 and S3) (48). Our scRNA-seq data suggested increased cell abundance of cluster 4 in response to Fxr mutation (fig. S6A). To validate this in vivo, we performed fluorescence-activated cell sorting (FACS) of anterior enterocytes collected from double transgenic fxr wt or mutant fish harboring the anterior enterocyte reporter Tg(-4.5fabp2:DsRed) and the pan-IEC reporter TgBAC(cldn15la-GFP). Consistent with our scRNA-seq data, we observed a significant increase in the relative abundance of anterior enterocytes in the fxr mutant compared to wt (Fig. 7A), confirming the role of Fxr in regulating the abundance of these cells in zebrafish. Fxr mutation also led to altered expression of over 250 genes in cluster 4 cells (fig. S6B and dataset S4). Notably, majority (~86%) of these differentially expressed genes were down-regulated. Functional categorization analysis revealed that these down-regulated genes in fxr mutant cells in cluster 4 were enriched for pathways involved in energy metabolism of diverse substrates (fig. S9A). This includes aspects of lipid metabolism such as lipid biosynthesis [Gene Ontology (GO) term: sterol biosynthetic process], trafficking (GO term: plasma lipoprotein particle assembly), and regulation [dre: peroxisome proliferator–activated receptor (PPAR) signaling pathway], amino acid metabolism (GO terms: peptide metabolic process; cellular modified amino acid metabolic process; creatine metabolism), and xenobiotic metabolism (GO terms: drug metabolic process; response to xenobiotic stimulus). To confirm that Fxr activity promotes expression of lipid metabolism genes in anterior enterocytes, we treated wt and fxr mutant larval zebrafish with the Fxr agonist GW4046 and observed Fxr-dependent induction of apoc2 (fig. S9B) which is expressed in cluster 4 (dataset S1). Because these pathways represent key functions of differentiated anterior enterocytes, we speculated that these gene expression differences in fxr mutant cells in cluster 4 may be due to reduced differentiation of these enterocytes. We therefore compared the zebrafish genes differentially regulated in cluster 4 in fxr mutants against defined sets of signature genes for intestinal stem cells (ISCs) and differentiated enterocytes in the small intestinal epithelium of adult mice (Fig. 7B) (49). This revealed an overlap of 102 one-to-one gene orthologs between the down-regulated genes of the fxr mutant cells in cluster 4 in this study and the genes preferentially expressed in either ISCs or differentiated enterocytes in mice. Approximately two-thirds of these genes (64 of 102) are preferentially expressed in differentiated enterocytes, suggesting that Fxr inactivation in cluster 4 preferentially reduces enterocyte differentiation programs. In support, we observed that the most enriched TFBS within accessible chromatin near the genes down-regulated in fxr mutant anterior enterocytes is Hnf4α, a TF known to promote enterocyte differentiation (Fig. 7C) (50). Collectively, these results reveal a novel role of Fxr in promoting differentiation programs of anterior enterocytes.
Fig. 7. Fxr regulates differentiation and functions of anterior absorptive enterocytes in zebrafish.
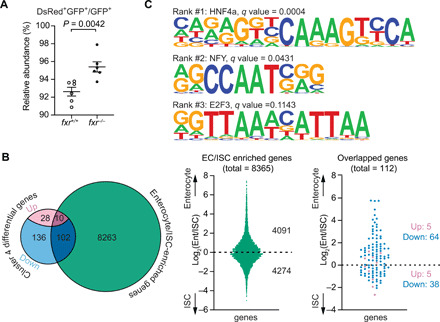
(A) FACS analysis comparing the relative abundance of anterior enterocytes in 7-dpf fxr+/+ and fxr−/− larvae expressing the Tg(-4.5fabp2:DsRed) and TgBAC(cldn15la-GFP) transgenes. The relative abundance was calculated by dividing the cell counts of DsRed+GFP+ double-positive cells by the cell counts of GFP+ single-positive cells (mean ± SEM). Statistical significance was calculated by unpaired t test. Representative data from two independent experiments are shown. (B) Comparisons between the genes that were differentially expressed in fxr−/− cells relative to fxr+/+ cells in cluster 4 and the mouse genes enriched in jejunal enterocytes/intestinal stem cells (ISCs) (49). Left: Venn diagram showing the number of overlapped genes between the two gene sets, of which the differentially expressed genes in fxr−/− cells were further classified based on the changes in their expression (up- or down-regulated) upon fxr mutation. Middle: Distributions of enterocyte- and ISC-enriched genes from the enterocyte/ISC dataset used in the current comparison. Right: Distribution of overlapped genes resulting from the comparison. (C) Top 3 HOMER-identified motifs enriched within accessible chromatin regions near genes that were down-regulated in the fxr−/− cells relative to fxr+/+ cells in cluster 4. The position weight matrices (PWMs) of the enriched nucleotide sequences are shown. The TF family that most closely matches the motif is indicated above the PWM.
DISCUSSION
The ability to synthesize bile salts and the bile salt–regulated TF Fxr are common features of all vertebrate classes, yet our knowledge of bile salt metabolism and bile salt–Fxr signaling is largely derived from mammals. Here, we characterize the bile salt–Fxr signaling axis in zebrafish by determining the bile salt composition and the key genetic components of Fxr signaling pathways. Furthermore, we elucidate the microbiota bile salt–Fxr relationships in zebrafish and highlight the importance of these interactions as they have been conserved over 420 million years since the last shared common ancestor between mammals and fishes. Collectively, we establish zebrafish as a valuable nonmammalian vertebrate model to study the bile salt–Fxr signaling axis and host-microbe coevolution. Using this model, we uncover novel functions of Fxr in modulating transcriptional programs controlling regional metabolic activities in the zebrafish intestine, including its role in repressing genes important for LRE functions in the ileum and promoting genes involved in enterocyte differentiation in the anterior intestine.
Our data show that zebrafish bile salts are composed predominantly of the evolutionarily “ancestral” C27 bile alcohol 5αCS, with only a small proportion of the evolutionarily recent C24 bile acid TCA that is commonly found in mammals (Fig. 2A). To our surprise, zebrafish 5αCS was not observed to undergo 7α-dehydroxylation, a common microbial modification of bile acids in mammals (Fig. 3B and fig. S4) (51). Furthermore, we did not observe 7α-dehydroxylation of CA by the zebrafish or carp microbiota, although CA is a suitable substrate for this modification in the mammalian gut environment (Figs. 2C and 3C). Although mammalian gut microbes have evolved numerous sulfatases that recognize and hydrolyze bile acid sulfates (52), we did not detect sulfatase activity toward 5αCS in zebrafish microbiota (Fig. 3B). Oxidation and epimerization of bile acids by microbial HSDH enzymes is well documented in mammalian gut microbiota (32, 52) and is now confirmed to occur in zebrafish (Fig. 3C). Deconjugation of TCA by bacterial BSHs was also observed in the present study, a function widespread among mammalian microbial taxa (Figs. 3C and 4A) (52), and important for regulation of lipid and cholesterol metabolism in diverse vertebrates (53). Together, these findings indicate that there may be top-down selection pressure in zebrafish to prevent evolution or acquisition of microbial enzymes that would recognize the side-chain sulfate and/or the 7α-hydroxyl group, therefore limiting secondary bile alcohol/acid production. Future study on how these primary and secondary bile salts contribute to digestive physiology and host-microbe interactions in different animals will shed light on understanding the evolutionary biology of vertebrate bile salts.
The binding pocket of FXR and the bile salt structures within a given vertebrate species are thought to have a coevolutionary relationship (7, 18). For example, C27 bile alcohol 5αCS, the major bile salt species in zebrafish, specifically binds and activates zebrafish Fxr but not mammalian FXR (7). Here, we show that the C24 bile acid TCA, a minor zebrafish bile salt, along with its derivative CA, can both stimulate zebrafish Fxr activity in vivo (Fig. 4, B and C). This raises the possibility that the zebrafish Fxr structure is able to bind both the ancestral bile alcohols and the derived bile acids, thereby representing an evolutionary transitional state. In addition, we find that key aspects of the Fxr signaling pathways remain conserved between zebrafish and mammals including Fxr-mediated induction of fabp6 and fgf19 (Fig. 1C). We further show that these zebrafish Fxr‑dependent genes, like their mammalian homologs, are more potently induced by the microbially derived deconjugated bile acid CA compared to its primary bile acid precursor TCA (Fig. 4, B and C) (33). Beyond these similarities, our analysis of intestinal genes regulated by Fxr function in zebrafish and mice also revealed substantial differences. An important example is the directionality of Fxr regulation of slc10a2. Unlike in mammals where FXR represses Slc10a2 (19), Fxr in zebrafish appears to induce slc10a2, as both the fxr and cyp7a1 mutants displayed reduced slc10a2 expression (Fig. 1C and fig. S2E). This suggests divergence of regulation of slc10a2 by Fxr since the common ancestor of fish and mammals. In fact, fxr-mediated regulation of Slc10a2 homologs appears to differ considerably even within mammals (54). For example, FXR negatively regulates intestinal Slc10a2 in mice but not in rats (55, 56). Furthermore, although Slc10a2 is repressed by FXR in both mice and humans, the underlying mechanisms are different (57). Future structure-function analyses are warranted to dissect the regulatory mechanisms responsible for such differential control of slc10a2 as well as the physiological consequences. Unlike mice where fxr mutation leads to increased inflammation (46), we did not observe significant changes in the inflammatory gene expression (fig. S7, F and G, and dataset S4) in fxr mutant zebrafish larvae. In accord, the bacterial load and the intestinal permeability were comparable between the two genotypes (fig. S7, D and E). This raises the question of functional diversification of Fxr signaling during the course of evolution and further highlights the critical need of using multiple animal models to decode bile salt–Fxr–microbiome interactions.
The zebrafish IEC scRNA-seq dataset reported here provides a useful new resource for zebrafish intestinal biology. The perspectives afforded by this scRNA-seq dataset allowed us to evaluate distinct regulatory roles of Fxr across different intestinal cell types. For example, we found that ileal epithelium (identified as cluster 17 in this dataset) is composed of multiple cell subtypes including ileocytes, LREs, and bifunctional cells expressing both bile transporter genes and lysosomal degradation markers (Fig. 6 and fig. S8, A to C). Close relationships between ileocytes and LREs have also been defined in mammals, suggesting that they are ancient cellular features of the vertebrate ileum. LREs develop in the mammalian ileum only during suckling stages before being replaced by ileocytes after weaning (44, 58). Expression of genes involved in lysosomal degradation declines during this transition, whereas the expression of genes associated with bile salt absorption increases, suggesting an inverse correlation between these two functions (44). Our results provide potential mechanistic insight into the regulation of these two functions by demonstrating that Fxr promotes the expression of bile salt absorption genes and concomitantly reduces lysosomal degradation genes in the zebrafish ileum (Fig. 6, A and B). In support, similar suppression of lysosomal genes by Fxr has been implicated in mouse studies examining Fxr influence in hepatic autophagy. In mice, Fxr trans-represses autophagy-related genes by competing for binding sites with transcriptional activators of these genes, such as CREB (cyclic adenosine monophosphate response element–binding protein) (59, 60). The binding motif of CREB (“TGACGT”) identified in the mouse study was the second most enriched binding motif near genes repressed by Fxr in zebrafish ileal epithelial cells (fig. S7C) (60), suggesting that Fxr may interact with a conserved transcriptional pathway to repress lysosomal functions across these vertebrate lineages. Other autophagy markers were expressed at similar levels between fxr mutant and wt cells in cluster 17 or any other clusters (dataset S4). Therefore, the increased expression of lysosomal genes in fxr mutant LREs is likely not due to increased autophagy in these cells. In accord, previous bulk RNA-seq comparing LREs to non-LRE enterocytes in zebrafish showed no enrichment of autophagosome pathways in LREs (44).
Whereas our data establish roles for zebrafish Fxr on ileocyte and LRE gene expression, we find that Fxr is not required for morphology of the ileal region similar to the observations from Fxr knockout mice (34). Loss of Fxr function did not overtly affect the relative abundance of ileal epithelial cells (cluster 17) or the spatial boundaries separating the typical ileocyte region from the adjacent LRE and anterior enterocyte regions (Fig. 6, C to G). The abundance of LREs also appears unaffected in fxr mutants, indicating that the observed impacts on LRE gene expression represent altered physiology in those cells (Fig. 6, A and F, and fig. S8D). The impacts of fxr mutation on ileocyte fate are less clear. Ileocytes are stereotypically defined by their expression of bile salt transport genes, which are markedly reduced in fxr mutants as expected (Fig. 6A). The differentiation and physiology of the cells that develop in fxr mutants within the typical ileocyte region remain unclear and were not resolved by our scRNA-seq dataset due to the relatively small number of cells located in cluster 17 as well as their substantial heterogeneity (figs. S7A and S8). Our results do show that Fxr is involved in tuning distinct transcriptional and physiologic programs of these ileal epithelial cell types, while other transcriptional pathways likely determine ileal organization and differentiation. This is consistent with the notion that multiple TFs regulate the same intestinal enterocytes but target distinct cellular processes (50).
In contrast to our grasp on Fxr regulation of ileal epithelial cell functions, relatively little is known about the impacts of Fxr on other intestinal cell types. Using scRNA-seq, we show that Fxr exhibits different regulatory effects in anterior enterocytes compared to ileal epithelial cells. Mutation of Fxr led to a significant increase in the abundance of the anterior enterocyte population (Fig. 7A and fig. S6A). This is consistent with the observations from intestinal tumorigenesis studies, which show that FXR restricts abnormal stem cell expansion, thereby balancing the epithelial proliferative and apoptotic pathways (34, 61, 62). It is therefore possible that Fxr similarly affects stem cell dynamics in the zebrafish intestinal epithelium; however, such studies await the establishment of markers and tools to study intestinal epithelial stem cells in the zebrafish. Along with the abundance, we also found that the differentiation status of the anterior enterocytes in zebrafish is regulated by Fxr (Fig. 7B). Similar roles of Fxr in promoting cell differentiation programs have been reported in other cell types in mammals, including mesenchymal stem cells, adipocytes, and osteoblasts (63–65). While the mechanism underlying Fxr’s regulation of cell differentiation remains unclear, we speculate that Fxr may coordinate with Hnf4α to elicit such regulatory effects in zebrafish anterior enterocytes, as Hnf4α binding motif was highly enriched near Fxr-dependent genes (Fig. 7C). Fxr and Hnf4α can directly interact and cooperatively modulate gene transcription (66, 67), and Fxr positively regulates Hnf4α protein levels in mouse liver (66). Therefore, it is possible that Fxr increases Hnf4α protein expression or activity to promote enterocyte differentiation in the zebrafish intestine. Nonetheless, our findings reveal novel roles of Fxr in modulating the abundance and differentiation of zebrafish anterior enterocytes.
The molecular and physiologic mechanisms by which Fxr mediates these effects on distinct IEC types warrant further investigation. Fxr function affects hundreds of genes in the zebrafish (this study) and mouse intestine (38), but it remains unclear how many of those are due to primary autonomous roles for Fxr interacting with those gene loci as opposed to secondary systemic effects caused by Fxr mutation. For example, Fxr mutation in the intestine can disrupt endocrine hormone Fgf19 signaling and bile salt homeostasis, therefore producing systemic impacts on energy metabolism, tissue regeneration, and control of inflammation (68). Furthermore, Fxr also has autonomous roles in other organ systems (69), which may be impaired in the whole-animal fxr mutant zebrafish that we used here. The resulting extraintestinal and systemic changes may, in turn, feedback to the intestine to elicit secondary effects on intestinal gene expression and physiology. Consequently, the phenotypes reported here may be caused directly by Fxr deficiency in the intestine, indirectly by Fxr deficiency in extraintestinal tissues, or a combination. Tissue-specific and conditional mutant alleles could help distinguish between these different possibilities in the future.
MATERIALS AND METHODS
Zebrafish lines and husbandry
All zebrafish experiments were performed following protocols approved by the Duke University Medical Center Institutional Animal Care and Use Committee (protocol numbers A115-16-05 and A096-19-04). Zebrafish stocks were maintained on Ekkwill (EK), Tuebingen long fin (TL), or a mixed EK/TL background on a 14-hour/10-hour light/dark cycle at 28.5°C in a recirculating system. Maintenance of conventionally raised zebrafish was performed as described (70). Generation, colonization, maintenance, and sterility test of GF zebrafish larvae were conducted as detailed in our earlier work with the exception that an additional gentamicin (50 μg/ml) (Sigma-Aldrich, G1264) was supplemented in the antibiotic-containing gnotobiotic zebrafish medium (71). Genotyping of larval and adult zebrafish was performed with the corresponding primers (table S2) using standard procedures (70). The following engineered zebrafish lines were used in this study: fxrrdu81/rdu81 (generated in this study), fxrrdu82/rdu82 (generated in this study), cyp7a1rdu8/rdu833 (generated in this study), cyp7a1rdu84/rdu84 (generated in this study), Tg(-1.7fabp6:EGFP-pA-cryaa:mCherry) (generated in this study), slc10a2sa2486 (28), Tg(-4.5fabp2:DsRed) (47), TgBAC(cldn15la-GFP) (35), TgBAC(lamp2:RFP) (72), Tg(-0.258fabp6-cfos:GFP) (16), TgBAC(tnfa:GFP) (73), and Tg(NFkB:EGFP) (47).
Construction of mutant zebrafish lines
Mutant zebrafish lines were generated using CRISPR-Cas9 as described previously (44). Briefly, the guide RNAs (gRNAs) were designed using the “CRISPRscan” tool (www.crisprscan.org/) and synthesized using oligo-based in vitro transcription method (table S2). At the one-cell stage, wt zebrafish embryos (TL or EK strain) were injected with 1 to 2 nl of a cocktail consisting of Cas9 mRNA (150 ng/μl), gRNA (120 ng/μl), 0.05% phenol red, 120 mM KCl, and 20 mM Hepes (pH 7.0). Injected embryos were screened for mutagenesis with the corresponding primers (table S2) using MeltDoctor High-Resolution Melting Assay (Thermo Fisher Scientific, 4409535) following the manufacturer’s specifications. The mutations were further determined through Sanger sequencing of the region encompassing the gRNA targeting sites. The fxr mutants were generated through targeted deletion at the exon 4 encoding the DNA binding domain of Fxr. We identified two independent deletion alleles, fxr-10/−10 and fxr-11/−11 (allele designations rdu81 and rdu82, respectively), that each resulted in frameshift mutations and displayed significantly reduced fxr mRNA (fig. S1, A and B). Likewise, the cyp7a1 mutants, cyp7a1–7/−7 and cyp7a1–16/−16 (allele designations rdu83 and rdu84, respectively), were generated by targeting the exon 2 encoding the cytochrome P450 domain and were validated via phenotypic assessment and/or qRT-PCR (fig. S2, B to E). Only the fxr-10/−10 (rdu81) and cyp7a1–16/−16 (rdu84) mutants were used in this study.
Construction of transgenic zebrafish line
The 1.7-kb promoter fragment of the fabp6 gene was PCR-amplified from the genomic DNA of wt Tübingen zebrafish and cloned into p5E-Fse-Asc plasmid (table S2). The resulting clone (p5E-1.7fabp6), along with the pME-EGFP and p3E-polyA plasmids, was further recombined into pDestTol2pACrymCherry through multisite Gateway recombination to generate pDestTol2-1.7fabp6:EGFPpACrymCherry (74, 75). This recombinant plasmid carries two linked fluorescent marker genes, a GFP and a mCherry. The expression of GFP is driven by the 1.7-kb fabp6 promoter fragment and reflects the expression of fabp6, whereas the expression of mCherry is driven by the lens marker cryaa and serves as a constitutive transgene marker. At the one-cell stage, wt zebrafish embryos (EK strain) were injected with 1 to 2 nl of a cocktail containing pDestTol2-1.7fabp6:EGFPpACrymCherry (50 ng/μl), transposase mRNA (25 ng/μl), 0.3% phenol red, and 1× Tango buffer (Thermo Fisher Scientific, BY5). Two mosaic germline founders were identified, raised to adulthood, and screened to isolate lines with the transgene inserted at a single locus. Stable Tg(-1.7fabp6:EGFP-pA-cryaa:mCherry) (allele designations rdu80) lines were generated by outcrossing the founder to wt EK for at least three generations [abbreviated as Tg(-1.7fabp6:GFP) in the rest of the article]. This Tg(-1.7fabp6:GFP) reporter line displayed a pattern of GFP expression in the ileocyte region and the LRE region similar to our previous transgenic line Tg(-0.258fabp6 -cfos:GFP) (16), which expresses GFP under control of a smaller 258-bp fabp6 promoter region and a mouse Cfos minimal promoter. Compared to that line, the new Tg(-1.7fabp6:GFP) line using the larger 1.7-kb fabp6 promoter region expresses GFP more distally into the LRE region and also in rare cells within the anterior regions of the intestine. In situ hybridization of fabp6 shows that its expression appears to be highest in the ileocyte region with lower expression in the LRE region (76), suggesting that the larger 1.7-kb fabp6 promoter is sufficient to recapitulate the endogenous pattern of fabp6 expression.
Quantitative RT-PCR analysis
RNA isolation, deoxyribonuclease treatment, RT, and quantitative PCR were performed as described previously (70). Data were analyzed with the ∆∆Ct method. For whole larvae samples, 6- to 7-dpf larvae were collected for RNA isolation (15 to 30 larvae per replicate; four to eight replicates per condition). For larval digestive tissue samples, 6- to 7-dpf larval zebrafish digestive tracts were dissected under a stereomicroscope and pooled for RNA isolation (25 to 35 guts per replicate; four to six replicates per condition). For adult digestive tissue samples, 3- to 12-month-old gender- and size-matched adult zebrafish livers or guts were used for RNA isolation (one gut or liver per replicate; five to eight replicates per condition).
In vivo imaging and densitometry
To quantify fluorescence in transgenic reporter lines, live zebrafish larvae were anesthetized, embedded in 3% methylcellulose [w/v in gnotobiotic zebrafish media (GZM)], and imaged using a Leica M205 FA stereomicroscope with identical exposure time and magnification in the same experiment. GFP densitometry analysis was performed using Fiji software. For each experiment, the areas of interest were selected using the shape tools, recorded using the ROI Manager, and applied to all images. The background was calculated as the average fluorescence from three to five nontransgenic siblings of the transgenic zebrafish lines from the same experiment and was subtracted from all images using the threshold tools. The mean fluorescence intensity values of each image were determined and plotted using GraphPad Prism software.
Bile salt collection in zebrafish
Twenty 6- to 9-month-old wt adult zebrafish from four different stocks were starved for 48 hours to eliminate the potential contribution of exogenous bile salt from the zebrafish diet on zebrafish de novo bile salts. The gallbladders were dissected under a stereoscope using autoclaved forceps and immediately placed in 1 ml of prechilled isopropanol. The suspension was vortexed and centrifuged (13,000 rpm for 10 min), and the supernatant was evaporated under a stream of nitrogen at room temperature. The residues were resuspended in 1 ml of 100% methanol. For thin-layer chromatography (TLC), 20 μl of the methanol extract was spotted onto a silica TLC plate. The butanol:acetic acid:water (BAW; 85:10:15) mobile phase system was used to separate the bile components. In addition, a diluted (1:100) sample was subjected to LC-MS analysis. For bile salt analysis of zebrafish intestinal contents, wt adult zebrafish from three randomly selected tanks were used. The intestines from these zebrafish were pooled in groups (10 guts per group, one group per tank) and subjected to the same procedures as gallbladders above and diluted (1:100) for LC-MS. Adult zebrafish diet composed of a 1:1 mixture of Skretting GEMMA Micro 500 and Wean 0.5 (Bio-Oregon, B1473 and B2818) was used as a background control.
Flash column chromatography of crude carp bile
Asian grass carp gallbladders (n = 3) were collected from a local supermarket in Champaign, IL. Bile was collected from each gallbladder and pooled for extraction (45 ml). Crude bile was extracted using 9× isopropanol, and the isopropanol-soluble portion was collected for analysis. The isopropanol layer was concentrated to approximately 20 ml under nitrogen. Diluted (1:100) crude bile samples were used for TLC analysis with the BAW mobile phase. Purification of carp bile acids and alcohols was performed using flash column chromatography as described previously (27). The flash column (80 cm by 2 cm; 100 ml) was packed two-thirds full with 40 μM silica gel. It was assembled using chloroform:methanol (80:20; v/v) mobile phase. The concentrated isopropanol-bile mixture was placed on top of the packed silica for purification. The eluates of crude bile were collected in 50-ml fractions using a gradient of chloroform:methanol (80:20; 500 ml, 75:25; 500 ml, 70:30; 1000 ml, 65:35; 500 ml). The fractions were evaporated under nitrogen and resuspended in 100% methanol. A dilute sample of each fraction was spotted (30 μl) and examined on a TLC plate using BAW mobile phase. Select fractions were chosen for LC-MS analysis.
TLC visualization and extraction of bile compounds from TLC
Zebrafish and carp bile in methanol were examined using silica gel TLC plate (JT Baker, JT4449-4). Two mobile phases were used to separate bile alcohols and bile acids. BAW mobile phase consisted of butanol:acetic acid:water (85:10:15) mobile phase. Solvent 25 mobile phase used was n-propanol:isoamyl acetate:acetic acid:water (4:3:2:1). Plates were sprayed with 10% phosphomolybdic acid (w/v) in ethanol, and plates were baked at 100°C for 10 min. To extract bile compounds from the TLC plate, silica from replicate plates was extracted twice with 3 ml of butanol and 3 ml of water. The butanol layer was removed after each extraction, combined, and evaporated under nitrogen gas.
Extraction of carp intestinal contents
Whole intestines were removed from Asian carp and collected in 50-ml conical tubes. The contents were placed in a −80°C freezer overnight and lyophilized to remove all liquid. For LC-MS analysis, dry intestinal contents (0.14 g) were resuspended in 1 ml of 90% ethanol and sonicated for 30 min to completely dissolve soluble compounds. Furthermore, the intestinal content was centrifuged (10,000 rpm for 15 min) and the supernatant was filtered (0.45 μm) to remove additional precipitates. Diluted samples (1:100) of the filtered supernatant were spotted (30 μl) onto a TLC using BAW mobile phase and also injected onto LC-MS in untargeted full-scan mode to analyze metabolites.
NMR analysis of purified zebrafish bile alcohol
Pure flash column chromatography fractions and TLC spots matching the Rf (retention factor) value for 5αCS were validated using MS in negative ion mode. One milligram of pure bile alcohol in methanol was used on a Waters SynaptG2-Si ESI MS. The MS data were analyzed using Waters MassLynx 4.1 software. In addition, a 4-mg sample of the evaporated bile alcohol was resuspended in 750 μl of deuterated methanol and analyzed by nuclear magnetic resonance spectroscopy using an Agilent 600 MHz with a 14.1-T 54-mm bore Agilent PremiumCOMPACT Shield Superconducting Magnet. Data were visualized at University of Illinois using MNova.
Liquid chromatography–mass spectrometry
LC-MS for all samples was performed using a Waters AQUITY UPLC coupled with a Waters Synapt G2-Si ESI MS. Chromatography was performed using a Waters Cortecs UPLC C18 column (1.6-μm particle size) (2.5 mm by 50 mm) with a column temperature of 40°C. Samples were injected at 1 μl. Solvent A consisted of 95% water, 5% acetonitrile, and 0.1% formic acid. Solvent B consisted of 95% acetonitrile, 5% water, and 0.1% formic acid. The initial mobile phase was 90% solvent A and 10% solvent B and increased linearly until the gradient reached 50% solvent A and 50% solvent B at 7.5 min. Solvent B was increased linearly again until it was briefly 100% at 8.0 min until returning to the initial mobile phase (90% solvent A, 10% solvent B) over the next 2 min. The total run was 10 min with a flow rate of 10 μl/min. MS was performed in negative ion mode. Nebulizer gas pressure was maintained at 400°C, and gas flow was 800 liters/hour. The capillary voltage was set at 2000 V in negative mode. MassLynx was used to analyze chromatographs and MS data. The limit of detection was defined as a 3:1 signal-to-noise ratio using the LC peak data. The limit of quantification was defined as a 10:1 signal-to-noise ratio using the LC peak data. A mixture containing 10 μM of the following bile standards was injected onto LC-MS for analysis: D4-glycocholic acid (D4-GCA) (internal standard), TCA, 5αCS, and allocholic acid (ACA). The LC-MS method was validated once a single peak for each compound was identified with the respective m/z value in negative mode.
Bacterial enrichment for in vitro bile salt metabolism assay
To prepare complex zebrafish microbiota for the in vitro bile metabolism assay, we randomly selected four different wt tanks from our conventional zebrafish aquaculture system and dissected guts from adult fish in each tank (15 to 20 fish per tank). All adult fish were dissected between 11:00 a.m. and 1:30 p.m. with sterilized dissection tools. Fish guts were pooled in groups (six guts per group, one group per tank), and each group was homogenized in 500 μl of filter-sterilized phosphate-buffered saline (PBS) containing 1 mM dithiothreitol. Results from each group #1 to #4 are reported separately. For each group, the intestinal homogenate was split into both aerobic and anaerobic vials containing modified tryptic soy broth (TSB) (1:10 dilution). Aerobic cultures were incubated with 200 rpm shaking, while anaerobic cultures were incubated statically. Both aerobic and anaerobic cultures were incubated at 30°C for 24 hours before testing for bile salt metabolism activity. To prepare individual microbial strains, Pseudomonas sp. ZWU0006, Acinetobacter sp. ZOR0008, Shewanella sp. ZOR0012, Exiguobacterium acetylicum ZWU0009, and Chryseobacterium sp. ZOR0023 were grown at 30°C for 24 hours before testing for bile salt metabolism activity.
In vitro bile salt modification assay
Modified TSB at pH 7.0 was prepared aerobically and anaerobically for experimental use with the composition as follows: vegetable peptone (17.0 g/liter), soy peptone (3.0 g/liter), glucose (2.5 g/liter), sodium chloride (0.5 g/liter), dipotassium phosphate (2.5 g/liter), and cysteine (1.0 g/liter). The substrate testing medium was generated by supplemented freshly made modified TSB medium with 50 μM methanol vehicle, 5αCS, TCA, ACA, or combinations of these bile salts of interest. The enriched complex zebrafish microbiota or individual zebrafish gut microbe of interest was subcultured (1:10 dilution) into different substrate testing media, respectively. The subcultures were grown at 30°C for 48 hours before being subjected to solid-phase extraction to detect metabolism of the bile salt substrates. Two microbes with known bile salt conversion activities were selected as reference controls for bile acid and bile alcohol conversion: Lactobacillus salivarius JCM1046 for the BSH activity (77) and Clostridium scindens ATCC (American Type Culture Collection) 35704 for 7α-dehydroxylation activity (78). In vitro bile salt modification assay with bacterial reference controls was performed similarly except that they were cultured at 37°C for 48 hours and that 1% Tween 80 was supplemented to media for growth enrichment of L. salivarius (79). Each modification assay was performed twice.
Solid-phase extraction of bacterial culture
Culture medium (1 ml) containing 50 μM bile salt substrate was used for further solid-phase extraction (SPE). Once grown, the culture was centrifuged (10,000 rpm for 5 min) to remove bacterial cells and conditioned medium was removed. A 10 μM spike of D4-GCA internal standard was added to each sample before SPE. Waters tC18 vacuum cartridges (3 ml of reservoir, 500 mg of sorbent) were used for SPE. The method was adapted from Abdel-Khalik et al. (80). Cartridges were preconditioned with 100% hexanes (6 ml), 100% acetone (3 ml), 100% methanol (6 ml), and water adjusted to pH 3.0 (6 ml). Conditioned medium was adjusted to pH 3.0, applied to the cartridge, and pulled through dropwise using a vacuum chamber. The cartridge was washed with water adjusted to pH 3.0 (6 ml) and allowed to air-dry for 30 min before being washed with 3 ml of 40% methanol. The 40% methanol fraction was tested on TLC to ensure that no substrates were being washed off of the column. Products were eluted using 3 ml of 100% methanol. Final eluates were evaporated under a stream of nitrogen and resuspended in 200 μl of 100% methanol for analysis on TLC (using solvent 25) or LC-MS.
Serial bile salt exposure and pharmacological manipulations
For serial bile salt exposure in conventionally raised larvae, embryos were collected from natural mating between cyp7a1+/−16 and cyp7a1+/−16; Tg(-1.7fabp6:GFP) and incubated in GZM at 28.5°C. At 3 dpf, larvae were randomly assigned into untreated group or groups treated with either 1 mM TCA or CA in GZM in six-well plates. The density of larvae in each well is maintained as 10 larvae in 10 ml of media, and the media in each well were changed daily (80% v/v) with fresh GZM or GZM supplemented with either 1 mM TCA or CA. At 7 dpf, larvae were sorted for mCherry under a fluorescence microscope, after which positive larvae were subjected to in vivo imaging and genotyping. Serial bile salt exposure in GF larvae was performed similarly except that GF larvae were maintained in T25 tissue flasks and that sterile TCA or CA was used for treatment. For pharmacological manipulations, (Z)-guggulsterone (Sigma-Aldrich, G5168) and GW4064 (Selleck Chemicals, S2782) were prepared in dimethyl sulfoxide (DMSO), aliquoted, and stored at −20°C. For guggulsterone treatment, Tg(-1.7fabp6:GFP) larvae were exposed to 2 μM guggulsterone in GZM daily from 3 to 5 dpf and were subjected to in vivo imaging at 6 dpf. For GW4064 treatment, Tg(-1.7fabp6:GFP) larvae were exposed to 1 μM GW4064 in GZM at 5 dpf for 16 hours and were imaged at 6 dpf. Matching DMSO-treated sibling larvae were used as vehicle control for each experiment.
Total bile salt quantification of zebrafish larvae
Zebrafish larvae (6 dpf) were euthanized and then rinsed with GZM for three times to remove carryover of food or debris (20 to 50 larvae per replicate; four to six replicates per condition). The pooled larvae were then placed in 1 ml of 1:1 chloroform:methanol and sonicated for 3 min with 2/1 ON/OFF cycle and 70% amplitude using a QSONICA Q700-MPX-110 cup horn sonicator. The homogenate of each replicate was spun at room temperature at 20,000g for 30 min. The supernatant was collected, air-dried in a fume hood overnight, and subjected to bile salt quantification using either the bile acid detection kit (Diazyme, DZ042A) as described (13) or LC-MS.
Gavage and intestinal barrier function assay
To label LREs in larval zebrafish, 6-dpf larvae were gavaged with 4 nl of Alexa Fluor 647 dextran (1.25 mg/ml) in PBS as described previously (44). After gavage, zebrafish were recovered in GZM for 4 hours before being subjected to in vivo imaging. To label LREs in adult zebrafish, size-matched fxr+/+ and fxr−/− adult siblings were fasted for 48 hours to clear luminal contents, gavaged with 50 μl of Alexa Fluor 488 dextran (1.25 mg/ml), and recovered in system water for 6 hours. Afterward, the zebrafish were euthanized and their intestines were dissected. The intestines were then fillet open, rinsed in PBS extensively to eliminate residual dextran, and imaged with a Leica M205 FA stereomicroscope. Intestinal permeability was assayed as described with some modifications (44). Briefly, 5- to 6-dpf fxr+/+ and fxr−/− larvae were gavaged with 4 nl of Alexa Fluor 568 dextran (1.25 mg/ml), recovered in GZM for 30 min, and re-gavaged with 8 nl of PBS to flush out the luminal dextran. The fluorescence is measured in the trunk for each zebrafish as a proxy for intestinal barrier leakage. As a positive control, 20 mM EDTA is gavaged along with dextran into wt larvae to disrupt epithelial tight junctions.
Fluorescence-activated cell sorting
Approximately 600 to 700 of 6-dpf TgBAC(cldn15la-GFP) zebrafish larvae of the fxr+/+ and fxr−/− genotypes were collected for the FACS experiment, respectively. The parental zebrafish used to generate the wt or mutant embryos were stock-matched siblings from heterozygous incrosses. Dissociation of the larvae was performed as previously described (70), after which the fxr+/+; TgBAC(cldn15la-GFP) and fxr−/−; TgBAC(cldn15la-GFP) cells were immediately subjected to FACS at the Duke Cancer Institute Flow Cytometry Shared Resource and were sorted side by side with two identical Beckman Coulter Astrios instruments. Nontransgenic and single transgenic controls (pools of 50 fish per genotype) were prepared as above and used for gating and compensation. Approximately 120,000 GFP-positive 7-aminoactinomycin D–negative cells per genotype were collected in 1.5 ml of Dulbecco’s modified Eagle medium/nutrient mixture F-12 supplemented with 10% heat-inactivated fetal bovine serum and 10 μM Y-27632 ROCK1 inhibitor and were immediately subjected to the downstream experiments.
Single-cell RNA sequencing
Each scRNA-seq library was generated from 10,000 FACS-sorted TgBAC(cldn15la-GFP) IECs of the indicated genotype following the 10× Genomics Single Cell 3′ protocol by the Duke Molecular Genomics Core. The sequencing ready libraries were cleaned with both Silane Dynabeads and solid phase reversible immobilization (SPRI) beads, and quality-controlled for size distribution and yield with the Agilent D5000 ScreenTape assays using the Agilent 4200 TapeStation system. Illumina P5 and P7 sequences, a sample index, and TruSeq Read 2 primer sequence were ligated for Illumina bridge amplification. Sequence was generated using paired-end sequencing on the NovaSeq SP flow cell sequencing platform at a minimum of 40,000 reads per cell.
Cell barcodes and unique molecular identifier (UMI) barcodes were demultiplexed, and reads were aligned to the reference genome, danRer11, following the CellRanger pipeline recommended by 10× Genomics. For quality control, we first performed UMI filtering by only including UMIs with <3000 detected genes. Next, we removed low-quality cells, which we define as cells that contain >25% transcript counts derived from mitochondrial genes. Furthermore, we removed the putative doublets by excluding cells that contain more than 30,000 UMIs. Through these steps, a total of 2625 low-quality or potential doublet cells were removed, after which 9918 cells passed the requirement, including 4710 cells from fxr wt and 5208 cells from fxr mutant samples. The genotype of fxr wt and mutant samples was confirmed by visualization of reads spanning the −10/−10 lesion (fig. S5A).
Clustering and statistical analysis of the scRNA-seq data were performed using the R package Seurat (version 3.1). Count matrices from both the fxr wt and mutant libraries were log-normalized, and highly variable genes were found in each library using the FindVariableFeatures() function. Afterward, these data were integrated together using the wt library as the reference dataset through the FindIntegrationAnchors (dims = 1:35) and IntegrateData (dims = 1:35) functions. The integrated expression matrix was then renormalized using the NormalizeData() function for visualization purposes. To mitigate the effects of unwanted sources of cell-to-cell variation in the integrated dataset, we used the ScaleData() function before running a principal components analysis. Jackstraw analysis revealed that the first 54 principal components significantly accounted for the variation in our data and were thus used as input to the FindClusters() function with the resolution parameter set to 0.82. Using the shared nearest neighbor algorithm within the FindNeighbors() function, cells were grouped into 27 distinct clusters and were visualized by uniform manifold approximation and projection (UMAP), which reduces the information captured in the selected significant principal components to two dimensions. The UMAP visualization was generated using the RunUMAP() function with the “n_neighbors” parameter set to 30.
To resolve putative distinct functional cell types in cluster 17 cells in fxr wt zebrafish, we performed subclustering of the cluster 17 using a similar strategy as described above with the exception that we used eight principal components following JackStraw analysis and a resolution of 0.5 in the FindClusters() function. This resulted in two subclusters: 17_0 and 17_1 (fig. S8A).
To identify marker genes of fxr wt cells in each cluster, we used two methods with different stringency standards. First, we used the FindAllMarkers() function using a Wilcoxon rank sum test to determine genes that are significantly up-regulated in each cluster compared to all other clusters combined as one group. These genes were further filtered based on an adjusted P value below 0.05 and an average loge fold change value over 0.25, resulting in a set of marker genes that we designated as “cluster markers” (dataset S2). Second, we performed pairwise comparisons between the cluster of interest and each and every other clusters using FindMarkers() function and only selected genes that showed higher expression, defined as an average loge fold change value over 0.25, in the cluster of interest in all comparisons. This pairwise comparison–based filtering step resulted in a set of more stringent marker genes, designated as “cluster-enriched markers,” that represented the most highly expressed genes in the cluster of interest (dataset S3). The expression and the distribution of relevant cluster markers or any gene of interest were visualized using FeaturePlot(), DotPlot(), and VlnPlot() functions.
To identify genes that were differentially expressed between the fxr wt and mutant cells in each cluster, we used the FindMarkers() function using a Wilcoxon rank sum test. Differentially expressed genes were arbitrarily defined as those that showed an average loge fold change value over 0.25.
TF binding motif enrichment analysis
We used FAIRE-Seq data from adult zebrafish intestinal epithelium (16) to identify accessible chromatin regions at genes that are differentially regulated in either cluster 17 or cluster 4. Using GALAXY, each FAIRE-Seq peak was associated with the nearest gene, including its surrounding regulatory regions (including 10 kb from the gene transcription start site, the gene body, and 10 kb from transcription termination sequence). We generated a BED file containing this information for every gene that could be filtered based on gene symbol identifier later based on whether a particular gene was differentially expressed in cluster 17 or 4. To identify enriched TFBS, we used “findMotifsGenome.pl” function of the HOMER software (http://homer.ucsd.edu/homer/) with foreground and background set of genomic coordinates. Specifically, genes that were differentially expressed between fxr wt and mutant cells in the cluster of interest were used as the foreground, and the ones that were not differentially expressed in the cluster of interest but exhibited expression in at least one of the IEC clusters were used as background.
Statistical methods
For the scRNA-seq experiment, statistical analyses for determination of the cluster markers, cluster-enriched markers, and differentially expressed genes of each clusters were calculated using the FindMarkers() function of the Seurat package in R with a Wilcoxon rank sum test. For all other experiments, statistical analysis was performed using unpaired t test, or one-way or two-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test with GraphPad Prism. P < 0.05 was defined as statistically significant.
Acknowledgments
We thank J. E. Park and L. Childers for technical assistance in LRE assays. We thank C. Scahill and E. Busch-Nentwich for sharing slc10a2 mutant zebrafish. We extend our gratitude to F. Sun, director of the Illinois Mass Spectrometry Core, for providing assistance with LC-MS analysis. We would also like to acknowledge the assistance of the Duke Molecular Physiology Institute Molecular Genomics Core for the generation of scRNA-seq data. Funding: We gratefully acknowledge support for this work to J.M.R. from USDA Hatch ILLU-538-916 and to J.F.R. from NIH grants R01-DK093399, R01-DK081426, and R01-DK121007, and an Innovation Grant from the Pew Charitable Trusts. H.L.D. is supported by the David H. and Norraine A. Baker Graduate Fellowship in Animal Sciences. C.K. was supported by NIH Ruth L. Kirschstein National Research Service Award Individual Predoctoral Fellowship F31-DK121392. J.L.C. was supported by NIH Ruth L. Kirschstein National Research Service Award Individual Postdoctoral Fellowship F32-DK094592. Author contributions: J.W., J.M.R., and J.F.R. designed research; J.W. and A.V. performed research; G.K., C.K., and J.L.C. contributed new reagents/analytic tools; J.W., G.P.M., A.V., H.L.D., C.R.L., T.C., G.K., J.M.R., and J.F.R. analyzed data; and J.W., G.P.M., A.V., H.L.D., C.R.L., J.M.R., and J.F.R. drafted and revised the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw and processed 10× Genomics scRNA-seq data can be found on GEO using accession number GSE173570. R scripts used to analyze the data can be found at https://github.com/gpadillamercado/scRNAseq_rawls5532.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/30/eabg1371/DC1
REFERENCES AND NOTES
- 1.Hofmann A. F., Hagey L. R., Krasowski M. D., Bile salts of vertebrates: Structural variation and possible evolutionary significance. J. Lipid Res. 51, 226–246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaap F. G., Trauner M., Jansen P. L., Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol. 11, 55–67 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., Lehmann J. M., Bile acids: Natural ligands for an orphan nuclear receptor. Science 284, 1365–1368 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre P., Cariou B., Lien F., Kuipers F., Staels B., Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89, 147–191 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Ridlon J. M., Kang D. J., Hylemon P. B., Bajaj J. S., Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 30, 332–338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winston J. A., Theriot C. M., Diversification of host bile acids by members of the gut microbiota. Gut Microbes 11, 158–171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reschly E. J., Ai N., Ekins S., Welsh W. J., Hagey L. R., Hofmann A. F., Krasowski M. D., Evolution of the bile salt nuclear receptor FXR in vertebrates. J. Lipid Res. 49, 1577–1587 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo G. L., Chiang J. Y. L., Is CYP2C70 the key to new mouse models to understand bile acids in humans? J. Lipid Res. 61, 269–271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGlone E. R., Tan T., Bloom S. R., Walters J. R. F., What can we learn from mouse models about bile acid-mediated changes after bariatric surgery? Gastroenterology 157, 4–8 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Carten J. D., Bradford M. K., Farber S. A., Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev. Biol. 360, 276–285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis J. L., Bove K. E., Schuetz E. G., Leino D., Valencia C. A., Schuetz J. D., Miethke A., Yin C., Zebrafish abcb11b mutant reveals strategies to restore bile excretion impaired by bile salt export pump deficiency. Hepatology 67, 1531–1545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S., Miller S. R., Ober E. A., Sadler K. C., Making it new again: Insight into liver development, regeneration, and disease from zebrafish research. Curr. Top. Dev. Biol. 124, 161–195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pham D. H., Yin C., Zebrafish as a model to study cholestatic liver diseases. Methods Mol. Biol. 1981, 273–289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alves-Costa F. A., Denovan-Wright E. M., Thisse C., Thisse B., Wright J. M., Spatio-temporal distribution of fatty acid-binding protein 6 (fabp6) gene transcripts in the developing and adult zebrafish (Danio rerio). FEBS J. 275, 3325–3334 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Enya S., Kawakami K., Suzuki Y., Kawaoka S., A novel zebrafish intestinal tumor model reveals a role for cyp7a1-dependent tumor-liver crosstalk in causing adverse effects on the host. Dis. Model. Mech. 11, dmm032383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lickwar C. R., Camp J. G., Weiser M., Cocchiaro J. L., Kingsley D. M., Furey T. S., Sheikh S. Z., Rawls J. F., Genomic dissection of conserved transcriptional regulation in intestinal epithelial cells. PLOS Biol. 15, e2002054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farber S. A., Pack M., Ho S. Y., Johnson I. D., Wagner D. S., Dosch R., Mullins M. C., Hendrickson H. S., Hendrickson E. K., Halpern M. E., Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science 292, 1385–1388 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Hagey L. R., Moller P. R., Hofmann A. F., Krasowski M. D., Diversity of bile salts in fish and amphibians: Evolution of a complex biochemical pathway. Physiol. Biochem. Zool. 83, 308–321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinal C. J., Tohkin M., Miyata M., Ward J. M., Lambert G., Gonzalez F. J., Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Plass J. R., Mol O., Heegsma J., Geuken M., Faber K. N., Jansen P. L., Muller M., Farnesoid X receptor and bile salts are involved in transcriptional regulation of the gene encoding the human bile salt export pump. Hepatology 35, 589–596 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., Gerard R. D., Repa J. J., Mangelsdorf D. J., Kliewer S. A., Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2, 217–225 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Gavalda-Navarro A., Pastor J. J., Mereu A., Villarroya F., Ipharraguerre I. R., Developmental regulation of the intestinal FGF19 system in domestic pigs. Am. J. Physiol. Gastrointest. Liver Physiol. 314, G647–G654 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Peng L., Piekos S., Guo G. L., Zhong X. B., Role of farnesoid X receptor in establishment of ontogeny of phase-I drug metabolizing enzyme genes in mouse liver. Acta Pharm. Sin. B 6, 453–459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheng L., Jena P. K., Liu H. X., Kalanetra K. M., Gonzalez F. J., French S. W., Krishnan V. V., Mills D. A., Wan Y. Y., Gender differences in bile acids and microbiota in relationship with gender dissimilarity in steatosis induced by diet and FXR inactivation. Sci. Rep. 7, 1748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Brolosy M. A., Kontarakis Z., Rossi A., Kuenne C., Gunther S., Fukuda N., Kikhi K., Boezio G. L. M., Takacs C. M., Lai S. L., Fukuda R., Gerri C., Giraldez A. J., Stainier D. Y. R., Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trauner M., Halilbasic E., Nuclear receptors as new perspective for the management of liver diseases. Gastroenterology 140, 1120–1125.e1–12 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Goto T., Holzinger F., Hagey L. R., Cerre C., Ton-Nu H. T., Schteingart C. D., Steinbach J. H., Shneider B. L., Hofmann A. F., Physicochemical and physiological properties of 5alpha-cyprinol sulfate, the toxic bile salt of cyprinid fish. J. Lipid Res. 44, 1643–1651 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Kettleborough R. N., Busch-Nentwich E. M., Harvey S. A., Dooley C. M., de Bruijn E., van Eeden F., Sealy I., White R. J., Herd C., Nijman I. J., Fenyes F., Mehroke S., Scahill C., Gibbons R., Wali N., Carruthers S., Hall A., Yen J., Cuppen E., Stemple D. L., A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496, 494–497 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urizar N. L., Liverman A. B., Dodds D. T., Silva F. V., Ordentlich P., Yan Y., Gonzalez F. J., Heyman R. A., Mangelsdorf D. J., Moore D. D., A natural product that lowers cholesterol as an antagonist ligand for FXR. Science 296, 1703–1706 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Maloney P. R., Parks D. J., Haffner C. D., Fivush A. M., Chandra G., Plunket K. D., Creech K. L., Moore L. B., Wilson J. G., Lewis M. C., Jones S. A., Willson T. M., Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med. Chem. 43, 2971–2974 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Une M., Matsumoto N., Kihira K., Yasuhara M., Kuramoto T., Hoshita T., Bile salts of frogs: A new higher bile acid, 3 alpha, 7 alpha, 12 alpha, 26-tetrahydroxy-5 beta-cholestanoic acid from the bile Rana plancyi. J. Lipid Res. 21, 269–276 (1980). [PubMed] [Google Scholar]
- 32.Doden H., Sallam L. A., Devendran S., Ly L., Doden G., Daniel S. L., Alves J. M. P., Ridlon J. M., Metabolism of oxo-bile acids and characterization of recombinant 12α-hydroxysteroid dehydrogenases from bile acid 7α-dehydroxylating human gut bacteria. Appl. Environ. Microbiol. 84, e00235-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiang J. Y., Kimmel R., Weinberger C., Stroup D., Farnesoid X receptor responds to bile acids and represses cholesterol 7α-Hydroxylase gene (CYP7A1) transcription. J. Biol. Chem. 275, 10918–10924 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Maran R. R., Thomas A., Roth M., Sheng Z., Esterly N., Pinson D., Gao X., Zhang Y., Ganapathy V., Gonzalez F. J., Guo G. L., Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J. Pharmacol. Exp. Ther. 328, 469–477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvers A. L., Ryan S., Scherz P. J., Huisken J., Bagnat M., Single continuous lumen formation in the zebrafish gut is mediated by smoothened-dependent tissue remodeling. Development 141, 1110–1119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parikh K., Antanaviciute A., Fawkner-Corbett D., Jagielowicz M., Aulicino A., Lagerholm C., Davis S., Kinchen J., Chen H. H., Alham N. K., Ashley N., Johnson E., Hublitz P., Bao L., Lukomska J., Andev R. S., Bjorklund E., Kessler B. M., Fischer R., Goldin R., Koohy H., Simmons A., Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567, 49–55 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Fang S., Suh J. M., Reilly S. M., Yu E., Osborn O., Lackey D., Yoshihara E., Perino A., Jacinto S., Lukasheva Y., Atkins A. R., Khvat A., Schnabl B., Yu R. T., Brenner D. A., Coulter S., Liddle C., Schoonjans K., Olefsky J. M., Saltiel A. R., Downes M., Evans R. M., Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat. Med. 21, 159–165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jadhav K., Xu Y., Xu Y., Li Y., Xu J., Zhu Y., Adorini L., Lee Y. K., Kasumov T., Yin L., Zhang Y., Reversal of metabolic disorders by pharmacological activation of bile acid receptors TGR5 and FXR. Mol. Metab. 9, 131–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt D. R., Schmidt S., Holmstrom S. R., Makishima M., Yu R. T., Cummins C. L., Mangelsdorf D. J., Kliewer S. A., AKR1B7 is induced by the farnesoid X receptor and metabolizes bile acids. J. Biol. Chem. 286, 2425–2432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Claudel T., Sturm E., Duez H., Torra I. P., Sirvent A., Kosykh V., Fruchart J. C., Dallongeville J., Hum D. W., Kuipers F., Staels B., Bile acid-activated nuclear receptor FXR suppresses apolipoprotein A-I transcription via a negative FXR response element. J. Clin. Invest. 109, 961–971 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim I., Ahn S. H., Inagaki T., Choi M., Ito S., Guo G. L., Kliewer S. A., Gonzalez F. J., Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 48, 2664–2672 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Wang Z., Du J., Lam S. H., Mathavan S., Matsudaira P., Gong Z., Morphological and molecular evidence for functional organization along the rostrocaudal axis of the adult zebrafish intestine. BMC Genomics 11, 392 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park J., Levic D. S., Sumigray K. D., Bagwell J., Eroglu O., Block C. L., Eroglu C., Barry R., Lickwar C. R., Rawls J. F., Watts S. A., Lechler T., Bagnat M., Lysosome-rich enterocytes mediate protein absorption in the vertebrate gut. Dev. Cell 51, 7–20.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harper J., Mould A., Andrews R. M., Bikoff E. K., Robertson E. J., The transcriptional repressor Blimp1/Prdm1 regulates postnatal reprogramming of intestinal enterocytes. Proc. Natl. Acad. Sci. U.S.A. 108, 10585–10590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inagaki T., Moschetta A., Lee Y. K., Peng L., Zhao G., Downes M., Yu R. T., Shelton J. M., Richardson J. A., Repa J. J., Mangelsdorf D. J., Kliewer S. A., Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 3920–3925 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanther M., Sun X., Muhlbauer M., Mackey L. C., Flynn E. J. III, Bagnat M., Jobin C., Rawls J. F., Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology 141, 197–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haber A. L., Biton M., Rogel N., Herbst R. H., Shekhar K., Smillie C., Burgin G., Delorey T. M., Howitt M. R., Katz Y., Tirosh I., Beyaz S., Dionne D., Zhang M., Raychowdhury R., Garrett W. S., Rozenblatt-Rosen O., Shi H. N., Yilmaz O., Xavier R. J., Regev A., A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kazakevych J., Sayols S., Messner B., Krienke C., Soshnikova N., Dynamic changes in chromatin states during specification and differentiation of adult intestinal stem cells. Nucleic Acids Res. 45, 5770–5784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.San Roman A. K., Aronson B. E., Krasinski S. D., Shivdasani R. A., Verzi M. P., Transcription factors GATA4 and HNF4A control distinct aspects of intestinal homeostasis in conjunction with transcription factor CDX2. J. Biol. Chem. 290, 1850–1860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ridlon J. M., Harris S. C., Bhowmik S., Kang D. J., Hylemon P. B., Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerard P., Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 3, 14–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joyce S. A., MacSharry J., Casey P. G., Kinsella M., Murphy E. F., Shanahan F., Hill C., Gahan C. G., Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. U.S.A. 111, 7421–7426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsubara T., Li F., Gonzalez F. J., FXR signaling in the enterohepatic system. Mol. Cell. Endocrinol. 368, 17–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arrese M., Trauner M., Sacchiero R. J., Crossman M. W., Shneider B. L., Neither intestinal sequestration of bile acids nor common bile duct ligation modulate the expression and function of the rat ileal bile acid transporter. Hepatology 28, 1081–1087 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Chen F., Ma L., Dawson P. A., Sinal C. J., Sehayek E., Gonzalez F. J., Breslow J., Ananthanarayanan M., Shneider B. L., Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J. Biol. Chem. 278, 19909–19916 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Neimark E., Chen F., Li X., Shneider B. L., Bile acid-induced negative feedback regulation of the human ileal bile acid transporter. Hepatology 40, 149–156 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Muncan V., Heijmans J., Krasinski S. D., Buller N. V., Wildenberg M. E., Meisner S., Radonjic M., Stapleton K. A., Lamers W. H., Biemond I., van den Bergh Weerman M. A., O’Carroll D., Hardwick J. C., Hommes D. W., van den Brink G. R., Blimp1 regulates the transition of neonatal to adult intestinal epithelium. Nat. Commun. 2, 452 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee J. M., Wagner M., Xiao R., Kim K. H., Feng D., Lazar M. A., Moore D. D., Nutrient-sensing nuclear receptors coordinate autophagy. Nature 516, 112–115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seok S., Fu T., Choi S. E., Li Y., Zhu R., Kumar S., Sun X., Yoon G., Kang Y., Zhong W., Ma J., Kemper B., Kemper J. K., Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 516, 108–111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Modica S., Murzilli S., Salvatore L., Schmidt D. R., Moschetta A., Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 68, 9589–9594 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Fu T., Coulter S., Yoshihara E., Oh T. G., Fang S., Cayabyab F., Zhu Q., Zhang T., Leblanc M., Liu S., He M., Waizenegger W., Gasser E., Schnabl B., Atkins A. R., Yu R. T., Knight R., Liddle C., Downes M., Evans R. M., FXR regulates intestinal cancer stem cell proliferation. Cell 176, 1098–1112.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rizzo G., Disante M., Mencarelli A., Renga B., Gioiello A., Pellicciari R., Fiorucci S., The farnesoid X receptor promotes adipocyte differentiation and regulates adipose cell function in vivo. Mol. Pharmacol. 70, 1164–1173 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Id Boufker H., Lagneaux L., Fayyad-Kazan H., Badran B., Najar M., Wiedig M., Ghanem G., Laurent G., Body J. J., Journe F., Role of farnesoid X receptor (FXR) in the process of differentiation of bone marrow stromal cells into osteoblasts. Bone 49, 1219–1231 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Sawitza I., Kordes C., Gotze S., Herebian D., Haussinger D., Bile acids induce hepatic differentiation of mesenchymal stem cells. Sci. Rep. 5, 13320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y., Yin L., Anderson J., Ma H., Gonzalez F. J., Willson T. M., Edwards P. A., Identification of novel pathways that control farnesoid X receptor-mediated hypocholesterolemia. J. Biol. Chem. 285, 3035–3043 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas A. M., Hart S. N., Li G., Lu H., Fang Y., Fang J., Zhong X. B., Guo G. L., Hepatocyte nuclear factor 4 alpha and farnesoid X receptor co-regulates gene transcription in mouse livers on a genome-wide scale. Pharm. Res. 30, 2188–2198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Somm E., Jornayvaz F. R., Fibroblast growth factor 15/19: From basic functions to therapeutic perspectives. Endocr. Rev. 39, 960–989 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Cariello M., Piccinin E., Garcia-Irigoyen O., Sabba C., Moschetta A., Nuclear receptor FXR, bile acids and liver damage: Introducing the progressive familial intrahepatic cholestasis with FXR mutations. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 1308–1318 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Murdoch C. C., Espenschied S. T., Matty M. A., Mueller O., Tobin D. M., Rawls J. F., Intestinal Serum amyloid A suppresses systemic neutrophil activation and bactericidal activity in response to microbiota colonization. PLOS Pathog. 15, e1007381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pham L. N., Kanther M., Semova I., Rawls J. F., Methods for generating and colonizing gnotobiotic zebrafish. Nat. Protoc. 3, 1862–1875 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodriguez-Fraticelli A. E., Bagwell J., Bosch-Fortea M., Boncompain G., Reglero-Real N., Garcia-Leon M. J., Andres G., Toribio M. L., Alonso M. A., Millan J., Perez F., Bagnat M., Martin-Belmonte F., Developmental regulation of apical endocytosis controls epithelial patterning in vertebrate tubular organs. Nat. Cell Biol. 17, 241–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marjoram L., Alvers A., Deerhake M. E., Bagwell J., Mankiewicz J., Cocchiaro J. L., Beerman R. W., Willer J., Sumigray K. D., Katsanis N., Tobin D. M., Rawls J. F., Goll M. G., Bagnat M., Epigenetic control of intestinal barrier function and inflammation in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 112, 2770–2775 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C. B., The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Berger J., Currie P. D., 503unc, a small and muscle-specific zebrafish promoter. Genesis 51, 443–447 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Oehlers S. H., Flores M. V., Okuda K. S., Hall C. J., Crosier K. E., Crosier P. S., A chemical enterocolitis model in zebrafish larvae that is dependent on microbiota and responsive to pharmacological agents. Dev. Dyn. 240, 288–298 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Raftis E. J., Forde B. M., Claesson M. J., O’Toole P. W., Unusual genome complexity in Lactobacillus salivarius JCM1046. BMC Genomics 15, 771 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marion S., Studer N., Desharnais L., Menin L., Escrig S., Meibom A., Hapfelmeier S., Bernier-Latmani R., In vitro and in vivo characterization of Clostridium scindens bile acid transformations. Gut Microbes 10, 481–503 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jacques N. A., Hardy L., Knox K. W., Wicken A. J., Effect of Tween 80 on the morphology and physiology of Lactobacillus salivarius strain IV CL-37 grown in a chemostat under glucose limitation. J. Gen. Microbiol. 119, 195–201 (1980). [DOI] [PubMed] [Google Scholar]
- 80.Abdel-Khalik J., Bjorklund E., Hansen M., Development of a solid phase extraction method for the simultaneous determination of steroid hormones in H295R cell line using liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 935, 61–69 (2013). [DOI] [PubMed] [Google Scholar]
- 81.Hall C. J., Boyle R. H., Sun X., Wicker S. M., Misa J. P., Krissansen G. W., Print C. G., Crosier K. E., Crosier P. S., Epidermal cells help coordinate leukocyte migration during inflammation through fatty acid-fuelled matrix metalloproteinase production. Nat. Commun. 5, 3880 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Phelps R. A., Chidester S., Dehghanizadeh S., Phelps J., Sandoval I. T., Rai K., Broadbent T., Sarkar S., Burt R. W., Jones D. A., A two-step model for colon adenoma initiation and progression caused by APC loss. Cell 137, 623–634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bedell V. M., Person A. D., Larson J. D., McLoon A., Balciunas D., Clark K. J., Neff K. I., Nelson K. E., Bill B. R., Schimmenti L. A., Beiraghi S., Ekker S. C., The lineage-specific gene ponzr1 is essential for zebrafish pronephric and pharyngeal arch development. Development 139, 793–804 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shu Y., Lou Q., Dai Z., Dai X., He J., Hu W., Yin Z., The basal function of teleost prolactin as a key regulator on ion uptake identified with zebrafish knockout models. Sci. Rep. 6, 18597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wingert R. A., Selleck R., Yu J., Song H. D., Chen Z., Song A., Zhou Y., Thisse B., Thisse C., McMahon A. P., Davidson A. J., The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLOS Genet. 3, 1922–1938 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shih L.-J., Lu Y.-F., Chen Y.-H., Lin C.-C., Chen J.-A., Hwang S.-P. L., Characterization of the agr2 gene, a homologue of X. laevis anterior gradient 2, from the zebrafish, Danio rerio. Gene Expr. Patterns 7, 452–460 (2007). [DOI] [PubMed] [Google Scholar]
- 87.Chen Y.-C., Lu Y.-F., Li I.-C., Hwang S.-P., Zebrafish Agr2 is required for terminal differentiation of intestinal goblet cells. PLOS ONE 7, e34408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park S. W., Zhen G., Verhaeghe C., Nakagami Y., Nguyenvu L. T., Barczak A. J., Killeen N., Erle D. J., The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc. Natl. Acad. Sci. U.S.A. 106, 6950–6955 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao F., Edwards R., Dizon D., Afrasiabi K., Mastroianni J. R., Geyfman M., Ouellette A. J., Andersen B., Lipkin S. M., Disruption of Paneth and goblet cell homeostasis and increased endoplasmic reticulum stress in Agr2−/− mice. Dev. Biol. 338, 270–279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Noah T. K., Kazanjian A., Whitsett J., Shroyer N. F., SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp. Cell Res. 316, 452–465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gregorieff A., Stange D. E., Kujala P., Begthel H., van den Born M., Korving J., Peters P. J., Clevers H., The ets-domain transcription factor Spdef promotes maturation of goblet and paneth cells in the intestinal epithelium. Gastroenterology 137, 1333–1345.e1-3 (2009). [DOI] [PubMed] [Google Scholar]
- 92.Wallace K. N., Akhter S., Smith E. M., Lorent K., Pack M., Intestinal growth and differentiation in zebrafish. Mech. Dev. 122, 157–173 (2005). [DOI] [PubMed] [Google Scholar]
- 93.Gerbe F., Sidot E., Smyth D. J., Ohmoto M., Matsumoto I., Dardalhon V., Cesses P., Garnier L., Pouzolles M., Brulin B., Bruschi M., Harcus Y., Zimmermann V. S., Taylor N., Maizels R. M., Jay P., Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schumacher M. A., Hsieh J. J., Liu C. Y., Appel K. L., Waddell A., Almohazey D., Katada K., Bernard J. K., Bucar E. B., Gadeock S., Maselli K. M., Washington M. K., Grikscheit T. C., Warburton D., Rosen M. J., Frey M. R., Sprouty2 limits intestinal tuft and goblet cell numbers through GSK3β-mediated restriction of epithelial IL-33. Nat. Commun. 12, 836 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kimura S., Kobayashi N., Nakamura Y., Kanaya T., Takahashi D., Fujiki R., Mutoh M., Obata Y., Iwanaga T., Nakagawa T., Kato N., Sato S., Kaisho T., Ohno H., Hase K., Sox8 is essential for M cell maturation to accelerate IgA response at the early stage after weaning in mice. J. Exp. Med. 216, 831–846 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Knoop K. A., Kumar N., Butler B. R., Sakthivel S. K., Taylor R. T., Nochi T., Akiba H., Yagita H., Kiyono H., Williams I. R., RANKL is necessary and sufficient to initiate development of antigen-sampling M cells in the intestinal epithelium. J. Immunol. 183, 5738–5747 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kimura S., Nakamura Y., Kobayashi N., Shiroguchi K., Kawakami E., Mutoh M., Takahashi-Iwanaga H., Yamada T., Hisamoto M., Nakamura M., Udagawa N., Sato S., Kaisho T., Iwanaga T., Hase K., Osteoprotegerin-dependent M cell self-regulation balances gut infection and immunity. Nat. Commun. 11, 234 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ye L., Mueller O., Bagwell J., Bagnat M., Liddle R. A., Rawls J. F., High fat diet induces microbiota-dependent silencing of enteroendocrine cells. eLife 8, e48479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Delporte F. M., Pasque V., Devos N., Manfroid I., Voz M. L., Motte P., Biemar F., Martial J. A., Peers B., Expression of zebrafish pax6b in pancreas is regulated by two enhancers containing highly conserved cis-elements bound by PDX1, PBX and PREP factors. BMC Dev. Biol. 8, 53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ng A. N., de Jong-Curtain T. A., Mawdsley D. J., White S. J., Shin J., Appel B., Dong P. D., Stainier D. Y., Heath J. K., Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev. Biol. 286, 114–135 (2005). [DOI] [PubMed] [Google Scholar]
- 101.Worthington J. J., Reimann F., Gribble F. M., Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol. 11, 3–20 (2018). [DOI] [PubMed] [Google Scholar]
- 102.Gross S., Garofalo D. C., Balderes D. A., Mastracci T. L., Dias J. M., Perlmann T., Ericson J., Sussel L., The novel enterochromaffin marker Lmx1a regulates serotonin biosynthesis in enteroendocrine cell lineages downstream of Nkx2.2. Development 143, 2616–2628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gribble F. M., Reimann F., Enteroendocrine cells: Chemosensors in the intestinal epithelium. Annu. Rev. Physiol. 78, 277–299 (2016). [DOI] [PubMed] [Google Scholar]
- 104.Moran-Ramos S., Tovar A. R., Torres N., Diet: Friend or foe of enteroendocrine cells--how it interacts with enteroendocrine cells. Adv. Nutr. 3, 8–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ye L., Bae M., Cassilly C. D., Jabba S. V., Thorpe D. W., Martin A. M., Lu H. Y., Wang J., Thompson J. D., Lickwar C. R., Poss K. D., Keating D. J., Jordt S.-E., Clardy J., Liddle R. A., Rawls J. F., Enteroendocrine cells sense bacterial tryptophan catabolites to activate enteric and vagal neuronal pathways. Cell Host Microbe 29, 179–196.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Verzi M. P., Shivdasani R. A., Epigenetic regulation of intestinal stem cell differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 319, G189–G196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang C., Meier U. T., Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 23, 1857–1867 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rao M. C., Physiology of electrolyte transport in the gut: Implications for disease. Compr. Physiol. 9, 947–1023 (2019). [DOI] [PubMed] [Google Scholar]
- 109.Plasschaert L. W., Zilionis R., Choo-Wing R., Savova V., Knehr J., Roma G., Klein A. M., Jaffe A. B., A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Montoro D. T., Haber A. L., Biton M., Vinarsky V., Lin B., Birket S. E., Yuan F., Chen S., Leung H. M., Villoria J., Rogel N., Burgin G., Tsankov A. M., Waghray A., Slyper M., Waldman J., Nguyen L., Dionne D., Rozenblatt-Rosen O., Tata P. R., Mou H., Shivaraju M., Bihler H., Mense M., Tearney G. J., Rowe S. M., Engelhardt J. F., Regev A., Rajagopal J., A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dubaissi E., Rousseau K., Lea R., Soto X., Nardeosingh S., Schweickert A., Amaya E., Thornton D. J., Papalopulu N., A secretory cell type develops alongside multiciliated cells, ionocytes and goblet cells, and provides a protective, anti-infective function in the frog embryonic mucociliary epidermis. Development 141, 1514–1525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Varsamos S., Nebel C., Charmantier G., Ontogeny of osmoregulation in postembryonic fish: A review. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 141, 401–429 (2005). [DOI] [PubMed] [Google Scholar]
- 113.Evans D. H., Teleost fish osmoregulation: What have we learned since August Krogh, Homer Smith, and Ancel Keys. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R704–R713 (2008). [DOI] [PubMed] [Google Scholar]
- 114.Ameen N. A., Ardito T., Kashgarian M., Marino C. R., A unique subset of rat and human intestinal villus cells express the cystic fibrosis transmembrane conductance regulator. Gastroenterology 108, 1016–1023 (1995). [DOI] [PubMed] [Google Scholar]
- 115.Jakab R. L., Collaco A. M., Ameen N. A., Characterization of CFTR High Expresser cells in the intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G453–G465 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gustafsson J. K., Ermund A., Ambort D., Johansson M. E., Nilsson H. E., Thorell K., Hebert H., Sjovall H., Hansson G. C., Bicarbonate and functional CFTR channel are required for proper mucin secretion and link cystic fibrosis with its mucus phenotype. J. Exp. Med. 209, 1263–1272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kayahara T., Sawada M., Takaishi S., Fukui H., Seno H., Fukuzawa H., Suzuki K., Hiai H., Kageyama R., Okano H., Chiba T., Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 535, 131–135 (2003). [DOI] [PubMed] [Google Scholar]
- 118.Fre S., Hannezo E., Sale S., Huyghe M., Lafkas D., Kissel H., Louvi A., Greve J., Louvard D., Artavanis-Tsakonas S., Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLOS ONE 6, e25785 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang J., Chan C. Y., Jiang B., Yu X., Zhu G. Z., Chen Y., Barnard J., Mei W., hnRNP I inhibits Notch signaling and regulates intestinal epithelial homeostasis in the zebrafish. PLOS Genet. 5, e1000363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Que J., Okubo T., Goldenring J. R., Nam K. T., Kurotani R., Morrisey E. E., Taranova O., Pevny L. H., Hogan B. L., Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development 134, 2521–2531 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Muncan V., Faro A., Haramis A. P., Hurlstone A. F., Wienholds E., van Es J., Korving J., Begthel H., Zivkovic D., Clevers H., T-cell factor 4 (Tcf7l2) maintains proliferative compartments in zebrafish intestine. EMBO Rep. 8, 966–973 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ito Y., Kobayashi S., Nakamura N., Miyagi H., Esaki M., Hoshijima K., Hirose S., Close association of carbonic anhydrase (CA2a and CA15a), Na+/H+ exchanger (Nhe3b), and ammonia transporter Rhcg1 in zebrafish ionocytes responsible for Na+ uptake. Front. Physiol. 4, 59 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.B. T. Thisse, C. Thisse, Fast release clones: a high throughput expression analysis, in ZFIN Direct Data Submission (2004); http://zfin.org.
- 124.Grosser T., Yusuff S., Cheskis E., Pack M. A., FitzGerald G. A., Developmental expression of functional cyclooxygenases in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 99, 8418–8423 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.G. J. Rauch, D.A. Lyons, I. Middendorf, B. Friedlander, N. Arana, T. Reyes, W.S. Talbot, Submission and curation of gene expression data, in ZFIN Direct Data Submission (2003); http://zfin.org.
- 126.B. Thisse, S. Pflumio, M. Fürthauer, B. Loppin, V. Heyer, A. Degrave, R. Woehl, A. Lux, T. Steffan, X. Q. Charbonnier, C. Thisse, Expression of the zebrafish genome during embryogenesis, in ZFIN Direct Data Submission (2001); http://zfin.org.
- 127.Wilm T. P., Solnica-Krezel L., Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development 132, 393–404 (2005). [DOI] [PubMed] [Google Scholar]
- 128.Parkin C. A., Allen C. E., Ingham P. W., Hedgehog signalling is required for cloacal development in the zebrafish embryo. Int. J. Dev. Biol. 53, 45–57 (2009). [DOI] [PubMed] [Google Scholar]
- 129.Sun X. J., Xu P. F., Zhou T., Hu M., Fu C. T., Zhang Y., Jin Y., Chen Y., Chen S. J., Huang Q. H., Liu T. X., Chen Z., Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLOS ONE 3, e1499 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.B. Thisse, G.J. Wright, C. Thisse, Embryonic and larval expression patterns from a large scale screening for novel low affinity extracellular protein interactions, in ZFIN Direct Data Submission (2008); http://zfin.org. [DOI] [PMC free article] [PubMed]
- 131.Lepage S. E., Bruce A. E., Characterization and comparative expression of zebrafish calpain system genes during early development. Dev. Dyn. 237, 819–829 (2008). [DOI] [PubMed] [Google Scholar]
- 132.Jevtov I., Samuelsson T., Yao G., Amsterdam A., Ribbeck K., Zebrafish as a model to study live mucus physiology. Sci. Rep. 4, 6653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Laue K., Daujat S., Crump J. G., Plaster N., Roehl H. H., Tubingen Screen C., Kimmel C. B., Schneider R., Hammerschmidt M., The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development 135, 1935–1946 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sperber S. M., Dawid I. B., barx1 is necessary for ectomesenchyme proliferation and osteochondroprogenitor condensation in the zebrafish pharyngeal arches. Dev. Biol. 321, 101–110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bakkers J., Hild M., Kramer C., Furutani-Seiki M., Hammerschmidt M., Zebrafish ΔNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev. Cell 2, 617–627 (2002). [DOI] [PubMed] [Google Scholar]
- 136.Lee H., Kimelman D., A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev. Cell 2, 607–616 (2002). [DOI] [PubMed] [Google Scholar]
- 137.Dodd M. E., Hatzold J., Mathias J. R., Walters K. B., Bennin D. A., Rhodes J., Kanki J. P., Look A. T., Hammerschmidt M., Huttenlocher A., The ENTH domain protein Clint1 is required for epidermal homeostasis in zebrafish. Development 136, 2591–2600 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kimelman D., Smith N. L., Lai J. K. H., Stainier D. Y., Regulation of posterior body and epidermal morphogenesis in zebrafish by localized Yap1 and Wwtr1. eLife 6, e31065 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Reischauer S., Levesque M. P., Nusslein-Volhard C., Sonawane M., Lgl2 executes its function as a tumor suppressor by regulating ErbB signaling in the zebrafish epidermis. PLOS Genet. 5, e1000720 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.C. Thisse, B. Thisse, High throughput expression analysis of zf-models consortium clones, in ZFIN Direct Data Submission (2005); http://zfin.org.
- 141.Oliver V. F., van Bysterveldt K. A., Cadzow M., Steger B., Romano V., Markie D., Hewitt A. W., Mackey D. A., Willoughby C. E., Sherwin T., Crosier P. S., McGhee C. N., Vincent A. L., A COL17A1 splice-altering mutation is prevalent in inherited recurrent corneal erosions. Ophthalmology 123, 709–722 (2016). [DOI] [PubMed] [Google Scholar]
- 142.Eisenhoffer G. T., Slattum G., Ruiz O. E., Otsuna H., Bryan C. D., Lopez J., Wagner D. S., Bonkowsky J. L., Chien C. B., Dorsky R. I., Rosenblatt J., A toolbox to study epidermal cell types in zebrafish. J. Cell Sci. 130, 269–277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Siddiqui M., Sheikh H., Tran C., Bruce A. E., The tight junction component Claudin E is required for zebrafish epiboly. Dev. Dyn. 239, 715–722 (2010). [DOI] [PubMed] [Google Scholar]
- 144.Lee R. T., Asharani P. V., Carney T. J., Basal keratinocytes contribute to all strata of the adult zebrafish epidermis. PLOS ONE 9, e84858 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lyons P. J., Ma L. H., Baker R., Fricker L. D., Carboxypeptidase A6 in zebrafish development and implications for VIth cranial nerve pathfinding. PLOS ONE 5, e12967 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tao L., DeRosa A. M., White T. W., Valdimarsson G., Zebrafish cx30.3: Identification and characterization of a gap junction gene highly expressed in the skin. Dev. Dyn. 239, 2627–2636 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Abbas L., Whitfield T. T., Nkcc1 (Slc12a2) is required for the regulation of endolymph volume in the otic vesicle and swim bladder volume in the zebrafish larva. Development 136, 2837–2848 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Winata C. L., Korzh S., Kondrychyn I., Zheng W., Korzh V., Gong Z., Development of zebrafish swimbladder: The requirement of Hedgehog signaling in specification and organization of the three tissue layers. Dev. Biol. 331, 222–236 (2009). [DOI] [PubMed] [Google Scholar]
- 149.Fang F., Li Y., Bumann M., Raftis E. J., Casey P. G., Cooney J. C., Walsh M. A., O’Toole P. W., Allelic variation of bile salt hydrolase genes in Lactobacillus salivarius does not determine bile resistance levels. J. Bacteriol. 191, 5743–5757 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ishibashi S., Schwarz M., Frykman P. K., Herz J., Russell D. W., Disruption of cholesterol 7alpha-hydroxylase gene in mice. I. Postnatal lethality reversed by bile acid and vitamin supplementation. J. Biol. Chem. 271, 18017–18023 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/30/eabg1371/DC1


