A malaria parasite plasma membrane phospholipid flippase protects ookinetes in mosquito midgut epithelium for transmission.
Abstract
Mosquito midgut epithelium traversal is essential for malaria parasite transmission. Phospholipid flippases are eukaryotic type 4 P-type adenosine triphosphatases (P4-ATPases), which, in association with CDC50, translocate phospholipids across the membrane lipid bilayers. In this study, we investigated the function of a putative P4-ATPase, ATP7, from the rodent malaria parasite Plasmodium yoelii. Disruption of ATP7 blocks the parasite infection of mosquitoes. ATP7 is localized on the ookinete plasma membrane. While ATP7-depleted ookinetes are capable of invading the midgut, they are eliminated within the epithelial cells by a process independent from the mosquito complement-like immunity. ATP7 colocalizes and interacts with the flippase cofactor CDC50C. Depletion of CDC50C phenocopies ATP7 deficiency. ATP7-depleted ookinetes fail to uptake phosphatidylcholine across the plasma membrane. Ookinete microinjection into the mosquito hemocoel reverses the ATP7 deficiency phenotype. Our study identifies Plasmodium flippase as a mechanism of parasite survival in the midgut epithelium that is required for mosquito transmission.
INTRODUCTION
Malaria is among the deadliest parasitic diseases and numbered an estimated 219 million cases and 435,000 deaths in humans in 2018 (1). Malaria is caused by protozoan parasites of the genus Plasmodium that are transmitted between humans by female anopheline mosquitoes. Mosquito acquisition of Plasmodium starts when a mosquito ingests an infected human blood containing gametocytes. Once in the mosquito midgut lumen, female and male gametocytes immediately transform into extracellular gametes. Fertilization of gametes leads to a spherical zygote that, within 12 to 20 hours, further differentiates into a crescent-shaped motile ookinete. The ookinetes invade and traverse the single-layer midgut epithelium before reaching the subepithelial basal space where they transform into stationary oocysts. Thousands of sporozoites form within the oocyst over a period of 10 to 18 days. Upon release into the hemocoel, sporozoites migrate to mosquito salivary glands from where they are released when the mosquito bites a new host (2). Upon midgut traversal, parasite numbers decrease markedly because of the complement-like immune responses mediated by mosquito TEP1 (thioester-containing protein 1; the homolog of human complement factor C3) and two TEP1-interacting proteins LRIM1 (leucine-rich immune molecule 1) and APL1C (Anopheles Plasmodium-responsive Leucine-rich repeat protein 1) (3–9).
Granulocytes, a subtype of phagocytic hemocytes, have also been implicated in promoting the activation of the mosquito complement-like immunity against Plasmodium (10). In addition, ookinete invasion triggers apoptosis of the traversed cell, causing extrusion and clearance of the epithelium into the lumen (11). Plasmodium has also evolved mechanisms to avoid host attack. Parasite surface proteins have been implicated in conferring resistance to anti-Plasmodium immunity. A variety of ookinete surface proteins interact with the mosquito host, promoting parasite infection, survival, and transmission (12). The ookinete surface protein P47, a member of six-cysteine protein family (13, 14), suppresses mosquito c-Jun N-terminal kinase signaling in the invaded midgut cell, inhibiting ookinete nitration and destruction by complement-like reactions (15–17). Another parasite surface protein PIMMS43 (Plasmodium infection of the mosquito midgut screen 43) was recently shown to protect ookinetes from the mosquito complement-like immunity (18). It is generally accepted that both P47 and PIMMS43 play critical roles to enable ookinete evade mosquito complement-like attack upon arriving at the subepithelial basal space. However, mosquito anti-Plasmodium immunity and ookinete defenses remain incompletely understood.
Asymmetric distribution of lipid molecules across the two leaflets of biological membranes is an important feature of eukaryotic cells. To establish asymmetry, eukaryotic organisms ranging from yeast to human encode a specific type of membrane transporters known as flippases that translocate phospholipids to the cytosolic side of membranes in a reaction driven by adenosine triphosphate (ATP) (19). These flippases belong to the P4-ATPase subfamily of P-type adenosine triphosphatases (ATPases) (19), and CDC50 proteins act as essential P4-ATPase cofactors (19). Primate malaria parasites encode four putative canonical P4-ATPases, while rodent malaria parasites encode three. However, the precise roles of these P4-ATPases in malaria parasite infection, development, and transmission have not been established.
Recently, we performed a CRISPR-Cas9–based screen of a large number of Plasmodium yoelii membrane protein-encoding genes to search for genes critical for development in the mosquito (20). Notably, one gene (PY17X_0809500) was determined to be essential for P. yoelii parasite development in the mosquito. This gene encodes a putative P4-ATPase protein (designated as ATP7 in this study) that is conserved among Plasmodium species. In this study, we performed in-depth in vitro and in vivo analyses of the P. yoelii P4-ATPase (PY17X_0809500) using gene disruption. We found an essential role for ATP7 and its cofactor CDC50C in ookinete survival and evasion from mosquito midgut immunity.
RESULTS
P4-ATPase ATP7 is specifically expressed in the mosquito stages of Plasmodium
The P. yoelii PY17X_0809500 gene encodes a 1764–amino acid protein with 10 predicted transmembrane helixes with extensive identity to key P4-ATPase subdomains (Fig. 1A). To investigate the temporal expression and subcellular localization of ATP7 in the parasite, we tagged endogenous ATP7 with a sextuple hemagglutinin (HA) epitope (6HA) at the C terminus in the P. yoelii 17XNL strain by homologous double crossover using a CRISPR-Cas9–based approach (fig. S1) (21, 22). The resulting atp7::6HA parasite showed normal development throughout its life cycle. Immunoblot and immunofluorescence assay (IFA) showed that ATP7 was expressed in gametocytes, mosquito midgut oocysts, and salivary gland sporozoites but was not detected in asexual blood-stage parasites (Fig. 1, B and C). ATP7 was detected in the cytoplasm of gametocytes and oocysts but displayed a peripheral localization in sporozoites (Fig. 1C). Costaining of the atp7::6HA gametocytes with α-tubulin (male gametocyte specific) and HA antibodies showed that ATP7 was expressed only in female gametocytes (Fig. 1D). During zygote to ookinete differentiation in vitro, ATP7 was distributed in both the cytoplasm and the cell periphery from zygote to retort but was mostly localized to the cell periphery of mature ookinetes (Fig. 1E). Furthermore, we tagged the endogenous ATP7 with a quadruple Myc epitope (4Myc) (fig. S1) and obtained similar results in the atp7::4Myc parasites (Fig. 1, F and G). In summary, ATP7 is specifically expressed in female gametocytes and then throughout parasite development in the mosquito.
Fig. 1. The P4-ATPase ATP7 is expressed in Plasmodium mosquito stages.
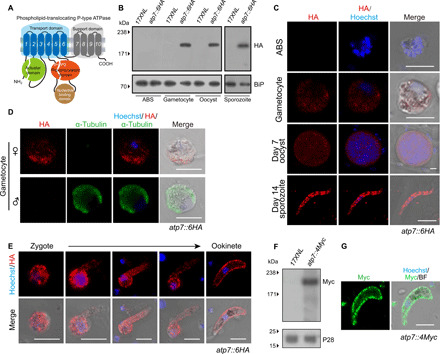
(A) Predicted protein topology of P. yoelii P4-type ATPase ATP7 (PY17X_0809500). Transmembrane helixes 1 to 10, actuator domain (green), phosphorylation domain (orange), and nucleotide binding domain (brown) are indicated. (B) Immunoblot of ATP7 at asexual blood stages (ABSs), gametocytes, midgut oocysts (7 days after blood feeding), and salivary gland sporozoites (14 days) of the 17XNL (WT parasite) and the tagged parasite atp7::6HA. Endoplasmic reticulum protein BiP was used as a loading control. (C) IFA of ATP7 expression in ABS, gametocytes, midgut oocysts, and salivary gland sporozoites of the atp7::6HA parasite. Nuclei are labeled with Hoechst 33342. (D) Costaining atp7::6HA gametocytes with HA and male gametocyte–specific α-tubulin II antibodies. (E) IFA for ATP7 expression dynamics during in vitro zygote to ookinete differentiation of the atp7::6HA parasite. (F) Immunoblot of ATP7 in the ookinetes of the tagged parasite atp7::4Myc. P28 protein was used as a loading control. (G) IFA of ATP7 expression in ookinetes of another tagged parasite atp7::4Myc. Scale bars, 5 μm for all images. Parasites were permeabilized by Triton X-100 in IFA analysis. All experiments in this figure were independently repeated three times with similar results, and the data are shown from one representative experiment.
ATP7 is essential for transmission of malaria parasites to the mosquito
To investigate the function of ATP7 in the parasite life cycle, we generated Cas9 constructs to disrupt the atp7 gene in P. yoelii (strain 17XNL) by homologous double crossover (fig. S1). This gene has a single exon with a 5295–base pair (bp) coding region. We successfully obtained a mutant clone ∆atp7c bearing a 1-kb deletion of the C-terminal coding region (Fig. 2A and fig. S1). The ∆atp7c clone exhibited normal development of asexual blood stages and gametocytes in the mice (Fig. 2, D and E).
Fig. 2. ATP7 is dispensable for asexual stages but essential for mosquito stages of parasites.
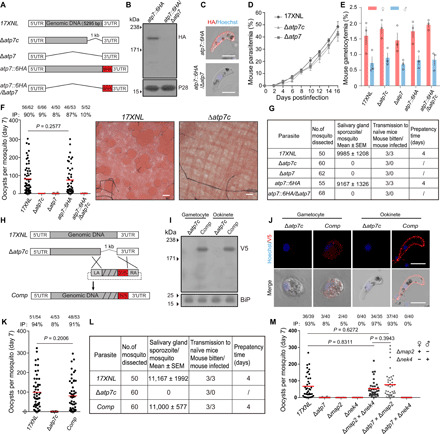
(A) Diagram showing genetic disruption (C terminus or full length) of the atp7 gene in the 17XNL and atp7::6HA parasite background (see fig. S1). UTR, untranslated region. (B) Immunoblot of ATP7 expression in atp7::6HA and atp7::6HA/Δatp7 ookinetes. P28, loading control. (C) IFA of ATP7 in atp7::6HA and atp7::6HA/Δatp7 ookinetes permeabilized by Triton X-100. Scale bars, 5 μm. (D) Asexual blood stage proliferation in mice. (E) Gametocyte formation in mice. (F) Midgut oocyst formation in mosquito 7 days after blood feeding. Midguts were stained with 0.5% Mercurochrome. Scale bars, 50 μm. (G) Salivary gland sporozoite counts and mouse infectivity. (H) Diagram of Δatp7c mutant complementation. A triple V5 (3V5) was fused to the gene. (I) Immunoblot of the 3V5-tagged ATP7 in gametocytes and ookinetes of the complemented parasite (Comp). BiP, loading control. (J) IFA of 3V5-tagged ATP7 in gametocytes and ookinetes of the Comp. Scale bars, 5 μm. (K) Midgut oocyst in mosquitoes infected with the Comp. (L) Salivary gland sporozoite counts and mouse infectivity of the Comp. (M) Midgut oocyst formation in mosquitoes infected with parasite genetic cross. △nek4 and △map2 are female and male gamete defective strains, respectively. Data are shown as means ± SD (D and E). n/n, the number of midguts carrying oocysts/the number of midguts dissected; IP, mosquito infection prevalence; red lines, mean value. Mann-Whitney test was applied (F, K, and M). Experiments in this figure were independently repeated three times except two times (M), and the data are shown from one representative experiment.
To evaluate the role of ATP7 in parasite development in the mosquito, Anopheles stephensi mosquitoes were fed on infected mice. The mosquitoes were dissected, and the formed oocysts in the midguts were counted after Mercurochrome staining. We found that ∆atp7c parasites did not produce any oocyst on day 7 postinfection (pi) (Fig. 2F), indicating failure of mosquito transmission in the absence of this gene. Consistent with this finding, no sporozoites were detected in the salivary glands on day 14 pi and no transmission from mosquitoes to mice was observed (Fig. 2G). To confirm that the defects were indeed caused by ATP7 deletion, we reintroduced the deleted 1.0-kb sequence fused with a triple V5 epitope (3V5) coding sequence back into the atp7 locus of the ∆atp7c parasite strain (Fig. 2H and fig. S1). Expression of V5-tagged ATP7 was detected in gametocytes and ookinetes from the complemented parasites using immunoblot (Fig. 2I) and IFA (Fig. 2J). In line with the localization of the endogenous ATP7 (Fig. 1E), the ATP7::3V5 fusion protein also exhibited a peripheral localization in the complemented ookinetes (Fig. 2J). Notably, oocyst and sporozoite formation as well as mouse infectivity were all restored (Fig. 2, K and L). To further confirm that the defects of the ∆atp7c strain resulted directly from atp7 deficiency, we generated two additional parasite mutants by deleting the entire coding region of the atp7 gene in the wild-type (WT) and atp7::6HA strains, respectively (Fig. 2, A to C). Consistent with the findings with the ∆atp7c mutant, these ∆atp7 and atp7::6HA/∆atp7 mutants were also able to generate gametocytes in mice (Fig. 2E) but did not produce oocysts or sporozoites in mosquitoes (Fig. 2, F and G). To confirm the female inheritance of ATP7 function, we performed a genetic cross of the ∆atp7 with either ∆map2 (male gamete-deficient) or ∆nek4 (female gamete-deficient) parasites (20). Normal numbers of midgut oocysts were observed in mosquitoes on day 7 pi in the ∆atp7 × ∆map2 but not the ∆atp7 × ∆nek4 cross (Fig. 2M), in agreement with the specific expression of ATP7 in female gametocytes (Fig. 1D).
ATP7-deficient P. yoelii produce ookinetes but not early oocysts
In the lumen of the mosquito midgut, malaria parasites undergo a series of distinct stage transformations (gametocyte to gamete to zygote and to ookinete) before traversing the midgut epithelium to initiate differentiation into oocysts in the subepithelial basal space. We performed experiments to investigate which stages are affected by ATP7 deficiency. The ∆atp7 parasites produced normal male and female gametes in vitro (Fig. 3A). Similarly, zygote to ookinete differentiation in vitro (Fig. 3, B and C) and in vivo were normal in the mutant parasites compared to controls (Fig. 3, D and E). Scanning electron microscopy showed crescent-shaped ∆atp7 ookinetes with an intact apex indistinguishable from that of WT ookinetes (Fig. 3F). Since gliding motility is a prerequisite for midgut invasion by ookinetes, we assessed their gliding capability using an in vitro Matrigel-based assay and observed no appreciable difference in gliding speed between the WT and ∆atp7 (Fig. 3G). In contrast, ookinetes lacking the ctrp gene, which is essential for ookinete gliding (23, 24), completely lost motility (Fig. 3G). Moreover, we tested whether ATP7 regulates microneme secretion, which is essential for ookinete gliding and invasion (24–26). Comparable levels of the secreted proteins from the microneme including CTRP (circumsporozoite- and TRAP-related protein), chitinase, and WARP (von Willebrand factor A domain-related protein) were detected in the supernatants of WT and mutant ookinete cultures (fig. S2A), suggesting normal microneme secretion. In addition, transmission electron microscopy (TEM) revealed intact micronemes, apical ring, and apical collar in the apex of ∆atp7 ookinetes (fig. S2, B and C). Further experiments showed that WT and the ATP7-depleted ookinetes exhibit similar cell morphology and survival under different stress conditions in vitro (fig. S2, D to I). Together, the ATP7-depleted parasite is able to develop mature and fully motile ookinetes.
Fig. 3. ATP7-deficient parasites develop ookinetes but no early oocysts.
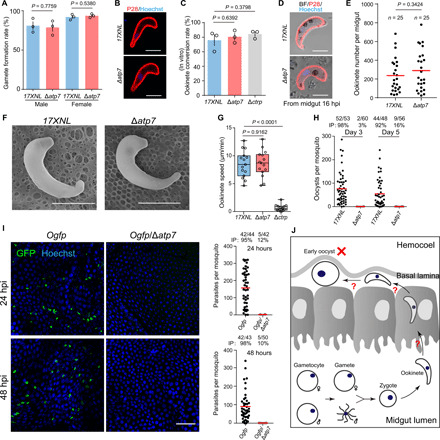
(A) Gametocyte activation to gamete in vitro. Male and female gamete formation rates are the percentage of male gametocytes showing exflagellation and the percentage of female gametocytes showing P28 expression after xanthurenic acid (XA) stimulation. (B) IFA of P28 in 17XNL and Δatp7 ookinetes in vitro. P28 is an ookinete plasma membrane protein. (C) Quantification of ookinete formation in (B). Ookinete conversion rate is the number of ookinete per 100 female gametocytes. Means ± SEM from three independent tests. (D) IFA of P28 in ookinetes from infected mosquito midguts 16 hours pi (hpi). (E) Quantification of the ookinetes in (D). n is the number of mosquitoes dissected. (F) Ookinete images by scanning electron microscopy. (G) Ookinete gliding motility measured using the in vitro Matrigel-based assay. (H) Midgut oocyst numbers at 3 and 5 days pi. (I) Fluorescence microscopy observation of ookinetes and early oocysts in mosquito midguts infected with the reporter strains Ogfp and Ogfp/Δatp7 at 24 and 48 hours pi. Right panels show the quantification of parasites. (J) Model for ATP7 deficiency in mosquito transmission. Scale bars, 5 μm (B, D, and F) and 50 μm (I). Data are shown as means ± SD (A and C). The numbers on the top are the number of mosquito carrying oocysts/the number of mosquito dissected. IP expressed as percentages. Red horizontal lines show mean oocyst number values (H and I). Two-tailed unpaired Student’s t test (A, C, E, and G). All experiments were repeated three times, and the data are shown from one representative experiment.
We next investigated the development of early oocysts. Compared to WT, no ∆atp7 oocysts were detected in mosquito midguts on both days 3 and 5 pi (Fig. 3H). Unlike late-stage oocysts, early-stage oocysts (days 1 and 2 pi) cannot be easily visualized with Mercurochrome staining. Since the soap gene is highly expressed in mature ookinetes and early oocysts (27), we engineered two parasite strains Ogfp and OmCherry, wherein the endogenous soap sequence is fused with the coding sequence of green fluorescent protein (GFP) and mCherry, respectively, at the C terminus to track early oocysts (fig. S1). A “ribosome skip” T2A peptide was inserted between the SOAP and GFP or mCherry to ensure the separate expression of GFP or mCherry and the endogenous SOAP. As expected, GFP and mCherry fluorescence was detected specifically in the mature ookinetes in vitro and in the ookinetes and early oocysts in vivo (fig. S3, A and B). Disruption of atp7 in the Ogfp parasites led to almost complete absence of GFP+ cells in mosquito midguts on both 24 and 48 hours pi (Fig. 3I), clearly indicating a lack of early oocysts (Fig. 3J).
ATP7-depleted ookinetes are capable of invading mosquito midgut epithelium
Four possibilities may account for the fact that ATP7-depleted parasites develop to motile ookinetes but fail to form early oocysts in the midgut (Fig. 3J): (i) The ookinetes fail to invade midgut epithelium, (ii) the ookinetes are developmentally arrested within the midgut epithelial cells, (iii) the ookinetes are eliminated by the mosquito immune system, and (iv) the ookinetes fail to transform into early oocysts after traversal. To address the first possibility, we analyzed the transcriptional activation of the A. stephensi gene AsSRPN6, a hallmark of ookinete invasion (28), in the midguts of infected mosquitoes at 24 hours pi. As expected, AsSRPN6 mRNA remained minimally expressed in mosquitoes that fed on naïve mouse blood (control), as determined by quantitative reverse transcription polymerase chain reaction (qRT-PCR), and was notably elevated in mosquitoes that fed on blood carrying WT and ∆atp7 but not ∆ctrp parasites (fig. S4A). Similar results were obtained when AsSRPN6 protein was assayed (fig. S4B). In addition, we examined tyrosine nitration and peroxidase activity, which are induced in epithelial cells invaded by ookinetes (29). Tyrosine nitration was detected at 24 hours pi in midguts infected with WT and ∆atp7 parasites but not in control midguts or midguts infected with ∆ctrp parasites (fig. S4C). Similarly, no differences were detected in peroxidase activity between midguts infected with WT and ∆atp7 parasites (fig. S4D).
To further confirm successful epithelium invasion by the atp7-deficient ookinetes, we examined the location of ookinetes in midguts using confocal microscopy. Parasite-infected midguts were dissected at 18 hours pi and visualized after staining with an antibody against P28. P28 is a plasma membrane protein of ookinetes and early oocysts (30). On average, 87% fewer P28+ atp7-deficient parasites were detected compared with WT (fig. S4, E and F), recapitulating the results of Ogfp/∆atp7 ookinetes (Fig. 3I). However, the percentage of ∆atp7 parasites (96%) localizing at the basal side is not notably different from that of the WT (97%) (fig. S4F), indicating successful epithelium invasion of the ∆atp7 ookinetes. As was previously reported, no ookinetes were detected at the basal side in the ∆ctrp and ∆pplp5 mutants that are motility defective and invasion defective, respectively (fig. S4, E and F) (24, 31). Together, these experiments show that ATPase7-depleted ookinetes are capable of invading the mosquito midgut epithelium (fig. S4G).
∆atp7 ookinetes are eliminated during epithelium traversal
We next asked whether the ATP7-deficient ookinetes are eliminated during midgut traversal. In parasite-infected midguts dissected at 24 hours pi, the number of P28+ ∆atp7 ookinetes (mean number: 9) substantially decreased compared with the WT ookinetes (mean number: 112), although the mosquito infection level is comparable (infection prevalence is 90% for the WT and 86% for the ∆atp7) (Fig. 4A). Notably, the ∆atp7 parasites in the midguts were morphologically aberrant. A total of 99% of the WT parasites had an intact shape (crescent or round), while 94% of the ∆atp7 mutants lost cell integrity and appeared to be lysed (Fig. 4B). In addition, ookinete nuclei were barely detected in ∆atp7 ookinetes in contrast with the intact nuclei observed in controls (Fig. 4B). Staining with antibody against GAP45, another pellicle protein in the parasite inner membrane complex (IMC), revealed similar defects (Fig. 4C). These results suggest drastic elimination of the ookinetes rather than developmental arrest within the epithelium. Moreover, deleting atp7 in the Ogfp strain resulted in only 0.5% (2 of 375) of the GFP+ cells among those were labeled by P28 in comparison to 90% (530 of 587) in the controls (Fig. 4, D and E). These results are consistent with the previous observations that GFP fluorescence rapidly fades away as the ookinetes are eliminated during midgut traversal, with P28 continuing to be on the cell surface (3, 15). Together, these results suggest that ATP7-depleted ookinetes are eliminated within the midgut epithelium (Fig. 4F).
Fig. 4. Δatp7 ookinetes are eliminated during midgut traversal.
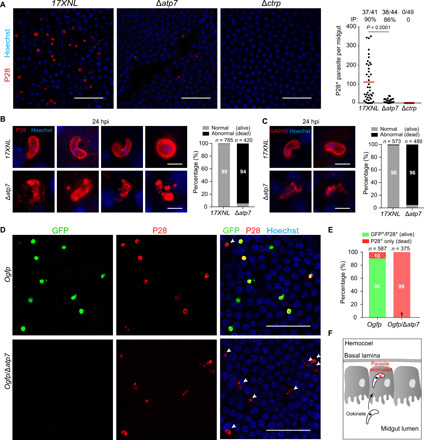
(A) Parasite plasma membrane (PPM) and nucleus staining in the infected midguts at 24 hours pi. P28 is a plasma membrane protein of ookinete. Right: Quantification of the number of P28+ parasites per midgut. The numbers on the top are the number of midguts containing parasite/the number of midguts measured; red horizontal lines show the mean value of parasite numbers. (B) Enlarged images of P28+ parasites in (A). Right: Percentage of normal (alive) and abnormal (dead) parasites. n is the number of parasites counted. (C) Parasite IMC and nucleus staining in the infected midguts at 24 hours pi. GAP45 is an IMC protein of ookinete. Right: Percentage of normal and abnormal parasites. n is the number of parasites counted. (D) P28 and Hoechst 33342 staining of midguts infected with fluorescent reporter strains Ogfp and Ogfp/Δatp7 at 24 hours pi. White arrows indicate dead parasites showing P28 staining but no GFP fluorescence. (E) Percentage of normal (alive) and abnormal (dead) parasites in (D). n is the number of parasites counted. (F) Diagram illustrating elimination of ATP7-depleted ookinetes within the epithelium. Scale bars, 50 μm (A and D) and 5 μm (B and C). Analyzed by Mann-Whitney test (A). All experiments in this figure were independently repeated three times, and the data are shown from one representative experiment.
∆atp7 ookinetes are eliminated within hours after midgut invasion
Following mosquito infection with blood containing gametocytes, ookinetes develop in the midgut lumen through an asynchronous process lasting 10 to 20 hours before midgut traversal. To allow synchronized ookinete invasion of epithelium, mosquitoes were infected by membrane feeding of ookinetes that were differentiated in vitro and purified using a Hemotek system (32). Mosquitoes were fed for 20 min with a mixture of equal numbers of Ogfp and OmCherry ookinetes, and midguts were dissected at 2.5, 4.5, and 8 hours postfeeding (pf) (Fig. 5A). At 2.5 hours, comparable numbers of GFP+ ookinetes (Ogfp) and mCherry+ ookinetes (OmCherry) were detected, although the number of ookinetes among the midguts varied (Fig. 5B). Similar results were also obtained at 4.5 and 8 hours pf (Figs. 5C and 6D). To delineate the precise timing of ookinete eradication, we disrupted the atp7 gene in the OmCherry parasites (fig. S3D) and performed cofeeding experiments with the resulting OmCherry/∆atp7 mutant and the Ogfp ookinetes. Equal numbers of the GFP+ and mCherry+ ookinetes were observed at 2.5 hours pf (Fig. 5, E and F), indicating successful epithelium invasion by the ATP7-depleted ookinetes. However, at 4.5 hours pf, the number of mCherry+ ookinetes was markedly lower compared with that of GFP+ ookinetes (GFP+, 13.8 ± 1.4; mCherry+, 0.8 ± 0.2) (Fig. 5G). By 8 hours pf, mCherry+ ookinetes completely disappeared (Fig. 5H). These results suggest that the ATP7-depleted ookinetes are eliminated within 2 to 4 hours after midgut invasion.
Fig. 5. Δatp7 ookinetes are eliminated within hours after midgut invasion.
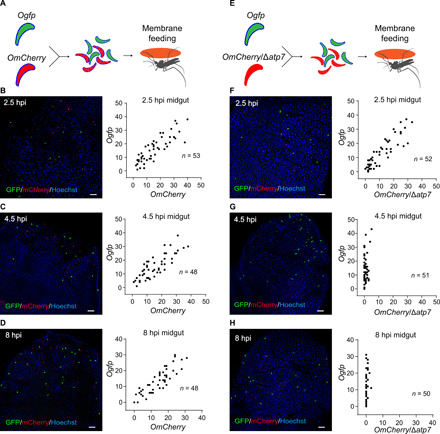
(A and E) Schematic of mosquito infection with in vitro purified ookinetes through membrane feeding. After cofeeding mosquitoes with equal number of ookinetes of Ogfp and OmCherry reporter strains (A) or Ogfp and OmCherry/Δatp7 strains (E), fluorescent ookinetes were counted in midguts dissected at 2.5, 4.5, and 8 hours after membrane feeding (pf). (B to D) Ogfp (green) and OmCherry (red) fluorescent ookinetes in coinfected midguts at 2.5 hours (B), 4.5 hours (C), and 8 hours (D). The right panels display the quantification of GFP and mCherry fluorescent ookinetes. (F to H) Ogfp (green) and OmCherry/Δatp7 (red) ookinetes in coinfected midguts at 2.5 hours (F), 4.5 hours (G), and 8 hours (H). The right panels display the quantification of GFP and mCherry fluorescent ookinetes. n, number of midguts measured. Scale bars, 50 μm. Experiments in this figure were independently repeated twice, and the data are shown from one representative experiment.
Fig. 6. ATP7 is localized on ookinete plasma membrane, and flippase activity is essential for its function.
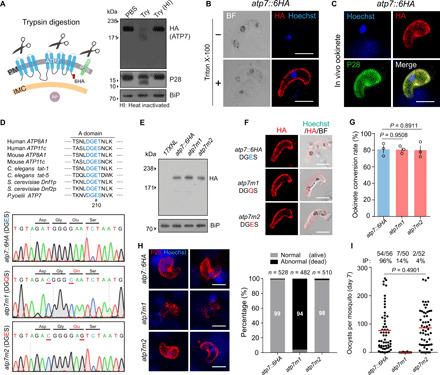
(A) Immunoblot of ATP7, P28, and BiP from atp7::6HA ookinetes treated with PBS, trypsin (Try), or heat-inactivated (HI) trypsin. Left: Schematic of plasma membrane (PM) and IMC in ookinetes. (B) IFA of ATP7 in the atp7::6HA ookinetes with or without cell permeabilization. (C) ATP7 and P28 staining of atp7::6HA ookinetes from infected mosquito midguts. (D) Generation of mutant parasites with impaired flippase activity of ATP7. Conserved motif residues (DGE) in actuator domain of eukaryotic P4-ATPases are indicated, and E210 is marked with #. DNA sequencing results confirming E210Q substitution in atp7m1 and E210E in atp7m2 mutants. (E) Immunoblot of ATP7 in ookinetes of atp7::6HA, atp7m1, and atp7m2 parasites. BiP, loading control. (F) IFA of ATP7 in the ookinetes permeabilized by Triton X-100. (G) In vitro ookinete differentiation. Data are shown as means ± SEM of three independent experiments, two-tailed unpaired Student’s t test. (H) PPM and nucleus staining from infected mosquito midguts at 24 hours pi. Right: Percentage of normal and abnormal parasites. n is the number of parasites counted. (I) Day 7 midgut oocysts in mosquitoes infected with the modified parasites. n/n, the number of midguts carrying oocysts/the number of midguts dissected; red lines indicate mean values of parasite numbers. Scale bars, 5 μm in all images. Experiments in this figure were repeated three times, and the data are shown from one representative experiment.
Elimination of ∆atp7 ookinetes is independent of mosquito complement system
We next asked how the ∆atp7 ookinetes get eliminated in the midgut. Mosquito complement-like immunity plays a major role in eliminating parasites during midgut traversal (3). Is this immunity responsible for the elimination of ∆atp7 ookinetes? We infected A. stephensi mosquitoes in which the gene encoding a key component of the complement-like system, TEP1 (3), was silenced by injection with double-stranded RNA (dsRNA) (fig. S5A). Compared to the control GFP dsRNA, TEP1 silencing slightly increased (but not significantly) the number of WT oocysts. However, ∆atp7 oocyst formation was not restored in midguts neither at 2 nor 7 days pi after TEP1 silencing (fig. S5, B and C). Silencing of LRIM1 or APL1, two other critical complement components (8), also failed to reverse ∆atp7 ookinete elimination (fig. S5, D to G). These data suggest that the elimination of ∆atp7 ookinetes in the midgut is independent of mosquito complement immunity.
To test whether mosquito hemocytes are involved in ∆atp7 ookinete elimination, we depleted phagocytic hemocytes in mosquito hemocoel using clodronate (CLD) liposomes (LPs) (fig. S5, H and I) (10). CLD-treated mosquitoes were infected with blood containing Ogfp or Ogfp/∆atp7 gametocytes. As expected (10), phagocyte depletion increased the number of Ogfp midgut parasites 24 hours pi (fig. S5J). However, it did not rescue the Ogfp/∆atp7 defect (fig. S5J). These results suggest that ∆atp7 ookinete elimination is not mosquito hemocyte mediated.
Ookinete microinjection into the mosquito hemocoel rescues the ∆atp7 defects
∆atp7 ookinetes are completely eliminated during midgut traversal, and they fail to further develop into oocysts and sporozoites. However, it is not clear whether ATP7 deficiency also affects the intrinsic developmental program of oocyst and sporozoite differentiation. By bypassing the physical barrier of the midgut, WT ookinetes injected into the hemocoel can develop into oocysts and salivary gland sporozoites in mosquitoes (33, 34). At 24 and 48 hours after injection of 690 cultured Ogfp and Ogfp/∆atp7 ookinetes (fig. S6A), comparable numbers of spherical GFP+ oocysts were detected in the hemocoel of the abdominal and thoracic cavities (fig. S6, B and C). Notably, the injected Ogfp/∆atp7 ookinetes developed into approximately equal numbers of salivary gland sporozoites as the Ogfp ookinetes at 13 days after injection (fig. S6D).
We also evaluated ookinete-to-oocyst transformation using an in vitro culture system (35). As early as 5 hours, GFP+ crescent-shaped Ogfp ookinetes protruded onto the outer convex edge, forming a snail-like stage described as a transforming ookinete (36), and further expanded to a spherical oocyst with a diameter of 7 to 8 μm at 24 hours and 10 μm at 48 hours (fig. S6E). Depletion of ATP7 had no effect on this oocyst transformation process (fig. S6F). Together, these experiments show that ATP7 is not required for differentiation into oocysts and sporozoites. Rather, it is required for protection of the ookinete from destruction in the midgut epithelial cells.
ATP7 is localized in the ookinete plasma membrane, and flippase activity is essential for its function
The ookinete pellicle harbors two membrane structures: the parasite plasma membrane (PPM) and, beneath this, the IMC. We sought to determine whether ATP7 is localized to either of these membrane structures. We reasoned that if ATP7 is localized in the PPM, then trypsin digestion of the extracellular segment is capable of degrading ATP7. Immunoblotting with an HA antibody detected a band corresponding to the HA-tagged ATP7 (~205 kDa) in cell lysates prepared from atp7::6HA ookinetes treated with phosphate-buffered saline (PBS)– or heat-inactivated trypsin. Trypsin treatment completely abolished the immunoblot signal (Fig. 6A). It is worth noting that the PPM-residing protein P28 (but not the endoplasmic reticulum protein BiP) was efficiently cleaved, which validated our approach (Fig. 6A). In agreement with its predicted topology, the 6HA-tagged ATP7 could only be labeled if the ookinetes are permeabilized by Triton X-100 (Fig. 6B). ATP7 was also colocalized with P28 in the ookinetes (Fig. 6C). Together, these results indicate that ATP7 is localized in the ookinete plasma membrane.
To test whether a conserved flippase motif within ATP7 is required for its function, we generated parasites carrying atp7 mutations that are predicted to compromise its flippase activity but not subcellular localization. It is known that the conserved motif Asp-Gly-Glu-Ser/Thr (DGES/T) in the actuator domain are critical for the catalytic activity of P4-ATPases (Fig. 6D) and that E to Q mutation in these residues abolish the flippase activity (37, 38). Accordingly, we replaced E210 with Q in the atp7::6HA strain in an attempt to generate an enzymatically inactive mutant designated as atp7m1 (Fig. 6D). A control strain (atp7m2) was also generated with a silent mutation still encoding “DGES” (Fig. 6D). The E210Q substitution had no effect on the protein level or localization of ATP7 in atp7m1 ookinetes compared to the parental strain (Fig. 6, E and F). The atp7m1 parasites produced comparable levels of ookinetes as the atp7::6HA strain (Fig. 6G) but were eradicated during midgut traversal (Fig. 6H) and therefore developed no oocysts in the mosquitoes (Fig. 6I). These results suggest that ATP7 is a functional flippase and that its activity is required for midgut traversal of ookinetes.
Phosphatidylcholine uptake defect of ∆atp7 ookinetes
To investigate the possible defect of ookinete plasma membrane caused by ATP7 disruption, we compared the uptake of labeled phospholipids in WT and ATP7-depleted ookinetes. Using a phospholipid uptake assay established previously for use with mammalian P4-ATPases (39), we analyzed the uptake activity of ookinetes toward four phospholipids, including phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylcholine (PC), and sphingomyelin (SM), which were labeled with fluorescent NBD (7-nitrobenz-2-oxa-1,3-diazol-4-yl). Ookinetes were incubated in the presence of NBD-PS, PE, PC, or SM for 60 min at 22°C, followed by the removal of fluorescent unincorporated phospholipids and those attached to the ookinete surfaces. Uptake of PS, PE, PC, or SM occurred in the WT ookinetes in the normal medium but did not occur in medium with NaN3 to deplete cellular ATP (fig. S7, A and B) (40). This is in agreement with the fact that P-type ATPases use energy from ATP hydrolysis to transport substrates across biological membranes (41). Notably, only uptake of PC is impaired in the ATP7-depleted ookinetes compared to the WT ookinetes (fig. S7, C and D). Furthermore, we tested the uptake of PC in ookinetes of the atp7m1 and atp7m2 parasites. The enzymaticlly inactive mutant atp7m1 had impaired PC uptake, similar to that of ∆atp7 ookinetes, while the atp7m2 ookinetes behaved normally (fig. S7, E and F). These results indicate that ATP7-decifient ookinetes had the defect in uptake of extracellular phospholipid PC but not PS, PE, and SM.
It is not currently possible to explore whether PC is exposed at the PPM surface in the ATP7-depleted ookinetes because no commercial probe is available. However, staining living ookinetes with either annexin V (PS probe) or Duramycin (PE probe) detected no fluorescent signal in either WT or ∆atp7 parasites (fig. S8, A and B), excluding the surface exposure of PS and PE in ATP7-deficient ookinetes.
ATP7 colocalizes and interacts with the CDC50C cofactor in ookinetes
P4-ATPase flippase function is reliant on its interaction with a CDC50 cofactor (Fig. 7A) (42). We investigated whether any of the three CDC50 paralogs in malaria parasites interact with ATP7. In P. yoelii, these are CDC50A, PY17X_0619700; CDC50B, PY17X_0916600; and CDC50C, PY17X_0514500 (23). Of these, only CDC50C showed peripheral localization in ookinete (23). CDC50C is predicted to contain two transmembrane helices and a large exocytosolic loop (Fig. 7B). Similar to ATP7, CDC50C is sensitive to ookinete trypsin treatment and permeabilization (Fig. 7, C and D), indicating the PPM localization of CDC50C in ookinetes of the tagged parasite cdc50c::6HA. To determine whether CDC50C interacts with ATP7, we generated a double-tagged strain atp7::6HA/50c::3V5 (fig. S1), wherein ATP7 and CDC50C were tagged with 6HA and 3V5, respectively. ATP7 and CDC50C showed marked colocalization at the cell periphery in the mature ookinetes (Fig. 7E). This colocalization was also observed in another independent double-tagged strain atp7::6HA/50c::4Myc (Fig. 7F). Furthermore, immunoprecipitation using an HA antibody indicated that ATP7 is bound to CDC50C in the atp7::6HA/50c::4Myc ookinete lysates (Fig. 7G). Last, we performed a proximity ligation assay (PLA), an immunohistochemical method to detect protein interaction in situ (43). A robust PLA signal was detected at the ookinete periphery in the atp7::6HA/50c::4Myc parasites when both anti-HA and anti-Myc primary antibodies were present (Fig. 7H), indicative of ATP7 and CDC50C interaction. Noteworthy, no signal was observed in either atp7::6HA or atp7::6HA/50c::3V5 ookinetes. These results demonstrate that ATP7 colocalizes and interacts with CDC50C in the PPM of ookinetes.
Fig. 7. CDC50C is an ATP7 cofactor for flippase function.
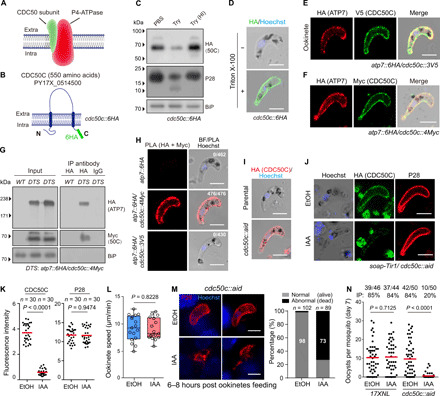
(A) Diagram of the eukaryotic flippase made of a P4-ATPase and a CDC50. (B) Topology of the P. yoelii CDC50C protein (PY17X_0514500) in ookinete plasma membrane of the tagged parasite cdc50c::6HA. (C) Immunoblot of CDC50C, P28, and BiP of cdc50c::6HA ookinetes treated with PBS, trypsin, or heat-inactivated trypsin. (D) IFA of ATP7 in atp7::6HA ookinetes with or without cell permeabilization. (E and F) IFA of ATP7 and CDC50C in ookinetes of two double-tagged parasite strains: atp7::6HA/cdc50c::3V5 (E) and atp7::6HA/cdc50c::4Myc (F). (G) Coimmunoprecipitation of ATP7 and CDC50C. (H) PLA detecting protein interaction between ATP7 and CDC50C in atp7::6HA/cdc50c::4Myc ookinetes. Number of cell displaying signal/number of cell counted is shown. (I) Generation of the cdc50c::aid strain with endogenous CDC50C tagged with an AID::6HA motif. The parental is an engineered strain soap-Tir1 stably expressing the Tir1. (J) IAA-induced depletion of CDC50C in the cdc50c::aid ookinetes. Ethanol treatment as control. (K) Quantification of IFA fluorescent signal in (J). Two-tailed unpaired Student’s t test. (L) Ookinete gliding in vitro. (M) PPM and nucleus staining in midguts infected with in vitro purified and IAA-treated ookinetes through membrane feeding. Midguts were dissected at 6 to 8 hours pf. (N) Day 7 midgut oocyst numbers in mosquitoes infected with the modified parasites. Scale bars, 5 μm in all images. Experiments were repeated three times in (B) to (F), (H) to (J), and (L) and twice in (M) and (N). The data are shown from one representative experiment.
CDC50C depletion in ookinetes phenocopies ATP7 deficiency during mosquito transmission
To investigate the potential function(s) of CDC50C in mosquito transmission, we attempted to disrupt the cdc50c gene but failed to obtain mutants, suggesting an essential role of CDC50C in parasite asexual blood stages. This observation is in agreement with the recent global knockout screening results in Plasmodium falciparum and Plasmodium berghei (44, 45). Therefore, we applied an auxin-inducible degron (AID)–based protein degradation system we recently adapted in P. yoelii (46), which allows rapid depletion of a target protein fused to an AID motif with the aid of auxin/indole-3-acetic acid (IAA) (47). The C terminus of the endogenous cdc50c locus was tagged with the sequence encoding the AID::6HA in the soap-Tir1 strain (fig. S1) (46). The resulting cdc50c::aid parasite exhibited the expected cell periphery localization of CDC50C in ookinetes (Fig. 7I), indicating no marked effect of AID tagging on CDC50C localization. IAA treatment (1 mM for 1 hour) markedly reduced the CDC50C protein abundance but had little impact on P28 levels (Fig. 7, J and K). Of note, in vitro gliding motility of the IAA-treated ookinetes is comparable to controls (Fig. 7L). Furthermore, we used a membrane feeding experiment using ookinetes treated with either IAA or EtOH to test the effect of CDC50C depletion on parasite transmission. Notably, IAA-treated ookinetes were lysed during midgut epithelium traversal (Fig. 7M). Consistent with these observations, IAA-treated ookinetes developed a much lower number of oocysts in mosquito midguts compared to the controls at 7 days pf (Fig. 7N). Together, these results strongly suggest a critical role of CDC50C in safeguarding ookinetes during midgut traversal, similar to that of its binding partner ATP7.
DISCUSSION
In this study, we identified a Plasmodium flippase complex comprising a catalytic P4-ATPase ATP7 (α subunit) and a CDC50C (β subunit) cofactor in the rodent malaria parasite P. yoelii. ATP7 and CDC50C are localized in the plasma membrane of mature ookinetes, facilitating ookinete survival during its traversal of the mosquito midgut epithelium. While ATP7- or CDC50C-depleted ookinetes are motile and capable of midgut invasion, they are quickly eliminated by the midgut epithelium, leading to failure of parasite transmission. It is noted that successful gene disruption of atp7 orthologs have been reported in genome-wide gene knockout screens of P. falciparum and P. berghei (44, 45). However, an attempt to disrupt atp7 ortholog in another gene knockout screen of P. berghei was not successful using conventional homologous recombination method (48), possibly resulting from variable rate of recombination at this locus among parasite species and strains tested.
As a putative flippase based on protein sequence homology, ATP7 was predicted to transport phospholipids from the outer leaflet to the inner leaflet of the lipid bilayer, but the identity of the lipid substrate was unknown. Our experiments revealed that ATP7 is a functional P4-ATPase essential for midgut traversal of malaria parasite. Furthermore, in the phospholipid uptake assay, WT ookinetes were able to take all four phospholipids tested (PS, PE, PC, and SM) from the medium into the ookinetes. However, the uptake of only PC is impaired in the ATP7-depleted ookinetes, suggesting that PC is the possible substrate for ATP7. Consistent with these data, the uptake of PC was abrogated by a point mutation in its enzymatic active site of ATP7. Therefore, we speculate thas in the absence of APT7, PC is exposed at the outer plasma membrane of the ookinete. Direct demonstration of PC exposure in the ∆atp7 ookinetes is not currently possible for lack of a PC-specific chemical probe.
How does a defect in ATP7/CDC50C-mediated phospholipids translocation across plasma membrane results in ookinete elimination during midgut traversal? We proposed three possibilities (fig S9). First, the P4-type ATPases are involved in the maintenance of phospholipid asymmetry that might directly or indirectly protect the parasites from the environmental stress imposed by the hosts. Conceivably, an altered distribution of phospholipids may compromise the structure and permeability of the plasma membrane and, therefore, cell viability. However, our data show that WT and the ATP7-depleted ookinetes exhibit similar cell morphology and survival under different stress conditions (fig. S2, D to I). Moreover, ∆atp7 ookinetes are also indistinguishable from WT regarding microneme secretion, cell pellicle and apical structures, and gliding motility. The second possibility is that, in the absence of ATP7/CDC50C flippase complex, impaired asymmetry of phospholipid distribution in ookinete plasma membrane lipid bilayer enables ookinete to invade the midgut epithelium but fails to egress out. The “time bomb model” of epithelial responses to ookinete invasion proposes that ookinete invasion causes irreversible cell damage and triggers a strong nitration response that is deleterious to the parasite (11). As a result, the ookinete has a limited time window to complete cell traversal before it is damaged by intracellular high levels of reactive nitrogen species and reactive oxygen species. The outcome depends on the balance between how fast the ookinete traverses the epithelium and how fast the epithelium mount a strong nitration response. In this scenario, the egress-defected ∆atp7 ookinetes could be trapped and quickly lysed inside the midgut epithelium. This would also imply that the mechanism of ookinetes to invade the epithelium is different from that to egress. While this is an intriguing hypothesis, currently, it is technically challenging to be tested. The third possibility is that once the ∆atp7 ookinetes invade the midgut epithelium, the PC exposed on the ookinete surface possibly functions as a pathogen-associated molecular pattern, which is recognized by the pattern recognition receptors of the mosquito host (49), inducing rapid ookinete elimination. Notably, the surface display of PS due to the absence of flippase activity marks apoptotic cells for consumption by phagocytes (50). However, the exact mechanisms underlying ATP7-deficient ookinete elimination within mosquito midgut epithelium requires further investigation.
During the Plasmodium life cycle, three invasion events need to occur: sporozoite invasion of hepatocytes, merozoite invasion of erythrocytes in the verterbrate host, and ookinete invasion of the midgut in the mosquito. All three invasive forms share cellular machinery that drives these processes (51). Merozoite and sporozoite invasion are accompanied by invagination of the host cell plasma membrane, resulting in the formation of a parasitophorous vacuole (PV). The parasites undergo asexual proliferation inside the PV, where the PV membrane functions as a barrier between the parasite and the host cell cytoplasm and protects the parasite from anti-Plasmodium responses by the host cells (52, 53). However, no PV formation occurs during ookinete invasion of the midgut epithelium (54). This scenario of direct contact between the invading ookinete and epithelium cytoplasm raises the possibility that ookinetes could be recognized and targeted by the mosquito intracellular immunity. Consistent with this hypothesis, coinfection experiments showed that ATP7 on the surface of WT ookinetes does not confer protection to the coinvading ATP7-deficient ookinetes. Therefore, protection of the ookinete by ATP7 is not due to a systemic mechanism in the midgut but an intrinsic mechanism to the individual ookinetes within the epithelium.
It is well established that the ookinete surface protein P47 is a key molecule that protects the Plasmodium ookinetes from mosquito complement-dependent immune attack during the midgut traversal (5). P47 inhibits mosquito Janus kinase signaling as the ookinetes come in close contact with the mosquito hemolymph (17). However, in our case, genetic and chemical ablation of either mosquito complement or hemocyte cellular immunity failed to rescue the aberrant eradication of ∆atp7 ookinetes. Therefore, Plasmodium ATP7/CDC50C-mediated protection of ookinetes likely represents a new mechanism of malaria parasite evasion from the mosquito host immune system.
MATERIALS AND METHODS
Mice and mosquitoes usage and ethics statement
All animal experiments were performed in accordance with approved protocols (XMULAC20140004) by the Committee for Care and Use of Laboratory Animals of Xiamen University. ICR mice (female, 5 to 6 weeks old) were purchased from the animal care center of Xiamen University and used for parasite propagation, drug selection, parasite cloning, and mosquito infection. The mosquito of A. stephensi (Hor strain) was reared at 28°C, 80% relative humidity, and at a 12-hour light/12-hour dark cycle in a standard insect facility. Adult mosquitoes were maintained on a 10% sucrose solution.
Plasmid construction and parasite transfection
CRISPR-Cas9 plasmid pYCm was used for all the parasite genetic modification. To construct vectors for gene editing, we amplified 5′ and 3′ genomic sequence (400 to 500 bp) of target genes as homologous arms using specific primers (table S2) and inserted the sequences into specific restriction sites in pYCm. Oligonucleotides for single guide RNAs (sgRNAs) (table S2) were mixed in pairs, denatured at 95°C for 3 min, annealed at room temperature for 5 min, and ligated into pYCm. The sgRNAs were designed to target the coding region of a gene (table S2) using the online program EuPaGDT. DNA fragments encoding 6HA, 4Myc, 3V5, and GFP or mCherry were inserted between the left and right arms in frame with the gene of interest. For each gene, two sgRNAs were designed to target sites close to the C- or N-terminal part of the coding region. Infected red blood cells (iRBCs) were electroporated with 5 to 10 μg of plasmid DNA using the Lonza Nucleofector described previously (21). Transfected parasites were immediately intravenously injected into a naïve mouse and were exposed to pyrimethamine (6 mg/ml) 24 hours after transfection.
Genotypic analysis of genetic modified parasites
All transgenic parasites were generated from the P. yoelii 17XNL strain and are listed in table S1. Blood samples from infected mice were collected from the orbital sinus, and blood cells were lysed using 1% saponin in PBS. Parasite genomic DNAs were isolated from blood-stage parasites using DNeasy blood kits (#69504, QIAGEN). For each parasite, both 5′ and 3′ homologous recombination events were detected using specific PCR primers (see fig. S1). The PCR results for parasite transfection, selection, cloning, and verification of genetic modifications are summarized in fig. S1. PCR products from some modified parasites were DNA-sequenced. All the primers used in this study are listed in table S2. Parasite clones with targeted modifications were obtained after limiting dilution. At least two clones for each gene-modified parasite were used for phenotype analysis.
Negative selection with 5-fluorouracil
Parasites subjected to sequential modification were negatively selected with 5-fluorouracil (5FC; Sigma-Aldrich, F6627) to remove episomal plasmid. 5FC (2 mg/ml) in drinking water in a dark bottle was provided to mice for 8 days with a replacement on day 4. Clearance of episomal plasmid in parasites after negative selection was confirmed by checking the parasite survival after reapplying pyrimethamine pressure (6 μg/ml) in the new infected mice.
Parasite proliferation assay in asexual mouse blood stage
Parasites (1.0 × 105) were injected into the tail vein of each of the five naïve ICR mice in each group. Parasitemia is monitored every 2 days through Giemsa staining of the thin mouse blood smears from days 2 to 16 pi. The parasitemia is calculated as the ratio of parasitized erythrocytes over total erythrocytes.
Gametocyte induction in mouse
ICR mice were treated with phenylhydrazine (80 μg/g mouse body weight) through intraperitoneal injection. Three days after injection, each mouse was infected with 4.0 × 106 parasites through intravenous injection. Gametocytemia usually peaks at day 3 pi. Male and female gametocytes were counted after Giemsa staining of blood smears. Male and female gametocytemia were calculated as the ratio of male and female gametocytes over the parasitized erythrocytes. These experiments were repeated three times independently.
Gametocyte activation to gamete in vitro
Two and a half microliters of mouse tail blood with 4 to 6% gametocytemia was added to 100 μl of exflagellation medium [RPMI 1640, 10% fetal calf serum (FCS), and 100 μM xanthurenic acid (XA) (pH 7.4)] containing 1 μl of heparin (200 U/ml) for 10 min at 22°C. For male gametocyte activation, the exflagellation centers (ECs) and total RBCs were counted in a hemocytometer under light microscope. The percentage of RBCs containing male gametocytes was calculated from Giemsa-stained smears, and the number of ECs per 100 male gametocytes was calculated as male gamete formation rate. For female gametocyte activation, P28-positive female gametocytes after staining with P28 antiserum and RBC were counted. The percentage of RBCs containing female gametocytes was calculated from Giemsa-stained smears, and the number of P28-positive female gametocytes per 100 female gametocytes was calculated as female gamete formation rate.
In vitro ookinete differentiation
In vitro ookinete differentiation was performed as described previously (23). Briefly, 1 ml of mouse blood with 4 to 6% gametocytemia was collected via orbital sinus and immediately transferred to a 10-cm cell culture dish (Corning, catalog no. 801002) containing ookinete culture medium [RPMI 1640, 10% FCS, 100 μM XA, and 25 mM Hepes (pH 8.0)]. The cultures were incubated at 22°C for 12 to 14 hours to allow gametogenesis, gamete fertilization, and ookinete differentiation. Ookinete formation was monitored by Giemsa staining of culture smears. Ookinete conversion rate was calculated as the number of ookinetes per 100 female gametocytes. For time course analysis of ookinete differentiation, parasites from 2-, 4-, 6-, 8-, and 12-hour culture were collected. Ookinetes were purified using an ammonium-chloride-potassium (ACK) lysing method (23). Briefly, the cultured ookinetes were collected by centrifugation and transferred into ACK lysing buffer (A1049201, Thermo Fisher Scientific) on ice for 8 min. After erythrocytes lysis, the remaining ookinetes were isolated via centrifugation and washed twice with PBS. The ookinetes were examined and counted on the hemocytometer under 40× objective lens. Samples containing >80% ookinete population were used for further analysis.
In vitro ookinete motility assay
Ookinete gliding motility was tested as previously described (23). All procedures were performed in a temperature-controlled room at 22°C. Briefly, 20 μl of mature ookinete cultures was mixed with 20 μl of Matrigel (#356234, BD) on ice. The mixtures were transferred onto a slide, covered with a coverslip, and sealed with nail varnish. The slide was placed at 22°C for 30 min before observation under a microscope. After tracking a gliding ookinete under view of 40× objective lens, time-lapse videos (1 frame/20 s) were taken to record ookinete movement for 20 min using a Nikon ECLIPSE E100 microscope fitted with an ISH500 digital camera and ISCapture v3.6.9.3N software (Tucsen). Time-lapse movies were analyzed with Fiji software and the Manual Tracking plugin. The motility speed was calculated by dividing the gliding distance of ookinete by the time. The experiments were repeated three times independently with 15 to 20 ookinetes tested for each parasite in each time.
Mosquito feeding and transmission assay
Thirty female mosquitoes were allowed to feed on an anesthetized mouse carrying 8 to 10% gametocytemia for 30 min at 22°C. Mosquito midguts were dissected at the indicated time (1, 2, 3, 5, or 7 days) after blood feeding and stained with 0.1% Mercurochrome for visualizing the midgut oocysts. Mosquito salivary glands were dissected on day 14 after blood feeding, and the number of sporozoites per mosquito was calculated. For parasite transmission from mosquito to mouse, 20 infected mosquitoes were allowed to feed on a naïve mouse, and the parasites emerging in mouse blood were examined.
Ookinete microneme secretion
Ookinete microneme secretion was analyzed as previously reported (47). Briefly, about 2.0 × 106 of ookinetes were purified from in vitro culture using the LS magnetic column (#130-042-401, Miltenyi Biotec) and incubated in 500 μl of PBS for 4 hours at 22°C. The cells were spun down at 750g, and the supernatant was collected, filtered through a filter (0.45 μm; SLHP033RS, Millipore), and subjected to Western blot assay.
Parasite genetic cross
The phenylhydrazine-pretreated mice were coinfected with same amount (2.0 × 106) of each parasite for genetic crosses (∆nek4 × ∆map2, ∆atp7 × ∆map2, and ∆atp7 × ∆nek4). On day 3 pi, 50 female mosquitoes were allowed to feed on gametocyte-containing mice for 30 min. Mosquito midguts were dissected on day 7 after blood feeding and stained with 0.1% Mercurochrome for oocyst counting.
Mosquito membrane feeding with ookinetes
Purified ookinetes (2.0 × 106) from in vitro culture were suspended in 1 ml of naïve mouse blood. The blood mix was added to the membrane feeder and fed to about 60 female mosquitoes for 30 min using the Hemotek (6W1, Hemotek Limited, England). After the feed, fully engorged mosquitoes were transferred to the new container and maintained under standard conditions. The mosquito midguts were dissected and observed at indicated times (2.5, 4.5, and 6 hours pf). These experiments were repeated three times. To test the mosquito transmission of ookinetes with chemical depletion of CDC50C, the ookinetes treated with IAA or EtOH were immediately added to the membrane feeder and fed to the mosquitoes.
In vitro culture for oocyst transformation
The procedure and culture conditions were followed, as previous study with some modifications (36). The in vitro–differentiated ookinetes were seeded into the eight-well Lab-Tek chamber slide or the 48-well plate at a density of 1 × 104 ookinetes per well and cultured in oocyst culture medium [85% Schneider’s medium, 15% fetal bovine serum, 23.8 mM sodium bicarbonate, 44 mM para-aminobenzoic acid (PABA), and 3.68 mM hypoxanthine (pH 8.0)]. Parasites were maintained at 22°C for 2 days, and the oocysts were monitored using the fluorescent microscope. These experiments were repeated three times with six replicate cells for each parasite in each test.
Ookinete microinjection into mosquito
After ookinete purification from in vitro culture described above, each of 40 mosquitoes (4 to 5 days old) was injected intrathoracically with 138 nl of PBS containing 690 ookinetes using a Nanoject II injector (Drummond Scientific, USA). The hemocoel oocysts within the mosquitoes were observed under the stereoscopic fluorescence microscope (M165FC, Leica) at indicated times after injection.
Quantitative real-time PCR
Thirty mosquito midguts are pooled in each group per condition per experiment. Total RNA was isolated from homogenized midgut using the RNeasy Mini Kit (74106, QIAGEN) and reverse-transcribed to complementary DNA (cDNA) using the kit (KR116-02, TIANGEN). Transcript expression of target mosquito genes was quantified using the SYBR Green Supermix (#1708882, Bio-Rad) and Bio-Rad iCycler iQ system (Bio-Rad, USA). The cycling conditions were as follows: 95°C for 20 s followed by 40 cycles at 95°C for 3 s and 72°C for 30 s. The samples were run in triplicate and normalized to the A. Stephensi rps7 gene (ASTE004816) using a ΔΔ cycle threshold–based algorithm. Arbitrary unit is provided to represent relative expression levels. PCR primers used are listed in table S2.
dsRNA synthesis and mosquito gene silencing
dsRNAs were synthesized by in vitro transcription using the MEGAscript RNAi Kit (AM1626, Ambion). The DNA templates for transcription were PCR-amplified from the mosquito cDNA using the primer pairs containing the sequence of T7 polymerase promoter at the 5′ end (table S2). For gene silencing, each of 50 mosquitoes (4 to 6 days old) was injected intrathoracically with 69 nl of dsRNA solution (3 μg/μl) using a Nanoject II injector. Ten mosquitoes were collected on 2 or 3 days after injection, and the gene knockdown efficiency was evaluated using the real-time quantitative PCR before parasite infection. The experiments were repeated three times.
Mosquito phagocyte depletion using CLD LP
Mosquito phagocyte depletion was performed according to a method recently developed (10). Each naïve female mosquito was injected intrathoracically with 69 nl of control LP or CLD (#8901, Encapsula NanoSciences) using the Nanoject II injector. LP (100%) and different concentrations (100, 50, and 20%) of CLD diluted with 1x PBS were initially tested to determine the efficacy on phagocyte depletion. The CLD (20%) was used in the subsequent experiments. Injection was performed on naïve mosquitoes 1 day before the infected-mouse feeding.
Mosquito hemolymph perfusion and hemocyte counting.
Mosquito hemolymph was collected using the perfusion method (10). Two days after injection with LP, the mosquito was injected intrathoracically with 10 μl of the anticoagulant solution [60% Schneider’s insect medium, 10% FCS, 30% citrate buffer, 98 mM NaOH, 186 mM NaCl, 1.7 mM EDTA, and 41 mM citric acid (pH 4.5)] and was perfused through an incision made in the lateral abdomen. About 10 μl of hemolymph was collected from one mosquito.
Mosquito hemocyte staining using CM-DiI
Hemocytes were visualized by staining with Vybrant CM-DiI (V22888, Life Technologies), a lipophilic dye for specifically labeling mosquito hemocytes (10). Mosquitoes pretreated with LP were infected with parasites. Forty-eight hours pi, the mosquitoes were injected with 138 nl of 100 μM CM-DiI and incubated for 20 min at 22°C. Perfused hemolymph (10 μl) was collected onto a cover glass (#801010, NEST, China), and hemocytes were allowed to adhere to the slide for 30 min. Without washing, 4% paraformaldehyde was added to each well for fixation. The cells were incubated at room temperature for 30 min, washed three times with PBS, and observed under the fluorescence microscope (DM4B, Leica, Germany). Hemocytes per mosquito were quantified after counting 15 fields (40×) of microscope.
Antibodies and antiserum
The primary antibodies used in this study include the following: rabbit anti-HA [Western blot, 1:1000 dilution; IFA, 1:500 dilution; 3724S, Cell Signaling Technology (CST)], rabbit anti-Myc (Western blot, 1:1000; IFA, 1:500; 2276S, CST), mouse anti-V5 (Western blot, 1:1000; IFA, 1:500; A01724-100, GenScript), mouse anti-HA (IFA, 1:200; sc-57592, Santa Cruz Biotechnology), mouse anti-Myc (IFA, 1:200; sc-40, Santa Cruz Biotechnology), mouse anti–α-tubulin II (IFA, 1:1000; T6199, Sigma-Aldrich), mouse anti-nitrotyrisine (IFA, 1:1000; SAB5200009, Sigma-Aldrich), and Biotin-LC-Duramycin (IFA, 1:1000; 25690-100, Polysciences). The secondary antibodies include the following: goat anti-rabbit immunoglobulin G (IgG) horseradish peroxidase (HRP)–conjugated and goat anti-mouse IgG HRP-conjugated secondary antibodies (1:5000; Abcam); the Alexa Fluor 555–labeled goat anti-rabbit IgG, Alexa Fluor 555–labeled goat anti-mouse IgG, and Alexa Fluor 488–labeled goat anti-mouse IgG secondary antibodies from Thermo Fisher Scientific (1:500); and streptavidin-488 (IFA, 1:500; Biosciences). Antiserum of rabbit anti-P28 (Western blot, 1:1000; IFA, 1:1000), rabbit anti-GAP45 (IFA, 1:1000), and rabbit anti-BiP (Western blot, 1:1000) were prepared previously (23). Other polyclonal antiserum were prepared by immunization of rabbit with synthetic peptides or recombinant protein as follow: α-enolase (1:1000; KTYDLDFKTPNNDK), α-chitinase (1:1000; HTEKQYKSLSHVDALC), α-CTRP (1:1000; LNGGETPHNSNMEFENVENNDGIIEEENEDFEVIDANDPMW), α-WARP (1:1000; CNKNNPSSLTSERKTTIKN), and α-SRPN6 (1:1000; RRDQWRRQTCPHDED).
Parasite IFA
Parasites of different stages were fixed using freshly prepared 4% paraformaldehyde (P6148, Sigma-Aldrich) in PBS for 15 min at room temperature and transferred to a 24-well cell plate containing a poly-l-lysine (#E607015, Sangon Biotech)–pretreated coverslip at the bottom. The fixed cells were then immobilized on the coverslip via centrifuging the plate at 2000 rpm for 10 min and washed twice with PBS. The fixed cells were permeabilized with 0.1% Triton X-100 PBS solution for 10 min at room temperature, washed with PBS three times, blocked in 5% bovine serum albumin (BSA) solution for 60 min at room temperature, and incubated with the primary antibodies diluted in 5% BSA-PBS at 4°C for 12 hours. The coverslip was incubated with fluorescent-conjugated secondary antibodies for 1 hour at room temperature and washed with PBS three times. Cells were stained with Hoechst 33342 (#62249, Thermo Fisher Scientific), mounted in 90% glycerol solution, and sealed with nail polish. All images were captured and processed using identical settings on a Zeiss LSM 780 confocal microscope.
Mosquito midgut IFA
Mosquitoes fed on parasite-infected mice were dissected, and the blood bolus contents were removed. Midguts were fixed with 4% paraformaldehyde for 1 hour in PBS at room temperature, permeabilized with 100% methanol for 20 min at −20°C, and then blocked in 5% BSA solution for 1 hour at room temperature. Next, midguts were incubated overnight with the primary antibodies or antisera (1:1000 dilution in 5% BSA) and then with the Alexa Fluor 555– or Alexa Fluor 488–conjugated secondary antibodies (1:1000 diluted in 5% BSA). Midguts were then stained with Hoechst 33342, mounted in 90% glycerol solution, and sealed with nail polish. All images were captured and processed using identical settings on a Zeiss LSM 880 confocal microscope. To visualize the midgut in a Z-stack manner, midguts were fixed in 4% paraformaldehyde in PBS for 1 hour at room temperature and stained with anti-P28 antiserum. Then, the midguts were incubated with 6.6 μM Alexa Fluor 488–conjugated phalloidin (#21833, Invitrogen) for 20 min at room temperature. Z-stacks were approximately 20 μm in length to encompass the interface of the epithelial layer and the basal lamina, and images were taken at intervals of 1 μm. The final images were obtained and analyzed using Zeiss ZEN 3.0 software.
Protein extraction and immunoblot
Proteins were extracted from asexual blood parasites, gametocytes, and ookinete using buffer A [0.1% SDS, 1 mM dithiothreitol (DTT), 50 mM NaCl, and 20 mM tris-HCl (pH 8.0)] and from mosquitoes using a mosquito protein extraction buffer [8 M urea, 2% SDS, 5% β-mercaptoethanol, and 125 Mm tris-HCl (pH8.0)] containing protease inhibitor cocktail and phenylmethylsulfonyl fluoride. After ultrasonication, the protein solution was kept on ice for 15 min before centrifugation at 14,000g for 10 min at 4°C. The supernatant was lysed in Laemmli sample buffer stored at 4°C or immunoblotting. The protein samples were separated in SDS-PAGE and transferred to polyvinylidene difluoride membrane that was blocked in TBST (tris-buffered saline with Tween 20) buffer with 5% skim milk and then incubated with primary antibodies. After incubation, the membrane was washed three times with TBST and incubated with HRP-conjugated secondary antibodies. The membrane was washed four times in TBST before enhanced chemiluminescence detection.
Proximity ligation assay
The PLA assay detecting in situ protein interaction was performed using the kit (DUO92008, 92001, 92005, and 82049, Sigma-Aldrich). Purified ookinetes were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked with a blocking solution overnight at 4°C. The primary antibodies were diluted in the Duolink Antibody Diluent, added to the cells, and then incubated in a humidity chamber overnight at 4°C. The primary antibodies were removed, and the slides were washed with wash buffer A twice. The PLUS and MINUS PLA probe were diluted in the Duolink Antibody Diluent, added to the cells, and incubated in a preheated humidity chamber for 1 hour at 37°C. Next, cells were washed with wash buffer A and incubated with the ligation solution for 30 min at 37°C. Then, cells were washed with wash buffer A twice and incubated with the amplification solution for 100 min at 37°C in the dark. Cells were washed with 1× wash buffer B twice and 0.01× wash buffer B once. Last, cells were incubated with Hoechst 33342 and washed with PBS. Images were captured and processed using identical settings on a Zeiss LSM 880 confocal microscope.
Protein immunoprecipitation
Ookinetes (1.0 × 107) were lysed in 1 ml of protein extraction buffer A plus [0.01% SDS, 1 mM DTT, 50 mM NaCl, and 20 mM Tris-HCl (pH 8.0)]. After ultrasonication, the protein solution was incubated on ice for 15 min before centrifugation at 14,000g at 4°C for 10 min. One microgram of rabbit anti-HA antibody (#3724S, CST) and control IgG antibody (#2729, CST) was added to the supernatant, respectively, and the solution was incubated on a vertical mixer at 4°C for 15 hours. After incubation, 20 μl of buffer A plus prebalanced protein A/G beads (#20423, Pierce) was added and incubated for 5 hours. The beads were washed three times with buffer A plus before elution with Laemmli buffer.
Scanning electron microscopy analysis
The purified ookinetes were fixed in 2.5% glutaraldehyde solution in 0.1 M phosphate buffer overnight, rinsed three times with PBS, and then postfixed with 1% osmium tetroxide for 2 hours. The fixed samples were dehydrated using a graded acetone series, CO2- dried in a critical-point drying device (K850, Emitech, USA), and gold-coated in a sputter coater (JFC-1600, JEOL, USA) as detailed previously (55). The samples were imaged in a JSM-6390LV scanning electron microscope.
TEM analysis
TEM experiments were performed using the protocol as described previously (56). Purified ookinetes were prefixed with 4% glutaraldehyde in 0.1 M phosphate buffer at 4°C overnight, rinsed three times with PBS, postfixed with 1% osmium acid for 2 hours, and rinsed three times with PBS. The samples were dehydrated with concentration-gradient acetone. After embedding and slicing, thin sections were stained with uranyl acetate and lead citrate before imaging. All samples were imaged under a HT-7800 electron microscope (USA).
Peroxidase activity detection of mosquito midgut
Midgut staining with DAB (3,3-diaminobenzidine) was performed as described (29). Briefly, infected mosquito midguts were dissected at 24 hours pf. Midguts were fixed in 0.5% glutaraldehyde for 10 min at room temperature, washed in PBS, and developed for DAB activity. Samples were incubated at room temperature with 2.5 mM DAB (D8001, Sigma-Aldrich) and 1 mM H2O2 (18304, Sigma-Aldrich) in PBS (pH 6.5) and continuously observed under the microscope.
NBD-lipid uptake
1-oleoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoserine (NBD-PS, #810194C), 1-oleoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoethanolamine (NBD-PE, #810155C), 1-oleoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphocholin (NBD-PC, #810132C), and N-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-sphingosine-1-phosphocholine (NBD-SM, #810218C) were purchased from Avanti Polar Lipids (Birmingham, USA). NBD-phospholipid uptake experiments were performed according to the procedures (57). Ookinetes (1.0 × 105) were collected from the in vitro culture, washed (2000 rpm for 5 min at 22°C), and equilibrated at 22°C for 15 min in 500 μl of Hanks’ balanced salt solution (pH 7.4) containing glucose (1 g/liter) (HBSS-glucose). A 500 μl of 3 mM NBD-phospholipid in HBSS-glucose was added to the ookinete suspension and incubated at 22°C for 1 hour. After centrifugation (2000 rpm for 5 min at 22°C) and discarding the supernatant, the ookinetes were mixed with 500 μl of ice-cold HBSS-glucose containing 5% fatty acid–free BSA (#9048-46-8, Sigma-Aldrich) to extract NBD-lipids incorporated into the exoplasmic leaflet of the ookinete plasma membrane, as well as unincorporated ones. The ookinete suspension was then placed in a petri dish for measuring the fluorescence signal of NBD-lipids inside the ookinetes. The fluorescence signals were captured using the Zeiss LSM 780 confocal microscope, and the signal intensity per cell was calculated using the ImageJ software. To test the effect of ATP depletion on the phospholipid uptake, ookinetes were preincubated in ATP depletion medium (HBSS without glucose but containing 20 mM NaN3) for 1 hour at 22°C before the NBD-lipid uptake assay (40).
Phospholipid staining
For annexin V staining, the live ookinetes or dead ookinetes (after 60°C treatment for 5 min) were incubated in 1:100 Alexa Fluor 488–annexin V (#A13201, Invitrogen) and 1:100 propidium iodide (#P3566, Invitrogen) for 15 min, washed in PBS (2000 rpm at 22°C for 5 min), and mounted on slides. For Duramycin staining, the live or dead ookinetes were incubated in biotinylated Duramycin (0.25 μg/ml) (58) for 30 min, stained with Alexa Fluor 488–streptavidin (1 μg/ml) (#S32354, Invitrogen) and Hoechst 33342 for 30 min, washed in PBS (2000 rpm at 22°C for 5 min), and mounted on slides. The ookinetes were analyzed using a Zeiss LSM 780 confocal microscope.
Auxin-induced protein depletion
A stock solution of 250 mM auxin/IAA (Sigma-Aldrich, I2886) was prepared using the 100% EtOH. Mock treatment includes an equivalent volume of 100% EtOH. To determine the degradation efficiency of CDC50C-AID fusing protein, ookinetes (1.0 × 105) were incubated with 1 mM IAA dissolved in ookinete media [RPMI 1640, 10% FCS, 100 μM XA, and 25 mM Hepes (pH 8.0)] at 37°C for 1 hour. After washing for three times in PBS (2000 rpm for 5 min at room temperature), the cells were immediately fixed with 4% paraformaldehyde and transferred onto the slide for IFA. After imaging by Zeiss LSM 780 confocal microscope, the intensities of fluorescence signal of targeting proteins were quantitatively analyzed using the ImageJ software.
Bioinformatics analysis and tools
The genomic sequences of Plasmodium genes were downloaded from the Plasmodium database of PlasmoDB (http://plasmodb.org). The genomic sequences of mosquito genes were downloaded from the A. Stephensi database (www.vectorbase.org/). The sgRNAs of target gene were designed using EuPaGDT (http://grna.ctegd.uga.edu/). The transmembrane domains of proteins were identified using the PROTTER Server (http://wlab.ethz.ch/protter/start/) (59). Multiple sequence alignments were performed by ClustalW in MEGA 7.0 (60).
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0. Data collected as raw values are shown as means ± SEM or means ± SD. Two-tailed t test or Mann-Whitney test was used to compare differences between treated groups and their paired controls. Details of statistical methods are reported in the figure legend. n represents the number of mosquitos or parasite cells tested in each group or experimental replications.
Acknowledgments
We thank B. Wang, D. Baker, and M. Jacobs-Lorena for the comments on this manuscript. Funding: This work was supported by the National Natural Science Foundation of China (31772443, 31872214, and 31970387), the Natural Science Foundation of Fujian Province (2019J05010), and the 111 Project sponsored by the State Bureau of Foreign Experts and Ministry of Education of China (BP2018017). Author contributions: Z.Y., Y.S., and H.G. generated the modified parasites and conducted the phenotype analysis, IFA assay, image analysis, mosquito experiments, and biochemical experiments. S.Y. conducted the mosquito experiments. J.Y. and H.C. supervised the work. Z.Y., H.C., and J.Y. analyzed the data, and J.Y. wrote the manuscript. Competing interests: The authors declare that have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/30/eabf6015/DC1
REFERENCES AND NOTES
- 1.World Health Organization, World malaria report 2018 (WHO, 2018).
- 2.Sinden R. E., Molecular interactions between Plasmodium and its insect vectors. Cell. Microbiol. 4, 713–724 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Blandin S., Shiao S. H., Moita L. F., Janse C. J., Waters A. P., Kafatos F. C., Levashina E. A., Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116, 661–670 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Simoes M. L., Caragata E. P., Dimopoulos G., Diverse host and restriction factors regulate mosquito-pathogen interactions. Trends Parasitol. 34, 603–616 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Molina-Cruz A., Canepa G. E., Barillas-Mury C., Plasmodium P47: A key gene for malaria transmission by mosquito vectors. Curr. Opin. Microbiol. 40, 168–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blandin S. A., Marois E., Levashina E. A., Antimalarial responses in Anopheles gambiae: From a complement-like protein to a complement-like pathway. Cell Host Microbe 3, 364–374 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Smith R. C., Vega-Rodriguez J., Jacobs-Lorena M., The Plasmodium bottleneck: Malaria parasite losses in the mosquito vector. Mem. Inst. Oswaldo Cruz 109, 644–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraiture M., Baxter R. H., Steinert S., Chelliah Y., Frolet C., Quispe-Tintaya W., Hoffmann J. A., Blandin S. A., Levashina E. A., Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe 5, 273–284 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Povelones M., Waterhouse R. M., Kafatos F. C., Christophides G. K., Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science 324, 258–261 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon H., Smith R. C., Chemical depletion of phagocytic immune cells in Anopheles gambiae reveals dual roles of mosquito hemocytes in anti-Plasmodium immunity. Proc. Natl. Acad. Sci. U.S.A. 116, 14119–14128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Y. S., Thompson J., Kafatos F. C., Barillas-Mury C., Molecular interactions between Anopheles stephensi midgut cells and Plasmodium berghei: The time bomb theory of ookinete invasion of mosquitoes. EMBO J. 19, 6030–6040 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennink S., Kiesow M. J., Pradel G., The development of malaria parasites in the mosquito midgut. Cell. Microbiol. 18, 905–918 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Annoura T., van Schaijk B. C., Ploemen I. H., Sajid M., Lin J. W., Vos M. W., Dinmohamed A. G., Inaoka D. K., Rijpma S. R., van Gemert G. J., Chevalley-Maurel S., Kielbasa S. M., Scheltinga F., Franke-Fayard B., Klop O., Hermsen C. C., Kita K., Gego A., Franetich J. F., Mazier D., Hoffman S. L., Janse C. J., Sauerwein R. W., Khan S. M., Two Plasmodium 6-Cys family-related proteins have distinct and critical roles in liver-stage development. FASEB J. 28, 2158–2170 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Arredondo S. A., Kappe S. H. I., The s48/45 six-cysteine proteins: Mediators of interaction throughout the Plasmodium life cycle. Int. J. Parasitol. 47, 409–423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ukegbu C. V., Giorgalli M., Yassine H., Ramirez J. L., Taxiarchi C., Barillas-Mury C., Christophides G. K., Vlachou D., Plasmodium berghei P47 is essential for ookinete protection from the Anopheles gambiae complement-like response. Sci. Rep. 7, 6026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molina-Cruz A., Garver L. S., Alabaster A., Bangiolo L., Haile A., Winikor J., Ortega C., van Schaijk B. C., Sauerwein R. W., Taylor-Salmon E., Barillas-Mury C., The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. Science 340, 984–987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramphul U. N., Garver L. S., Molina-Cruz A., Canepa G. E., Barillas-Mury C., Plasmodium falciparum evades mosquito immunity by disrupting JNK-mediated apoptosis of invaded midgut cells. Proc. Natl. Acad. Sci. U.S.A. 112, 1273–1280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ukegbu C. V., Giorgalli M., Tapanelli S., Rona L. D. P., Jaye A., Wyer C., Angrisano F., Blagborough A. M., Christophides G. K., Vlachou D., PIMMS43 is required for malaria parasite immune evasion and sporogonic development in the mosquito vector. Proc. Natl. Acad. Sci. U.S.A. 117, 7363–7373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi T., Menon A. K., Transbilayer lipid asymmetry. Curr. Biol. 28, R386–R391 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y., Wei J., Cui H., Liu C., Zhi Y., Jiang Z., Li Z., Li S., Yang Z., Wang X., Qian P., Zhang C., Zhong C., Su X. Z., Yuan J., An intracellular membrane protein GEP1 regulates xanthurenic acid induced gametogenesis of malaria parasites. Nat. Commun. 11, 1764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C., Xiao B., Jiang Y., Zhao Y., Li Z., Gao H., Ling Y., Wei J., Li S., Lu M., Su X. Z., Cui H., Yuan J., Efficient editing of malaria parasite genome using the CRISPR/Cas9 system. MBio 5, e01414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C., Gao H., Yang Z., Jiang Y., Li Z., Wang X., Xiao B., Su X. Z., Cui H., Yuan J., CRISPR/Cas9 mediated sequential editing of genes critical for ookinete motility in Plasmodium yoelii. Mol. Biochem. Parasitol. 212, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao H., Yang Z., Wang X., Qian P., Hong R., Chen X., Su X. Z., Cui H., Yuan J., ISP1-anchored polarization of GCbeta/CDC50A complex initiates malaria ookinete gliding motility. Curr. Biol. 28, 2763–2776.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Dessens J. T., Beetsma A. L., Dimopoulos G., Wengelnik K., Crisanti A., Kafatos F. C., Sinden R. E., CTRP is essential for mosquito infection by malaria ookinetes. EMBO J. 18, 6221–6227 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langer R. C., Vinetz J. M., Plasmodium ookinete-secreted chitinase and parasite penetration of the mosquito peritrophic matrix. Trends Parasitol. 17, 269–272 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Yuda M., Yano K., Tsuboi T., Torii M., Chinzei Y., von Willebrand Factor A domain-related protein, a novel microneme protein of the malaria ookinete highly conserved throughout Plasmodium parasites. Mol. Biochem. Parasitol. 116, 65–72 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Dessens J. T., Siden-Kiamos I., Mendoza J., Mahairaki V., Khater E., Vlachou D., Xu X. J., Kafatos F. C., Louis C., Dimopoulos G., Sinden R. E., SOAP, a novel malaria ookinete protein involved in mosquito midgut invasion and oocyst development. Mol. Microbiol. 49, 319–329 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Abraham E. G., Pinto S. B., Ghosh A., Vanlandingham D. L., Budd A., Higgs S., Kafatos F. C., Jacobs-Lorena M., Michel K., An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 102, 16327–16332 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta L., Kumar S., Han Y. S., Pimenta P. F., Barillas-Mury C., Midgut epithelial responses of different mosquito-Plasmodium combinations: The actin cone zipper repair mechanism in Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 102, 4010–4015 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grotendorst C. A., Kumar N., Carter R., Kaushal D. C., A surface protein expressed during the transformation of zygotes of Plasmodium gallinaceum is a target of transmission-blocking antibodies. Infect. Immun. 45, 775–777 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ecker A., Pinto S. B., Baker K. W., Kafatos F. C., Sinden R. E., Plasmodium berghei: Plasmodium perforin-like protein 5 is required for mosquito midgut invasion in Anopheles stephensi. Exp. Parasitol. 116, 504–508 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Childs L. M., Cai F. Y., Kakani E. G., Mitchell S. N., Paton D., Gabrieli P., Buckee C. O., Catteruccia F., Disrupting mosquito reproduction and parasite development for malaria control. PLOS Pathog. 12, e1006060 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weathersby A. B., The role of the Stolviach wall in the exogenous development of Plasmodium Galllnaceum as studied by means of Haelviocoel injections of susceptible and refractory mosquitoes. J. Infect. Dis. 91, 198–205 (1952). [DOI] [PubMed] [Google Scholar]
- 34.Sinden R. E., Butcher G. A., Beetsma A. L., Maintenance of the Plasmodium berghei life cycle. Methods Mol. Med. 72, 25–40 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Al-Olayan E. M., Beetsma A. L., Butcher G. A., Sinden R. E., Hurd H., Complete development of mosquito phases of the malaria parasite in vitro. Science 295, 677–679 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Carter V., Nacer A. M., Underhill A., Sinden R. E., Hurd H., Minimum requirements for ookinete to oocyst transformation in Plasmodium. Int. J. Parasitol. 37, 1221–1232 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takada N., Naito T., Inoue T., Nakayama K., Takatsu H., Shin H. W., Phospholipid-flipping activity of P4-ATPase drives membrane curvature. EMBO J. 37, 29599178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coleman J. A., Vestergaard A. L., Molday R. S., Vilsen B., Andersen J. P., Critical role of a transmembrane lysine in aminophospholipid transport by mammalian photoreceptor P4-ATPase ATP8A2. Proc. Natl. Acad. Sci. U.S.A. 109, 1449–1454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takatsu H., Tanaka G., Segawa K., Suzuki J., Nagata S., Nakayama K., Shin H. W., Phospholipid flippase activities and substrate specificities of human type IV P-type ATPases localized to the plasma membrane. J. Biol. Chem. 289, 33543–33556 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grant A. M., Hanson P. K., Malone L., Nichols J. W., NBD-labeled phosphatidylcholine and phosphatidylethanolamine are internalized by transbilayer transport across the yeast plasma membrane. Traffic 2, 37–50 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Palmgren M. G., Nissen P., P-type ATPases. Annu. Rev. Biophys. 40, 243–266 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Paulusma C. C., Folmer D. E., Ho-Mok K. S., de Waart D. R., Hilarius P. M., Verhoeven A. J., Oude Elferink R. P., ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology 47, 268–278 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Bagchi S., Fredriksson R., Wallen-Mackenzie A., In situ proximity ligation assay (PLA). Methods Mol. Biol. 1318, 149–159 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Bushell E., Gomes A. R., Sanderson T., Anar B., Girling G., Herd C., Metcalf T., Modrzynska K., Schwach F., Martin R. E., Mather M. W., McFadden G. I., Parts L., Rutledge G. G., Vaidya A. B., Wengelnik K., Rayner J. C., Billker O., Functional profiling of a plasmodium genome reveals an abundance of essential genes. Cell 170, 260–272.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M., Wang C., Otto T. D., Oberstaller J., Liao X., Adapa S. R., Udenze K., Bronner I. F., Casandra D., Mayho M., Brown J., Li S., Swanson J., Rayner J. C., Jiang R. H. Y., Adams J. H., Uncovering the essential genes of the human malaria parasite Plasmodium falciparumby saturation mutagenesis. Science 360, eaap7847 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu C., Yang Z., Cai M., Shi Y., Cui H., Yuan J., Generation of Plasmodium yoelii malaria parasite for conditional degradation of proteins. Mol. Biochem. Parasitol. 241, 111346 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Philip N., Waters A. P., Conditional degradation of plasmodium calcineurin reveals functions in parasite colonization of both host and vector. Cell Host Microbe 18, 122–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kenthirapalan S., Waters A. P., Matuschewski K., Kooij T. W., Functional profiles of orphan membrane transporters in the life cycle of the malaria parasite. Nat. Commun. 7, 10519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar A., Srivastava P., Sirisena P., Dubey S. K., Kumar R., Shrinet J., Sunil S., Mosquito innate immunity. Insects 9, 95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segawa K., Kurata S., Yanagihashi Y., Brummelkamp T. R., Matsuda F., Nagata S., Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 344, 1164–1168 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Frenal K., Dubremetz J. F., Lebrun M., Soldati-Favre D., Gliding motility powers invasion and egress in Apicomplexa. Nat. Rev. Microbiol. 15, 645–660 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Spielmann T., Montagna G. N., Hecht L., Matuschewski K., Molecular make-up of the Plasmodium parasitophorous vacuolar membrane. Int. J. Med. Microbiol. 302, 179–186 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Agop-Nersesian C., Niklaus L., Wacker R., Theo Heussler V., Host cell cytosolic immune response during Plasmodium liver stage development. FEMS Microbiol. Rev. 42, 324–334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinden R. E., Billingsley P. F., Plasmodium invasion of mosquito cells: Hawk or dove? Trends Parasitol. 17, 209–211 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Orfano A. S., Nacif-Pimenta R., Duarte A. P., Villegas L. M., Rodrigues N. B., Pinto L. C., Campos K. M., Pinilla Y. T., Chaves B., Barbosa Guerra M. G., Monteiro W. M., Smith R. C., Molina-Cruz A., Lacerda M. V., Secundino N. F., Jacobs-Lorena M., Barillas-Mury C., Pimenta P. F. P., Species-specific escape of Plasmodium sporozoites from oocysts of avian, rodent, and human malarial parasites. Malar. J. 15, 394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferguson D. J., Henriquez F. L., Kirisits M. J., Muench S. P., Prigge S. T., Rice D. W., Roberts C. W., McLeod R. L., Maternal inheritance and stage-specific variation of the apicoplast in Toxoplasma gondii during development in the intermediate and definitive host. Eukaryot. Cell 4, 814–826 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naito T., Takatsu H., Miyano R., Takada N., Nakayama K., Shin H. W., Phospholipid flippase ATP10A translocates phosphatidylcholine and is involved in plasma membrane dynamics. J. Biol. Chem. 290, 15004–15017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wehman A. M., Poggioli C., Schweinsberg P., Grant B. D., Nance J., The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr. Biol. 21, 1951–1959 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Omasits U., Ahrens C. H., Muller S., Wollscheid B., Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Stecher G., Liu L., Sanderford M., Peterson D., Tamura K., Kumar S., MEGA-MD: Molecular evolutionary genetics analysis software with mutational diagnosis of amino acid variation. Bioinformatics 30, 1305–1307 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/30/eabf6015/DC1


