Abstract
COVID-19 exhibits a global health threat among the elderly and the population with underlying health conditions. During infection, the host's innate immune response acts as a frontline of defense by releasing cytokines such as type I interferon (IFN α and β) thereby initiating antiviral activity. However, this particular interferon response is interrupted by factors such as SARS-CoV-2 non-structural proteins, aging, diabetes, and germ-line errors eventually making the host more susceptible to illness. Therefore, enhancing the host's innate immune response by administering type I IFN could be an effective treatment against COVID-19. Here, we highlight the importance of innate immune response and the role of IFN β monotherapy against COVID-19.
Keywords: Coronavirus disease 2019, Severe acute respiratory syndrome, Type I Interferon, Innate immunity, Interferon β monotherapy
Abbreviations
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- CTSB/L
cathepsin B/L
- TMPRSS2
transmembrane protease, serine 2
- ACE2
angiotensin-converting enzyme 2 receptor
- AEC
airway epithelial cell
- PAMPS
pathogen-associated molecular patterns
- ssRNA
single-stranded RNA
- RLRs
RIG-I like receptor
- CARD
caspase activator, and recruitment domain
- MAVS
mitochondrial antiviral signaling
- TBK 1
TANK-binding kinase 1
- TRAF3
TNF receptor-associated factor 3
- IRF 3 & 7
interferon regulatory factor 3 and 7
- IFNAR
interferon-alpha/beta receptor
- Tyk 2
tyrosine kinase 2
- Jak 1
Janus kinase 1
- STAT 1 & 2
signal transducer and activator of transcription 1 and 2
- ISGs
interferon-stimulated genes
- IFITM 3
interferon-induced transmembrane protein 3
- PAI-1
plasminogen activator inhibitor
- ssRNA
single-stranded RNA
- PKR
protein kinase R
- IKK
IκB kinase complex
- NF κB
nuclear factor kappa-light-chain-enhancer of an activated B cell
- eIFα2
eukaryotic initiation factor α 2
- IFN β
interferon β
- IRF 9
interferon regulatory factor 9
- IFN
interferon
- HAM
HTLV-1 associated myelopathy
- CHC
chronic hepatitis C
- COPD
chronic obstructive pulmonary disease
- IM
intramuscular
- SC
subcutaneous
- NEB
nebulization
1. Introduction
In December 2019, the epidemic in Wuhan, China, was designated as a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing coronavirus disease (COVID-19). In March 2020, the COVID-19 outbreak was announced as a pandemic [1]. As of mid-July 2021, COVID- 19 has infected more than 180 million and killed more than 4 million people worldwide [2]. With the lack of specific therapeutic interventions, the COVID-19 exhibits a potential threat of fatality among the elderly and others with underlying health ailments [3].
SARS-CoV-2 is a zoonotic virus with a bilipid membrane coated with spike proteins, membrane proteins, and envelope proteins providing a stronger protective layer for viral single-stranded RNA (ssRNA) [4]. SARS-CoV-2 enters the airway and infects airway epithelial cells (AECs) initially causing mild pneumonia followed by dyspnea and finally leading to respiratory failure [5]. Simultaneously, SARS-CoV-2 transmits from the infected individual to the non-infected primarily through respiratory droplets [6]. During infection, the host's innate immune response acts as a frontline of defense by releasing cytokines such as type I interferon (IFN α and IFN β). The type I IFN induces hundreds of interferon-stimulated genes (ISGs) which set up the antiviral state thereby protecting the host from the infection [7]. However, the type I IFN production is impaired by SARS-CoV-2 non-structural proteins (nsp), aging, diabetes, and germ-line errors [[8], [9], [10], [11], [12]]. Thus, the deficiency of type I IFN leads to poor innate immune responses in the host against SARS-CoV-2 infection, eventually leading to severe illness. Therefore, enhancing the host's innate immune response by administering type I IFN could be a key to prevent the host from COVID-19 severity. We evaluated here the importance of the innate immune response during SARS-CoV-2 infection and the molecular mechanism of IFN β-mediated antiviral activity against SARS-CoV-2 infection and concluded that the IFN β monotherapy as an attractive potent treatment option against SARS-CoV-2.
2. Host-virus interaction
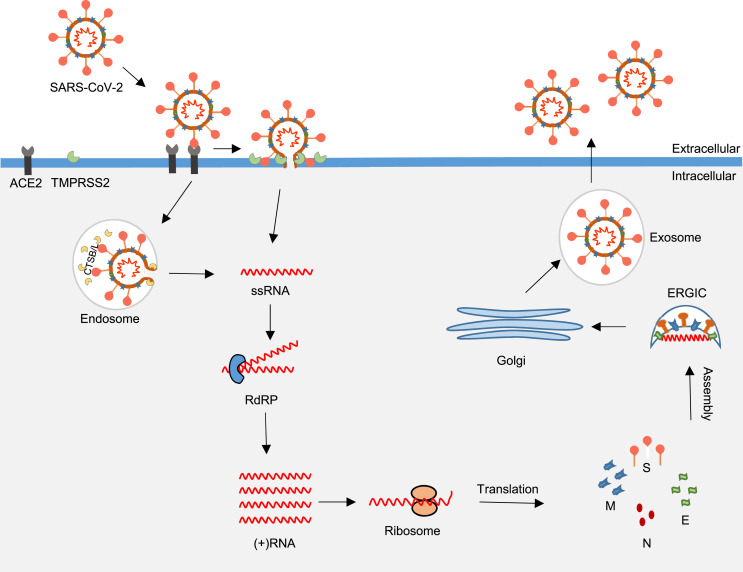
When the SARS-CoV-2 enters the airway, the virus spike protein (S) recognizes and binds the angiotensin-converting enzyme-2 receptor (ACE-2) on the surface of airway epithelial cells (AECs) [13]. The receptor-bound spike protein is cleaved by the transmembrane protease serine 2 (TMPRSS2) followed by membrane-fusion or endosomal entry into the AECs [14]. After entering the host cell, the virion releases viral ssRNA into the cytoplasm. The viral ssRNA replicates in the cytoplasm via RNA-dependent RNA polymerase (RdRP), a viral polymerase, followed by translation using cellular machinery [15]. The replicated viral particles are assembled in the endoplasmic reticulum-Golgi apparatus intermediate compartment (ERGIC) followed by exocytosis [14].
In the host-virus interaction, ACE-2 and TMPRSS2 play a huge role in recognizing and cleaving the S protein eventually leading to viral entry. Unlike SARS-CoV-1, SARS-CoV-2 S protein has a unique four amino acid (proline-arginine-arginine-alanine) insertion at the S1/S2 site which can be primed by furin. In the in vitro, the S1/S2 priming plays a crucial role in subsequent cleavage activation by TMPRSS2 in the membrane-fusion entry (early) in Calu-3 cells, on the other hand, the S1/S2 priming is not essential in the endosomal entry (late) in Vero E6 cells [16]. Notably, SARS-CoV-2 is shown to be more dependent on TMPRSS2 mediated membrane-fusion entry than SARS-CoV-1 in TMPRSS2+ 293T-ACE2 cells [17]. In the in-vitro study, E−64d and camostat, a cathepsin B/L and TMPRSS2 inhibitor, completely inhibited entry of PV SARS-S and PV SARS-2-S into TMPRSS2+ Caco-2 cells. On the other hand, when the TMPRSS2+ Caco-2 cells were treated with either E−64d or camostat, the cells showed only partial inhibition against viral entry [18]. Similar effects had been observed in the TMPRSS2+ 293T-ACE2 cells when treated with hydroxychloroquine, an interferer of endosomal acidification, and camostat [17]. These findings show that both the membrane-fusion and endosomal entry should be targeted to inhibit the host cell from SARS-CoV-2 infection. Therefore, we hypothesize that the combination of TMPRSS2 and Cathepsin B/L inhibitors would be an effective treatment option against COVID-19 which warrants further animal studies and clinical trials.
Similarly, Vero cells pre-incubated with anti-ACE2 antibodies significantly inhibited entry of pseudovirions (PV) harboring SARS-S and SARS-2-S [18]. Of note, ACE2 plays an important role in the renin-angiotensin-aldosterone system (RAAS). The ACE converts angiotensin I to angiotensin II thereby stimulating inflammation, vasoconstriction, fibrosis, apoptosis, and fluid retention. Concurrently, ACE2 converts angiotensin I & II into angiotensin-(1–9) & angiotensin-(1–7) and stimulates the opposite effect eventually counterbalancing the ACE effect. As the SARS-CoV-2 infection disrupts the ACE/ACE2 physiological balance, it leads to RAAS hyperactivation eventually causing acute lung injury, pulmonary edema, high blood pressure, and fibrosis [19]. Therefore, anti-ACE2 antibodies would disrupt the cellular homeostasis eventually supporting COVID-19 progression, hence, we hypothesize anti-ACE2 antibodies would be an inappropriate treatment option against COVID-19. Intriguingly, innate immune response plays a central role in controlling both the membrane fusion and endosomal viral entry with undisrupted cellular homeostasis and safeguards the non-infected cells from pathogenicity followed by initiating adaptive immunity. Hence, the innate immune system acts as the first line of defense in viral infections by preventing the viral invasion or replication in the host (see Fig. 1 ) [7].
Fig. 1.
The host-virus interaction in the airway epithelial cell. The SARS-CoV-2 spike protein binds ACE-2 followed by TMPRSS2-mediated proteolytic cleavage of the receptor-bound spike protein. The virus enters the host through the endosomal-mediated or membrane-fusion entry. After entering the host, the virion releases ssRNA into the cytoplasm. The ssRNA replicates via RdRP and translates using cellular machinery. The replicated viral particles (S, E, M and N) are assembled in the ERGIC followed by exocytosis. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; CTSB/L, cathepsin B/L; TMPRSS2, transmembrane protease, serine 2; ACE2, angiotensin-converting enzyme 2 receptor; RdRP, RNA-dependent RNA polymerase; ERGIC, Endoplasmic Reticulum-Golgi apparatus Intermediate Compartment; E, envelope proteins; S, spike proteins; M, membrane proteins; and N, nucleocapsid proteins.
3. Innate immune response
In the innate immune response, when a pathogen-associated molecular pattern (PAMP) is generated in the host cell, it is recognized by an intracellular pathogen recognition receptor (PRR) such as retinoic acid-inducible gene I (RIG-I) like receptor (RLR). Activated RLR undergoes ubiquitination by E3 ligase and the CARD domain of ubiquitinated RLR interacts with the CARD domain of mitochondrial antiviral signaling protein (MAVS). This process is followed by MAVS interaction with nuclear factor-kB (NF-kB) and interferon regulatory factor (IRF) leading to the expression of pro-inflammatory cytokines, chemokines, and type I and type III interferon (IFN α/β and IFN λ) [20]. The pro-inflammatory cytokines and chemokines recruit lymphocytes and leukocytes to the site of infection thereby initiating an inflammatory response. On the other hand, type I interferon induces interferon-stimulated genes (ISGs) and anti-inflammatory cytokines through the JAK-STAT signaling pathway (Fig. 2 ) where ISGs inhibit viral replication while the anti-inflammatory cytokines compensate the inflammatory response. This innate immune response acts as a frontline of defense in preventing the host from viral infections and severe inflammation [21].
Fig. 2.
A proposed model of host innate immunity induced by PAMPs. (A) The ssRNA enters the AEC and is recognized by intracellular receptors such as RLRs eventually transforming to an active form. The activated RIG-1 undergoes ubiquitination by E3 ligases and the CARD domain of ubiquitinated RIG-I interacts with the CARD domain of MAVS. The MAVS activates TBK1 and NF-kB through TRAF3 and IKK complex. The TBK1 phosphorylates IRF 7 and IRF 3 thereby stimulating type I IFN production; On the other hand, NF kB induces pro-inflammatory cytokine production. Aging is associated with the downregulation of proteins such as RLR, E3, and IRFs which impairs type I IFN production. Furthermore, nsp 6, N protein, nsp 13, nsp 14 and ORF 6 impair type I IFN production by inhibiting viral RNA sensing, TBK1 phosphorylation, and IRF phosphorylation. Conversely, N protein directly interacts with NF-kB and promotes cytokine release. (B) The interferon binds the IFNAR induces STAT homodimerization and heterodimerization thereby promoting the expression of ISGs and anti-inflammatory cytokines. Intriguingly, nsp 1, nsp 6, N protein, and ORF 6 inhibit STAT 1/STAT 2 phosphorylation and nuclear translocation eventually impairing interferon signaling. AEC, airway epithelial cell; PAMPS, pathogen-associated molecular patterns; ssRNA, single-stranded RNA; RLRs, RIG-I like receptor; CARD, caspase activator, and recruitment domain; MAVS, mitochondrial antiviral signaling; TBK 1, TANK-binding kinase 1; TRAF3, TNF receptor-associated factor 3; IRF 3 & 7, interferon regulatory factor 3 and 7; IFNAR, interferon-alpha/beta receptor; Tyk1, tyrosine kinase 1; Jak 2, Janus kinase 2; STAT 1 & 2, signal transducer and activator of transcription 1 and 2; ISGs, interferon-stimulated genes; nsp, non-structural proteins; ORF, open reading frame proteins; N, SARS-CoV-2 N protein.
Recently, several studies conducted on severe COVID-19 patients reported an elevated level of proinflammatory cytokines such as interleukin 6 (IL-6), IL-1β, IL-2, interleukin 1 receptor antagonist (IL-1RA), and tumor necrosis factor (TNF α) in the blood. Furthermore, chemokines such as C-X-C motif ligand 8 (CXC8), CXCL9, CXCL16, C–C motif ligand (CCL8), interferon-induced protein (IP-10), monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory proteins (MIP-1α), MIP-1β, CXCL8, granulocyte-macrophage colony-stimulating factor (GM-CSF) are found in the bronchoalveolar lavage fluid (BALF) [22]. Notably, an in vitro study conducted on SARS-CoV-1 reported that the SARS-CoV-1 nucleocapsid (N) protein can directly interact with NF-kB and facilitates nuclear translocation, eventually manipulating cytokine release [23]. Similarly, human monocytes and macrophages cultured in the presence of SARS-CoV-2 N protein and S protein expressed high levels of cytokines such as IL-6, IL1B, IL-10 and TNF [24]. Supportively, SARS-CoV-2 N protein promotes NF-kB signaling by targeting IKK complex eventually initiating cytokine release in vitro [25]. Additionally, SARS-CoV-2 N protein antagonizes interferon signaling by impairing phosphorylation and nuclear translocation of STAT 1 and STAT 2 in 293T cells [26]. However, the studies were conducted in the non-infected cells either expressing or being treated with SARS-CoV-2 N and S protein. Therefore, the role of SARS-CoV-2 N protein and S protein in NFkB signaling and cytokine storm during infection warrants further investigation.
Concurrently, lower levels of type I and type III IFN in the blood had also been reported among COVID-19 severity than the control [27,28]. Similar effects had also been observed in coronaviruses such as SARS-CoV-1 and middle eastern respiratory syndrome coronavirus (MERS-CoV). The interferon antagonistic effect may be due to SARS-CoV-2 N protein, non-structural proteins (nsp) and open reading frame proteins (ORF). In vitro studies reported that among 27 viral proteins of SARS-CoV-2, such as nsp 1, nsp 6, nsp 13, nsp 14, nsp 15, and ORF 6 can be potent interferon antagonists (Fig. 2) [11,12]. Specifically, N protein, nsp 1, nsp 6 and ORF 6 impair ISGs expression by inhibiting STAT 1/STAT 2 phosphorylation and nuclear translocation in both SARS-CoV-1 and SARS CoV-2 [11,29,30]. Notably, nsp 14, a guanine-N7-methyltransferase, methylates the 5’ cap structure of the viral RNA to mimic the host RNA eventually leading to virus immune evasion. These findings point at the ability of SARS CoV-2 in impairing interferon production and signaling leading to a muted innate immune response. However, the role of SARS-CoV-2 nsp and ORF in virus immune evasion, interferon production, and interferon signaling needs further evaluation. Additionally, the type I interferon production is also impaired by, i) the downregulation of proteins such as RLRs, E3 ligases, and IRFs due to aging (11), ii) germline errors in toll-like receptors 3 (TLR3) and IRF 7 [10], and iii) enhanced glucose levels (diabetes) in the blood [8] (Fig. 2). These host immanent factors render a host more susceptible to SARS-CoV-2 infection and severity.
In innate immunity, type I and III interferon acts as a mediator of the antiviral activity and anti-inflammatory response thereby preventing viral infection and severe tissue damage. However, coronaviruses such as SARS-CoV-1, MERS-CoV, and SARS-CoV-2 are known to manipulate the innate immune response by targeting interferon production and signaling. Unlike SARS-CoV-1, the SARS-CoV-2 is more sensitive to type I IFN treatment in vitro [31]. Hence, it could be beneficial to enhance the innate immune response in the infected individual to control the severity of COVID-19, and type I IFN therapy could be an effective treatment option against COVID -19 [32].
4. Pathogenesis of COVID-19
As mentioned earlier, COVID-19 severity has been associated with uncontrolled cytokine release leading to cytokine storm syndrome. Comparatively, the cytokines such as IL-2, IL-4, IL-6, IL-1β, and TNF α were significantly elevated especially in severe patients. In the clinic, an elevated level of IL-6 was associated with disease progression and poor prognosis among severe COVID-19 patients [33]. In SARS-CoV-1, N protein directly promotes IL-6 expression through NF-kB in vitro [23]. Similarly, SARS-CoV-2 N protein may exhibit a potential mechanism in IL-6 elevation, as it shares a 90 % amino acid identity with SARS-CoV-1 N protein [34]. Excessive IL-6 enhances vascular endothelial growth factor (VEGF) and destabilizes vascular endothelial cadherin (VE-cadherin) thereby causing high vascular permeability in vitro [35]. The increased vascular permeability infiltrates excessive macrophages and monocytes thus contributing to severe inflammation, as well as increases fluid influx into the lungs leading to pulmonary edema [36]. Additionally, elevated IL-6 signaling increases fibrinogen synthesis and platelet hyperactivity in vivo and promotes microvascular thrombosis, a condition observed among severe COVID-19 patients [37,38]. Therefore, IL-6 may play a crucial role in the pathogenesis of COVID-19. In the clinic, tocilizumab, an anti-IL 6 receptor drug, is shown to be effective against moderate to severe forms of COVID-19 with a reduced mortality rate than the control group [[39], [40], [41], [42]]. Conversely, a few clinical studies observed that Tocilizumab therapy is neither effective nor reduced mortality among moderate to severe COVID-19 patients [43,44]. Of note, tocilizumab administration in COVID-19 patients was associated with adverse side effects such as late-onset infection, elevated liver function test, neutropenia, and hypertension [45]. A small cohort study reported that early tocilizumab therapy is safe and effective against COVID-19 [46]. Although anti-IL 6 therapy is a key component of a strategy to control COVID-19 pathogenesis, current knowledge of anti-IL6 therapy such as tocilizumab against COVID-19 is controversial. Therefore, anti-IL 6 therapy requires further clinical studies which may reveal the complete safety and efficacy.
Notably, T cell response mediates B cell activation, pathogen clearance, and apoptosis of the infected cells thereby controlling pathogenesis. However, severe COVID-19 patients exhibit a dysregulated T cell response. Like MERS and SARS-COV-1, COVID-19 severity was also associated with reduced T cell count in the peripheral blood, a condition called lymphopenia [47]. In a transcriptome analysis, the elevated expression of TP53, a T-cell apoptosis signaling factor, in the BALF of severe COVID-19 was reported. It explicates that the lymphopenia among severe COVID-19 patients may indeed be due to apoptosis [48,49]. On the other hand, persistent activation of CD8+ and CD4+ T cells leads to an exhausted state of the surviving T cells thereby impairing adaptive immunity which correlates with the elevated cytokines such as IL-6, IL-10, and TNF α [50]. Conversely, robust CD8 T cell clonal expansion was observed among mild and recovered COVID-19 patients [49]. Therefore, the role of dysregulated T cell response in COVID-19 pathogenesis warrants further investigation. Additionally, the immune cells such as antigen-presenting cells (APC), dendritic cells (DC), and B cells also exhibit a dysregulated response among COVID-19 patients (Table 2). In summary, the dysregulated immune response in COVID-19 severity causes acute respiratory distress syndrome (ARDS) followed by multiple organ failure eventually leading to mortality [36].
Table 2.
IFN β therapy against viral diseases in clinical trials.
| Clinical Trial No. | Disease | Drug and Dosing | Adm. | Clinical Phase |
|---|---|---|---|---|
| NCT04350281 | COVID-19 | Interferon β-1b (25 μg 3x weekly) Hydroxychloroquine (400 mg daily) |
SC | Phase 3 |
| NCT04350671 | COVID-19 | Interferon β-1a (30 μg weekly) Lopinavir/Ritonavir (400/100 mg 2x daily) Hydroxychloroquine (400 mg daily) |
IM | Phase 2 |
| NCT04385095 | COVID-19 | Interferon β-1a (150 μg × 2 weeks) | NEB | Phase 2 |
| NCT04449380 | COVID-19 | Interferon β-1a (44 μg 3x weekly) | SC | Phase 2 |
| NCT02845843 | MERS | Interferon β-1b (250 μg 3 x weekly) Lopinavir/Ritonavir (400/100 mg 2x daily) |
SC | Phase 2 |
| NCT00001785 | HAM | Interferon β-1a (30 μg weekly) | IM | Phase 2 |
| NCT00249860 | CHC | Interferon β-1a (44 μg 3x weekly) | SC | Phase 3 |
| NCT03570359 | COPD | Interferon β-1a (150 μg × 2 weeks) | NEB | Phase 2 |
IFN, interferon; HAM, HTLV-1 associated myelopathy; CHC, chronic hepatitis C; COPD, chronic obstructive pulmonary disease; IM, intramuscular; SC, subcutaneous; NEB, nebulization.
5. Therapeutic potential of IFN β
For decades, type I interferon such as IFN α and IFN β have been a promising treatment option against the hepatitis B virus (HBV) and multiple sclerosis (MS). In a clinical study, IFN β monotherapy against chronic hepatitis C (CHC) is shown to be more effective with the sustained viral response (SVR) than IFN α and ribavirin combination therapy [51]. IFN β therapy is under clinical trials against viral diseases such as HTLV-1 associated myelopathy (HAM) [52], chronic hepatitis C (CHC) [53], chronic obstructive pulmonary disease (COPD) [54] (Table 1 ). Recently, an in vitro study reported that SARS-CoV-2 is more sensitive to IFN β than IFN α [55]. In the clinic, autoantibodies against IFN α (not against IFN β) have been found in more severe COVID-19 patients than mild and asymptomatic patients [56]. In transcriptome analysis, IRF 3 expression, a key regulator of IFN β production, was significantly lower in COVID-19 patients than in the control group [57]. Therefore, we hypothesize that COVID-19 specifically targets IFN β production, as well as stimulates IFN α antagonists eventually evading the innate immune response. Hence, IFN β therapy could be an appropriate treatment option against COVID-19 compared with IFN α in terms of specificity, efficacy, and associated side effects [58]. In the following section, we discuss the antiviral activity and the immunomodulatory effect of IFN β.
Table 1.
Immunomodulatory effect of IFN β
| Dysregulation in COVID-19 | Effect of IFN β administration | |
|---|---|---|
| APC |
↓ pDC impairs antigen presentation and reduces type I IFN production. ↓ mTOR levels in pDC [76] |
↑ DC differentiation from CD14+ monocytes and CD34+ progenitors [77] ↓ naive T cell activation in draining lymph nodes by inhibiting DC migration [72]. ↓ Antigen presentation by downregulating IFN-γ induced MHC II molecule expression among non-professional APC [78]. |
| T cells |
↓ CD4+ T cell and CD8+ T cell leading to pathogen persistence. ↑ Tex exhibits dysregulated function. ↑ CD4+ CCR6+ (Th17) secretes IL-17 which recruits neutrophils and induces pro-inflammatory cytokines such as IL-1β and TNF α. ↑ CD4+ CXCR3+ (Th1) secretes GM-CSF and IFN-γ. IFN-γ increases MHC I and II antigen presentation, chemokine secretion, macrophage activation, and phagocytosis, on the other hand, GM-CSF activates CD14+ CD16+ monocytes that secretes high levels of IL-6 [49]. |
↑ CD4+ CD25+ Foxp3+ (T reg) population secretes IL-10 and TGF β which inhibits T cell activation and Th1/Th2 differentiation. ↑ Bcl2 expression among the T reg population. ↑ CD4+ CCR4+ (Th2) secretes IL-4 which actively inhibits activated macrophage and pro-inflammatory cytokines such as IL-1 and TNF. ↓ IL-17 through T reg population [79] |
| B cell | ↓ CD3− CD19+ B cell impairs pathogen clearance [80]. |
↑ CD69, CD86, and MHC II on the B cell surface which enhances B cell function. ↑ Bcl2 expression leading to B cell survival [81] |
| Cytokines |
↑ IL-2, IL-6, IL-1β, IL-1Ra, TNF α, and IFN-γ leading to cytokine storm syndrome. ↓Type I IFN and type III IFN impairs innate immune response [82] |
↑ IL-10 and IL-4 actively inhibit immune cell activation and recruitment. ↓ IL-1β maturation by inhibiting inflammasome activation [83] |
| Others |
↑ Neutrophils secrete IL-1β and IL-1Ra. ↓ Monocytes, eosinophil, and basophil [82] |
↑ Neutrophils apoptosis and efferocytosis [74] |
pDC, plasmacytoid dendritic cell; Th, T helper cells; T reg, T regulatory cells; T ex, exhausted state T cell; IL, interleukins; MHC, major histocompatibility complex; GM-CSF, granulocyte-macrophage colony-stimulating factor; TNF, tumor necrosis factor; IFN, interferon; mammalian target of rapamycin (mTOR);
5.1. I. IFN β and antiviral activity
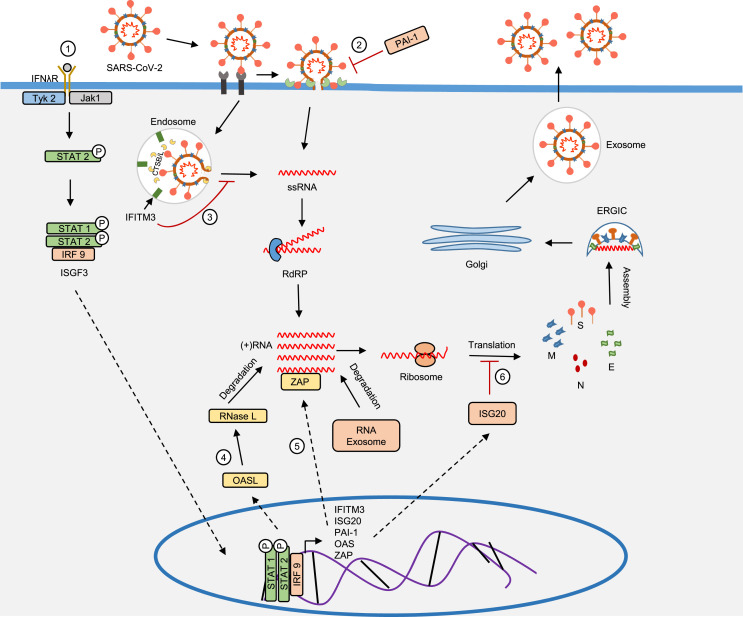
In principle, IFN β binds the IFNAR and initiates the JAK-STAT signaling pathway thereby inducing several hundred ISGs, notably the interferon-induced transmembrane protein 3 (IFITM3), plasminogen activator inhibitor (PAI-1), protein kinase R (PKR), and 2′-5′-oligoadenylate synthetase (OAS) [59]. The IFITM 3, a transmembrane protein present on the late endosome inhibits the release of viral particles into the cytoplasm, as well, initiates lysosomal degradation of the virus [60]. Concurrently, PAI-1, an inhibitor protein, also inhibits viral entry through TMPRSS2 [61,62]. OAS inhibits viral replication by degrading viral RNA through latent ribonuclease (RNase L) [63]. Upon viral RNA sensing, the zinc-finger anti-viral protein (ZAP) recruit RNA exosomes, eventually initiating viral RNA degradation [64]. Simultaneously, the interferon-induced protein 20 (ISG20), an RNA exonuclease inhibits viral replication by impairing protein translation ( Fig. 3 ) [65].
Fig. 3.
The role of IFN β in antiviral activity. 1, The IFN binds the IFNAR on the cell surface and initiates the JAK-STAT signaling pathway inducing ISGS which encodes IFITM, PAI-1, OASL, ZAP and ISG20, etc.; 2, PAI-1 inhibits TMPRSS2 that cleaves ACE-2 bound spike protein of SARS- COV-2; 3, IFITM3 inhibits the viral ssRNA release into the cytoplasm through endosome and initiates lysosomal degradation of the virus; 4, OAS activates RNase L that effectively degrades viral RNA; 5, ZAP senses and attaches to viral RNA, followed by recruiting RNA exosome that eventually degrades viral RNA; 6, The ISG20 inhibits viral replication by impairing protein translation. IFITM 3, interferon-induced transmembrane protein 3; PAI-1, plasminogen activator inhibitor; CTSB/L cathepsin B/L; ssRNA, single-stranded RNA; PKR, protein kinase R; OASL, 2′-5′-oligoadenylate synthetase like; IKK, IκB kinase complex; NF κB, nuclear factor kappa-light-chain-enhancer of an activated B cell; eIFα2, eukaryotic initiation factor α 2; IFN β, interferon β; IFNAR, interferon-alpha/beta receptor; Jak 1, Janus kinase 1; Tyk 2, tyrosine kinase 2; STAT 1 and 2, signal transducer and activator of transcription 1 and 2; IRF 9, interferon regulatory factor 9.
Therefore, IFN β signaling actively inhibits viral entry, and replication thereby setting up an effective antiviral state in the infected and the non-infected cells.
In the in-vitro study, IFN β administration in SARS-CoV-2 infected cells effectively downregulated viral proteins such as E protein, nsp 14, and S protein [66]. Conversely, blocking the interferon signaling by ruxolitinib, a JAK kinase inhibitor, had no significant effect on viral reads. These data indicate that IFN β controls viral protein translation through ISGs. Previous studies reported that both SARS-CoV-1 and SARS-CoV-2 effectively downregulated ISGs eventually leading to viral progression [28,67]. Hence, it would be a logical approach to up-regulate these ISGs through IFN β administration to retain the antiviral activity in the host.
5.2. II. IFN β and immunomodulatory effect
In addition to antiviral activity, IFN β also exhibits potent anti-inflammatory properties. IFN β induces intracellular anti-inflammatory proteins such as tristetraprolin (TTP) and suppressor of cytokine signaling (SOCS) through the JAK-STAT signaling cascade. TTP destabilizes the mRNA thereby inhibiting pro-inflammatory cytokine expression [68] and the suppressor of cytokine signaling (SOCS) inhibits JAK thereby impairing the IL-6 signaling cascade in vitro [69]. Furthermore, IFN β induces anti-inflammatory cytokines such as IL-10 and IL-4 (Fig. 2). While IL-10 inhibits T helper cell 1 (Th1) differentiation, inflammatory APC priming, and macrophage activation, IL-4 promotes Th2 differentiation and inhibits lipopolysaccharide (LPS) induced cytokine production [70]. Of note, sustained IL-10 and IL-4 production after immune cell priming has a pathogenic role which has been observed among severe COVID-19 patients [71]. Therefore, the timing of IFN β therapy is crucial in COVID-19.
Additionally, IFN β controls immunopathogenesis by modulating the immune responses of cells such as T cell, dendritic cell (DC), B cell, etc (Table 1). In CD4+ T cells, IFN β treatment upregulates Bcl2 expression, an anti-apoptotic protein, thereby preventing apoptosis and promoting survival in vitro. It also promotes Treg differentiation from the CD4+ population, which actively inhibits Th1/Th17 differentiation [70]. In DC, IFN β administration downregulates chemokine receptor 7 (CCR7) and matrix metallopeptidase (MMP) thereby inhibiting migration and T cell activation in the draining lymph nodes [72]. It also promotes DC differentiation from CD14+ monocytes and CD34+ progenitors thereby promoting the production of cytokines such as IL-10, and TGF β [73]. In B cells, IFN β promotes survival by upregulating Bcl2 and enhances function by inducing surface markers such as CD69, CD86, and MHC II. In neutrophils, IFN β promotes apoptosis thereby controlling excessive neutrophils at the site of inflammation and promotes the efferocytosis of apoptotic cells [74]. Thus, IFN β exhibits a central role in driving towards the resolution phase from chronic inflammation. However, a delayed IFN β response has pathogenic consequences [75]; hence, the timing of IFN β therapy in COVID-19 is crucial.
6. Clinical interventions
Recently, drugs such as remdesivir, lopinavir/ritonavir, and hydroxychloroquine are clinically approved therapeutic interventions against COVID-19. However, clinical studies conducted on these drugs reported neither significant clinical improvement nor reduction in mortality rate among COVID-19 patients, which may be due to their non-specificity [[84], [85], [86]]. It explicates the importance of more targeted therapeutic interventions such as the administration of IFN β. IFN β is shown to be effective against several viral diseases by enhancing the host's innate immune response (Table 2 ). Of note, in a clinical study, IFN β monotherapy against CHC exhibited a potent antiviral activity with sustained viral response among Asian patients, and, intriguingly, IFN β and ribavirin combination therapy increased the proportion of patients who fully recovered [87]. In vivo, early IFN β administration showed significantly reduced viral RNA levels and an increased survival rate than the delayed IFN β treatment among the MERS-CoV infected mice [88]. Supportively, a pilot study reported that IFN β and lopinavir/ritonavir combination therapy significantly lowered mortality when administered within 7 days after symptom onset among hospitalized MERS patients [89]. In vitro study, SARS-CoV-2 was more sensitive to IFN β than SARS-CoV-1 and the effective concentration (EC 50) of IFN β was less than that of IFN α [55]. In a recent clinical study, the triple combination therapy (interferon beta-1b, lopinavir-ritonavir, and ribavirin) for COVID-19 patients significantly reduced the recovery time compared to the control (lopinavir-ritonavir) group [90]. Similarly, the IFN β with hydroxychloroquine plus lopinavir-ritonavi showed improved odds of recovery and death than the control. However, the IFN β did not change the time to reach clinical response than the control [91]. Taken together, we hypothesize that IFN β could be the key component for reaching efficiency in triple and double combination therapy against COVID-19.
Conversely, the SOLIDARITY randomized trial reported that 2000 COVID-19 patients, who received subcutaneous interferon beta 1-a or interferon beta-1a plus lopinavir-ritonavir, had little or no effect in clinical improvement [92]. The result of the SOLIDARITY trial is in complete contrast to the above-mentioned interferon beta combination therapy. Recently the nebulized interferon beta-1a (SNG001) treated COVID-19 patients showed improved odds of recovery and death than the control group. Of note, improvement of patient-reported breathlessness was greater among the SNG001 group than the control [93].
Comparatively, the nebulized therapy directly delivers the interferon in the airway than the subcutaneous route, used in the SOLIDARITY trial, eventually initiating anti-viral response in the mucosa. On the other hand, the patients recruited in the nebulized therapy are less severe than the patients in the SOLIDARITY trial. Furthermore, in a transcriptomic analysis IFN β expression in the nasal mucosa, independent of viral road, predicted the clinical outcome of severe COVID-19 patients [94]. Of note, the INF β has a detrimental effect in the severe stages of COVID-19. Therefore, the stage of COVID-19 and route of administration is crucial during interferon therapy against COVID-19. Once timing, dosing, and immunogenicity issues have been thoroughly investigated, INF β could be a potential early-stage treatment option against COVID-19 [95,96]. Currently, IFN β monotherapy and combination therapy (Table 2) are under clinical trials [[97], [98], [99]], which will reveal the safety and efficacy of IFN β against COVID-19.
7. Conclusions
Several viral diseases primarily target the innate immune response to replicate and progress in the host. Intriguingly, COVID-19 severity was associated with a significantly low innate immune response. IFN β monotherapy would be an effective early-stage treatment option to prevent COVID-19 severity. For instance, SNG001, an IFN β product, is in phase 3 clinical trial against COVID-19 and its final data will reveal the efficiency of IFN β monotherapy. Therefore, IFN β monotherapy-mediated enhancement of innate immune response could be a key component in future strategies to prevent large-scale viral disease outbreaks.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Prof. Adhikarla Rao for his useful suggestions. We thank Mr. Nachiyappan Venkatachalam for his digital support. Without them, it would have been a completely different review. This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CORONAVIRUS RESOURCE CENTER, Johns Hopkins Univ. Med. 2020. https://coronavirus.jhu.edu/map.html (accessed June 1, 2020)
- 3.Jean S.S., Lee P.I., Hsueh P.R. Treatment options for COVID-19: the reality and challenges. J. Microbiol. Immunol. Infect. 2020;53:436–443. doi: 10.1016/j.jmii.2020.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacKenzie J.S., Smith D.W. COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don't. Microbiol. Aust. 2020;41:45–50. doi: 10.1071/MA20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Z., McGoogan J.M. Characteristics of and important Lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. J. Am. Med. Assoc. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 6.Who Modes of transmission of virus causing COVID-19 : implications for IPC precaution recommendations. Sci. Br. 2020:1–3. doi: 10.1056/NEJMoa2001316.5. [DOI] [Google Scholar]
- 7.Koyama S., Ishii K.J., Coban C., Akira S. Cytokine Innate immune response to viral infection. 2008;43:336–341. doi: 10.1016/j.cyto.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Ronghua Hu M.C.-S., Xia Chang-Qing, Butfiloski Edward. Effect of high glucose on cytokine production by human peripheral blood immune cells and type I interferon signaling in monocytes: implications for the role of Hyperglycemia in the diabetes inflammatory process and host defense against infection. Physiol. Behav. 2019;176:139–148. doi: 10.1016/j.physbeh.2017.03.040. and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molony R.D., Nguyen J.T., Kong Y., Montgomery R.R., Shaw A.C., Iwasaki A. Aging impairs both primary and secondary RIG-I signaling for interferon induction in human monocytes. Sci. Signal. 2017;10:1–12. doi: 10.1126/scisignal.aan2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A.A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W.M., Razooky B.S., Hoffmann H.-H., Michailidis E., Moens L., Han J.E., Lorenzo L., Bizien L., Meade P., Neehus A.-L., Ugurbil A.C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schlüter A., Le Voyer T., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M.F., Mansouri D., Al-Saud H., Al-Mulla F., Almourfi F., Al-Muhsen S.Z., Alsohime F., Al Turki S., Hasanato R., van de Beek D., Biondi A., Bettini L.R., D'Angio M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A.J., Tompkins M.F., Alba C., Vandernoot I., Goffard J.-C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P.-E., Keles S., Çölkesen F., Ozcelik T., Yasar K.K., Senoglu S., Karabela Ş.N., Gallego C.R., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouénan C., Snow A.L., Dalgard C.L., Milner J., Vinh D.C., Mogensen T.H., Marr N., Spaan A.N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., García-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R., Zhang S.-Y., Gorochov G., Béziat V., Jouanguy E., Sancho-Shimizu V., Rice C.M., Abel L., Notarangelo L.D., Cobat A., Su H.C., Casanova J.-L. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;80– doi: 10.1126/science.abd4570. eabd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuen C.-K., Lam J.-Y., Wong W.-M., Mak L.-F., Wang X., Chu H., Cai J.-P., Jin D.-Y., To K.K.-W., Chan J.F.-W., Yuen K.-Y., Kok K.-H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microb. Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., Menachery V.D., Rajsbaum R., Shi P.-Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33(1) doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;(80-):782. doi: 10.1126/science.abb7498. eabb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang T., Jaimes J.A., Bidon M.K., Straus M.R., Daniel S., Whittaker G.R. Proteolytic activation of SARS-CoV-2 spike at the S1/S2 boundary: potential role of proteases beyond furin. ACS Infect. Dis. 2021;7:264–272. doi: 10.1021/acsinfecdis.0c00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou T., Mou H., Zhang L., Ojha A., Choe H., Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beyerstedt S., Casaro E.B., Rangel É.B. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:905–919. doi: 10.1007/s10096-020-04138-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wieczorek M., Abualrous E.T., Sticht J., Álvaro-Benito M., Stolzenberg S., Noé F., Freund C. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 2017;8:1–16. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.García L.F. Immune response, inflammation, and the clinical spectrum of COVID-19. Front. Immunol. 2020;11:1441. doi: 10.3389/fimmu.2020.01441. https://www.frontiersin.org/article/10.3389/fimmu.2020.01441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Wu K., Wang D., Yue X., Song D., Zhu Y., Wu J. Nucleocapsid protein of SARS-CoV activates interleukin-6 expression through cellular transcription factor NF-κB. Virology. 2007;365:324–335. doi: 10.1016/j.virol.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karwaciak I., Sałkowska A., Karaś K., Dastych J., Ratajewski M. Nucleocapsid and spike proteins of the coronavirus SARS-CoV-2 induce IL6 in monocytes and macrophages-potential implications for cytokine storm syndrome. Vaccines. 2021;9:54. doi: 10.3390/vaccines9010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y., Ma L., Cai S., Zhuang Z., Zhao Z., Jin S., Xie W., Zhou L., Zhang L., Zhao J., Cui J. RNA-induced liquid phase separation of SARS-CoV-2 nucleocapsid protein facilitates NF-κB hyper-activation and inflammation. Signal Transduct. Target. Ther. 2021;6:167. doi: 10.1038/s41392-021-00575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mu J., Fang Y., Yang Q., Shu T., Wang A., Huang M., Jin L., Deng F., Qiu Y., Zhou X. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020;6:65. doi: 10.1038/s41421-020-00208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuda Y., Homma T., Inoue H., Onitsuka C., Ikeda H., Goto Y., Sato Y., Kimura T., Hirai K., Ohta S., Yamamoto M., Kusumoto S., Suzuki S., Tanaka A., Sagara H. Downregulation of type III interferons in patients with severe COVID-19. J. Med. Virol. 2021;93:4559–4563. doi: 10.1002/jmv.26993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pène F., Marin N., Roche N., Szwebel T.-A., Merkling S.H., Treluyer J.-M., Veyer D., Mouthon L., Blanc C., Tharaux P.-L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kernéis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;(80-):369. doi: 10.1126/science.abc6027. 718 LP – 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/golgi membrane. J. Virol. 2007;81:9812. doi: 10.1128/JVI.01012-07. LP – 9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548. doi: 10.1128/JVI.01782-06. LP – 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Interactions V., Lokugamage K.G., Hage A., De Vries M., Valero-jimenez A.M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V.D. Type I interferon susceptibility distinguishes SARS-CoV-2 from. SARS-CoV. 2020;94:1–13. doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sallard E., Lescure F.-X., Yazdanpanah Y., Mentre F., Peiffer-Smadja N. Type 1 interferons as a potential treatment against COVID-19. Antivir. Res. 2020;178:104791. doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herold T., Jurinovic V., Arnreich C., Lipworth B.J. 2020. Since January 2020 Elsevier Has Created a COVID-19 Resource Centre with Free Information in English and Mandarin on the Novel Coronavirus COVID- 19 . The COVID-19 Resource Centre Is Hosted on Elsevier Connect , the Company ’ S Public News and Information. [Google Scholar]
- 34.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence Homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayakabe K., Kuroiwa T., Sakurai N., Ikeuchi H., Kadiombo A.T., Sakairi T., Matsumoto T., Maeshima A., Hiromura K., Nojima Y. Interleukin-6 promotes destabilized angiogenesis by modulating angiopoietin expression in rheumatoid arthritis. Rheumatology. 2012;51:1571–1579. doi: 10.1093/rheumatology/kes093. [DOI] [PubMed] [Google Scholar]
- 36.Levy H.R., Sanchez C.I.S. COVID-19 and cytokine storm syndrome. MLO. Med. Lab. Obs. 2020;52 http://search.ebscohost.com/login.aspx?direct=true&AuthType=athens&db=cin20&AN=143565168&site=ehost-live 8–14. [Google Scholar]
- 37.Senchenkova E.Y., Komoto S., Russell J., Almeida-Paula L.D., Yan L.-S., Zhang S., Granger D.N. Interleukin-6 mediates the platelet abnormalities and thrombogenesis associated with experimental colitis. Am. J. Pathol. 2013;183:173–181. doi: 10.1016/j.ajpath.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iba T., Levy J.H., Levi M., Thachil J. Coagulopathy in COVID-19. J. Thromb. Haemostasis. 2020;18:2103–2109. doi: 10.1111/jth.14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castelnovo L., Tamburello A., Lurati A., Zaccara E., Marrazza M.G., Olivetti M., Mumoli N., Mastroiacovo D., Colombo D., Ricchiuti E., Vigano’ P., Paola F., Mazzone A. Anti-IL6 treatment of serious COVID-19 disease: a monocentric retrospective experience. Medicine (Baltim.) 2021:100. doi: 10.1097/MD.0000000000023582. https://journals.lww.com/md-journal/Fulltext/2021/01080/Anti_IL6_treatment_of_serious_COVID_19_disease__A.8.aspx [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salama C., Han J., Yau L., Reiss W.G., Kramer B., Neidhart J.D., Criner G.J., Kaplan-Lewis E., Baden R., Pandit L., Cameron M.L., Garcia-Diaz J., Chávez V., Mekebeb-Reuter M., Lima de Menezes F., Shah R., González-Lara M.F., Assman B., Freedman J., Mohan S.V. Tocilizumab in patients hospitalized with covid-19 pneumonia. N. Engl. J. Med. 2021;384:20–30. doi: 10.1056/nejmoa2030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guaraldi G., Meschiari M., Cozzi-Lepri A., Milic J., Tonelli R., Menozzi M., Franceschini E., Cuomo G., Orlando G., Borghi V., Santoro A., Di Gaetano M., Puzzolante C., Carli F., Bedini A., Corradi L., Fantini R., Castaniere I., Tabbì L., Girardis M., Tedeschi S., Giannella M., Bartoletti M., Pascale R., Dolci G., Brugioni L., Pietrangelo A., Cossarizza A., Pea F., Clini E., Salvarani C., Massari M., Viale P.L., Mussini C. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2 doi: 10.1016/S2665-9913(20)30173-9. e474–e484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon A.C., Mouncey P.R., Al-beidh F., Kathryn M., Nichol A.D., Arabi Y.M., Annane D. Interleukin-6 receptor antagonists in critically ill patients with covid-19 – writing Committee : corresponding author, MedRxiv. 2021. 1-12. [DOI]
- 43.Stone J.H., Frigault M.J., Serling-Boyd N.J., Fernandes A.D., Harvey L., Foulkes A.S., Horick N.K., Healy B.C., Shah R., Bensaci A.M., Woolley A.E., Nikiforow S., Lin N., Sagar M., Schrager H., Huckins D.S., Axelrod M., Pincus M.D., Fleisher J., Sacks C.A., Dougan M., North C.M., Halvorsen Y.-D., Thurber T.K., Dagher Z., Scherer A., Wallwork R.S., Kim A.Y., Schoenfeld S., Sen P., Neilan T.G., Perugino C.A., Unizony S.H., Collier D.S., Matza M.A., Yinh J.M., Bowman K.A., Meyerowitz E., Zafar A., Drobni Z.D., Bolster M.B., Kohler M., D'Silva K.M., Dau J., Lockwood M.M., Cubbison C., Weber B.N., Mansour M.K. Efficacy of tocilizumab in patients hospitalized with covid-19. N. Engl. J. Med. 2020;383:2333–2344. doi: 10.1056/nejmoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosas I.O., Bräu N., Waters M., Go R.C., Hunter B.D., Bhagani S., Skiest D., Aziz M.S., Cooper N., Douglas I.S., Savic S., Youngstein T., Del Sorbo L., Cubillo Gracian A., De La Zerda D.J., Ustianowski A., Bao M., Dimonaco S., Graham E., Matharu B., Spotswood H., Tsai L., Malhotra A. Tocilizumab in hospitalized patients with severe covid-19 pneumonia. N. Engl. J. Med. 2021:1–14. doi: 10.1056/NEJMoa2028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettit N.N., Nguyen C.T., Mutlu G.M., Wu D., Kimmig L., Pitrak D., Pursell K. Late onset infectious complications and safety of tocilizumab in the management of COVID-19. J. Med. Virol. 2021;93:1459–1464. doi: 10.1002/jmv.26429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guillén L., Padilla S., Fernández M., Agulló V., García J.A., Telenti G., García-Abellán J., Botella Á., Gutiérrez F., Masiá M. Preemptive interleukin-6 blockade in patients with COVID-19. Sci. Rep. 2020;10:16826. doi: 10.1038/s41598-020-74001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin L., Lu L., Cao W., Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection–a review of immune changes in patients with viral pneumonia. Emerg. Microb. Infect. 2020;9:727–732. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Liu Y., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients, Emerg. Microb. Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. https://www.frontiersin.org/article/10.3389/fimmu.2020.00827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barbaro G.D.L.G., Soldini M., Giancaspro G., Pellicelli A., Grisorio B., Barbarini G. Intravenous recombinant interferon-beta versus interferon-alpha-2b and ribavirin in combination for short-term treatment of chronic hepatitis C patients not responding to interferon-alpha. Scand. J. Gastroenterol. 1999;34:928–933. doi: 10.1080/003655299750025426. [DOI] [PubMed] [Google Scholar]
- 52.Ninds . ClinicalTrials.Gov; 2008. Recombinant Human Interferon Beta-1a (Avonex) for the Treatment of Patients with HTLV-1-Associated Myelopathy (HAM)https://clinicaltrials.gov/ct2/show/study/NCT00001785?recrs=e [Google Scholar]
- 53.Emd&Merck A. ClinicalTrials.Gov.; 2013. Multicenter phase 3 study of interferon-beta-1a for the treatment of chronic hepatitis C in asian subjects. [Google Scholar]
- 54.Synairgen A. ClinicalTrials.Gov; 2018. Study to Test a Potential New Treatment for COPD Patients Suffering from the Common Cold or Influenza.https://clinicaltrials.gov/ct2/show/study/NCT03570359?term=interferon+beta [Google Scholar]
- 55.Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., Manry J., Shaw E., Haljasmägi L., Peterson P., Lorenzo L., Bizien L., Trouillet-Assant S., Dobbs K., de Jesus A.A., Belot A., Kallaste A., Catherinot E., Tandjaoui-Lambiotte Y., Le Pen J., Kerner G., Bigio B., Seeleuthner Y., Yang R., Bolze A., Spaan A.N., Delmonte O.M., Abers M.S., Aiuti A., Casari G., Lampasona V., Piemonti L., Ciceri F., Bilguvar K., Lifton R.P., Vasse M., Smadja D.M., Migaud M., Hadjadj J., Terrier B., Duffy D., Quintana-Murci L., van de Beek D., Roussel L., Vinh D.C., Tangye S.G., Haerynck F., Dalmau D., Martinez-Picado J., Brodin P., Nussenzweig M.C., Boisson-Dupuis S., Rodríguez-Gallego C., Vogt G., Mogensen T.H., Oler A.J., Gu J., Burbelo P.D., Cohen J.I., Biondi A., Bettini L.R., D'Angio M., Bonfanti P., Rossignol P., Mayaux J., Rieux-Laucat F., Husebye E.S., Fusco F., Ursini M.V., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Castagnoli R., Montagna D., Licari A., Marseglia G.L., Duval X., Ghosn J., Tsang J.S., Goldbach-Mansky R., Kisand K., Lionakis M.S., Puel A., Zhang S.-Y., Holland S.M, Gorochov G., Jouanguy E., Rice C.M., Cobat A., Notarangelo L.D, Abel L., Su H.C., Casanova J.-L. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;80(370) doi: 10.1126/science.abd4585. eabd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bizzotto J., Sanchis P., Abbate M., Lage-Vickers S., Lavignolle R., Toro A., Olszevicki S., Sabater A., Cascardo F., Vazquez E., Cotignola J., Gueron G. SARS-CoV-2 infection boosts <em>MX1</em> antiviral effector in COVID-19 patients. IScience. 2020;23 doi: 10.1016/j.isci.2020.101585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hosseini-Moghaddam S.M., Mousavi A., Alavian S.M. Is β-interferon a promising therapeutic option for the management of hepatitis C? J. Antimicrob. Chemother. 2009;63:1097–1103. doi: 10.1093/jac/dkp092. [DOI] [PubMed] [Google Scholar]
- 59.García-Sastre A. Ten strategies of interferon evasion by viruses. Cell Host Microbe. 2017;22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spence J.S., He R., Hoffmann H.-H., Das T., Thinon E., Rice C.M., Peng T., Chandran K., Hang H.C. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat. Chem. Biol. 2019;15:259–268. doi: 10.1038/s41589-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen L.W., Mao H.J., Wu Y.L., Tanaka Y., Zhang W. vol. 142. 2017. pp. 1–10. (TMPRSS2: A Potential Target for Treatment of Influenza Virus and Coronavirus Infections, Biochimie). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/jvi.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi U.Y., Kang J.-S., Hwang Y.S., Kim Y.-J. Oligoadenylate synthase-like (OASL) proteins: dual functions and associations with diseases. Exp. Mol. Med. 2015;47 doi: 10.1038/emm.2014.110. e144–e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nchioua R., Kmiec D., Müller J.A., Conzelmann C., Groß R., Swanson C.M., Neil S.J.D., Stenger S., Sauter D., Münch J., Sparrer K.M.J., Kirchhoff F. SARS-CoV-2 is restricted by zinc finger antiviral protein despite preadaptation to the low-CpG environment in humans. mBio. 2020;11 doi: 10.1128/mBio.01930-20. e01930-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu N., Nguyen X.-N., Wang L., Appourchaux R., Zhang C., Panthu B., Gruffat H., Journo C., Alais S., Qin J., Zhang N., Tartour K., Catez F., Mahieux R., Ohlmann T., Liu M., Du B., Cimarelli A. The interferon stimulated gene 20 protein (ISG20) is an innate defense antiviral factor that discriminates self versus non-self translation. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1008093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu W., Yen Y.-T., Singh S., Kao C.-L., Wu-Hsieh B.A. SARS-CoV regulates immune function-related gene expression in human monocytic cells. Viral Immunol. 2012;25:277–288. doi: 10.1089/vim.2011.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sauer I., Schaljo B., Vogl C., Gattermeier I., Kolbe T., Müller M., Blackshear P.J., Kovarik P. Interferons limit inflammatory responses by induction of tristetraprolin. Blood. 2006;107:4790–4797. doi: 10.1182/blood-2005-07-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whitmarsh R.J., Gray C.M., Gregg B., Christian D.A., May M.J., Murray P.J., Hunter C.A. A critical role for SOCS3 in innate resistance to <em>Toxoplasma gondii</em>. Cell Host Microbe. 2011;10:224–236. doi: 10.1016/j.chom.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Opal S.M., DePalo V.A. Anti-inflammatory cytokines. Chest. 2000;117:1162–1172. doi: 10.1378/chest.117.4.1162. [DOI] [PubMed] [Google Scholar]
- 71.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease, Cytokine Growth Factor Rev. 2020. 54, 62-75. [DOI] [PMC free article] [PubMed]
- 72.Yen J.-H., Kong W., Ganea D. IFN-beta inhibits dendritic cell migration through STAT-1-mediated transcriptional suppression of CCR7 and matrix metalloproteinase 9. J. Immunol. 2010;184:3478–3486. doi: 10.4049/jimmunol.0902542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gessani S., Conti L., Del Cornò M., Belardelli F. Type I interferons as regulators of human antigen presenting cell functions, Toxins (Basel) 2014. [DOI] [PMC free article] [PubMed]
- 74.Kumaran Satyanarayanan S., El Kebir D., Soboh S., Butenko S., Sekheri M., Saadi J., Peled N., Assi S., Othman A., Schif-Zuck S., Feuermann Y., Barkan D., Sher N., Filep J.G., Ariel A. IFN-β is a macrophage-derived effector cytokine facilitating the resolution of bacterial inflammation. Nat. Commun. 2019;10:3471. doi: 10.1038/s41467-019-10903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park A., Iwasaki A. Type I and type III interferons- induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., Wagh D., Coller J., Pellegrini K.L., Kazmin D., Alaaeddine G., Leung W.S., Chan J.M.C., Chik T.S.H., Choi C.Y.C., Huerta C., Paine McCullough M., Lv H., Anderson E., Edupuganti S., Upadhyay A.A., Bosinger S.E., Maecker H.T., Khatri P., Rouphael N., Peiris M., Pulendran B. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;80:369. doi: 10.1126/science.abc6261. 1210 LP – 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang Y.-M., Hussien Y., Yarilin D., Xiao B.-G., Liu Y.-J., Link H. INTERFERON-BETA induces the development OF type 2 dendritic cells. Cytokine. 2001;13:264–271. doi: 10.1006/cyto.2000.0835. [DOI] [PubMed] [Google Scholar]
- 78.Lu H.T., Riley J.L., Babcock G.T., Huston M., Stark G.R., Boss J.M., Ransohoff R.M. Interferon (IFN) beta acts downstream of IFN-gamma-induced class II transactivator messenger RNA accumulation to block major histocompatibility complex class II gene expression and requires the 48-kD DNA-binding protein, ISGF3-gamma. J. Exp. Med. 1995;182:1517–1525. doi: 10.1084/jem.182.5.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martín-Saavedra F.M., González-García C., Bravo B., Ballester S. Beta interferon restricts the inflammatory potential of CD4+ cells through the boost of the Th2 phenotype, the inhibition of Th17 response and the prevalence of naturally occurring T regulatory cells. Mol. Immunol. 2008;45:4008–4019. doi: 10.1016/j.molimm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 80.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.-S. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in wuhan, China, Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kiefer K., Oropallo M.A., Cancro M.P., Marshak-Rothstein A. Role of type I interferons in the activation of autoreactive B cells, Immunol. Cell Biol. 2012;90:498–504. doi: 10.1038/icb.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang L., Liu S., Liu J., Zhang Z., Wan X., Huang B., Chen Y., Zhang Y. COVID-19: immunopathogenesis and Immunotherapeutics. Signal Transduct. Target. Ther. 2020;5:128. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guarda G., Braun M., Staehli F., Tardivel A., Mattmann C., Förster I., Farlik M., Decker T., Du Pasquier R.A., Romero P., Tschopp J. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 84.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir–ritonavir in adults hospitalized with severe covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Magagnoli J., Narendran S., Pereira F., Cummings T.H., Hardin J.W., Sutton S.S., Ambati J. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with COVID-19. Med. 2020;1:114–127. doi: 10.1016/j.medj.2020.06.001. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan H.L.Y., Ren H., Chow W.C., Wee T., I. beta-1a H.C.S. Group Randomized trial of interferon beta-1a with or without ribavirin in Asian patients with chronic hepatitis C. Hepatology. 2007;46:315–323. doi: 10.1002/hep.21683. [DOI] [PubMed] [Google Scholar]
- 88.Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Jr., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arabi Y.M., Asiri A.Y., Assiri A.M., Balkhy H.H., Al Bshabshe A., Al Jeraisy M., Mandourah Y., Azzam M.H.A., Bin Eshaq A.M., Al Johani S., Al Harbi S., Jokhdar H.A.A., Deeb A.M., Memish Z.A., Jose J., Ghazal S., Al Faraj S., Al Mekhlafi G.A., Sherbeeni N.M., Elzein F.E., Al-Hameed F., Al Saedi A., Alharbi N.K., Fowler R.A., Hayden F.G., Al-Dawood A., Abdelzaher M., Bajhmom W., AlMutairi B.M., Hussein M.A., Alothman A. Interferon beta-1b and lopinavir–ritonavir for Middle East respiratory syndrome. N. Engl. J. Med. 2020;383:1645–1656. doi: 10.1056/NEJMoa2015294. [DOI] [PubMed] [Google Scholar]
- 90.Hung I.F.-N., Lung K.-C., Tso E.Y.-K., Liu R., Chung T.W.-H., Chu M.-Y., Ng Y.-Y., Lo J., Chan J., Tam A.R., Shum H.-P., Chan V., Wu A.K.-L., Sin K.-M., Leung W.-S., Law W.-L., Lung D.C., Sin S., Yeung P., Yip C.C.-Y., Zhang R.R., Fung A.Y.-F., Yan E.Y.-W., Leung K.-H., Ip J.D., Chu A.W.-H., Chan W.-M., Ng A.C.-K., Lee R., Fung K., Yeung A., Wu T.-C., Chan J.W.-M., Yan W.-W., Chan W.-M., Chan J.F.-W., Lie A.K.-W., Tsang O.T.-Y., Cheng V.C.-C., Que T.-L., Lau C.-S., Chan K.-H., To K.K.-W., Yuen K.-Y. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Davoudi-Monfared E., Rahmani H., Khalili H., Hajiabdolbaghi M., Salehi M., Abbasian L., Kazemzadeh H., Yekaninejad M.S. A randomized clinical trial of the efficacy and safety of interferon β-1a in treatment of severe COVID-19, antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.01061-20. e01061-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Repurposed antiviral drugs for covid-19 — interim WHO solidarity trial results. N. Engl. J. Med. 2020;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peiffer-Smadja N., Yazdanpanah Y. Nebulised interferon beta-1a for patients with COVID-19. Lancet Respir. Med. 2021;9:122–123. doi: 10.1016/S2213-2600(20)30523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Menezes S.M., Braz M., Llorens-Rico V., Wauters J., Van Weyenbergh J. Endogenous IFNβ expression predicts outcome in critical patients with COVID-19. The Lancet Microbe. 2021;2 doi: 10.1016/S2666-5247(21)00063-X. e235–e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Haji Abdolvahab M., Mofrad M.R.K., Schellekens H. Elsevier Inc.; 2016. Interferon Beta: from Molecular Level to Therapeutic Effects. [DOI] [PubMed] [Google Scholar]
- 96.Creeke P.I., Farrell R.A. Clinical testing for neutralizing antibodies to interferon-β in multiple sclerosis. Ther. Adv. Neurol. Disord. 2013;6:3–17. doi: 10.1177/1756285612469264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.S.B.U. of M.S. Seyed Sina Naghibi Irvani, Md, Mph, Mba, Senior Researcher Interferon beta 1a in hospitalized COVID-19 patients (IB1aIC), ClinicalTrials.Gov. 2020. https://clinicaltrials.gov/ct2/show/study/NCT04350671?term=interferon&cond=Covid-19&draw=2&rank=1
- 98.T.U. of H.K. Ivan Fn Hung Md . ClinicalTrials.Gov; 2020. Double Therapy with IFN-Beta 1b and Hydroxychloroquine.https://clinicaltrials.gov/ct2/show/study/NCT04350281?term=interferon&cond=Covid-19&draw=2&rank=17 [Google Scholar]
- 99.Synairgen Research Ltd . ClinicalTrials.Gov; 2020. Trial of Inhaled Anti-viral (SNG001) for SARS-CoV-2 (COVID-19) Infection.https://clinicaltrials.gov/ct2/show/NCT04385095?term=interferon&cond=Covid-19&draw=2&rank=12 [Google Scholar]