Abstract
Coronavirus Disease-2019 (COVID-19), a viral disease caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was declared a global pandemic by WHO in 2020. In this scenario, SARS-CoV-2 main protease (COVID-19 Mpro), an enzyme mainly involved in viral replication and transcription is identified as a crucial target for drug discovery. Traditionally used medicinal plants contain a large amount of bioactives and pave a new path to develop drugs and medications for COVID-19. The present study was aimed to examine the potential of Emblica officinalis (amla), Phyllanthus niruri Linn. (bhumi amla) and Tinospora cordifolia (giloy) bioactive compounds to inhibit the enzymatic activity of COVID-19 Mpro. In total, 96 bioactive compounds were selected and docked with COVID-19 Mpro and further validated by molecular dynamics study. From the docking and molecular dynamics study, it was revealed that the bioactives namely amritoside, apigenin-6-C-glucosyl7-O-glucoside, pectolinarin and astragalin showed better binding affinities with COVID-19 Mpro. Drug-likeness, ADEMT and bioactivity score prediction of best drug candidates were evaluated by DruLiTo, pkCSM and Molinspiration servers, respectively. Overall, the in silico results confirmed that the validated bioactives could be exploited as promising COVID-19 Mpro inhibitors.
Keywords: COVID-19, SARS-CoV-2, Main protease, Medicinal plants, Molecular docking, Molecular dynamics
1. Introduction
Coronavirus disease-2019 (COVID-19) is an infectious respiratory disease caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). COVID-19 started spreading worldwide since December 2019 from Wuhan city of China and has currently affected nearly 230 countries. The World Health Organization (WHO) has declared this outbreak as a global pandemic [1]. COVID-19 related deaths have crossed around 3 million worldwide and nearly 0.6 million new cases emerge everyday as of May 2021 [2]. The main symptoms of COVID-19 include fever, dry cough, tiredness, body ache, chills with shivering, breathing trouble or shortness in breath, chest pain or pressure, and loss of speech or movement. SARS-CoV-2 bind to the lung endothelial cells and induce immune response and cytokine storm syndrome thereby causing respiratory failure, which is being considered the main cause of death in COVID-19 patients [3]. SARS-CoV-2 is a positive strand RNA virus, mostly in spherical structure. It has the potential to change its morphology corresponding to varying environmental conditions [4]. The size of the SARS-CoV-2 genome is around 30 kb, enclosed with 5′-cap and 3′-poly(A) tail and the structure is comprised of spike, envelope, membrane and nucleocapsid proteins [5]. Currently, there is no specific antiviral drug available for the treatment of COVID-19 and the infected patients are primarily treated only by supportive therapy [6]. As of date, several vaccine candidates such as Tozinameran, AZD1222, Covishield, Ad26.COV2·S, Vero Cell, InCoV, mRNA-1273, Sputnik V, Ad5-nCoV, EpiVacCorona, Covaxin are available for COVID-19 prevention and more than ten candidates of vaccines are under clinical trials. More than 200 FDA approved drugs and vaccines have been registered for clinical trials. Several antiviral drugs (remdesivir, lopinavir, ritonavir, oseltamivir, favipiravir), anti-inflammatory agents (chloroquine, hydroxychloroquine (HCQ), glucocorticoids) and antimicrobials (azithromycin and antiprotozoals) have been attempted for management of COVID-19 patients [7]. However, the use of antiviral, anti-inflammatory and antimicrobial drugs can lead to toxicity associated health issues [8].
Ayurveda and Siddha medicines are often employed as alternative therapies for allopathy treatment. Medicinal plants are considered a rich source of bioactives for natural drug development and have played a crucial role in controlling numerous diseases [7,8]. Now-a-days, researchers are focused on the development of antiviral bioactive compounds from medicinal plants with high curative effect and no toxicity [8,9]. Medicinal plant-based bioactives have been traditionally used to treat many infectious and respiratory diseases [10,11]. In earlier reports, numerous computational screening studies have mapped out the potential bioactives from medicinal plants against the targets of SARS-CoV-2 [[12], [13], [14]]. These plant bioactives such as flavonoids, phenolics, polyphenols, tannins, saponins, alkaloids possess promising antiviral activities against SARS-CoV-2 and may guide the development of novel anti-COVID-19 prophylactics [10,15]. Emblica officinalis (Amla), Phyllanthus niruri Linn. (Bhumi amla) and Tinospora cordifolia (Giloy) are important medicinal plants native to India and are widely used in many ayurvedic formulations for treating several diseases [[16], [17]]. The bioactive compounds of these medicinal plants can act as immunomodulators and reduce cytokine storm against viral infections [[18], [19], [20]]. The present study was aimed to perform computational screening against the SARS-CoV-2 main protease (COVID-19 Mpro) using amla, bhumi amla and giloy derived bioactives.
2. Materials and methods
2.1. Database and software
DruLiTo 1.0.0 software, Open Babel v2.30, Discovery studio 2017R2, PubMed Database (https://pubmed.ncbi.nlm.nih.gov/), PubChem (https://pubchem.ncbi.nlm.nih.gov/), RCSB-Protein Data Bank (https://www.rcsb.org/), pkCSM – pharmacokinetics online server (http://biosig.unimelb.edu.au/pkcsm/), AutoDock Vina inbuild PyRx 0.8, GLIDE, Schrodinger, LLC, New York, NY, 2014, AMBER 2018 and AMBER 18 tools, University of California, San Francisco were used for molecular docking and simulation studies.
2.2. Blind docking protocol using autodock vina
2.2.1. Preparation of protein
The crystal structure of COVID-19 Mpro (PDB ID: 6LU7) was retrieved from the RCSB PDB site (www.rcsb.org). The structure contained a total of 306 amino acids with a 2.16 Å resolution (single chain). The water molecules, heteroatoms and undesired ligands were removed from the COVID-19 Mpro structure using Discovery studio 2017R2. The polar hydrogens were added to the COVID-19 Mpro structure to stabilize the charges. The PDB file of Mpro structure was converted into PDBQT file format using PyRx [21].
2.3. Preparation of ligand
Totally, 96 bioactive compounds of amla, bhumi amla and giloy were retrieved from recent literature [18,19,22]. The 2D and/or 3D structure data file (SDF) format of each ligand (bioactives) were collected from NCBI PubChem public database. All bioactive compound structures were subjected to energy minimization process using mmff94 force field algorithm and 3D structures of ligand were converted into PDBQT format using PyRx, before performing molecular docking analysis [23].
2.3.1. Virtual screening
Screening of bioactives against the COVID-19 Mpro was done using PyRx software. Initially the target protein of COVID-19 Mpro and bioactive ligands were loaded using AutoDock Wizard. The grid box for COVID-19 Mpro was generated using the PyRx software (X = −26.28, Y = 12.60, Z = 58.97) and the dimensions were set as 25.00 × 25.00 × 25.00 Å. The docking conformation of protein-ligand interactions was predicted with the energy value in kcal/mol wherein lowest binding energies (most negative) represented highest binding affinities [24,25].
2.4. Molecular docking and refining using GLIDE and prime
The protein and bioactive compounds were prepared using optimized potential for liquid simulations (OPLS) force field and three steps of docking were implemented using GLIDE Schrodinger, LLC, New York, NY, 2014 [26,27]. The binding affinity of protein-inhibitor complexes were estimated using Prime MM-GBSA module [20,28].
The protocol included removal of the water molecules, heteroatoms and peptide like inhibitors from the X-ray crystal structure of COVID-19 MPro. Hydrogen atoms are added to the structure after assigning them with suitable protonation states of acidic, basic and histidine residues of COVID-19 Mpro. using Maestro Protein Preparation Wizard [28]. The added hydrogen atoms were optimized to enhance the hydrogen bond network of the protein structure. Further, all the ligand SDF files were prepared through Maestro ligand preparation wizard using OPLS 2005 force field that analyzed various plausible 3D stereoisomers and protonation states through EpiK. The lower energy 3D conformer of bioactive compounds was considered for docking. Initially, the 96 compounds along with different protonation states for a few compounds (104) were subjected to high throughput virtual screening (HTVS) with OPLS force field where the grid center of receptor (Mpro) was defined as the coordinates of the peptide like inhibitor (X = −11.8762, Y = 11.5144, Z = 70.5154) with specified inner box (15, 15, 15) and outer box (27.9878, 27.9878, 27.9878) dimensions. To obtain accuracy on binding poses of these molecules, they were subjected to standard precision (SP) with OPLS 2005 force field [29]. Further, Extra Precision sampling (XP) was performed to remove the false positives and the advanced scoring function that enables inclusion of non-covalent interactions, penalty of entropy effect and penalty of restriction of ligands, as Glide G-score or XP glide score to validate the binding docking poses [20,26,30].
| XP glide Score = 0.065 * Van derWaals energy +0.130 * Coulomb energy + Lipo + Hbond + Metal + BuryP + RotB + Site |
Where Lipo represents hydrophobic interactions, Hbond represents hydrogen bonding interactions, Metal signifies metal binding, BuryP represents buried polar group penalty, RotB defines the penalty for freezing rotatable bonds and Site represents polar interactions present in the active site [31]. After removal of false positives, a total of 12 bioactive compounds-protein complexes were minimized using OPLS force field and the free energy (Gbind) of binding was estimated using molecular mechanics generalized born surface area (MM-GBSA) method of Prime with other default parameters.
| ΔGbind = ΔGcomplex – (ΔGprotein+ ΔGligand) |
The binding energy estimation includes generalized born molecular mechanics energies (EMM), surface generalized Born (SGB) solvation model for polar solvation (GSGB) and a non-polar solvation term (GNP).
2.5. Molecular simulation study
Although the rigid receptor approximation in GLIDE docking protocol has substantially considered the inductive effect of the protein conformation, the stability of binding poses was still unclear. To understand the stability of protein-inhibitor complexes and estimation of binding energies from an ensemble of conformations, a molecular dynamic run of 50ns for each of the 12 protein-bioactive complexes was performed. Quantum mechanical calculations of remdesivir, amritoside, pectolinarin, astragalin, apigenin-6-C-glucosyl7-O-glucoside, 7-keto sitosterol, 20a hydroxy ecdysone, chlorogenic acid, ellagic acid, cyanidin, tinosporine B, quercetin and epicatechin, at HF and 6–31 G* level of theory using Gaussian 16 [32] were performed to obtain the missing generalized atomic force field (GAFF) parameters for the bioactive compounds. The missing parameters were obtained through antechamber module of AMBER 18 [33]. All docking complexes were neutralized with sodium ions. A solvated octahedron box of TIP3P waters from the periphery of protein to the 10 Å, were added to each complex. The solvated water was initially minimized with steepest descent for 5000 steps and conjugate gradient for 5000 steps to optimize the hydrogen bond network in order to remove poor water clashes with the protein. The protein was employed with AMBER ff14SB [34] whereas the bioactive compounds were employed with GAFF force field. The complexes were annealed from 10 to 300 K under NVT conditions for 50ps and subjected to NPT ensemble at 1 atm pressure and 300K temperature for 500ps to obtain uniform density. Berendsen barostat and Langevin thermostat were implemented to maintain constant pressure and temperature for NPT ensemble. Later the complexes were equilibrated for ~3ns, and 50ns productive MD run was performed for each of these complexes using AMBER 18 GPU implementation [35]. Analysis of MD trajectories such as hierarchical agglomerative approach of cluster analysis, hydrogen bond analysis, root mean square fluctuations was executed using CPPTRAJ as mentioned in previous methodology [36]. The binding energies of the complexes were obtained from an ensemble of 5000 frames of 50ns MD run using MM-GBSA method in AMBER 18.
| ΔG0bind, solv = ΔG0bind, vacuum + ΔG0solv, complex– (ΔG0solv, ligand + ΔG0solv, receptor) |
where, ΔG0 bind, solv is the solvation free energy difference between the bound and unbound forms of receptor and ligand. It is comprised of ΔG0 bind, vacuum, the free energy difference between the bound and unbound forms of ligand and receptor in vacuum. ΔG0 solv, complex, ΔG0 solv, ligand, and ΔG0 solv, receptor are the differences between solvation free energies of the complex, ligand and receptor in solvent and vacuum, respectively. The individual binding energy contributions of ligand, receptor and complex in solvent and vacuum can be calculated as,
| ΔG0solv = ΔG0electrostatic+ ΔG0hydrophobic |
| ΔG0vaccum = ΔE0MM - T ΔS0 |
Where, ΔG0 solv is the solvation free energy which is the combination of electrostatic energy (ΔG0 electrostatic) and non-electrostatic energy or hydrophobic energy (ΔG0 hydrophobic) of the system. ΔE0 MM is the molecular mechanical energy contribution from the gas phase which is comprised of the average interaction energy between receptor and ligand, and the entropy change upon binding at 300K (T ΔS0).
| ΔEMM = ΔEelec + ΔEvdW + ΔGpolar + ΔGnon-polar |
Where Eelec and EvdW are the electrostatic and van der Waal's contributions, and Gploar and Gnon-polar are the polar and non-polar solvation terms (EGB and ESURF), respectively. It was noted that similar ranking order of binding poses was observed from MM-GBSA calculations from Prime and AMBER 18. All the graphics were prepared using the Maestro, Schrodinger LLC NY 2021.
2.6. Determination of drug-likeness properties
Drug-likeness properties of selected bioactive compounds were analyzed using DruLiTo software. Lipinski's rule (molecular mass (MM) less than 500 Da, no more than 5 hydrogen bond donors (HBD), no more than 10 hydrogen bond acceptors (HBA), and partition coefficient (log P) not greater than 5) were used to filter the bioactive compounds based on their physicochemical properties [37]. In addition, Atom Molar Refractivity (PSA), atom molar refractivity (AMR) and number of rotable ond (nRB) were also studied using DruLiTo [30].
2.7. Bioactivity score prediction
The bioactivity scores (ion channel modulation (ICM), G protein-coupled receptor (GPCR), nuclear receptor ligand (NRL) and enzyme inhibitors: protease, kinase) of the filtered bioactive compounds were predicted by using Molinspiration Cheminformatics online server [38].
2.8. ADMET/pharmacokinetic properties analysis
ADMET (Absorption, Distribution, Metabolism, Excretion, And Toxicity) properties analysis of the selected bioactive ligands was performed by pkCSM server. The compound structure SMILES was retrieved from NCBI PubChem database and was used as the input file for the pkCSM online tool [39]. The absorption (gastrointestinal absorption, bioavailability, water solubility (log S), Caco-2 and skin permeability), distribution (blood–brain barrier (BBB), central nervous system (CNS) permeability, volume of distribution (VDss) unbound state), metabolism (various metabolic enzymes of Cytochromes P450 (CYP)), excretion (drug and renal clearance) and toxicity (AMES, acute and chronic hepatotoxicity, Lethal Dose (LD50) values, Skin Sensitization, and etc) properties of the selected bioactives were predicted.
3. Results and discussion
3.1. Molecular docking and dynamics studies
The computational screening approach is mainly used for identifying the potential drug candidates from the chemical libraries. The present study employed virtual screening of compounds from amla, bhumi amla and giloy against COVID-19 Mpro. The flow diagram of the computational work is given in Figure-1 . COVID-19 Mpro is a class of viral protease, which is considered as a functional therapeutic target protein due to its important role in processing of viral polyproteins and viral maturation inside the infected host cells. Initially, 96 bioactive compounds were subjected to molecular docking analysis using blind docking mode in Autodock vina. These molecules were more bound to domain II and domain III residues rather than the substrate binding pocket. Further, virtual screening using the receptor grid docking at the cleft of domain I and domain II active site [40,41] slightly altered the relative ranking of the bioactive molecules (Table 1, Table 2, Table 3 ). The refining of docking poses using extensive sampling and advanced scoring function under the extra precision method of Glide and MM-GBSA enabled to finalize the most suitable bioactive molecules for binding affinity prediction (Table 4 ). The docked complex of COVID-19 Mpro with bioactive drug candidates was subjected to molecular simulations (MD) to understand the protein flexibility influencing the orientation of binding poses and non-covalent interaction patterns that contribute to binding free energy [20,26,30,31].
Fig. 1.
Flowchart of stepwise methodology used in docking studies to identify the potential bioactive compounds-based inhibitors for COVID-19 Mpro.
Table 1.
Binding energies (kcal/mol) obtained during blind docking (Autodock Vina), high through put screening (HTVS) and standard precision (SP) docking score of selected bioactives from Giloy to the substrate binding cleft (GLIDE) of COVID-19 Mpr.o.
| S. No |
Pubchem ID | Compound name | Binding energy (kcal/mol) | HTVS Docking score |
SP Docking score |
|---|---|---|---|---|---|
| 1 | 122,206,356 | Tinosporine B | −7.4 | −4.2 | – |
| 2 | 73,981,613 | Amritoside | −7.2 | −5.63 | −7.61 |
| 3 | 15,215,479 | Tinosponone | −7.1 | −6.4 | – |
| 4 | 72,276 | Epicatechin | −7.1 | −6.3 | −6.76 |
| 5 | 100,926,541 | Tinocordifolioside | −6.9 | −4.92 | −5.44 |
| 6 | 100,926,540 | tinocordifolin | −6.8 | −5.27 | −6.12 |
| 7 | 46,173,925 | Isocolumbin | −6.8 | −5.90 | −5.70 |
| 8 | 101,916,313 | Tinocordioside | −6.7 | −5.36 | – |
| 9 | 2353 | Berberine | −6.5 | −6.06 | −6.06 |
| 10 | 72,323 | Jatrorrhizine | −6.4 | −5.75 | −6.0 |
| 11 | 44,257,772 | Apigenin-6-C-glucosyl7-O-glucoside | −6.4 | −6.7 | −8.05 |
| 12 | 21,636,215 | Borapetoside | −6.3 | −3.72 | −3.85 |
| 13 | 439,653 | Reticuline | −6.2 | −5.79 | −7.04 |
| 14 | 167,718 | tembetarine | −6.1 | −6.3 | −6.82 |
| 15 | 637,775 | Sinapic acid | −6.1 | −4.91 | −5.24 |
| 16 | 101,915,817 | cordioside | −5.9 | −2.2 | −5.16 |
| 17 | 72,301 | Tetrahydropalmatine | −5.9 | −4.86 | −5.34 |
| 18 | 3638 | Hydrastinine | −5.8 | −5.03 | −5.62 |
| 19 | 73,337 | Magnoflorine | −5.7 | −6.28 | −6.18 |
| 20 | 19,009 | palmatine | −5.7 | −5.69 | – |
| 21 | 11,168,362 | Pinoresinol-di-O-glucoside | −5.6 | −5.42 | −5.17 |
| 22 | 5,459,840 | 20a-Hydroxy ecdysone | −5.5 | −4.47 | −5.41 |
| 23 | 5,316,860 | Syringin | −5.4 | −4.03 | −7.1 |
| 24 | 10,153 | Corydine | −5.4 | −6.49 | −6.16 |
| 25 | 8468 | Vanillic acid | −5.4 | −5.14 | – |
| 26 | 122,206,355 | Tinosporine A | −5.3 | −4.49 | – |
| 27 | 30,358 | Menisperine | −5.2 | −6.72 | −5.94 |
| 28 | 12,312,690 | makisterone | −5.1 | −4.62 | −4.67 |
| 29 | 45,359,937 | Cordifolioside A | −5.1 | −6.27 | −6.48 |
| 30 | 11,081,347 | Ecdysterone | −5 | −4.47 | −5.41 |
| 31 | 305 | Choline | −3.3 | −4.06 | −4.5 |
Table 2.
Binding energies (kcal/mol) obtained during blind docking (Autodock Vina), high through put screening (HTVS) and standard precision (SP) docking score of selected bioactives from Bhumi amla to the substrate binding cleft (GLIDE) of COVID-19 Mpr.o.
| S. No | Pubchem ID | Compound name | Binding energy (kcal/mol) | HTVS Docking score | SP Docking score |
|---|---|---|---|---|---|
| 1 | 5,281,855 | Ellagic acid | −8.4 | −5.9 | −6.32 |
| 2 | 128,861 | Cyanidin | −6.9 | −7.02 | −7.13 |
| 3 | 1,794,427 | Chlorogenic acids | −6.9 | −5.22 | −5.21 |
| 4 | 193,552 | Phyllanthine | −6.8 | −5.3 | −5.25 |
| 5 | 44,257,151 | Quercetol | −6.8 | −5.69 | −5.79 |
| 6 | 5,280,343 | Isoquercetin | −6.8 | −6.25 | −6.87 |
| 7 | 68,071 | Pinocambrin | −6.8 | −6.0 | −7.33 |
| 8 | 122,173,234 | Kaempferol-3- O-rutinoside | −6.7 | −7.44 | −8.57 |
| 9 | 135,403,798 | Theaflavin | −6.7 | −6.39 | −7.76 |
| 10 | 439,246 | Naringenin | −6.7 | −6.69 | – |
| 11 | 92,158 | Lupenone | −6.7 | −4.3 | −4.0 |
| 12 | 637,760 | Chalcone | −6.6 | −5.82 | −6.69 |
| 13 | 68,079 | Isopimpinellin | −6.6 | −6.21 | −5.76 |
| 14 | 10,151,874 | Valoneic acid dilactone | −6.5 | −4.94 | −5.37 |
| 15 | 5,281,672 | Myricetin | −6.5 | −6.08 | – |
| 16 | 129,720,117 | Glycolic acid | −6.4 | −5.55 | −5.67 |
| 17 | 442,428 | Naringin | −6.3 | −5.62 | – |
| 18 | 164,893 | 5-pcoumaroyl quinic acid | −6.2 | −6.96 | – |
| 19 | 442,872 | securinine | −6.2 | −4.7 | −5.59 |
| 20 | 131,752,343 | procyanidin dimer | −6.1 | −5.57 | −6.17 |
| 21 | 5,320,835 | Quercetin-3, 4-O-diglucoside | −6.1 | −7.52 | −7.50 |
| 22 | 689,043 | Caffeic acid | −6.1 | −4.56 | −5.03 |
| 23 | 72,193,643 | 4-sinapoyl quinic acid | −6 | −4.15 | −5.73 |
| 24 | 121,225,501 | 1-caffeoyl-5-feruloylquinic acid | −5.9 | −3.4 | −5.03 |
| 25 | 168,849 | Pectolinarin | −5.8 | −4.23 | −6.76 |
| 26 | 5,282,102 | Astragalin | −5.8 | −7.22 | −7.31 |
| 27 | 338 | Salicylic acid R1 | −5.7 | −5.26 | −4.85 |
| 28 | 5,317,238 | Ethyl caffeate | −5.7 | −5.4 | −5.59 |
| 29 | 323 | Coumarin | −5.6 | −5.4 | −5.32 |
| 30 | 5,274,585 | Quercetin-3-O-glucuronide | −5.6 | −6.84 | −6.89 |
| 31 | 5,280,459 | Quercitrin | −5.6 | −6.32 | −6.21 |
| 32 | 5,281,613 | Diosmin | −5.5 | −5.17 | −6.98 |
| 33 | 72 | Protocatechuic acid | −5.3 | −5.53 | −5.29 |
| 34 | 462,192 | Malvidin3,5-O-diglucoside | −5 | −6.79 | −7.24 |
| 35 | 4133 | Methyl salicylate | −4.9 | −4.91 | – |
Table 3.
Binding energies (kcal/mol) obtained during blind docking (Autodock Vina), high through put screening (HTVS) and standard precision (SP) docking score of selected bioactives from amla to the substrate binding cleft (GLIDE) of COVID-19 Mpr.o.
| S. No | Pubchem ID | Name | Binding energy (kcal/mol) | HTVS docking score | SP docking score |
|---|---|---|---|---|---|
| 1 | 5,281,855 | Ellagic acid | −8.4 | −5.89 | −6.32 |
| 2 | 1,794,427 | Chlorogenic acid | −6.9 | −5.22 | −5.21 |
| 3 | 5,280,343 | Quercetin | −6.9 | −6.25 | −6.87 |
| 4 | 72,277 | Epigallocatechin | −6.9 | −4.65 | −6.65 |
| 6 | 10,914,547 | Phyllaemblic acid C | −6.9 | −5.41 | −6.71 |
| 5 | 5,280,443 | Apigenin | −6.7 | −7.42 | −7.34 |
| 7 | 181,681 | Medioresinol | −6.5 | −6.27 | −4.95 |
| 8 | 5,280,863 | Kaempferol | −6.5 | −7.45 | −7.13 |
| 9 | 5,281,672 | Myricetin | −6.5 | −6.08 | – |
| 10 | 102,039,055 | Mucic acid 2-O-gallate | −6.4 | −5.06 | −5.29 |
| 11 | 124,375 | Glucogallin | −6.4 | −5.77 | −5.34 |
| 12 | 44,258,098 | luteolin-4 neohesperidoside | −6.4 | −6.83 | −5.93 |
| 13 | 13,917,513 | Isostrictinin | −6.3 | −4.72 | −4.62 |
| 14 | 5,089,687 | Prodelphinidin | −6.3 | −4.55 | −7.15 |
| 15 | 11,057,167 | Phyllaemblic acid B | −6.2 | −6.05 | −5.34 |
| 16 | 689,043 | Caffeic acid | −6.1 | −4.56 | −5.03 |
| 17 | 444,539 | Cinnamic acid | −6 | −4.97 | −4.11 |
| 18 | 637,542 | Coumaric acid | −6 | −5.03 | −4.43 |
| 19 | 5,280,805 | Rutin | −5.9 | −7.38 | −7.41 |
| 20 | 100,067 | Lirioresinol A | −5.8 | −4.48 | −5.89 |
| 21 | 5,280,536 | Coniferyl aldehyde | −5.6 | −5.1 | −5.25 |
| 22 | 7428 | Methyl gallate | −5.6 | −4.80 | −5.31 |
| 23 | 160,608 | 7-Ketositosterol | −5.5 | −2.59 | −5.55 |
| 24 | 73,568 | Corilagin | −5.5 | −3.16 | −4.93 |
| 25 | 370 | Gallic acid | −5.3 | −5.50 | −5.61 |
| 26 | 3,037,582 | Galactaric acid | −5.1 | −4.86 | −3.86 |
| 27 | 54,670,067 | vitamin-C | −5.1 | −3.65 | −4.57 |
| 28 | 1057 | Pyrogallol | −4.9 | −5.23 | −6.75 |
| 29 | 7456 | Methyl-4-hydroxybenzoate | −4.9 | −5.19 | – |
| 30 | 289 | Catechol | −4.6 | −4.78 | −5.62 |
Table 4.
Top hits of bioactives of Giloy, Bhumi amla and Amla with COVID-19 Mprobased on extra precision method (XP) of Glide.
| Bioactive name | Docking score | Glide emodel | XP GScore | MMGBSA ΔGBind | Molecular simulations |
||
|---|---|---|---|---|---|---|---|
| ΔG Bind | EEL | Vdw | |||||
| Remedesivir | −9.27 | −89.55 | −9.27 | −63.50 | −47.59 | −60.85 | −30.30 |
| Amritoside | −11.28 | −83.05 | −11.33 | −60.35 | −34.21 | −36.16 | −47.54 |
| Pectolinarin | −9.55 | −84.07 | −9.56 | −54.02 | −32.14 | −24.73 | −44.63 |
| Astragalin | −7.87 | −72.75 | −7.90 | −50.08 | −31.19 | −33.89 | −37.14 |
| Apigenin-6-C-glucosyl7-O-glucoside | −11.34 | −90.28 | −11.34 | −50.50 | −34.68 | −65.08 | −36.92 |
| 7-Ketositosterol | −5.75 | −40.67 | −5.75 | −46.67 | −29.81 | −10.60 | −39.51 |
| 20a-Hydroxy ecdysone | −7.52 | −55.15 | −7.52 | −44.45 | −12.90 | −15.13 | −20.33 |
| Chlorogenic acid | −9.07 | −57.89 | −9.07 | −42.53 | −22.64 | −28.67 | −28.56 |
| Ellagic acid | −6.42 | −53.03 | −6.42 | −40.69 | −13.91 | −13.35 | −21.82 |
| cyanidin | −6.76 | −54.88 | −6.76 | −39.92 | – | – | – |
| Tinosporine B | −5.05 | −50.62 | −5.05 | −39.69 | −19.47 | −13.80 | −29.93 |
| Quercetin | −8.27 | −51.81 | −8.27 | −38.77 | −22.51 | −18.07 | −31.02 |
| Epicatechin | −7.19 | −51.55 | −7.19 | −38.00 | −21.50 | −19.48 | −26.96 |
| Apigenin | −7.05 | −45.03 | −7.09 | −32.88 | – | – | – |
| Epigallocatechin | −7.62 | −50.05 | −7.62 | −32.69 | – | – | – |
| Phyllaemblic acid C | −6.04 | −33.94 | −6.04 | −11.98 | – | – | – |
| Paracetmol | −6.23 | −32.21 | −6.23 | −27.05 | – | – | – |
(Docking score, glide emodel, XP Gscore and binding energies (kcal/mol) from ΔG bind from Prime MM/GBSA. MM/GBSA binding energy (ΔG bind, kcal/mol), electrostatic (EEEL) and Van der Waals (EVdw) energy contribution of EMM from molecular simulations).
3.1.1. Phytochemicals of giloy
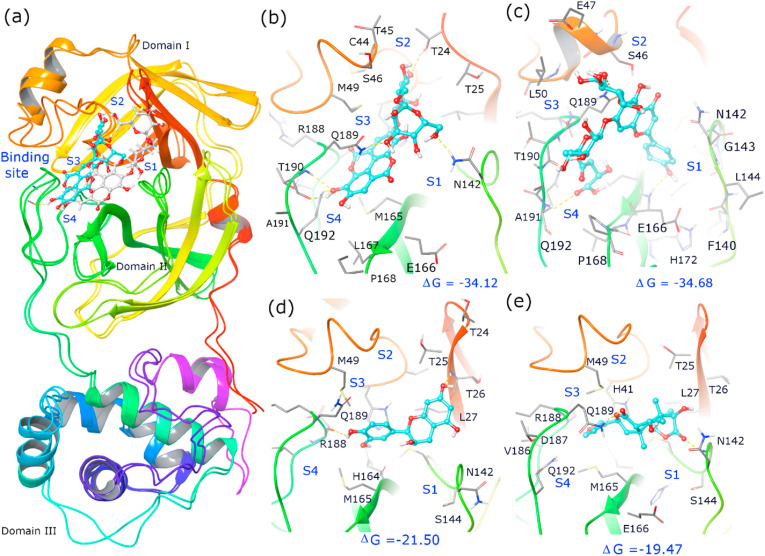
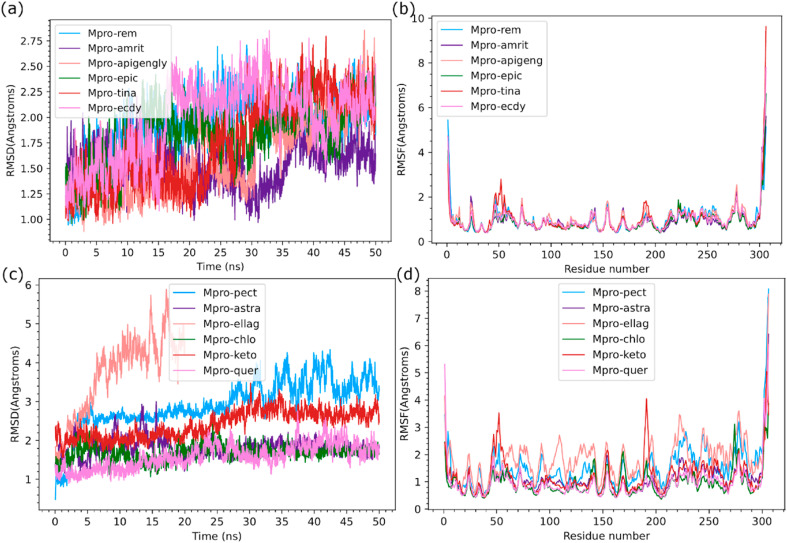
Giloy is a familiar medicinal plant containing various biological and therapeutic properties to treat conditions like skin diseases, anemia, inflammation, rheumatism, etc. Giloy bioactives assist the immune system to resist infections and maintain leucocyte functioning [[42], [43]]. Also, giloy formulations are commercially used for digestive problems and as immunomodulators. The previous reports of Kapil and Sharma (1997) and Bishayi et al. (2002) have suggested that giloy bioactives namely cordifolioside A, magnoflorine, syringin and tinocordiside showed cytotoxic and immunomodulatory properties [27,28]. These giloy secondary metabolites enhanced the phagocytic activity of macrophage cells. Upadhyaya and coworkers (2011) reported that aqueous extracts of giloy influenced the stimulation of immune cells along with cytokine production [44]. The crude extracts of giloy with a polyclonal B cell mitogen showed enhancement of immune response. along with macrophage activation and prevention of oxidative damage [45]. In the present study, 32 bioactive candidates were chosen from giloy for the docking analysis. Among the 32 bioactives, amritoside (−60.35 kcal/mol), apigen-6-C-glucosyl7-O-glucoside (−50.50 kcal/mol), 20a hydroxy ecdysone (−44.45 kcal/mol), tinosporine B (−39.69 kcal/mol) and epicatechin (−38.0 kcal/mol) showed higher binding affinity compared to other compounds (Table-4). These bioactives also revealed interactions with crucial active site residues observed in acetoamide and peptide inhibitors of Mpro (Table-5 ). Figure-2 and Figure-3 depict the binding pose observed in the populated cluster of the molecular simulations. The docking and binding pose of amritoside during MD simulation has root mean square deviation (RMSD) of Cα atoms below 2 Å (Figure-4a ). The root mean square fluctuation (RMSF) of protein residues indicated flexibility at N- and C-terminal residues and minor perturbation of active site residues. It is noted that tinosporine B, apigen-6-C-glucosyl7-O-glucoside and 20a hydroxy ecdysone have increased flexibility of loops in the regions connecting the domains (38–52 and 176–200) and observed to have slightly higher RMSD (2.50–2.75 Å). A detailed analysis of noncovalent interactions of MD simulation trajectory with respect to the initial docking pose indicated that except for 20a hydroxy ecdysone the other bioactive molecule retained their initial docking pose in the active site pocket (Figure-3a). Based on previous reports [[33], [34], [35]], the crucial active site residues were defined to have four sub sites namely, S1 (F140, N142, E166, H163, H172), S2 (H41, M49, Y54), S3 (M165, L167, F185, Q192), and S4 (P168, T190, A191). Interaction with these active site residues is observed in the docking pose and throughout the MD run (Table-5). However, there was a slight tumbling and reorientation of the moieties of bioactive compounds to S1 and S2 pockets, that caused an alteration in the number of hydrogen bonds and van der Waals interactions in the populated cluster of tinasporine B and apigen-6-C-glucosyl7-O-glucoside. The binding free energy of these complexes implied amritoside ~ apigen-6-C-glucosyl7-O-glucoside > epicatechin > tinasporine B > 20a hydroxy ecdysone. A slight difference in a binding affinity order was obtained, while performing the minimized estimation of docking poses. This conformational ensemble changes indicate that the protein flexibility is varied in each pose. The diterpene glucoside derivatives of amritoside present in the active site of COVID-19 Mpro indicated strong hydrogen bond interaction with the backbone atoms of T190 of S4 sub site and the side chain of N142 of S1. Additionally, one glycoside is embedded in S2 pocket forming hydrogen bond with side chain of T24 and also has hydrophobic interactions with catalytic residue H41 (Figure-2b). The apigen-6-C-glucosyl7-O-glucoside has two interactions with the catalytic residues. However, it forms crucial hydrogen bonds with back bone atoms of E166, Q192 and side chain of Q189 along with hydrophobic interactions with N142 of S1, M165 of S3 and S4 sub site residues namely, P168, A191 and T190 (Figure-2c). The phenolic compound of epicatechin is retained in the active site pocket through three H-bonds with side chain of T26 and back bone residues of D187 and H164 along with hydrophobic residue of S1, S2 and S3 sub sites (Table-5, Figure-2d). Tinosporine B is mainly accommodated to S1 and S3 pocket using hydrogen bonding interactions with side chains of N142 and Q189 respectively along with hydrophobic interactions with catalytic residue C145 (Figure-2e). 20a Hydroxy ecdysone migrated away from the substrate binding cleft during equilibration and interacted only with the S3 site residues (Figure-3a). Amritoside ~ apigen-6-C-glucosyl7-O-glucoside seem to have higher binding affinity than the others as the former has higher contribution of van der Waals interactions and strong electrostatic interactions (Table-4).
Table 5.
Altered hydrogen bonding interactions in binding pose of docking and most populated cluster protein complexes during molecular simulation.
| Compound Name | Docking Pose |
Cluster binding pose |
||
|---|---|---|---|---|
| H-bond residues (Bond lengths Å) | Vdw Interactions | H-bond residues (Bond lengths Å) | Vdw Interactions | |
| Amritoside | R188 (1.99 Å), N142 (2.05 Å), G143 (2.73 Å), T24 (1.79 Å), S46 (1.77 Å) | T25, T26, L27, H41, M49, T45, M165, Q189, D187, E166, C145 | Q189 (1.81 Å), T190 (1.76 Å,2.02 Å) Q192 (2.08 Å), N142 (2.0 Å) C44 (1.54 Å,2.26 Å) T24 (2.00 Å) |
R188, M49, S46, D187, H41, S46, T45, N142 |
| Apigenin-6-C-glucosyl7-O-glucoside | N142 (2.09 Å), Q192 (2.31 Å), E166 (2.50 Å) | F140, L141, M165, L167, P168, C145, H163, H164, H41, Q189 | Q189 (2.45 Å), Q192 (2.39 Å) E166 (2.09 Å) | M165, H163, N142, L167, P168, A191, T190 |
| Epicatechin | T26 (1.83 Å), Q189 (1.96 Å) | T25, L27, G143, N142, M49, R188, D187, M165, H164, H41, | H164 (2.32 Å), D187 (2.35 Å, 1.86 Å), T26 (2.30 Å) | Q189, M165, L27, G143, T25, H41, M49, |
| Tinosporine B | G143 (2.04 Å) | H41, H164, E166, N142, C145, M165, N189, R188 | N189 (1.80 Å), G143 (2.82 Å) | N142, C145, M165, M49, D187, R188 |
| 20a-Hydroxy ecdysone | E166 (1.79 Å, 1.73 Å), L141 (1.84 Å) | N142, H163, C145, H164, F140, M165, P168, Q189 | R40 (2.08 Å, 1.85 Å) | - |
| Pectolinarin | E166 (1.84 Å), F140 (1.92, 1.93 Å), G143 (1.70 Å) | T26, H41, N142, M165, L167, P168, D187, R188, Q189 | R 188 (2.60 Å), Q192 (1.98 Å), E166 (2.30 Å) | M165, D187, H164, Q180, C145, L27, H41, Q189, L167 |
| Astragalin | T190 (1.80 Å, 1.77 Å), E166 (2.49 Å), F140 (1.99 Å) | P168, R188, Q192, T190, M165, Q189, D187, H164, H163, H41, L141 | T190 (1.59 Å, 1.52 Å), Q192 (1.82 Å,2.33 Å), D187 (1.66 Å) | A191, L167, E166, N142, H163, M165, M49, Q189, H41 |
| Chlorogenic acid | N142 (2.09 Å G143 (2.05 Å), T26 (1.67 Å, 1.97 Å) | C145, T25, R188, D187, H41, M165, H164, M49, Q189 | D187 (1.71 Å), H41 (2.00 Å), T26 (1.76 Å, 2.05 Å) | T25, C145, H164, N189, M49, R188, M165 |
| Ellagic acid | F140 (1.71 Å) S144 (2.55 Å), C145 (2.24 Å) | Q189, M165, E166, H164, H41, H163, L141, | F140 (1.93 Å), H172 (2.18 Å) | L141, N142, E166, H163 |
| cyanidin | E166 (2.4 Å), R188 (1.93 Å), T190 (2.11 Å), L141 (2.29 Å), | C145, F140, M165, Q189 | – | – |
| 7-Ketositosterol | T26 (1.68 Å, 2.78 Å), S 144 (2.51 Å) | T25, H41, M49, M165, Q189, E166, N142, G143, C145 | T25 (1.83 Å) | H41, R188, L167, P168, M149, D187 |
| Quercetin | T26 1.86 Å, 2.14 Å), D187 (2.14 Å) | T25, L27, H41, M49, M165, Q189, H164, G143 | R188 (2.11) C145 (2.28 Å) G143 Å), N142 (2.20 Å) | D187, H41, M165, V186, Q189, H164, L27 |
| Remedesivir | Q189 (1.72 Å), T24 (1.92 Å) | M49, T25, T26, L27, H41, C145, H164, D187, M165, GLU166, L167, Q192, P168, T190 | T25 (1.73 Å), H41 (2.01 Å) | V42, C44, T25, L27, T45, S46, M49, G143, Q192, H164, V186, R188, M165, P168, E166 |
Fig. 2.
(a) Cartoon representation of COVID-19 Mpro with superimposition of protein (6LU7) with docking pose and binding pose of amritoside in highest populated cluster of 50ns MD run. Binding pose and interactions of bioactive compounds of giloy in the active site pocket of highest populated cluster Mpro complex with (a) amritoside (b) apigenin-6-C-glucosyl7-O-glucoside, (c) epicatechin (d) tinosporine B. Note that bioactive compounds are shown in ball and stick model and cyan color indicates binding pose clustered in MD run and white color indicates docking pose. Crucial active site residues are labelled and shown in sticks along with key hydrogen bonds (dashed yellow lines) and subsites (S1, S2, S3, S4) and the relative binding free energy (ΔG) is given in kcal/mol (Maestro, Schrödinger).
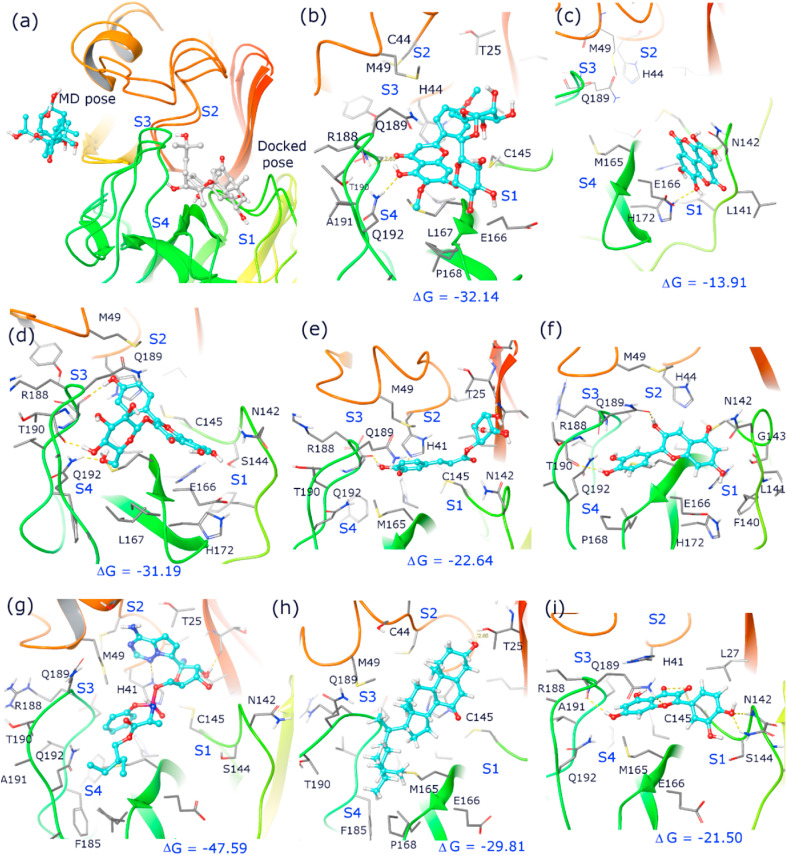
Fig. 3.
Binding pose and interactions of inhibitor and bioactive compounds of giloy, bhumi amla and amla in the active site pocket of Mpro during 50 ns MD run (a) 20ahydroxy ecdysone (b) pectolinarin, (c) ellagic acid (d) astragalin (e) chlorogenic acid (f) cyanidin (g) Remedesivir (h) 7-ketositosterol (i) quercetin. Note that bioactive compounds are shown in ball and stick model and cyan color indicates of MD run and white color indicates docking pose. Crucial active site residues are labelled and shown in sticks along with key hydrogen bonds (dashed yellow lines) and subunits (S1, S2, S3, S4) and the relative binding free energy (ΔG) is given in kcal/mol (Maestro, Schrödinger).
Fig. 4.
Root mean square deviation of C-alpha atoms (Å) of COVID-19 Mpro complexed with inhibitor and bioactive compounds during 50 ns MD run for (a) Remedisivir and giloy (amritoside, apigenin-6-C-glucosyl7-O-glucoside, epicatechin, tinosporine B, 20ahydroxy ecdysone); (b) Bhumi amla and Amla (pectolinarin, astragalin, chlorogenic acid, ellagic acid, cyanidin, 7-ketositosterol and quercetin); (c &d) Root mean square fluctuation per residue (Å) of COVID-19 Mpro complexed with inhibitor and bioactive compounds of giloy and bhumi amla and amla during 50ns MD. Note that ellagic acid has gradual increase in RMSD to 4.0 Å in 20ns indicating the weak association and also altered the protein conformation.
3.1.2. Phytochemicals of bhumi amla
Bhumi amla is widely used as a medication for many illnesses such as jaundice, kidney stones, dysentery, diabetes, etc. In addition, the bhumi amla extract stimulated both cellular and humoral immune responses and induced the activity of lymphocytes and macrophages [46]. The major bioactives of bhumi amla - catechin, quercetin and astragalin regulate the immune system via various signaling pathways such as mitogen-activated protein kinase (MAPK), Nuclear factor kappa B (NF-κB) and Interferon regulatory factors (IRF) pathways. Oral administration of bhumi amla extract was shown to regulate both the innate and adaptive immunity and modulate the levels of primary and secondary antibodies in the blood [47]. In an earlier report, clinical studies on consumption of bhumi amla extracts (50 mg, thrice a day, for 2 months) increased the Interleukin (IL-6), Tumour necrosis factor (TNF-α) production and regulated T-helper cells (Th 2) lymphocytes in tuberculosis (TB) patients [19]. Another report suggested that bhumi amla extracts stimulated the immune cells activity in TB patients [48]. Bhumi amla also promote the cell proliferation and enhance the phagocytosis of macrophage. In the present study, 35 bioactives were selected from bhumi amla as per the recent literature and docked with COVID-19 Mpro. Among these compounds, pectolinarin (−54.02 kcal/mol), astragalin (−50.50 kcal/mol), chlorogenic acid (−42.53 kcal/mol), ellagic acid (−40.69 kcal/mol), cyanidin (−39.92 kcal/mol), exhibited maximum binding affinities to the target COVID-19 Mpro (Table-4). The binding free energy ranking order from the MD simulations is similar to that obtained from Prime MMGBSA. However, there is tumbling and reorientation of binding poses for ellagic acid and pectolinarin in molecular simulations. Ellagic acid, present in both Amla and Bhumi Amla showed high RMSD and RMSF (Figure-4c and 4d), having similar binding pose till 10ns after which it slowly drifted away from the active site pocket at 20ns MD run, thereby indicating very weak association and low affinity towards Mpro (Table-5). Pectolinarin complex has also shown increased RMSD of Cα atom up to 3.5 Å (Figure-4c), where there is a reorientation of binding pose and higher penetration into S2 pocket, leading to van der Waals interactions with catalytic residues H41 and C145 (Figure-3b, Table-5). The complex also formed two hydrogen bonds with the backbone atoms of E166 and R188, and one with the side chain of Q192, belonging to S1 and S3 subsite residues. While astragalin and chlorogenic acid have low RMSD and RMSF, the docking binding pose is retained with increased noncovalent interactions towards the catalytic residues C145 and H41 (Table-5). Astragalin mostly resided in S1, S3 and S4 sub sites where it has formed hydrogen bonds with back bone atoms of T190 and side chain of Q192. It also developed crucial van der Waals interactions with H41, M49, N142, E166, L167, H163, M165, Q189 and A191 residues (Figure-3d). Chlorogenic acid (present in both amla and bhumi amla) has formed hydrogen bond with side chain of H41 and T26 with back bone of D187 and hydrophobic interactions with T25, M49, C145, H164, M165, R188 and N189 residues (Figure-3e). The high binding affinity of astragalin as compared to chlorogenic acid is attributed to the substantially higher electrostatic and van der Waals contributions. On the other hand, high binding energy of pectolinarin is due to the higher van der Waals contribution (Table-4).
3.1.3. Phytochemicals of amla
Amla has been widely used for medicinal and nutraceutical purposes. Every part of amla is useful for different pharmaceutical applications. Amla is often reported for its various biological properties such as immunomodulatory, anti-inflammatory, antioxidant, anticancer, antiviral, anti-diabetic, antimicrobial, etc. [22]. Amla is a rich source of vitamin C, which significantly increase the natural killer cell activity and reduce oxidative stress [49]. Srikumar et al. (2006) reported that amla extracts increased the immune response and regulated the activities of superoxide dismutase, catalase and glutathione S transferase [50]. In the present study, 30 bioactives of amla were selected from literature and docked with COVID-19 Mpro. Among these compounds, 7-ketositosterol (−46.67 kcal/mol), Quercetin (−38.77 kcal/mol), Epigallocatechin (−32.69 kcal/mol) and Phyllaemblic acid-C (−11.98 kcal/mol) were observed to fit the substate binding cleft of Mpro (Table-4). During the MD simulations, 7-ketositosteol showed slightly higher RMSD and RMSF compared to Quercetin, indicating its induced perturbation to protein conformation and tumbling of the molecule in the active site. Therefore 7-ketositosterol was observed to have higher binding energy (−29.81 kcal/mol) than Quercetin (−21.5 kcal/mol) due to the increased van der Waals contribution. The 7-ketositosterol was stretched to S4 pocket and held via one hydrogen bond with the side chain of T25 and via van der Waals interactions with catalytic residue H41, and other interactions with L167, M149, P168, D187 and R188 residues of S3 and S4 subsites (Figure-3h). On the other hand, Quercetin occupied S1 and S2 sub sites forming hydrogen bonds with catalytic residue C145, backbone of G143 and side chains of N142 and R188 of S1 subsite residues. It was also observed to have van der Waals interactions with H41, L27, M16, H164, V186, D187 and Q189 (Figure-3j).
3.2. Molecular docking for standard drugs
The molecular docking results of the lead bioactive candidates were compared to HCQ, remdesivir and paracetamol. Remdesivir is an antiviral medication, commonly used for the management of viral infections including recent COVID-19. Remdesivir and COVID-19 Mpro docked complex exhibited two crucial hydrogen bond interactions with side chains of T24 and Q189. During the MD run, the adenine ring stretched to the S2 sub site that resulted in the formation of hydrogen bond with catalytic residues H41 and T25 (Figure-3g, Table-5). The binding mode and nature of interactions are in line with the molecular docking studies of anti-viral drugs reported earlier [51,52]. The binding energy of remdesivir with minimized docked pose (−63.50 kcal/mol) and from the ensemble of conformations of MD run (−47.59 kcal/mol) was higher as compared to all the other bioactive compounds. HCQ and paracetamol are also under clinical trials as possible medications for COVID-19. HCQ with COVID-19 Mpro docked complex showed two hydrogen bonds (Q110, T111) and four hydrophobic interactions (I106, V101, V104) with significant binding energy (−6.6 kcal/mol). Similarly, the protein ligand interaction of paracetamol and COVID-19 Mpro formed three hydrogen bonds (T111, N151, D295) to the amino acid residues. It can be inferred from the above results that the traditional medicinal plants of E. officinalis, P. niruri and T. cordifolia derived bioactives also showed similar results as compared to the standard antiviral drugs (Table-5).
3.3. COVID-19 Mpro inhibitors from medicinal plants
Recently, Rajagopal et al. (2020) performed in silico studies for COVID-19 Mpro using Curcuma longa (turmeric). Cyclocurcumin, a curcumin derivative, showed significantly active interaction with COVID-19 Mpro (−6.77 kcal/mol). The docking results of cyclocurcumin displayed formation of two hydrogen bonds with amino acid residues of COVID-19 Mpro (T26, H41) [53]. Similarly, turmeric derivatives, curcuminoid and tetrahydroxycurcumin showed minimized binding affinity (−9.08 and −8.07 kcal/mol) and displayed hydrogen bonding (T25, G166, T190) [54]. Sesamum indicum (Sesame) is an oilseed crop used as a traditional medicine in India. S. indicum derived compounds, Sesami, Sesaminol and Sesamolin docked with COVID-19 Mpro showed minimum binding affinities (−8.2, −7.8, −7.7 kacl/mol). These compounds interacted with the COVID-19 Mpro amino acid residues T26, H41, G143, H163, M165 covalently [14]. Similarly, Natesh et al. (2021) performed the docking studies for COVID-19 Mpro using Ferula asafoetida bioactive compounds. Farnesiferol B, a F. asafoetida derivative, displayed minimized binding energy (−7.2 kacl/mol) with the COVID-19 Mpro amino acid residues and indicated H-bond (T26, D187) and hydrophobic interactions (T25, H41, T54, N142, C145, M165, Q189) [14]. Ghosh and coworkers reported that gallocatechin-3-gallate, a compound derived from Camellia sinensis (Green tea), showed significantly more activity (−9.0 kcal/mol) to the COVID-19 Mpro amino acids of E166, F140, H163, S144, C145, G113 (H-bond) and M49, L141, M165, E166, R188 (Hydrophobic interactions) [55]. Some of the herbaceous plant compounds with COVID-19 Mpro docking reports are summarized in Table-6. Based on the previous reports available in literature and the results of the present in silico study, it can be implied that the bioactives of E. officinalis, P. niruri and T. cordifolia could act as potential inhibitors for COVID-19 Mpro (Table-6 ).
Table 6.
Previous reports of molecular docking studies to identify COVID-19 Mpro inhibitors.
| Compound names | Binding energy (kcal/mol) | No of H-bond formation | source | Reference |
|---|---|---|---|---|
| Shogasulfonic acid A Gingerenone-A Isogingerenone-B |
−6.9 −6.5 −6.4 |
3 1 2 |
Zingiber officinale (Ginger) | [65] |
| Tinosponone Xanosporic acid Cardiofolioside B |
−7.7 −7.5 −7.3 |
2 3 5 |
Tinospora cordifolia (Giloy) | [15] |
| Assafoetidnol A Conferol Farnesiferol B |
−7.4 −7.6 −7.2 |
1 1 2 |
Ferula asafoetida (Asafoetida) | [14] |
| Sesami Sesaminol Sesamolin |
−8.2 −7.8 −7.7 |
2 5 4 |
Sesamum indicum (Sesame) | [14] |
| Cyclocurcumin Andrographolide Dihydroxy-dimethoxyflavone |
−6.77 −6.26 −6.23 |
2 3 3 |
Curcuma longa (Turmeric) | [53] |
| Curcuminoid Tetrahydroxycurcumin |
−9.08 −8.07 |
3 3 |
Curcuma longa (Turmeric) | [54] |
| Rutin Isorhamnetin-3-O-β-D Calendoflaside |
−8.8 −8.7 −8.6 |
Not clearly mentioned | Calendula officinalis (Pot marigold) | [66] |
| Gallocatechin-3-gallate Epicatechingallate Epigallocatechin gallate |
−9.0 −8.2 −7.6 |
5 6 7 |
Camellia sinensis (Green tea) | [55] |
| Kazinol A Broussochalcone A Broussoflavan A |
−8.2 −8.1 −8.1 |
5 5 3 |
Broussonetia papyrifera (Paper mulberry) | [55] |
| Withanoside II Withanoside IV Withanoside V |
−11.30* −11.02* −8.96* |
5 3 4 |
Withania somnifera (Ashwagandha) | [16] |
| Amentoflavone Bilobetin Ginkgetin |
−9.2 −9.1 −9.0 |
2 0 2 |
Torreya nucifera leaves | [67] |
| Withanoside V Somniferine |
10.32** 9.62** |
3 1 |
Withania somnifera (Ashwagandha) | [16] |
| Tinocordiside | 8.10** | 2 | Tinospora cordifolia (Giloy) | [16] |
| Vicenin IGHB Ursolic acid |
8.97** 8.55** 8.52** |
2 3 1 |
Ocimum sanctum (Tulsi) | [16] |
| Kaempferol Anthraquinone |
−6.2 −6.0 |
2 2 |
Moringa oleifera (Drumstick tree) | [68] |
| Andrographolide Neoandrographolide 14-deoxy 11,12-didehydro-andrographolide |
−7.2 −7.1 −7.0 |
Not clearly mentioned | Andrographis paniculata | [69] |
* glide score value; ** YASARA score.
3.4. Drug-likeness properties analysis
From the molecular docking studies, 12 drug candidates (amritoside, pectolinarin, astragalin, apigenin-6-C-glucosyl7-O-glucoside, 7-ketositosterol, 20a-hydroxy ecdysone, chlorogenic acid, ellagic acid, tinosporine B, quercetin, epicatechin and remdesivir) were subjected to drug-likeness analysis using DruLiTo (Table-7 ). The drug-likeness properties were evaluated by Pfizer's rule (also called as Lipinski's rule), wherein it states that those candidates that have values of log P ≤ 5, HBD ≤ 5, HBA ≤ 10, MW ≤ 500, TPSA (<140), and AMR (40–130), are considered to have passed the drug-likeness analysis [37]. These above parameters influence the bioavailability, absorption, receptor-drug interactions, metabolism and their toxicity [56]. The size of the molecule is also an important factor for drug candidates and it is useful for membrane transportation [37]. Drug-likeness study, based on the physicochemical nature of the bioactive compounds, is a preliminary criterion to assess its structural resemblance of an ideal drug, based on the Lipinski's rule of five [57]. However, it is not necessary for a drug to obey all the rules to be a potential drug candidate. As per the previous investigation by Bickerton et al. (2012), the oral bioavailability of the compounds did not affect the bioactivity or pharmacological potencies of a drug [58]. It was observed from this study that remdesivir obeyed two rules from the DruLiTo study, whereas all the other test compounds except apigenin-6-C-glucosyl7-O-glucoside obeyed ≥2 rule of drug-likeness. Among the selected drug candidates, tinosporin B, quercetin and epicatechin exhibited great structural properties to be an ideal drug.
Table 7.
Physiochemical properties of potential bioactive compounds from medicinal plants.
| Compound | MM | log P | Alog P | HBA | HBD | TSPA | AMR | nRB |
|---|---|---|---|---|---|---|---|---|
| Amritoside | 626.11 | −1.906 | −5.142 | 18 | 10 | 291.82 | 139.81 | 6 |
| Pectolinarin | 622.19 | −0.441 | −3.996 | 15 | 7 | 223.29 | 155.57 | 8 |
| Astragalin | 448.1 | −0.249 | −2.771 | 11 | 7 | 186.37 | 114.53 | 4 |
| Apigenin-6-C-glucosyl7-O-glucoside | 710.17 | −3.096 | −5.2 | 19 | 11 | 319.89 | 166.59 | 11 |
| 7-Ketositosterol | 428.37 | 9.499 | 1.029 | 2 | 1 | 37.3 | 125.1 | 6 |
| 20a-Hydroxy ecdysone | 480.31 | 1.049 | −1.198 | 7 | 6 | 138.45 | 126.18 | 5 |
| Chlorogenic acids | 354.1 | −0.7 | −1.194 | 9 | 6 | 164.75 | 85.8 | 5 |
| Ellagic acid | 608.17 | −0.762 | −4.06 | 15 | 8 | 234.29 | 150.53 | 7 |
| Tinosporine B | 374.14 | 1.379 | −1.134 | 7 | 2 | 102.29 | 93.42 | 1 |
| Quercetin | 302.04 | 1.834 | −1.244 | 7 | 5 | 127.45 | 83.44 | 1 |
| Epicatechin | 290.08 | 0.852 | −0.936 | 6 | 5 | 110.38 | 81.07 | 1 |
| Remdesivir | 602.23 | 0.336 | −3.217 | 14 | 4 | 211.13 | 151.49 | 14 |
MM: molecular mass, HBD: hydrogen bond donors, HBA hydrogen bond acceptors, PSA: polar surface area, AMR: Atom Molar Refractivity, nRB: number of Rotable Bond (MM less than 500 Da, no more than 5 HBD, no more than 10 HBA, and partition coefficient (log P) not greater than 5, TPSA no greater than140 Å2, AMR: 40 to 130, nRB: not more than 3 RB).
3.5. Bioactivity scores analysis
The five lead bioactives from the docking and MD results (7-Ketositosterol, astragalin, pectolinarin, amritoside and apigenin-6-C-glucosyl7-O-glucoside) were subjected to bioactivity scores analysis for different parameters including ICM, GPCR, NRL and enzyme inhibitors (protease, kinase). The compounds with score values more than 0.00 were determined highly active, values between −0.50 and 0.00 were considered abstemiously active and scores less than −0.50 were considered inactive [59]. All the five bioactive compounds exhibited promising bioactivity score and the results of summarized in Table-8 [60]. 7-ketositosterol, astragalin and amritoside were predicted with higher enzyme inhibitor activity. From the bioactivity score prediction results, it can be noted that the key compounds of 7-Ketositosterol, astragalin and amritoside could be used as COVID-19Mpro inhibitors for further drug development.
Table 8.
Bioactivity score prediction of selected bioactive compounds from Ayurvedic medicinal plants using Molinspiration cheminformatics online software.
| Compound Name | GPCR ligand | Ion channel modulator | Kinase inhibitor | Nuclear receptor ligand | Protease inhibitor | Enzyme inhibitor |
|---|---|---|---|---|---|---|
| Amritoside | −0.17 | −0.67 | −0.28 | −0.38 | −0.05 | 0.02 |
| Pectolinarin | −0.13 | −0.69 | −0.24 | −0.39 | −0.13 | −0.03 |
| Astragalin | 0.06 | −0.05 | 0.10 | 0.20 | −0.05 | 0.41 |
| Apigenin-6-C-glucosyl7-O-glucoside | −0.53 | −1.42 | −1.01 | −0.91 | −0.30 | −0.52 |
| 7-Ketositosterol | 0.11 | −0.03 | −0.69 | 0.72 | 0.12 | 0.56 |
| Remdesivir | 0.27 | −0.35 | 0.20 | −0.48 | 0.49 | 0.38 |
Highly active (more than 0.00); abstemiously active (between −0.50 and 0.00); inactive (less than −0.50).
3.6. Pharmacokinetic properties analysis
The pharmacokinetic profile analysis of a drug candidate is defined based on its ADMET properties. ADMET analysis is exceptionally useful in the early phase of drug discovery to facilitate significant reduction of clinical trial failures [39]. The five lead compounds were subjected to ADMET analysis. Aqueous solubility, GI absorption, skin and Caco2 permeability are important absorption parameters in the drug development process [61]. It is implied that a GI absorption value > 30% implies good absorbance. 7-Ketositosterol showed the highest percentage of absorption (95.8%) followed by astragalin (48.05%) and pectolinarin (41.84%) which showed good absorbance rates; whereas, amritoside (24.38%) and apigenin-6-C-glucosyl7-O-glucoside (8.03%) exhibited moderate absorption percentage (Table-9 ). A skin permeability value greater than −2.5 cm/h is deemed as low skin permeability and all drug compounds exhibited acceptable skin permeability. All the drug candidates had low Caco2 permeability (<0.9 cm/s) except 7-ketositosterol (1.293 cm/s). Another important factor during ADMET analysis was to predict the P-glycoprotein non-substrate candidature. All compounds were observed to be a substrate for P-glycoprotein except 7-ketositosterol (Table-9).
Table 9.
Pharmacokinetics, toxicities and receptor binding properties of potential bioactive compounds from medicinal plants using pkCSM web server.
| Model Name | Amritoside | Pectolinarin | Apigenin-6-C-glucosyl7-O-glucoside | 7-Ketositosterol | Astragalin | Unit | |
|---|---|---|---|---|---|---|---|
| Absorption | Water solubility | −2.839 | −2.986 | −2.828 | −6.292 | −2.863 | Numeric (log mol/L) |
| Caco2 permeability | −0.858 | 0.309 | −1.188 | 1.293 | 0.306 | Numeric (log Papp in 10−6 cm/s) | |
| Intestinal absorption (human) | 24.384 | 41.847 | 8.034 | 95.807 | 48.052 | Numeric (% Absorbed) | |
| Skin Permeability | −2.735 | −2.735 | −2.735 | −2.748 | −2.735 | Numeric (log Kp) | |
| P-glycoprotein substrate | Yes | Yes | Yes | No | Yes | Categorical (Yes/No) | |
| P-glycoprotein I inhibitor | No | No | No | Yes | No | Categorical (Yes/No) | |
| P-glycoprotein II inhibitor | No | No | No | Yes | No | Categorical (Yes/No) | |
| Distribution | VDss (human) | 0.736 | 0.684 | 0.134 | −0.182 | 1.444 | Numeric (log L/kg) |
| Fraction unbound (human) | 0.191 | 0.123 | 0.268 | 0 | 0.218 | Numeric (Fu) | |
| BBB permeability | −2.28 | −1.863 | −2.402 | −0.143 | −1.514 | Numeric (log BB) | |
| CNS permeability | −5.476 | −4.794 | −5.301 | −1.795 | −3.908 | Numeric (log PS) | |
| Metabolism | CYP2D6 substrate | No | No | No | No | No | Categorical (Yes/No) |
| CYP3A4 substrate | No | No | No | Yes | No | Categorical (Yes/No) | |
| CYP1A2 inhibitior | No | No | No | No | No | Categorical (Yes/No) | |
| CYP2C19 inhibitior | No | No | No | No | No | Categorical (Yes/No) | |
| CYP2C9 inhibitior | No | No | No | No | No | Categorical (Yes/No) | |
| CYP2D6 inhibitior | No | No | No | No | No | Categorical (Yes/No) | |
| CYP3A4 inhibitior | No | No | No | No | No | Categorical (Yes/No) | |
| Excretion | Total Clearance | −0.619 | 0.027 | −0.202 | 0.575 | 0.462 | Numeric (log ml/min/kg) |
| Renal OCT2 substrate | No | No | No | No | No | Categorical (Yes/No) | |
| Toxicity | AMES toxicity | No | No | No | No | No | Categorical (Yes/No) |
| Max. tolerated dose (human) | 0.405 | 0.543 | 0.349 | −0.65 | 0.582 | Numeric (log mg/kg/day) | |
| hERG I inhibitor | No | No | No | No | No | Categorical (Yes/No) | |
| hERG II inhibitor | Yes | Yes | No | Yes | No | Categorical (Yes/No) | |
| Oral Rat Acute Toxicity (LD50) | 2.479 | 2.521 | 2.479 | 2.664 | 2.546 | Numeric (mol/kg) | |
| Oral Rat Chronic Toxicity (LOAEL) | 5.171 | 3.382 | 5.317 | 2.351 | 4.53 | Numeric (log mg/kg_bw/day) | |
| Hepatotoxicity | No | No | No | No | No | Categorical (Yes/No) | |
| Skin Sensitization | No | No | No | No | No | Categorical (Yes/No) | |
| T.Pyriformis toxicity | 0.285 | 0.285 | 0.285 | 0.425 | 0.285 | Numeric (log ug/L) |
Intestinal absorption (30 % <), skin permeability (−2.5 cm/h <), Caco2 permeability (<0.9 cm/s), log VDss (0.45 <), BBB membrane permeability, log BB > 0.3 to < −1, CNS permeability, log PS > −2 to < −3, Cytochrome P450 enzymes (CYP2D6, CYP2A4, CYP2C9, CYP2C19, CYP2D6 and CYP3A4).
The VDss, CNS and BBB membrane permeability were used to study the drug distribution [62]. The log VDss greater-than 0.45 were considered to be relatively high. Astragalin (1.444), Amritoside (0.736) and pectolinarin (0.684) showed greater distribution volumes (Table-9). For BBB membrane permeability, log BB values > 0.3 but < −1 indicated that the drug molecules crossed the BBB membrane. For CNS permeability, range of log PS values > −2 to < −3 indicated impenetrability. All drug candidates were predicted to be neither capable of penetrating the CNS nor crossing the BBB membrane (Table-9).
The CYP450 plays an important role in drug metabolism in the liver system [63]. The metabolism scores showed that all the drug compounds except 7-ketositosterol did not affect/inhibit CYP2D6 and CYP3A4 enzymes, and also did not act as inhibitors for CYP2D6, CYP2A4, CYP2C9, CYP2C19 enzymes. The total drug clearance is measured by a combination of hepatic and renal clearance. Total clearance defines the concentration of drug in the body using its elimination rate [64]. The predicted results showed that the drug candidates’ excretion ranges from −0.6 to 1.1 mL/min/kg (Table-9).
In drug discovery, toxicity is an important criteria and plays a significant role in the selection of most suitable drug candidates [62]. All the drug compounds in this analysis have not expressed any skin allergic action and hepatotoxic effect (Table-9). hERG inhibition (I and II) is an important factor for toxicity analysis and it also involves cardiotoxicity. None of the compounds exhibited inhibitory actions for hERG-I. Astragalin, Amritoside and pectolinarin drug candidates were predicted to be hERG II inhibitors. All the drug candidates have not expressed any AMES toxicity and Tetrahymena Pyriformis toxicity. The LD50, lowest-observed-adverse-effect level (LOAEL) and maximum tolerated dosage range of drug candidates were predicted by the toxicity analysis server and the predicted scores are shown in Table-9. Thus, from these results, the present study concluded that these bioactive drug candidates could be used as potential protease inhibitor drugs against COVID-19.
4. Conclusion
Traditionally employed medicinal plants are gaining significant attention in recent years in the search for therapeutic solutions against emerging infectious diseases. Natural compounds from medicinal plants can be synergistically combined with pharmacological treatments in various disease pathologies. As biologists, we need to be more concerned and vigilant about the status of these medicinal plants and their secondary metabolites (phytoconstituents) in developing phyto-antiviral drugs and controlling pandemics like COVID-19. In the present study, we have portrayed the screening of 3 medicinal plants and their derivatives (96 bioactive compounds) in search for new, potential COVID-19 Mpro inhibitors. The lead candidates (10 ligands) were selected from computational screening approaches. The present study indicates that amritoside, pectolinarin, astragalin, apigenin-6-C-glucosyl7-O-glucoside, 7-ketositosterol and quercetin efficiently occupy the substrate binding cleft of COVID-19 Mpro. These bioactives non-covalently interacted with catalytic residues H41 and C145, thereby signifying them as plausible inhibitors for COVID-19 Mpro. Further experimental and clinical studies are highly warranted to transform these potential inhibitors into therapeutic drugs for COVID-19. We believe that the insights gained in the current in silico study may be highly valuable for discovering and developing novel natural COVID-19 therapeutic drugs in the future.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgement
The authors acknowledge the support of the MLP270 project funded by CSIR-CFTRI, Mysuru, India.
Dr. Dandamudi Usharani duly acknowledge IIT Guwahati Param Ishan High performance computing facility for computing facilities.
References
- 1.Chauhan S. Comprehensive review of coronavirus disease 2019 (COVID-19) Biomed. J. 2020;43:334–340. doi: 10.1016/j.bj.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chosidow O., Bernigaud C., Guillemot D., Giraudeau B., Lespine A., Changeux J.-P., Bourhy H., Lecuit M., Amoura Z. Ivermectin as a potential treatment for COVID-19? PLoS Neglected Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Geng X., Tan Y., Li Q., Xu C., Xu J., Hao L., Zeng Z., Luo X., Liu F., Wang H. New understanding of the damage of SARS-CoV-2 infection outside the respiratory system. Biomed. Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prathiviraj R., Kiran G.S., Selvin J. Phylogenomic proximity and comparative proteomic analysis of SARS-CoV-2. Gene Reports. 2020;20:100777. doi: 10.1016/j.genrep.2020.100777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prathiviraj R., Saranya S., Bharathi M., Chellapandi P. A hijack mechanism of Indian SARS-CoV-2 isolates for relapsing contemporary antiviral therapeutics. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh T.U., Parida S., Lingaraju M.C., Kesavan M., Kumar D., Singh R.K. Drug repurposing approach to fight COVID-19. Pharmacol. Rep. 2020;72:1479–1508. doi: 10.1007/s43440-020-00155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yildiz C., Ozsurekci Y., Gucer S., Cengiz A.B., Topaloglu R. Acute kidney injury due to acyclovir. CEN Case Reports. 2013;2:38–40. doi: 10.1007/s13730-012-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murugesan S., Kumar Lakshmanan D., Arumugam V., Alexander R.A. Nutritional and therapeutic benefits of medicinal plant Pithecellobium dulce (Fabaceae): a review ARTICLE INFO. J. Appl. Pharmaceut. Sci. 2019;9:130–139. doi: 10.7324/JAPS.2019.90718. [DOI] [Google Scholar]
- 10.Lakshmanan D.K., Ravichandran G., Elangovan A., Jeyapaul P., Murugesan S., Thilagar S. Cissus quadrangularis (veldt grape) attenuates disease progression and anatomical changes in mono sodium iodoacetate (MIA)-induced knee osteoarthritis in the rat model. Food Funct. 2020 doi: 10.1039/d0fo00992j. [DOI] [PubMed] [Google Scholar]
- 11.Venkateswaran M.R., Jayabal S., Murugesan S. Evaluation of antioxidant and antidiabetic potentials of a polyherbal formulation-Mehani. Nat. Prod. Res. 2019:1–5. doi: 10.1080/14786419.2019.1660978. 0. [DOI] [PubMed] [Google Scholar]
- 12.Siddiqui A.J., Danciu C., Ashraf S.A., Moin A., Singh R., Alreshidi M., Patel M., Jahan S., Kumar S., Alkhinjar M.I.M., Badraoui R., Snoussi M., Adnan M. Plants-derived biomolecules as potent antiviral phytomedicines: new insights on ethnobotanical evidences against coronaviruses. Plants. 2020;9:1–41. doi: 10.3390/plants9091244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alamgeer W., Younis H., Asif A., Sharif H., Riaz I.A., Bukhari A.M., Assiri Traditional medicinal plants used for respiratory disorders in Pakistan: a review of the ethno-medicinal and pharmacological evidence. Chin. Med. 2018;13:48. doi: 10.1186/s13020-018-0204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natesh J., Mondal P., Penta D., Abdul Salam A.A., Meeran S.M. Culinary spice bioactives as potential therapeutics against SARS-CoV-2: computational investigation. Comput. Biol. Med. 2021;128 doi: 10.1016/j.compbiomed.2020.104102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krupanidhi S., Abraham Peele K., Venkateswarulu T.C., Ayyagari V.S., Nazneen Bobby M., John Babu D., Venkata Narayana A., Aishwarya G. Screening of phytochemical compounds of Tinospora cordifolia for their inhibitory activity on SARS-CoV-2: an in silico study. J. Biomol. Struct. Dyn. 2020:1–5. doi: 10.1080/07391102.2020.1787226. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shree P., Mishra P., Selvaraj C., Singh S.K., Chaube R., Garg N., Tripathi Y.B. Targeting COVID-19 (SARS-CoV-2) main protease through active phytochemicals of ayurvedic medicinal plants–Withania somnifera (Ashwagandha), Tinospora cordifolia (Giloy) and Ocimum sanctum (Tulsi)–a molecular docking study. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1810778. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh Y.D., Jena B., Ningthoujam R., Panda S., Priyadarsini P., Pattanayak S., Panda M.K., Singh M.C., Satapathy K.B. Potential bioactive molecules from natural products to combat against coronavirus. Adv. Tradit. Med. 2020 doi: 10.1007/s13596-020-00496-w. [DOI] [Google Scholar]
- 18.Sharma P., Dwivedee B.P., Bisht D., Dash A.K., Kumar D. The chemical constituents and diverse pharmacological importance of Tinospora cordifolia. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e02437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jantan I., Haque M.A., Ilangkovan M., Arshad L. An insight into the modulatory effects and mechanisms of action of Phyllanthus species and their bioactive metabolites on the immune system. Front. Pharmacol. 2019;10:878. doi: 10.3389/fphar.2019.00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yadav T.C., Kumar N., Raj U., Goel N., Vardawaj P.K., Prasad R., Pruthi V. Exploration of interaction mechanism of tyrosol as a potent anti-inflammatory agent. J. Biomol. Struct. Dyn. 2020;38:382–397. doi: 10.1080/07391102.2019.1575283. [DOI] [PubMed] [Google Scholar]
- 21.Murugesan S., Ravichandran D., Lakshmanan D.K., Ravichandran G., Arumugam V., Raju K., Geetha K., Thilagar S. Evaluation of anti rheumatic activity of Piper betle L. (Betelvine) extract using in silico, in vitro and in vivo approaches. Bioorg. Chem. 2020;103 doi: 10.1016/j.bioorg.2020.104227. [DOI] [PubMed] [Google Scholar]
- 22.Variya B.C., Bakrania A.K., Patel S.S. Emblica officinalis (Amla): a review for its phytochemistry, ethnomedicinal uses and medicinal potentials with respect to molecular mechanisms. Pharmacol. Res. 2016;111:180–200. doi: 10.1016/j.phrs.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Murugesan S., Venkateswaran M.R., Jayabal S., Periyasamy S. Evaluation of the antioxidant and anti-arthritic potential of Zingiber officinale Rosc. by in vitro and in silico analysis. South Afr. J. Bot. 2020;130:45–53. doi: 10.1016/j.sajb.2019.12.019. [DOI] [Google Scholar]
- 24.Kumar A., Mehta V., Raj U., Varadwaj P.K., Udayabanu M., Yennamalli R.M., Singh T.R. Computational and in-vitro validation of natural molecules as potential acetylcholinesterase inhibitors and neuroprotective agents. Curr. Alzheimer Res. 2019;16:116–127. doi: 10.2174/1567205016666181212155147. [DOI] [PubMed] [Google Scholar]
- 25.Dallakyan S., Olson A.J. In: Chem. Biol. Methods Protoc. Hempel J.E., Williams C.H., Hong C.C., editors. Springer; New York, New York, NY: 2015. Small-molecule library screening by docking with PyRx; pp. 243–250. [DOI] [PubMed] [Google Scholar]
- 26.Kar P., Sharma N.R., Singh B., Sen A., Roy A. Natural compounds from Clerodendrum spp. as possible therapeutic candidates against SARS-CoV-2: an in silico investigation. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1780947. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 28.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 29.Banks J.L., Beard H.S., Cao Y., Cho A.E., Damm W., Farid R., Felts A.K., Halgren T.A., Mainz D.T., Maple J.R., Murphy R., Philipp D.M., Repasky M.P., Zhang L.Y., Berne B.J., Friesner R.A., Gallicchio E., Levy R.M. Integrated modeling program, applied chemical theory (IMPACT) J. Comput. Chem. 2005;26:1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aier I., Varadwaj P., Raj U. Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci. Rep. 2016;6:34984. doi: 10.1038/srep34984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for Protein−Ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 32.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Petersson G.A., Nakatsuji H. Gaussian. 2016;16 [Google Scholar]
- 33.Case D.A., Walker R.C., Cheatham T.E., Simmerling C., Roitberg A., Merz K.M., Luo R., Darden T., Amber . Vol. 2018. Univ. California; 2018. pp. 1–923.http://ambermd.org/contributors.html%0Ahttp://ambermd.org/doc12/Amber18.pdf San Fr. 2018. [Google Scholar]
- 34.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Götz A.W., Williamson M.J., Xu D., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J. Chem. Theor. Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abduldileep S., Narayanasamy R., Usharani D., Singh A., Rajasekharan R. A bioactive polypeptide from sugarcane selectively inhibits intestinal sucrase. Int. J. Biol. Macromol. 2020;156:938–948. doi: 10.1016/j.ijbiomac.2020.03.085. [DOI] [PubMed] [Google Scholar]
- 37.Lipinski C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Khan T., Dixit S., Ahmad R., Raza S., Azad I., Joshi S., Khan A.R. Molecular docking, PASS analysis, bioactivity score prediction, synthesis, characterization and biological activity evaluation of a functionalized 2-butanone thiosemicarbazone ligand and its complexes. J. Chem. Biol. 2017;10:91–104. doi: 10.1007/s12154-017-0167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.V Pires D.E., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58:4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. BioRxiv. 2020 doi: 10.1101/2020.02.26.964882. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;80–(368):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapil A., Sharma S. Immunopotentiating compounds from Tinospora cordifolia. J. Ethnopharmacol. 1997;58:89–95. doi: 10.1016/s0378-8741(97)00086-x. [DOI] [PubMed] [Google Scholar]
- 43.Bishayi B., Roychowdhury S., Ghosh S., Sengupta M. Hepatoprotective and immunomodulatory properties of Tinospora cordifolia in CCl4 intoxicated mature albino rats. J. Toxicol. Sci. 2002;27:139–146. doi: 10.2131/jts.27.139. [DOI] [PubMed] [Google Scholar]
- 44.Upadhyaya R., Pandey R.P., Sharma V., Verma Anita K. Assessment of the multifaceted immunomodulatory potential of the aqueous extract of Tinospora cordifolia. Res. J. Chem. Sci. 2011;1:71–79. [Google Scholar]
- 45.Raghu R., Sharma D., Ramakrishnan R., Khanam S., Chintalwar G.J., Sainis K.B. Molecular events in the activation of B cells and macrophages by a non-microbial TLR4 agonist, G1-4A from Tinospora cordifolia. Immunol. Lett. 2009;123:60–71. doi: 10.1016/j.imlet.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Nworu C.S., Akah P.A., Okoye F.B.C., Proksch P., Esimone C.O. The effects of Phyllanthus niruri aqueous extract on the activation of murine lymphocytes and bone marrow-derived macrophages. Immunol. Invest. 2010;39:245–267. doi: 10.3109/08820131003599585. [DOI] [PubMed] [Google Scholar]
- 47.Eze C.O. Immunomodulatory activities of methanol extract of the whole aerial part of Phyllantus niruri L. J. Pharmacogn. Phyther. 2014;6:41–46. [Google Scholar]
- 48.Putri D.U., Rintiswati N., Soesatyo M.H.N.E., Haryana S.M. Immune modulation properties of herbal plant leaves: Phyllanthus niruri aqueous extract on immune cells of tuberculosis patient - in vitro study. Nat. Prod. Res. 2018;32:463–467. doi: 10.1080/14786419.2017.1311888. [DOI] [PubMed] [Google Scholar]
- 49.Suresh K., Vasudevan D.M. Augmentation of murine natural killer cell and antibody dependent cellular cytotoxicity activities by Phyllanthus emblica, a new immunomodulator. J. Ethnopharmacol. 1994;44:55–60. doi: 10.1016/0378-8741(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 50.Srikumar R., Parthasarathy N.J., Manikandan S., Narayanan G.S., Sheeladevi R. Effect of Triphala on oxidative stress and on cell-mediated immune response against noise stress in rats. Mol. Cell. Biochem. 2006;283:67–74. doi: 10.1007/s11010-006-2271-0. [DOI] [PubMed] [Google Scholar]
- 51.Naik V.R., Munikumar M., Ramakrishna U., Srujana M., Goudar G., Naresh P., Kumar B.N., Hemalatha R. Remdesivir (GS-5734) as a therapeutic option of 2019-nCOV main protease – in silico approach. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1781694. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmed S., Mahtarin R., Ahmed S.S., Akter S., Islam M.S., Al Mamun A., Islam R., Hossain M.N., Ali M.A., Sultana M.U.C., Parves M.D.R., Ullah M.O., Halim M.A. Investigating the binding affinity, interaction, and structure-activity-relationship of 76 prescription antiviral drugs targeting RdRp and Mpro of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1796804. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajagopal K. Activity of phytochemical constituents of Curcuma longa (turmeric) against SARS-CoV-2 main protease (Covid19): anin-silico approach. Int. J. Pharm. \& Life. 2020;6(1):1–10. doi: 10.1186/s43094-020-00126-x. http://search.ebscohost.com/login.aspx?direct=true%5C&profile=ehost%5C&scope=site%5C&authtype=crawler%5C&jrnl=09767126%5C&AN=144984858%5C&h=QAqQvHezAEUEL9ZvD%2FuFS5sVr4iOQ9vhMlAZZq0ecmvKVKBlHtW3QFaTZ3Fypkg2SdiuWwdD1s6lbIQE26WrJg%3D%3D%5C&crl=c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta S., Singh A.K., Kushwaha P.P., Prajapati K.S., Shuaib M., Senapati S., Kumar S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1776157. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors–an in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1779818. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider G. Madame Curie Biosci. Database. Landes Bioscience; 2013. Prediction of drug-like properties. [Internet] [Google Scholar]
- 57.Bei Y. 乳鼠心肌提取 HHS public access. Physiol. Behav. 2017;176:139–148. doi: 10.1016/j.addr.2016.05.007. BDDCS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bickerton G.R., V Paolini G., Besnard J., Muresan S., Hopkins A.L. Quantifying the chemical beauty of drugs europe PMC funders group. Nat Chem. 2012;4:90–98. doi: 10.1038/nchem.1243.Quantifying. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lakshmanan D.K., Murugesan S., Rajendran S., Ravichandran G., Elangovan A., Raju K., Prathiviraj R., Pandiyan R., Thilagar S. Brassica juncea (L.) Czern. leaves alleviate adjuvant-induced rheumatoid arthritis in rats via modulating the finest disease targets - IL2RA, IL18 and VEGFA. J. Biomol. Struct. Dyn. 2021:1–14. doi: 10.1080/07391102.2021.1907226. 0. [DOI] [PubMed] [Google Scholar]
- 60.Selvakumar M., Palanichamy P., Arumugam V., Venkatesan M., Aathmanathan S., Krishnamoorthy H., Pugazhendhi A. In silico potential of nutraceutical plant of Pithecellobium dulce against GRP78 target protein for breast cancer. Appl. Nanosci. 2021 doi: 10.1007/s13204-021-01840-5. [DOI] [Google Scholar]
- 61.Dahlgren D., Lennernäs H. Intestinal permeability and drug absorption: predictive experimental, computational and in vivo approaches. Pharmaceutics. 2019;11 doi: 10.3390/pharmaceutics11080411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han Y., Zhang J., Hu C.Q., Zhang X., Ma B., Zhang P. In silico ADME and toxicity prediction of ceftazidime and its impurities. Front. Pharmacol. 2019;10:434. doi: 10.3389/fphar.2019.00434. https://www.frontiersin.org/article/10.3389/fphar.2019.00434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 64.Horde G.W., Gupta V. StatPearls. StatPearls Publishing; 2020. Drug clearance. [Internet] [PubMed] [Google Scholar]
- 65.Prasanth D.S.N.B.K., Panda S.P., Lakshmana Rao A., Chakravarti G., Teja N., Naga Vani V.B., Sandhya T. In-silico strategies of some selected phytoconstituents from zingiber officinale as sars cov-2 main protease (COVID-19) inhibitors. Indian J. Pharm. Educ. Res. 2020;54:S552–S559. doi: 10.5530/ijper.54.3s.154. [DOI] [Google Scholar]
- 66.Das P., Majumder R., Mandal M., Basak P. In-Silico approach for identification of effective and stable inhibitors for COVID-19 main protease (Mpro) from flavonoid based phytochemical constituents of Calendula officinalis. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1796799. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Computer aided identification of potential SARS CoV-2 main protease inhibitors from diterpenoids and biflavonoids of Torreya nucifera leaves. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1841680. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamza M., Ali A., Khan S., Ahmed S., Attique Z., Ur Rehman S., Khan A., Ali H., Rizwan M., Munir A., Khan A.M., Siddique F., Mehmood A., Nouroz F., Khan S. nCOV-19 peptides mass fingerprinting identification, binding, and blocking of inhibitors flavonoids and anthraquinone of Moringa oleifera and hydroxychloroquine. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1778534. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murugan N.A., Pandian C.J., Jeyakanthan J. Computational investigation on Andrographis paniculata phytochemicals to evaluate their potency against SARS-CoV-2 in comparison to known antiviral compounds in drug trials. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1777901. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]