Abstract
The emergence and subsequent global outbreak of the novel coronavirus SARS-CoV-2 prompted our laboratory to launch efforts to develop methods for SARS-CoV-2 antigen detection and quantification. We present an isotope dilution mass spectrometry method (IDMS) for rapid and accurate quantification of the primary antigens, spike and nucleocapsid proteins. This IDMS method utilizes liquid chromatography-tandem mass spectrometry (LC-MS/MS) to analyze sample tryptic digests for detection and quantification of selected conserved peptides of SARS-CoV-2 spike and nucleocapsid proteins. The IDMS method has the necessary attributes to be successfully utilized for accurate quantification in SARS-CoV-2 protein-based vaccines and as targets of rapid diagnostic tests. Absolute quantification was achieved by quantifying and averaging 5 peptides for spike protein (3 peptides in the S1 subunit and 2 peptides in the S2 subunit) and 4 peptides for nucleocapsid protein. The overall relative standard deviation of the method was 3.67% for spike protein and 5.11% for nucleocapsid protein. IDMS offers speed (5 h total analysis time), sensitivity (LOQ; 10 fmol/µL) and precision for quantification of SARS-CoV-2 spike and nucleocapsid proteins.
Keywords: Coronavirus, COVID-19, SARS-CoV-2, Mass spectrometry, Spike, Nucleocapsid, Quantification, Epidemic, Pandemic, Outbreak, Vaccine
1. Introduction
Mass spectrometry (MS) plays an important role in many biomedical applications including clinical diagnostics [1], [2], biomarker discovery [3], [4], drug metabolism [5] and pharmaceutical development and testing [6]. Vaccine development, characterization, and quality control also rely heavily on MS techniques [7]. Isotope dilution liquid chromatography tandem mass spectrometry (IDMS) has become the gold standard for the quantification of small molecules [8], [9], [10] and proteins [11], [12], [13], [14], [15]. Several reference methods employ IDMS. Protein quantification by IDMS involves enzymatic digestion of viral proteins, addition of 13C and 15N labeled internal standards, and the selective detection of target labeled and native peptides by mass spectrometry. The 13C and 15N labeled peptides behave chromatographically the same as the native peptides but are distinguishable by mass allowing for accurate and precise quantification. Some examples of antigens that have been quantified in different vaccines using IDMS include the quantification of the viral envelope glycoprotein in Ebola virus-like particle vaccine [16], the two porins, PorA, the major antigenic protein, and PorB, the most abundant membrane protein in the meningococcal vaccine [17], and hemagglutinin (HA), the primary antigenic component of the influenza virus and main constituent of the influenza vaccine [18], [19].
The CDC, in collaboration with the World Health Organization (WHO) Essential Regulatory Laboratories (ERL), developed the IDMS methods to quantify influenza hemagglutinins (HA) [18], [19]. These HA IDMS methods are now used to value assign the cell-derived influenza primary liquid standards that calibrate and standardize the single radial immunodiffusion (SRID) assay. SRID is the accepted regulatory potency method for the quantification of HA in seasonal and potential pandemic influenza vaccines. The IDMS methods are also being used to selectively quantify, in a single analytical run, the individual HA contents of H1, H3, B (Yamagata) and B (Victoria) in trivalent [18] and tetravalent vaccines and to explore alternative potency assays [20], [21], [22], [23]. Additionally, IDMS has successfully been used to quantify neuraminidase (NA) [24], matrix protein, and nucleoprotein to support vaccine development by identifying faster growing seed strains [25], [26] and support influenza diagnostics [27].
The novel coronavirus SARS-CoV-2 is a zoonotic respiratory virus, the causative agent of the COVID-19 illnesses [28], and the first coronavirus to spark a pandemic [29]. SARS-CoV-2 is a positive-sense single-stranded RNA virus that has 16 predicted non-structural proteins along with 4 structural proteins found in all coronaviruses that include the envelope, the matrix or membrane protein, spike, and nucleocapsid [30]. The coronavirus spike protein, the primary antigen of COVID-19 vaccines, is a transmembrane envelope protein that is a member of the class I viral membrane fusion proteins that also includes the influenza glycoprotein HA [31]. The spike glycoprotein, consisting of S1 and S2 domains that are responsible for viral binding and fusion, respectively [32], is characteristically similar to influenza’s major surface protein HA and its HA1 and HA2 subunits [33]. Both spike and HA are the most important targets for acquired protective immunity in populations for their respective viruses and are the primary antigens of which vaccines are comprised [34], [35].
Multiple strategies for SARS-CoV-2 vaccine development are currently being pursued, including inactivated virus, recombinant protein, mRNA, DNA, and viral vector vaccines [22]. While the spike glycoprotein is the main antigenic target for vaccine development, the nucleocapsid protein, a multifunctional protein that primarily functions for binding to the viral RNA genome and packing it into a long helical nucleocapsid structure, has also been found to be highly immunogenic and expressed abundantly during coronavirus infections [36]. Additionally, the nucleocapsid gene is more highly conserved and stable than the spike gene [37], [38]. T-cells also specifically target the nucleocapsid protein in SARS-CoV-1 infection and have been found to be highly immunogenic, eliciting important IgG antibodies against SARS-CoV-1 in human patients during the immune response [39]. Currently, most viral antigen-based detection of SARS-CoV-2, including molecular-based testing, rapid fluorescent immunoassays (FIA) [40] and rapid fluorescence immunochromatographic (FIC) assays [41], target nucleocapsid protein to detect the SARS-CoV-2.
Here, we describe an IDMS method utilizing liquid chromatography-tandem mass spectrometry (LC-MS/MS) for quantification of SARS-CoV-2 spike and nucleocapsid proteins. The IDMS method offers high precision and specificity of spike and nucleocapsid measurements to aid in the development and quality control of vaccines and diagnostics.
2. Materials and methods
2.1. Synthesis of native and isotopically labeled SARS-CoV-2 peptide standards
Custom synthetic peptides for SARS-CoV-2 spike protein were synthesized using different strategies and vendors to assess timeliness, cost, and quality of standard preparations. Peptides were chosen based on general characteristic rules that have been described elsewhere that lend themselves as good peptides for protein quantification [24]. In brief, peptides were chosen that were 6 to 12 amino acids long to ensure selectivity of the method. Peptides that contain tryptophan and methionine are avoided since these amino acids are prone to oxidation in vivo or during sample preparation and handling, which would change the peptide’s molecular weight. Peptides that contain cysteine are avoided because they can form disulfide bonds with distant areas of the protein and would be difficult to measure accurately. Peptides were also chosen in both the S1 and S2 regions of the spike protein. The peptides selected for quantification are shown in Table 1 .
Table 1.
Target peptides and their 13C15N isotopically labeled counterparts employed for quantification of SARS-CoV-2 spike protein (S) and nucleocapsid protein (N). Underlined amino acids correspond to those that we 13C15N - labeled.
| Target peptide | SARS-CoV-2 protein | Precursor ion m/z | Target ion m/z | Confirmation ion m/z | Confirmation ion m/z |
|---|---|---|---|---|---|
| GVYYPDK | S/S1 | 421.2 (+2) | 685.3 (y5) | 359.2 (y3) | 522.3 (y4) |
| GVYYPDK | S/S1 | 425.2 (+2) | 693.3 (y5) | 367.2 (y3) | 530.3 (y4) |
| GIYQTSNFR | S/S1 | 543.3 (+2) | 624.3 (y5) | 752.4 (y6) | 915.4 (y7) |
| GIYQTSNFR | S/S1 | 548.3 (+2) | 634.3 (y5) | 762.4 (y6) | 925.4 (y7) |
| FLPFQQFGR | S/S1 | 570.3 (+2) | 879.5 (y7) | 635.3 (y5) | 782.4 (y6) |
| FLPFQQFGR | S/S1 | 575.3 (+2) | 889.5 (y7) | 645.3 (y5) | 792.4 (y6) |
| VTLADAGFIK | S/S2 | 517.8 (+2) | 721.4 (y7) | 650.4 (y6) | 535.3 (y5) |
| VTLADAGFIK | S/S2 | 521.8 (+2) | 729.4 (y7) | 658.4 (y6) | 543.3 (y5) |
| ASANLAATK | S/S2 | 423.7 (+2) | 688.4 (y7) | 617.4 (y6) | 503.3 (y5) |
| ASANLAATK | S/S2 | 427.4 (+2) | 696.4 (y7) | 625.4 (y6) | 511.3 (y5) |
| DHIGTR | N | 349.7 (+2) | 446.3(y4) | 333.2 (y3) | 583.3(y5) |
| DHIGTR | N | 354.7 (+2) | 456.7(y4) | 343.2 (y3) | 593.3 (y5) |
| GFYAEGSR | N | 443.7 (+2) | 682.3 (y6) | 519.3 (y5) | 448.2 (y4) |
| GFYAEGSR | N | 448.7 (+2) | 692.3 (y6) | 529.3 (y5) | 458.2 (y4) |
| LNQLESK | N | 416.2 (+2) | 604.3 (y5) | 718.4 (y6) | 476.3 (y4) |
| LNQLESK | N | 420.2 (+2) | 612.3 (y5) | 726.4 (y6) | 484.3 (y4) |
| DQVILLNK | N | 471.8 (+2) | 699.5 (y6) | 600.4 (y5) | 487.3 (y4) |
| DQVILLNK | 475.8 (+2) | 707.5 (y6) | 608.4 (y5) | 495.3 (y4) |
New England Peptide Inc. (NEP), recently rebranded as Vivitide, (Gardner, MA, USA) was chosen to generate peptides for our IDMS method utilizing two different synthesis strategies. NEP was first asked to synthesize twelve SARS-CoV-2 spike protein peptides (6 native and 6 labeled (13C/15N on C terminus R or K)). Five mg (gross weight) of each SARS-CoV-2 spike protein peptide was synthesized with > 95% purity and shipped to our laboratory for in-house processing. Each 5 mg lyophilized peptide was reconstituted in 8 mL of 10% formic acid in water (FA (aq)) followed by 42 mL of 0.1% FA (aq) to make a 50 mL solution. 200 µL of each peptide was then dispensed into 216 MAXYMum Recovery™ 1.5-mL screw cap tubes (Axygen Scientific, Union City, CA, USA) using a Biomek FXP Laboratory Automation Workstation (Beckman Coulter, Brea, CA, USA) with a 54-well plate configuration. All 2,592 vials (216 vials × 6 native peptides × 6 labeled peptides) were lyophilized using a Labconco™ Refrigerated Centrivap Concentrator (Labconco Corporation, Kansas City, MO, USA) and stored below -20 °C until use. Peptide standards were then analyzed by amino acid analysis (AAA) to confirm peptide sequence, quantify peptide amount and ensure vial-to-vial reproducibility. Amino acid analysis is a well-established technique for quantifying peptides. Three vials of each native lyophilized peptide standard were randomly selected, shipped, and analyzed by both NEP and AAA Services Laboratory, Inc. (Damascus, OR, USA). Additionally, for each labeled lyophilized peptide standard, one vial was randomly selected and quantified using duplicate AAA with NIST-certified amino acid standards by NEP and one vial was randomly selected and quantified by AAA Services Laboratory, Inc.
To prepare the in-house standards mixtures to contain 0.5 nmol of each peptide, a vial for each of the individual native peptides was reconstituted in 0.1% FA (aq) based on the AAA value. Once reconstituted, 100 µL of each native peptide was pooled to make the standard mixture. The native vial, containing 600 µL total of peptide material was then lyophilized and stored at −80 °C. An identical means of preparation was utilized for the labeled mixtures.
Additional peptide standards, native mixtures, and labeled mixtures of target peptides, prepared by NEP, were purchased and carefully compared to the standards prepared in-house. NEP synthesized 10 nmols of each (7 native and 7 labeled) SARS-CoV-2 spike peptide with > 95% purity, tested in duplicate by AAA, and vialed as 2 mixtures—one containing 7 native spike peptides and one containing 7 labeled spike peptides. NEP also synthesized 10 nmols of each (5 native and 5 labeled) SARS-CoV-2 nucleocapsid peptide with > 95% purity, tested in duplicate by AAA, and vialed as 2 mixtures--one containing 5 nucleocapsid native peptides and one containing 5 nucleocapsid labeled peptides. Both spike and nucleocapsid peptide mixtures were provided as 10 vials each containing 1 nmol of each peptide.
All target peptide standard preparations were stored at −80 °C until thawed prior to analysis.
2.2. SARS-CoV-2 recombinant proteins
SARS-CoV-2 recombinant spike S (S1 + S2) protein, rabbit Fc tag was received from Euprotein Inc. (Wuhan virus - Cat. No. EPY275672) (Euprotein Inc., North Brunswick Township, NJ, USA). SARS-CoV-2 nucleocapsid recombinant protein (1 mg in PBS; Cat. No. RP87665) was obtained from Life Technologies Corporation (Carlsbad, CA, USA). All samples were aliquoted into smaller volumes to eliminate multiple freeze–thaw cycles and stored at -80 °C until thawed prior to analysis.
2.3. Preparation of working stock, calibration, and labeled solutions
The in-house prepared 6-peptide native and labeled spike protein mixtures contained 0.5 nmol of each of the six peptides. The peptide mixture was reconstituted in 1 mL of 0.1% (v/v) FA (aq) to yield a 0.5 pmol/µL solution of each peptide. Seven 0.5-mL stock calibration standard mixtures, ranging from 10 to 250 fmol/µL, were prepared by adding 10, 30, 50, 70, 90, 180 and 250 µL of the native working stock peptide standard mixture, 50 µL of the labeled working stock peptide standard mixture, and a sufficient volume of 0.1% FA (aq) to make the final volume 0.5 mL. Ten µL of the 0.5–pmol/µL solution of the labeled peptides was used as the internal standard cocktail for the analysis of unknowns.
The NEP prepared native and labeled SARS-CoV-2 spike protein peptide mixtures contained 1 nmol of each of the 7 peptides. The peptide mixture was reconstituted in 1 mL of 0.1% FA (aq) to yield 1.0 pmol/ µL native and labeled working stock standard mixtures. Seven 1.0-mL stock calibration standard mixtures, ranging from 10 to 250 fmol/µL, were prepared by adding 10, 30, 50, 70, 90, 180 and 250 µL of the native peptide standard mixture, 50 µL of the labeled standard mixture, and a sufficient volume of 0.1% FA (aq) to make the final volume 1.0 mL. The internal standard cocktail for the analysis of unknown samples was prepared by adding 5 µL of the 1.0 pmol/µL spiked solution of the labeled peptides.
NEP also prepared the native and labeled nucleocapsid peptide mixtures that contain 1 nmol of 5 peptides (LNQLESK, GFYAEGSR, DHIGTR, DQVILLNK and ADETQALPQR). The procedure for preparing calibration curves for nucleocapsid and spike protein are identical and each provide a range from 10 to 250 fmol/µL.
Mean area ratios (native/labeled) were plotted against concentrations for each standard. Linear regression without weighting was applied to the data sets and calibration curves were generated for each peptide. Regression analysis of the calibration curves showed a linear relationship with R2 values between 0.997 and 0.999 for each spike and nucleocapsid peptide using each set of standards.
2.4. Preparation of SARS-CoV-2 recombinant protein digests and IDMS quantification
SARS-CoV-2 recombinant spike S (S1 + S2) protein, rabbit Fc tag (Euprotein Inc.) was used to quantify total spike content. Euprotein’s recombinant spike was first diluted 1:2 in 1X PBS and a 10 µL volume was used as our starting volume. Life Technologies’ SARS-CoV-2 nucleocapsid recombinant protein (RP-87665) was diluted 1:20 in 1X PBS and 10 µl was used as our starting volume. The recombinant samples were then diluted in 10 µL of a 0.1% solution of Rapigest™ SF Surfactant. (Waters Corporation, Milford, MA) in 50 mM ammonium bicarbonate to solubilize proteins and improve tryptic protein digestion [42], [43]. The samples were heated for 5 min at 100 °C and allowed to cool to RT. After cooling, 5 µL (~86 pmol) of frozen Promega Sequencing Grade Modified Trypsin (V5113, Promega, Madison, WI, USA) was added to each vial, and samples were incubated at 37 °C for 2 h to achieve complete in-solution digestion. After digests were cooled, 10 µL of a 0.45 M HCl solution was added to reduce the pH, and digests were incubated at RT for 30 min to cleave the acid-labile surfactant. After incubation, 10 µL of the 0.5-pmol/µL labeled spike (in-house prepared) internal standard (ISTD) working stock mixture, 5 µL of the 1.0 pmol/µL labeled spike (NEP prepared), or 5 µL of the 1.0 pmol/µL labeled nucleocapsid (NEP prepared) ISTD working stock mixture was added to each sample depending upon the sample contents. 0.1% FA (aq) was used to dilute the final sample volume to 100 µL. The digested samples were mixed, centrifuged, and transferred to LC autosampler vials for analysis.
2.5. IDMS instrumentation parameters
An Agilent 1200 series LC system (Agilent Technologies, Inc., Santa Clara, CA, USA) was configured for separation of target peptides. The analytical columns utilized were 150 mm × 1 mm i.d. Symmetry300™ reverse phase C18 (3.5 µm particle size, Waters Corporation). The injection volume was 5 µL. The aqueous mobile phase (A) consisted of 0.1% FA in HPLC-grade water, while the organic mobile phase (B) was 0.1% FA in acetonitrile. A gradient profile for the analysis column was utilized at a flow rate of 50 µL/min. Initially, the mobile phase consisted of 98% A and 2% B. At 3 min the gradient was stepped to 80% A and 20% B over the next 7 min. After 10 min the gradient was stepped to 75% A and 25% B over the next 5 min and then held constant for 2 min. After 17 min total the gradient was stepped to 2% A and 98% B for 7 min to clean the column, then stepped to 98% A and 2% B for the next 2 min to begin equilibrating the column to initial conditions. The isocratic gradient for the regeneration column utilized a 50 µL/min flow rate and consisted of a constant eluent composition of 98% A and 2% B. The total analysis run time was 57 min.
The column eluent was introduced into a Thermo Scientific TSQ Altis™ triple quadrupole tandem mass spectrometer with an electrospray interface. The instrument was operated in positive ion multiple reaction monitoring mode. For each peptide, a quantification transition and two additional transitions were monitored for confirmation purposes. All quantitative and confirmation transitions are listed in Table 1. Instrument parameters were as follows: spray voltage 4000 V, sheath gas 4, auxiliary gas 2, capillary tube temperature 270 °C, and collision gas pressure of 1.5 mTorr. Collision energies and tube lens settings were optimized for each peptide. Instrument control and data processing was performed via the Thermo Scientific TraceFinder™.
2.6. Data analysis
Mean area ratios (native/labeled) were plotted against concentrations for each standard. Linear regression without weighting was applied to the data sets and calibration curves were generated for each peptide. Regression analysis of the calibration curves showed a linear relationship with R2 values between 0.997 and 0.999 for each spike and nucleocapsid peptide using each set of standards.
Two independent IDMS digest preparations were analyzed in duplicate. General practice for selecting peptides and peptide standards for protein quantification by mass spectrometry and the standard rules and guidance for optimization of proteolytic MS-based assays have been described by others [44]. Five unique peptides were quantified independently to ensure completeness of digestion in our targeted spike regions and four unique peptides were quantified independently to ensure completeness of digestion in our targeted nucleocapsid regions. Data acquired on the triple quadrupole mass spectrometer were analyzed and processed by Thermo Scientific TraceFinder™ software. Genesis peak integration settings included an area noise factor of 10, a peak noise factor of 5, and a tailing factor of 2. Boxcar smoothing with 7 points averaged was utilized. Peak integrations were reviewed manually, and transitions from analyte peptides were confirmed by having the same retention times of the heavy stable isotope-labeled peptides. Linear regression was performed without weighting in TraceFinder on the native:labeled peak area ratio versus expected spike and nucleocapsid concentrations to construct response curves. The most abundant transition for each peptide was selected as the quantitative transition to be used in quantitative and statistical analyses. Spike and nucleocapsid limits of quantification (LOQ), both established at 10 fmol/µL, correspond to our lowest calibrant (10 fmol/µL). The values obtained using the peptides’ quantitative responses were then averaged. Percent coefficient of variation is expressed as percent relative standard deviation (standard deviation divided by the mean multiplied by 100). Once the mean value has been calculated using all quantitative peptides (spike = 5; nucleocapsid n = 4), peptide agreement was evaluated to report final IDMS results. Peptide agreement RSD’s must be within 20% for an IDMS run to be accepted. The RSD for the peptide agreement was 12.71% for the 5 spike peptides and 7.78% for the 4 nucleocapsid peptides. The overall method RSD was 3.67% for spike protein and 5.11% for nucleocapsid protein.
3. Results and discussion
We have developed a targeted IDMS method to quantify spike and nucleocapsid proteins from SARS-CoV-2. The total time for analysis is approximately 5 h. This includes sample preparation, a 2 h tryptic digestion, 30 min quenching step and a 57 min LC-MS/MS run time followed by data analysis.
The SARS-CoV-2 spike glycoprotein is 1,273 amino acids in length (SARS-CoV-2 reference virus hCoV-19/Wuhan/IVDC-HB-01/2019). Predicted tryptic digestion sites were identified by database analysis. At the time of in-house standard preparation, the availability of SARS-CoV-2 spike reference materials for analysis was limited, therefore candidate spike peptides were chosen based solely on the sequence of the protein and the absence of methionine, cysteine, and tryptophan residues. We initially selected peptides GVYYPDK (S1), FQTLLALHR (S1), GIYQTSNFR (S1), FLPFQQFGR (S1), VTLADAGFIK (S2) and VEAEVQIDR (S2) for our standard preparation, spanning the S1 and S2 regions of the protein. Of these six candidates, we eliminated VEAEVQIDR from the S2 region because while the VEAEVQIDR calibration curve was linear and accurate, analysis of unknowns showed lower than expected results and discordance with the other peptides. These results may be due to incomplete digestion in the region of the peptide or due to the protein conformation at this site. Additionally, in vaccine candidates, mutations have been made in this region to stabilize the trimer in a pre-fusion structure. Two proline substitutes (2P) at residues K986 and V987 were used to retain the protein in its antigenically optimal pre-fusion confirmation and the new S-2P design is being used as the target antigen in current vaccines [45]. Following our initial investigation and preparation of in-house standard mixtures, we utilized NEP’s services for synthesis of spike standard mixtures. The NEP mixture contained seven peptides, two new peptides, SFIEDLLFNK and ASANLAATK, both in the S2 region, as well as GVYYPDK, GIYQTSNFR, FLPFQQFGR, FQTLLALHR and VTLADAGFIK. The peptide FQTLLALHR in the S1 region was not as robust during analysis and we eliminated this peptide from further investigation. Of the NEP spike tryptic peptides selected, five candidates for MRM analysis were selected for quantitative analyses, GVYYPDK (S1), GIYQTSNFR (S1), FLPFQQFGR (S1), ASANLAATK (S2), and VTLADAGFIK (S2).
The Wuhan reference virus spike protein sequence was then aligned with representative lineages of the known SARS-CoV-2 variants of concern Alpha (first detected in the United Kingdom), Beta (first detected in South Africa), Gamma (first detected in Brazil/Japan), Delta (first detected in India), [46] and variants of interest Epsilon (first detected in USA/California), Zeta (first detected in Brazil), Eta (first detected in United Kingdom/Nigeria), Theta (first detected in the Philippines), Iota (first detected in USA/New York), Kappa (first detected in India), and Lambda (first detected in Peru) (www.who.int/en/activities/tracking-SARS-CoV-2-variants/) to identify if these peptides were located in conserved regions of the spike protein. All of these variants contain the D614G mutation except for the Wuhan wild type virus. Fig. 1 , illustrates cthat the target spike peptides are conserved in the amino acid sequence of the Wuhan reference strain, as well as, that of the variants of concern (Alpha, Beta, Gamma and Delta). While not shown in Fig. 1, the target peptides were also conserved in all of the varients of interest (Epsilon, Zeta, Eta, Theta, Iota, Kappa, and Lambda). Thus, the IDMS method can be used to quantify all viral lineages. While the Gamma hCoV-19/Brazil/AM-987/2020 lineage does have a mutation from ASANLAATK to ASANLAAIK, the other four peptides can be used to quantify the spike protein. The redundancy of having five target peptides, which are generally conserved among the viral lineages, was designed to help ensure that multiple peptides were available to provide accurate protein quantification. Anticipating the possibility of peptide mutations occurring is a key reason for choosing multiple peptide sequences within the protein of interest. Should it be necessary to have this specific peptide, or if a novel variant or lineage emerges that is not covered by our selected target peptides, it is a simple matter to have the variant peptide and isotopically labeled peptide synthesized in a few weeks as previously described.
Fig. 1.
The spike protein sequence of the reference Wuhan virus is aligned with representative variants of concern Alpha, Beta, Gamma, and Delta to show that the target peptides are conserved and can be used to quantify all strains. The Gamma lineage has a mutation from ASANLAATK to ASANLAAIK in one of the S2 target peptides. All peptide targets that were tested are shown in color. The peptides in green were selected for the final IDMS method.
Similarly, while peptide ADETQALPQR in the NEP nucleocapsid peptide mixture, containing five candidate peptides, was comparable in signal intensity for our peptide standards, this peptide yielded lower than expected results when analyzed in our recombinant matrix. This variation is likely attributed to a change in protein conformation or incomplete digestion and was excluded from our final IDMS analyses. Target peptide sequences with corresponding peptide internal standards used for quantification are listed in Table 1.
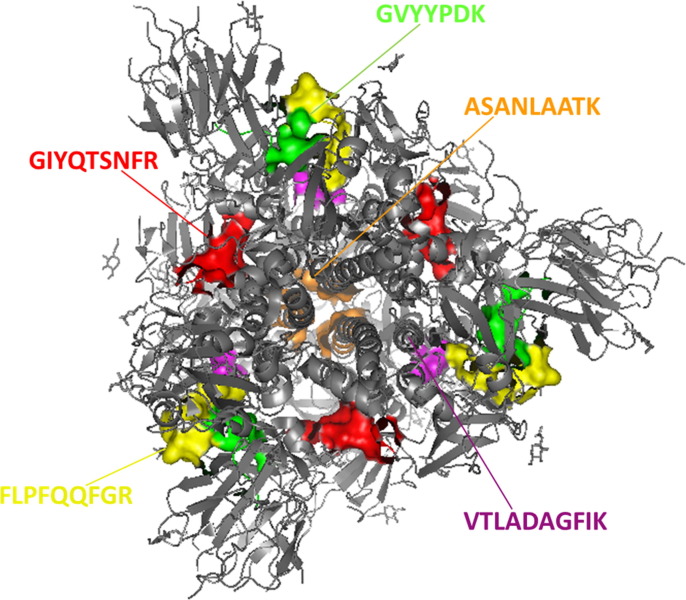
The amino acid sequence of the spike protein in Fig. 2 a indicates targeted spike peptides highlighted and in red, bold font, while the peptides that were initially chosen, but determined not to be appropriate for IDMS analysis of this protein are simply underlined. The location within the trimeric spike structure of the five final peptides used for analysis is shown in Fig. 3 ’s 3-D illustration. These five tryptic peptides from both S1 and S2 subunits were selected as stoichiometric representatives of the spike protein and quantified against the corresponding spiked isotopologue internal standard to yield a measure of protein concentration under the assumption that one mole of peptide equals one mole of protein. The SARS-CoV-2 nucleocapsid protein amino acid sequence seen in Fig. 2b (419 aa in length) indicates our four target MRM tryptic peptides LNQLESK, GFYAEGSR, DHIGTR and DQVILLNK highlighted and in red, bold font, while the peptide that was initially chosen but not used in our final IDMS analysis is underlined.
Fig. 2.
Amino acid sequences of the (A) spike protein and (B) nucleocapsid protein of hCoV-19/Wuhan/WIV04/2019. Target peptides for quantification are highlighted and indicated in red bold font. Candidate peptides chosen, but not included in final quantitative results, are underlined. The protein sequences for this reference virus were obtained from GISAID. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Prefusion 2019-nCoV trimeric spike glycoprotein. PDB:6VSB. https://doi.org/10.2210/pdb6VSB/pdb. Targeted peptides are color-illustrated.
Peptide synthesis options were evaluated by timeliness, cost, and quality of standards. In general, three formats are commercially available for synthesis of custom peptide sequences for quantification: 1) Standard bulk format – a single peptide standard is synthesized, purified (>95%), lyophilized, and received as a 5 mg gross weight deliverable; 2) a single peptide standard is synthesized, purified (>95%), tested for the peptide content’s accuracy by amino acid analysis (AAA), aliquoted as a specific concentration into separate vials, and partially or wholly lyophilized; 3) a single peptide standard is synthesized, purified (>95%), tested for the peptide content’s accuracy by AAA, pooled to create a peptide standard mixture, and aliquoted as specific concentrations into separate vials lyophilized. Here, we are reporting on peptide mixtures made by procedures 1 and 3. Format 1 has been in place in our laboratory since 2008, and while this approach is cost-effective and provides large quantities of high quality and accurate standards, this approach also involves a significant amount of effort from the end user. Peptide standards synthesized by formats 2 and 3 are received as final deliverables. Format 3 involves NEP to generate a native mixture and a label mixture of target peptides to quantify a protein of interest. This strategy would be the most convenient packaging for the end user. Because our laboratory had not previously evaluated a commercial entity’s ability to make high quality peptide mixtures to use as standards, how long it would take to synthesize the mixtures for pandemic preparedness and how accurate the mixtures would be was not clear. For these reasons, we chose to also prepare our mixtures in-house via format 1 for comparative purposes. While format 3′s strategy requires the least effort on the end user and is very popular among the proteomics community, the accuracy of the standards relies solely on the company, the price per standard is the most costly, and the small batch quantity of material produced makes inter-lab distribution of standards difficult for comparative purposes. NEP typically provides peptide mixtures in 4–6 weeks, however we requested expedited service, which added significantly to the cost but also allowed us to receive the material within 2–3 weeks. Format 2 is also well known among the proteomics community, but we proceeded with format 3 so that NEP’s synthesis of peptide mixtures eliminated the need for handling multiple vials of target peptides to analyze a single protein.
Peptides received in 1 standard bulk format (5 mg; spike protein peptides (6 native; 6 labeled) from NEP were further processed in-house and sent to NEP and AAA Service Laboratories, Inc. for AAA testing to evaluate turn-around time, accuracy and precision of peptide contents.
Following in-house processing, 3 vials of each individual spike native peptide and 1 vial of each labeled spike peptide were randomly selected and sent to AAA Service Laboratories, Inc. and NEP for AAA testing. AAA results were received electronically within a few days of receipt confirming peptide contents. Following AAA, the single peptide standards were prepared in-house as peptide mixtures. Individual native peptides were reconstituted in FA according to the specific AAA result and 100 µL of each peptide (600 µL total) was pooled into one vial and lyophilized so that each finalized mixture vial contained 0.5 nmol of each peptide for use as research grade peptide standards for quantitative analyses. Preparation of the labeled pool was performed identically to the native. The total time for preparation of the in-house mixtures was approximately 3 weeks, similar to NEP’s expeditated synthesis of format 3′s mixtures.
Nucleocapsid protein peptide standard mixtures (5 native; 5 labeled) and spike protein peptide standard mixtures (7 native; 7 labeled) were ordered via format 3 from NEP. Each peptide was synthesized, purified (>95%), tested by AAA in duplicate, aliquoted using a liquid handler into two separate mixtures (one native pool and one labeled pool), vialed in 1 nmol quantities, dried and provided as 1 nmol/peptide/vial dry aliquots ready for quantitative analyses.
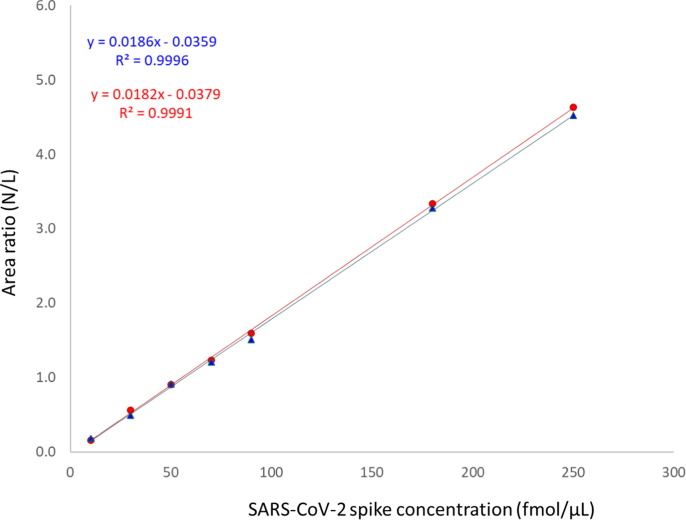
The two spike peptide synthesis formats (in-house mixture preparation (format 1) and NEP mixture preparation (format 3)) were evaluated and their ability to provide accurate quantitative results was determined (Fig. 4 )
Fig. 4.
Comparison of Standard curves for in-house peptide synthesis and NEP peptide synthesis for CoV-2 spike peptide GIYQTSNFR. The similarity of the in-house calibration curve’s slope and y-intercept in comparison to the slope and y-intercept of the NEP-prepared calibration curve shows the agreement of measurements for unknowns regardless of preparation format. Blue triangles = in-house standard preparation; Red circles = NEP standard preparation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
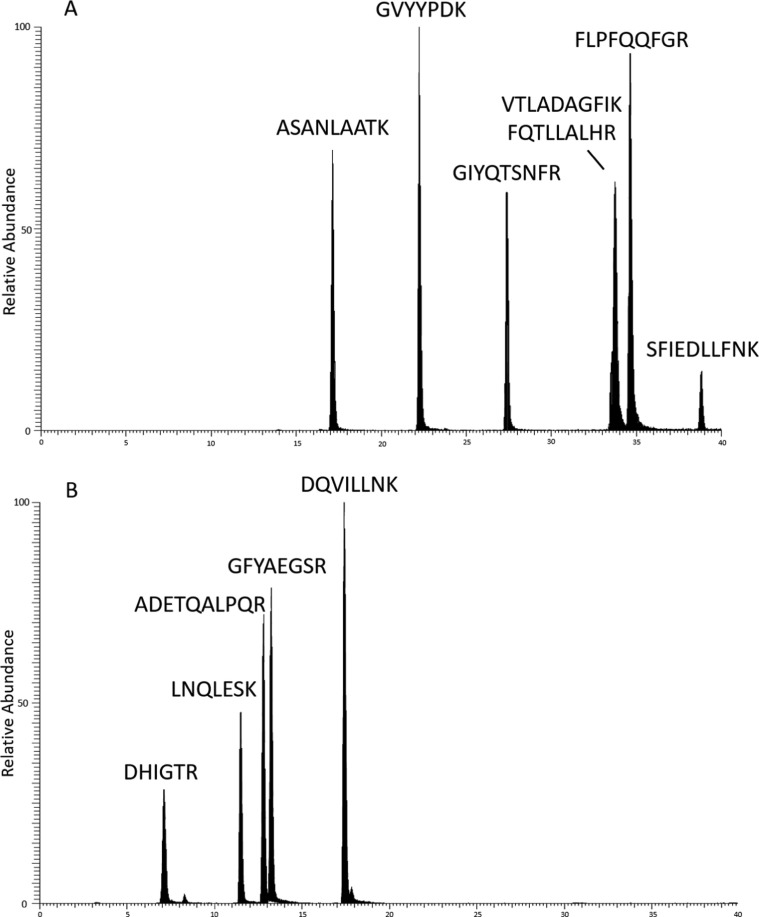
All targeted spike and nucleocapsid peptides were analyzed using the MRM transitions described in Table 1. Typical total ion chromatograms (TIC) are shown in Fig. 5 a, 5b. The resulting calibration curves from the dilution series confirmed that the instrument responses were proportional to spike and nucleocapsid peptide concentrations for the MRM quantitative transitions. Reported spike and nucleocapsid LOQs, both 10 fmol/µL, correspond to the lowest standard on our calibration curves (10 fmol/µL). Recombinant spike and nucleocapsid protein measurements reported here were significantly higher than 10 fmol/µL. However, if quantification below our lowest standard is needed, our LOQ is not limited to 10 fmol/µL (based on the S/N of our lowest standard).
Fig. 5.
Total ion chromatogram showing targeted peptides for (A) SARS-CoV-2 spike protein and (B) nucleocapsid protein quantification. Data were acquired on a Thermo Scientific TSQ Altis™ in SRM mode.
We evaluated the performance of our method and these peptide mixtures for SARS-CoV-2 spike and nucleocapsid quantification using commercially available recombinant protein. Table 2 presents fmol/µL amounts as analyzed by IDMS, summarizing each peptide’s ability to quantify spike and nucleocapsid proteins in the recombinant protein solutions. The mean, standard deviation and % relative standard deviation in Table 2 shows agreement of our spike and nucleocapsid target peptides for a typical IDMS preparation analyzed on our Thermo Scientific TSQ Altis in SRM mode (spike protein n = 5 peptides; nucleocapsid protein n = 4 peptides). To evaluate precision of the LC-MS/MS peptide measurements, two independent digestions were prepared on five separate days (biological replicates) and each digest was analyzed in duplicate (technical replicates). The mean, method standard deviation and % relative standard deviation (RSD) for the SARS-CoV-2 samples, presented in Table 2, shows the performance of the method for both the spike protein and nucleocapsid protein (n = 20). The RSD for the peptide agreement was 12.71% for the 5 spike peptides and 7.78% for the 4 nucleocapsid peptides. Peptide agreement is used to report final IDMS results. Peptide agreement RSD’s must be within 20% for an IDMS run to be accepted. The overall method RSD was 3.67% for spike protein and 5.11% for nucleocapsid protein.
Table 2.
Precision and accuracy of SARS-CoV-2 spike (S) and nucleocapsid (N) quantitative measurements.
| SPIKE (fmol/µL) | NUCLEOCAPSID (fmol/µL) | ||
|---|---|---|---|
| Euprotein Inc. (Cat no. EPY275672) | Life Technologies Corp. (Cat no. RP-87665) | ||
| Target peptide | Target peptide | ||
| GVYYPDK (S1) | 92.02 | DHIGTR (N) | 1979.60 |
| GIYQTSNFR(S1) | 78.22 | LNQLESK (N) | 1941.00 |
| FLPFQQFGR (S1) | 85.60 | GFYAEGSR (N) | 1986.00 |
| ASANLAATK (S2) | 85.80 | DQVILLNK (N) | 1676.60 |
| VTLADAGFIK (S2) | 108.54 | ||
| peptide agreement mean (n = 5)a | 90.04 | peptide agreement mean (n = 4)a | 1895.80 |
| StDev. (n = 5) | 11.44 | StDev. (n = 4) | 147.48 |
| %RSD | 12.71 | %RSD | 7.78 |
| Inter-day mean (n = 20)b | 87.96 | Inter-day mean (n = 20)b | 1830.80 |
| StDev. (n = 20) | 3.22 | StDev. (n = 20) | 93.60 |
| %RSD | 3.67 | %RSD | 5.11 |
4. Conclusions
Mass spectrometry-based protein quantification methods play a number of significant roles in biomedical applications including vaccine development and quality control. Isotope dilution methods can be rapidly developed, and targeted MRM detection of selective peptides yields highly specific methods that have been applied to diverse range of proteins in a vast array of complex matrices. For several years, IDMS analytical techniques, developed and validated in our laboratory for seasonal and potentially pandemic influenza strains, have been approved by the FDA and are currently being performed in several vaccine manufacturers’ laboratories for QA/QC confirmatory testing on whole virus and recombinant protein vaccines. We have adapted these analytical techniques for potential quantification of SARS-CoV-2 spike and nucleocapsid proteins in whole virus and recombinant protein vaccines and in materials to be used as standards for diagnostic tests. Spike protein is the primary antigen in SARS-CoV-2 vaccines and nucleocapsid protein is the target for a majority of SARS-CoV-2 rapid antigen tests. The choice of peptides used to quantify the proteins is a critical step in IDMS method development to provide a selective, accurate and precise analytical method. The IDMS method can be used in the development and quality control of future vaccines and diagnostics by rapidly yielding accurate and precise protein quantification.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This project was supported in part by an appointment to the Research Participation Program at the Centers for Disease Control and Prevention, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and CDC. We gratefully acknowledge the following Authors from the Originating laboratories responsible for obtaining specimens and the Submitting laboratories where genetic sequence data were generated and shared via GISAID Initiative, on which this research is based.
1. hCoV-19/Wuhan/IVDC-HB-01/2019; Accession No. EPI_ISL_402119; Collected 2019-12-30; Originating laboratory National Institute for Viral Disease Control and Prevention; Submitting laboratory Chinese Center for Disease Control and Prevention; Authors Wenjie Tan, Xiang Zhao, Wenling Wang, Xuejun Ma, Yongzhong Jiang, Roujian Lu, Ji Wang, Weimin Zhou, Peihua Niu, Peipei Liu, Faxian Zhan, Weifeng Shi, Baoying Huang, Jun Liu, Li Zhao, Yao Meng, Xiaozhou He, Fei Ye, Na Zhu Yang Li, Jing Chen, Wenbo Xu, George F. Gao, Guizhen Wu.
2. hCoV-19/Brazil/AM-987/2020 (P.1); Accession No. EPI_ISL_833167; Collected 2020-12-16; Originating laboratory DB Diagnosticos do Brasil; Submitting laboratory Instituto Adolfo Lutz, Interdisciplinary Procedures Center, Strategic Laboratory; Authors Claudio Tavares Sacchi, Claudia Regina Goncalves, Erica Valessa Ramos Gomes, Karoline Rodrigues Campos.
3. hCoV-19/England/205090260/2020 (B.1.1.7); Accession No. EOI_ISL_728343; Collected 2020-12-22; Originating laboratory Respiratory Virus Unit, National Infection Service, Public Health England; Submitting laboratory COVID-19 Genomics UK (COG-UK) Consortium; Authors PHE Covid Sequencing Team.
4. hCov-19/South Africa/KRISP-K006851/2020; (B.1.351); Accession No. EPI_ISL_825115; Collected 2020-11-24; Originating laboratory NHLS-IALCH; Submitting laboratory KRISP, KZN Research Innovation and Sequencing Platform; Authors Giandhari J, Pillay, S Lessels R, Mdlalose K, York D, Khan S, Tegally H, Wilkinson E, de Oliveira T.
5. hCoV-19/India/TG-CDFD-OMC11/2021; Accession No. EPI_ISL_2924251; Collected 2021-05-24; Originating laboratory Osmania Medical College; Submitting laboratory CDFD; Authors Bashyam M, Gupta A, Basu R, Donipadi V, Kammili N, Vashisht D, Dalal A.
References in this paper to any specific commercial products, process, service, manufacturer, or company do not constitute an endorsement or a recommendation by the U.S. Government or the Centers for Disease Control and Prevention. The findings and conclusions reported in this article are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
References
- 1.Banerjee S. Empowering clinical diagnostics with mass spectrometry. ACS Omega. 2020;5(5):2041–2048. doi: 10.1021/acsomega.9b03764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seger C., Salzmann L. After another decade: LC-MS/MS became routine in clinical diagnostics. Clin Biochem. 2020;82:2–11. doi: 10.1016/j.clinbiochem.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Geyer P.E., et al. Revisiting biomarker discovery by plasma proteomics. Mol Syst Biol. 2017;13(9):942. doi: 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker C.E., Borchers C.H. Mass spectrometry based biomarker discovery, verification, and validation–quality assurance and control of protein biomarker assays. Mol Oncol. 2014;8(4):840–858. doi: 10.1016/j.molonc.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross D.H., Xu L. Determination of drugs and drug metabolites by ion mobility-mass spectrometry: A review. Anal Chim Acta. 2021;1154 doi: 10.1016/j.aca.2021.338270. [DOI] [PubMed] [Google Scholar]
- 6.Loos G., Van Schepdael A., Cabooter D. Quantitative mass spectrometry methods for pharmaceutical analysis. Philos Trans A Math Phys Eng Sci. 2016;374(2079):20150366. doi: 10.1098/rsta.2015.0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma V.K., Sharma I., Glick J. The expanding role of mass spectrometry in the field of vaccine development. Mass Spectrom Rev. 2020;39(1-2):83–104. doi: 10.1002/mas.21571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botelho J.C., et al. Evaluation of an isotope dilution HPLC tandem mass spectrometry candidate reference measurement procedure for total 17-beta estradiol in human serum. Anal Chem. 2016;88(22):11123–11129. doi: 10.1021/acs.analchem.6b03220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vesper H.W., Botelho J.C. Standardization of testosterone measurements in humans. J Steroid Biochem Mol Biol. 2010;121(3-5):513–519. doi: 10.1016/j.jsbmb.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 10.Mineva E.M., Schleicher R.L., Chaudhary-Webb M., Maw K.L., Botelho J.C., Vesper H.W., et al. A candidate reference measurement procedure for quantifying serum concentrations of 25-hydroxyvitamin D(3) and 25-hydroxyvitamin D(2) using isotope-dilution liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2015;407(19):5615–5624. doi: 10.1007/s00216-015-8733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J., Zhu W., Sun H., Song D., Xiao P., Xu B., et al. Development of a primary reference material of natural C-reactive protein: verification of its natural pentameric structure and certification by two isotope dilution mass spectrometry. Anal Methods. 2021;13(5):626–635. doi: 10.1039/d0ay02289f. [DOI] [PubMed] [Google Scholar]
- 12.Barr JR, et al. Isotope dilution--mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem, 1996. 42(10): p. 1676-82. [PubMed]
- 13.Brun V, et al. Isotope dilution strategies for absolute quantitative proteomics. J Proteomics, 2009. 72(5): p. 740-9. [DOI] [PubMed]
- 14.Cobbaert CM, et al. Towards an SI-traceable reference measurement system for seven serum apolipoproteins using bottom-up quantitative proteomics: conceptual approach enabled by cross-disciplinary/cross-sector collaboration. Clin Chem, 2021. 67(3): p. 478-489. [DOI] [PubMed]
- 15.Ruhaak LR, et al. MS-based proteomics: a metrological sound and robust alternative for apolipoprotein E phenotyping in a multiplexed test. Clin Chem Lab Med, 2019. 57(5): p. e102-e104. [DOI] [PubMed]
- 16.Cazares LH, et al., Development of a liquid chromatography high resolution mass spectrometry method for the quantitation of viral envelope glycoprotein in Ebola virus-like particle vaccine preparations. Clin Proteomics, 2016. 13(1): p. 18. [DOI] [PMC free article] [PubMed]
- 17.Whiting G., Vipond C., Facchetti A., Chan H., Wheeler J.X. Measurement of surface protein antigens, PorA and PorB, in Bexsero vaccine using quantitative mass spectrometry. Vaccine. 2020;38(6):1431–1435. doi: 10.1016/j.vaccine.2019.11.082. [DOI] [PubMed] [Google Scholar]
- 18.Williams T.L., Luna L., Guo Z., Cox N.J., Pirkle J.L., Donis R.O., et al. Quantification of influenza virus hemagglutinins in complex mixtures using isotope dilution tandem mass spectrometry. Vaccine. 2008;26(20):2510–2520. doi: 10.1016/j.vaccine.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Santana W.I., Williams T.L., Winne E.K., Pirkle J.L., Barr J.R. Quantification of viral proteins of the avian H7 subtype of influenza virus: an isotope dilution mass spectrometry method applicable for producing more rapid vaccines in the case of an influenza pandemic. Anal Chem. 2014;86(9):4088–4095. doi: 10.1021/ac4040778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierce C.L., Williams T.L., Moura H., Pirkle J.L., Cox N.J., Stevens J., et al. Quantification of immunoreactive viral influenza proteins by immunoaffinity capture and isotope-dilution liquid chromatography-tandem mass spectrometry. Anal Chem. 2011;83(12):4729–4737. doi: 10.1021/ac2006526. [DOI] [PubMed] [Google Scholar]
- 21.Pierce C.L., Williams T.L., Santana W.I., Levine M., Chen L.-M., Cooper H.C., et al. Immunocapture isotope dilution mass spectrometry in response to a pandemic influenza threat. Vaccine. 2017;35(37):5011–5018. doi: 10.1016/j.vaccine.2017.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgenstern K., Xie Y., Palladino G., Barr J.R., Settembre E.C., Williams T.L., et al. Reference antigen-free and antibody-free LTD-IDMS assay for influenza H7N9 vaccine in vitro potency determination. Vaccine. 2018;36(41):6144–6151. doi: 10.1016/j.vaccine.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper H.C., Xie Y., Palladino G., Barr J.R., Settembre E.C., Wen Y., et al. Limited tryptic digestion-isotope dilution mass spectrometry (LTD-IDMS): A reagent-free analytical assay to quantify hemagglutinin of A(H5N1) vaccine material. Anal Chem. 2020;92(17):11879–11887. doi: 10.1021/acs.analchem.0c02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams T.L., Pirkle J.L., Barr J.R. Simultaneous quantification of hemagglutinin and neuraminidase of influenza virus using isotope dilution mass spectrometry. Vaccine. 2012;30(14):2475–2482. doi: 10.1016/j.vaccine.2011.12.056. [DOI] [PubMed] [Google Scholar]
- 25.Johnson A., Chen L.-M., Winne E., Santana W., Metcalfe M.G., Mateu-Petit G., et al. Identification of influenza A/PR/8/34 donor viruses imparting high hemagglutinin yields to candidate vaccine viruses in eggs. PLoS ONE. 2015;10(6):e0128982. doi: 10.1371/journal.pone.0128982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ridenour C., et al. Development of influenza A(H7N9) candidate vaccine viruses with improved hemagglutinin antigen yield in eggs. Influenza Other Respir Viruses. 2015;9(5):263–270. doi: 10.1111/irv.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bose M.E., Sasman A., Mei H., McCaul K.C., Kramp W.J., Chen L.-M., et al. Analytical reactivity of 13 commercially available rapid influenza diagnostic tests with H3N2v and recently circulating influenza viruses. Influenza Other Respir Viruses. 2014;8(4):474–481. doi: 10.1111/irv.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Client CN. New coronavirus! Preliminary determination of “culprit” of unexplained pneumonia in Wuhan; 2020.
- 29.Prevention, C.D.C. Coronavirus Disease 2019 (COVID-19) Situation Summary. COVID Data Tracker 2020 [cited 2020 April 9]; Available from: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/summary.html
- 30.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., et al. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69(1):531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 34.Soema P.C., Kompier R., Amorij J.-P., Kersten G.F.A. Current and next generation influenza vaccines: Formulation and production strategies. Eur J Pharm Biopharm. 2015;94:251–263. doi: 10.1016/j.ejpb.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 35.Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F., et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol. 2020;94(4) doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marra M.A., et al. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 38.Holmes K.V., Enjuanes L. Virology. The SARS coronavirus: a postgenomic era. Science. 2003;300(5624):1377–1378. doi: 10.1126/science.1086418. [DOI] [PubMed] [Google Scholar]
- 39.Leung D., Tam F., Ma C., Chan P., Cheung J., Niu H., et al. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J Infect Dis. 2004;190(2):379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beck E.T., Paar W., Fojut L., Serwe J., Jahnke R.R., Miller M.B. Comparison of the Quidel Sofia SARS FIA Test to the Hologic Aptima SARS-CoV-2 TMA Test for Diagnosis of COVID-19 in Symptomatic Outpatients. J Clin Microbiol. 2021;59(2) doi: 10.1128/JCM.02727-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diao B, et al. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect, 2021. 27(2): p. 289 e1-289 e4. [DOI] [PMC free article] [PubMed]
- 42.Yu YQ, et al. Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal Chem, 2003. 75(21): p. 6023-8. [DOI] [PubMed]
- 43.Suder P., Bierczynska A., König S., Silberring J. Acid-labile surfactant assists in-solution digestion of proteins resistant to enzymatic attack. Rapid Commun Mass Spectrom. 2004;18(7):822–824. doi: 10.1002/rcm.1411. [DOI] [PubMed] [Google Scholar]
- 44.Hoofnagle AN, et al. Recommendations for the generation, quantification, storage, and handling of peptides used for mass spectrometry-based assays. Clin Chem, 2016. 62(1): p. 48-69. [DOI] [PMC free article] [PubMed]
- 45.Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol, 2021. 21(2): p. 73-82. [DOI] [PMC free article] [PubMed]
- 46.CDC SARS-CoV-2 Variant Classifications and Definitions; 2021.