Abstract
Extraction-based real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR) is currently the “gold standard” in SARS-CoV-2 diagnostics. However, some extraction-free RT-qPCR techniques have recently been developed. In this study, we compared the sensitivity of traditional extraction-based, heated extraction-free, and unheated extraction-free RT-qPCR methods for SARS-CoV-2 detection in nasopharyngeal swabs from symptomatic individuals. The unheated extraction-free method showed perfect agreement with the standard extraction-based RT-qPCR. By contrast, the heat-treated technique was associated with an 8.2% false negativity rate. Unheated extraction-free RT-qPCR for the molecular diagnosis of SARS-CoV-2 is a valuable alternative to the traditional extraction-based methods and may accelerate turnaround times by about two hours.
Introduction
The ongoing pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has had an unprecedented socioeconomic and public health impact worldwide. The availability of accurate COVID-19 diagnostic tests is one of the cornerstones of tackling the pandemic [1–5].
Today, real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR) is considered the “gold standard” assay for the diagnosis of both symptomatic and asymptomatic cases [6] and is included in the protocols issued by several international institutional bodies [7, 8]. These commonly used protocols involve a ribonucleic acid (RNA) extraction step, which may be seen as a major bottleneck in the routine laboratory testing process [9].
Since March 2020, some manufacturers have reported supply shortages of RNA extraction kits, owing to the sudden increase in their global demand. This situation has called for alternative protocols with similarly high diagnostic accuracy, in order to ensure the continuity of testing [10–13]. Consequently, various direct approaches that avoid RNA extraction have been suggested, including heat-processed methods [9, 14].
The objective of this study was to evaluate the diagnostic performance of two different extraction-free RT-qPCR techniques and to compare their performance with that of the traditional extraction-based method in a real-world setting.
Materials and methods
In this study, a total of 98 fresh nasopharyngeal swabs routinely and prospectively collected in October 2020 from COVID-19 symptomatic individuals were analyzed. All these samples were deemed positive according to the traditional extraction-based (EB) RT-qPCR technique. During an internal discussion, it was decided that a sample size of 100 positive samples would be sufficiently powered to determine a sensitivity of 90% [15, 16] provided by the RNA extraction-free (EF) methods. It was assumed that the specificity of the EF method was 100%. During execution of the study, two samples from previously known positive subjects yielded invalid results (i.e., the internal control was not amplified) in the EB RT-qPCR and were therefore excluded from the analysis.
Each sample analyzed was eluted in universal transport medium (UTM™, Copan Diagnostics Inc., USA) and processed at the regional reference laboratory for COVID-19 diagnosis of the San Martino Policlinico Hospital (Genoa, Liguria, Northwest Italy).
All samples were processed on the same day using three methods: (i) a standard EB method performed with STARMag 96x4 Viral DNA/RNA 200C Kit (Seegene Inc., South Korea), (ii) an RNA extraction-free (EF) method with a heating step (EFh+), and (iii) an unheated EF technique (EFh–).
For the EB method, total RNA was extracted using a STARMag Universal Cartridge Kit (Seegene Inc., South Korea) on an automated Nimbus IVD (Seegene Inc., South Korea) platform. As recommended by the manufacturer, a total of 200 µl of each sample was extracted and eluted with 100 µl of elution buffer and set up for RT-qPCR [17].
For the EFh– technique, an input volume of 5 µl of each sample diluted 1:3 with molecular-grade water was used directly for RT-qPCR. By contrast, for the EFh+ method, thermolysis was first performed at 95°C for 3 min and at 4°C for 5 min on a Bio-Rad CFX96™ thermal cycler (Bio-Rad Laboratories, USA).
RT-qPCR was performed on a CFX96™ instrument (Bio-Rad Laboratories, USA) using an Allplex™ 2019-nCoV assay (Seegene Inc., South Korea). This is a multiplex RT-qPCR assay that is able to simultaneously detect three different genes, targeting the nucleoprotein region (N), the RNA-dependent RNA-polymerase (RdRp)/spike (S) region, and the envelope (E) region. Amplification was performed at 50°C for 20 min, followed by 95°C for 15 min and 45 cycles at 95°C for 10 s, 60°C for 15 s with first acquisition, and 72°C for 10 s with second acquisition on a CFX96™ thermal cycler. For each reaction, 5 µl of the extracted RNA in a final volume of 20 µl was used. According to the manufacturer, the analytical specificity of this method is 100% [17].
In the analysis, samples yielding a cycle threshold (Ct) value <40 for at least two genes were considered positive.
The standard EB technique was considered a reference method against both EFh+ and EFh–. A sensitivity value with 95% confidence intervals (CIs) was calculated. As the observed Ct values were approximately normally distributed, the average target-specific differences in Ct values among the three techniques were computed and compared by applying repeated-measures analysis of variance (ANOVA) and the follow-up Tukey honestly significant difference post-hoc test. Data were analyzed in R stats packages, v. 4.0.3 [18].
Results
When using the traditional EB technique as the “gold standard”, the EFh– method displayed perfect accordance, with no false negative results (sensitivity of 100% [95% CI: 95.3–100%]). By contrast, a total of eight (8.2%) swabs treated as per the EFh+ methodology were deemed false negative, giving a sensitivity of 91.8% (95% CI: 84.1–96.2%). As per the EB method, all of these specimens had Ct values >30. These discordant samples were retested, and the initial result was confirmed.
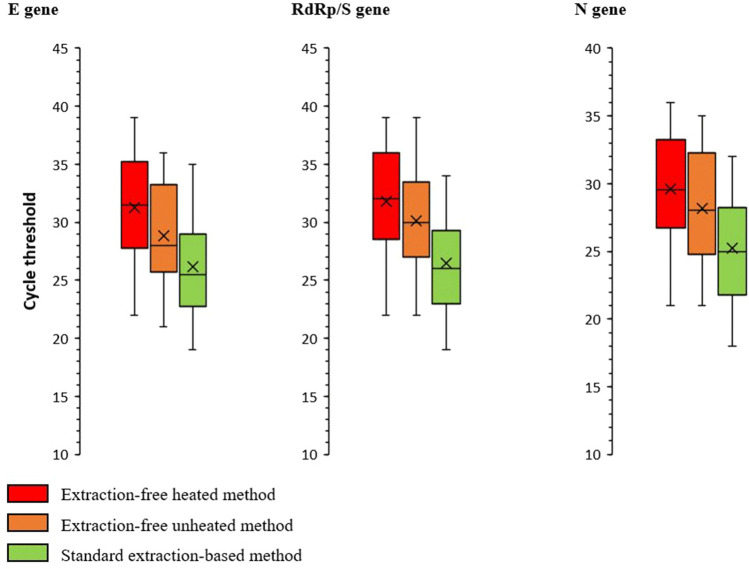
Figure 1 and Table 1 report the summary distributions and mean differences of Ct values according to the technique used and gene target. The omnibus ANOVA test rejected (P < 0.001) the null hypothesis that Ct values yielded by the three methods had identical means; moreover, all of the pairwise post-hoc comparisons were also statistically significant (P < 0.01). Indeed, the average Ct value for any gene was lowest in the EB group and highest in the EFh+ group. Moreover, the highest between-method difference concerned the RdRp/S gene, and the lowest, the N gene (Table 1). Of note, all eight false negatives determined by the EFh+ method had a Ct value >30, regardless of the gene target (average of 34.1, 34.5, and 32.9 for the E, RdRp/S, and N gene, respectively).
Fig. 1.
Distribution of the cycle threshold values by method used and gene target
Table 1.
Mean absolute difference in cycle threshold values by method used and gene target*
| Method | Genet target | ||
|---|---|---|---|
| E | RdRp/S | N | |
| EFh+ vs EB | 4.92 (4.44–5.40) | 5.29 (4.74–5.85) | 4.10 (3.71–4.48) |
| Efh– vs EB | 2.26 (1.89–2.63) | 2.87 (2.48–3.26) | 2.53 (2.13–2.93) |
| Efh+ vs Efh– | 2.66 (2.34–2.98) | 2.42 (1.99–2.86) | 1.56 (1.23–1.90) |
*Results are reported as mean difference (95% CIs for paired samples)
Discussion
In the context of the COVID-19 pandemic, it is crucial to obtain rapid and reliable results in order to diagnose patients and take timely public health decisions. Here, we demonstrated that EF methods may be valuable alternatives to traditional EB techniques. This study adds to the growing body of evidence of the utility of extraction-free methods of detecting SARS-CoV-2.
Several EF protocols have recently been described and validated [9, 15, 19–24]. Across these studies, the relative sensitivity of EF techniques has been seen to vary from 55% to >95%. In our study, the RT-qPCR readout yielded by the direct EFh– method was in complete agreement with the traditional EB technique. Although there were significant differences between the Ct values obtained by these two methods, no disagreement in the interpretation of the overall output was found. On the other hand, we found some negative effect of heat inactivation on virus detection. This, however, affected only samples with low viral loads (Ct >30). Cameron et al. [19] observed a relative sensitivity of EFh+ (heat treatment at 95°C for 10 min) vs. EB of 100% for specimens with Ct values ≤ 25. This value, however, dropped to 22–33% for specimens with Ct values >35 [19]. Similarly, Zou et al. [20] reported that, following heat treatment, about 13.0% of positive swabs became negative; all of these false negatives had a Ct value ≥37. Grant et al. [21] also observed lower Ct values in unheated samples. Our results are also consistent with those of a recent study by Burton et al. [22]. Indeed, these authors observed that, in comparison with unheated samples, heat treatment at 95°C for 5 min increased the average Ct by 4.5–6 cycles. The application of lower heating regimens may therefore be useful in order to increase the sensitivity of the EFh+ method and should be investigated further. For instance, Burton et al. [22] did not register any significant differences on implementing heating regimens of 56/60°C. In sum, in line with the available evidence [9, 15, 19–24], our results support the use of EF techniques in routine practice. The heat treatment procedure introduced in order to lyse epithelial cells may, however, reduce sensitivity. The differences in the sensitivity values reported are most probably attributable to different EF protocols adopted in single studies, including, for example, the relative distribution of specimens with different Ct values, sample storage, heat inactivation regimen, etc. Therefore, a well-established and recognized EF protocol is warranted.
At least two study limitations may affect the overall interpretation of our findings. First, we assumed that the specificity of EB RT-qPCR was 100%; therefore, no EB-based negative samples were analyzed in the present study. Second, for the fully concordant samples (91.8%, 90/98), only one replicate for each RT-qPCR technique was used.
In conclusion, our data suggest that EF methods (especially unheated) are a useful tool in providing RT-qPCR results in a shorter time, thereby speeding up clinical diagnostics and the subsequent burden on molecular labs and hospitals. Indeed, the average processing times in our study were 270, 163, and 156 min for the standard EB, EFh+, and EFh–, respectively. These EF methods therefore urgently require an internationally recognized protocol.
Acknowledgments
The authors would like to thank Chessa Valerio, Qosjia Rexhina, Randazzo Nadia, and Theimer Matteo for performing RT-qPCR tests.
Funding
This study received no external funding.
Availability of data and materials
The whole raw dataset used may be available from the corresponding author on reasonable request and following approval by San Martino Policlinico Hospital.
Declarations
Conflict of interest
The authors declare no conflict of interest regarding this publication.
Ethics statement
The study was conducted according to the guidelines of the Declaration of Helsinki. Ethical review and approval were waived for this analysis, as it was based on routine COVID-19 testing.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Park M, Cook AR, Lim JT, Sun Y, Dickens BL. A systematic review of COVID-19 epidemiology based on current evidence. J Clin Med. 2020;9:967. doi: 10.3390/jcm9040967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pradhan D, Biswasroy P, Kumar Naik P, Ghosh G, Rath G. A review of current interventions for COVID-19 prevention. Arch Med Res. 2020;51:363–374. doi: 10.1016/j.arcmed.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang YW, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID-19: Current issues and challenges. J Clin Microbiol. 2020;58:e00512–e520. doi: 10.1128/jcm.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McArthur L, Sakthivel D, Ataide R, Chan F, Richards JS, Narh CA. Review of burden, clinical definitions, and management of COVID-19 cases. Am J Trop Med Hyg. 2020;103:625–638. doi: 10.4269/ajtmh.20-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venter M, Richter K. Towards effective diagnostic assays for COVID-19: A review. J Clin Pathol. 2020;73:370–377. doi: 10.1136/jclinpath-2020-206685. [DOI] [PubMed] [Google Scholar]
- 6.Böger B, Fachi MM, Vilhena RO, Cobre AF, Tonin FS, Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am J Infect Control. 2021;49:21–29. doi: 10.1016/j.ajic.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization (WHO) (2021) Laboratory testing for coronavirus disease (COVID-19) in suspected human cases: Interim guidance, 19 March 2020. https://apps.who.int/iris/handle/10665/331501. Accessed 5 Mar 2021
- 8.European Centre for Disease Prevention and Control (ECDC) (2021) COVID-19 testing strategies and objectives. https://www.ecdc.europa.eu/sites/default/files/documents/TestingStrategy_Objective-Sept-2020.pdf. Accessed 5 Mar 2021
- 9.Smyrlaki I, Ekman M, Lentini A, Rufino de Sousa N, Papanicolaou N, Vondracek M, Aarum J, Safari H, Muradrasoli S, Rothfuchs AG, Albert J, Högberg B, Reinius B. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat Commun. 2020;11:4812. doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhoads DD, Cherian SS, Roman K, Stempak LM, Schmotzer CL, Sadri N. Comparison of Abbott ID Now, DiaSorin Simplexa, and CDC FDA emergency use authorization methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J Clin Microbiol. 2020;58:e00760–e820. doi: 10.1128/jcm.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Yao H, Xu X, Zhang P, Zhang M, Shao J, Xiao Y, Wang H. Limits of detection of 6 approved RT-PCR kits for the novel SARS-Coronavirus-2 (SARS-CoV-2) Clin Chem. 2020;66:977–979. doi: 10.1093/clinchem/hvaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran A, Beavis KG, Matushek SM, Ciaglia C, Francois N, Tesic V, Love N. Detection of SARS-CoV-2 by use of the cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 assays. J Clin Microbiol. 2020;58:e00772–e820. doi: 10.1128/jcm.00772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waggoner JJ, Stittleburg V, Pond R, Saklawi Y, Sahoo MK, Babiker A, Hussaini L, Kraft CS, Pinsky BA, Anderson EJ, Rouphael N. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26:1633–1635. doi: 10.3201/eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fomsgaard AS, Rosenstierne MW. An alternative workflow for molecular detection of SARS-CoV-2—escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Euro Surveill. 2020;25:2000398. doi: 10.2807/1560-7917.es.2020.25.14.2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byrnes SA, Gallagher R, Steadman A, Bennett C, Rivera R, Ortega C, Motley ST, Jain P, Weigl BH, Connelly JT. Multiplexed and extraction-free amplification for simplified SARS-CoV-2 RT-PCR tests. Anal Chem. 2021;93:4160–4165. doi: 10.1021/acs.analchem.0c03918. [DOI] [PubMed] [Google Scholar]
- 16.Brown JR, Atkinson L, Shah D, Haris K (2021) Validation of an extraction-free RT-PCR protocol for detection of SARS-CoV-2 RNA. 10.1101/2020.04.29.20085910v1.full.pdf. Accessed 20 May 2021
- 17.Seegene Inc (2021) Allplex™ 2019-nCoV Assay. http://www.seegene.com/upload/product/IFU_FDA_COVID19_Seegene.pdf Accessed 20 May 2021
- 18.R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. Accessed 5 Mar 2021
- 19.Cameron A, Pecora ND, Pettengill MA. Extraction-free methods for the detection of SARS-CoV-2 by reverse transcription-PCR: a comparison with the Cepheid Xpert Xpress SARS-CoV-2 assay across two medical centers. J Clin Microbiol. 2021;59:e02643–e2720. doi: 10.1128/JCM.02643-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou J, Zhi S, Chen M, Su X, Kang L, Li C, Su X, Zhang S, Ge S, Li W. Heat inactivation decreases the qualitative real-time RT-PCR detection rates of clinical samples with high cycle threshold values in COVID-19. Diagn Microbiol Infect Dis. 2020;98:115109. doi: 10.1016/j.diagmicrobio.2020.115109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant PR, Turner ME, Shin GY, Nastouli E, Levett LJ (2021) Extraction-free COVID-19 (SARS-CoV-2) diagnosis by RT-PCR to increase capacity for national testing programmes during a pandemic. 10.1101/2020.04.06.028316v1.full.pdf. Accessed 5 Mar 2021
- 22.Burton J, Love H, Richards K, Burton C, Summers S, Pitman J, Easterbrook L, Davies K, Spencer P, Killip M, Cane P, Bruce C, Roberts ADG. The effect of heat-treatment on SARS-CoV-2 viability and detection. J Virol Methods. 2021;290:114087. doi: 10.1016/j.jviromet.2021.114087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visseaux B, Collin G, Houhou-Fidouh N, Le Hingrat Q, Ferré VM, Damond F, Ichou H, Descamps D, Charpentier C. Evaluation of three extraction-free SARS-CoV-2 RT-PCR assays: a feasible alternative approach with low technical requirements. J Virol Methods. 2021;291:114086. doi: 10.1016/j.jviromet.2021.114086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruce EA, Huang ML, Perchetti GA, Tighe S, Laaguiby P, Hoffman JJ, Gerrard DL, Nalla AK, Wei Y, Greninger AL, Diehl SA, Shirley DJ, Leonard DGB, Huston CD, Kirkpatrick BD, Dragon JA, Crothers JW, Jerome KR, Botten JW. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. PLoS Biol. 2020;18:e3000896. doi: 10.1371/journal.pbio.3000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole raw dataset used may be available from the corresponding author on reasonable request and following approval by San Martino Policlinico Hospital.