Abstract
Simple Summary
Multiparametric magnetic resonance imaging (mpMRI) has become an important diagnostic tool in the assessment of clinically significant prostate cancer. The disadvantages of this technique are primarily related to its long examination time and limited availability. This prospective study presents a novel accelerated deep learning image reconstructed T2-weighted turbo spin echo (TSE) sequence providing an acquisition time reduction of more than 60%. The study results show that the acquisition of three imaging planes of the prostate is feasible within 3:50 min, using deep learning image reconstruction, compared to 10:21 min in standard imaging. Additionally, image quality parameters were evaluated to be superior in deep learning imaging.
Abstract
Multiparametric MRI (mpMRI) of the prostate has become the standard of care in prostate cancer evaluation. Recently, deep learning image reconstruction (DLR) methods have been introduced with promising results regarding scan acceleration. Therefore, the aim of this study was to investigate the impact of deep learning image reconstruction (DLR) in a shortened acquisition process of T2-weighted TSE imaging, regarding the image quality and diagnostic confidence, as well as PI-RADS and T2 scoring, as compared to standard T2 TSE imaging. Sixty patients undergoing 3T mpMRI for the evaluation of prostate cancer were prospectively enrolled in this institutional review board-approved study between October 2020 and March 2021. After the acquisition of standard T2 TSE imaging (T2S), the novel T2 TSE sequence with DLR (T2DLR) was applied in three planes. Overall, the acquisition time for T2S resulted in 10:21 min versus 3:50 min for T2DLR. The image evaluation was performed by two radiologists independently using a Likert scale ranging from 1–4 (4 best) applying the following criteria: noise levels, artifacts, overall image quality, diagnostic confidence, and lesion conspicuity. Additionally, T2 and PI-RADS scoring were performed. The mean patient age was 69 ± 9 years (range, 49–85 years). The noise levels and the extent of the artifacts were evaluated to be significantly improved in T2DLR versus T2S by both readers (p < 0.05). Overall image quality was also evaluated to be superior in T2DLR versus T2S in all three acquisition planes (p = 0.005–<0.001). Both readers evaluated the item lesion conspicuity to be superior in T2DLR with a median of 4 versus a median of 3 in T2S (p = 0.001 and <0.001, respectively). T2-weighted TSE imaging of the prostate in three planes with an acquisition time reduction of more than 60% including DLR is feasible with a significant improvement of image quality.
Keywords: deep learning, multiparametric magnetic resonance imaging, prostatic neoplasms, diagnostic imaging
1. Introduction
Multiparametric magnetic resonance imaging (mpMRI) of the prostate has become the standard of care in prostate cancer imaging during the last decade [1,2]. mpMRI plays an important role in biopsy planning, local staging, as well as active surveillance [2,3,4,5,6]. The technical development and increasing experience with mpMRI reporting has finally led to the establishment of the prostate imaging reporting and data system (PI-RADS). The currently recommended protocol of the European Society of Urology consists of the following sequences: T2-weighted turbo spin echo (TSE) imaging in three planes with a slice thickness of 3 mm without gaps and with high morphological resolution. Furthermore, diffusion-weighted imaging (DWI) including apparent diffusion coefficient maps (ADC) as well as dynamic contrast-enhanced imaging should be acquired, to provide further parameters for analysis. Additionally, a precontrast T1-weighted sequence of the pelvis with a larger field of view should be acquired, for lymph node staging as well as for assessment of pelvic bones. A disadvantage of this comprehensive protocol consists of rather long acquisition times, ranging approximately from 30 to 45 min. This is especially problematic as prostate cancer commonly affects elderly men who may have difficulties remaining motionless during long MRI examinations. Furthermore, long examination times are problematic due to the growing demand for mpMRI of the prostate, resulting from extended life expectancy on the one hand and increasing importance of the examination modality to prevent significant prostatic cancers on the other hand. Especially the role in active surveillance might determine a significant contribution for increasing demands regarding mpMRI in future.
Established methods to shorten the acquisition time include parallel imaging techniques in TSE imaging or compressed sensing (CS) in gradient echo imaging. However, a significant disadvantage of parallel imaging is the loss of signal-to-noise ratio (SNR) proportional to the square root of the scan acceleration. Therefore, in clinical protocols, parallel imaging acceleration does usually not exceed a factor of two for two-dimensional acquisitions and a factor of four for three-dimensional acquisitions.
One of the newest techniques applied for MRI acquisition acceleration is based on deep learning (DL). While DL techniques have been primarily applied to facilitate and support diagnosis so far, recent applications concentrate on the acceleration of MRI protocols [7,8,9,10,11]. In previous studies, it could be shown that DL, e.g., via variational networks, is able to significantly accelerate MRI protocols of the abdomen, knee or of the pituitary [12,13,14,15]. However, literature regarding the clinical impact of DL strategies in accelerated MRI protocols is still sparse.
Due to the increasing importance of prostate MRI that is hampered by the long examination times, a thorough, systematic investigation of the potential benefits of DL reconstruction techniques in mpMRI seems worthwhile. Therefore, the aim of this study was to investigate the impact of DL reconstruction in accelerated T2 TSE imaging of the prostate in three orthogonal planes on image quality, lesion conspicuity, and diagnostic confidence, compared to standard T2 TSE imaging.
2. Materials and Methods
2.1. Study Design
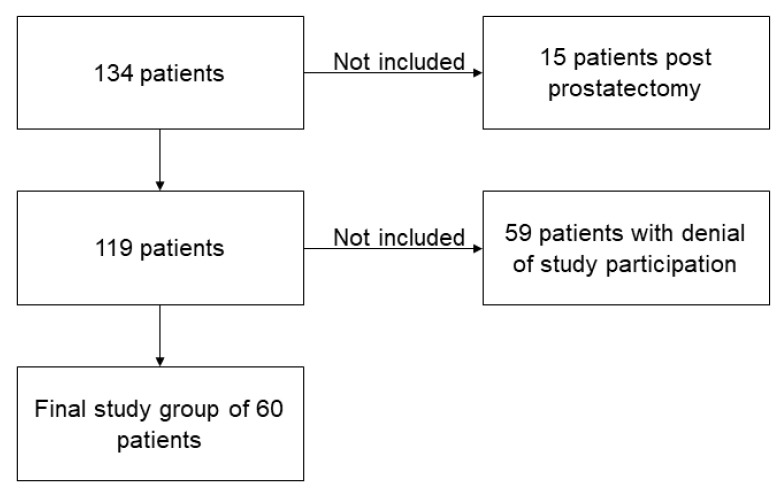
This prospective study was approved by the institutional review board. Inclusion criteria were: examination for evaluation of prostate cancer on appropriate 3T MRI scanners with installed DL reconstruction algorithm. Exclusion criteria were: non-conditional implants, severe claustrophobia, and status post prostatectomy. Figure 1 shows the flowchart of this study (Figure 1). Sixty patients who underwent 3T MRI of the prostate for evaluation of carcinoma between October 2020 and March 2021 were finally included (Table 1). Study participation was voluntary, and all patients gave informed consent to participate in this study. All study procedures were in line with the declaration of Helsinki and its later amendments.
Figure 1.
Flowchart of study participants.
Table 1.
Patients’ characteristics.
| Characteristics | Values |
|---|---|
| Number of patients | n = 60 |
| Age, mean ± standard deviation | 69 ± 9 years |
| Sex | 100% male |
| PSA, median (interquartile range) | 7.2 ng/mL (5.9–9.7 ng/mL) |
2.2. Deep Learning Image Reconstruction Technique
An unrolled variational network was employed for reconstruction [16,17]. Inspired by the iterative optimization used in CS, the trainable network alternated between data consistency steps and image regularization steps using a convolutional network. From this viewpoint, the approach can be considered a natural development from CS in that the regularization is not postulated but instead determined on representative data. Similar to CS, the input to the network consists of undersampled k-space data, coil sensitivity maps estimated from reference lines, and for the network used in the present study also a normalization field for image homogenization. Conventional sampling patterns also used in parallel imaging were used for acceleration. As the reconstruction was not designed to modify the image contrast, but instead mainly focused on signal-to-noise enhancement, this procedure had the additional advantage that the effect of acquisition parameters, such as echo time, repetition time, and echo train length was identical to conventional reconstructions.
The parameters of the model were determined through supervised training using about 10,000 slices from volunteer TSE acquisitions on various clinical 1.5T and 3T scanners (MAGNETOM scanners, Siemens Healthcare). The fully sampled training data was acquired in different body regions (head, pelvis, and knee) and with different image contrasts. The input data was gained via retrospective downsampling by a factor of 4 and usage of a phase resolution of 75%. An L1-norm and multiscale version of the structural similarity (SSIM) content between network prediction and ground truth images encompassed the loss function. The training was integrated in PyTorch and performed on a commercially available GPU cluster with 32 GB of memory. Finally, the trained network was converted for use in a scanner-integrated inference framework. Inference time for a single slice in the actual deployment was about 3 s for CPU on average and 0.5 s for GPU.
2.3. Imaging Protocol
All MRI studies were performed in clinical routine using three different scanners (MAGNETOM Skyra, MAGNETOM Prismafit, and MAGNETOM Vida, Siemens Healthcare). Butylscopolamine bromide was applied prior to the examination, given no present contraindications (e.g., cardiac arrhythmia or glaucoma). Patients were scanned in dorsal position using a setup of 12 elements of a 32-channel spine coil as well as an 18-channel body coil. No endorectal coil was applied in our imaging center. The institution’s standard mpMRI protocol consisted of the following sequences: T2w TSE imaging in three planes; DWI with three different acquired b-values in axial plane (50 s/mm², 500 s/mm², and 1000 s/mm²) as well as one calculated b-value of 2000 s/mm² and ADC mapping; for evaluation of possible bone lesions and lymph node status, a T1w precontrast TSE imaging with a larger field of view was obtained. Furthermore, after application of contrast media (0.1 mmol/kg body weight gadobutrol; Gadovist, Bayer Healthcare) using a flow rate of 1.5 mL/s followed by a saline flush of 20 mL, dynamic contrast-enhanced gradient echo imaging was acquired in axial direction followed by an additional post-contrast gradient-echo axial sequence.
After completion of standard T2w TSE imaging (T2S), the novel T2w TSE imaging with deep learning image reconstruction (T2DLR) was acquired using a prototype sequence.
Imaging parameters are displayed in Table 2. Acquisition time of axial T2S was 4:37 min as compared to 1:38 min of T2DLR (65% reduction of acquisition time), of coronal T2S was 3:07 min versus 1:10 min of T2DLR (63% reduction of acquisition time), and of sagittal T2S was 2:37 min versus 1:02 min of T2DLR (61% reduction of acquisition time). Overall, acquisition time for T2S resulted in 10:21 min versus 3:50 min for T2DLR.
Table 2.
MRI acquisition parameters.
| Axial | Coronal | Sagittal | ||||
|---|---|---|---|---|---|---|
| T2S | T2DLR | T2S | T2DLR | T2S | T2DLR | |
| TR (ms) | 4470 | 4470 | 7480 | 7760 | 7480 | 6900 |
| TE (ms) | 104 | 104 | 101 | 101 | 101 | 101 |
| Averages | 3 | 1 | 3 | 1 | 2 | 1 |
| Voxel size (mm) | 0.3 × 0.3 × 3.0 | 0.3 × 0.3 × 3.0 | 0.3 × 0.3 × 3.0 | 0.3 × 0.3 × 3.0 | 0.3 × 0.3 × 3.0 | 0.3 × 0.3 × 3.0 |
| Field of view (mm) | 200 | 200 | 200 | 200 | 200 | 200 |
| Slice thickness (mm) | 3 | 3 | 3 | 3 | 3 | 3 |
| Parallel imaging factor | 3 | 3 | 3 | 3 | 2 | 3 |
| Acquisition time (min:sec) | 4:37 | 1:38 | 3:07 | 1:10 | 2:37 | 1:02 |
2.4. Image Analysis
Image analysis was performed independently by two radiologists with five and eight years of experience. Reading sessions were carried out in a blinded random order consisting of datasets of T2 imaging (DLR or standard), dynamic contrast-enhanced imaging, as well as DWI, including ADC maps. One hundred and twenty imaging sets were created (60 datasets with T2S and 60 datasets with T2DRL). Both readers were blinded to the description of sequence names, to patient data as well as to the official radiological report.
T2 score and PI-RADS score according to PI-RADS v.2.1 were assessed for all datasets. The exact location and lesion size were reported for the most suspicious lesions given a PI-RADS score ≥ 3.
Additionally, image quality parameters were assessed for the following items using an ordinal Likert scale ranging from 1 to 4 (1 = non-diagnostic; 4 = excellent): noise levels, lesion conspicuity, magnitude of artifacts, diagnostic confidence of the readers and image quality overall.
Furthermore, T2S and T2DLR sequences of each patient were presented to both radiologists mentioned above and were evaluated regarding the naturality of image impression using a Likert scale ranging from 1 to 4 (1 = unnatural; 4 = very natural).
2.5. Statistical Evaluation
Statistical evaluation was performed using commercially available statistical software (SPSS Statistics Version 26; IBM). Continuous variables are shown using mean ± standard deviation (std), ordinal scaled variables are shown using median and interquartile range (IQR). The Wilcoxon signed-rank test was used for paired data of ordinal structure and non-normally-distributed parametric variables. p-values were adjusted using Bonferroni procedure. Intra- and inter-reader variability were assessed using Cohen’s kappa. The significance level alpha was set at 0.05.
3. Results
3.1. Patients’ Characteristics
The mean patient age was 69 ± 9 years (range, 49–85 years). The median prostate specific antigen (PSA) level was 7.2 ng/mL (IQR 5.9–9.7) (Table 1).
The results of the more experienced reader 1 are described in the following. All results are available within the tables of this manuscript.
3.2. Evaluation of Qualitative Imaging Parameters
Analysis of the inter-reader agreement between both readers regarding image quality parameters using Cohen’s kappa resulted in 0.76 for T2S and 0.79 for T2DLR. The impact and extent of image noise was rated to be significantly less in T2DLR compared to T2S with the following ratings: median of 4 (IQR 4–4) for T2DLR versus a median of 3 (IQR 3–3) for T2S in axial imaging, a median of 4 (IQR 4–4) for T2DLR versus a median of 3 (IQR 3–3) for T2S in coronal imaging, and a median of 4 (IQR 3–4) for T2DLR versus a median of 3 (IQR 3–3) for T2S in sagittal imaging (all p < 0.001). The extent of artifacts was also rated to be less in T2DLR versus T2S in axial imaging with a median of 4 (IQR 4–4) compared to 3 (3–4) (p = 0.003). The extent of artifacts in T2DLR was also significantly less in coronal (p = 0.002) and sagittal imaging (p = 0.002). There was no significant difference regarding the natural appearance of both sequences in all planes with a median of 4 for T2S and T2DLR (p > 0.05). Overall image quality was rated higher in axial T2DLR (median of 4 (IQR 4–4)) as compared to T2S (median of 3 (IQR 3–4)) (p < 0.001). Similar results regarding image quality were obtained for T2DLR versus T2S in coronal (p = 0.002) and sagittal imaging (p = 0.002). Diagnostic confidence was evaluated to be higher in T2DLR with a median of 4 (4–4) as compared to T2S with a median of 4 (3–4) (p = 0.03). Table 3 shows the image quality results of both readers.
Table 3.
Image quality in standard T2-weighted imaging (T2S) and deep learning reconstructed T2-weighted imaging (T2DLR).
| Characteristics | Reader 1 | Reader 2 | ||||
|---|---|---|---|---|---|---|
| T2S | T2DLR | p-Value | T2S | T2DLR | p-Value | |
| Image noise axial | 3 (3–3) | 4 (4–4) | <0.001 | 3 (3–3) | 4 (4–4) | <0.001 |
| Image noise coronal | 3 (3–3) | 4 (4–4) | <0.001 | 3 (3–4) | 4 (4–4) | <0.001 |
| Image noise sagittal | 3 (3–3) | 4 (3–4) | <0.001 | 4 (3–4) | 4 (3–4) | 0.005 |
| Artifacts axial | 3 (3–4) | 4 (4–4) | 0.003 | 4 (3–4) | 4 (4–4) | 0.003 |
| Artifacts coronal | 3 (3–4) | 4 (4–4) | 0.002 | 3 (3–4) | 4 (4–4) | 0.014 |
| Artifacts sagittal | 3 (3–4) | 4 (3–4) | 0.002 | 3 (3–4) | 4 (3–4) | 0.011 |
| Natural appearance axial | 4 (4–4) | 4 (4–4) | 1 | 4 (4–4) | 4 (4–4) | 0.630 |
| Natural appearance coronal | 4 (4–4) | 4 (4–4) | 1 | 4 (4–4) | 4 (4–4) | 1 |
| Natural appearance sagittal | 4 (3–4) | 4 (3–4) | 1 | 4 (3–4) | 4 (3–4) | 1 |
| Overall image quality axial | 3 (3–4) | 4 (4–4) | <0.001 | 3 (3–4) | 4 (4–4) | <0.001 |
| Overall image quality coronal | 3 (3–4) | 4 (4–4) | 0.002 | 3 (3–4) | 4 (4–4) | <0.001 |
| Overall image quality sagittal | 3 (3–4) | 4 (3–4) | 0.002 | 3 (3–4) | 4 (3–4) | 0.005 |
| Diagnostic confidence | 4 (3–4) | 4 (4–4) | 0.03 | 4 (3–4) | 4 (4–4) | 0.06 |
3.3. PI-RADS Scoring and Lesion Conspicuity
In no case was a different scoring affecting patient management (<3 versus ≥3) found between the readers and between the sequences. Table 4 shows the detailed results of the T2 and PI-RADS scoring of both readers. In 39 cases, a PI-RADS score ≥3 was found as the most suspicious lesion. In eight of these 39 cases, the most suspicious lesions were found in the transition zone, and in 31 cases in the peripheral zone. Inter-reader agreement regarding T2-scoring was 0.823 for T2S and 0.932 for T2DLR (Table 5). Cohen’s kappa for inter-reader agreement of PI-RADS scoring was 0.905 for T2S and 0.946 for T2DLR (Table 5). Lesion conspicuity was rated superior in T2DLR by both readers compared to T2S with a median of 4 (4–4) versus a median of 3 (3–4) for both readers (p = 0.001 and p < 0.001, respectively). There was no significant difference between lesion size measurements between T2S and T2DLR (Table 6). Figure 2, Figure 3, Figure 4 and Figure 5 show examples of T2S and T2DLR.
Table 4.
T2 and PI-RADS scoring in standard T2-weighted imaging (T2S) and deep learning reconstructed T2-weighted imaging (T2DLR).
| T2 and PI-RADS Scoring | Reader 1 | Reader 2 | ||
|---|---|---|---|---|
| T2S | T2DLR | T2S | T2DLR | |
| T2 score | ||||
| 1 | 0 | 0 | 0 | 0 |
| 2 | 21 | 21 | 21 | 21 |
| 3 | 14 | 13 | 13 | 14 |
| 4 | 15 | 16 | 16 | 15 |
| 5 | 10 | 10 | 10 | 10 |
| PI-RADS score | ||||
| 1 | 0 | 0 | 0 | 0 |
| 2 | 21 | 21 | 21 | 21 |
| 3 | 4 | 5 | 5 | 5 |
| 4 | 25 | 24 | 24 | 24 |
| 5 | 10 | 10 | 10 | 10 |
Table 5.
Intra- and inter-reader agreement.
| Intra-Reader Agreement | |
| Reader 1 T2 score | 0.931 |
| Reader 2 T2 score | 0.905 |
| Reader 1 PI-RADS score | 0.960 |
| Reader 2 PI-RADS score | 0.946 |
| Inter-Reader Agreement | |
| T2 score T2S | 0.823 |
| T2 score T2DLR | 0.932 |
| PI-RADS T2S | 0.905 |
| PI-RADS T2DLR | 0.946 |
Table 6.
Lesion size and conspicuity in standard T2-weighted imaging (T2S) and deep learning reconstructed T2-weighted imaging (T2DLR).
| Characteristics | Reader 1 | Reader 2 | ||||
|---|---|---|---|---|---|---|
| T2S | T2DLR | p-Value | T2S | T2DLR | p-Value | |
| Lesion size (mm) | 13 (10–16) | 13 (11–16) | 1 | 12 (10–16) | 13 (10–17) | 0.840 |
| Lesion conspicuity | 3 (3–4) | 4 (4–4) | <0.001 | 3 (3–4) | 4 (4–4) | 0.001 |
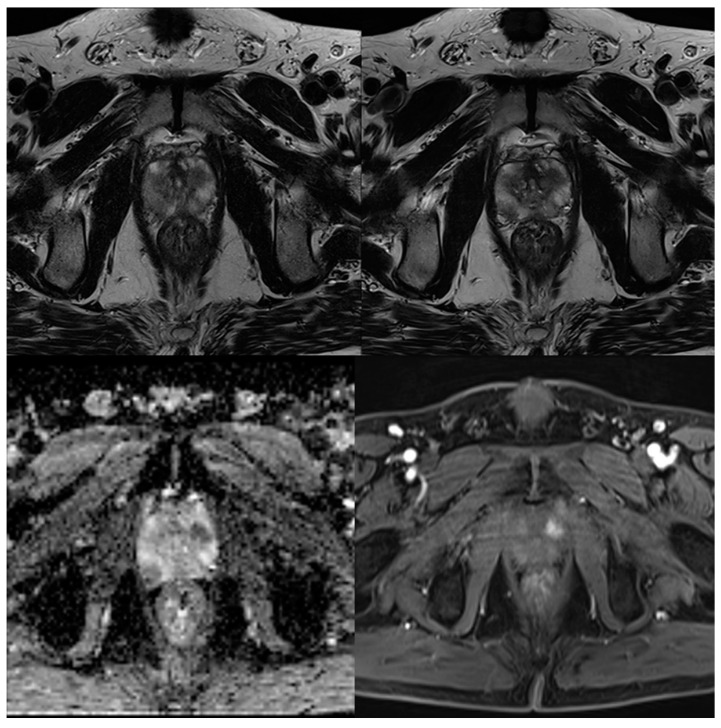
Figure 2.
A 59-year-old male patient with suspicion of prostate cancer. Example of axial standard T2-weighted TSE imaging (T2S) on the top left-hand side and deep learning–reconstructed (T2DLR) imaging on the top right-hand side. The bottom row shows apparent diffusion coefficient (ADC) map on the left-hand side and dynamic contrast-enhanced (DCE) imaging on the right-hand side. PI-RADS 5 lesion in the transition zone was found by both readers in both sequences. Motion artifacts are especially reduced in T2DLR demonstrating the advantages of reduced acquisition time.
Figure 3.
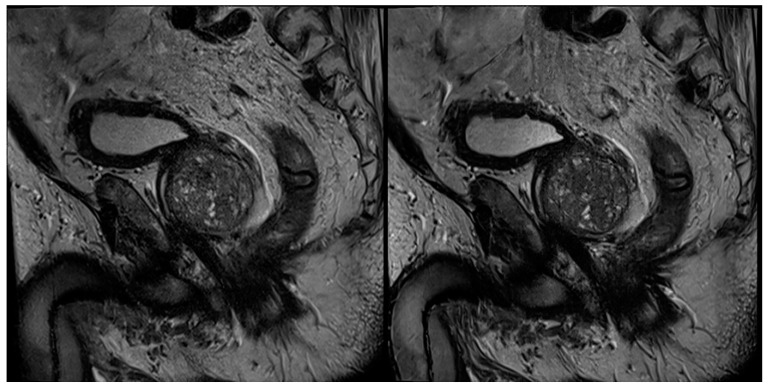
A 72-year-old male patient with suspicion of prostate cancer. Example of axial standard T2-weighted TSE imaging (T2S) on the left-hand side and deep learning–reconstructed (T2DLR) imaging on the right-hand side. Similar to Figure 2, less motion artifacts occurred in T2DLR with sharper depiction of the prostate.
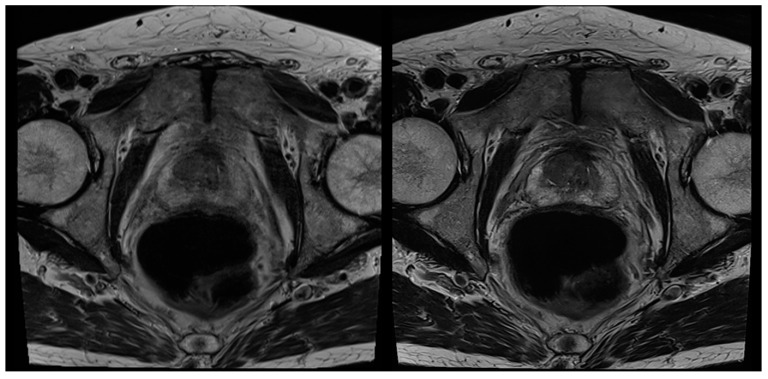
Figure 4.
A 67-year-old male patient with suspicion of prostate cancer. Example of coronal standard T2-weighted TSE imaging (T2S) on the left-hand side and deep learning–reconstructed (T2DLR) imaging on the right-hand side. Advantages of T2DLR are demonstrated, regarding less image noise and improved delineation of anatomic structures.
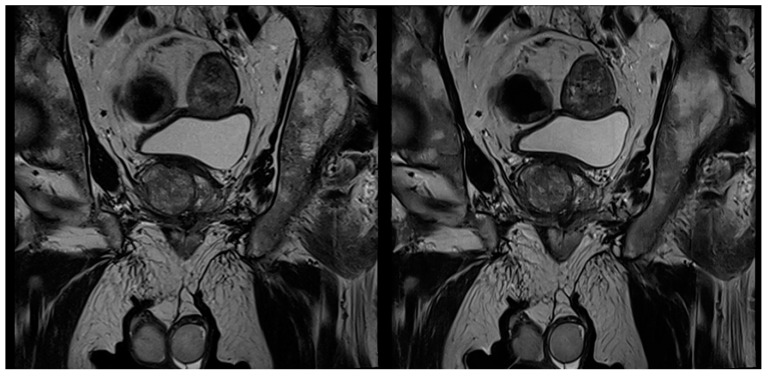
Figure 5.
A 76-year-old male patient with suspicion of prostate cancer. Example of sagittal standard T2-weighted TSE imaging (T2S) on the left-hand side and deep learning–reconstructed (T2DLR) imaging on the right-hand side. This figure demonstrates the advantages of deep learning image reconstruction, regarding image noise and sharpness of organ structures.
4. Discussion
This prospective study investigated the feasibility and impact of DL image reconstruction in accelerated T2w TSE imaging of the prostate in three planes. The results demonstrate that the application of DL image reconstruction in T2-weighted TSE imaging of the prostate is robust and ready to be clinically implemented, as well as being able to achieve a reduction of acquisition time above 60% and superior image quality, compared to a standard T2 acquisition. No significant difference between PI-RADS scoring and T2 scoring could be observed between the sequences.
The potential of DL techniques has been previously demonstrated in several studies [18,19,20,21]. However, most studies have primarily investigated DL methods regarding diagnosis support, e.g., regarding the detection of cancer or analysis of lung tissue [8,18,22]. It was shown in recent studies on prostate MRI that DL can be applied for cancer detection and classification and also for registration with histopathological images [10,11,23,24]. Wang et al. demonstrated that DL could also be applied for the omission of endorectal coils in mpMRI without compromising the image quality regarding noise [25]. However, one of the most important recent developments regarding DL is related to MRI acquisition times. DL-based reconstructions can be used for the acceleration of MRI sequences via the increased undersampling of the acquired data, e.g., via a variational neural network [16]. The clinical applicability of this technique was already demonstrated in knee MRI examinations by Recht et al. [13]. However, in this study, the authors used retrospectively undersampled data. Meanwhile, the potential of DL-based reconstructions for the acceleration and/or improvement of image quality has also been demonstrated in studies with “real” undersampled data, e.g., in prostate MRI and MRI of the pituitary gland [12,17]. However, in a previous prostate DL MRI investigation only a small number of patients were examined in only one imaging plane, limiting the generalizability of the results [17]. The present study demonstrates the first systematic prospective approach with the analysis of three imaging planes in a much larger patient cohort.
Conventional methods for shortening acquisition times involve parallel imaging or CS [26,27,28,29]. Although by using parallel imaging, a drastic reduction of acquisition time is theoretically possible, in clinical routine only factors of 2–4 are routinely applied. The reasons for this are due to SNR loss proportional to the square root of the scan acceleration and typical artifacts as noise bands [30]. CS is an established method for MRI acquisition acceleration that is based on redundancy of imaging data; however, a common disadvantage of this technique is the unnatural appearance of these images, as well as the long processing time [26]. In the present study, the DL-reconstructed images were evaluated as superior, regarding both image quality and noise. The very low degree of noise in some T2DRL images may cause an unrealistic image perception. Despite this issue, both readers evaluated the appearance of T2DLR images to be very natural, without significant difference to the T2S.
Despite the drastic shortening of acquisition time and improved image quality, the question arises as to which clinical benefits might be drawn from these results. MRI of the prostate has undergone an extraordinary evolution during the last decade, nowadays leading to a structured and standardized analysis via the PI-RADS classification system [2]. mpMRI is an important tool for biopsy planning and also for prostate cancer detection [4,31]. Furthermore, in the future, mpMRI might play an important role for patients under active surveillance [32]. However, an increase in demand for an examination does not automatically lead to an increase in supply, due to the restricted availability of MRI scanners and comparably long examination times using standard techniques. This is especially problematic due to the long examination times necessary for mpMRI of the prostate, to align with current guidelines and to achieve sufficient image and report quality. Therefore, the demonstrated approach of a drastic reduction of T2 TSE acquisition time above 60% provides enormous potential for the increase of scanner availability. T2 TSE imaging in three planes in less than 4 min might even allow ultra-short screening protocols for prostate cancer to be created. This is even more important, due to the updated PI-RADS version 2.1 with the possibility of acquiring only two planes of T2 TSE imaging and further research regarding biparametric MRI with omission of dynamic contrast-enhanced imaging [33].
Limitations
This study investigated only DL reconstructions of T2w TSE imaging. No other sequences were reconstructed using DL technology. However, due to the time-consuming acquisition process of high-resolution morphological T2 images, this sequence was preferred for initial DL implementation. Further studies will be necessary to investigate the impact on other sequences. Furthermore, it is possible to change the image perception via a change of DL networks or artificial increase of noise (dithering). However, these approaches were beyond the scope of our study and, therefore, not further analyzed. Additionally, as many patients were examined in an outpatient setting, possible biopsy results were not available and therefore diagnostic accuracy regarding histopathologic correlation was not part of this study.
5. Conclusions
This prospective study demonstrates the potential of a new DL reconstruction algorithm in accelerated T2w TSE imaging of the prostate in three planes with a reduction of acquisition times above 60% combined with an improvement of image quality and a reduction of artifacts, compared to standard T2w imaging. Concluding, DL reconstructions may be of great importance for establishing ultrafast screening protocols in prostate MRI.
Author Contributions
Conceptualization, S.G. and A.E.O.; data curation, S.G.; formal analysis, S.A. and A.E.O.; investigation, S.G., S.A. and A.E.O.; methodology, S.G., S.A. and A.E.O.; project administration, K.N. and A.E.O.; resources, K.N. and A.E.O.; software, M.D.N. and M.M.; supervision, K.N. and A.E.O.; validation, K.N. and A.E.O.; visualization, S.G., J.H. and H.A.; writing—original draft, S.G.; writing—review and editing, S.A., M.D.N., M.M., J.H., H.A., K.N. and A.E.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the University of Tuebingen (055/2017BO2 and 23 September 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
Dominik Nickel and Mahmoud Mostapha are employees of Siemens Healthineers.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Weinreb J.C., Barentsz J.O., Choyke P.L., Cornud F., Haider M.A., Macura K.J., Margolis D., Schnall M.D., Shtern F., Verma S., et al. PI-RADS Prostate Imaging—Reporting and Data System: 2015, Version 2. Eur. Urol. 2016;69:16–40. doi: 10.1016/j.eururo.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giganti F., Rosenkrantz A.B., Villeirs G., Panebianco V., Stabile A., Emberton M., Moore C.M. The Evolution of MRI of the Prostate: The Past, the Present, and the Future. AJR Am. J. Roentgenol. 2019;213:384–396. doi: 10.2214/AJR.18.20796. [DOI] [PubMed] [Google Scholar]
- 3.Cornud F., Delongchamps N.B., Mozer P., Beuvon F., Schull A., Muradyan N., Peyromaure M. Value of multiparametric MRI in the work-up of prostate cancer. Curr. Urol. Rep. 2012;13:82–92. doi: 10.1007/s11934-011-0231-z. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed H.U., Bosaily A.E.S., Brown L.C., Gabe R., Kaplan R., Parmar M.K., PROMIS Study Group Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. Lancet. 2017;389:815–822. doi: 10.1016/S0140-6736(16)32401-1. [DOI] [PubMed] [Google Scholar]
- 5.Kasivisvanathan V., Rannikko A.S., Borghi M., Panebianco V., Mynderse L.A., Vaarala M.H., Moore C.M. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N. Engl. J. Med. 2018;378:1767–1777. doi: 10.1056/NEJMoa1801993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickinson L., Ahmed H.U., Allen C., Barentsz J.O., Carey B., Futterer J.J., Emberton M. Clinical applications of multiparametric MRI within the prostate cancer diagnostic pathway. Urol. Oncol. 2013;31:281–284. doi: 10.1016/j.urolonc.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McBee M.P., Awan O.A., Colucci A.T., Ghobadi C.W., Kadom N., Kansagra A.P., Auffermann W.F. Deep Learning in Radiology. Acad. Radiol. 2018;25:1472–1480. doi: 10.1016/j.acra.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 8.Walsh S.L.F., Calandriello L., Silva M., Sverzellati N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: A case-cohort study. Lancet Respir. Med. 2018;6:837–845. doi: 10.1016/S2213-2600(18)30286-8. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y.D., Wang J., Wu C.J., Bao M.L., Li H., Wang X.N., Shi H.B. An imaging-based approach predicts clinical outcomes in prostate cancer through a novel support vector machine classification. Oncotarget. 2016;7:78140–78151. doi: 10.18632/oncotarget.11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong X., Cao R., Shakeri S., Scalzo F., Lee Y., Enzmann D.R., Sung K. Deep transfer learning-based prostate cancer classification using 3 Tesla multi-parametric MRI. Abdom. Radiol. 2019;44:2030–2039. doi: 10.1007/s00261-018-1824-5. [DOI] [PubMed] [Google Scholar]
- 11.Padhani A.R., Turkbey B. Detecting Prostate Cancer with Deep Learning for MRI: A Small Step Forward. Radiology. 2019;293:618–619. doi: 10.1148/radiol.2019192012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim M., Kim H.S., Kim H.J., Park J.E., Park S.Y., Kim Y.H., Lebel M.R. Thin-Slice Pituitary MRI with Deep Learning-based Reconstruction: Diagnostic Performance in a Postoperative Setting. Radiology. 2021;298:114–122. doi: 10.1148/radiol.2020200723. [DOI] [PubMed] [Google Scholar]
- 13.Recht M.P., Zbontar J., Sodickson D.K., Knoll F., Yakubova N., Sriram A., Zitnick C.L. Using Deep Learning to Accelerate Knee MRI at 3T: Results of an Interchangeability Study. AJR Am. J. Roentgenol. 2020 doi: 10.2214/AJR.20.23313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann J., Gassenmaier S., Nickel D., Arberet S., Afat S., Lingg A., Othman A.E. Diagnostic Confidence and Feasibility of a Deep Learning Accelerated HASTE Sequence of the Abdomen in a Single Breath-Hold. Investig. Radiol. 2020 doi: 10.1097/RLI.0000000000000743. [DOI] [PubMed] [Google Scholar]
- 15.Herrmann J., Nickel D., Mugler J.P., III, Arberet S., Gassenmaier S., Afat S., Othman A.E. Development and Evaluation of Deep Learning-Accelerated Single-Breath-Hold Abdominal HASTE at 3 T Using Variable Refocusing Flip Angles. Investig. Radiol. 2021 doi: 10.1097/RLI.0000000000000785. [DOI] [PubMed] [Google Scholar]
- 16.Hammernik K., Klatzer T., Kobler E., Recht M.P., Sodickson D.K., Pock T., Knoll F. Learning a variational network for reconstruction of accelerated MRI data. Magn. Reson. Med. 2018;79:3055–3071. doi: 10.1002/mrm.26977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gassenmaier S., Afat S., Nickel D., Mostapha M., Herrmann J., Othman A.E. Deep learning-accelerated T2-weighted imaging of the prostate: Reduction of acquisition time and improvement of image quality. Eur. J. Radiol. 2021;137:109600. doi: 10.1016/j.ejrad.2021.109600. [DOI] [PubMed] [Google Scholar]
- 18.Becker A.S., Marcon M., Ghafoor S., Wurnig M.C., Frauenfelder T., Boss A. Deep Learning in Mammography: Diagnostic Accuracy of a Multipurpose Image Analysis Software in the Detection of Breast Cancer. Investig. Radiol. 2017;52:434–440. doi: 10.1097/RLI.0000000000000358. [DOI] [PubMed] [Google Scholar]
- 19.Fayad L.M., Parekh V.S., de Castro Luna R., Ko C.C., Tank D., Fritz J., Jacobs M.A. A Deep Learning System for Synthetic Knee Magnetic Resonance Imaging: Is Artificial Intelligence-Based Fat-Suppressed Imaging Feasible? Investig. Radiol. 2021;56:357–368. doi: 10.1097/RLI.0000000000000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kart T., Fischer M., Küstner T., Hepp T., Bamberg F., Winzeck S., Gatidis S. Deep Learning-Based Automated Abdominal Organ Segmentation in the UK Biobank and German National Cohort Magnetic Resonance Imaging Studies. Investig. Radiol. 2021;56:401–408. doi: 10.1097/RLI.0000000000000755. [DOI] [PubMed] [Google Scholar]
- 21.Almansour H., Gassenmaier S., Nickel D., Kannengiesser S., Afat S., Weiss J., Othman A.E. Deep Learning-Based Superresolution Reconstruction for Upper Abdominal Magnetic Resonance Imaging: An Analysis of Image Quality, Diagnostic Confidence, and Lesion Conspicuity. Investig. Radiol. 2021 doi: 10.1097/RLI.0000000000000769. [DOI] [PubMed] [Google Scholar]
- 22.Chartrand G., Cheng P.M., Vorontsov E., Drozdzal M., Turcotte S., Pal C.J., Tang A. Deep Learning: A Primer for Radiologists. Radiographics. 2017;37:2113–2131. doi: 10.1148/rg.2017170077. [DOI] [PubMed] [Google Scholar]
- 23.Schelb P., Kohl S., Radtke J.P., Wiesenfarth M., Kickingereder P., Bickelhaupt S., Bonekamp D. Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology. 2019;293:607–617. doi: 10.1148/radiol.2019190938. [DOI] [PubMed] [Google Scholar]
- 24.Shao W., Banh L., Kunder C.A., Fan R.E., Soerensen S.J., Wang J.B., Rusu M. ProsRegNet: A deep learning framework for registration of MRI and histopathology images of the prostate. Med. Image Anal. 2021;68:101919. doi: 10.1016/j.media.2020.101919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Ma J., Bhosale P., Rovira J.J.I., Qayyum A., Sun J., Szklaruk J. Novel deep learning-based noise reduction technique for prostate magnetic resonance imaging. Abdom. Radiol. 2021 doi: 10.1007/s00261-021-02964-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng L., Benkert T., Block K.T., Sodickson D.K., Otazo R., Chandarana H. Compressed sensing for body MRI. J. Magn. Reson. Imaging. 2017;45:966–987. doi: 10.1002/jmri.25547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griswold M.A., Jakob P.M., Heidemann R.M., Nittka M., Jellus V., Wang J., Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn. Reson. Med. 2002;47:1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 28.Lustig M., Donoho D., Pauly J.M. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn. Reson. Med. 2007;58:1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 29.Yanasak N.E., Kelly M.J. MR imaging artifacts and parallel imaging techniques with calibration scanning: A new twist on old problems. Radiographics. 2014;34:532–548. doi: 10.1148/rg.342135051. [DOI] [PubMed] [Google Scholar]
- 30.Yang R.K., Roth C.G., Ward R.J., deJesus J.O., Mitchell D.G. Optimizing abdominal MR imaging: Approaches to common problems. Radiographics. 2010;30:185–199. doi: 10.1148/rg.301095076. [DOI] [PubMed] [Google Scholar]
- 31.De Rooij M., Hamoen E.H., Futterer J.J., Barentsz J.O., Rovers M.M. Accuracy of multiparametric MRI for prostate cancer detection: A meta-analysis. AJR Am. J. Roentgenol. 2014;202:343–351. doi: 10.2214/AJR.13.11046. [DOI] [PubMed] [Google Scholar]
- 32.Giganti F., Stabile A., Stavrinides V., Osinibi E., Retter A., Orczyk C., Moore C.M. Natural history of prostate cancer on active surveillance: Stratification by MRI using the PRECISE recommendations in a UK cohort. Eur. Radiol. 2021;31:1644–1655. doi: 10.1007/s00330-020-07256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turkbey B., Rosenkrantz A.B., Haider M.A., Padhani A.R., Villeirs G., Macura K.J., Weinreb J.C. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 2019;76:340–351. doi: 10.1016/j.eururo.2019.02.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.