Abstract
Investigation of changes in the skin microbiome following treatment of atopic dermatitis (AD) with dupilumab may provide valuable insights into the skin microbiome as a therapeutic target. The aim of this study is to assess changes in the AD skin microbiome following treatment of AD with dupilumab (n = 27). E-swabs were collected from nose, lesional, and nonlesional skin before and after 16 weeks of dupilumab therapy, and the microbiome was analyzed by 16S rRNA and tuf gene sequencing. Data for 17 patients with milder disease receiving treatment with non-targeted therapies are also presented. The results show that both groups experienced clinical improvement (p < 0.001) following dupilumab therapy and that Shannon diversity increased and bacterial community structure changed. The relative abundance of the genus Staphylococcus (S.) and S. aureus decreased, while that of S. epidermidis and S. hominis increased. No significant changes were observed for patients receiving non-targeted treatments. The increases in S. epidermidis and S. hominis and the decrease in S. aureus correlated with clinical improvement. Furthermore, changes in S. hominis and S. epidermidis correlated inversely with S. aureus. In conclusion, treatment with dupilumab significantly changed the skin microbiome and decreased S. aureus. Our results suggest a favorable role of commensal staphylococci in AD.
Keywords: atopic dermatitis, skin microbiome, staphylococcus, dupilumab
1. Introduction
The pathogenesis of atopic dermatitis (AD) is characterized by the interplay between activation of the Th2/Th22-skewed immune response and epidermal barrier impairment [1,2]. This is paralleled by changes in the skin microbiome, with decreased bacterial diversity and increased colonization with Staphylococcus aureus (S. aureus), which correlates with disease severity [3,4,5,6]. Conditions in AD skin promote S. aureus colonization [7,8], and S. aureus expresses a broad spectrum of virulence factors that aggravate the disease [9,10,11]. Commensal staphylococcal species, such as S. epidermidis and S. hominis, work in concert with host immunity to limit pathogen colonization [12,13,14,15], and were recently shown to be reduced in AD [16]. Accordingly, deficiency of commensal staphylococci may be related to the increased S. aureus colonization and disease severity in AD.
Dupilumab, a fully human monoclonal antibody targeting IL-4 and IL-13 by blocking IL-4Rα signaling [17], has been shown to improve the clinical disease activity, immune abnormalities, and epidermal barrier function, thereby highlighting IL-4Rα signaling as a key driver of the AD phenotype [18,19,20]. One previous study of changes in the skin microbiome and S. aureus colonization following dupilumab therapy showed that bacterial diversity increased and the absolute S. aureus abundance decreased [21]. However, the study did not assess treatment-related changes in other staphylococcal species and nasal microbiome.
Further understanding of the complex interactions within the bacterial communities and their cross-talk with host immunity is necessary to explore the potential of manipulating the skin microbiome in the management of AD. Accordingly, investigation of changes in the skin microbiome and staphylococcal species that are associated with successful responses to targeted and non-targeted treatments may lead to identification of genera and species that need to be further evaluated with regard to their link to AD. Therefore, the aim of the study was to investigate changes in the skin and nasal microbiome associated with successful response to dupilumab. By applying tuf gene sequencing, changes in the relative abundance of all staphylococcal species were assessed, revealing interesting new data on the potential interplay between different staphylococcal species following treatment.
2. Materials and Methods
2.1. Study Design
Patients were recruited from the Department of Dermatology, Bispebjerg Hospital, Copenhagen, from March 2018 to September 2019. Inclusion criteria comprised a diagnosis of AD according to UK criteria, age ≥ 18 years, and no pregnant or lactating women. Patients were scheduled to be evaluated at treatment initiation, after 16 weeks of systemic treatments (including dupilumab), and after four weeks of topical treatment. Data on the three most common loss-of-function mutations in the filaggrin gene (FLG) were collected [22]. Patients were instructed not to apply emollients within 24 h of each visit.
2.2. Sample Collection and Disease Severity Assessment
Swabs (e-Swab, Copan, Brescia, Italy) were collected from anterior nares and lesional and nonlesional skin from representative areas on the upper or lower extremities. Disease severity was assessed with the Eczema Area and Severity Index (EASI). Blood was collected for total serum IgE quantification.
2.3. DNA Extraction and Microbiome Characterization
DNA was extracted from swabs using an enzymatic prelysis step (30 min incubation at 37 °C with an enzyme solution containing 4 U lysostaphine (SAE0091), 25 U mutanolysin (sae0092), and 3 mg lysozyme (L4919) (Sigma-Aldrich, St. Louis, MO, USA); then 30 min incubation at 56 °C with 20 µL protein kinase K (RPROTKSOL-RO, Sigma-Aldrich, St. Louis, MO, USA), followed by DNA extraction on a MagNa-Pure 96 robot using a DNA and Viral NA Small Volume Kit (Roche, Mannheim, Germany).
The V3-V4 region of the 16S rRNA gene and that of the tuf gene were amplified in two separate PCRs (95 °C for 3 min; 25 cycles of 98 °C for 20 s, 60 °C for 15 s, 72 °C for 45 s; 72 °C for 5 min), using primers (16SrRNA: 341F: 5′- CCTACGGGNGGCWGCAG -3′; 805R: 5′- GACTACHVGGGTATCTAATC-3′; tuf: F: 5′- CAGAAGAAAAAGAACGTGG-3′; R: 5′- GTCCTCAACWGGCATCA-3′) with preceding heterogeneity spacers [23,24]. Amplicon libraries were constructed using nextera indexing primers (Illumina Inc., San Diego, CA, USA) (PCR program used: 95 °C for 3 min; 20 cycles of 98 °C for 20 s, 55 °C for 15 s, and 72 °C for 45 s; 72 °C for 5 min) and sequenced on a MiSeq instrument using a 600 cycle V3 kit (Illumina Inc., San Diego, CA, USA).
2.4. Sequence Pre-Processing
Demultiplexing of raw reads was performed by using the bcl2fastq Conversion Software (Illumina Inc., San Diego, CA, USA). Subsequently, cutadapt (version 2.3) [25] was used on 16S rRNA and tuf gene reads for heterogeneity spacer and primer trimming at an 8% error rate (corresponding to one mismatch per primer) in paired-end mode. Trimmed reads were subjected to quality filtering and amplicon sequence variant (ASV) inference with dada2 (version 1.12.1) [26]. The dada2 pipeline was utilized run-wise with default settings, except for truncation length. 16S rRNA gene reads were truncated at 270 (forward reads) and 210 bp (reverse reads), while tuf gene reads were truncated at 270 (forward reads) and 241 bp (reverse reads). We performed consensus chimera removal. In cases when a sample had a read count <5000 after quality filtering, it was re-sequenced. Taxonomic assignment of 16S rRNA gene sequence-derived ASVs was performed with dada2′s ‘assignTaxonomy’ and ‘addSpecies’ functions, using the Silva reference database and species-level training set (version 132) formatted for dada2, respectively. A staphylococcal-specific taxonomic database was used to classify tuf gene sequence-derived ASVs with the ‘assignTaxonomy’ function [24]. Using the R package phyloseq v, ASV count tables, taxonomic tables, and patient data were integrated [27]. Computational identification and removal of putative contaminants from the 16S rRNA gene sequencing data were performed for skin and nasal samples separately by using the decontam package and by additional manual filtering [28].
Twenty-five ASVs identified with the frequency method (threshold 0.01) were excluded from the skin data set, and 2041 ASVs were manually filtered out in addition. From the nasal data set, 29 ASVs were excluded based on the frequency method (threshold 0.05) and an additional 292 ASVs based on manual filtering.
Manual filtering concerned the following ASVs in both nasal and skin samples: ASVs classified no further than class-level, ASVs not classified as Bacteria (kingdom), and ASVs belonging to the phyla Cyanobacteria, Plantomycetes, Chloroflexi, and Deinococcus-Thermus, the class Rhodothermia, or the orders Rhizobiales, Rhodobacterales, Oceanospirillales, Azospirillales, and Rhodospirillales.
Read counts of re-sequenced samples were merged. One skin sample and one nasal sample, each with <4900 read counts after quality filtering, contaminant removal and merging, were excluded from downstream analyses. The final skin data set comprised 9364 ASVs, and the final nasal data set comprised 2240 ASVs.
2.5. Statistical Analysis
Statistical analyses and visualizations were performed using the statistical software R (version 4.0.1). The R package phyloseq was used for microbiota analysis and the ggplot2 package was used for visualization [27]. Alpha diversity was measured using Shannon’s diversity index through the phyloseq package on raw data. Shannon’s diversity index takes into account both the richness (number of different ASVs) and evenness (how evenly the ASVs are distributed) of the bacterial community. The Kruskal–Wallis test for unpaired samples and Wilcoxon’s signed rank test for paired samples were used to test for differences in Shannon diversity between visits in each group. To compare overall differences between groups, principal coordinate analysis (PCoA) plots were used for visualization, together with permutational multivariable analysis of variance, using distance matrices (PERMANOVA) tests based on Bray–Curtis distances for statistical analysis (function adonis in package vegan) [29]. This analysis was performed on Hellinger-transformed data. Homogeneity in within-group variations was tested for with the betadiper function (package vegan) as a prerequisite to applying PERMANOVA.
For barplots, all ASVs were merged at genus level, and counts were transformed to relative abundance. To assess staphylococcal species distribution, ASVs derived from tuf gene sequencing were transformed to relative abundance, and the 10 species with the highest percentage across all samples were plotted. Partitioning around medoid (PAM) clustering based on the Jensen–Shannon distance was used for grouping samples into an optimal number of clusters, pre-assessed by consensus of the gap statistic, silhouette width, and the elbow method (R packages cluster and factoextra) [30,31] (Maechler et al., 2019, Kassambara and Mundt). Temporal cluster dynamics were visualized in Sankey plots by utilizing the ggalluvial package [32] (Brunson, 2020). For each skin and nose site, differential abundance analyses were performed using DEseq2 [33] for the identification of differentially abundant genera, and for staphylococci differentially abundant species in relation to treatment groups, FLG mutation status, pre-versus-post treatment, and to identify changes in genera and species related to treatment response (i.e., change in EASI). Spearman’s correlation was calculated between the EASI score and Shannon diversity at baseline, as well as between change in the EASI score and Shannon diversity from baseline to post-treatment. Moreover, Spearman’s correlation was calculated between baseline relative abundances of staphylococcal species, their change in abundance from baseline to post-treatment, as well as their correlation with the EASI score. Benjamini–Hochberg multiple testing correction was used following all statistical tests when applicable. To assess changes in clinical parameters following treatment (i.e., EASI score, serum IgE, itch and sleep score, TEWL, and skin pH), Wilcoxon’s matched-pair signed rank sum test was used.
3. Results
3.1. Patient Characteristics and Clinical Response
Twenty-seven patients (mean age (range): 41.6 years (18–65); female: 14.8%; median EASI score (range): 16.3 (6.4–34.9)) were included in the study after initiating treatment with 300 mg dupilumab every second week. Patients additionally treated with topical treatment were instructed not to use topical treatments 7 days prior to each evaluation. The mean percentage reduction in the EASI score following dupilumab therapy was 70.0% (p < 0.001) (Table 1).
Table 1.
Demographic and clinical parameters at baseline and following treatment in the two treatment groups.
| Baseline | Follow-Up | p-Value 1 | |
|---|---|---|---|
| Dupilumab treatment group 2 | |||
| Age (y) mean (range) | 41.6 (18–65) | ||
| Gender, women % (n/ ntotal) | 14.8 (4/27) | ||
| FLG-mutation carrier, % (n/ntotal) | 23.8 (5/21) | ||
| EASI score Median (range) |
(n = 27) 16.3 (6.4–34.9) |
(n = 27) 3.4 (0.2–31.1) |
<0.001 |
| Percentage change in EASI score from baseline, % (sd) | 70.0 (26.7) | ||
| EASI-75, % (n/ntotal) | 55.5 (15/27) | ||
| Serum total IgE Median (min-max) |
(n = 27) 1920 (50–23,400) |
(n = 27) 1120 (23–12,300) |
<0.001 |
| Non-targeted treatment group | |||
| Age (y) mean (range) | 35.0 (20–55) | ||
| Gender, women % (n/ ntotal) | 47.1 (8/17) | ||
| FLG-mutation carrier, % (n/ntotal) | 33.3 (5/15) | ||
| EASI score Median (min-max) |
(n = 17) 7.1 (3.5–41.1) |
(n = 17) 4.7 (0.8–29.0) |
<0.001 |
| Percentage change in EASI score from baseline, % (sd) | 24.4 (53.4) | ||
| EASI-75, % (n/ntotal) | 23.5 (4/17) | ||
| Serum total IgE Median (min-max) |
(n = 15) 456 (11–20,600) |
(n = 11) 355 (23–12,300) |
0.27 |
1 Missing data were excluded for each analysis. 2 Abbreviations: EASI; Eczema Area and Severity index, sd; standard deviation IgE; immunoglobulin E, EASI-75; proportion of patients experiencing at least a 75-percentage reduction from baseline EASI score NRS; numeric range scale (0–10 with 10 being most severe). Statistically significant p-values are shown in bold.
An additional 17 patients (mean age: 35.0 years (20–55); female: 47.1%; median baseline EASI score (range): 7.1 (0.8–29.1)) with milder disease receiving non-targeted treatments (TCS (n = 13), TCI (n = 1), methotrexate (n = 2) and azathioprine (n = 1)) were included in the study. The mean percentage reduction in EASI score following non-targeted therapy was 24.4% (p < 0.001) (Table 1).
3.2. Dupilumab Changed Bacterial Diversity and Staphylococci Abundance
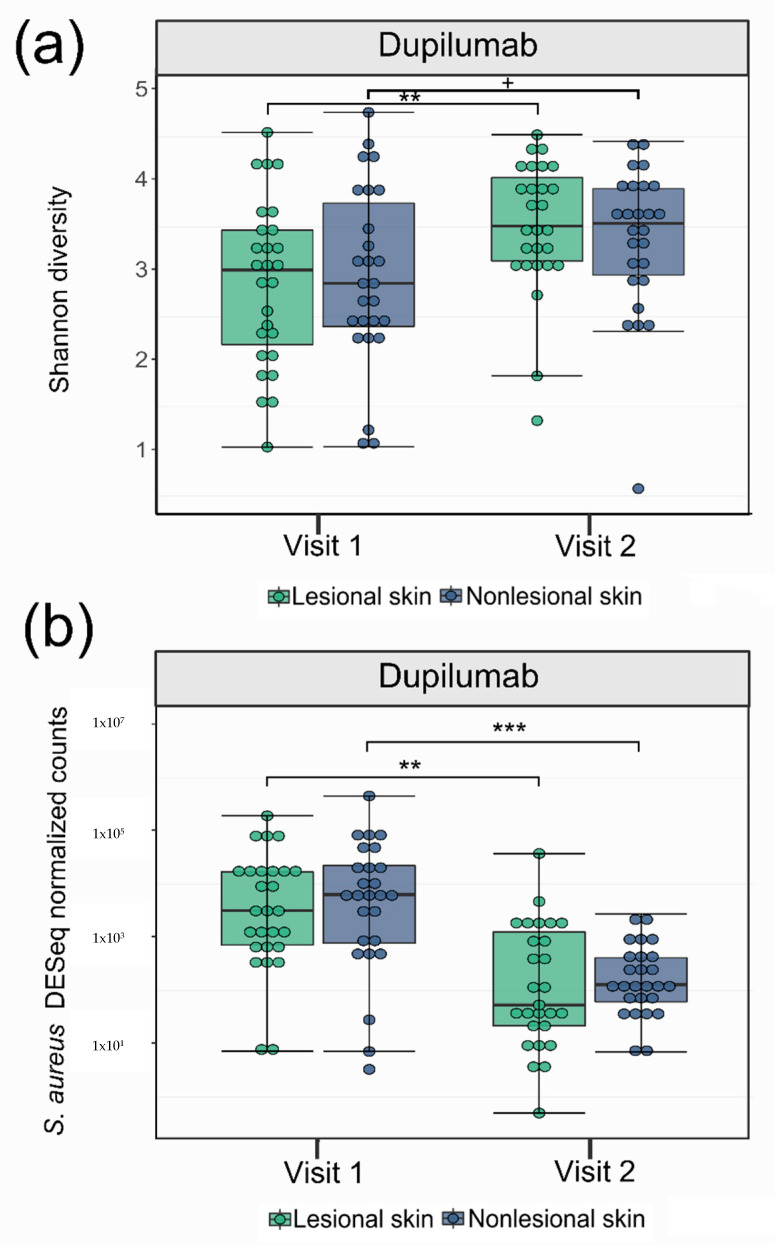
Following dupilumab therapy, Shannon diversity significantly increased on lesional skin (p = 0.005), and the bacterial community structure changed on both lesional and nonlesional skin (p < 0.001) (Figure 1a and Figure S1a,b, Table 2). The proportion of Staphylococcus on lesional skin (p = 0.02) and of S. aureus on lesional and nonlesional skin (p = 0.001, p < 0.001, respectively) was reduced (Figure 1b and Figure S2, Table 2), while the proportions of S. epidermidis, S. hominis, and S. saprophyticus increased on nonlesional skin (p < 0.001) (Figure S3a, Table 2).
Figure 1.
Shannon diversity and S. aureus relative abundance before and after treatment of atopic dermatitis patients with dupilumab. Boxplots showing (a) Shannon diversity and (b) Deseq2 normalized abundance of S. aureus in lesional and nonlesional skin at visit 1 and visit 2 for patients treated with dupilumab + = p < 0.1, ** = p < 0.01, *** = p < 0.001.
Table 2.
Treatment related changes in microbiome and staphylococcal species in the two treatment groups 1.
| Dupilumab (n = 27) | Non-Targeted Treatment Group (n = 17) | |||||
|---|---|---|---|---|---|---|
| Significant Changes Following Treatment | ||||||
| NLS | LS | Nose | NLS | LS | Nose | |
| Shannon Diversity | - | +↑ | +↑* | - | - | - |
| Bacterial community structure | + | + | + | - | - | - |
| Bacterial genera | - | ↓Staphylococcus |
↑Rothia
↑Corynebacterium ↑Haemophilus ↑Veillonella |
- | - | - |
| staphylococcal species | ↓S. aureus ↑S. epidermidis ↑S. hominis ↑ S. saprophyticus |
↓S. aureus |
- | - | - | - |
1 Abbreviations: LS: lesional skin; NLS; nonlesional skin; +: significant change; -: no significant change; * not significant after adjusting for multiple testing; ↑: increase; ↓: decrease.
Following non-targeted therapy, no significant changes in Shannon diversity, bacterial community structure, and relative abundance of genera and staphylococcal species were observed (Figures S5–S7), although the community structure of nonlesional skin showed a trend for significant change (p = 0.06) (Table 2).
Despite comparable post-treatment EASI scores, the bacterial community structures of lesional and nonlesional skin were significantly different between the two treatment groups, and the proportion of S. aureus in both lesional and nonlesional skin was significantly lower in patients treated with dupilumab (p < 0.001) (Table 2).
3.3. Shift in Composition of Bacterial Genera and Staphylococcal Species Following Treatment
Clustering on all samples revealed three clusters (Figure S4a,b). Cluster 1 was dominated by Staphylococcus, while cluster 2 and 3 were more diverse. Regarding staphylococcal species, cluster 1 was dominated by S. aureus and S. capitis, cluster 2 by S. hominis and S. epidermidis, and cluster 3 by a higher abundance of S. saprophyticus and S. epidermidis. The majority of patients in cluster 1 had moderate-to-severe AD (mean EASI: 17.8), while most patients in cluster 2 and 3 had mild eczema (mean EASI: 8.0 and 9.8, respectively).
Prior to dupilumab therapy, 47% of lesional and nonlesional samples were of the S. aureus dominated cluster 1, and the majority of these patients shifted to cluster 2 or cluster 3 following treatment (Figure S4c). A greater proportion of the patients experiencing at least 75% improvement in their EASI scores (EASI-75) shifted from cluster 1 to cluster 2 or 3. Shifts in clusters following non-targeted treatment are shown in Figure S8.
3.4. Correlations between the Skin Microbiome, Disease Severity and Treatment-Related Clinical Improvement
Overall, when grouping samples from the two treatment groups, bacterial Shannon diversity on lesional and nonlesional skin correlated inversely with the EASI score at baseline (p < 0.001), and an increase in Shannon diversity following treatment correlated with EASI improvement (p = 0.005 and p = 0.02). The proportion of S. aureus on nonlesional skin correlated positively with the EASI score at baseline (p = 0.01).
Furthermore, EASI improvement correlated significantly with a decrease in the proportion of nonlesional S. aureus (p = 0.001) and with an increased proportion of S. hominis on lesional and nonlesional skin (p = 0.003) and S. epidermidis on nonlesional skin (p = 0.003). Changes in the proportions of S. aureus were significantly inversely correlated with changes in S. epidermidis and S. hominis on both lesional and nonlesional skin (p = 0.02, p = 0.003 and p < 0.001, p = 0.02, respectively) (Table 3).
Table 3.
Correlations between the skin microbiome and severity, clinical improvement, and changes in S. aureus.
| Correlations with Change in EASI Score | ||
|---|---|---|
| Lesional skin | r | p-value |
| Change in Shannon diversty | −0.44 | 0.005 |
| Change in S. aureus relative abundance | 0.29 | 0.06 |
| Change in S. hominis relative abundance | −0.44 | 0.003 |
| Nonlesional skin | r | p-value |
| Change in Shannon diversity | −0.34 | 0.02 |
| Change in S. aureus relative abundance | 0.51 | 0.001 |
| Change in S. hominis relative abundance | −0.44 | 0.003 |
| Change in S. epidermidis relative abundance | −0.49 | 0.002 |
| Correlations with change in S. aureus relative abundance | ||
| Lesional skin | r | p-value |
| Change in S. hominis relative abundance | −0.43 | 0.003 |
| Change in S. epidermidis relative abundance | −0.32 | 0.02 |
| Nonlesional skin | r | p-value |
| Change in S. hominis relative abundance | −0.32 | 0.02 |
| Change in S. epidermidis relative abundance | −0.54 | <0.001 |
Samples from the dupilumab group (n = 27) and non-targeted group (n = 17) was merged into one group for correlation analyses. Abundance of S. aureus corresponds to the relative abundance (1 = 100%) in the staphylococcal species community. Correlations, r, were tested using spearman correlation test. Statistically significant p-values are shown in bold.
3.5. Relation between the Skin Microbiome and FLG Mutation and Serum IgE
Bacterial Shannon diversity on lesional and nonlesional skin correlated inversely with the total serum IgE at baseline (r = −0.29, p = 0.01). There was no significant difference in the skin microbiome between patients with and without FLG null mutation.
3.6. Characterization of the Nasal Microbiome in Relation to Treatment and Correlations with Skin Microbiome
Following dupilumab therapy, the nasal community structure changed (Table 2). Shannon diversity and community structure did not change in patients treated with non-targeted treatments.
Overall, staphylococcal species distribution did not change following treatment (Figure S3), but patients achieving EASI-75 experienced a significant increase in S. hominis compared to patients that did not (p = 0.02). However, subgroup analyses revealed that this was primarily driven by a few patients. Furthermore, relative S. hominis abundance in the nose correlated with the abundance of S. hominis on both lesional and nonlesional skin at visit 2 and on nonlesional skin at visit 1 (p = 0.005 and 0.02, respectively), although subgroup analysis revealed that this was likewise primarily driven by a few patients.
4. Discussion
In this study, we investigated changes in the skin and anterior nasal microbiome, including staphylococcal species in AD patients treated with dupilumab. Our study showed that dupilumab therapy was related to significant changes in the skin microbiome, with increased Shannon diversity and changes in bacterial community structure. Additionally, a decreased proportion of the genus Staphylococcus and S. aureus and an increase in S. hominis and S. epidermidis was observed.
Further insight into the temporal dynamics of the skin microbiome in relation to different treatments and clinical improvement is essential for exploring the potential of targeting the skin microbiome in the therapeutic management of AD [34]. At present, mechanistic evidence suggests that S. aureus is mutually promoted by and aggravated by Th2-mediated inflammation and skin barrier impairment in AD [10,35]. However, previous studies that evaluated the efficacy of topical antibiotics showed conflicting results [36,37], and it has been hypothesized that this may be due to recolonization by nasal S. aureus and eradication of beneficial bacteria [38,39]. Ongoing research is working with manipulating the skin microbiome in the therapeutic management of AD [40,41], and initial pilot trials showed that topical application of coagulase negative staphylococci (CoNS) [40] and Roseomonas mucosa [42] decreased disease severity.
Only one previous study has investigated changes in the skin microbiome and S. aureus colonization following dupilumab therapy. In line with our findings, this study demonstrated an increased Shannon diversity and a reduced absolute abundance of S. aureus that correlated with clinical improvement [21]. Tuf gene sequencing assessing the relative abundance of all staphylococcal species, not just S. aureus, was applied in our study. Thus, our study adds to these previous findings by elucidating the interplay between different staphylococcal species in relation to treatment, revealing a significant decrease in S. aureus and an increase in S. epidermidis and S. hominis proportions that correlate with clinical improvement. Furthermore, the change in proportion of S. aureus correlates inversely with changes in S. epidermidis and S. hominis. These findings suggest that the treatment-induced increase in CoNS might promote a sustained decrease in S. aureus, thereby limiting S. aureus re-colonization and clinical worsening of the disease. Our findings are in line with the results of previous studies demonstrating that S. hominis and S. epidermidis can produce S. aureus-selective antimicrobial peptides that work in concert with LL-37, a host antimicrobial peptide [15,16]. Moreover, a possible protective role of S. hominis in relation to the development of AD was suggested by Meylan et al. [43], showing that high S. hominis abundance was related to a reduced risk of developing AD in infants. Likewise, high abundance of S. epidermidis has been related to decreased severity of AD [4]; however, conflicting results exist regarding the role of S. epidermidis in AD [44].
The significant correlation between nasal and skin S. hominis abundance, together with a relationship between nasal S. hominis and clinical improvement, suggest that nasal microbiota may influence the skin microbiome by serving as a reservoir for bacteria that are transferred between nose and skin. This is supported by a previous study showing an association between nasal S. aureus and severity of AD [45]. The finding is interesting since it may suggest that targeting the nasal and skin microbiome simultaneously is beneficial for sustained change in the skin microbiome in some patients.
Our study showed that treatment with dupilumab was related to pronounced changes in the microbiome in both lesional and nonlesional skin and in the nose, with significant changes in bacterial community structure. Thus, our results highlight the close relationship between Th2 immune activation and global bacterial dysbiosis in AD and emphasize how blocking IL-4Rα signaling reverses the vicious circle in which S. aureus further promotes skin inflammation and the breakdown of the skin barrier. Interestingly, we found even stronger correlations between clinical improvement and change in relative abundance of staphylococcal species in nonlesional skin compared to lesional skin, highlighting the theoretic therapeutic potential of also including nonlesional skin in topical microbiome-based treatment strategies.
Contrary to some prior studies [5,42,43], non-targeted treatments were not related to significant changes in the skin microbiome. This may be due to the relatively few included patients, low baseline EASI scores, short follow-up time, and low relative abundance of S. aureus at baseline in these patients. Interestingly, despite comparable post-treatment EASI scores between the two treatment groups, patients treated with dupilumab had a significantly different community structure and lower abundance of S. aureus on lesional and nonlesional skin, suggesting that the impact of dupilumab on the skin microbiome extends beyond the effect of clinical improvement. A high proportion of patients treated with dupilumab reached EASI-75, supporting the promising results on the effectiveness of dupilumab.
The strength of this study is the homogenous patient cohort and the complimentary use of both 16S rRNA and tuf amplicon sequencing data, allowing an in-depth characterization of changes in relative abundance of staphylococcal species. Limitations include the size of the study population and lack of a control group; however, we decided to present some data from patients receiving other treatments, since treatment-related changes in general are sparsely investigated.
5. Conclusions
In conclusion, our study showed that targeting IL-4Rα signaling reverses the microbial dysbiosis in AD. The proportional increase in CoNS in both lesional and nonlesional skin may be particularly important for sustained treatment response by reducing S. aureus-induced inflammation. Furthermore, our results suggest that future microbiome-based therapeutic approaches should comprise the nasal and nonlesional, as well as the lesional, microbiome.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9071487/s1, Figure S1: Changes in bacterial community structure on lesional, nonlesional, and nasal skin following dupilumab therapy, Figure S2: Relative abundance of Staphylococcus on lesional and nonlesional skin before and after dupilumab therapy, Figure S3: Changes in relative abundance of staphylococcal species following dupilumab therapy stratified by lesional, nonlesional, and nasal skin, Figure S4: Genera and staphylococci species distribution grouped in clusters and shift in clusters following treatment with dupilumab, Figure S5: Shannon diversity and S. aureus relative abundance before and after non-targeted treatment, Figure S6: Changes in bacterial community structure on lesional, nonlesional, and nasal skin following non-targeted treatment, Figure S7: Relative abundance of Staphylococcus on lesional and nonlesional skin before and after non-targeted treatment, Figure S8: Shift in clusters following non-targeted treatment.
Author Contributions
Conceptualization, C.M.O., M.-L.C., and T.A.; methodology, C.M.O., M.-L.C., and T.A.; software, A.C.I.; validation, A.C.I.; formal analysis, A.C.I., S.M.E., and P.S.A.; investigation, C.M.O. and Y.T.Y.; resources, S.F.T., E.G.-Y., and T.A.; data curation, C.M.O. and Y.T.Y.; writing—original draft preparation, C.M.O.; writing—review and editing, all authors; visualization, C.M.O. and A.C.I.; supervision, S.F.T., P.S.A., E.G.-Y., and T.A.; project administration, C.M.O. and T.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by The Augustinus Foundation and Aage Bangs Foundation.
Institutional Review Board Statement
The study was approved by the local ethics committee (project number: H-1-2014-039) and by the Danish Data Protection Agency.
Informed Consent Statement
Oral and written information was provided and written informed consent was obtained from all subjects.
Data Availability Statement
The 16S rRNA and tuf sequences are available through the European Nucleotide Archive (ENA) at the European Bioinformatics Institute (EBI) under accession number PRJEB41628. Due to national data protection regulations regarding personally identifiable information, only a limited number of variables are included in the sample data file.
Conflicts of Interest
C.M.O., A.C.I., M.-L.C., S.M.E., Y.T.Y., and P.S.A.: none declared, S.F.T.: has served on advisory boards, been a paid speaker and received research grants from Sanofi; E.G.-Y.: has been an investigator and served as consultant for Regeneron Pharmaceuticals and Sanofi, and has received research grants from Regeneron Pharmaceuticals and Sanofi; T.A.: has served on advisory boards, been a paid speaker and received research grants from Sanofi.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ungar B., Garcet S., Gonzalez J., Dhingra N., Correa da Rosa J., Shemer A., Krueger J.G., Suarez-Farinas M., Guttman-Yassky E. An Integrated Model of Atopic Dermatitis Biomarkers Highlights the Systemic Nature of the Disease. J. Investig. Dermatol. 2017;137:603–613. doi: 10.1016/j.jid.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 2.Suarez-Farinas M., Tintle S.J., Shemer A., Chiricozzi A., Nograles K., Cardinale I., Duan S., Bowcock A.M., Krueger J.G., Guttman-Yassky E. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J. Allergy Clin. Immunol. 2011;127:954–964.e4. doi: 10.1016/j.jaci.2010.12.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clausen M.L., Agner T., Lilje B., Edslev S.M., Johannesen T.B., Andersen P.S. Association of Disease Severity With Skin Microbiome and Filaggrin Gene Mutations in Adult Atopic Dermatitis. JAMA Dermatol. 2018;154:293–300. doi: 10.1001/jamadermatol.2017.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrd A.L., Deming C., Cassidy S.K.B., Harrison O.J., Ng W.I., Conlan S., Belkaid Y., Segre J.A., Kong H.H. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong H.H., Oh J., Deming C., Conlan S., Grice E.A., Beatson M.A., Nomicos E., Polley E.C., Komarow H.D., Murray P.R., et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamazaki Y., Nakamura Y., Nunez G. Role of the microbiota in skin immunity and atopic dermatitis. Allergol. Int. 2017;66:539–544. doi: 10.1016/j.alit.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Clausen M.L., Slotved H.C., Krogfelt K.A., Andersen P.S., Agner T. In vivo expression of antimicrobial peptides in atopic dermatitis. Exp. Dermatol. 2016;25:3–9. doi: 10.1111/exd.12831. [DOI] [PubMed] [Google Scholar]
- 8.Geoghegan J.A., Irvine A.D., Foster T.J. Staphylococcus aureus and Atopic Dermatitis: A Complex and Evolving Relationship. Trends Microbiol. 2018;26:484–497. doi: 10.1016/j.tim.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura Y., Oscherwitz J., Cease K.B., Chan S.M., Munoz-Planillo R., Hasegawa M., Villaruz A.E., Cheung G.Y., McGavin M.J., Travers J.B., et al. Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature. 2013;503:397–401. doi: 10.1038/nature12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams M.R., Nakatsuji T., Gallo R.L. Staphylococcus aureus: Master Manipulator of the Skin. Cell Host Microbe. 2017;22:579–581. doi: 10.1016/j.chom.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwamoto K., Moriwaki M., Niitsu Y., Saino M., Takahagi S., Hisatsune J., Sugai M., Hide M. Staphylococcus aureus from atopic dermatitis skin alters cytokine production triggered by monocyte-derived Langerhans cell. J. Dermatol. Sci. 2017;88:271–279. doi: 10.1016/j.jdermsci.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Lai Y., Cogen A.L., Radek K.A., Park H.J., Macleod D.T., Leichtle A., Ryan A.F., Di Nardo A., Gallo R.L. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J. Investig. Dermatol. 2010;130:2211–2221. doi: 10.1038/jid.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wanke I., Steffen H., Christ C., Krismer B., Götz F., Peschel A., Schaller M., Schittek B. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J. Investig. Dermatol. 2011;131:382–390. doi: 10.1038/jid.2010.328. [DOI] [PubMed] [Google Scholar]
- 14.Meisel J.S., Sfyroera G., Bartow-McKenney C., Gimblet C., Bugayev J., Horwinski J., Kim B., Brestoff J.R., Tyldsley A.S., Zheng Q., et al. Commensal microbiota modulate gene expression in the skin. Microbiome. 2018;6:20. doi: 10.1186/s40168-018-0404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cogen A.L., Yamasaki K., Sanchez K.M., Dorschner R.A., Lai Y., MacLeod D.T., Torpey J.W., Otto M., Nizet V., Kim J.E., et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Investig. Dermatol. 2010;130:192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakatsuji T., Chen T.H., Narala S., Chun K.A., Two A.M., Yun T., Shafiq F., Kotol P.F., Bouslimani A., Melnik A.V., et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moyle M., Cevikbas F., Harden J.L., Guttman-Yassky E. Understanding the immune landscape in atopic dermatitis: The era of biologics and emerging therapeutic approaches. Exp. Dermatol. 2019;28:756–768. doi: 10.1111/exd.13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman-Yassky E., Bissonnette R., Ungar B., Suarez-Farinas M., Ardeleanu M., Esaki H., Suprun M., Estrada Y., Xu H., Peng X., et al. Dupilumab progressively improves systemic and cutaneous abnormalities in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2019;143:155–172. doi: 10.1016/j.jaci.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton J.D., Suárez-Fariñas M., Dhingra N., Cardinale I., Li X., Kostic A., Ming J.E., Radin A.R., Krueger J.G., Graham N., et al. Dupilumab improves the molecular signature in skin of patients with moderate-to-severe atopic dermatitis. J. Allergy Clin. Immunol. 2014;134:1293–1300. doi: 10.1016/j.jaci.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 20.He H., Olesen C.M., Pavel A.B., Clausen M.L., Wu J., Estrada Y., Zhang N., Agner T., Guttman-Yassky E. Tape-Strip Proteomic Profiling of Atopic Dermatitis on Dupilumab Identifies Minimally Invasive Biomarkers. Front. Immunol. 2020;11:1768. doi: 10.3389/fimmu.2020.01768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callewaert C., Nakatsuji T., Knight R., Kosciolek T., Vrbanac A., Kotol P., Ardeleanu M., Hultsch T., Guttman-Yassky E., Bissonnette R., et al. IL-4Rα Blockade by Dupilumab Decreases Staphylococcus aureus Colonization and Increases Microbial Diversity in Atopic Dermatitis. J. Investig. Dermatol. 2020;140:191–202.e7. doi: 10.1016/j.jid.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drucker A.M., Ellis A.G., Bohdanowicz M., Mashayekhi S., Yiu Z.Z.N., Rochwerg B., Di Giorgio S., Arents B.W.M., Burton T., Spuls P.I., et al. Systemic Immunomodulatory Treatments for Patients With Atopic Dermatitis: A Systematic Review and Network Meta-analysis. JAMA Dermatol. 2020 doi: 10.1001/jamadermatol.2020.0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iversen S., Johannesen T.B., Ingham A.C., Edslev S.M., Tevell S., Månsson E., Nilsdotter-Augustinsson Å., Söderquist B., Stegger M., Andersen P.S. Alteration of Bacterial Communities in Anterior Nares and Skin Sites of Patients Undergoing Arthroplasty Surgery: Analysis by 16S rRNA and Staphylococcal-Specific tuf Gene Sequencing. Microorganisms. 2020;8:1977. doi: 10.3390/microorganisms8121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17 doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 26.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMurdie P.J., Holmes S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O’Hara R.B., Simpson G.L., Solymos P. R Package Vegan: Community Ecology Package. [(accessed on 9 July 2021)];2019 Available online: https://github.com/vegandevs/vegan.
- 30.Maechler M., Rousseeuw P., Struyf A., Hubert M., Hornik K. Cluster: Cluster Analysis Basics and Extensions. [(accessed on 9 July 2021)];2019 Available online: https://cran.r-project.org/web/packages/cluster/index.html.
- 31.Kassambara A., Mundt F. R Package Factoextra. [(accessed on 9 July 2021)];2020 Available online: https://CRAN.R-project.org/package=factoextra.
- 32.Brunson J.C., Read Q.D. R Package Ggalluvial. [(accessed on 9 July 2021)];2020 Available online: https://corybrunson.github.io/ggalluvial/
- 33.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olesen C.M., Clausen M.-L., Andersen P.S., Agner T. The Skin Microbiome in Atopic Dermatitis—A Potential Treatment Target? Curr. Dermatol. Rep. 2018;7:199–208. doi: 10.1007/s13671-018-0245-6. [DOI] [Google Scholar]
- 35.Leyva-Castillo J.M., McGurk A., Geha M.D.R. Allergic skin inflammation and S. aureus skin colonization are mutually reinforcing. Clin. Immunol. 2020;218:108511. doi: 10.1016/j.clim.2020.108511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bath-Hextall F.J., Birnie A.J., Ravenscroft J.C., Williams H.C. Interventions to reduce Staphylococcus aureus in the management of atopic eczema: An updated Cochrane review. Br. J. Dermatol. 2010;163:12–26. doi: 10.1111/j.1365-2133.2010.09743.x. [DOI] [PubMed] [Google Scholar]
- 37.George S.M., Karanovic S., Harrison D.A., Rani A., Birnie A.J., Bath-Hextall F.J., Ravenscroft J.C., Williams H.C. Interventions to reduce Staphylococcus aureus in the management of eczema. Cochrane Database Syst. Rev. 2019;2019 doi: 10.1002/14651858.CD003871.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wróbel J., Tomczak H., Jenerowicz D., Czarnecka-Operacz M. Skin and nasal vestibule colonisation by Staphylococcus aureus and its susceptibility to drugs in atopic dermatitis patients. Ann. Agric. Environ. Med. 2018;25:334–337. doi: 10.26444/aaem/85589. [DOI] [PubMed] [Google Scholar]
- 39.Huang J.T., Abrams M., Tlougan B., Rademaker A., Paller A.S. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics. 2009;123:e808–e814. doi: 10.1542/peds.2008-2217. [DOI] [PubMed] [Google Scholar]
- 40.Paller A.S., Kong H.H., Seed P., Naik S., Scharschmidt T.C., Gallo R.L., Luger T., Irvine A.D. The microbiome in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2019;143:26–35. doi: 10.1016/j.jaci.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grice E.A. The skin microbiome: Potential for novel diagnostic and therapeutic approaches to cutaneous disease. Semin. Cutan. Med. Surg. 2014;33:98–103. doi: 10.12788/j.sder.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myles I.A., Earland N.J., Anderson E.D., Moore I.N., Kieh M.D., Williams K.W., Saleem A., Fontecilla N.M., Welch P.A., Darnell D.A., et al. First-in-human topical microbiome transplantation with Roseomonas mucosa for atopic dermatitis. JCI Insight. 2018;3 doi: 10.1172/jci.insight.120608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meylan P., Lang C., Mermoud S., Johannsen A., Norrenberg S., Hohl D., Vial Y., Prod’hom G., Greub G., Kypriotou M., et al. Skin Colonization by Staphylococcus aureus precedes the Clinical Diagnosis of Atopic Dermatitis in Infancy. J. Investig. Dermatol. 2017;137:2497–2504. doi: 10.1016/j.jid.2017.07.834. [DOI] [PubMed] [Google Scholar]
- 44.Hon K.L., Tsang Y.C., Pong N.H., Leung T.F., Ip M. Exploring Staphylococcus epidermidis in atopic eczema: Friend or foe? Clin. Exp. Dermatol. 2016;41:659–663. doi: 10.1111/ced.12866. [DOI] [PubMed] [Google Scholar]
- 45.Totté J.E.E., Pardo L.M., Fieten K.B., Vos M.C., van den Broek T.J., Schuren F.H.J., Pasmans S. Nasal and skin microbiomes are associated with disease severity in paediatric atopic dermatitis. Br. J. Dermatol. 2019;181:796–804. doi: 10.1111/bjd.17755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA and tuf sequences are available through the European Nucleotide Archive (ENA) at the European Bioinformatics Institute (EBI) under accession number PRJEB41628. Due to national data protection regulations regarding personally identifiable information, only a limited number of variables are included in the sample data file.