Abstract
Angelman syndrome (AS) is a rare neurodevelopmental disease that is caused by the loss of function of the maternal copy of ubiquitin–protein ligase E3A (UBE3A) on the chromosome 15q11–13 region. AS is characterized by global developmental delay, severe intellectual disability, lack of speech, happy disposition, ataxia, epilepsy, and distinct behavioral profile. There are four molecular mechanisms of etiology: maternal deletion of chromosome 15q11–q13, paternal uniparental disomy of chromosome 15q11–q13, imprinting defects, and maternally inherited UBE3A mutations. Different genetic types may show different phenotypes in performance, seizure, behavior, sleep, and other aspects. AS caused by maternal deletion of 15q11–13 appears to have worse development, cognitive skills, albinism, ataxia, and more autistic features than those of other genotypes. Children with a UBE3A mutation have less severe phenotypes and a nearly normal development quotient. In this review, we proposed to review genotype–phenotype correlations based on different genotypes. Understanding the pathophysiology of the different genotypes and the genotype–phenotype correlations will offer an opportunity for individualized treatment and genetic counseling. Genotype–phenotype correlations based on larger data should be carried out for identifying new treatment modalities.
Keywords: Angelman syndrome, genotype, phenotype, imprinting, intellectual disability, neurodevelopment
1. Introduction
Angelman syndrome (AS, OMIM #105830) is an incurable neurodevelopmental disease caused by the loss of function of the maternal copy of ubiquitin–protein ligase E3A (UBE3A) and other genes on chromosome 15q11–13 region [1,2]. AS was first described in 1965 by Harry Angelman following a study of three children with similar symptoms [3,4]. During the past decades, our understanding of AS’s clinical phenotypes and genetic pathology has improved. As a genomic imprinting disorder, AS has featured presentations that include global developmental delay, severe intellectual disability, lack of speech, happy disposition, ataxia, epilepsy, and distinct behavioral profile [1,5,6,7]. This rare neurodevelopment disorder has a prevalence of 1 in 10,000–24,000 births [8].
AS has four molecular mechanisms of etiology: (1) deletion of the maternal copy of chromosome 15q11–q13 (del15q11–13, 70%) [9], (2) paternal uniparental disomy of chromosome 15q11–q13 (UPD, 2–7%) [10], (3) imprinting defects within chromosome 15q11–q13 that disrupt the expression of maternally inherited UBE3A (3–5%) [11], and (4) maternally inherited UBE3A mutations (10%) [12]. The clinical picture is variable in different genotypes. Generally, patients with a deletion type have a more severe phenotype, and those with UPD or imprinting defects have a less severe phenotype [7,13,14,15,16,17]. Genotype–phenotype correlation should be highlighted for accurate prediction and genetic consultation.
Children with AS usually have normal prenatal and birth history, normal metabolic and hematologic, and chemical laboratory results. The following four frequent clinical characteristics can be seen in all patients: severe developmental delay, movement or balance disorder, behavioral abnormality, and speech disorder (Table 1). Developmental delay often becomes evident by 6 months of age [18]. Movement or balance disorders usually include ataxia of gait and/or tremulous movement of the limbs. Most individuals lack speech completely, and few can speak a few words [19]. Receptive language is less impaired and is better than expressive language [14]. Frequent clinical characteristics, including microcephaly and seizures, occur in >80% of children with AS, often developing by 3 years of age [17]. A characteristic electroencephalogram (EEG) “signature” can also be found in 80% of children with AS [20,21], which may be an important hint for diagnosis [22]. Children with AS have several associated clinical features (Table 1). These children are easily excited and have an apparent happy demeanor associated with frequent laughter, hyperactivity, stereotypes, and proactive social contact [23,24]. Most children have sleep disturbance with reduced need for sleep and with long or frequent periods of wakefulness during the night [25,26,27,28,29].
Table 1.
Clinical characteristics of Angelman syndrome (adapted from Williams et al. [30]).
| Consistent (100%) | Frequent (>80%) | Associated (<80%) |
|---|---|---|
|
|
|
2. Molecular Genetics and Diagnostics
Human chromosome 15q11–q13 contains a cluster of imprinted genes. In the imprinted region, methylation and gene expression are regulated by a bipartite imprinting center located in the small nuclear ribonucleoprotein associated protein N (SNRPN) region [31]. Genes in the imprinted center are clustered into paternally expressed only genes, maternally expressed only genes, and biallelically expressed genes (Figure 1). Differential expression of these maternal and paternal expression genes causes sister imprinting disorders: AS and Prader–Willi syndrome (PWS) [32].
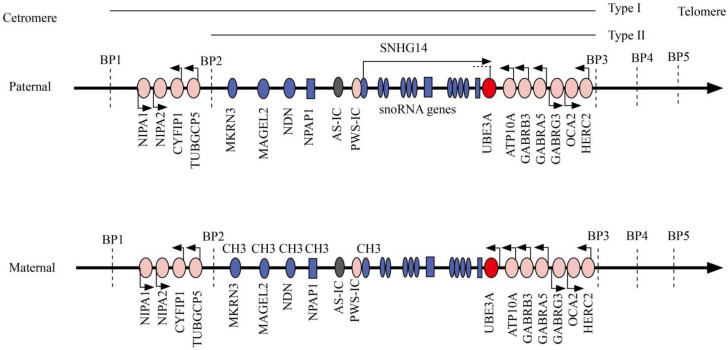
Figure 1.
Human chromosome 15q11–13 region. Paternal and maternal chromosome 15q11–13 regions around the Angelman syndrome imprinting center (AS-IC) and Prader–Willi syndrome imprinting center (PWS-IC) are presented. Paternally expressed genes are indicated as deep blue, maternally expressed genes are indicated in red, and genes expressed from both parental alleles are indicated as pink. Transcription orientation is noted with arrows. Class I and class II deletions are indicated as horizontal lines. BP, breakpoint cluster region; CH3, methylation; snoRNA, small nucleolar RNA; SNRPN, small nuclear ribonucleoprotein-associated protein N; UBE3A, ubiquitin–protein ligase E3A.
MKRN3, MAGEL2, NDN, PWRN1, NPAP1, SNURF-SNRPN, and several C/D box small nucleolar RNA (snoRNA) genes are paternally expressed genes [33]. The CpG islands in the promoter regions are differentially imprinted: the paternal allele is unmethylated and is expressed, whereas the maternal allele is methylated and repressed [34,35]. Loss of expression of these paternally expressed only genes causes PWS.
The pathogenesis of AS is similar to PWS. UBE3A is a maternally expressed gene: the unmethylated maternal allele is expressed, and the methylated paternal allele is repressed. Imprinted expression of UBE3A is regulated by small nucleolar RNA host gene 14 (SNHG14) with a noncoding antisense transcript as the product which is started at the paternal SNRPN promoter [36] (Figure 1). In neuronal cells of normal individuals, the paternal region lacks methylation, and the paternally derived UBE3A gene is silenced [37]; the maternal region is methylated, SNHG14 is not expressed, resulting in UBE3A gene transcription [38,39]. Any genetic causes leading to a non-functional UBE3A protein will result in a knockout of neuronal UBE3A and lead to AS [40].
Lack of UBE3A protein expression in the brain of children with AS can lead to abnormal ubiquitination in Purkinje cells in the cerebellum [37,41,42]. Abnormal ubiquitination is attributed to the abnormality in the nigrostriatal pathway along with the cerebellum in animal models, which produce phenotypes that include motor impairments, synaptic plasticity, and repaired memory in AS [43,44,45]. Major aspects of the core clinical phenotypes of AS, including cognitive, language and motor deficits, are results of impaired long-range connection between the cerebellar and cortical networks [46].
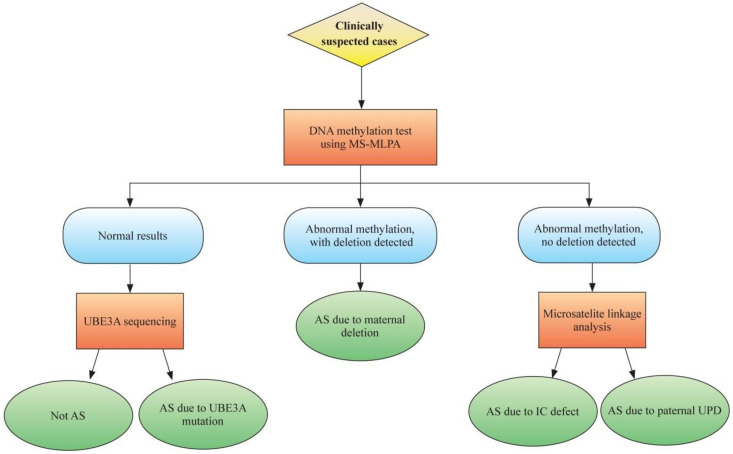
Although the criteria for diagnosis of AS were defined in 2005 based on the common phenotypes of AS, a definitive diagnosis still depends on molecular testing. Differential methylation of chromosome 15q11–q13 provides the basis for molecular diagnosis (Figure 1). For those suspected with AS clinically, the first diagnostic modality is methylation analysis of the chromosome 15q11–13 region, methylation-sensitive multiplex ligation-dependent probe amplification (MS-MLPA). For those with normal methylation results, UBE3A gene sequencing is recommended to detect the UBE3A gene mutation. If UBE3A gene mutation is not detected, AS is excluded, and other diseases should be considered. For those with abnormal methylation results and deletion detected, AS due to del15q11–13 is diagnosed; for those without deletion, microsatellite linkage analysis can be done to differentiate UPD or imprinting defects (Figure 2).
Figure 2.
Molecular diagnostics for Angelman syndrome (AS). For those suspected with AS clinically, methylation analysis of the chromosome 15q11–13 region can be performed using methylation-sensitive multiplex ligation-dependent probe amplification (MS-MLPA). For those with normal methylation results, UBE3A gene sequencing is recommended to detect the UBE3A gene mutation. If UBE3A gene mutation is not detected, AS is excluded, and other diseases should be considered. For those with abnormal methylation results and deletion detected, AS due to del15q11–13 is diagnosed; for those without deletion, microsatellite linkage analysis can be done to differentiate paternal uniparental disomy (UPD) or imprinting defects.
It is necessary to differentiate several microdeletion syndromes and single-gene disorders that resemble AS. Chromosome microarray testing can be used to detect microdeletion syndromes, including Phelan–McDermid syndrome (22q13.3 deletion), MBD5 haploinsufficiency syndrome (2q23.1 deletion), and KANSL1 haploinsufficiency syndrome (17q21.31 deletion) [18,47]. If the results of microarray analysis are normal, single-gene disorders should be considered, which include Pitt–Hopkins syndrome (TCF4 haploinsufficiency), Christianson syndrome (SLC9A6 mutation), Mowat–Wilson syndrome (ZEB2 haploinsufficiency), and Rett syndrome (MECP2 mutations) [18].
Several pilot newborn screening programs have been undertaken for the early diagnosis of AS. Mahmoud et al. carried out a pilot newborn screening program and established a cost-effective newborn screening test for early diagnosis of PWS [48], which is also applicable to early diagnosis of AS. Ferreira et al. used a methylation-sensitive high-resolution melting method to analyze DNA extracted from dried blood spots. This method provides an accurate approach for genetic screening of imprinting-related disorders (PWS and AS) in newborns [49]. Early identification by newborn screening will not only achieve early efficient intervention but also will decrease costly medical evaluations.
Defining the exact molecular mechanism of AS is beneficial for genetic consultations. The risk of recurrence in parents of different genotypes is variable. In AS cases due to maternal deletion, the risk of recurrence is less than 1% [50]; for cases with paternal UPD, the risk of recurrence is less than 1/200; however, for those rare cases of structural defects of chromosome 15, including Robertsonian translocation, the risk is as high of 100%. For those due to IC defect, the risk of occurrence is 50% or 1% for those with or without deletion in maternal chromosome 15 [50]. For those with de novo UBE3A mutation, the risk of occurrence is near to 0; and for those with UBE3A mutation in both cases and their mothers, the risk is 50% [50]. Therefore, genetic analysis should be undertaken in the cases together with parents for genetic consultation.
Exploring the disease mechanism will also benefit targeted gene, and molecular therapies for AS. UBE3A critically impacts early brain development, and reactivation of UBE3A gene expression can prevent the onset of behavioral deficits [51]. Researchers are identifying novel therapeutics aimed at correcting pathophysiological deficits of AS and restoring the loss of UBE3A expression in the brain [52]. Restoring the function of UBE3A is the most promising therapeutic modality for AS.
3. Genotype–Phenotype Correlation in Angelman Syndrome
The severity of the phenotype depends on the molecular etiology. Individuals with a deletion commonly present with a more severe phenotype, whereas those with non-deletion have slightly milder and more variable presentations [7,53,54]. Intrauterine and postnatal growth, neurobehavioral and neuropsychiatric development phenotypes in children with AS depend on the genotype. Previous findings have revealed that patients with AS caused by del15q11–13 appear to have worse development, cognitive skills, albinism, ataxia, and more autistic features than individuals with other genotypes [13,53,55,56,57,58]. Children with UBE3A mutation appear to have less severe phenotypes with a nearly normal development quotient [58]. Below we review genotype–phenotype correlations based on different genotypes (Table 2).
Table 2.
Comparison of major phenotypes of different subtypes.
| Major Aspects | AS Due to Maternal del15q11–13 | AS Due to Non-Deletion | ||
|---|---|---|---|---|
| Paternal Uniparental Disomy for Chromosome15q11–q13 (UPD) |
Imprinting Defect | Pathogenic UBE3A Mutation | ||
| Development | More delayed across all development domains than other types Cognitive skills lower than other types Delayed gross and fine motor skills more severe Reduced developmental age regarding visual perception, receptive language, and expressive language |
Higher overall age equivalent scores and growth score equivalents than deleion type but lower than UBE3A mutation subtype; Better development and expressive language ability in patients with UPD and imprinting defect Higher scores and greater rates of skill attainment in all development domains in patients UBE3A mutation |
||
| Seizures | More common and severe in the deletion group | Lower prevalence of epilepsy, and more with late-onset seizures. UPD subtype has the lowest frequency of epilepsy and exhibits the least severe epilepsy phenotype The severity of epilepsy in the UBE3A mutation subtype ranks second after the deletion subtype |
||
| Behavior | Lower response rates to the social reinforcement paradigm than other types |
The imprinting defect a high rate of reinforcement by social stimuli. Patients with UBE3A mutations | ||
| Sleep | Common in all subtypes but Sleep problems are more prevalent in children with UPD and UBE3A mutations | |||
| Others | Higher rate of hypopigmentation | UPD and imprinting defects have a higher risk of obesity than deletion type | ||
3.1. AS Due to Maternal del15q11–13
Most patients with AS have maternal 15q11–13 deletions at a length of 5–6 Mb. The deletion is classified into two types based on the deletion length: Class I patients have breakpoints at BP1 and BP3 with various noncoding regions deleted (~6 Mb, ~16 genes), and Class II have breakpoints at BP2 and BP3 with a deletion of ~12 genes at a length of 5 Mb [8] (Figure 1). Classes I and II deletions represent 95% of AS patients with del15q11–13. Four evolutionarily conserved genes (NIPA-1, NIPA-2, CYF1P1, and GCP5) are located between BP1 and BP2, which are involved in central nervous system development and functioning [59]. Deletion of these genes may result in speech impairment and developmental delay. Genes causing the difference also include three GABAA receptor subunit genes (GABRB3, GABRG3, GABRA5) that are single-copies for the deletion genotypes but are intact for all non-deletion genotypes [8].
Developmental studies indicate that children with AS caused by a deletion are developmentally more delayed across all domains compared with those due to a UBE3A pathogenic variant or UPD [14,54,60]. Cognitive skills are much lower in the deletion group than in the non-deletion group [14,60]. Delayed gross and fine motor skills are more severe in the deletion group [14,53,60,61,62]. Seizure and microcephaly have been reported to be more common and severe in the deletion group [53,63,64,65]. The deletion subtype is associated with the most severe epilepsy phenotype; in contrast, non-deletion patients may have relatively late-onset seizures [64,66]. Mertz et al. found that children with a deletion type had significantly reduced developmental age regarding visual perception, receptive language, and expressive language compared with those having a UBE3A mutation and pUPD [58]. Therefore, children with deletion type had lower response rates to the social reinforcement paradigm than those with non-deletion type [67]; in other words, children with the deletion type are more difficult to treat using social intervention methods. Hypopigmentation was also significantly more prevalent in the deletion group [64].
For different types of deletion, children with Class I deletion are reported to have lower expressive and total language abilities than those having Class II deletion [55,57]. Children with BP1—BP3 deletion have more daily, disabling seizures, a higher frequency of seizures, and recurrent seizures aggravated by fever [16]. Class II deletion is associated with >50% intermittent rhythmic theta activity and normal posterior rhythm, whereas Class I deletion is associated with <50% intermittent rhythmic theta activity and epileptiform discharges during wakefulness [68]. Burnside et al. found that with more deleted genes and with locations nearer to the centromere, patients may have a higher prevalence of microcephaly, epilepsy, ataxia, speech disorders, and autistic symptoms [59]. The study results are conflicting on the difference of clinical severity in different deletion subtypes. Some studies found no difference in language abilities and cognitive function between Class I and Class II patients [8,13,14,56,58]. Additional studies should be carried out to delineate genotype–phenotype correlations of Class I and II deletion types.
3.2. AS Due to Paternal Uniparental Disomy for Chromosome15q11–q13
For children with paternal UPD of chromosome 15q11–q13, the structure and number of chromosomes are normal. However, there are paternally imprinted (maternal expressed) genes within the chromosomal region 15q11–q13, and maternally imprinted genes have been elevated but with very little expression of UBE3A. The developmental profiles are generally similar for patients due to UPD and imprinting defects. Both subtypes generally had slightly lower overall age equivalent scores and growth score equivalents and had slower rates of growth than the UBE3A mutation subtype across all domains. However, children with UPD or imprinting defects had higher overall age equivalent scores and growth score equivalents than those with both deletion classes [54]. Compared to the patients with deletion type, patients with paternal UPD have a much lower prevalence of epilepsy, better development and expressive language ability; some patients may even speak 2–7 words [58,59]. The UPD subtype is also associated with the lowest frequency of epilepsy and exhibits the least severe epilepsy phenotype [66]. Varela et al. found that swallowing disorders, hypotonia, and microcephaly in the UPD type are less severe than those in the deletion type [57].
Children with UPD presented significantly more severe hyperphagic behavior, hyperphagic severity, and hyperphagic drive than children in the other genetic groups [69]. These children also have significantly higher birth weight and birth length, being taller at five years of age than children with other genetic types [69]. Children with UPD and imprinting defects may have a higher risk of obesity than those with 15q11.2–q13 deletions and UBE3A mutations [15,70]. Sleep problems are more prevalent in children with UPD and UBE3A mutations compared with other types [64].
3.3. AS Due to Imprinting Defect
The imprinting control region 15q11–q13 cluster has been designated the “imprinting center” (IC). The IC can control the differential expression of alleles in varying tissues. The mechanism of the imprinting defect is related to failed imprinting or lacking submicroscopic structure [71]. Only maternal-of-origin-specific UBE3A genes can be found in neuronal cells. The maternally active UBE3A gene lacks differential DNA methylation. Therefore, no UBE3A gene is expressed in the brain of AS patients due to the imprinting defect. The phenotypes in patients with the imprinting defect are similar to those with UPD. They have better language and intellectual intelligence than the deletion type. Almost all patients with imprinting defect or UPD will become overweight before 44 months of age. The imprinting defect group may benefit more from behavioral interventions because they showed a high rate of reinforcement by social stimuli [67].
3.4. AS Due to Pathogenic UBE3A Mutation
Patients with UBE3A mutations have the mildest clinical presentations among the four genetic types. The developmental quotient is the highest among different types, and children due to UBE3A mutation have higher scores and greater rates of skill attainment in all development domains than other types [54]. These patients have relatively normal adaptive behaviors and are without obesity or overweight. Some patients can present symptoms of tremor in the head, limbs, and trunk [72]. However, attention should be paid to the severity of epilepsy in the UBE3A mutation subtype, which ranks second after the deletion subtype [64].
4. Conclusions
The current studies revealed that AS patients with different genetic types may have different phenotypes in performance, seizure, behavior, sleep, and other aspects. However, the genotype–phenotype correlation atlas has been drawn based on numerous studies with sample sizes varying in different genotypes. Many studies are limited owing to the small number of patients in a certain genotype, particularly the non-deletion types. Other genotype–phenotype correlations will be delineated if a larger number of patients are studied. Another limitation is that patients with AS from different regions have not been compared due to under-registration and underreporting. More data on carefully phenotyped patients from a global Angelman registry and multi-center study will benefit the development of genotype–phenotype correlations of AS.
Further studies should focus on several main issues, including epilepsy and neurodevelopmental outcomes that need further precise interventions. Understanding the pathophysiology of the different genotypes and the genotype–phenotype correlations will offer an opportunity for individualized treatment, genetic counseling, and better outcomes. Genotype–phenotype correlations based on larger data sets should be carried out to identify new treatment modalities, including gene therapy.
Acknowledgments
We want to thank Robert D. Dorazio for his language editing of this paper.
Author Contributions
L.Y. and X.S. wrote the first draft of the paper. L.Y., X.S., S.M., Y.W., X.D. and C.Z. revised the paper and approved the final version for publication. All authors have read and agreed to the published version of the manuscript.
Funding
This project is supported by the Key R & D Projects of Zhejiang Provincial Department of Science and Technology (2021C03094), National Natural Science Foundation of China (81371215 and 81670786).
Institutional Review Board Statement
Not applicable for this review article.
Informed Consent Statement
Not required for the review article.
Data Availability Statement
No data are available for this review article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Williams C.A., Driscoll D.J., Dagli A.I. Clinical and genetic aspects of Angelman syndrome. Genet. Med. 2010;12:385–395. doi: 10.1097/GIM.0b013e3181def138. [DOI] [PubMed] [Google Scholar]
- 2.Maranga C., Fernandes T.G., Bekman E., Da Rocha S.T. Angelman syndrome: A journey through the brain. FEBS J. 2020;287:2154–2175. doi: 10.1111/febs.15258. [DOI] [PubMed] [Google Scholar]
- 3.Buonfiglio D., Hummer D.L., Armstrong A., Ehlen J.C., DeBruyne J.P. Angelman syndrome and melatonin: What can they teach us about sleep regulation. J. Pineal Res. 2020;69:12697. doi: 10.1111/jpi.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angelman H. “Puppet children”. A report of three cases. Dev. Med. Child. Neurol. 1965;7:688. doi: 10.1111/j.1469-8749.1965.tb07844.x. [DOI] [PubMed] [Google Scholar]
- 5.Williams C.A. Neurological aspects of the Angelman syndrome. Brain Dev. 2005;27:88–94. doi: 10.1016/j.braindev.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Thibert R.L., Larson A.M., Hsieh D.T., Raby A.R., Thiele E.A. Neurologic Manifestations of Angelman Syndrome. Pediatr. Neurol. 2013;48:271–279. doi: 10.1016/j.pediatrneurol.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Bird L. Angelman syndrome: Review of clinical and molecular aspects. Appl. Clin. Genet. 2014;7:93–104. doi: 10.2147/TACG.S57386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keute M., Miller M.T., Krishnan M.L., Sadhwani A., Chamberlain S., Thibert R.L., Tan W.-H., Bird L.M., Hipp J.F. Angelman Syndrome Genotypes Manifest Varying Degrees of Clinical Severity and Developmental Impairment. [(accessed on 13 August 2020)]; doi: 10.1038/s41380-020-0858-6. Available online: https://www.nature.com/articles/s41380-020-0858-6. [DOI] [PMC free article] [PubMed]
- 9.Kaplan L.C., Wharton R., Elias E., Mandell F., Donlon T., Latt S.A., Opitz J.M., Reynolds J.F. Clinical heterogeneity associated with deletions in the long arm of chromosome 15: Report of 3 new cases and their possible genetic significance. Am. J. Med. Genet. 1987;28:45–53. doi: 10.1002/ajmg.1320280107. [DOI] [PubMed] [Google Scholar]
- 10.Nicholls R.D. Genomic imprinting and uniparental disomy in Angelman and Prader-Willi syndromes: A review. Am. J. Med. Genet. 1993;46:16–25. doi: 10.1002/ajmg.1320460106. [DOI] [PubMed] [Google Scholar]
- 11.Buiting K., Saitoh S., Gross S., Dittrich B., Schwartz S., Nicholls R.D., Horsthemke B. Inherited microdeletions in the Angelman and Prader–Willi syndromes define an imprinting centre on human chromosome 15. Nat. Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- 12.Kishino T., Lalande M., Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 13.Peters S.U., Horowitz L., Barbieri-Welge R., Taylor J.L., Hundley R.J. Longitudinal follow-up of autism spectrum features and sensory behaviors in Angelman syndrome by deletion class. J. Child Psychol. Psychiatry. 2011;53:152–159. doi: 10.1111/j.1469-7610.2011.02455.x. [DOI] [PubMed] [Google Scholar]
- 14.Gentile J.K., Tan W.-H., Horowitz L.T., Bacino C.A., Skinner S.A., Barbieri-Welge R., Bauer-Carlin A., Beaudet A.L., Bichell T.J., Lee H.-S., et al. A Neurodevelopmental Survey of Angelman Syndrome with Genotype-Phenotype Correlations. J. Dev. Behav. Pediatr. 2010;31:592–601. doi: 10.1097/DBP.0b013e3181ee408e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan W.-H., Bacino C.A., Skinner S.A., Anselm I., Barbieri-Welge R., Bauer-Carlin A., Beaudet A.L., Bichell T.J., Gentile J.K., Glaze D.G., et al. Angelman syndrome: Mutations influence features in early childhood. Am. J. Med. Genet. Part A. 2011;155:81–90. doi: 10.1002/ajmg.a.33775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valente K.D., Varela M.C., Koiffmann C.P., Andrade J.Q., Grossmann R., Kok F., Marques-Dias M.J. Angelman syndrome caused by deletion: A genotype–phenotype correlation determined by breakpoint. Epilepsy Res. 2013;105:234–239. doi: 10.1016/j.eplepsyres.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Margolis S.S., Sell G.L., Zbinden M.A., Bird L.M. Angelman Syndrome. Neurotherapeutics. 2015;12:641–650. doi: 10.1007/s13311-015-0361-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buiting K., Williams C., Horsthemke K.B.B. Angelman syndrome—Insights into a rare neurogenetic disorder. Nat. Rev. Neurol. 2016;12:584–593. doi: 10.1038/nrneurol.2016.133. [DOI] [PubMed] [Google Scholar]
- 19.Andersen W.H., Rasmussen R.K., Strømme P. Levels of cognitive and linguistic development in Angelman syndrome: A study of 20 children. Logoped. Phoniatr. Vocol. 2001;26:2–9. doi: 10.1080/14015430117324. [DOI] [PubMed] [Google Scholar]
- 20.Boyd S., Harden A., Patton M.A. The EEG in early diagnosis of the Angelman (Happy Puppet) syndrome. Eur. J. Nucl. Med. Mol. Imaging. 1988;147:508–513. doi: 10.1007/BF00441976. [DOI] [PubMed] [Google Scholar]
- 21.Dan B., Boyd S. Angelman Syndrome Reviewed from a Neurophysiological Perspective. The UBE3A-GABRB3 Hypothesis. Neuropediatrics. 2003;34:169–176. doi: 10.1055/s-2003-42213. [DOI] [PubMed] [Google Scholar]
- 22.Born H.A., Martinez L.A., Levine A.T., Harris S.E., Mehra S., Lee W.L., Dindot S.V., Nash K.R., Silverman J.L., Segal D.J., et al. Early Developmental EEG and Seizure Phenotypes in a Full Gene Deletion of Ubiquitin Protein Ligase E3A Rat Model of Angelman Syndrome. Eneuro. 2021;8:ENEURO.0345-20.2020. doi: 10.1523/ENEURO.0345-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Micheletti S., Palestra F., Martelli P., Accorsi P., Galli J., Giordano L., Trebeschi V., Fazzi E. Neurodevelopmental profile in Angelman syndrome: More than low intelligence quotient. Ital. J. Pediatr. 2016;42:1–8. doi: 10.1186/s13052-016-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dan B., Pelc K., Cheron G. Behavior and neuropsychiatric manifestations in Angelman syndrome. Neuropsychiatr. Dis. Treat. 2008;4:577–584. doi: 10.2147/NDT.S2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruni O., Ferri R., D’Agostino G., Miano S., Roccella M., Elia M. Sleep disturbances in Angelman syndrome: A questionnaire study. Brain Dev. 2004;26:233–240. doi: 10.1016/S0387-7604(03)00160-8. [DOI] [PubMed] [Google Scholar]
- 26.Didden R., Korzilius H., Smits M.G., Curfs L.M.G. Sleep Problems in Individuals with Angelman Syndrome. Am. J. Ment. Retard. 2004;109:275–284. doi: 10.1352/0895-8017(2004)109<275:SPIIWS>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 27.Walz N.C., Beebe D., Byars K. Sleep in Individuals with Angelman Syndrome: Parent Perceptions of Patterns and Problems. Am. J. Ment. Retard. 2005;110:243–252. doi: 10.1352/0895-8017(2005)110[243:SIIWAS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 28.Pelc K., Cheron G., Boyd S., Dan B. Are there distinctive sleep problems in Angelman syndrome? Sleep Med. 2008;9:434–441. doi: 10.1016/j.sleep.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Takaesu Y., Komada Y., Inoue Y. Melatonin profile and its relation to circadian rhythm sleep disorders in Angelman syndrome patients. Sleep Med. 2012;13:1164–1170. doi: 10.1016/j.sleep.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 30.Williams C.A., Beaudet A.L., Clayton-Smith J., Knoll J.H., Kyllerman M., Laan L.A., Magenis R.E., Moncla A., Schinzel A.A., Summers J.A., et al. Angelman syndrome 2005: Updated consensus for diagnostic criteria. Am. J. Med. Genet. Part A. 2006;140A:413–418. doi: 10.1002/ajmg.a.31074. [DOI] [PubMed] [Google Scholar]
- 31.Nakao M., Sutcliffe J.S., Durtschl B., Mutlrangura A., Ledbetter D.H., Beaudet A.L. Imprinting analysis of three genes in the Prader—Willi/Angelman region: SNRPN, E6-associated protein, and PAR-2 (D15S225E) Hum. Mol. Genet. 1994;3:309–315. doi: 10.1093/hmg/3.2.309. [DOI] [PubMed] [Google Scholar]
- 32.Smith E.Y., Futtner C.R., Chamberlain S.J., Johnstone K.A., Resnick J.L. Transcription Is Required to Establish Maternal Imprinting at the Prader-Willi Syndrome and Angelman Syndrome Locus. PLoS Genet. 2011;7:e1002422. doi: 10.1371/journal.pgen.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beygo J., Buiting K., Ramsden S.C., Ellis R., Clayton-Smith J., Kanber D. Update of the EMQN/ACGS best practice guidelines for molecular analysis of Prader-Willi and Angelman syndromes. Eur. J. Hum. Genet. 2019;27:1326–1340. doi: 10.1038/s41431-019-0435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa R.A., Ferreira I.R., Cintra H.A., Gomes L.H.F., Guida L.D.C. Genotype-Phenotype Relationships and Endocrine Findings in Prader-Willi Syndrome. Front. Endocrinol. 2019;10:864. doi: 10.3389/fendo.2019.00864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glenn C.C., Saitoh S., Jong M.T., Filbrandt M.M., Surti U., Driscoll D.J., Nicholls R.D. Gene structure, DNA methylation, and imprinted expression of the human SNRPN gene. Am. J. Hum. Genet. 1996;58:335–346. [PMC free article] [PubMed] [Google Scholar]
- 36.Georgieva B., Atemin S., Todorova A., Todorov T., Miteva A., Avdjieva-Tzavella D., Mitev V. Molecular-Genetic Diagnostics of Angelman Syndrome—The Bulgarian Experience. Acta Med. Bulg. 2020;47:9–16. doi: 10.2478/amb-2020-0002. [DOI] [Google Scholar]
- 37.Judson M.C., Sosa-Pagan J.O., Del Cid W.A., Han J.E., Philpot B.D. Allelic specificity of Ube3a Expression In The Mouse Brain During Postnatal Development. J. Comp. Neurol. 2014;522:1874–1896. doi: 10.1002/cne.23507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chamberlain S.J., Chen P.-F., Ng K.Y., Bourgois-Rocha F., Lemtiri-Chlieh F., Levine E., Lalande M. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader–Willi syndromes. Proc. Natl. Acad. Sci. USA. 2010;107:17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaSalle J.M., Reiter L.T., Chamberlain S.J. Epigenetic regulation ofUBE3Aand roles in human neurodevelopmental disorders. Epigenomics. 2015;7:1213–1228. doi: 10.2217/epi.15.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heussler H.S. Emerging Therapies and challenges for individuals with Angelman syndrome. Curr. Opin. Psychiatry. 2021;34:123–128. doi: 10.1097/YCO.0000000000000674. [DOI] [PubMed] [Google Scholar]
- 41.Zaaroor-Regev D., de Bie P., Scheffner M., Noy T., Shemer R., Heled M., Stein I., Pikarsky E., Ciechanover A. Regulation of the polycomb protein Ring1B by self-ubiquitination or by E6-AP may have implications to the pathogenesis of Angelman syndrome. Proc. Natl. Acad. Sci. USA. 2010;107:6788–6793. doi: 10.1073/pnas.1003108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daily J., Smith A.G., Weeber E.J. Spatial and temporal silencing of the human maternal UBE3A gene. Eur. J. Paediatr. Neurol. 2012;16:587–591. doi: 10.1016/j.ejpn.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulherkar S.A., Jana N.R. Loss of dopaminergic neurons and resulting behavioural deficits in mouse model of Angelman syndrome. Neurobiol. Dis. 2010;40:586–592. doi: 10.1016/j.nbd.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Moreira-De-Sá A., Gonçalves F.Q., Lopes J.P., Silva H.B., Tomé Ângelo R., Cunha R.A., Canas P.M. Motor Deficits Coupled to Cerebellar and Striatal Alterations in Ube3am−/p+ Mice Modelling Angelman Syndrome Are Attenuated by Adenosine A2A Receptor Blockade. Mol. Neurobiol. 2021;58:2543–2557. doi: 10.1007/s12035-020-02275-9. [DOI] [PubMed] [Google Scholar]
- 45.Moreira-De-Sá A., Gonçalves F.Q., Lopes J.P., Silva H.B., Tomé Ângelo R., Cunha R.A., Canas P.M. Adenosine A2A receptors format long-term depression and memory strategies in a mouse model of Angelman syndrome. Neurobiol. Dis. 2020;146:105137. doi: 10.1016/j.nbd.2020.105137. [DOI] [PubMed] [Google Scholar]
- 46.Cheron G., Mã¡rquez-Ruiz J., Kishino T., Dan B. Disruption of the LTD dialogue between the cerebellum and the cortex in Angelman syndrome model: A timing hypothesis. Front. Syst. Neurosci. 2014;8:221. doi: 10.3389/fnsys.2014.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madaan M., Mendez M.D. Angelman Syndrome. StatPearls Publishing; Treasure Island, FL, USA: 2020. [PubMed] [Google Scholar]
- 48.Mahmoud R., Singh P., Weiss L., Lakatos A., Oakes M., Hossain W., Butler M.G., Kimonis V. Newborn screening for Prader–Willi syndrome is feasible: Early diagnosis for better outcomes. Am. J. Med. Genet. Part A. 2018;179:29–36. doi: 10.1002/ajmg.a.60681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferreira I.R., Costa R.A., Gomes L.H.F., Cunha W.D.D.S., Tyszler L.S., Freitas S., Junior J.C.L., De Vasconcelos Z.F.M., Nicholls R.D., Guida L.D.C. A newborn screening pilot study using methylation-sensitive high resolution melting on dried blood spots to detect Prader-Willi and Angelman syndromes. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-020-69750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stalker H.J., Williams C.A. Genetic counseling in Angelman syndrome: The challenges of multiple causes. Am. J. Med. Genet. 1998;77:54–59. doi: 10.1002/(SICI)1096-8628(19980428)77:1<54::AID-AJMG12>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 51.Sonzogni M., Hakonen J., Kleijn M.B., Silva-Santos S., Judson M.C., Philpot B.D., Van Woerden G.M., Elgersma Y. Delayed loss of UBE3A reduces the expression of Angelman syndrome-associated phenotypes. Mol. Autism. 2019;10:1–9. doi: 10.1186/s13229-019-0277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elgersma Y., Sonzogni M. UBE3A reinstatement as a disease-modifying therapy for Angelman syndrome. Dev. Med. Child Neurol. 2021;63:802–807. doi: 10.1111/dmcn.14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lossie A.C., Whitney M.M., Amidon D., Dong H.J., Chen P., Theriaque D., Hutson A., Nicholls R.D., Zori R.T., Williams C.A., et al. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J. Med. Genet. 2001;38:834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadhwani A., Wheeler A., Gwaltney A., Peters S.U., Barbieri-Welge R.L., Horowitz L.T., Noll L.M., Hundley R.J., Bird L.M., Tan W.-H. Developmental Skills of Individuals with Angelman Syndrome Assessed Using the Bayley-III. [(accessed on 30 January 2021)]; doi: 10.1007/s10803-020-04861-1. Available online: http://link.springer.com/article/10.1007/s10803-020-04861-1. [DOI] [PMC free article] [PubMed]
- 55.Sahoo T., Peters S.U., Madduri N.S., Glaze D.G., German J.R., Bird L.M., Barbieri-Welge R., Bichell T.J., Beaudet A.L., Bacino C.A. Microarray based comparative genomic hybridization testing in deletion bearing patients with Angelman syndrome: Genotype-phenotype correlations. J. Med. Genet. 2006;43:512–516. doi: 10.1136/jmg.2005.036913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahoo T., Bacino C.A., German J.R., Shaw C., Bird L.M., Kimonis V., Anselm I., Waisbren S., Beaudet A.L., Peters S.U. Identification of novel deletions of 15q11q13 in Angelman syndrome by array-CGH: Molecular characterization and genotype–phenotype correlations. Eur. J. Hum. Genet. 2007;15:943–949. doi: 10.1038/sj.ejhg.5201859. [DOI] [PubMed] [Google Scholar]
- 57.Varela M.C., Kok F., Otto P.A., Koiffmann C.P. Phenotypic variability in Angelman syndrome: Comparison among different deletion classes and between deletion and UPD subjects. Eur. J. Hum. Genet. 2004;12:987–992. doi: 10.1038/sj.ejhg.5201264. [DOI] [PubMed] [Google Scholar]
- 58.Mertz L.G.B., Thaulov P., Trillingsgaard A., Christensen R., Vogel I., Hertz J.M., Østergaard J.R. Neurodevelopmental outcome in Angelman syndrome: Genotype–phenotype correlations. Res. Dev. Disabil. 2014;35:1742–1747. doi: 10.1016/j.ridd.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 59.Burnside R.D., Pasion R., Mikhail F.M., Carroll A.J., Robin N.H., Youngs E.L., Gadi I.K., Keitges E., Jaswaney V.L., Papenhausen P.R., et al. Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: A susceptibility region for neurological dysfunction including developmental and language delay. Qual. Life Res. 2011;130:517–528. doi: 10.1007/s00439-011-0970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams D., Roche L., Heussler H. Parent perceptions, beliefs, and fears around genetic treatments and cures for children with Angelman syndrome. Am. J. Med. Genet. Part A. 2020;182:1716–1724. doi: 10.1002/ajmg.a.61631. [DOI] [PubMed] [Google Scholar]
- 61.Moncla A., Malzac P., Livet M., Voelckel M., Mancini J., Delaroziere J.C., Philip N., Mattei J.F. Angelman syndrome resulting from UBE3A mutations in 14 patients from eight families: Clinical manifestations and genetic counselling. J. Med. Genet. 1999;36:554–560. [PMC free article] [PubMed] [Google Scholar]
- 62.Peters S.U., Goddard-Finegold J., Beaudet A.L., Madduri N., Turcich M., Bacino C.A. Cognitive and adaptive behavior profiles of children with Angelman syndrome. Am. J. Med. Genet. 2004;128A:110–113. doi: 10.1002/ajmg.a.30065. [DOI] [PubMed] [Google Scholar]
- 63.Moncla A., Malzac P., Voelckel M.-A., Auquier P., Girardot L., Mattei M.-G., Philip N., Mattei J.-F., Lalande M., Livet M.-O. Phenotype–genotype correlation in 20 deletion and 20 non-deletion Angelman syndrome patients. Eur. J. Hum. Genet. 1999;7:131–139. doi: 10.1038/sj.ejhg.5200258. [DOI] [PubMed] [Google Scholar]
- 64.Luk H., Lo I.F. Angelman syndrome in Hong Kong Chinese: A 20 years’ experience. Eur. J. Med. Genet. 2016;59:315–319. doi: 10.1016/j.ejmg.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 65.Shaaya E.A., Grocott O.R., Laing O., Thibert R.L. Seizure treatment in Angelman syndrome: A case series from the Angelman Syndrome Clinic at Massachusetts General Hospital. Epilepsy Behav. 2016;60:138–141. doi: 10.1016/j.yebeh.2016.04.030. [DOI] [PubMed] [Google Scholar]
- 66.Samanta D. Epilepsy in Angelman syndrome: A scoping review. Brain Dev. 2021;43:32–44. doi: 10.1016/j.braindev.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heald M., Adams D., Walls E., Oliver C. Refining the Behavioral Phenotype of Angelman Syndrome: Examining Differences in Motivation for Social Contact Between Genetic Subgroups. Front. Behav. Neurosci. 2021;15:618271. doi: 10.3389/fnbeh.2021.618271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vendrame M., Loddenkemper T., Zarowski M., Gregas M., Shuhaiber H., Sarco D.P., Morales A., Nespeca M., Sharpe C., Haas K., et al. Analysis of EEG patterns and genotypes in patients with Angelman syndrome. Epilepsy Behav. 2012;23:261–265. doi: 10.1016/j.yebeh.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 69.Mertz L.G., Christensen R., Vogel I., Hertz J.M., Østergaard J.R. Eating behavior, prenatal and postnatal growth in Angelman syndrome. Res. Dev. Disabil. 2014;35:2681–2690. doi: 10.1016/j.ridd.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 70.Brennan M.-L., Adam M.P., Seaver L.H., Myers A., Schelley S., Zadeh N., Hudgins L., Bernstein J.A. Increased body mass in infancy and early toddlerhood in Angelman syndrome patients with uniparental disomy and imprinting center defects. Am. J. Med. Genet. Part A. 2015;167:142–146. doi: 10.1002/ajmg.a.36831. [DOI] [PubMed] [Google Scholar]
- 71.Horsthemke B., Buiting K. Imprinting defects on human chromosome 15. Cytogenet. Genome Res. 2006;113:292–299. doi: 10.1159/000090844. [DOI] [PubMed] [Google Scholar]
- 72.Goto M., Saito Y., Honda R., Saito T., Sugai K., Matsuda Y., Miyatake C., Takeshita E., Ishiyama A., Komaki H., et al. Episodic tremors representing cortical myoclonus are characteristic in Angelman syndrome due to UBE3A mutations. Brain Dev. 2015;37:216–222. doi: 10.1016/j.braindev.2014.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data are available for this review article.