Abstract
Simple Summary
We examined the relevance of immunoglobulin kappa C (IGKC), an important part of the humoral immune system, in early breast cancer. To our knowledge, our results confirm for the first time previous retrospective findings of a cancer recurrence protective role of IGKC in a large cohort of early breast cancer patients who were treated in the prospective, randomized FinHer clinical trial. We show that an increased amount of IGKC in the tumor is linked to longer distant metastasis-free survival, especially in patients whose breast cancer does not express hormone receptors or human epidermal growth factor receptor-2. This type of breast cancer often has poor prognosis. Since an improved outcome is associated with the presence of tumor-infiltrating IGKC expressing immune cells, this may be a further argument for the use of immunotherapies in these patients.
Abstract
We studied the prognostic impact of tumor immunoglobulin kappa C (IGKC) mRNA expression as a marker of the humoral immune system in the FinHer trial patient population, where 1010 patients with early breast cancer were randomly allocated to either docetaxel-containing or vinorelbine-containing adjuvant chemotherapy. HER2-positive patients were additionally allocated to either trastuzumab or no trastuzumab. Hormone receptor-positive patients received tamoxifen. IGKC was evaluated in 909 tumors using quantitative real-time polymerase chain reaction, and the influence on distant disease-free survival (DDFS) was examined using univariable and multivariable Cox regression and Kaplan–Meier estimates. Interactions were analyzed using Cox regression. IGKC expression, included as continuous variable, was independently associated with DDFS in a multivariable analysis also including age, molecular subtype, grade, and pT and pN stage (HR 0.930, 95% CI 0.870–0.995, p = 0.034). An independent association with DDFS was also found in a subset analysis of triple-negative breast cancers (TNBC) (HR 0.843, 95% CI 0.724–0.983, p = 0.029), but not in luminal (HR 0.957, 95% CI 0.867–1.056, p = 0.383) or HER2-positive (HR 0.933, 95% CI 0.826–1.055, p = 0.271) cancers. No significant interaction between IGKC and chemotherapy or trastuzumab administration was detected (Pinteraction = 0.855 and 0.684, respectively). These results show that humoral immunity beneficially influences the DDFS of patients with early TNBC.
Keywords: triple-negative breast cancer, prognosis, immune system, immunoglobulin kappa C
1. Introduction
During the last decade, numerous, largely retrospective, analyses showed that tumor-infiltrating lymphocytes or transcripts of immune cells play an important prognostic and predictive role in breast cancer. We have previously reported a strong beneficial prognostic impact of T cell as well as B cell metagenes for breast cancer prognosis [1]. The strong protective impact of a B cell/plasma cell signature were later confirmed by others [2,3]. Tumor-infiltrating plasmablasts and plasma cells were identified as the source of immunoglobulin kappa C (IGKC) expression using confocal microscopy [4]. In this study, co-staining with anti-human IgG showed that IGKC was expressed in IgG-positive cells, a well-known feature of B cell maturation and plasma cell differentiation after antigen encounter. IGKC was associated with favorable prognosis in patients who had not been treated with systemic therapy, and also with response to anthracycline-containing neoadjuvant chemotherapy in early breast cancer. Indeed, Gentles et al. confirmed that plasma cell signatures and also plasma cells expressing IGKC were associated with improved survival in a comprehensive analysis of the prognostic landscape of genes and infiltrating immune cells across human cancers [3]. Taken together, these and other results suggest that humoral immunity might be as important as cellular immunity in eliminating cancer [5]. For example, B cell-attracting C-X-C motif chemokine ligand 13 (CXCL13)-positive, CD4-positive T follicular helper T (Tfh) cells were independently associated with distant disease-free survival (DDFS) in patients with triple-negative breast cancer (TNBC) in the FinHer trial patient population [6]. Recently, Garaud and co-workers examined tumor-infiltrating B cells (TIL-B) in TNBC from the BIG 02-98 clinical trial and showed a correlation between Tfh TILs and antibody secretion [7].
Even though Tfh cells play an important role in humoral immune responses, antibody-secreting immune cells are the most obvious and definite proof of humoral immunity. A strong association between IGKC, tumor-associated plasma cell infiltration and improved prognosis was found by Yeong and co-workers in a retrospective cohort of 269 TNBC samples [8]. However, all of these results were obtained analyzing retrospectively collected breast cancer samples, which carries the risk of a selection bias. Thus, in order to avoid the potential biases that may arise in such retrospective series, we studied the prognostic impact of IGKC in the patient population of the prospective FinHer trial that evaluated the inclusion of vinorelbine and trastuzumab to the standard adjuvant treatments in early breast cancer [9]. To our knowledge, the results presented here are the first to confirm an independent prognostic impact of IGKC in patients with early TNBC in an exploratory analysis of a large randomized trial.
2. Materials and Methods
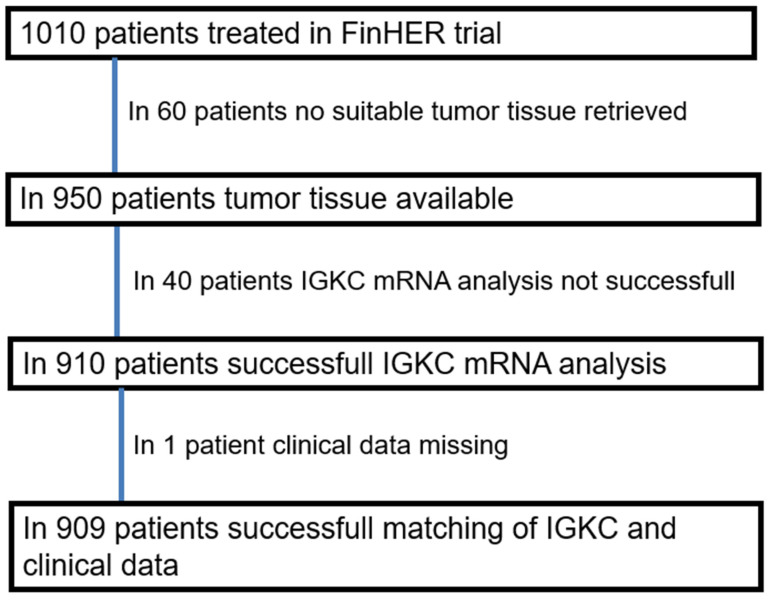
In the FinHer trial (International Standard Randomized Controlled Trial number, ISRCTN76560285) 1010 node-positive or high-risk node-negative breast cancer patients were randomized. Patients received either three cycles of adjuvant docetaxel or vinorelbine, followed by three cycles of fluorouracil, epirubicin, and cyclophosphamide [9]. Patients with human epidermal growth factor receptor 2 (HER2)-positive cancer were additionally randomized between nine trastuzumab infusions administered at one-week intervals concomitantly with chemotherapy (either with vinorelbine or docetaxel) and no trastuzumab. Steroid hormone receptor-positive patients received tamoxifen. The patients were recruited from 17 study centers from October 2000 to September 2003. One patient was excluded from the analysis due to presence of overt distant metastases already at the time of study entry (Figure 1).
Figure 1.
CONSORT diagram showing patient selection for the study.
Ethical approval was obtained from an ethics committee at the Helsinki University Central Hospital. Study participants provided signed informed consent to allow further research analyses on their tumor tissue. This exploratory biomarker study is reported according to the REMARK (Reporting Recommendations for Tumor Marker Prognostic Studies) criteria [10]. The characteristics of the patients and the tumors are shown in Table 1.
Table 1.
Association of IGKC mRNA content in the tumor with clinicopathological characteristics. IGKC low was defined as <median (n = 452) and IGKC high as ≥median (n = 457). The p value from Fisher’s exact test is given. IGKC, immunoglobulin kappa C; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; TNBC, triple negative breast cancer.
| Characteristic | N | IGKC Low n (%) | IGKC High n (%) | p | |
|---|---|---|---|---|---|
| Age | <50 years | 399 | 195 (49) | 204 (51) | |
| ≥50 years | 510 | 257 (50) | 253 (50) | 0.688 | |
| Grade 1 | Grade I | 133 | 73 (55) | 60 (45) | |
| Grade II | 365 | 202 (55) | 163 (45) | ||
| Grade III | 371 | 158 (43) | 213 (57) | 0.001 | |
| ER status | Positive | 250 | 93 (37) | 157 (63) | |
| Negative | 659 | 359 (54) | 300 (46) | <0.001 | |
| PR status 1 | Positive | 382 | 160 (42) | 222 (58) | |
| Negative | 526 | 292 (56) | 234 (44) | <0.001 | |
| HER2 status | Positive | 706 | 367 (52) | 339 (48) | |
| Negative | 203 | 85 (41) | 118 (59) | 0.013 | |
| Ki67 1 | ≤20% | 412 | 236 (57) | 176 (43) | |
| >20% | 397 | 168 (42) | 229 (58) | <0.001 | |
| Molecular subtype | Luminal | 574 | 321 (56) | 253 (44) | |
| TNBC | 132 | 46 (35) | 86 (65) | ||
| HER2+ | 203 | 85 (41) | 118 (59) | <0.001 | |
| pT stage 1 | pT1 | 381 | 189 (50) | 192 (50) | |
| pT2 | 447 | 226 (51) | 221 (49) | ||
| pT3 or pT4 | 80 | 36 (45) | 44 (55) | 0.356 | |
| pN stage | pN0 | 96 | 42 (44) | 54 (56) | |
| pN1 | 786 | 392 (50) | 394 (50) | ||
| pN2 or pN3 | 27 | 18 (67) | 9 (33) | 0.113 | |
| Breast surgery | Mastectomy | 540 | 260 (48) | 280 (52) | |
| Breast conserving | 369 | 192 (52) | 177 (48) | 0.252 | |
| Lymph node dissection 1 |
Axillary dissection | 877 | 435 (50) | 442 (50) | |
| Sentinel node | 31 | 16 (52) | 15 (48) | 0.857 | |
1 Missing values: grade N = 40, PR status N = 1, Ki67 N = 100, pT stage N = 1.
Determination of steroid hormone receptor status and HER2 expression by immunohistochemistry (IHC) was performed according to the guidelines of each institution [9]. When HER2 expression was scored 2+ or 3+, the number of copies of HER2 was centrally confirmed by means of chromogenic in situ hybridization (CISH) [11]. The cancers were considered hormone receptor positive when ≥10% of cancer cells expressed estrogen receptor (ER) and/or progesterone receptor PR). Patients with ER-positive or PR-positive tumor were scheduled to receive 5 years of tamoxifen. Breast cancer subtypes were classified using IHC as previously described as either luminal (ER+ and/or PR+, HER2−), HER2-positive (HER2+, irrespective of the steroid hormone receptor status), or TNBC (ER−, PR−, HER2−) [12].
Total RNA was extracted from 5-μm thick tumor formalin-fixed paraffin-embedded (FFPE) tissue sections with ≥30% of the section surface area consisting of cancer as previously described [6]. In 950 (94.1%) out of the 1010 cases, tumor tissue was available for RNA extraction. IGKC could be successfully analyzed from 910 (90.0%) tumors. After xylene-free deparaffinization driven by heat, the RNA extraction was done using a commercially available kit (XTRAKT-R02; STRATIFYER, Cologne, Germany). The DNase digestion was done using Ambion RNase-free DNase I. The same quality measures were applied as were previously used for the successful clinical development of RNXtract® and MammaTyper® IVD [13,14]. Each extraction was checked for mRNA quantity and amplificability by using clinically developed reference genes demonstrating sufficient fragment length of total RNA extract for the amplification. Samples with low mRNA content (raw CT value of CALM2 > 30) were excluded from analysis. DNA content after DNase digestion was checked by PCR using DNA specific, intron-spanning PAEP gene as quality control gene [15]. Assay performance and specificity of the IGKC assay was validated by using commercially available universal human reference RNA (Stratagene, La Jolla, CA, USA) and human genomic DNA (Roche Diagnostics, Mannheim, Germany). The RNA quality was assessed by the ability to amplify a 65 bp fragment of RPL37A and the Cq value was used as a surrogate marker for the mRNA yield, as described in Bohmann et al. [16]. One-step qRT-PCR to measure tumor IGKC content was done using a custom-designed gene-specific TaqMan-based assay. IGKC and the reference gene CALM2 mRNA content were assessed in triplicates using the SuperScript III Platinum One-Step Quantitative RT-PCR System with ROX (Invitrogen, Karlsruhe, Germany) in a Versant kPCR system (Siemens, Erlangen, Germany). The thermal profile included 30 min at 50 °C, 20.5 min at 8 °C, and 2 min at 95 °C followed by 40 cycles of 15 s at 95 °C, and 30 s at 60 °C. Forty amplification cycles were applied and the cycle quantification threshold (Ct) values of IGKC and one reference gene for each sample were estimated as the median of the triplicate measurements. The primer and probe sequences used for IGKC mRNA quantification were for the probe AGCAGCCTGCAGCCTGAAGATTTTGC, the forward primer was GATCTGGGACAGAATTCACTCTCA and the reverse primer GCCGAACGTCCAAGGGTAA. For CALM2, the probe TCGCGTCTCGGAAACCGGTAGC, forward primer GAGCGAGCTGAGTGGTTGTG and reverse primer TGTGGTTCCTGCATGAAGACA were used. For RPL37A, the probe TGGCTGGCGGTGCCTGGA, forward primer TGTGGTTCCTGCATGAAGACA and reverse primer GTGACAGCGGAAGTGGTATTGTAC were used. The relative mRNA expression levels of IGKC were calculated as delta cycle threshold (ΔCt) values ΔCt = 40 − [Ct GOI (mean of gene of interest) − Ct REF (mean of CALM2)]. The final expression values were generated by subtracting ΔCT from the total number of cycles (40 – ΔCT) to ensure that the normalized gene expression obtained by the test was proportional to the corresponding mRNA expression.
The primary objective of this explorative study was to evaluate the association of tumor IGKC content with distant disease-free survival (DDFS), which was the survival endpoint in the final analysis of the FinHer trial [9]. The secondary objectives were to study the influence of IGKC expression in defined breast cancer molecular subtypes (i.e., luminal, HER2-positive, triple-negative), and the associations with the type of adjuvant therapy administered. DDFS was defined as the time interval between the date of randomization and the date of first cancer recurrence outside of the ipsilateral locoregional region or the date of death, whenever death occurred before distant recurrence. Patients alive without documented evidence of distant metastases were censored at the time of the latest contact. DDFS rates were determined using the Kaplan-Meier analyses. The log-rank test was used to compare survival between groups. We stratified the samples using the median as well as the top quartile IGKC mRNA expression as the cut-off value for each molecular subgroup; 66 triple-negative cancers had high and 66 low IGKC expression (median, 35.76), 289 luminal cancers had high and 285 low IGKC expression (median, 34.29), and 102 HER2-positive cancers had high and 101 low IGKC expression (median, 35.07). Frequency tables were analyzed using the Fisher’s exact test.
Univariable and multivariable Cox proportional hazards models were fitted to investigate the association of tumor IGKC expression as a continuous variable, as well as dichotomized using either the median or the top quartile as cutoff, with DDFS for each of the three molecular subtypes separately as well as combined. Other co-variables in the multivariate models were age at the time of study entry (≤50 vs. >50 years), breast tumor size (pT1 vs. pT2-4), axillary nodal status (pN0 vs. pN1-3), histological grade of differentiation (grade I vs. II-III for the entire cohort and the luminal subgroup, and I + II vs. III for the TNBC- and HER2-positive subgroups, respectively, due to the small number of grade I tumors in these subgroups), HER2 status (positive vs. negative), ER status (positive vs. negative), PR status (positive vs. negative), and molecular subtype (luminal vs. HER2-positive vs. triple-negative). A Cox proportional hazards model containing the treatment group (docetaxel vs. vinorelbine, or trastuzumab vs. no trastuzumab when the tumor was HER2-positive), IGKC expression (high [≥median] vs. low [<median]), and the treatment-by-biomarker interaction term was used to study the potential interactions between tumor IGKC content and the treatment assigned.
All p values were two-sided, and p < 0.05 was considered statistically significant. All analyses were explorative and not adjusted for multiple testing. Therefore, p values should be interpreted with caution and in connection with the effect estimates. Statistical analyses were performed using the Statistical Package for Social Science (SPSS) (SPSS Inc., version 27, Chicago, IL, USA) and the statistical programming language R version 4.0.3.
3. Results
3.1. IGKC mRNA Expression Depends on the Molecular Subtype
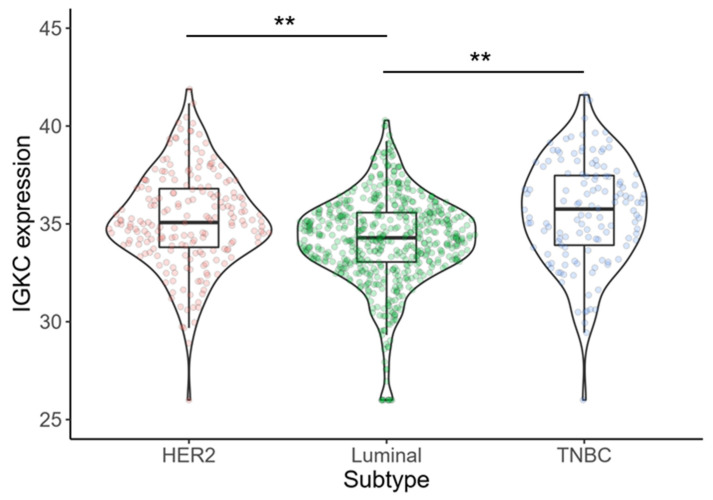
Of the 909 cancers, 574 (63.1%) were luminal, 203 (22.3%) HER2-positive, and 132 (14.5%) triple-negative. IGKC expression was significantly associated with molecular subtype (TNBC vs. luminal, p < 0.001; TNBC vs. HER2-positive, p = 0.327; HER2-positive vs. luminal, p < 0.001). Triple-negative cancers showed the highest expression of IGKC (median 35.76; interquartile range [IQR] 33.89–37.52) followed by HER2-positive tumors (median 35.07; IQR 33.73–36.82) and luminal cancers (median 34.29; IQR 33.05–35.58) (Figure 2).
Figure 2.
Statistically significant association of IGKC expression and molecular subtype (p < 0.001; one-way ANOVA). Triple-negative cancers and HER2-positive cancers show higher expression of IGKC mRNA compared to luminal cancers. ** p < 0.001.
3.2. IGKC Expression Is Associated with Adverse Histopathological Characteristics
First, we examined an association between cancer IGKC expression (<median vs. ≥median) and clinicopathological characteristics using Fisher’s exact test. High levels of IGKC were significantly associated with poor histological grade of differentiation (p < 0.001), negative ER (p < 0.001) and PR (p < 0.001) status, positive HER2 status (p = 0.013), high (>20%) Ki-67 expression (p < 0.001), and HER2-positive subtype as well as TNBC molecular subtype (p < 0.001). Tumor IGKC mRNA content was not associated with age at diagnosis (p = 0.688), tumor size (p = 0.356), axillary nodal status (p = 0.113), surgery of the breast (p = 0.252) or lymph node dissection (p = 0.857) (Table 1). Radiotherapy was given according to each institution’s guidelines; it was mandated after breast-conserving surgery and was given to most patients with node-positive disease.
3.3. Association between IGKC Expression and Distant Disease-Free Survival
In a univariable Cox analysis, tumor IGKC expression did not show a significant association with DDFS (HR 0.982, 95% CI 0.920–1.048, p = 0.589); whereas, the standard prognostic factors were as expected significantly associated with DDFS (Table S1). In a multivariable analysis of the whole series, IGKC was significantly associated with DDFS (HR 0.930, 95% CI 0.870–0.995, p = 0.034) when included as continuous variable (Table 2). Furthermore, in the multivariable analysis, cancer ER expression, PR expression, HER2 expression, Ki-67 expression, pT stage, pN stage, histological grade of differentiation, and the molecular subtypes were also independently associated with DDFS. When we instead stratified the patients using the median IGKC mRNA expression as the cut-off value, IGKC did not reach independent significance (HR 0.815, 95% CI 0.580–1.145, p = 0.238) (Table S2a). However, using the upper quartile, IGKC retained its independent significance for DDFS (HR 0.619, 95% CI 0.410–0.934, p = 0.022) (Table S2b).
Table 2.
Multivariable Cox analysis for DDFS. 868 patients had complete data for all variables and were included in the multivariable analysis. IGKC expression was included as a continuous variable. The results of the univariable analysis are shown in Table S1. CI, confidence interval; IGKC, immunoglobulin kappa C; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer.
| Variable | Hazard Ratio | 95% CI | p |
|---|---|---|---|
| IGKC | 0.930 | 0.870–0.995 | 0.034 |
| Age | |||
| ≤50 years | 1.000 | ||
| >50 years | 0.899 | 0.643–1.256 | 0.532 |
| Molecular subtype | |||
| Luminal | 1.000 | ||
| TNBC | 2.661 | 1.667–4.249 | <0.001 |
| HER2+ | 2.122 | 1.425–3.159 | <0.001 |
| pT stage | |||
| pT1 | 1.000 | ||
| pT2-4 | 1.605 | 1.111–2.319 | 0.012 |
| pN stage | |||
| pN0 | 1.000 | ||
| pN1-3 | 4.369 | 1.993–9.581 | <0.001 |
| Grade | |||
| Grade I | 1.000 | ||
| Grade II–III | 2.871 | 1.235–6.675 | 0.014 |
3.4. Significance of IGKC in Triple-Negative Breast Cancer
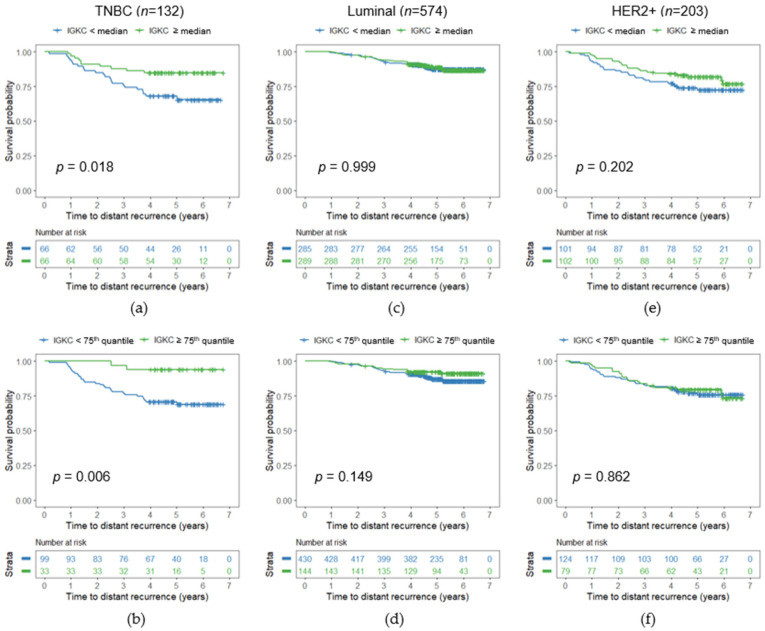
We next analyzed the association between IGKC expression and DDFS in each molecular subtype separately. In TNBC, IGKC was not significantly associated with DDFS in the univariable Cox analysis when included as a continuous variable (HR 0.907, 95% CI 0.806–1.022, p = 0.109) (Table 3). However, when using the median as a cut-off, high IGKC expression was associated with favorable DDFS (HR 0.418, 95% CI 0.198–0.882, p = 0.022) (Table S3a) (Figure 3a). This association was even stronger when the top quartile was used to stratify the patients into groups with high and low IGKC expression (HR 0.172, 95% CI 0.041–0.719, p = 0.016) (Table S3b) (Figure 3b).
Table 3.
Univariable and multivariable Cox analyses for DDFS in patients with TNBC. 132 patients had data for IGKC and were included in the univariable analysis. 129 patients had complete data for all variables and were included in the multivariable analysis. IGKC expression was included as a continuous variable. The corresponding results for IGKC dichotomized using the median or top-quartile expression as a cutoff are shown in Table S3. CI, confidence interval; IGKC, immunoglobulin kappa C; HER2, human epidermal growth factor receptor 2.
| Variable | Univariable Analysis | Multivariable Analysis | ||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | p | Hazard Ratio | 95% CI | p | |
| IGKC | 0.907 | 0.806–1.022 | 0.109 | 0.843 | 0.724–0.983 | 0.029 |
| Age | ||||||
| ≤50 years | 1.000 | 1.000 | ||||
| >50 years | 0.427 | 0.209–0.874 | 0.020 | 0.319 | 0.143–0.708 | 0.005 |
| pT stage | ||||||
| pT1 | 1.000 | 1.000 | ||||
| pT2-4 | 0.839 | 0.388–1.813 | 0.654 | 1.421 | 0.610–3.310 | 0.416 |
| pN stage | ||||||
| pN0 | 1.000 | 1.000 | ||||
| pN1-3 | 2.796 | 1.076–7.262 | 0.035 | 4.025 | 1.327–12.209 | 0.014 |
| Grade 1 | ||||||
| Grade I-II 2 | 1.000 | 1.000 | ||||
| Grade III | 0.642 | 0.286–1.442 | 0.283 | 0.778 | 0.332–1.823 | 0.563 |
1n = 129; 2 grade was dichotomized as I-II vs. III since there was only one TNBC patient with a grade I tumor.
Figure 3.
Influence of cancer IGKC expression on DDFS illustrated by Kaplan–Meier plots for (a,b) triple-negative (TNBC), (c,d) luminal, and (e,f) HER2+ cancers in the FinHer trial, with IGKC dichotomized as low (<median of the corresponding subtype) and high (≥ median of the corresponding subtype) (upper panels) as well as dichotomized as low (<75th quantile of the corresponding subtype) and high (≥75th quantile of the corresponding subtype) (lower panels). The p-value from the log-rank test is given in the figure.
In the multivariable analysis of TNBC, adjusted for the key prognostic clinical parameters age, pT stage, pN stage, and grade, IGKC expression was independently associated with DDFS as a continuous variable (HR 0.843, 95% CI 0.724–0.983, p = 0.029) (Table 3) and also when using the median (HR 0.322, 95% CI 0.146–0.712, p = 0.005) or top quartile (HR 0.197, 95% CI 0.045–0.852, p = 0.030) as cutoff to define high and low IGKC expression (Table S3). IGKC expression was not significantly associated with DDFS in the uni- or multivariate analysis in the luminal subtype (Table S4) (Figure 3c,d) nor in the HER2-positive subtype (Table S5) (Figure 3e,f).
3.5. Cancer IGKC mRNA Content Shows No Association with Systemic Treatment Effects
An examination of the potential interactions between IGKC expression and the systemic treatments given with DDFS revealed no significant interaction with the type of chemotherapy or whether or not trastuzumab was administered (Pinteraction = 0.855 and 0.684, respectively).
4. Discussion
In this retrospective analysis of a prospective trial, breast cancer IGKC mRNA content was associated with DDFS in the total cohort of patients treated in the FinHer trial in multivariable but not in univariable analysis. When we analyzed the molecular subtypes separately, cancer IGKC content was independently associated with longer DDFS in TNBC, while it was not significant in luminal or HER2-positive breast cancer.
The lack of an association between cancer IGKC expression and survival differs from our previous study in cohorts of node-negative breast cancer patients who did not receive systemic therapies [4]. A possible explanation for this is that the prognostic effect of cancer IGKC content may be obscured by the effects of the systemic cancer treatments administered, since all hormone receptor-positive patients in the FinHer trial received chemotherapy followed by tamoxifen. Besides chemotherapy, tamoxifen may also have immune-modulatory effects, potentially interfering with the prognostic effects of tumor-infiltrating immune cells, as anticipated when we developed a prognostic gene expression signature for ER-positive, HER2-negative patients treated with adjuvant endocrine therapy (EndoPredict™) [17]. In fact, not a single gene from our previously defined B cell metagene was selected for the EndoPredict™ test [17]. In breast cancer patients who did not receive adjuvant systemic therapy, a strong prognostic effect of a B cell metagene including IGKC was seen in patients with rapidly proliferating, node-negative, ER-positive, HER2-negative breast cancer [1]. In contrast, this prognostic effect was not observed in the present study investigating IGKC in ER-positive, HER2-negative patients treated with adjuvant tamoxifen.
Moreover, we recently reported a significant interaction between the prognostic impact of IGKC and tamoxifen treatment in a retrospective and non-randomized analysis of patients treated more than two decades ago with adjuvant chemotherapy with or without tamoxifen [18]. The positive prognostic impact of high cancer IGKC expression was most pronounced in patients who did not receive tamoxifen as endocrine treatment compared with those breast cancer patients treated with tamoxifen. The interaction test confirmed a significant interaction between tamoxifen treatment and the prognostic impact of IGKC expression (Pinteraction = 0.04). Indeed, there is a growing body of evidence demonstrating immunomodulatory effects of tamoxifen [19]. These authors proposed that tamoxifen leads to a shift away from Th1 to Th2 immunity. In addition, Li and co-workers profiled differentially expressed intratumoral cytokines as a signature to evaluate the immune-polarizing side effects of tamoxifen. They could show that patients with low immune-polarizing side effects of tamoxifen (low Th2 polarization) had a lower risk of distant metastasis in a cohort of 608 breast cancer patients. In addition, in vitro data revealed that tamoxifen impaired differentiation of dendritic cells and reduced their immunostimulatory capacity [20]. These authors even speculated that tamoxifen may depress immunity and potentially interfere with immunotherapeutic strategies to improve antitumor immunity in breast cancer patients. Furthermore, a recent exploratory analysis of a prospective trial showed that CD8+ TILs have a considerably stronger prognostic impact in ER-positive patients not treated with tamoxifen compared to patients treated with tamoxifen [21]. The interaction test between CD8 status and tamoxifen treatment for relapse-free interval showed a trend (Pinteraction = 0.082).
Taken together, the proposed interaction between tamoxifen and the immune response may explain why IGKC loses its prognostic relevance in patients treated with tamoxifen. On the other hand, one has to consider the relationship of immune-related markers such as IGKC with trastuzumab. Currently, it is well accepted that the host immune system contributes significantly to trastuzumab efficacy [22]. Supporting this, the baseline percentage of TILs was not only associated with pathologic complete response (pCR) but also provided independent prognostic information in patients treated with trastuzumab/pertuzumab-based neoadjuvant chemotherapy [23]. Conversely, Loi and co-workers failed to find a significant association between TILs and distant disease-free survival of HER2-positive patients in the FinHer study [24]. This is in line with our findings for IGKC in HER2-positive FinHer patients. Instead, they detected a statistically significant interaction between higher TILs and increased trastuzumab benefit in HER2-positive disease (DDFS Pinteraction = 0.025). However, this positive association between TILs and benefit from trastuzumab is disputable. Perez et al. reported conflicting results with respect to an association between TILs and trastuzumab benefit in HER2-positive patients randomized to trastuzumab or no trastuzumab within the N9831 trial [25]. A meta-analysis in early HER2-positive breast cancer showed that high baseline TILs were associated with increased pCR probability [26]. However, this meta-analysis failed to show an interaction between TILs and response to trastuzumab. Consistent with these results, we could not detect a significant interaction of IGKC and trastuzumab benefit.
Using the B cell/plasma cell associated transcript IGKC, we could confirm the beneficial prognostic effects of tumor-infiltrating immune cells in TNBC. Our results in this particular molecular subtype support previous findings [24,27,28,29,30] of an independent prognostic association of TILs with improved survival in TNBC. Considering that the total mutational burden is highest in TNBC, this significant association of tumor-infiltrating immune cells and TNBC is not surprising [31]. In addition, these authors described that the mutational burden was highly correlated with the neoepitope load (R2 = 0.86). A higher neoepitope load renders immunotherapy more efficacious. Indeed, clinical results using immune checkpoint inhibition with monoclonal antibodies against programmed cell death protein 1 (PD-1) or its ligand programmed cell death 1 ligand 1 (PD-L1) in patients with early as well as advanced TNBC showed encouraging results [32,33,34,35]. Conversely, and consistent with our findings of a lesser role for the immune system in luminal breast cancer, a recent randomized trial in advanced hormone receptor-positive patients showed that the addition of the PD-1 antibody pembrolizumab to eribulin did not improve survival compared with eribulin alone [36].
A potential limitation of our study is that it is exploratory and that the different molecular subtypes received different therapies in addition to the randomized adjuvant chemotherapy (e.g., tamoxifen in hormone receptor-positive and trastuzumab in HER2-positive subtypes, respectively) potentially obscuring the prognostic effect of IGKC in these molecular subtypes. A strength of the study, however, is that we report the prognostic significance of IGKC in a large, randomized trial with approximately 90% of the formalin-fixed, paraffin-embedded tumor tissue available for the IGKC mRNA analysis.
5. Conclusions
We confirm the independent prognostic significance of cancer IGKC content in a prospective-retrospective study. A multivariate analysis supported the favorable DDFS associated with high cancer IGKC content in the subset of TNBC patients treated in the FinHer trial. This highlights the importance of the humoral immune system in early TNBC.
Acknowledgments
We are grateful to all patients who participated in the FinHer study. We thank Rosemarie Marchan for critical reading of the manuscript and language editing.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13143626/s1, Table S1: Univariable Cox analysis for DDFS; Table S2: Multivariable Cox analysis for DDFS; Table S3: Uni- and multivariable Cox analysis for DDFS in the subset of patients with TNBC; Table S4: Uni- and multivariable Cox analysis for DDFS in the subset of patients with luminal cancer; Table S5: Uni- and multivariable Cox analysis for DDFS in the subset of patients with HER2-positive cancer.
Author Contributions
Conceptualization, M.S., K.E., H.J., J.G.H. and R.M.W.; data curation, K.E., M.S., H.J., J.G.H., J.R., A.-S.H. and R.M.W.; formal analysis, K.E., M.S., H.J., J.G.H., J.R., A.-S.H. and R.M.W.; funding acquisition, J.G.H., R.M.W., H.J. and M.S.; investigation, M.S., K.E., J.G.H., R.M.W., H.J. and P.-L.K.-L.; methodology, M.S., K.E., J.G.H., R.M.W., H.J. and P.-L.K.-L.; project administration, M.S., R.M.W. and H.J.; writing—original draft, M.S., K.E., H.J., J.G.H. and R.M.W.; writing—review and editing, M.S., K.E., J.G.H., A.-S.H., K.A., S.K., A.L., M.J.B., W.B., A.H., J.R., M.G., P.-L.K.-L., R.M.W. and H.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Federal Ministry of Education and Research (BMBF, NGFN project Oncoprofile, number 01GR0816). The funder had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of the Helsinki University Central Hospital (Ethics Committee approval number HUS 124/13/03/02/2014, approved on 9 April 2014).
Informed Consent Statement
Written informed consent was obtained from all subjects involved in the study to allow research assays to be carried out on their tumor tissue.
Data Availability Statement
The dataset analyzed during the current study is available from the corresponding author on reasonable request.
Conflicts of Interest
M.S. reports personal fees from AstraZeneca, Eisai, Lilly, MSD, Novartis, Pantarhei Bioscience, Pfizer, Roche, and SeaGen outside the submitted work. Institutional research funding from AstraZeneca, BioNTech, Eisai, Genentech, German Breast Group, Novartis, Palleos, Pantarhei Bioscience, Pierre-Fabre, and Roche. Travel reimbursement from Pfizer and Roche. In addition, M.S. is named inventor on patent EP 2390370 B1 and patent EP 2951317 B1 issued. K.A. reports personal fees from AstraZeneca, Pfizer, Roche. M.J.B. received honorarium and travelling expenses from Roche Pharma AG, Tesaro Bio GmbH, Glaxo Smith Kline, Clovis Oncology, Astra Zeneca, Pharma Mar GmbH, Celgene, Pierre Fabre Pharma GmbH. S.K. received speaker honoraria from Roche Pharma AG. He received research funding from Novartis and travel reimbursement from PharmaMar. A.H. reports personal fees from AstraZeneca, Celgen, GSK, LEO Pharma, MedConcept GmbH, Med update GmbH, Medicultus, PharmaMar, Pfizer, Promedicis GmbH, Softconsult, Roche Pharma AG, Streamedup! GmbH, Tesaro Bio Germany GmbH. M.G. is named inventor on patents for breast cancer prognosis and prediction (EP1954830 B1, EP2066805 B1, EP2390370 B1, EP2553119 B1, EP2737081 B1, EP2845911 B1 and EP 2951317 B1. P.-L.K.-L. received honorarium and travelling expenses from BMS and Sanofi. R.M.W. is founder & employee of STRATIFYR Molecular Pathology. H.J. is the Chair of the Scientific Advisory Board at Orion Pharma and at Neutron Therapeutics Ltd. and owns stock of Orion Pharma and Sartar Therapeutics. The other authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schmidt M., Bohm D., von Torne C., Steiner E., Puhl A., Pilch H., Lehr H.-A., Hengstler J.G., Kolbl H., Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 2.Bianchini G., Qi Y., Alvarez R.H., Iwamoto T., Coutant C., Ibrahim N.K., Valero V., Cristofanilli M., Green M.C., Radvanyi L., et al. Molecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancers. J. Clin. Oncol. 2010;28:4316–4323. doi: 10.1200/JCO.2009.27.2419. [DOI] [PubMed] [Google Scholar]
- 3.Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D., Nair V.S., Xu Y., Khuong A., Hoang C.D., et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt M., Hellwig B., Hammad S., Othman A., Lohr M., Chen Z., Boehm D., Gebhard S., Petry I., Lebrecht A., et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin kappa C as a compatible prognostic marker in human solid tumors. Clin. Cancer Res. 2012;18:2695–2703. doi: 10.1158/1078-0432.CCR-11-2210. [DOI] [PubMed] [Google Scholar]
- 5.Whiteside T.L., Ferrone S. For breast cancer prognosis, immunoglobulin kappa chain surfaces to the top. Clin. Cancer Res. 2012;18:2417–2419. doi: 10.1158/1078-0432.CCR-12-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt M., Weyer-Elberich V., Hengstler J.G., Heimes A.-S., Almstedt K., Gerhold-Ay A., Lebrecht A., Battista M.J., Hasenburg A., Sahin U., et al. Prognostic impact of CD4-positive T cell subsets in early breast cancer: A study based on the FinHer trial patient population. Breast Cancer Res. 2018;20:15. doi: 10.1186/s13058-018-0942-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garaud S., Buisseret L., Solinas C., Gu-Trantien C., de Wind A., van den Eynden G., Naveaux C., Lodewyckx J.-N., Boisson A., Duvillier H., et al. Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI Insight. 2019;5 doi: 10.1172/jci.insight.129641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeong J., Lim J.C.T., Lee B., Li H., Chia N., Ong C.C.H., Lye W.K., Putti T.C., Dent R., Lim E., et al. High Densities of Tumor-Associated Plasma Cells Predict Improved Prognosis in Triple Negative Breast Cancer. Front. Immunol. 2018;9:1209. doi: 10.3389/fimmu.2018.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joensuu H., Bono P., Kataja V., Alanko T., Kokko R., Asola R., Utriainen T., Turpeenniemi-Hujanen T., Jyrkkio S., Moykkynen K., et al. Fluorouracil, epirubicin, and cyclophosphamide with either docetaxel or vinorelbine, with or without trastuzumab, as adjuvant treatments of breast cancer: Final results of the FinHer Trial. J. Clin. Oncol. 2009;27:5685–5692. doi: 10.1200/JCO.2008.21.4577. [DOI] [PubMed] [Google Scholar]
- 10.McShane L.M., Altman D.G., Sauerbrei W., Taube S.E., Gion M., Clark G.M. REporting recommendations for tumor MARKer prognostic studies (REMARK) Breast Cancer Res. Treat. 2006;100:229–235. doi: 10.1007/s10549-006-9242-8. [DOI] [PubMed] [Google Scholar]
- 11.Joensuu H., Kellokumpu-Lehtinen P.-L., Bono P., Alanko T., Kataja V., Asola R., Utriainen T., Kokko R., Hemminki A., Tarkkanen M., et al. Adjuvant docetaxel or vinorelbine with or without trastuzumab for breast cancer. N. Engl. J. Med. 2006;354:809–820. doi: 10.1056/NEJMoa053028. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z., Gerhold-Ay A., Gebhard S., Boehm D., Solbach C., Lebrecht A., Battista M., Sicking I., Cotarelo C., Cadenas C., et al. Immunoglobulin kappa C predicts overall survival in node-negative breast cancer. PLoS ONE. 2012;7:e44741. doi: 10.1371/journal.pone.0044741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laible M., Schlombs K., Kaiser K., Veltrup E., Herlein S., Lakis S., Stöhr R., Eidt S., Hartmann A., Wirtz R.M., et al. Technical validation of an RT-qPCR in vitro diagnostic test system for the determination of breast cancer molecular subtypes by quantification of ERBB2, ESR1, PGR and MKI67 mRNA levels from formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer. 2016;16:398. doi: 10.1186/s12885-016-2476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varga Z., Lebeau A., Bu H., Hartmann A., Penault-Llorca F., Guerini-Rocco E., Schraml P., Symmans F., Stoehr R., Teng X., et al. An international reproducibility study validating quantitative determination of ERBB2, ESR1, PGR, and MKI67 mRNA in breast cancer using MammaTyper®. Breast Cancer Res. 2017;19:55. doi: 10.1186/s13058-017-0848-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kostadima L., Pentheroudakis G., Fountzilas G., Dimopoulos M., Pectasides D., Gogas H., Stropp U., Christodoulou C., Samantas E., Wirtz R., et al. Survivin and glycodelin transcriptional activity in node-positive early breast cancer: mRNA expression of two key regulators of cell survival. Breast Cancer Res. Treat. 2006;100:161–167. doi: 10.1007/s10549-006-9240-x. [DOI] [PubMed] [Google Scholar]
- 16.Bohmann K., Hennig G., Rogel U., Poremba C., Mueller B.M., Fritz P., Stoerkel S., Schaefer K.-L. RNA extraction from archival formalin-fixed paraffin-embedded tissue: A comparison of manual, semiautomated, and fully automated purification methods. Clin. Chem. 2009;55:1719–1727. doi: 10.1373/clinchem.2008.122572. [DOI] [PubMed] [Google Scholar]
- 17.Filipits M., Rudas M., Jakesz R., Dubsky P., Fitzal F., Singer C.F., Dietze O., Greil R., Jelen A., Sevelda P., et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 2011;17:6012–6020. doi: 10.1158/1078-0432.CCR-11-0926. [DOI] [PubMed] [Google Scholar]
- 18.Heimes A.-S., Krämer H., Härtner F., Almstedt K., Krajnak S., Battista M.J., Brenner W., Hasenburg A., Schmidt M. Prognostic Impact of Immunoglobulin Kappa C in Breast Cancer Patients Treated with Adjuvant Chemotherapy. Breast Care. 2020 doi: 10.1159/000511992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B., Li Y., Wang X.-Y., Yan Z.-Q., Liu H., Liu G.-R., Liu S.-L. Profile of differentially expressed intratumoral cytokines to predict the immune-polarizing side effects of tamoxifen in breast cancer treatment. Am. J. Cancer Res. 2015;5:726–737. [PMC free article] [PubMed] [Google Scholar]
- 20.Nalbandian G., Paharkova-Vatchkova V., Mao A., Nale S., Kovats S. The selective estrogen receptor modulators, tamoxifen and raloxifene, impair dendritic cell differentiation and activation. J. Immunol. 2005;175:2666–2675. doi: 10.4049/jimmunol.175.4.2666. [DOI] [PubMed] [Google Scholar]
- 21.Sobral-Leite M., Salomon I., Opdam M., Kruger D.T., Beelen K.J., van der Noort V., van Vlierberghe Ronald L.P., Blok E.J., Giardiello D., Sanders J., et al. Cancer-immune interactions in ER-positive breast cancers: PI3K pathway alterations and tumor-infiltrating lymphocytes. Breast Cancer Res. 2019;21:90. doi: 10.1186/s13058-019-1176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bianchini G., Gianni L. The immune system and response to HER2-targeted treatment in breast cancer. Lancet Oncol. 2014;15:68. doi: 10.1016/S1470-2045(13)70477-7. [DOI] [PubMed] [Google Scholar]
- 23.Ignatiadis M., van den Eynden G., Roberto S., Fornili M., Bareche Y., Desmedt C., Rothe F., Maetens M., Venet D., Holgado E., et al. Tumor-Infiltrating Lymphocytes in Patients Receiving Trastuzumab/Pertuzumab-Based Chemotherapy: A TRYPHAENA Substudy. J. Natl. Cancer Inst. 2019;111:69–77. doi: 10.1093/jnci/djy076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loi S., Michiels S., Salgado R., Sirtaine N., Jose V., Fumagalli D., Kellokumpu-Lehtinen P.-L., Bono P., Kataja V., Desmedt C., et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann. Oncol. 2014;25:1544–1550. doi: 10.1093/annonc/mdu112. [DOI] [PubMed] [Google Scholar]
- 25.Perez E.A., Ballman K.V., Tenner K.S., Thompson E.A., Badve S.S., Bailey H., Baehner F.L. Association of Stromal Tumor-Infiltrating Lymphocytes With Recurrence-Free Survival in the N9831 Adjuvant Trial in Patients with Early-Stage HER2-Positive Breast Cancer. JAMA Oncol. 2016;2:56–64. doi: 10.1001/jamaoncol.2015.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solinas C., Ceppi M., Lambertini M., Scartozzi M., Buisseret L., Garaud S., Fumagalli D., de Azambuja E., Salgado R., Sotiriou C., et al. Tumor-infiltrating lymphocytes in patients with HER2-positive breast cancer treated with neoadjuvant chemotherapy plus trastuzumab, lapatinib or their combination: A meta-analysis of randomized controlled trials. Cancer Treat. Rev. 2017;57:8–15. doi: 10.1016/j.ctrv.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Adams S., Gray R.J., Demaria S., Goldstein L., Perez E.A., Shulman L.N., Martino S., Wang M., Jones V.E., Saphner T.J., et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J. Clin. Oncol. 2014;32:2959–2966. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hida A.I., Watanabe T., Sagara Y., Kashiwaba M., Sagara Y., Aogi K., Ohi Y., Tanimoto A. Diffuse distribution of tumor-infiltrating lymphocytes is a marker for better prognosis and chemotherapeutic effect in triple-negative breast cancer. Breast Cancer Res. Treat. 2019;178:283–294. doi: 10.1007/s10549-019-05390-x. [DOI] [PubMed] [Google Scholar]
- 29.Ibrahim E.M., Al-Foheidi M.E., Al-Mansour M.M., Kazkaz G.A. The prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancer: A meta-analysis. Breast Cancer Res. Treat. 2014;148:467–476. doi: 10.1007/s10549-014-3185-2. [DOI] [PubMed] [Google Scholar]
- 30.Loi S., Sirtaine N., Piette F., Salgado R., Viale G., van Eenoo F., Rouas G., Francis P., Crown J.P.A., Hitre E., et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. 2013;31:860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 31.Narang P., Chen M., Sharma A.A., Anderson K.S., Wilson M.A. The neoepitope landscape of breast cancer: Implications for immunotherapy. BMC Cancer. 2019;19:200. doi: 10.1186/s12885-019-5402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cortes J., Cescon D.W., Rugo H.S., Nowecki Z., Im S.-A., Yusof M.M., Gallardo C., Lipatov O., Barrios C.H., Holgado E., et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): A randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. 2020;396:1817–1828. doi: 10.1016/S0140-6736(20)32531-9. [DOI] [PubMed] [Google Scholar]
- 33.Mittendorf E.A., Zhang H., Barrios C.H., Saji S., Jung K.H., Hegg R., Koehler A., Sohn J., Iwata H., Telli M.L., et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): A randomised, double-blind, phase 3 trial. Lancet. 2020;396:1090–1100. doi: 10.1016/S0140-6736(20)31953-X. [DOI] [PubMed] [Google Scholar]
- 34.Schmid P., Adams S., Rugo H.S., Schneeweiss A., Barrios C.H., Iwata H., Dieras V., Hegg R., Im S.-A., Shaw Wright G., et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 35.Schmid P., Cortes J., Pusztai L., McArthur H., Kümmel S., Bergh J., Denkert C., Park Y.H., Hui R., Harbeck N., et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020;382:810–821. doi: 10.1056/NEJMoa1910549. [DOI] [PubMed] [Google Scholar]
- 36.Tolaney S.M., Barroso-Sousa R., Keenan T., Li T., Trippa L., Vaz-Luis I., Wulf G., Spring L., Sinclair N.F., Andrews C., et al. Effect of Eribulin With or Without Pembrolizumab on Progression-Free Survival for Patients With Hormone Receptor-Positive, ERBB2-Negative Metastatic Breast Cancer: A Randomized Clinical Trial. JAMA Oncol. 2020;6:1598–1605. doi: 10.1001/jamaoncol.2020.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset analyzed during the current study is available from the corresponding author on reasonable request.